Background: Rift Valley fever virus is a single-stranded RNA virus that causes disease in humans and livestock.
Results: Rift Valley fever virus infection activates the host NFκB signaling cascade.
Conclusion: NFκB inhibitors, particularly curcumin, down-regulate virus in both in vitro and in vivo models.
Significance: Novel versions of host components resulting from an infection make them ideal therapeutic targets.
Keywords: Animal Viruses, Cell Signaling, Infectious Diseases, Molecular Biology, NF-κB, Curcumin, IKK, Inhibitor, P65, RVFV
Abstract
Rift Valley fever virus (RVFV) is an arbovirus that is classified as a select agent, an emerging infectious virus, and an agricultural pathogen. Understanding RVFV-host interactions is imperative to the design of novel therapeutics. Here, we report that an infection by the MP-12 strain of RVFV induces phosphorylation of the p65 component of the NFκB cascade. We demonstrate that phosphorylation of p65 (serine 536) involves phosphorylation of IκBα and occurs through the classical NFκB cascade. A unique, low molecular weight complex of the IKK-β subunit can be observed in MP-12-infected cells, which we have labeled IKK-β2. The IKK-β2 complex retains kinase activity and phosphorylates an IκBα substrate. Inhibition of the IKK complex using inhibitors impairs viral replication, thus alluding to the requirement of an active IKK complex to the viral life cycle. Curcumin strongly down-regulates levels of extracellular infectious virus. Our data demonstrated that curcumin binds to and inhibits kinase activity of the IKK-β2 complex in infected cells. Curcumin partially exerts its inhibitory influence on RVFV replication by interfering with IKK-β2-mediated phosphorylation of the viral protein NSs and by altering the cell cycle of treated cells. Curcumin also demonstrated efficacy against ZH501, the fully virulent version of RVFV. Curcumin treatment down-regulated viral replication in the liver of infected animals. Our data point to the possibility that RVFV infection may result in the generation of novel versions of host components (such as IKK-β2) that, by virtue of altered protein interaction and function, qualify as unique therapeutic targets.
Introduction
Rift Valley fever virus (RVFV)3 is a RNA virus that belongs to the genus Phlebovirus, family Bunyaviridae (1). It infects humans and livestock and causes Rift Valley fever. RVFV is classified as an emerging infectious agent and as a category A select agent. RVFV is included as an agricultural pathogen by the USDA, as it causes 100% abortion in cattle and extensive death of newborns. In humans, RVFV infection causes fever, ocular damage, liver damage, hemorrhagic fever, and death in some cases (2). There are currently limited options for vaccine candidates, which include the MP-12 and clone 13 versions of RVFV (3). Ribavirin may be used as a treatment option, albeit with suboptimal efficacy (2, 4). Overall, we are lacking in our knowledge of host factors that contribute to RVFV-induced pathogenesis, which is vital not only to understanding the disease progression but also to designing better therapeutics.
RVFV is a single-stranded RNA virus with a tripartite genome. The L (large) segment encodes the RNA dependent RNA polymerase. The M (medium) segment codes for the glycoproteins Gn and Gc. Additionally, the M segment encodes a 78-kDa protein of unknown function and a small nonstructural protein, NSm. NSm has been demonstrated to have an anti-apoptotic function in infected cells (5, 6). The S (small) segment encodes the N protein and a second nonstructural protein, NSs. NSs, a viral virulence factor, is a transcriptional repressor critical to the down-regulation of the host interferon response (7–16).
Our earlier reverse-phase protein microarray (RPMA) studies reveal that infection of human small airway lung epithelial cells (HSAECs) by the virulent ZH501 strain of RVFV elicit multiple host phospho-signaling events relevant to diverse pathological manifestations such as oxidative stress, activation of stress response MAPKs, and DNA damage (17, 18). Multiple ongoing follow-up studies indicate that the signaling responses detected in HSAECs is also observed in many other cell types such as HepG2 cells, HeLa cells, and 293T cells following RVFV infection (Refs. 17 and 48; data not shown). Therefore, our results support the use of HSAECs as a model cell line to study host signaling events after RVFV infection.
One host signaling event that is connected to multiple manifestations, including cellular stress and DNA damage following RVFV infection, is the p65/NFκB signaling cascade (19–24). In fact, Billecocq et al. (7), as part of a study demonstrating the involvement of NSs in interferon suppression, show the nuclear presence and DNA binding function of NFκB after RVFV infection. Activation of the NFκB response is a multistep process that originates at the plasma membrane in the form of receptor activation and terminates in the nuclear activation of NFκB-responsive genes (25). In the classical NFκB activation cascade, a heterotrimeric IκB kinase (IKK) complex consisting of IKK-α, IKK-β, and IKK-γ (NFκB essential modulator or NEMO) induces phosphorylation of IκBα, which is then degraded by the host proteasome. Degradation of IκBα exposes the nuclear localization signal on p65, which is then translocated to the nucleus. Once within the nucleus, p65 forms dimers on κB elements of NFκB-responsive genes. Transcription of these genes determines the cell fate by regulating numerous host cell events such as apoptosis, survival, and cell cycle progression.
We demonstrated previously that inhibition of the host signaling kinase components such as JNK and MEK inhibits viral replication (18). Along these lines, recent publications by our colleagues have provided evidence that regulation of the host factors in the context of RVFV infection is a viable and attractive therapeutic strategy to down-regulate virus replication (26, 27). In this study, we sought to expand on the activation of the NFκB-signaling cascade following infection by MP-12 virus. Our experiments have resulted in the identification of a novel low molecular form of IKK-β that is enzymatically active and unique only to infected cells. We have labeled this novel complex IKK-β2. Additionally, our results suggest that the IKK complex may play a role in the viral life cycle, because inhibitors that target the IKK complex also result in the down-regulation of extracellular virus. We have identified curcumin as a candidate inhibitor that displays effective inhibition of virus, in the case of both pre-exposure and post-exposure treatment. We provide evidence suggesting that curcumin may exert its inhibitory effect on RVFV replication by influencing cell cycle progression of the host cell. Additionally, we demonstrate that IKK-β2 may phosphorylate NSs; this could enhance the ability of NSs to interact with host proteins such as mSin3A, which is critical for NSs-induced down-regulation of the host transcription function. We provide evidence that curcumin prevents phosphorylation of NSs by IKK-β2, thus providing an additional mechanistic explanation for curcumin-mediated viral inhibition. Experiments carried out using the virulent ZH501 strain demonstrate that curcumin can inhibit replication of the fully virulent virus as well. Finally, our experiments using the INFAR−/− murine model (28, 29) provide preliminary proof-of-concept validation that curcumin can down-regulate virus in the livers of infected animals as well, thus paving the way for further development of novel curcumin-based therapeutic options.
EXPERIMENTAL PROCEDURES
Viruses
The MP-12 strain of RVFV is a live attenuated vaccine derivative of the ZH548 strain. ZH548 was isolated from a patient with uncomplicated RVFV infection in 1977. MP-12 was generated by 12 serial passages in MRC5 cells in the presence of 5-fluorouracil, which induced a total of 25 nucleotide changes across the three viral genome segments. arMP-12-del21/384 has a large deletion in the pre-Gn region of the M segment and as a result does not express NSm or 78-, 75-, or 73-kDa proteins encoded by this region. rMP-12-NSdel completely lacks the NSs ORF. The ZH501 strain of RVFV is a fully virulent strain of RVFV.
Cell Culture, Viral Infection, and Extract Preparation
HSAECs were cultured and infected with RVFV, and whole cell extracts were prepared as described previously (18). Briefly, HSAECs were grown in Ham's F12 medium and infected with MP-12 (m.o.i. = 3). In the case of infections with the NSs and NSm mutant viruses (6, 28), cells were infected with the appropriate mutant constructs (m.o.i. = 3). When infections were carried out in 6-well plates, the existing medium was removed (and stored as “conditioned medium”), and cells were washed with PBS and overlaid with 400 μl of medium with virus. After incubation for 1 h at 37 °C, the overlay was removed, and wells were washed twice with PBS and replaced with the conditioned medium. When infections were carried out in 96-well plates, a similar approach was used with the only exception being the utilization of an overlay medium (with virus) of 50 μl. HepG2, HeLa, and A549 cells were also infected following similar methods. To prepare whole cell extracts, the supernatant was removed from the wells, and cells were lysed in lysis buffer (1:1 mixture of T-PER reagent (Pierce), 2× Tris-glycine SDS sample buffer (Novex, Invitrogen), 2.5% β-mercaptoethanol, and a protease and phosphatase inhibitor mixture (1× Halt mixture, Pierce)) and boiled for 10 min prior to electrophoresis.
Western Blot Analysis
Whole cell lysates were separated in 4–20% Tris-glycine gels and transferred to nitrocellulose membranes (iBlot gel transfer system, Invitrogen). The membranes were blocked with a 1% dry milk solution in PBS-T (PBS + Tween 20) at room temperature. Primary antibodies to RVFV (ProSci, catalog No. 4519), Total p65 (Abcam, catalog No. ab7970), phospho-p65 (Ser-536; Santa Cruz Biotechnology, Inc., catalog No. 33020), phospho-IκBα (Santa Cruz Biotechnology, catalog No. sc-21869), HRP-conjugated actin (Abcam, catalog No. ab49900), IKK-α (Santa Cruz Biotechnology, catalog No. sc-7182), IKK-β (Santa Cruz Biotechnology, catalog No. sc-7329), and IKK-γ (Cell Signaling, catalog No. 2685P) were used according to manufacturer's instructions, and the blots were incubated overnight at 4 °C. The blots were then washed three times with PBS-T and incubated with secondary HRP-coupled goat anti-rabbit antibody (Cell Signaling, catalog No. 7074). The blots were visualized by chemiluminescence using a Super Signal West Femto maximum sensitivity substrate kit (Thermo Fisher Scientific) and a molecular imager ChemiDoc XRS system (Bio-Rad). Band intensities were calculated using Quantity One 4.6.5 software (Bio-Rad).
Inhibitor Studies
For inhibitor studies, HSAECs were seeded at 50,000 cells/well in a 96-well plate, and viral infections were carried out (m.o.i. = 0.1). The inhibitors used in this study are as follows: curcumin (Santa Cruz Biotechnology, catalog No. sc-200509), synthetic curcumin (Santa Cruz Biotechnology, catalog No. sc-294110), dimethoxycurcumin (Santa Cruz Biotechnology, catalog No. 205217), lactacystin (Calbiochem, catalog No. 426100), genistein (Sigma, catalog No. G6649), resveratrol (Sigma, catalog No. R5010), 17-Dimethylaminoethylamino-17-demethoxygeldanamycin (Santa Cruz Biotechnology, catalog No. sc-202005), SC-514 (Santa Cruz Biotechnology, catalog No. sc-205504), arctigenin (Santa Cruz Biotechnology, catalog No. sc-202957), IKK2 compound IV (Santa Cruz Biotechnology, catalog No. sc-203083), BAY-11-7082 (Sigma, catalog No. B5556), BAY-11–7085 (Sigma, catalog No. B5681), RO-106-9920 (Santa Cruz Biotechnology, catalog No. sc-203240), CAPE (Santa Cruz Biotechnology, catalog No. sc-200800), 5,7-dihydroxy-4-methylcoumarin (Santa Cruz Biotechnology, catalog No. sc-254863), o-phenanthroline (Santa Cruz Biotechnology, catalog No. 202256). The inhibitors were dissolved in 100% DMSO and added to the cells at a final inhibitor concentration of 10 μm (0.1% DMSO final concentration).
Quantitative RT-PCR (qRT-PCR) Analysis
HSAECs were grown at a density of 50,000 cells/well in 96-well plates. Viral RNA from cell culture supernatants was extracted using Ambion's MagMAXTM-96 viral RNA isolation kit, and RNA was analyzed by qRT-PCR. The primers and probe used for amplification of viral RNA were originally described by Drosten et al. (29). qRT-PCR assays were performed using the ABI Prism 7000 and Invitrogen's RNA UltraSenseTM one-step quantitative RT-PCR system. Cycling conditions were as follows: 1 cycle at 50 °C for 15 min, 1 cycle at 95 °C for 2 min, and 40 cycles at 95 °C for 15 s and 60 °C for 30 s. The absolute quantification was calculated based on the threshold cycle (Ct) relative to the standard curve.
Plaque Assays
Neutral red plaque assays were performed by standard procedures. Briefly, Vero cells were plated in 6-well plates (106 cells/well). Supernatants were diluted in DMEM (102-106) and used to infect the cells in duplicate. After a 1-h infection, the medium was removed and the wells overlaid with a 2× E-MEM and 0.5% agarose solution. After the overlay solidified, the plates were incubated for 48 h. Plaques were visualized with neutral red, E-MEM, and an 0.5% agarose solution by overlaying neutral red on top of the first layer. After the overlay had solidified, the plates were placed in the incubator for an additional 24 h after which plaques were counted and viral titers determined.
Flow cytometry Analysis
HSAECs were prepared for flow cytometry analysis by standard procedures. Briefly, the cells were washed twice in 1× PBS (without calcium and magnesium) and trypsinized. Trypsin was neutralized by adding back cold medium with 10% serum, and the cells were spun down at 2000 rpm for 10 min in a refrigerated microcentrifuge. The cell pellet was washed twice with 1× PBS and resuspended in 70% ice-cold ethanol. The cells were rehydrated using 1× PBS (without calcium and magnesium) for 15 min and pelleted. Cells were then stained with 1 ml of propidium iodide solution, and cell cycle analysis was carried out on an Accuri C6 flow cytometer. Data analysis was performed with Multicycle AV and FCS Express.
Cell Viability Assays
HSAECs were seeded in 96-well plates at 50,000 cells/well, and cell viability was measured using a CellTiter-Glo luminescent cell viability kit (Promega) as per the manufacturer's instructions. Briefly, an equal volume of room temperature medium and CellTiter-Glo reagent was added to the cells. The plate was shaken for 2 min on an orbital shaker, and after incubation for 10 min at room temperature luminescence was detected using the DTX 880 multimode detector (Beckman Coulter).
Size-exclusion Chromatography
HSAECs were infected with MP-12 virus (m.o.i. = 10), and cells were pelleted for analysis at ∼20 h post-infection. In the case of TNFα treatment, HSAECs were treated with TNFα (10 and 50 ng/ml). BCBL-1 cells are Kaposi sarcoma herpesvirus (KSHV)-infected cells, and BJAB cells are their uninfected counterparts. In all cases, cell pellets were washed twice with PBS without Ca2+ and Mg2+, resuspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 120 mm NaCl, 5 mm ethylenediaminetetraacetic acid, 0.5% Nonidet P-40, 50 mm NaF, 0.2 mm Na3VO4, 1 mm DTT, and one Complete protease mixture tablet/50 ml), and incubated on ice for 20 min with gentle vortexing every 5 min. Lysates were then centrifuged at 4 °C at 10,000 rpm for 10 min. Supernatants were transferred to a fresh tube, and protein concentrations were determined using the Bradford protein assay (Bio-Rad). Two milligrams of protein from each treatment was equilibrated and degassed in chromatography running buffer (0.2 m Tris-HCl (pH 7.5), 0.5 m NaCl, and 5% glycerol). The lysates were run on a Superose 6 HR 10/30 size-exclusion chromatography column using the AKTA purifier system (GE Healthcare). The samples were injected over a 1-ml loop into the column, and the columns in this study were used for multiple runs prior to fractionation of the actual virus-infected or uninfected control extracts. This is an essential step to ensure that the column is conditioned and the flow rates and elution patterns are reproducible. Flow-through was collected at 4 °C at a flow rate of 0.3 ml/min at 0.5 ml for ∼70 fractions. Every fifth fraction was acetone-precipitated using 4 volumes of ice-cold 100% acetone and incubated for 15 min on ice. Lysates were centrifuged at 4 °C for 10 min at 12,000 rpm, supernatants were removed, and the pellets were allowed to dry for a few minutes at room temperature. The pellets were resuspended in Laemmli buffer and analyzed by immunoblotting for IKK-α, IKK-β, IKK-γ, and β-actin.
Immunoprecipitation and in Vitro Kinase Assay
Immunoprecipitation (IP) and in vitro kinase assays were carried out as described previously (30). Briefly, for immunoprecipitation, low molecular weight complex fractions from MP-12- or UV-MP-12-infected cells were immunoprecipitated at 4 °C overnight with IKK-β antibody. The next day, complexes were precipitated with A/G beads (Calbiochem) for 2 h at 4 °C. Immunoprecipitated samples were washed twice with appropriate TNE buffer (Tris (pH 7.5), NaCl, EDTA) and kinase buffer. The reaction mixtures (20 μl) contained the following final concentrations: 40 mm β-glycerophosphate (pH 7.4), 7.5 mm MgCl2, 7.5 mm EGTA, 5% glycerol, [γ-32P]ATP (0.2 mm, 1 μCi), 50 mm NaF, 1 mm orthovanadate, and 0.1% (v/v) β-mercaptoethanol. Phosphorylation reactions were performed with immunoprecipitated material and [γ-32P]-labeled GST-IκBα as a substrate in TTK kinase buffer containing 50 mm HEPES (pH 7.9), 10 mm MgCl2, 6 mm EGTA, and 2.5 mm dithiothreitol. When using a GST-NSs substrate, a similar reaction setup was employed. For reactions involving curcumin, synthetic curcumin (Santa Cruz Biotechnology, catalog No. sc-294110) was included in the reaction at 0.1 and 1 μm concentrations. Reactions were incubated at 37 °C for 1 h, stopped by the addition of 1 volume of Laemmli sample buffer containing 5% β-mercaptoethanol, and run on a 4–20% SDS-PAGE. Gels were subjected to autoradiography and quantification using Amersham Biosciences PhosphorImager software (Amersham Biosciences).
Mass Spectrometry
Fractions corresponding to the medium molecular weight IKK-β complex and the low molecular weight IKK-β2 complex were immunoprecipitated with an anti-IKKβ antibody. The immunoprecipitated material was separated on a gel, and multiple bands were cut out that spanned the entire length of the gel. The separated material was then subjected to in-gel tryptic digestion (trypsin, Promega) overnight at 37 °C. The digested peptides were eluted using ZipTip purification (Millipore), and identification of the peptides was performed by LTQ-MS/MS equipped with a reverse-phase liquid chromatography nanospray (Thermo Fisher Scientific). The reverse-phase column was slurry-packed in-house with 5 μm, 200 Å-pore size, C18 resin (Michrom Bioresources) in a 100 μm × 10 cm fused silica capillary (Polymicro Technologies) with a laser-pulled tip. After a sample injection, the column was washed for 5 min at 200 nl/min with 0.1% formic acid; peptides were eluted using a 50-min linear gradient from 0 to 40% acetonitrile and an additional step of 80% acetonitrile (all in 0.1% formic acid) for 5 min. The LTQ-MS was operated in a data-dependent mode in which each full MS scan was followed by five MS/MS scans where the five most abundant molecular ions were dynamically selected and fragmented by collision-induced dissociation using normalized collision energy of 35%. Tandem mass spectra were matched against the National Center for Biotechnology Information mouse database by SEQUEST BioWorks software (Thermo Fisher Scientific) with full tryptic cleavage constraints and static cysteine alkylation by iodoacetamide. For a peptide to be considered legitimately identified, it had to be the top number one matched and had to achieve cross-correlation scores of 1.9 for [M+H]1+, 2.2 for [M+2H]2+, and 3.5 for [M+3H]3+ with ΔCn > 0.1 and a maximum probability of randomized identification of 0.01.
Curcumin-Bead Binding Assay
Curcumin immobilized in trimethoxysilane-based nanoparticles (patent pending) or nanoparticles without curcumin were synthesized as follows. 30 ml of 50 mm PBS buffer (pH 7.5), 1 ml of PEG 200, and 4 ml of curcumin (1 mg/ml in DMSO) were added sequentially and vortexed thoroughly. Next, 4 ml of previously hydrolyzed trimethoxysilane was added to the tube, and the contents were vortexed for about 2 min. The tube was allowed to sit undisturbed for gelation (2–5 h). The gel was then lyophilized for 24 to 48 h. The resulting particles were ball-milled at 150 rpm for 6.5 h. The particles were incubated with fractions 18–21 (medium molecular weight complex) and 33–36 (low molecular weight complex) from MP-12-infected and UV-MP-12-infected cells at 4 °C overnight. The next day, the particles were washed twice with TNE-50 (0.1% Nonidet P-40) buffer, resuspended in 20 μl of Laemmli sample buffer containing 5% β-mercaptoethanol, and run on 4–20% Tris-glycine gels.
Animal Studies
Six-to-eight-week-old INFAR−/− mice were obtained from the National Center for Biodefense and Infectious Disease breeding colony (George Mason University). All experiments were carried out in bio-safety level 2 (BSL-2) facilities and in accordance with the Guide for the Care and Use of Laboratory Animals (Committee on Care And Use of Laboratory Animals, Institute of Laboratory Animal Resources, National Research Council, National Institutes of Health Publication 86-23, revised 1996). For infection experiments, all animals were treated subcutaneously with DMSO alone or with curcumin every day for 4 days. Animals were pretreated with curcumin 24 h prior to infection and infected with 104 pfu of MP-12 virus by intraperitoneal injection. Mice were weighed daily and monitored for morbidity and mortality, including lethargy and ruffled fur. Liver tissue was dissected from DMSO- and curcumin-treated animals and analyzed for viral load by standard plaque assay.
Statistical Analysis
All quantifications indicated are based on data obtained from triplicate experiments. The p values were calculated by using Student's t test.
RESULTS
The NFκB Subunit p65 Is Phosphorylated at Serine 536 in MP-12-infected Cells
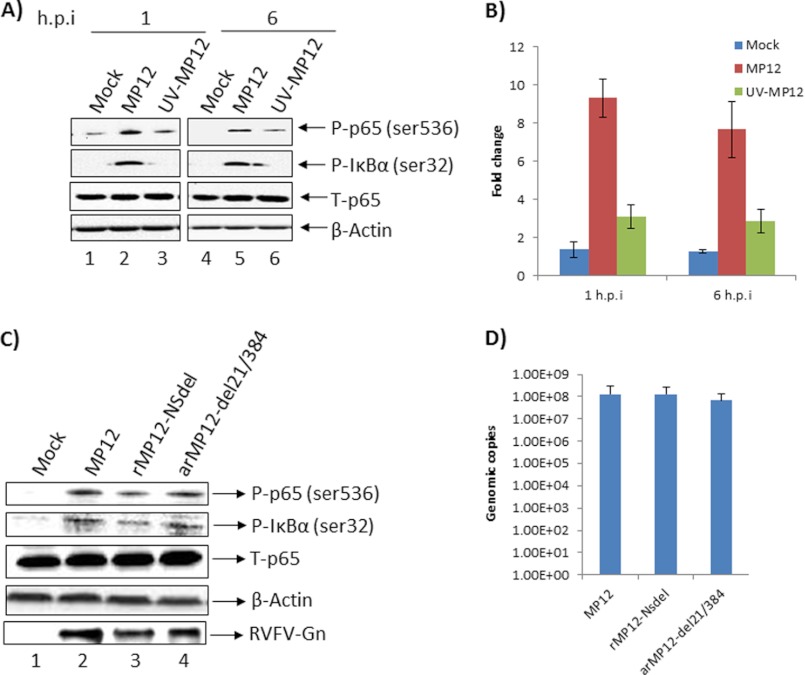
Using a RPMA approach, we demonstrated previously that infection of HSAECs with the virulent strain of RVFV (ZH501) causes phosphorylation and activation of multiple signal transduction events including p38 MAPK, JNK, ERK, and their downstream transcription targets including p65 (NFκB) in the host cell (18). Specific to the NFκB-signaling pathway, RPMA studies indicate that following infection by ZH501 virus, p65 is phosphorylated at serine 536 (18). In the current study, we first asked the question whether the attenuated strain (MP-12) also caused phosphorylation of p65 at serine 536 in HSAECs. We carried out Western blot analysis of MP-12-infected and mock-infected cell extracts obtained at 1 and 6 h post-infection. Activation of the NFκB cascade and p65 phosphorylation (serine 536) has been demonstrated to occur at similar time points after viral infection (31). During the course of these experiments, care was taken to maintain cells in “conditioned medium” (the same medium that cells were maintained in prior to infection was returned to the cells post-infection) to avoid any p65 phosphorylation event due to medium/serum components. The results demonstrated that p65 is phosphorylated at serine 536 at both 1 and 6 h post-infection when compared with the mock-infected control samples (Fig. 1A, compare lane 1 with 2 and lane 4 with 5). Phosphorylation of p65, however, was not due to an increase in total p65 levels (Fig. 1A). Phosphorylation of IκBα was also detectable at 1 and 6 h post-infection (Fig. 1A) suggesting that the activation of p65 occurred by the classical pathway. We performed similar experiments with a UV-inactivated virus to address the issue of specificity of the phosphorylation event. Quantification of signal intensities of the phosphorylated p65 observed in multiple experiments revealed that MP-12 infection resulted in an approximate 8–9-fold increase in phospho-p65 when compared with the mock-infected control (Fig. 1B). This fold increase in phosphorylation was significantly higher than that observed in the case of infection by the UV-inactivated virus, which did not change over the time frame tested. This observation provided specificity to the observed phosphorylation of p65 following MP-12 infection.
FIGURE 1.
MP-12 infection caused phosphorylation of p65 by the classical pathway. A, HSAECs were infected with MP-12 or UV-inactivated MP-12 virus and analyzed for phosphorylation of p65 and IκBα by Western blot analysis. Mock-infected cells were maintained alongside as negative controls. Total p65 and β-actin Western blots were carried out as controls. B, extent of phosphorylation of p65 was quantified by averaging signal intensities observed in three different experiments after infection by MP-12 or UV-MP-12 virus. Phosphorylation is represented as -fold increase over that of uninfected cells. h.p.i., (hours post-infection). C, phosphorylation of p65 and IκBα after infection by wild type MP-12 virus, NSs mutant (rMP-12-NSdel), and NSm mutant (arMP-12-del21/384) were analyzed by Western blot. Total p65 and β-actin levels were analyzed as controls. D, intracellular RNA levels of MP-12 virus or the NSs and NSm mutant viruses were determined by qRT-PCR using total RNA extracted from infected cells at 6 h post-infection.
We performed plaque assays with the UV-inactivated virus in comparison with wild type MP-12 and were unable to observe any plaques with the inactivated virus (supplemental Fig. 1A). We also tested whether p65 was phosphorylated on additional residues (serine 276) and were unable to detect significant changes in the phosphorylation of serine 276 at the same time points (data not shown).
We then asked whether any viral protein component played a role in the phosphorylation of p65. We infected HSAECs with the wild type MP-12 virus or mutant viruses that lacked the NSs (rMP-12-NSdel) or the NSm (arMP-12-del21/384) coding sequences and checked for p65 phosphorylation. The results demonstrated a modest influence of NSs on the phosphorylation of p65 (an average drop of about 34% as determined from two experiments) suggesting that the viral protein may play a role in inducing p65 phosphorylation (Fig. 1C). We then asked whether the NSs and the NSm mutant viruses displayed any inherent replication differences in our cell system, to verify that the observed effects on p65 phosphorylation were not indirect. To that end, we performed qRT-PCR analysis of intracellular viral RNA at 6 h post-infection. We reasoned that a quantitative analysis of the intracellular RNA of the wild type and mutant viruses performed at the time point when phosphorylation differences were observed would demonstrate whether it was an indirect consequence of decreased replication kinetics of the NSs mutant. The data are shown in Fig. 1D; we did not observe any significant difference between replication of the wild type or mutants at the time point tested, suggesting that the observed differences in the p65 phosphorylation levels following infection by MP-12 and mutant viruses were not merely a reflection of altered replication kinetics.
We next asked whether phosphorylation of p65 on serine 536 was a cell type-specific event. HeLa and HepG2 cells were infected with MP-12 virus (m.o.i. = 3), and cell extracts were analyzed by Western blot. These experiments revealed that p65 was indeed phosphorylated in both cell types following viral infection without any change in total p65 levels (supplemental Fig. 1B). Cumulatively, these experiments revealed that p65 is phosphorylated on serine 536 following infection by MP-12 virus.
Presence of a Novel IKK Complex in RVFV-infected Cells
The IKK complex is the upstream component in the NFκB cascade that leads to the phosphorylation of IκBα and p65 (31, 32). The IKK complex that functions as the IκBα and p65 kinase is typically a heterotrimer that consists of IKK-α, IKK-β, and IKK-γ (NEMO) proteins. IKK-α and IKK-β possess kinase activity, whereas IKK-γ (NEMO) is required to stabilize the heterotrimeric complex.
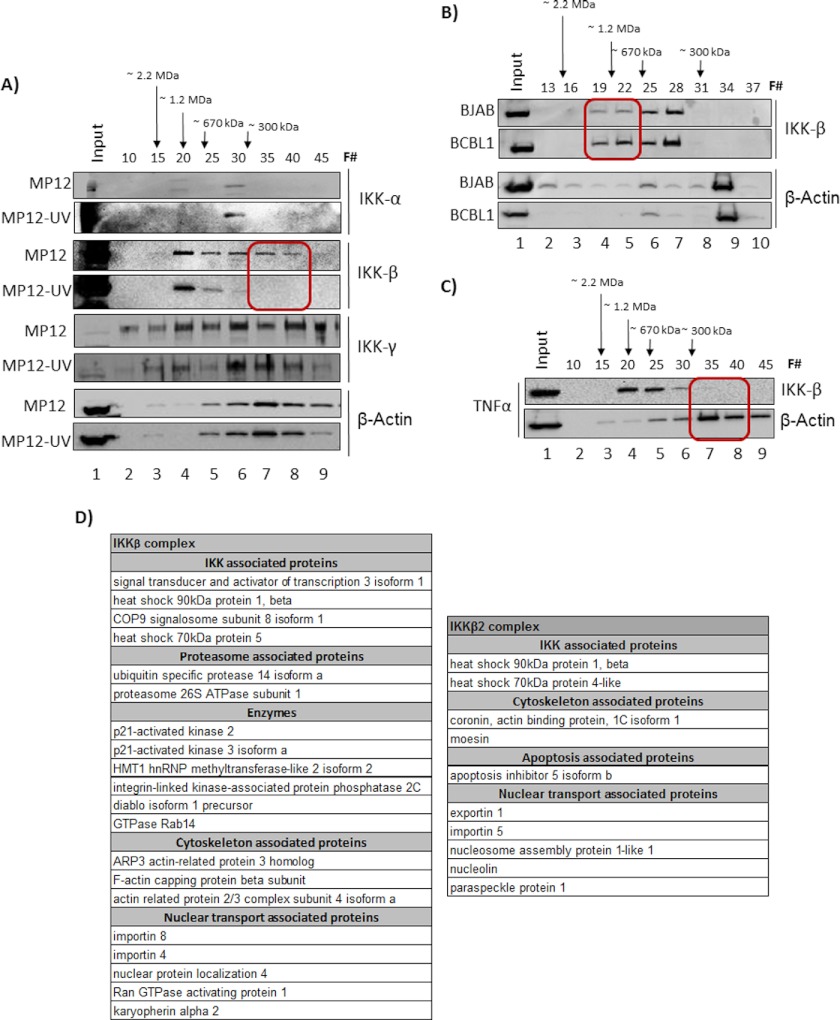
First, we asked whether the IKK complex is altered because of MP-12 infection. When no major changes in total IKK levels could be detected upon infection (data not shown), we asked whether changes could be manifested as alterations of IKK higher order protein complexes. To answer that question, we performed conventional chromatography separation of MP-12-infected cell extracts using a Superose 6 sizing column and analyzed fractions for distribution of individual IKK components by Western blots. The Superose 6 sizing column is designed to separate higher order molecular complexes based on cumulative molecular weight, as indicated in Fig. 2A. Therefore, if individual components of the IKK complex display alterations in higher order protein association, the cumulative molecular weight of that component will be expected to change. This will result in a different elution profile, which can then be detected by methods such as Western blotting. As a control, we utilized extracts from cells infected with the UV-inactivated virus. Whole cell lysates from infected cells were fractionated, and every fifth fraction (Fig. 2A, F#) was analyzed by Western blot for IKK-α, IKK-β, and IKK-γ (NEMO) proteins. β-Actin was analyzed as a control. The results showed that there was an alteration in the distribution profile of IKK-β in MP-12-infected cells that was not observable in the UV-inactivated virus-infected cells (Fig. 2A). Among the changes, IKK-β reproducibly displayed unique low molecular weight complexes in MP-12-infected cells, which we will refer to henceforth as IKK-β2 (Fig. 2A, red squares). Immunoblotting the same extracts with actin demonstrated a comparable distribution of actin in fractions 30–40 in both MP-12- and UV-MP-12-infected cell extracts.
FIGURE 2.
IKK complex components were altered after MP-12 infection. A, HSAECs were infected with MP-12 and UV-inactivated MP-12, and cell lysates were analyzed for IKK complex components at ∼20 h post-infection. Whole cell lysates were fractionated in a Superose 6 HR 10/30 size-exclusion chromatography column using the AKTA purifier system. A total of 70 fractions were obtained, and every fifth fraction was analyzed for IKK-α,β,γ complexes and for β-actin by Western blots. The fraction numbers analyzed are indicated above each panel by F#. The red square in the IKK-β blot indicates the lower molecular weight complexes (IKK-β2) that eluted in the case of MP-12 infection. B, total protein extracts from BCBL1 cells (KSHV-infected) and BJAB (uninfected control) cells were fractionated using a new, conditioned Superose 6 size-exclusion column. Fraction numbers for subsequent analysis were determined based on the elution profiles of known standards. Accordingly, the fractions indicated in B match with fractions indicated in A and C in terms of elution profiles. C, total protein extracts from HSAECs treated with TNFα were fractionated, and every fifth fraction was analyzed for IKK-β and β-actin.
IKK-β subunit is one of the kinases involved in phosphorylation of p65 at serine 536 (34). The native IKK complex exhibits a molecular mass of ∼700 to 900 kDa. The IKK complex that is common to both MP-12 virus- and UV-MP-12 virus-infected cells migrated at an approximate molecular range of 600 to 900 kDa in our sizing columns (Fig. 2A). The novel IKK-β2 complex eluted around 300 kDa in the case of MP-12-infected cells (Fig. 2A, fractions 35–40). The existence of the IKK-β2 complex after infection by MP-12 has been confirmed by us in multiple independent experiments. We did not observe any significant or reproducible alterations in the case of the IKK-α or IKK-γ subunits in the MP-12- and UV-MP-12-infected cells.
We then asked the question of whether a similar alteration in IKK-β could be observed in the case of an infection by a DNA virus. To that end, we utilized KSHV/HHV-8-infected cells (BCBL-1) and their uninfected counterparts, BJAB (33), to perform a fractionation experiment similar to that with the RVFV-infected cells. KSHV infection is known to activate the host NFκB signaling cascade with involvement of the IKK complex (34–36). Interestingly, as observed in Fig. 2B, when infected with a DNA virus, IKK-β distribution shifted toward the high molecular weight range (> 670 kDa) (Fig. 2B, compare lanes 4 and 5 between BCBL1 and BJAB samples). This suggested an altered regulation of IKK-β in the case of at least one DNA virus that was different from that of RVFV infection. Additionally, we also tested whether activation of the NFκB pathway by inducers such as TNFα would result in the formation of IKK-β2. To that end, HSAECs were treated with TNFα. Cells were analyzed 24 h later by a similar fractionation scheme. As seen in Fig. 2C, TNFα treatment did not result in any significant change to the IKK-β profile, with almost all detectable IKK-β eluting between fractions 20 and 30. We tested the fractionation profile of TNFα at 5× higher concentration (50 ng/ml) and were still unable to detect any difference in the IKK-β distribution profile between untreated and treated cells (data not shown). Therefore, the data from the fractionation studies revealed that a low molecular weight version of IKK-β (IKK-β2) may be a distinctive occurrence during RVFV infection; this was not observed during infection by a DNA virus or during NFκB activation by inducers such as TNFα.
We carried out proteomic analysis of the IKK-β and IKK-β2 complex by immunoprecipitating the kinase using an anti-IKK-β antibody from pooled fractions. Specifically, we pooled fractions 19–23 (medium molecular weight complex) and fractions 34–38 (low molecular weight complex) and subjected the immunoprecipitated complexes to LC-MS/MS analysis. The data, as shown in Fig. 2D, demonstrated multiple common and unique aspects of the IKK-β and IKK-β2 complexes. As for the commonalities, in both cases we observed interaction of chaperone proteins including Hsp70 and Hsp90. Hsp90 is also known to be a protein that interacts strongly with IKK-γ. Fig. 2A shows that IKK-γ co-fractionated with both IKK-β and IKK-β2 in the Superose column. Therefore, both complexes are likely to contain IKK-γ. In fact, based on data shown in Fig. 2A, we suspect that the IKK-β2 complex may contain slightly more IKK-γ when compared with IKK-β. Additionally, we observed that IKK-β and IKK-β2 contained actin-related proteins and proteins involved in nuclear-cytoplasmic transport. The fact that both complexes contained nuclear export/import-related proteins suggested that both complexes are likely to be present in the nuclear and cytoplasmic compartments of the cell; however, it would be of great interest to determine the targets of the two kinase complexes in the nuclear and cytoplasmic compartments. In terms of aspects unique to either complex, our results suggested that specific proteasome components may be associated with IKK-β, which were not found to be associated with IKK-β2. This indicates interesting regulatory possibilities with altered stability kinetics between the two enzymatic complexes. The association of proteasome components may extend to the inclusion of Diablo in this complex as well. It was also interesting to observe protein phosphatase 2C associated with IKK-β. This may also contribute to differences in kinase-active states of IKK-β versus IKK-β2, with IKK-β2 possibly being more kinase-active than IKK-β. The association of a methyltransferase enzyme component may be suggestive of a nuclear inhibitory function for IKK-β, which is absent in the case of IKK-β2, again alluding to IKK-β2 having different kinetic rates and targets from the IKK-β enzyme. Therefore, analysis of the proteomic composition of IKKβ and IKKβ2, although not indicating any noteworthy changes in intracellular distribution, pointed to differences in regulation that will affect function and targets. Overall, our results indicated that MP-12 infection resulted in the formation of a novel IKK-β2 complex without any major alterations in the other components of the IKK complex, which may be a distinctive consequence of RNA virus infection.
Inhibition of the IKK Complex Resulted in Decreased Viral Replication
We then investigated whether activation of the NFκB cascade was essential for viral replication. We utilized well established inhibitors of the NFκB cascade including those that inhibit the IKK complex, IκBα degradation, and p65 nuclear translocation (Fig. 3A). Briefly, the inhibitors utilized include IKK inhibitors (geldanamycin, SC514, curcumin, arctigenin, and IKK2 compound IV), IkBa inhibitors (BAY-11-7082, BAY-11-7085, and RO-106-9920), a nuclear translocation inhibitor (CAPE), and those that prevent NFκB-dependent transcriptional activation (5,7-dihydroxy-4-methylcoumarin and o-phenanthroline). HSAECs were pretreated with inhibitors for 2 h followed by infection with MP-12 virus. Infected cells were treated with DMSO alone as a negative control. After infection, cells were post-treated with the inhibitor or DMSO for 24 h, and supernatants were analyzed for infectious virus by plaque assays. The results of our inhibitor studies demonstrated that activation of the IKK complex and nuclear translocation was necessary for viral replication (Fig. 3B and supplemental Fig. 2). Untreated cells (Fig. 3, UT) and DMSO- or inhibitor-treated cells were also analyzed by CellTiter-Glo (luminescence units) at 24 h post-treatment to determine the cytotoxic effects. The toxicity studies confirmed that the inhibitors were not toxic to the cells at these concentrations. Among the inhibitors tested, curcumin was the strongest and down-regulated RVFV by 3–4 logs (Fig. 3B). Arctigenin, IKK-IV, SC-514, and CAPE were able to down-regulate viral replication by about 1 log (Fig. 3B and supplemental Fig. 2). Other inhibitors that interfered with Iκbα phosphorylation (BAY-11-7082, BAY-11-7085, and RO-106-9920) and transcription activation (5,7-dihydroxy-4-methylcoumarin and o-phenanthroline) did not exert any inhibitory effect on RVFV replication (supplemental Fig. 2), suggesting that the influence of an activated NFκB cascade in the context of RVFV replication is likely to transcend a mere transcription regulatory function.
FIGURE 3.
Inhibitors of the IKK complex down-regulated MP-12 replication. A, diagrammatic representation of the NFκB activation cascade and the interface of tested inhibitors with the cascade. B, HSAECs were pretreated with inhibitors of the NFκB cascade for 2 h. Pretreated cells were infected with MP-12 and continued to be post-treated with the inhibitors for up to 24 h. Supernatants were collected from infected, untreated cells, infected, DMSO-treated cells, and infected, inhibitor-treated cells. All supernatants were quantified for infectious progeny virus by plaque assays. Inhibitor toxicity was evaluated by measuring survival of inhibitor treated cells in comparison with DMSO treated and untreated (UT) cells.
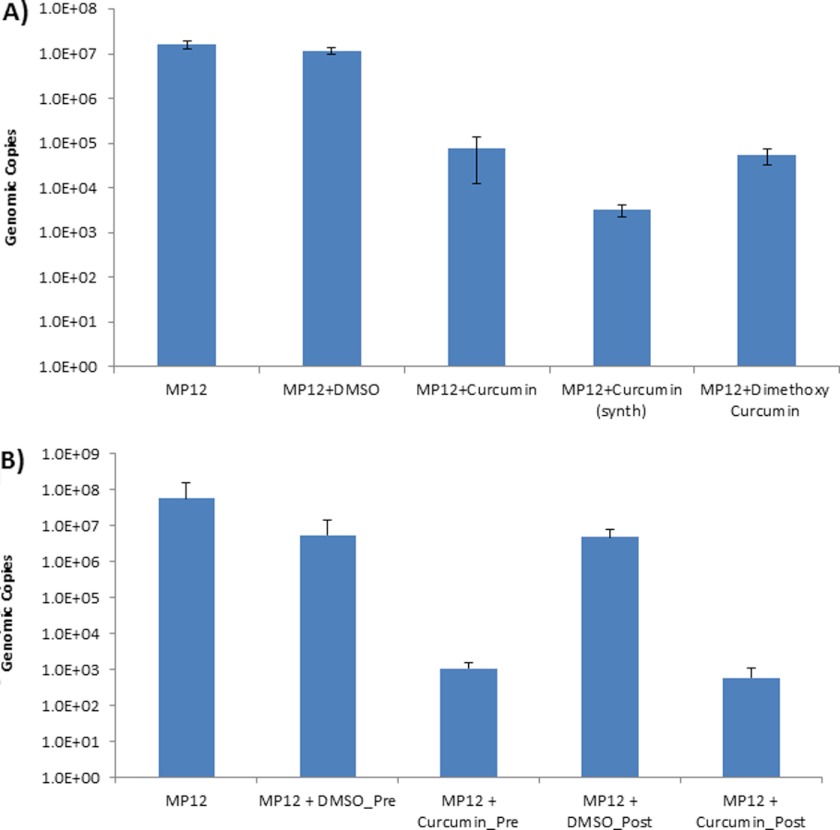
Next, we asked whether the observed down-regulation of extracellular virus after curcumin treatment was due to low amounts of virus being released or to infection-defective viruses. To determine the extracellular viral genomic copy numbers after treatment with curcumin, we carried out qRT-PCR studies of culture supernatants with a RVFV-specific primer directed against the G2 region of the RVFV genome (31). Our studies revealed that curcumin treatment decreased total genomic copies by more than 3 logs (Fig. 4A, compare columns 1 and 2 with column 3). When we treated cells with a dimethoxy derivative of curcumin, we found that the level of inhibition of viral replication by the dimethoxy derivative was comparatively lower than that of synthetic curcumin (Fig. 4A, compare columns 4 and 5).
FIGURE 4.
Down-regulation of extracellular genomic RNA by curcumin. A, HSAECs were pre- and post-treated with curcumin, and supernatants were analyzed for viral genomic RNA copies by qRT-PCR in comparison with untreated and DMSO-treated, MP-12-infected cells. Viral genomic copy numbers in the context of inhibition by synthetic curcumin or dimethoxycurcumin were determined. Supernatants were obtained from infected, inhibitor-treated cells at 24 h post-infection and analyzed by qRT-PCR. B, HSAECs were either pretreated (2 h) or post-treated (3 h) with curcumin, and the viral genomic copy number in the supernatant was determined by qRT-PCR.
We then determined whether the time of addition of curcumin would have an influence on the extent of inhibition. We compared extracellular viral genomic copies following pretreatment (2 h prior to infection) with curcumin and post-treatment (3 h post-infection) using qRT-PCR. HSAECs were infected with MP-12, and supernatants were collected 24 h post-exposure. DMSO treatments were included as controls. As shown in Fig. 4B, treatment of infected cells 3 h post-exposure with curcumin continued to down-regulate extracellular virus in a manner comparable with pretreatment with curcumin (compare columns 3 and 5).
As indicated earlier, we observed a unique low molecular weight IKK-β2 complex in MP-12-infected extracts (Fig. 2A). We next asked whether the IKK-β2 complex exhibited kinase activity similar to IKK-β and could be inhibited by curcumin. To answer that question, we immunoprecipitated the IKK-β2 complex from fractions 33–36 and performed an in vitro kinase assay using a GST-IκBα as substrate (Fig. 5A). An IKK-β immunoprecipitation from HTLV-1-infected cells was used as a positive control (Fig. 5A, C81) in the kinase assay. The immunoprecipitated IKK-β2 complex demonstrated kinase activity, whereas comparable fractions from UV-MP-12-infected extracts showed no activity (Fig. 5A, compare lanes 4 and 6). We performed similar kinase assays from the IKK-β complex that was observed in both MP-12 and UV-MP-12 extracts (fractions 18–21) and detected comparable kinase activity (data not shown). When the kinase assay was carried out using immunoprecipitated IKK-β2 in the presence of increasing concentrations of curcumin, we observed down-regulation of kinase activity at a low concentration of curcumin and a complete loss of activity at higher concentrations (Fig. 5A, compare lane 4 with lanes 7 and 8), suggesting that curcumin inhibits the IKK-β2 complex in infected cells. As curcumin is known to inhibit the conventional IKK-β complex, we utilized IKK-β immunoprecipitated from fractions 18–21 in the kinase assay in the presence of increasing concentrations of curcumin (Fig. 5A, lanes 9 and 10). Interestingly, we found that while both IKK-β and IKK-β2 were susceptible to curcumin at high concentrations (Fig. 5A, lanes 8 and 10) IKK-β2 was more susceptible to curcumin than IKK-β at lower concentrations (compare lanes 7 and 9).
FIGURE 5.

IKK-β2 derived from MP-12-infected cells retained kinetic function and was bound by curcumin. A, fractions 33–36 from MP-12-infected and UV-MP-12-infected cell extracts were pooled and immunoprecipitated with anti-IKK-β antibody. Phosphorylation reactions were carried out with the immunoprecipitated material and a GST-IκBα substrate. After incubation, samples were separated on a 4–20% Tris-glycine gel, dried, and analyzed by a PhosphorImager (Amersham Biosciences). In lane 2 (C81), total C81 cell extract was used as a positive control for IKK-β activity. In lane 1, GST alone without the IκBα substrate was used as a negative control. In lanes 7 and 8, increasing concentrations of curcumin were added to the phosphorylation reaction. In lanes 9 and 10, fractions 18–21 were used as the source of IKK-β, and the effect of curcumin on its kinase activity was determined. B, fractions 18–21 and 33–36 were pooled independently and subjected to a pulldown assay by incubation with immobilized curcumin beads. The washed beads were resuspended in Laemmli buffer, separated by SDS-PAGE, and analyzed by Western blot using an anti-IKK-β antibody.
Finally, we asked whether immobilized curcumin could bind to IKK-β2 in vitro. To answer that question, we immobilized curcumin in trimethoxysilane-based nanoparticles and incubated the particles with medium (fractions 18–21) and low molecular weight fractions (fractions 33–36) overnight at 4 °C. Next day, bound samples were separated in a 4–20% Tris-glycine gel, and Western blot analysis was carried out using IKK-β antibody (Fig. 5B). Interestingly, immobilized curcumin did not bind to IKK-β in the medium molecular weight complex from the MP-12-infected cell fractions (Fractions 18–21); however, we observed binding of curcumin to the low molecular weight IKK-β2 complex (fractions 33–36) (Fig. 5B, compare lanes 2 and 4). We did not observe similar interactions with immobilized curcumin and IKK-β2 in UV-MP-12-infected cells. This suggested a unique interaction of curcumin with the IKK-β2 complex that is present in infected cells. Taken together, our results demonstrated for the first time that inhibitors of the IKK complex such as curcumin cause down-regulation of extracellular virus. Additionally, our functional assays indicated that the kinase activity of the IKK-β2 complex is inhibited by curcumin.
Curcumin, in addition to being an inhibitor of the NFκB cascade, is also a well documented inhibitor of the host proteasome (37). Relevant to the critical role played by the host proteasome pathway in RVFV biology, Ikegami et al. (10–12) have demonstrated that two proteasome inhibitors, lactacystin and MG-132, are capable of reversing the NSs-induced post-transcriptional down-regulation of PKR. We extended the studies to see whether proteasome inhibitors other than curcumin could down-regulate extracellular virus levels. To answer that question, we pretreated HSAECs with three different proteasome inhibitors: lactacystin (38), resveratrol (39), and genistein (40). Lactacystin is a specific and irreversible inhibitor of the 26S proteasome and has been demonstrated to down-regulate the trypsin-like, chymotrypsin-like, and peptidyl glutamyl hydrolase-like proteasomal activity of the proteasome (41). Resveratrol has been demonstrated to suppress cytokine-induced proteasome function and degradation of IkBa (42). Genistein is thought to interact with the proteasomal β5 subunit and result in the inhibition of the chymotrypsin-like activity of the proteasome (43). After 2 h of pretreatment, cells were infected with MP-12 virus. DMSO-treated cells were maintained as controls. Supernatants were obtained from inhibitor-treated and DMSO-treated cells at 24 h post-infection and analyzed for extracellular virus by plaque assays. Our results demonstrated that some of the inhibitors could modestly down-regulate extracellular virus levels by approximately 1 log (supplemental Fig. 3A). Although treatment with lactacystin down-regulated extracellular virus, we observed some level of cytotoxicity associated with inhibitor treatment. We then determined whether the observed down-regulation of infectious virus was a reflection of low extracellular genomic copy numbers. We carried out qRT-PCR studies with virus-specific primers (29). Our studies revealed that treatment with various proteasome inhibitors resulted in comparable decrease in genomic copies (supplemental Fig. 3B). Taken together, these data confirm prior studies that the host proteasomal pathway is a critical component of RVFV-host interaction.
Reduced Viral Replication Correlates with Rescue of RVFV-induced Cell Cycle Arrest
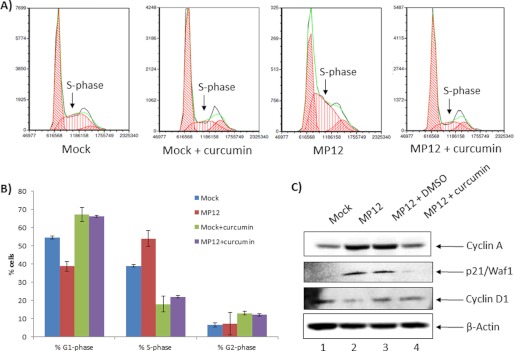
We have consistently observed that RVFV infection induces a strong S-phase arrest of the infected cells in diverse cell types, and this arrest is dependent on the viral protein NSs (44) (Fig. 6, A and B). Here we asked whether inhibition of viral replication could be sufficient to rescue the cells from the infection-induced S-phase arrest. Therefore, we pretreated synchronized cells with curcumin for 2 h and then infected the cells with MP-12 virus. Cells were maintained in curcumin post-infection. Untreated, infected cells were maintained alongside as controls. We performed FACS analysis 24 h post-infection to evaluate cell cycle progression and observed that treatment of cells with curcumin rescued the infected cells from S-phase arrest. The pronounced S-phase peak that is observed in the third histogram (Fig. 6A) is reduced to a level that can be compared with the peak observed in the uninfected panel, thus suggesting that the infection induced S-phase arrest may be relieved upon curcumin treatment. This is quantitatively demonstrated in Fig. 6B. We also observed that treatment of uninfected cells with curcumin decreased the population of cells that were at S phase (Fig. 6A, compare S-phase peaks between histograms 1 (Mock) and 2 (Mock + curcumin); Fig. 6B, compare first and third bars in the % S-phase panel). Therefore, it may be possible that S-phase cells are better hosts for supporting RVFV replication, and curcumin may inhibit RVFV by lowering the population of susceptible host cells. Additionally, Western blot analyses revealed that marker proteins of cell cycle arrest such as cyclin A and p21/Waf1 accumulated in the MP-12-infected cells, whereas curcumin treatment restored cyclin A and p21/Waf1 levels to those seen in control mock-infected cells (Fig. 6C). Rescue of cyclin A and p21/Waf1 phenotypes suggested that the population of cells that responded to curcumin were likely to be in late G1/early S phase of the cell cycle. Cyclin D1, which is an early G1 phase marker, did not appear to be influenced significantly by curcumin treatment (in comparison with the DMSO control), thus arguing against curcumin being effective in early G1 phase of the cell cycle. Collectively these experiments suggested that inhibition of viral replication by curcumin may be able to reverse infection-induced host cell phenotypes. Importantly, the efficacy of curcumin treatment may be a cell cycle-regulated phenomenon, with curcumin exerting its antiviral activity and cell cycle rescue in the late G1/early S-phase cells.
FIGURE 6.
Curcumin reversed S-phase arrest induced by MP12 infection. A, HSAECs were synchronized by serum starvation for 72 h prior to infection. After infection, cells were analyzed for cell cycle phenotypes by FACS. Each histogram demonstrates relative distributions of cells in the G1, S, and G2 phases of the cell cycle. In each histogram, the y axis refers to collected events (cell counts), and the x axis shows fluorescence from propidium iodide stain. The three histograms show the relative distributions of cells in G1, S, and G2 phases of the cell cycle in mock-infected HSAECs, MP-12-infected HSAECs, and curcumin-treated HSAECs infected with MP-12. B, relative percentages of cells in G1, S, and G2/M phases were quantified. C, changes in total protein levels of p21/Waf1, cyclin A, and cyclin D were determined by Western blot analysis of serum-starved, MP-12-infected, and curcumin-treated cells in comparison with DMSO-treated or untreated cells. β-actin was determined as a control.
Curcumin-based Down-regulation of RVFV Replication Is Likely due to Phosphorylation of NSs and Cell Cycle Changes
We sought to arrive at mechanistic explanations for the inhibition of RVFV replication by curcumin. We approached this question by using two independent assays. We first addressed the important issue of whether IKK-β2 could function as a kinase for any of the viral proteins. Accordingly, we asked whether IKK-β2 phosphorylated NSs protein. We chose NSs because there is evidence in the literature that the nuclear and cytoplasmic forms of NSs can be phosphorylated at two different serine residues, 252 and 256 (16). We performed in vitro kinase assays using immunoprecipitated IKK-β2 (fractions from MP-12- and UV-MP-12-infected cells as the source of the kinase) and a GST-NSs substrate similar to that used in the studies carried out in Fig. 5A. The results shown in Fig. 7A demonstrate that NSs can be phosphorylated by IKK-β2 in vitro. Additionally, we observed that this phosphorylation event was sensitive to curcumin, as even low concentrations of curcumin could inhibit phosphorylation (Fig. 7A, compare lanes 4 and 5 with lane 3). Therefore, it is possible that curcumin exerts its inhibitory influence on RVFV replication by interfering with the IKK-β2-mediated phosphorylation of NSs.
FIGURE 7.
Inhibition by curcumin may be due to its influence on NSs phosphorylation and cell cycle progression. A, phosphorylation of a GST-NSs substrate by IKK-β2 was determined by in vitro kinase assay. Specifically, fractions 34–38 from MP-12-infected cell extracts were pooled and immunoprecipitated with anti-IKK-β antibody. Phosphorylation reactions were carried out with the immunoprecipitated material and a GST-NSs substrate. To determine the effects of curcumin on the phosphorylation reaction, the reaction was carried out in the presence of increasing concentrations of curcumin (0.1 and 1 μm). After incubation, samples were separated on a 4–20% Tris-glycine gel, dried, and analyzed by PhosphorImager (Amersham Biosciences). B, efficacy of NSs phosphorylation by IKK-β (obtained from pooled fractions 20–25) and IKK-β2 (obtained from pooled fractions 35–40) was compared by titrating increasing concentrations of both enzymes (5, 50, and 500 ng) in the presence of a constant amount of GST-NSs substrate. After incubation of the substrate with increasing concentrations of enzymes, the substrate was electrophoresed and analyzed by a PhosphorImager. C, relative binding of phosphorylated NSs with mSin3A and HDAC3 was evaluated by incubating uninfected whole cell extract with phosphorylated GST-NSs (using IKK-β2 and ATP). The same reaction without ATP (lanes 2 and 4) was set up alongside as a negative control. The next day, the reaction was washed three times with TNE-50 (0.1% Nonidet P-40) buffer and run on a 4–20% Tris-glycine gel. The proteins were then transferred to a nitrocellulose membrane and Western-blotted for mSin3A and HDAC3. D, HSAECs were synchronized at either G0/G1 phase (Sync-G1) by serum starvation or at late G1/early S phase (Sync-S) by treatment with hydroxyurea for up to 48 h and then infected with MP-12 (m.o.i. = 0.1). Cells were maintained in serum starvation conditions or in hydroxyurea for 24 h post-infection, and infectious progeny were determined by plaque assays. UT, untreated. E, similar experiment as described in D, except that following infection cells were maintained in complete medium for 24 h after which supernatants were analyzed for infectious virus by plaque assays.
We performed a comparative phosphorylation assay of NSs using IKK-β (fractions 20–24) and IKK-β2 (fractions 34–38) to determine the relative efficacies of either kinase on the NSs substrate. To that end, GST-NSs was phosphorylated using increasing concentrations (5, 50, and 500 ng) of either immunoprecipitated IKK-β or IKK-β2. It was striking to note that there was a strong increase in phosphorylated NSs in the presence of increasing amounts of IKK-β2 (Fig. 7B, lanes 5 and 6). In comparison, even though we could detect a band corresponding to phosphorylated NSs in the presence of highest amount of IKK-β (Fig. 7B, lane 3), the extent of phosphorylation with IKK-β2 was 6-fold higher (compare lanes 3 and 6), thus suggesting that IKK-β2 is a better kinase when it comes to phosphorylating NSs. The band corresponding to phosphorylated NSs was not observed when GST alone was used as a substrate for IKK-β2 or IKK-β at the highest concentration of enzyme (Fig. 7B, lanes 1 and 2). This experiment provided critical evidence that the phosphorylation of NSs may be more strongly influenced by the IKK-β2 enzyme.
Next, we performed an experiment to address the consequences of NSs phosphorylation. Specifically, we asked the question of whether phosphorylated NSs would be able to interact with and bind better to other inhibitory proteins such as mSin3A and HDAC3, which are known to co-localize with NSs at the interferon promoter (16). To perform this in vitro binding assay, we utilized IKK-β2 immunoprecipitated from fractions 33–37 in a kinase assay to phosphorylate GST-NSs in the presence of cold ATP. We performed the same reaction with [γ-32P]ATP alongside to ensure that GST-NSs was indeed phosphorylated by IKK-β2 (data not shown). As a negative control, we carried out the same reaction in the absence of ATP. We then incubated the phosphorylated NSs protein (or unphosphorylated control NSs protein) with HSAEC whole cell extracts (as the source of mSin3A and HDAC3) overnight and then washed the beads three times with TNE-50 (0.1% Nonidet P-40) buffer. The material was then electrophoresed on a 4–20% gel, and proteins were transferred to nitrocellulose membranes and Western-blotted using anti-mSin3A and HDAC3 antibodies to compare the relative interaction of mSin3A and HDAC3 with either phosphorylated NSs or unphosphorylated NSs. The data shown in Fig. 7C indicate that a higher amount of mSin3A was pulled down along with NSs when NSs was phosphorylated (Fig. 7C, compare lanes 4 and 5). Interestingly, phosphorylated NSs did not appear to have any significant enhancement of binding with HDAC3 (Fig. 7C, HDAC3 panel, compare lanes 4 and 5), suggesting that the influence of phosphorylated NSs may be specific to some proteins such as mSin3A.
As the second step, to correlate with the observation that the late G1/early S-phase cells were effective responders to curcumin, we asked if these cells were effective hosts for viral infection as well. To answer that question, we arrested cells in late G1/early S phase using hydroxyurea. As controls, we maintained unsynchronized cells and cells that were arrested in G0/G1 using serum starvation. We infected the unsynchronized and synchronized cells with MP-12 virus, maintained them in a serum-starved or hydroxyurea-treated condition for 24 h, and then collected supernatants for plaque assays. As seen in Fig. 7D, infection of cells synchronized in late G1/early S phase resulted in a modest increase of virus. Therefore, as suggested earlier, one way by which curcumin may inhibit the virus could be by lowering the population of cells in the late G1/early S transition stage (Fig. 6, A and B).
As a control, we performed the same experiment as outlined above, but released the cells in complete medium after infection for 24 h, after which we collected the supernatants for plaque assays. As seen in Fig. 7E, an increased level of virus can be detected in the cells that were arrested originally at G0/G1 stage. This may be the result of the arrested cells re-entering the cell cycle, specifically, entering into early S phase now due to the addition of complete medium. Along similar lines, cells that were originally arrested at early S phase by hydroxyurea treatment are likely to have proceeded into later stages of the cell cycle, such as G2/M or early G1, and therefore are not ideal hosts for RVFV replication as evidenced by the low levels of virus released (Fig. 7E). Hydroxyurea treatment affects the cellular nucleoside and nucleotide pools, which could influence viral replication. This may explain the modest change in virus levels seen in Fig. 7D, whereas removal of hydroxyurea (Fig. 7E) resulted in a more robust change.
Therefore, collectively our data point to the possibility that IKK-β2 may phosphorylate NSs, which will influence the ability of NSs to bind with specific host proteins such as mSin3A to shut down host transcription. One of the consequences of this phenomenon may be a stalled cell cycle (S-phase arrest) that is NSs-dependent.
Curcumin Inhibits ZH501 Replication
We then asked whether curcumin can exert its inhibitory effect against ZH501, the fully virulent form of RVFV. To test that possibility, we adopted a similar approach as with the attenuated strain (MP-12). Briefly, A549 cells were pretreated with curcumin (10 μm) for 2 h and then infected with ZH501 virus (m.o.i. = 0.1). The infected cells that were not treated with curcumin were maintained as controls alongside. Supernatants were obtained at 24 h post-infection and analyzed by plaque assays for extracellular virus levels. As shown in Fig. 8A, there was a four log drop in the levels of extracellular virus in the presence of curcumin. We carried out validation studies alongside using MP-12 virus in the same cell type and were able to observe a robust inhibition of MP-12 as well (Fig. 8B). Based on toxicity studies (Fig. 8C), we determined that the inhibition of extracellular virus by curcumin was not a consequence of cell death. Cumulatively, the experiments performed with ZH501 virus demonstrated that curcumin treatment can down-regulate replication of virulent virus.
FIGURE 8.
Curcumin inhibits ZH501 replication. A, A549 cells were infected with ZH501 at m.o.i. = 0.1 and were either treated with curcumin or left untreated (UT) for 24 h. Culture supernatants were then analyzed for infectious virus by plaque assays. B, A549 cells were infected with MP-12 virus at m.o.i. = 0.1, and the extent of inhibition was determined by plaque assays. C, inhibitor toxicity was evaluated by measuring survival of inhibitor-treated A549 cells after 24-h treatment with curcumin.
Treatment of INFAR−/− Mice with Curcumin Decreases Hepatic Viral Load
Our cell culture studies revealed that curcumin is a strong inhibitor of MP-12 and ZH501 replication. We next asked whether curcumin can exert a similar influence on viral replication in vivo. We used the INFAR−/− mouse model, as this is the only model that has been documented to manifest disease and mortality following infection by MP-12 virus (8, 45), and performed a preliminary proof-of-concept experiment.
Our specific question was whether curcumin treatment can down-regulate virus in the liver of MP-12-infected mice, as the liver is a prominently affected organ in RVFV infection. INFAR−/− mice were pretreated with curcumin 24 h prior to infection by the subcutaneous route. Mice infected by the intraperitoneal route with MP-12 virus (104 pfu/animal) continued to be treated with curcumin once every 24 h for a period of up to 4 days. All of the control untreated, infected animals were dead by 3 days post-infection, whereas 60% of the treated animals survived for up to 4 days after infection (Fig. 9A). We dissected out the liver from the infected animals and measured the hepatic viral load by plaque assays. In agreement with our in vitro data, the plaque assays revealed a strong reduction of viral titers in the livers (up to 90% decrease) of treated animals in comparison with the untreated controls (Fig. 9B). Cumulatively, our in vivo experiments using curcumin provide preliminary evidence that curcumin can down-regulate viral replication in vivo.
FIGURE 9.
Curcumin treatment reduces hepatic viral load in INFAR−/− mice. A, INFAR−/− mice (n = 5) were treated subcutaneously with DMSO or curcumin (60 mg/kg) every day. Concentrations of curcumin up to 300 mg/kg have been used by others in murine models (in some cases, for up to periods of 21 days with daily administration) with no apparent toxicity effects observed (59–63). All animals were infected with MP-12 virus at a concentration of 104 pfu/animal. All mice were pretreated with inhibitor at 24 h prior to infection and were post-treated for a total of 4 days post-infection. Percentage of survivors is shown in the y axis, whereas the number of days post-infection is shown in the x axis. B, liver tissue from infected, DMSO-treated and infected, and curcumin-treated mice was isolated, and infectious viral titers were determined by plaque assays.
DISCUSSION
RVFV is a category A select agent, an emerging infectious virus, and an agricultural pathogen that infects humans and livestock. Whereas in the case of human RVFV infections mortality rates up to 45% have been reported in certain instances, the infection of cattle and other livestock with RVFV results in an extreme 100% abortion rate. Treatment with generic antivirals such as ribavirin and supportive therapy are the only options currently available to treat Rift Valley fever. Understanding the interactions between the virus and the host is an important step toward designing effective therapeutics. The pursuit of host-based therapeutic intervention is an advantageous approach for multiple reasons, which include positive influences on the host mean survival time, decreased potential for evolution of resistant viruses, and broad-spectrum application potential.
Earlier RPMA studies using both ZH501 and MP-12 viruses revealed that the host NFκB signaling subunit, p65, is phosphorylated upon infection (18). This phosphorylation of p65 agrees with the reported nuclear migration and DNA binding of activated NFκB following RVFV infection as reported by Billecocq et al. (7). The term NFκB refers to a family of ubiquitously expressed, structurally related transcription factors, namely p65 (RelA), RelB, c-Rel, p50, and p52 (46, 47). These five proteins can form homo- or heterodimers that are usually held in the cytosol by the IκB family of inhibitory proteins. The p65/p50 dimer, the most abundant and well studied heterodimer, regulates a wide array of NFκB-responsive genes by binding to the κB site on the promoter. p65 is retained in the cytoplasm by IκBα, which, in the classical pathway, is phosphorylated by IKK-β and proteasomally degraded; this in turn leads to nuclear translocation of p65. Our in vitro studies with the MP-12 strain of RVFV demonstrated phosphorylation of p65 on serine 536 in multiple cell types (Fig. 1 and supplemental Fig. 1B). This phosphorylation event on p65 is known to increase its transactivation potential (46). We have also demonstrated that p65 is phosphorylated on serine 536 via the classical NFκB activation pathway. Similar results have been demonstrated for Rous sarcoma virus in a recent publication by Yoboua et al. (31). We were unable to detect alternate phosphorylation events on p65, such as phosphorylation of serine 276, within the time frames analyzed. Relevant to that observation, our RPMA studies with MP-12-infected HSAECs revealed that MSK1, one of the upstream kinases that phosphorylates p65 on serine 276, does not display significant changes in phosphorylation at such early time points (data not shown).
The IKK complex is one of the kinase complexes that phosphorylates p65 on serine 536 (47). IKK is a macromolecular signaling complex or the signalosome that consists of two related kinases, IKK-α and IKK-β, and a third regulatory subunit, IKK-γ (NEMO), of which the exact mechanistic role in the trimeric complex is still unclear. Although both the IKK-α and IKK-β kinases can phosphorylate IκBα, IKK-β-mediated phosphorylation is more rapid, dramatically more efficient, and corresponds to the classical activation cascade, which is what we observed relevant to RVFV infection. Considering that the entire viral life cycle is completed within a matter of a few hours (∼8–10 h between entry and exit), a rapid phosphorylation and activation would be more plausible, in contrast to a delayed IKK-α-mediated alternative activation cascade. Under stringent isolation parameters, the IKK complex demonstrates an apparent molecular mass of ∼700–900 kDa. The IKK complex that is common to both MP-12 virus- and UV-MP-12 virus-infected cells migrated at an approximate molecular range of 600–900 kDa in our sizing columns (Fig. 2A). A novel IKK-β-containing low molecular weight complex, which we refer to as IKK-β2, eluted around 300 kDa in the case of MP-12-infected cells (Fig. 2A). Interestingly, such a phenomenon does not happen in the case of infection by a DNA virus or by other NFκB inducers such as TNFα (Fig. 2, B and C). In contrast to our observation following RVFV infection, the IKK-β profile was shifted toward the higher molecular weight range in the case of infection by at least one DNA virus (Fig. 2B), suggesting that IKK-β2 may be a phenomenon that is associated with RVFV infection. It would be interesting to determine whether IKK-β2 is associated with infection by other RNA viruses as well. Our proteomic analysis of the composition of IKK-β2 indicated that this complex contains multiple chaperones, including Hsp70 and Hsp90, that are also associated with the IKK-β complex (Fig. 2D). However, interestingly, the IKK-β2 complex appeared to be missing certain regulatory components, such as inhibitory enzymes and proteasome units, that were seen in association with IKK-β, thus suggesting altered regulation (Fig. 2D). IKK-α, in some instances, has been suggested to down-regulate the kinase activity of IKK-β (48). Additionally, the absence of proteasome subunits and inhibitory enzymatic components hints at the possibility that the IKK-β2 complex may respond to a different set of activator-signaling components and have more of an increased and/or constitutive kinase activity than the regular IKK-α-, IKK-β-, and IKK-γ-containing signalosome. An in vitro kinase assay using immunoprecipitated IKK-β2 demonstrated kinase activity, suggesting that this complex may be functional in RVFV-infected cells (Fig. 5A). In support of such a possibility, the screening of multiple inhibitors of the NFκB cascade singled out inhibitors of the IKK-β component as being most effective in the down-regulation of RVFV replication (Figs. 3 and 4). Thus, it is possible that the novel IKK-β2 complex observed in RVFV-infected cells is a hyperactive enzyme that lacks the control that could be exerted normally on the IKK-β complex (43).
The IKK component of the NFκB signaling cascade is a critical node that is often exploited by multiple viruses to either suppress the host innate immune response or enhance viral replication potential. Murine γ-herpesvirus (gamma-HV68), a model system for KSHV and EBV, is known to activate IKK-β. Interestingly, activated, phosphorylated IKK-β promotes viral replication, because the viral protein RTA (replication and transcription activator) is a phosphorylation target for IKK-β kinase activity (49). In contrast to this situation, in the case of vaccinia virus infection, the viral protein B14 modulates IKK-β kinase activity in a way that inhibits the phosphorylation of its downstream target IκBα without exerting any influence on IKK-α activity (50, 51). In a similar manner, enterovirus 71 2C protein is known to target IKK-β activity by interacting with IKK-β, which is a critical event in the infectious process (52). In the case of EBV infection, the viral protein EBNA1 negatively regulates IKK activation by inhibiting phosphorylation of IKK-α/β (53). There are some interesting lines of evidence in the literature indicating that viruses can influence the functionality of IKK-β by altering the composition of the canonical IKK-β complex. A herpes simplex virus, virulence factor γ(49) 34.5, is known to block dendritic cell maturation and hence influence the host innate immune response by associating with IKK-α/β. Importantly, the association of γ(49) 34.5 with the canonical IKK-α/β complex now alters the protein composition by recruiting protein phosphatase 1, which by dephosphorylating IKK-β influences the host innate immune response (54). Bovine foamy virus, infection by which results in a persistent activation of the NFκB cascade, uses its transactivator protein BTas to keep the NFκB cascade persistently active. To achieve this end, the viral transactivator interacts with the IKK-α/β complex as demonstrated by co-immunoprecipitation experiments (55).
At this juncture, we hypothesized that the kinase function of IKK-β2 may play a role in the viral replication cycle beyond influencing host transcription. This hypothesis was also based on the observation by Ikegami et al. (12) that inhibition of host basal transcription by generic inhibitors such as α-amanitin does not result in down-regulation of virus. It would be interesting to determine whether any of the viral proteins are phosphorylated by IKK-β2. To that effect, our in vitro kinase studies with a GST-NSs substrate revealed that NSs may be phosphorylated by IKK-β2 (Fig. 7, A and B). Additionally, phosphorylated NSs appeared to interact better in an in vitro binding assay with specific inhibitory host components such as mSin3A (Fig. 7C), whereas certain other components of transcriptional repressive complexes such as HDAC3 remained unaffected by a phosphorylated NSs. The interaction of NSs with transcription repressor proteins such as mSin3A is a key step in the viral suppression of the host innate immune response. Therefore, we suspect that a critical aspect of an activated NFκB cascade in the context of RVFV replication is the activation of the IKK-β complex, which is likely to exert its effect on NSs in the nucleus. This speculation is supported by our observation that inhibitors that interfered with these two functions (Fig. 3B and supplemental Fig. 2) negatively influenced RVFV replication, whereas those that did not influence these two functions had no effect on viral replication. Additionally, although we focused on the interaction between NSs and IKK-β2, it may be possible that such a phenomenon may extend to other viral proteins as well and have influences on virus-induced pathology. With regards to the formation of IKK-β2, although it is interesting to speculate whether the viral protein NSs may have any role to play in the formation of IKK-β2, qRT-PCR studies do not demonstrate any significant difference in replication competencies between the wild type MP-12 virus and the NSs mutant virus at early time points (Fig. 1C). We suspect that formation of IKK-β2 may be associated with infection by a replication-competent virus, and data shown with UV-inactivated MP-12 (Fig. 2A) provide support for this idea. Therefore, based on current data, we do not believe that the formation of IKK-β2 is linked directly to NSs function. However, as shown in Fig. 7A, NSs can be phosphorylated by IKK-β2 and is therefore a potential substrate for this kinase.
Among the multiple IKK inhibitors that showed efficacy against RVFV, curcumin emerged as the most potent, with about 3–4 logs of inhibition of virus in treated cells, which included reduced total genomic copies and lowered infectivity (Figs. 3 and 4). Our experiments also demonstrated that curcumin directly bound to IKK-β2 and suppressed its kinase function (Fig. 5). Curcumin is a well documented natural polyphenolic compound, which in recent years has been demonstrated to have extensive antiproliferative, antiviral, anti-arthritic, anti-amyloid, and anti-inflammatory properties (56–58).
We also observed that curcumin down-regulated viral replication in the liver of infected animals. Our current preliminary study was performed using well tolerated doses of curcumin in the context of other murine models (59–63). Our studies revealed that treatment of infected animals with curcumin resulted in the down-regulation of liver viremia (Fig. 9B). Further studies are ongoing to explore derivatives of curcumin that may confer better bioavailability at low doses and hence offer increased survival advantage to infected animals.
Collectively, our observations indicated that viral infections may cause alterations in macromolecular complexes such as IKK, which result in the presence of novel versions of enzymes, such as IKK-β2 observed in RVFV infection. These novel host components, by virtue of altered protein-protein interactions and function, may serve as therapeutic targets. There are multiple inhibitors of host signaling components that are currently FDA-approved and on the market for treatment of many cancers. The identification of altered host-signaling components will be a critical step in drug repositioning to utilize them in the treatment of infectious diseases as well.
Supplementary Material
Acknowledgments
We are grateful to Dr. Shinji Makino (University of Texas Medical Branch (at Galveston)) for generously providing us with the NSs (rMP-12-NSdel) and NSm (arMP-12-del21/384) strains. We also thank Dr. Sina Bavari (United States Army Medical Research Institute for Infectious Diseases) for the MP-12 strain of RVFV.
This work was supported by United States Department of Energy Grant DE-FC52-04NA25455 (to C. B. and F. K.).

This article contains supplemental Figs. 1–3.
- RVFV
- Rift Valley fever virus
- RPMA
- reverse-phase protein microarray
- HSAEC
- human small airway lung epithelial cell
- IKK
- IκB kinase
- NEMO
- NFκB essential modulator
- m.o.i.
- multiplicity of infection
- DMSO
- dimethyl sulfoxide
- qRT-PCR
- quantitative RT-PCR
- KSHV
- Kaposi sarcoma herpesvirus
- CAPE
- caffeic acid phenethyl ester.
REFERENCES
- 1. Flick R., Bouloy M. (2005) Rift Valley fever virus. Curr. Mol. Med. 5, 827–834 [DOI] [PubMed] [Google Scholar]
- 2. LaBeaud A. D., Kazura J. W., King C. H. (2010) Advances in Rift Valley fever research: insights for disease prevention. Curr. Opin. Infect. Dis. 23, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ikegami T., Makino S. (2009) Rift valley fever vaccines. Vaccine 27, Suppl. 4, D69–D72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouloy M., Flick R. (2009) Reverse genetics technology for Rift Valley fever virus: current and future applications for the development of therapeutics and vaccines. Antiviral Res. 84, 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Won S., Ikegami T., Peters C. J., Makino S. (2007) NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 81, 13335–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Won S., Ikegami T., Peters C. J., Makino S. (2006) NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J. Virol. 80, 8274–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Billecocq A., Spiegel M., Vialat P., Kohl A., Weber F., Bouloy M., Haller O. (2004) NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78, 9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouloy M., Janzen C., Vialat P., Khun H., Pavlovic J., Huerre M., Haller O. (2001) Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J. Virol. 75, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dasgupta A. (2004) Targeting TFIIH to inhibit host cell transcription by Rift Valley fever virus. Mol. Cell 13, 456–458 [DOI] [PubMed] [Google Scholar]
- 10. Habjan M., Pichlmair A., Elliott R. M., Overby A. K., Glatter T., Gstaiger M., Superti-Furga G., Unger H., Weber F. (2009) NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83, 4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikegami T., Narayanan K., Won S., Kamitani W., Peters C. J., Makino S. (2009) Rift Valley fever virus NSs protein promotes post-transcriptional down-regulation of protein kinase PKR and inhibits eIF2α phosphorylation. PLoS Pathog. 5, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ikegami T., Narayanan K., Won S., Kamitani W., Peters C. J., Makino S. (2009) Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional down-regulation of protein kinase PKR. Ann. N.Y. Acad. Sci. 1171, Suppl. 1, E75–E85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikegami T., Peters C. J., Makino S. (2005) Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79, 5606–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalveram B., Lihoradova O., Ikegami T. (2011) NSs protein of Rift Valley fever virus promotes posttranslational down-regulation of the TFIIH subunit p62. J. Virol. 85, 6234–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le May N., Dubaele S., Proietti De Santis L., Billecocq A., Bouloy M., Egly J. M. (2004) TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116, 541–550 [DOI] [PubMed] [Google Scholar]
- 16. Le May N., Mansuroglu Z., Leger P., Josse T., Blot G., Billecocq A., Flick R., Jacob Y., Bonnefoy E., Bouloy M. (2008) A SAP30 complex inhibits IFN-β expression in Rift Valley fever virus-infected cells. PLoS Pathog. 4, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narayanan A., Popova T., Turell M., Kidd J., Chertow J., Popov S. G., Bailey C., Kashanchi F., Kehn-Hall K. (2011) Alteration in superoxide dismutase 1 causes oxidative stress and p38 MAPK activation following RVFV infection. PLoS One 6, e20354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Popova T. G., Turell M. J., Espina V., Kehn-Hall K., Kidd J., Narayanan A., Liotta L., Petricoin E. F., 3rd, Kashanchi F., Bailey C., Popov S. G. (2010) Reverse-phase phosphoproteome analysis of signaling pathways induced by Rift Valley fever virus in human small airway epithelial cells. PLoS One 5, e13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahmed K. M., Li J. J. (2007) ATM-NF-κB connection as a target for tumor radiosensitization. Curr. Cancer Drug Targets 7, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dent P., Yacoub A., Contessa J., Caron R., Amorino G., Valerie K., Hagan M. P., Grant S., Schmidt-Ullrich R. (2003) Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 159, 283–300 [DOI] [PubMed] [Google Scholar]
- 21. Haddad J. J., Saadé N. E., Safieh-Garabedian B. (2002) Redox regulation of TNF-α biosynthesis: augmentation by irreversible inhibition of γ-glutamylcysteine synthetase and the involvement of an IκB-α/NF-κB-independent pathway in alveolar epithelial cells. Cell. Signal. 14, 211–218 [DOI] [PubMed] [Google Scholar]
- 22. Pan D., Pan L. Z., Hill R., Marcato P., Shmulevitz M., Vassilev L. T., Lee P. W. (2011) Stabilisation of p53 enhances reovirus-induced apoptosis and virus spread through p53-dependent NF-κB activation. Br. J. Cancer 105, 1012–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pise-Masison C. A., Mahieux R., Radonovich M., Jiang H., Duvall J., Guillerm C., Brady J. N. (2000) Insights into the molecular mechanism of p53 inhibition by HTLV type 1 Tax. AIDS Res. Hum. Retroviruses 16, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 24. Rahman I., Marwick J., Kirkham P. (2004) Redox modulation of chromatin remodeling: impact on histone acetylation and deacetylation, NF-κB and pro-inflammatory gene expression. Biochem. Pharmacol. 68, 1255–1267 [DOI] [PubMed] [Google Scholar]
- 25. Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Cross-talk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 26. Filone C. M., Hanna S. L., Caino M. C., Bambina S., Doms R. W., Cherry S. (2010) Rift Valley fever virus infection of human cells and insect hosts is promoted by protein kinase Cϵ. PLoS One 5, e15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panchal R. G., Reid S. P., Tran J. P., Bergeron A. A., Wells J., Kota K. P., Aman J., Bavari S. (2012) Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antiviral Res. 93, 23–29 [DOI] [PubMed] [Google Scholar]
- 28. Ikegami T., Won S., Peters C. J., Makino S. (2006) Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80, 2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Drosten C., S., Schilling S., Asper M., Panning M., Schmitz H., S. (2002) Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40, 2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agbottah E., Yeh W. I., Berro R., Klase Z., Pedati C., Kehn-Hall K., Wu W., Kashanchi F. (2008) Two specific drugs, BMS-345541 and purvalanol A, induce apoptosis of HTLV-1-infected cells through inhibition of the NF-κB and cell cycle pathways. AIDS Res. Ther. 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoboua F., Martel A., Duval A., Mukawera E., Grandvaux N. (2010) Respiratory syncytial virus-mediated NF-κB p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKKβ. J. Virol. 84, 7267–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douillette A., Bibeau-Poirier A., Gravel S. P., J. F., V., Moreau P., Servant M. J. (2006) The proinflammatory actions of angiotensin II are dependent on p65 phosphorylation by the IκB kinase complex. J. Biol. Chem. 281, 13275–13284 [DOI] [PubMed] [Google Scholar]
- 33. Sripadi P., Shrestha B., Easley R. L., Carpio L., Kehn-Hall K., Chevalier S., Mahieux R., Kashanchi F., Vertes A. (2010) Direct detection of diverse metabolic changes in virally transformed and Tax-expressing cells by mass spectrometry. PLoS One 5, e12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sgarbanti M., Arguello M., tenOever B. R., Battistini A., Lin R., Hiscott J. (2004) A requirement for NF-κB induction in the production of replication-competent HHV-8 virions. Oncogene 23, 5770–5780 [DOI] [PubMed] [Google Scholar]
- 35. Sun Q., Matta H., Lu G., Chaudhary P. M. (2006) Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene 25, 2717–2726 [DOI] [PubMed] [Google Scholar]
- 36. Caselli E., Fiorentini S., Amici C., Di Luca D., Caruso A., Santoro M. G. (2007) Human herpesvirus 8 acute infection of endothelial cells induces monocyte chemoattractant protein 1-dependent capillary-like structure formation: role of the IKK/NF-κB pathway. Blood 109, 2718–2726 [DOI] [PubMed] [Google Scholar]
- 37. Milacic V., Banerjee S., Landis-Piwowar K. R., Sarkar F. H., Majumdar A. P., Dou Q. P. (2008) Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 68, 7283–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fernandez-Garcia M. D., Meertens L., Bonazzi M., Cossart P., Arenzana-Seisdedos F., Amara A. (2011) Appraising the roles of CBLL1 and the ubiquitin/proteasome system for flavivirus entry and replication. J. Virol. 85, 2980–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wyke S. M., Russell S. T., Tisdale M. J. (2004) Induction of proteasome expression in skeletal muscle is attenuated by inhibitors of NF-κB activation. Br. J. Cancer 91, 1742–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith H. J., Tisdale M. J. (2003) Signal transduction pathways involved in proteolysis-inducing factor induced proteasome expression in murine myotubes. Br. J. Cancer 89, 1783–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pasquini L. A., Paez P. M., Moreno M. A., Pasquini J. M., Soto E. F. (2003) Inhibition of the proteasome by lactacystin enhances oligodendroglial cell differentiation. J. Neurosci. 23, 4635–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shakibaei M., Csaki C., Nebrich S., Mobasheri A. (2008) Resveratrol suppresses interleukin-1β-induced inflammatory signaling and apoptosis in human articular chondrocytes: potential for use as a novel nutraceutical for the treatment of osteoarthritis. Biochem. Pharmacol. 76, 1426–1439 [DOI] [PubMed] [Google Scholar]
- 43. Kazi A., Daniel K. G., Smith D. M., Kumar N. B., Dou Q. P. (2003) Inhibition of the proteasome activity, a novel mechanism associated with the tumor cell apoptosis-inducing ability of genistein. Biochem. Pharmacol. 66, 965–976 [DOI] [PubMed] [Google Scholar]
- 44. Baer A., Austin D., Narayanan A., Popova T., Kainulainen M., Bailey C., Kashanchi F., Weber F., Kehn-Hall K. (2012) Induction of DNA damage signaling upon Rift Valley fever virus infection results in cell cycle arrest and increased viral replication. J. Biol. Chem. 287, 7399–7410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lorenzo G., Martín-Folgar R., Hevia E., Boshra H., Brun A. (2010) Protection against lethal Rift Valley fever virus (RVFV) infection in transgenic IFNAR(−/−) mice induced by different DNA vaccination regimens. Vaccine 28, 2937–2944 [DOI] [PubMed] [Google Scholar]
- 46. Viatour P., Merville M. P., Bours V., Chariot A. (2005) Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30, 43–52 [DOI] [PubMed] [Google Scholar]
- 47. Häcker H., Karin M. (2006) Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 48. O'Mahony A., Lin X., Geleziunas R., Greene W. C. (2000) Activation of the heterodimeric IκB kinase α (IKKα)-IKKβ complex is directional: IKKα regulates IKKβ under both basal and stimulated conditions. Mol. Cell. Biol. 20, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong X., Feng P. (2011) Murine γ herpesvirus 68 hijacks MAVS and IKKβ to abrogate NFκB activation and antiviral cytokine production. PLoS Pathog. 7, e1002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benfield C. T., Mansur D. S., McCoy L. E., Ferguson B. J., Bahar M. W., Oldring A. P., Grimes J. M., Stuart D. I., Graham S. C., Smith G. L. (2011) Mapping the IκB kinase beta (IKKβ)-binding interface of the B14 protein, a vaccinia virus inhibitor of IKKβ-mediated activation of nuclear factor κB. J. Biol. Chem. 286, 20727–20735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen R. A., Ryzhakov G., Cooray S., Randow F., Smith G. L. (2008) Inhibition of IκB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 4, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng Z., Li H., Zhang Z., Meng J., Mao D., Bai B., Lu B., Mao P., Hu Q., Wang H. (2011) Enterovirus 71 2C protein inhibits TNF-α-mediated activation of NF-κB by suppressing IκB kinase β phosphorylation. J. Immunol. 187, 2202–2212 [DOI] [PubMed] [Google Scholar]
- 53. Valentine R., Dawson C. W., Hu C., Shah K. M., Owen T. J., Date K. L., Maia S. P., Shao J., Arrand J. R., Young L. S., O'Neil J. D. (2010) Epstein-Barr virus-encoded EBNA1 inhibits the canonical NF-kappaB pathway in carcinoma cells by inhibiting IKK phosphorylation. Mol. Cancer 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jin H., Yan Z., Ma Y., Cao Y., He B. (2011) A herpesvirus virulence factor inhibits dendritic cell maturation through protein phosphatase 1 and IκB kinase. J. Virol. 85, 3397–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang J., Tan J., Zhang X., Guo H., Zhang Q., Guo T., Geng Y., Qiao W. (2010) BFV activates the NF-κB pathway through its transactivator (BTas) to enhance viral transcription. Virology 400, 215–223 [DOI] [PubMed] [Google Scholar]
- 56. Aggarwal B. B., Harikumar K. B. (2009) Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune, and neoplastic diseases. Int. J. Biochem. Cell Biol. 41, 40–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou H., Beevers C. S., Huang S. (2011) The targets of curcumin. Curr. Drug Targets 12, 332–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wan S. B., Yang H., Zhou Z., Cui Q. C., Chen D., Kanwar J., Mohammad I., Dou Q. P., Chan T. H. (2010) Evaluation of curcumin acetates and amino acid conjugates as proteasome inhibitors. Int. J. Mol. Med. 26, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dattani J. J., Rajput D. K., Moid N., Highland H. N., George L. B., Desai K. R. (2010) Ameliorative effect of curcumin on hepatotoxicity induced by chloroquine phosphate. Environ. Toxicol. Pharmacol. 30, 103–109 [DOI] [PubMed] [Google Scholar]
- 60. Tu C. T., Han B., Yao Q. Y., Zhang Y. A., Liu H. C., Zhang S. C. (2012) Curcumin attenuates concanavalin A-induced liver injury in mice by inhibition of Toll-like receptor (TLR) 2, TLR4, and TLR9 expression. Int. Immunopharmacol. 12, 151–157 [DOI] [PubMed] [Google Scholar]
- 61. Tu C. T., Han B., Liu H. C., Zhang S. C. (2011) Curcumin protects mice against concanavalin A-induced hepatitis by inhibiting intrahepatic intercellular adhesion molecule-1 (ICAM-1) and CXCL10 expression. Mol. Cell. Biochem. 358, 53–60 [DOI] [PubMed] [Google Scholar]
- 62. Yu W. G., Xu G., Ren G. J., Xu X., Yuan H. Q., Qi X. L., Tian K. L. (2011) Preventive action of curcumin in experimental acute pancreatitis in mouse. Indian J. Med. Res. 134, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oh S. W., Cha J. Y., Jung J. E., Chang B. C., Kwon H. J., Lee B. R., Kim D. Y. (2011) Curcumin attenuates allergic airway inflammation and hyper-responsiveness in mice through NF-κB inhibition. J. Ethnopharmacol. 136, 414–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.