Background: Ketone bodies have been implicated not only in energy metabolism, but also in lipogenesis.
Results: Discovery and characterization of human extramitochondrial HMG-CoA lyase-like protein (HMGCLL1) has been accomplished.
Conclusion: Catalytically active HMGCLL1 is myristoylated and vesicle associated.
Significance: Extramitochondrial HMG-CoA lyase may be crucial to lipid biosynthesis or to energy metabolism in certain tissues and cancer cells.
Keywords: Acetoacetate, Acetyl Coenzyme A, Energy Metabolism, Enzymes, Ketone Body Metabolism, Lipid Synthesis, HMG-CoA
Abstract
3-Hydroxy-3-methylglutaryl-CoA lyase-like protein (HMGCLL1) has been annotated in the Mammalian Genome Collection as a previously unidentified human HMG-CoA lyase (HMGCL). To test the validity of this annotation and evaluate the physiological role of the protein, plasmids were constructed for protein expression in Escherichia coli and Pichia pastoris. Protein expression in E. coli produced insoluble material. In contrast, active HMGCLL1 could be recovered upon expression in P. pastoris. Antibodies were prepared against a unique peptide sequence found in the N terminus of the protein. In immunodetection experiments, the antibodies discriminated between HMGCLL1 and mitochondrial HMGCL. Purified enzyme was characterized and demonstrated to cleave HMG-CoA to acetoacetate and acetyl-CoA with catalytic and affinity properties comparable with human mitochondrial HMGCL. The deduced HMGCLL1 sequence contains an N-terminal myristoylation motif; the putative modification site was eliminated by construction of a G2A HMGCLL1. Modification of both proteins was attempted using human N-myristoyltransferase and [3H]myristoyl-CoA. Wild-type protein was clearly modified, whereas G2A protein was not labeled. Myristoylation of HMGCLL1 affects its cellular localization. Upon transfection of appropriate expression plasmids into COS1 cells, immunofluorescence detection indicates that G2A HMGCLL1 exhibits a diffuse pattern, suggesting a cytosolic location. In contrast, wild-type HMGCLL1 exhibits a punctate as well as a perinuclear immunostaining pattern, indicating myristoylation dependent association with nonmitochondrial membrane compartments. In control experiments with the HMGCL expression plasmid, protein is localized in the mitochondria, as anticipated. The available results for COS1 cell expression, as well as endogenous expression in U87 cells, indicate that HMGCLL1 is an extramitochondrial hydroxymethylglutaryl-CoA lyase.
Introduction
3-Hydroxy-3-methylglutaryl-CoA (HMG-CoA) lyase (EC 4.1.3.4; HMGCL)2 catalyzes a cation-dependent cleavage of substrate into acetyl-CoA and acetoacetate (1) (Scheme 1).
SCHEME 1.
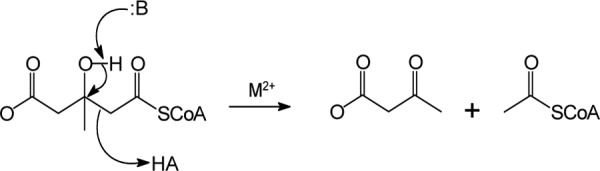
A series of functional and structural studies on human HMGCL, recently summarized in a paper by Fu et al. (2), have demonstrated that residues critical to reaction catalysis include Arg-41, Asp-42, and Cys-266. This reaction is a key step in ketogenesis, the products of which support energy production in nonhepatic animal tissues (e.g. muscle and, after adaptation, brain and nervous tissue (3)).
Ketogenesis is particularly important to human metabolism during the perinatal period and during fasting or starvation. In accordance with these physiological roles, it is not surprising that gene knock-out in mice results in embryonic lethality (4). The physiological importance of the enzyme in humans is underscored by the observation that many mutations that diminish HMGCL activity correlate with inherited metabolic disease. Outcome of this inherited disease can be lethal if uncontrolled (5). An extensive compilation of missense human HMGCL mutations has been included in a recent review article (6).
Other metabolic roles, e.g. a function in biosynthesis, have been suggested by work in transformed cells or tissues. In work with rat hepatoma cells, Shrago's lab (7) reported that ketone bodies are converted to cholesterol and fatty acids using a cytoplasmic pathway. In neuroblastoma and glioma cells, acetoacetate has been shown to be a preferred substrate for neural lipid synthesis (8). Recently, transcriptional profiling of human breast cancer tumor stroma (9) has indicated up-regulation effects for HMG-CoA synthase (HMGCS2) and the HMG-CoA lyase-like protein (HMGCLL1). The results prompted speculation that these proteins should be included as “druggable” candidates for chemotherapeutic targeting. The study also included metabolomic profiling work. The results were interpreted to suggest a role for ketone body production in tumor stromal cells as a mechanism to provide fuel for energy production in adjacent epithelial cancer cells.
Implication of HMG-CoA lyase-like protein (HMGCLL1) by transcriptional profiling may raise new interest in ketone body metabolism. The HMGCLL1 gene, which maps to human chromosome 6, is distinct from the HMGCL gene, which maps to chromosome 1 and encodes the mitochondrial enzyme traditionally associated with ketogenesis. It has not yet been established that expression of the HMGCLL1 gene produces an enzyme with any ketogenic function. Additionally, the possibility of extramitochondial acetoacetate biosynthesis has not been addressed in the context of this potential HMG-CoA lyase homolog. This report addresses these issues in work that includes the engineering of an expression plasmid containing the HMGCLL1 coding sequence and the consequent expression of the protein in Pichia pastoris. The protein has been isolated; characterization has provided a test of enzymatic functionality. Moreover, the possibility of post-translational modification of HMGCLL1 has been investigated to discover and evaluate additional contrasts between this enzyme and mitochondrial HMG-CoA lyase. A preliminary account of these studies has appeared (10).
EXPERIMENTAL PROCEDURES
Materials
Primers used for amplification and mutagenesis were synthesized by Integrated DNA Technologies. DNA sequencing was performed at the DNA Core Facility, University of Missouri, Columbia, MO. Edman analyses were performed at the Iowa State University Protein Core Facility. The Easy Select Pichia Expression Kit and Zeocin were purchased from Invitrogen. [9,10-3H]Myristic acid was obtained from PerkinElmer Life Sciences. Antibodies against Golgin 58 and GM-130 protein were generously provided by Prof. Carolyn Machamer (Johns Hopkins University). Antibody against PEX-14 was provided by Stephanie Mihalik (Children's Hospital of Pittsburgh).
Plasmid Construction
The HMGCLL1 coding sequence was amplified from IMAGE Clone ID 4818781 (11) by PCR using primers complimentary to the 5′ and 3′ ends of the gene. For insertion of the gene into the pPICZ A vector (Invitrogen), the 5′ primer (5′-AAAAAAGAATTCATAATGGGGAATGTGCCATCCGCG-3′) encoded an EcoRI site (underlined) and a partial yeast Kozak consensus sequence (12) (boldface), which includes the ATG start codon; the 3′ primer (5′-AAAAAACTCGAGTCAATGATGATGATGATGATGAGCATTGAAGG-3′) encoded six histidines (boldface) followed by a stop codon (italicized) and finally an XhoI site (underlined). A G2A HMGCLL1 mutant was also constructed by amplifying the gene using the 5′ primer (5′-AAAAAAGAATTCATAATGGCGAATGTGCCATCCGCG-3′), which includes the single base substitution G5C (italicized), coding for an alanine at position 2 instead of glycine, an EcoRI site (underlined) and a partial yeast Kozak consensus sequence (boldface), which includes the ATG start codon. For insertion of the gene into the pET23d vector (Novagen), the 5′ primer (5′-GCCGCCACAGGCTCCGCCA-3′) encoded a partial NcoI site (underlined); the 3′ primer (5′AAAAAAGGATCCTCAATGATGATGATGATGATGAGCATTGAAGG-3′) encoded six histidines (boldface) followed by a stop codon (italicized) and finally a BamHI site (underlined). The amplified DNA was digested with the restriction endonucleases EcoRI and XhoI or NcoI and BamHI and gel purified. The purified restriction fragment was ligated with a similarly digested and purified pPICZ A or pET23d vector, respectively. The ligations produced constructs pPICZ-HLL1-His6, pPICZ-HLL1-G2A-His6, and pET23-HLL1-His6, which encode full-length HMGCLL1 with a C-terminal His6 tag. DNA sequence analysis was used to verify the integrity of the final products.
Mutagenesis
A pET23-HLL1-G2A-His6 mutant was generated using full circle PCR according to the QuikChange site-directed mutagenesis protocol from Stratagene. The WT pET23-HLL-His6 construct was used as a template. The mutation was verified by DNA sequence analysis. Forward and reverse mutagenic primer sequences (mutagenic bases underlined) are as follows: CLL1-G2A-F, 5′-GGAGATATACCATGGCGAATGTGCCATCCGCG-3′; CLL1-G2A-R, 5′-CGCGGATGGCACATTCGCCATGGTATATCTCC-3′.
Solubilization of HMGCLL1 from Inclusion Bodies
Escherichia coli cell pellets containing HMGCLL1 expressed using pET-23-derived plasmids that contain the coding sequences for WT or G2A protein were prepared from 1 liter of ampicillin containing culture. Cell pellets were resuspended in 50 ml of ice-cold lysis buffer containing 50 mm Tris (pH 8.0), 1 mm EDTA, and 25% (w/v) sucrose. Immediately before cell disruption, 1 mm PMSF, 1 unit/ml of DNase I, and 5 mm mercaptoethanol were added. Cells were mechanically disrupted by passing twice through a microfluidizer at ∼17 kpsi. The lysate was centrifuged at 17,400 × g for 20 min at 4 °C to collect inclusion bodies. The remaining steps were carried out at room temperature. The pellet was resuspended by homogenization (∼5 strokes) in 40 ml of wash buffer containing 20 mm Tris (pH 8.0), 200 mm NaCl, 1% (w/v) sodium deoxycholate, and 2 mm EGTA. The suspension was centrifuged and the pellet was resuspended as above three times in 40 ml of wash buffer containing 10 mm Tris (pH 8.0), 0.25% (w/v) sodium deoxycholate, and 1 mm EGTA. The final pellet was resuspended by homogenization (∼10 strokes) in buffer containing 10 mm Tris (pH 8.0), 8 m urea (or 6 m guanidinium HCl). The suspension was diluted to 6 m urea, centrifuged as above, and the supernatant was stored at room temperature until use. Protein concentration was determined by the method of Bradford (13). A typical expression and solubilization yields about 100 mg of protein.
Antibody Production and Purification
A synthetic peptide corresponding to residues 19–37 of human HMGCLL1 (a region that is not conserved in mitochondrial HMG-CoA lyase; Fig. 1) was produced, conjugated to keyhole limpet hemocyanin, and used to raise antibodies in rabbits (Global Peptide Services, Fort Collins, CO). For immunofluorescence microscopy, antibodies were purified essentially as described by Pringle et al. (14). Protein solubilized from inclusion bodies (described above) was purified by immobilized metal affinity chromatography. A 12% SDS gel was prepared using a two-dimensional electrophoresis comb to produce a single 6.7-cm wide well. Sixty-seven μg of purified HMGCLL1 were loaded evenly across the well and the sample was run for 10 min at 110 V. This was repeated five more times to produce a total of 6 strips of protein. Following the final load, the gel was allowed to run until the dye front was ∼1 cm from the bottom of the gel. The protein was transblotted to nitrocellulose overnight. The blot was stained with Ponceau S, strips containing the protein bands were cut out, destained with TBS (20 mm Tris, pH 7.5, containing 500 mm NaCl), and stored dry until use. Crude, nondiluted antiserum was applied to the blocked strips and incubated for 2 h at room temperature, with agitation. The protein strips with bound antibody were washed 3 times in TBS, the antibodies were eluted with 0.2 m glycine (pH 2.5), 1 mm EGTA, and immediately neutralized with 1 m Tris (pH 8.0). The purified antibodies were buffer exchanged into PBS with 0.1% (w/v) BSA by centrifugal ultrafiltration to a final antibody concentration of about 1 mg/ml (as determined by A280; ϵ1% = 13.5) and stored in aliquots at −80 °C until use.
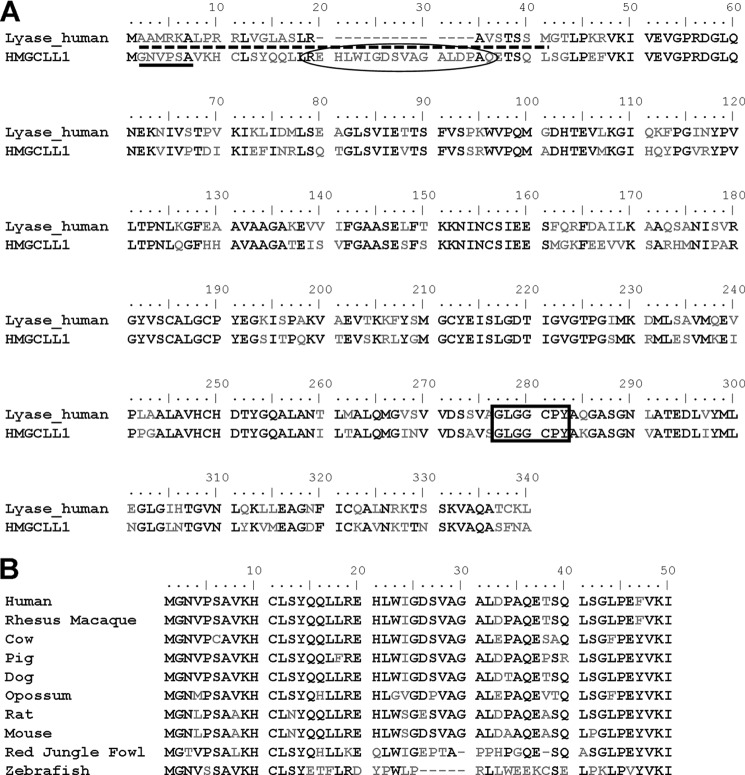
FIGURE 1.
Sequence comparisons of HMG-CoA lyase and HMGCLL1 proteins. A, alignment comparing the sequences of human HMG-CoA lyase and HMGCLL1; identical residues are boldfaced. A dashed underline indicates a mitochondrial leader sequence. A solid underline denotes residues consistent with an N-myristoylation motif. Boxed residues represent the HMG-CoA lyase signature sequence. Residues corresponding to the anti-HMGCLL1 antibody epitope are circled. B, sequence alignment comparing the deduced N-terminal sequences of HMGCLL1 from a variety of vertebrates. Highly conserved residues (>70% identity) are boldfaced. Sequences from the following organisms are included: Homo sapiens, Macaca mulatta, Bos taurus, Sus scrofa, Canis lupus familiaris, Monodelphis domestica, Rattus norvegicus, Mus musculus, Gallus gallus, and Danio rerio. Amino acid sequences were obtained from the NCBI Protein database using the following reference numbers: NP_001035865.1, XP_002803820.1, DAA16714.1, XP_003356653.1, XP_538973.2, XP_001371535.1, XP_001058331.2, NP_776092.1, XP_419903.2, and NP_001103870.1, respectively. Alignments were generated using Clustal W. Residue numbering corresponds to human HMGCLL1.
Native Protein Expression
pPICZ-HLL1-His6 and pPICZ-HLL1-G2A-His6 plasmids were propagated in E. coli JM109 cells grown in a 50-ml culture of LB + 25 μg/ml of Zeocin; yield of each plasmid after purification was ∼150–200 μg. Purified plasmids were linearized by digestion with SacI endonuclease and 3 μg of each were transformed into chemically competent P. pastoris KM71H cells using an Invitrogen EasyComp Transformation kit. Transformation reactions were plated on yeast extract peptone dextrose (YPD) + 100 μg/ml of Zeocin and incubated at 30 °C until colonies formed (3–5 days). Integration of HMGCLL1 was confirmed by direct PCR screening (15) using the primers described for plasmid construction. An isolated colony of each integrant was used to inoculate 4 ml of YPD for overnight growth. A glycerol stock of each confirmed integrant was made as described above. Small-scale cultures were produced and analyzed by Western blot to select an integrant with good HMGCLL1 expression. For full-scale expression, a 75-ml starter culture of minimal glycerol + histidine media (MGYH) was inoculated from glycerol stock and incubated at 25 °C with vigorous shaking until the A600 was about 5. Ten milliliters of the starter culture was used to inoculate each of three 1-liter cultures of MGYH, which were incubated as above until the A600 was about 6. Expression of HMGCLL1 was induced by harvesting the cells by centrifugation at 3,000 × g for 5 min and resuspending the combined pellets in 750 ml of minimal methanol + histidine media (MMH). Induced cultures were incubated at 25 °C with vigorous shaking and induction was maintained by the addition of 3.75 ml of 100% methanol every 24 h. At 72 h post-induction, cells were harvested by centrifugation at 5,000 × g for 6 min at room temperature and pellets (∼20 g of wet cells) were stored at −80 °C until protein purification. Similar conditions were used for expression of the HLL1-G2A-His6 mutant.
Native Enzyme Purification
All steps were carried out at 4 °C. Pichia cell pellets from 750 ml of induction culture were resuspended in 400 ml of cold lysis buffer containing 25 mm NaPi (pH 7.4), 300 mm NaCl, 10% (v/v) glycerol, 1% (v/v) Triton X-100, and 10 mm imidazole. Protease inhibitors (1 mm PMSF, 1 μm pepstatin A, and 10 μm leupeptin) and 5 mm mercaptoethanol were added immediately before cell disruption. Cells were mechanically disrupted in a Bead Beater (Bio Spec) chamber containing equal volumes of sample and ice-cold glass beads (0.5 mm) for 12 cycles of 30 s with a 60-s pause between each cycle. DNase I was added to the lysate at a final concentration of 1 unit/ml and incubated for 15 min at 4 °C. A gram of Chelex 100 was added to the lysate just prior to clarification by centrifugation at 3,000 × g (wt HMGCLL1) or 5,000 × g (G2A HMGCLL1) for 10 min. The supernatant was loaded onto Ni-Sepharose Fast Flow resin (0.2–0.4 ml). The column was washed with buffer containing 25 mm NaPi (pH 7.4), 250 mm NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 50 mm imidazole, and 5 mm mercaptoethanol until the A280 < 0.010. The protein was eluted slowly overnight with buffer containing 25 mm NaPi (pH 7.4), 250 mm NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 300 mm imidazole, and 5 mm mercaptoethanol. Fractions containing HMGCLL1 were pooled, concentrated by centrifugal ultrafiltration, and the final concentration was determined by the method of Bradford (13). Similar conditions were utilized for the purification of the G2A mutant. A typical expression and purification yielded 0.1–0.5 mg of WT or G2A HMGCLL1.
Enzyme Activity Measurement
Enzyme activity was determined using the method of Stegink and Coon (1) as modified by Kramer and Miziorko (16). HMG-CoA was synthesized using the method of Goldfarb and Pitot (17). Briefly, this spectrophotometric assay couples the acetyl-CoA produced upon the cleavage of HMG-CoA to the reactions of malate dehydrogenase and citrate synthase. For each acetyl-CoA that condenses with oxaloacetate to form citrate, one malate is oxidized to oxaloacetate, producing one NADH. The rate of NADH production is determined by measuring the increase in A340 and is proportional to the amount of HMG-CoA lyase added. Reactions are performed at 30 °C (pH 8.2). For estimates of maximum velocity (Vmax) and Michaelis constant (Km), reaction velocities at varying substrate concentrations were fitted to the Michaelis-Menten equation using GraphPad Prism 4.0 (GraphPad Software, San Diego, CA). For determination of the Km for Mg2+ and Mn2+, all assay components including buffers and enzymes were treated with Chelex 100 to remove trace metals. Assays for estimates of maximum velocity (Vmax) and Michaelis constant (Km) contained 3 nm HMGCLL1 or G2A HMGCLL1.
Expression and Purification of Human N-Myristoyltransferase
The pET15-MHL vector containing the gene for human NMT1 (obtained from Structural Genomics Consortium, Toronto, Canada; deposited by Cheryl Arrowsmith) was harbored in E. coli DH5α cells. The plasmid was purified and transformed into chemically competent E. coli BL21(DE3). Transformed cells were plated onto LB agar containing 100 μg/ml of ampicillin, and incubated overnight at 37 °C. A single colony was used to inoculate a small volume of LB/ampicillin for overnight growth. Two milliliters of the overnight culture was used to inoculate a 1-liter culture of LB/ampicillin. The culture was incubated at 37 °C until the A600 was 0.6–0.8 at which point protein expression was induced by the addition of sterile isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 1 mm and the temperature was reduced to 20 °C. After overnight incubation the induced cells were harvested by centrifugation, the pellet was resuspended in 40 ml of cold lysis buffer (50 mm HEPES (pH 7.5), 500 mm NaCl, 5 mm imidazole, 5% (v/v) glycerol, 2.5 mm PMSF, and 10 μm pepstatin), and the suspension was stored at −80 °C until protein purification. All purification steps were carried out at 4 °C. The cell suspension was thawed, the volume adjusted to 100 ml with lysis buffer, and DNase I (1 units/ml) was added. Cells were mechanically disrupted by passing twice through a microfluidizer at ∼17 kpsi. The lysate was clarified by centrifugation at 20,000 × g for 1 h and the supernatant was loaded onto Ni-Sepharose Fast Flow resin (∼0.5 ml). The column was washed with 50 mm HEPES (pH 7.5), 500 mm NaCl, 30 mm imidazole, and 10% (v/v) glycerol until the A280 of the effluent was about 0.075. Protein was eluted slowly overnight with 50 mm HEPES (pH 7.5), 500 mm NaCl, 250 mm imidazole, and 20% (v/v) glycerol. Fractions containing N-myristoyltransferase (NMT) were pooled and the concentration was determined by the method of Bradford (13). About 7 mg of homogeneous enzyme were produced.
In Vitro N-Myristoylation Assay
First, [3H]myristoyl-CoA was synthesized by adding 0.03 units of Pseudomonas sp. acyl-coenzyme A synthase (Sigma) to 100 μl of buffer (20 mm Tris (pH 7.4), 1 mm DTT, 10 mm MgCl2, and 0.1 mm EGTA) containing 6 mm coenzyme A, 12 mm ATP, and 3.3 μm [9,10-3H]myristic acid (30 Ci/mmol). The reaction was incubated at 37 °C for 2 h with agitation. Then, the NMT reaction was initiated by addition of 10 μl of the [3H]myristoyl-CoA reaction mixture to 10 μl of NMT reaction mixture containing 1 mm EDTA, 0.2% Triton X-100, 7.4 μg of HMGCLL1 (solubilized from inclusion bodies), and 6.2 μg of purified human NMT. This reaction was incubated overnight at 25 °C. NMT-treated and untreated proteins were subjected to electrophoresis on a 12% SDS gel, which was then soaked in FluoroHance, dried, and exposed to autoradiography film for 3 days at −80 °C.
Immunofluorescence Detection of HMGCLL1 or HMGCL in COS-I Cells
Full-length HMGCLL1 and G2A HMGCLL1 were amplified from PICZ-HLL1-His6 using the primers described under “Plasmid Construction.” The PCR products were digested with EcoRI and XhoI, gel purified, and ligated with a similarly digested and purified pcDNA3 (Invitrogen) vector backbone. Similarly, a restriction fragment from pTrc-HL encoding the HMGCL coding sequence was also ligated into the pcDNA3 vector. DNA sequence analysis was used to verify the integrity of the pcDNA3-HLL1-His6, pcDNA3-HLL1-G2A-His6, and pcDNA3-HMGCL expression plasmids. COS-1 (American Type Culture Collection) cells were transfected by electroporation with pcDNA3-HLL1-His6, pcDNA3-HLL1-G2A-His6, pcDNA3-HMGCL, or empty vector, and cultured to about 80% confluence at 37 °C in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) in a 5% CO2 atmosphere. Coverslips were removed from medium and washed 3 times in Dulbecco's phosphate-buffered saline (D-PBS). Cells were then fixed in 3% (w/v) formaldehyde in PBS for 20 min and rinsed 3 times with PBS. Fixed cells were permeabilized with 1% (v/v) Triton X-100 in PBS for 5 min and rinsed 3 times in PBS. Cells were incubated for 45 min with rabbit anti-HMGCLL1, rabbit anti-HMGCL, mouse anti-ATP synthase (Santa Cruz), or mouse anti-GM130, rinsed 7 times with PBS and then incubated for 45 min with Alexa Fluor 488-labeled goat anti-rabbit IgG or Cy3-labeled goat anti-mouse IgG (Santa Cruz Biotechnology). The coverslips were rinsed 7 times with PBS and mounted in 100 mm Tris (pH 8.7), 1 mg/ml of phenylenediamine. Cells were photographed using a digital camera at 1–10-s exposures with fluorescent light of appropriate wavelengths.
Rat Organ Lysate Blots
Six 60-day-old white rats (3 male, 3 female) were euthanized and the major organs were harvested and homogenized on ice (3 cycles of 1 min on/1 min pause; Brinkman/Kinematica Polytron; speed 3) in 3 volumes of homogenization buffer (25 mm HEPES (pH 7.4), 5 mm EDTA, 1 mm DTT, and 0.1% (v/v) Triton X-100). PMSF and pepstatin (1 mm and 1 μm, respectively) were added immediately prior to homogenization. Homogenates were clarified by centrifugation at 800 × g for 10 min at 25 °C and the supernatants were filtered through a 0.2-μm centrifugal device. Ten-μg protein samples (1–6 μl) from each filtrate were diluted to 100 μl in homogenization buffer without Triton X-100 and applied to nitrocellulose using a Hybri-Slot manifold (Invitrogen). Casein blocked, peroxidase-suppressed blots were incubated separately in 5.6 μg/ml of affinity purified anti-HMGCLL1 or anti-HMG-CoA lyase antibodies. Rinsed blots were incubated in a 1:5000 dilution of goat anti-rabbit IgG-horseradish peroxidase-conjugated antibodies. Finally, rinsed blots were incubated in West Pico ECL Western blotting substrate and exposed to autoradiography film.
Subcellular Fractionation of U87 Glioblastoma Cells
Human U87 glioblastoma cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells from three confluent 10-cm culture dishes were harvested by gentle trypsinization, homogenized, and a postnuclear supernatant was prepared as described previously (25). The supernatant was loaded onto a linear gradient of increasing Nycodenz (Accudenz; Accurate Chemical and Scientific Corp.) concentration (15–30%) and decreasing sucrose concentration (0.25–0 m); 10 mm Tris (pH 7.5) and 0.1 mm EDTA were present throughout the gradient. A cushion of Purdens (Accurate Chemical and Scientific Corp.) was present below the gradient. Centrifugation at 77,000 × g for 30 min was performed in a Beckman VTi 65.1 rotor. Fractions of ∼1 ml were collected from the bottom of the centrifuge tube and subjected to Western blotting as described above. Marker antibodies used were Golgin-58 (Golgi; gift from C. Machamer), Cop-1 (Golgi-derived vesicles; Sigma), manganese-superoxide dismutase (mitochondria; Stressgen), and PEX14 (peroxisomes; gift from S. Mihalik).
RESULTS
Comparison of Human HMGCL and Human HMGCLL1 Amino Acid Sequences
Based on sequence homology, the National Institute of Health's Mammalian Gene Collection Program (MGC) has recently identified the protein encoded by the gene 3-hydroxymethyl-3-methylglutaryl-coenzyme A lyase-like 1 (HMGCLL1) as being a potential HMG-CoA lyase. The HMGCLL1 gene-encoded protein contains 340 residues and is 64% identical and 78% similar to HMG-CoA lyase (Fig. 1A). HMGCLL1 contains the highly conserved HMG-CoA lyase signature sequence G(L/A)GGCP(Y/F) (residues 277–283; Fig. 1A), which is found in all HMG-CoA lyase proteins identified to date. HMGCLL1 lacks the basic and hydrophobic residues typically found in a N-terminal mitochondrial leader sequence (e.g. as found in precursor HMGCL) but instead the N-terminal MGNVPSA sequence represents a potential N-myristoylation motif. Target recognition by N-myristoyltransferase absolutely requires a terminal glycine (produced upon methionine cleavage by methionine aminopeptidase) followed by an uncharged residue. Residues 3 and 4 may be any residue. The 5th residue is a small uncharged residue but serine is favored. The 6th residue may not be proline (18). Although the N terminus of HMGCLL1 is not conserved in HMGCL, it is well conserved among the predicted sequences for higher vertebrate HMGCLL1 (Fig. 1B). At the C terminus, HMGCLL1 also lacks the peroxisomal targeting sequence C(K/R)L present in HMGCL. Mammalian HMG-CoA lyases contain 8 highly conserved cysteines. Interestingly, although HMGCLL1 also contains 8 cysteines it lacks the C-terminal cysteine implicated in thiol/disulfide exchange (19). Instead, the 8th cysteine in HMGCLL1 is located in its nonconserved leader sequence. These sequence features and their contrasts with the mitochondrial HMGCL sequence suggest that HMGCLL1 may be an active, extramitochondrial HMG-CoA lyase and may be subject to post-translational myristoylation.
Expression and Purification of Wild-type Human HMGCLL1 and a G2A Mutant
Because several E. coli expression systems failed to produce substantial quantities of soluble protein,3 the methylotrophic yeast P. pastoris was tested as an expression system. Although HMGCLL1 was expressed as only a small fraction of the total P. pastoris lysate, from 750 ml of induction culture about 0.5 mg of >95% homogenous protein was recovered after purification by immobilized metal affinity chromatography (Fig. 2A). During initial attempts at purification it was observed that upon high-speed centrifugation, some HMG-CoA lyase activity could be detected in the pellet fraction. The addition of Triton X-100 to purification buffers and minimization of centrifugation of the crude lysate improved the yield of protein in the supernatant. HMGCLL1 recovered from the nickel column eluate remains in the supernatant fraction even upon extended high-speed centrifugation. Expression and purification of a G2A mutant was performed using similar methodology. Although a variety of protease inhibitors are present during purification, the protein band is often observed as a doublet upon SDS-PAGE (e.g. Fig. 2A). Edman sequencing of purified proteins revealed that substantial proteolyic cleavage primarily occurs after residue 17 (leucine) in E. coli preparations and after residue 18 (leucine) in P. pastoris preparations. The extent of this cleavage did not correlate with any apparent major change in the enzymatic function of the protein (see below).
FIGURE 2.
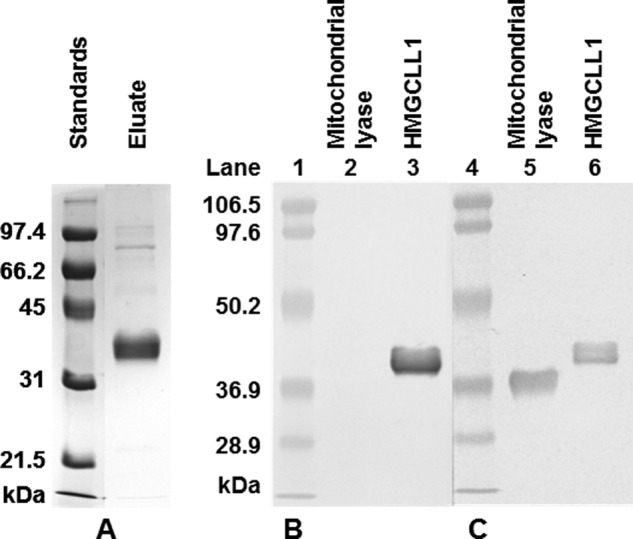
Purification of HMGCLL1 and specificity of antibody against HMGCLL1. A, left panel, HMGCLL1 protein was expressed in P. pastoris. 5 μg of protein from a Ni-Sepharose eluate were subjected to electrophoresis on a 10% SDS gel. Protein bands were detected using a Coomassie stain. Molecular weight markers (left lane) include (highest to lowest): phosphorylase b, 97.4 kDa; bovine serum albumin, 66.2 kDa; ovalbumin, 45 kDa; carbonic anhydrase, 31 kDa; and trypsin inhibitor, 21.5 kDa. Right panel, duplicate samples of purified human mitochondrial HMG-CoA lyase and wild-type HMGCLL1 (1 μg each) were subjected to electrophoresis on an SDS gel and transblotted to nitrocellulose. The duplicate halves were separated and treated individually with either (B) rabbit anti-HMGCLL1 antibody (whole serum) or (C) rabbit anti-avian HMG-CoA lyase antibody (whole serum). The blots were incubated with alkaline phosphatase-conjugated secondary antibody. Protein bands were colorimetrically detected using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium. Prestained molecular mass markers (Bio-Rad low range; lot number 310001516) include: phosphorylase b, 106.5 kDa; bovine serum albumin, 97.6 kDa; ovalbumin, 50.2 kDa; carbonic anhydrase, 36.9 kDa; trypsin inhibitor, 28.9 kDa.
Antibody Reactivity with HMGCLL1 Versus HMGCL
To facilitate detection of HMGCLL1 protein in complex samples from cell lysates or fractions and to distinguish it from mitochondrial or peroxisomal HMGCL, an antibody was raised in rabbit against a synthetic peptide corresponding to residues 19–37 of the nonconserved HMGCLL1 N terminus (Fig. 1). When used to probe Western blots of purified HMGCLL1, and HMGCL proteins, the anti-HMGCLL1 antibody detects only the HMGCLL1 protein (Fig. 2B). In contrast, the anti-avian HMGCL antibody detects both proteins (Fig. 2C). When used to probe Western blots of unfractionated whole cell lysates of E. coli or P. pastoris expressing HMGCLL1, the anti-HMGCLL1 antibody detects only a single band at the molecular mass expected for HMGCLL1 protein.
Functional Characterization of HMGCLL1
To determine whether HMGCLL1 is a functional HMG-CoA lyase, properties that are well characterized for other HMG-CoA lyases were examined using HMGCLL1. Wild-type and G2A HMGCLL1 were assayed for HMG-CoA lyase activity as described under “Experimental Procedures.” Specific activities measured for wild-type and G2A HMGCLL1 enzymes were determined to be 150 and 117 units/mg, respectively. The specific activities of these enzymes are reasonably comparable with that of WT recombinant human HMGCL (159 units/mg; Table 1; see Roberts et al. (20)). Activity of HMGCLL1 is markedly stimulated by divalent cations. Wild-type and G2A HMGCLL1 exhibit Km values for Mg2+ (49 and 88 μm; Table 1) that are somewhat tighter than reported for recombinant human HMGCL (233 μm), whereas Km values for Mn2+ are comparable (0.18, 0.24, and 0.34 μm, respectively). The Km values for (S)HMG-CoA measured for WT and G2A HMGCLL1 (28 and 24 μm; Table 1) are equivalent to the value reported for recombinant human HMGCL (24 μm). A comparison of these parameters measured for HMGCLL1 and those reported for recombinant human (20) and avian HMGCL (16, 21) is presented in Table 1. The comparability between specific activity estimates, as well as substrate and divalent cation Michaelis constants, for HMGCLL1 and the previously characterized eukaryotic HMG-CoA lyases indicate that HMGCLL1 is a catalytically functional HMG-CoA lyase.
TABLE 1.
Kinetic characterization of wild-type and G2A human HMGCLL1
| Property | HMGCLL1 |
Recombinant human HMGCLa |
Avian HMGCLb | ||
|---|---|---|---|---|---|
| Wild-type | G2A | Wild-type | C323S | ||
| Specific activity (μmol min−1 mg−1)c | 150 ± 6d | 117 ± 5d | 159 | 348 | 350 |
| Km (S-HMG-CoA, μm) | 28 ± 3 | 24 ± 2 | 24 | 45 | 8 |
| Km (Mg2+, μm) | 49 ± 7 | 88 ± 9 | 233 | 322 | 50 |
| Km (Mn2+, μm) | 0.18 ± 0.06 | 0.24 ± 0.06 | 0.34 | 0.37 | 10 |
a Properties of human wild-type and C323S HMG-CoA lyase have been described by Roberts et al. (20).
b Properties of avian HMG-CoA lyase have been described by Kramer and Miziorko (16) and Hruz and Miziorko (21).
c Specific activities determined in the presence of 5 mm DTT.
d Maximum specific activity is observed at 2.5 mm Mg2+.
Myristoylation of HMGCLL1
The N-terminal amino acid sequence of HMGCLL1 is MGNVPS, a motif that suggests it may be a suitable substrate for N-myristoyltransferase (18). N-Myristoyltransferase absolutely requires an N-terminal glycine. Mutation of the N-terminal glycine to alanine as in the G2A mutant HMGCLL1 precludes myristoylation. Limitations in expression of HMGCLL1 protein in P. pastoris hindered attempts to determine whether HMGCLL1 was myristoylated by endogenous Pichia N-myristoyltransferase, which may not optimally process human HMGCLL1. Edman degradation of WT HMGCLL1 failed to identify the N terminus of full-length protein and only yielded sequences of the proteolyzed sample. Additionally, phenylthiohydantoin-derivative yields were only ∼20% of the expected value. These observations are consistent with the possibility that any unproteolyzed protein is modified at the N terminus (blocking Edman degradation). To further test the possibility of N-terminal modification, the following in vitro approach was used. Denatured HMGCLL1 and G2A HMGCLL1 proteins solubilized from E. coli inclusion bodies were incubated with recombinant human NMT and [3H]myristoyl-CoA. Comparable amounts of protein from each reaction were subjected to electrophoresis on an SDS gel and the dried gel exposed to autoradiography film (Fig. 3, top panel). In the control reaction (lane 1), HMGCLL1 was incubated with [3H]myristoyl-CoA but not NMT; no signal is detected. Upon reaction of NMT with wild-type HMGCLL1 (lane 2), a signal can be seen at the expected molecular mass of 37.1 kDa, indicating that HMGCLL1 is modified with [3H]myristic acid. In the G2A mutant reaction (lane 3), no signal is seen at the expected molecular weight, confirming that this modification occurs at the N-terminal glycine, which is lacking in the G2A mutant. In a control experiment, densitometry of the Coomassie-stained gel (Fig. 3, bottom panel) indicates that intensity of the G2A band is ∼72% of that measured for the wild-type band. Based on this observation of comparable amounts of protein target for the NMT reaction, the radiolabeling pattern shown in the top panel clearly indicates myristoylation of HMGCLL1.
FIGURE 3.
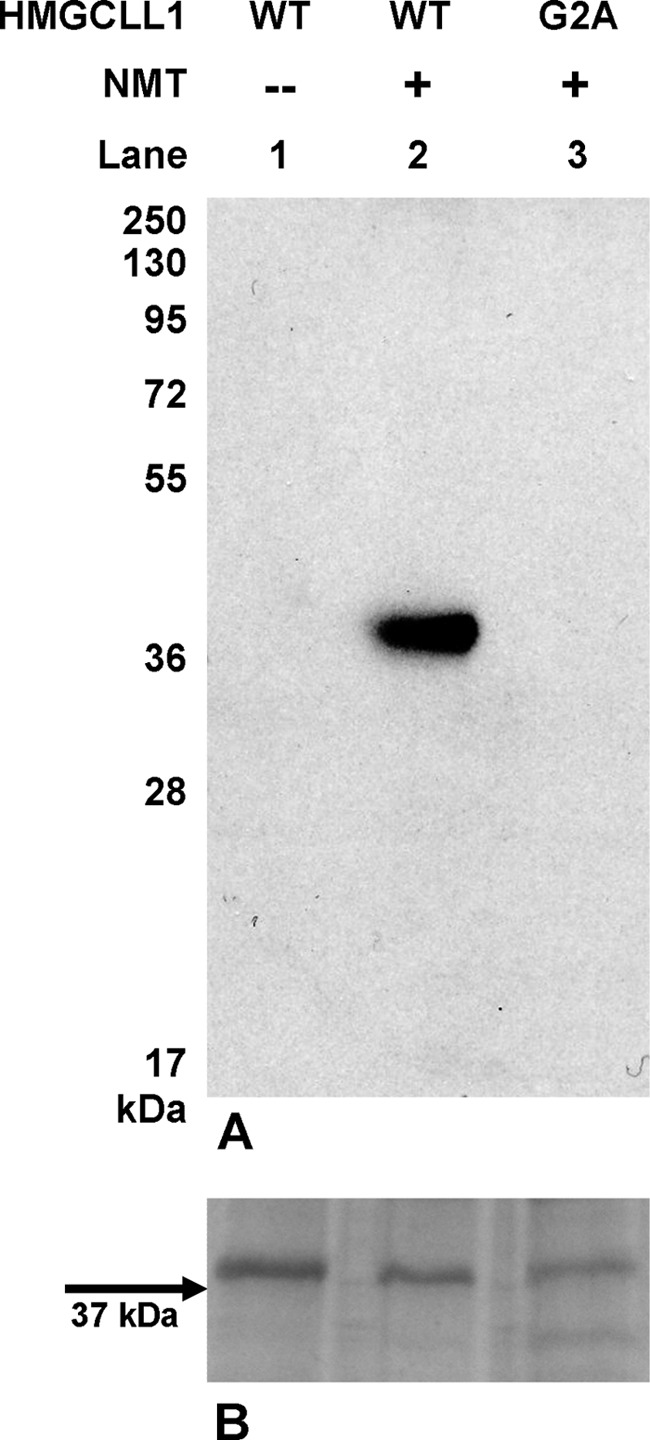
In vitro myristoylation of HMGCLL1. A, equal amounts (7 μg) of wild-type HMGCLL1 (lanes 1 and 2) and G2A mutant (lane 3) were incubated in separate reactions with 6.2 μg of human NMT as described under “Experimental Procedures.” NMT was omitted from the negative control reaction (lane 1). Protein from each reaction was subjected to electrophoresis on a 12% SDS gel. Any residual unincorporated [3H]myristoyl-CoA was allowed to run off with the dye front. The gel was dried, soaked in FluoroHance, and exposed to autoradiography film for 3 days at −80 °C. Positions indicated for pre-stained molecular weight standards (Fermentas Page Ruler Plus) were determined when exposed autoradiography film was overlaid on the dried gel. B, an SDS gel run in parallel with the gel described in panel A was Coomassie stained to demonstrate comparable loading of HMGCLL1 proteins.
Expression of HMGCLL1, G2A HMGCLL1, and HMGCL in COS1 Cells
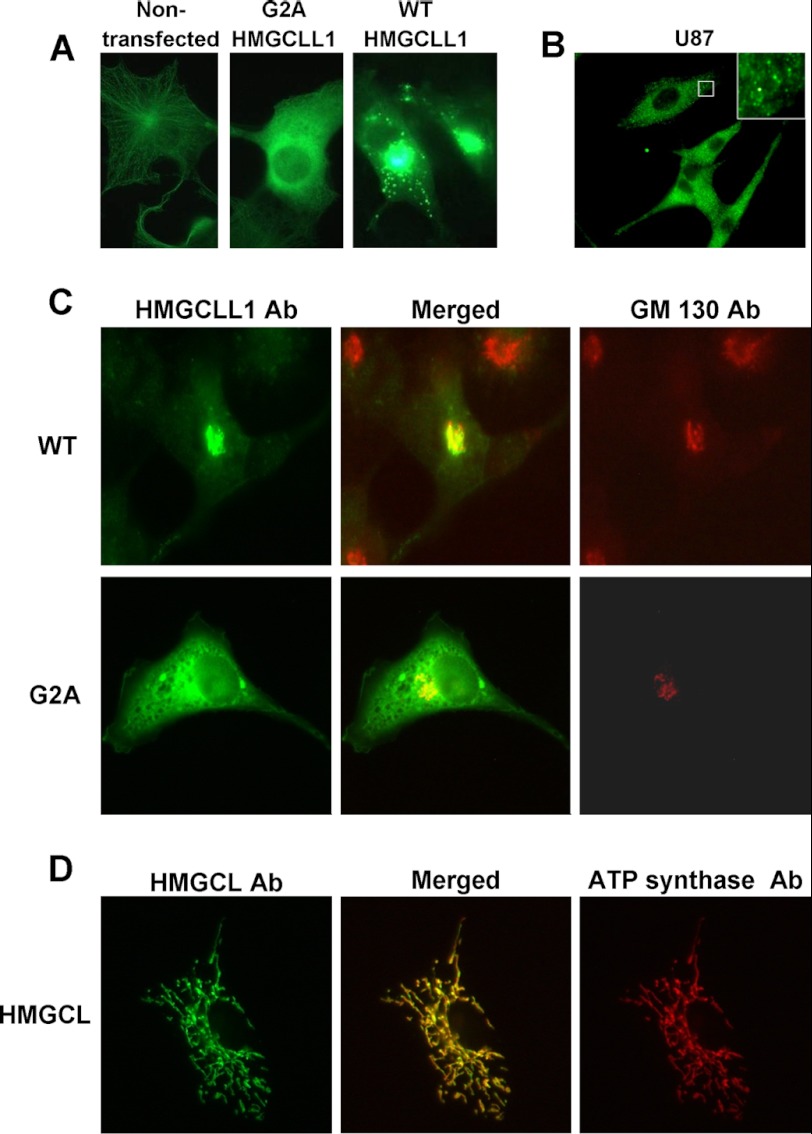
Immunofluorescence microscopy was used to determine the effect of myristoylation on the subcellular localization of HMGCLL1. Antibodies used in this experiment were affinity purified by incubation with HMGCL or HMGCLL1 proteins immobilized on nitrocellulose and then eluted for use in immunodetection. Wild-type HMGCLL1, G2A HMGCLL1, and HMGCL were cloned into the mammalian expression vector pcDNA3 and the proteins were overexpressed in COS1 cells. Immunofluorescence microscopy in nontransfected COS1 cells reveals minor background fluorescence due to cross-reaction of the anti-HMGCLL1 antibody with a cytoskeletal-like protein (Fig. 4A, left). Cells transfected with G2A mutant HMGCLL1 (Fig. 4A, center) demonstrate a diffuse pattern that is consistent with cytoplasmic localization. However, cells transfected with WT HMGCLL1 (Fig. 4A, right) had both a perinuclear and a punctate immunostaining pattern. Perinuclear HMGCLL1 (Fig. 4C, top row, left) colocalized with the cis-Golgi marker protein GM130 (Fig. 4C, top row, right) when the images were merged (Fig. 4C, top row, center). The differences in localization seen upon mutation of the N-terminal glycine to an alanine (Fig. 4C, second row panels) suggest that myristoylation promotes the association of HMGCLL1 with subcellular membranes. In contrast, control experiments with HMGCL expression (Fig. 4D) confirm the expected localization of the protein in mitochondria (Fig. 4D, left panel), as indicted by its colocalization (Fig. 4D, middle panel) with the mitochondrial marker protein, ATP synthase (Fig. 4D, right panel).
FIGURE 4.
Immunofluorescence microscopy detection of HMGCLL1 in COS-1 and U87 cells. Panel A indicates levels and localization of HMGCLL1 in COS-1 cells that are nontransfected (left) or transfected with expression plasmids encoding G2A HMGCLL1 (center) or wild-type HMGCLL1 (right). Panel B reflects endogenous expression of HMGCLL1 in U87 glioblastoma cells. The inset illustrates HMGCLL1 localization in punctate vesicles. Panel C (top panel) compares COS-1 cell localization of wild-type HMGCLL1 (left) and the Golgi marker protein GM130 (right); the center image results upon merging of the left and right images. The panels in the bottom row depict contrasting localization results when the G2A mutant protein is expressed. Panel D depicts the results of control experiments in which COS-1 cells, transfected with pcDNA3-HMGCL, express the HMGCL precursor. After expression and processing, the mature protein is detected in organelles (left). Upon comparison with localization of an ATP synthase mitochondrial marker protein (right), the mitochondrial localization of mature HMGCL (center; merged image) is confirmed.
Immunodetection of Endogenous HMGCLL1 Expression
The expression pattern of HMGCLL1 protein under a variety of metabolic conditions and/or during different developmental stages may be a useful tool in future investigation of the physiological function(s) of this novel HMG-CoA lyase. To screen for tissues that endogenously express HMGCLL1, duplicate arrays of rat organ lysates were probed with either anti-HMGCL or anti-HMGCLL1 antibody. To produce the arrays, the 800 × g supernatants of a variety of homogenized rat organs was passed through a 0.2-μm filter and the filtrates were transferred to nitrocellulose. In organ lysate blots probed with antibody against HMGCL (which detects both HMGCL and HMGCLL1), strong signals are observed for the gut (duodenum, caecum, small intestine, and large intestine) and liver (Fig. 5A). Weaker signals are observed for kidney and adrenal gland. These results are consistent with current literature describing HMG-CoA lyase activity in these organs (22). In contrast, for organ lysate blots probed with antibody against HMGCLL1 (which does not cross-react with HMGCL), strong signals are observed for the duodenum and small intestine (Fig. 5B). In interpreting these results, it is important to keep in mind that anti-HMGCL antibody recognizes both HMGCL and HMGCLL1 (Fig. 2). Therefore, the signals seen in duodenum and small intestine on the anti-HMGCL blot (Fig. 5A) may be partially or primarily due to HMCGLL1 protein. Although the experiment did not unequivocally reveal a tissue in which HMGCLL1, but not HMGCL, is expressed, it does demonstrate that HMGCLL1 is expressed in mammals in a pattern that is distinct from that of the traditional mitochondrial HMG-CoA lyase.
FIGURE 5.
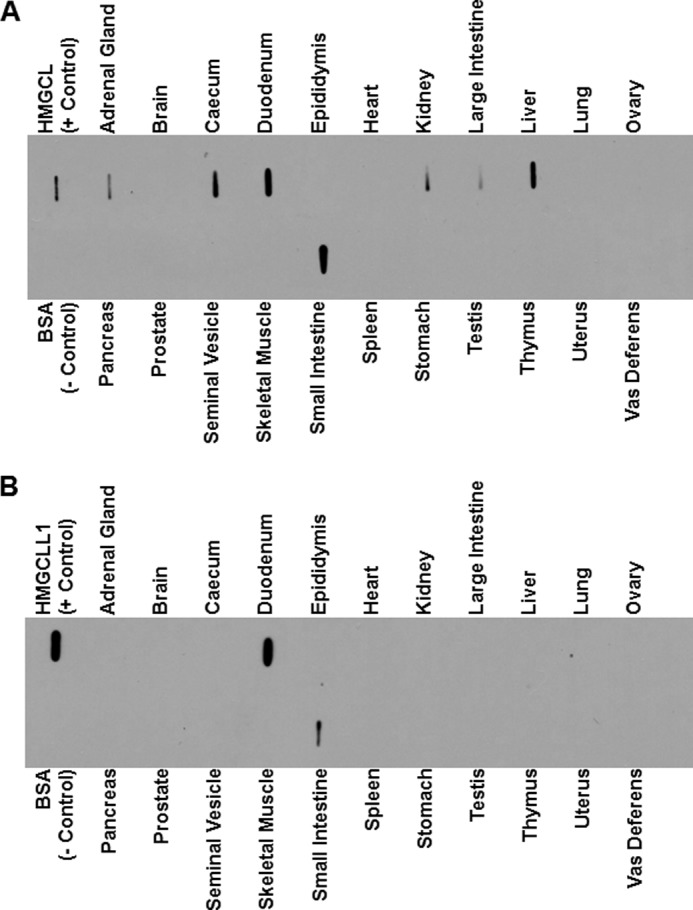
Endogenous tissue expression of HMGCLL1. Protein (10 μg) of clarified organ lysates from the indicated rat tissues was applied to nitrocellulose using a slot-blot manifold. Negative control lanes contained BSA (1 μg); positive control lanes contained 1 ng of purified HMGCL or HMGCLL1. Blocked, peroxidase suppressed blots were incubated separately with anti-HMG-CoA lyase (A) or anti-HMGCLL1 antibodies (B). The blots were incubated with goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody and developed using enhanced chemiluminescence. As demonstrated in Fig. 2, the antibody against HMG-CoA lyase recognizes both mitochondrial HMG-CoA lyase and HMGCLL1, whereas the antibody against HMGCLL1 does not recognize mitochondrial HMG-CoA lyase.
Endogenous Expression of HMGCLL1 in Glioblastoma Cells
The UniGene database expressed sequence tag (EST) profile for HMGCLL1 predicts the highest levels of transcripts to be in brain tissue (www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist = Hs.147054). In a preliminary test for expression of HMGCLL1 protein, protein lysate (30 μg) from cultured U87 (human glioblastoma) cells was subjected to SDS-PAGE and the proteins were transblotted to nitrocellulose. A strong 37-kDa signal was immunodetected in the human glioblastoma cell line U87 (data not shown). Immunofluorescence analysis revealed that endogenous HMGCLL1 localized to punctate vesicles (Fig. 4B) similar to those seen in COS-1 cells overexpressing the protein (Fig. 4, A and C). However, no perinuclear immunostaining was evident. To further extend these observations, U87 cell organelles were separated by centrifugation on a 15–30% Nycodenz gradient. Western blots containing aliquots of the various gradient fractions were tested for the presence of HMGCLL1; equivalent tests were performed to identify the location of subcellular organelles. The markers tested were: Golgin-58 (Golgi), Cop-1 (Golgi-derived vesicles), MnSOD (mitochondria), and PEX14 (peroxisomes). Results (Fig. 6) indicated that HMGCLL1 cleanly sediments in fractions 8 and 9, which coincide with the lanes in which both Golgin-58 and Cop-1 are detected. These results are consistent with the possibility that HMGCLL1 is overexpressed in certain types of human brain cancer cells and localized to Golgi-derived vesicles. Further study will be needed to characterize the specific nature of these vesicles.
FIGURE 6.
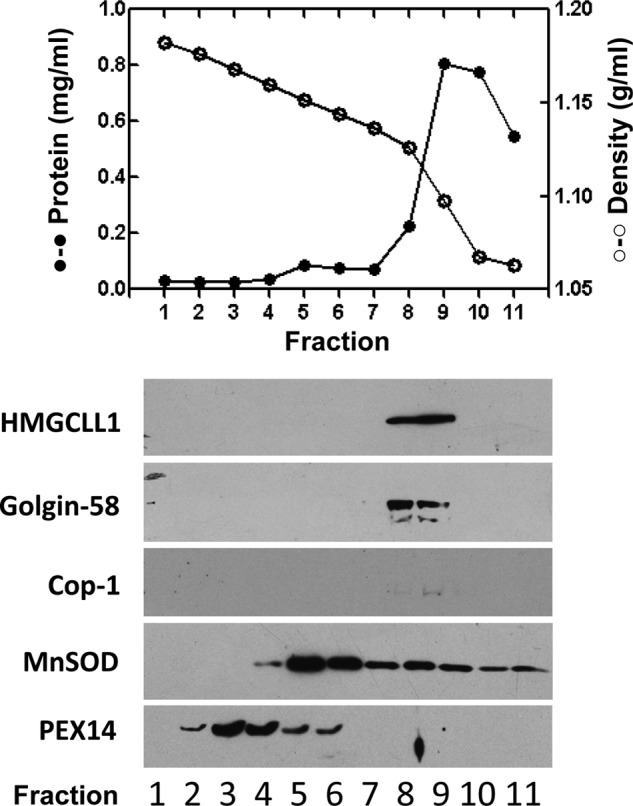
Subcellular localization of endogenously expressed HMGCLL1 in U87 glioblastoma cells. U87 cell organelles were separated by centrifugation on a 15–30% Nycodenz gradient as described under “Experimental Procedures.” Top panel, the protein concentration (closed circles) and density (open circles) of each fraction is shown. Bottom panel, Western blots of the various gradient fractions (16 μl/lane) were tested for the presence of HMGCLL1; equivalent tests were performed to identify the location of subcellular organelles. The organelle markers tested were: Golgin-58 (Golgi), Cop-1 (Golgi derived vesicles), MnSOD (mitochondria), and PEX14 (peroxisomes). HMGCLL1 sediments in fractions 8 and 9, which coincide with the fractions in which both Golgin-58 and Cop-1 are detected.
DISCUSSION
The documentation of protein encoding open reading frames in the mammalian gene collection (MGC) or other data bases affords the opportunities of identifying and retrieving clones from repositories or synthesizing DNA appropriate for construction of expression plasmids. For a protein that is newly documented in a database, any hypothesis of specific function should be confirmed by protein or enzymological techniques because sequence similarities between proteins that are members of a larger family can complicate accurate functional assignments. For example, the Pfam database indicates that the HMG-CoA lyase (HMGL) family of proteins (PF00682) includes several that catalyze acetyl-CoA linked aldol or Claisen condensation/cleavage reactions that are mechanistically similar to HMG-CoA cleavage. All are β barrel proteins and, for several family members that utilize similar chemistry during reaction catalysis, stretches of homologous amino acid sequence are quite common. For this reason, designation of a new human protein (HMGCLL1) as a previously undiscovered HMG-CoA lyase required validation by protein expression and characterization. Expression of HMGCLL1 in P. pastoris allowed recovery of this protein in sufficient amounts so that efficient purification and functional characterization could be accomplished. The kinetic characterization (Table 1) indicates good agreement between the Vm and Km parameters for HMGCLL1 and recombinant or tissue isolated HMGCL, making it reasonable to assign a function to HMGCLL1 in catalysis of a ketogenic reaction. In contrast, one member (DHRS6) of the large family of short chain dehydrogenases has been characterized as a novel human cytosolic hydroxybutyrate dehydrogenase (BDH2) (23), which catalyzes another important reaction in ketone body metabolism. This assignment was offered even though the available functional characterization indicates specific activity and substrate Km values that differ by an order of magnitude from comparable parameters for the well established mitochondrial enzyme (BDH1) (24). This cytosolic dehydrogenase may possess adequate BDH function to support extramitochondrial ketone body metabolism. However, its properties illustrate the point that, in making functional assignments to newly identified enzymes, the possibility that they may more efficiently catalyze other related metabolic reactions must not be overlooked. Likewise, the possibility that they have additional cellular functions unrelated to catalysis should not be dismissed.
In attempting to establish a physiological function for HMGCLL1, the possibility of post-translational modification needed to be addressed because the N-terminal sequence suggests a myristoylation site. Fortunately, an expression plasmid encoding human NMT had been made available by C. Arrowsmith (Structural Genomics Consortium). Expression and isolation of NMT made it possible to convincingly demonstrate [3H]myristic acid incorporation into wild-type HMGCLL1 but not into the G2A mutant, which lacks the myristoylation target residue. Although HMGCL loses its N-terminal leader sequence upon transport into the mitochondria, the N-terminal myristoylation of HMGCLL1 persists in the mature protein, accounting for its detection at an extramitochondrial location when expressed in COS1 cells. These contrasts in post-translational processing for the two homologous proteins underscore their distinct properties. Nonetheless, the precise physiological role for HMGCLL1 remains to be addressed. The endogenous HMGCLL1 detected as a punctate pattern in U87 cells resembles the punctate labeling observed in COS-1 overexpression. The Golgi staining detected in COS-1 cells may reflect an artificially high HMGCLL1 level upon overexpression. The identity of the vesicles that account for punctate labeling remains unclear, because these vesicles colocalize with Golgi and Golgi-derived vesicle markers upon density gradient centrifugation. However, the collected experimental results consistently indicate that HMGCLL1 is not targeted to mitochondria.
Although myristoylation of the N terminus of HMGCLL1 targets enzyme for a cellular membrane destination, it remains possible that both the lumen of its β barrel as well as the “signature” sequence containing loop that harbors the catalytic cysteine are accessible to cytoplasmic metabolites. Such a situation is well precedented based on results for the cholesterogenic enzyme, HMG-CoA reductase; whereas this protein is membrane associated, its catalytic site can access cytosolic substrate. This possibility suggests that HMGCLL1 can access soluble substrate, produced by HMGCS1, the cytosolic HMG-CoA synthase. HMGCLL1 catalysis would then make available an extramitochondrial pool of acetoacetate, which has been shown (7) to be incorporated into cholesterol and fatty acid. The observation that certain transformed brain cells contain elevated levels of HMGCLL1 is interesting based on the report (9) of elevated transcripts for HMGCLL1 in human breast cancer stroma. Those results suggest an alternative role for the reaction product of HMGCLL1, acetoacetate. In this context, the ability of acetoacetate to enter the mitochondria in either the stromal cells or the proximal epithelial cancer cells would allow subsequent activation to its acyl-CoA derivative by CoA transferase. Mitochondrial thiolase could then convert the acetoacetyl-CoA to acetyl-CoA for use in metabolic energy production upon TCA cycle metabolism in these cancer cells, supporting cell maintenance or possible growth. Thus, interpretation of our experimental data in the context of these literature precedents suggests plausible physiological roles for HMGCLL1 in either lipid biosynthesis and/or energy metabolism.
Acknowledgments
Dr. M. S. R. Sastry evaluated various methodologies for bacterial expression of HMGCLL1. We appreciate Dr. Peter Kramer's observations on a putative HMGCLL1 in the Mammalian Genome Collection.
This work was supported, in whole or in part, by National Institutes of Health Grants DK21491 and HD020461 and the Marion-Merrell-Dow foundation.
M. S. R. Sastry, unpublished observations.
- HMGCL
- 3-hydroxy-3-methylglutaryl-CoA lyase
- HMG-CoA
- 3-hydroxy-3-methylglutaryl-CoA
- HMGCLL1
- HMG-CoA lyase-like protein
- NMT
- N-myristoyltransferase
- BDH
- 3-hydroxybutyrate dehydrogenase
- MGC
- mammalian gene collection.
REFERENCES
- 1. Stegink L. D., Coon M. J. (1968) Stereospecificity and other properties of highly purified β-hydroxy-β-methylglutaryl coenzyme A cleavage enzyme from bovine liver. J. Biol. Chem. 243, 5272–5279 [PubMed] [Google Scholar]
- 2. Fu Z., Runquist J. A., Montgomery C., Miziorko H. M., Kim J. J. (2010) Functional insights into human HMG-CoA lyase from structures of acyl-CoA-containing ternary complexes. J. Biol. Chem. 285, 26341–26349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson A. M., Williamson D. H. (1980) Physiological roles of ketone bodies as substrates and signals in mammalian tissues. Physiol. Rev. 60, 143–187 [DOI] [PubMed] [Google Scholar]
- 4. Wang S. P., Marth J. D., Oligny L. L., Vachon M., Robert M. F., Ashmarina L., Mitchell G. A. (1998)3-Hydroxy-3-methylglutaryl-CoA lyase (HL). Gene targeting causes prenatal lethality in HL-deficient mice. Hum. Mol. Genet. 7, 2057–2062 [DOI] [PubMed] [Google Scholar]
- 5. Gibson K. M., Breuer J., Nyhan W. L. (1988) 3-Hydroxy-3-methylglutaryl-coenzyme A lyase deficiency. Review of 18 reported patients. Eur. J. Pediat. 148, 180–186 [DOI] [PubMed] [Google Scholar]
- 6. Pié J., López-Viñas E., Puisac B., Menao S., Pié A., Casale C., Ramos F. J., Hegardt F. G., Gómez-Puertas P., Casals N. (2007) Molecular genetics of HMG-CoA lyase deficiency. Mol. Genet. Metab. 92, 198–209 [DOI] [PubMed] [Google Scholar]
- 7. Hildebrandt L. A., Spennetta T., Elson C., Shrago E. (1995) Utilization and preferred metabolic pathway of ketone bodies for lipid synthesis by isolated rat hepatoma cells. Am. J. Physiol. 269, C22–27 [DOI] [PubMed] [Google Scholar]
- 8. Patel M. S., Russell J. J., Gershman H. (1981) Ketone body metabolism in glioma and neuroblastoma cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7214–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pavlides S., Tsirigos A., Migneco G., Whitaker-Menezes D., Chiavarina B., Flomenberg N., Frank P. G., Casimiro M. C., Wang C., Pestell R. G., Martinez-Outschoorn U. E., Howell A., Sotgia F., Lisanti M. P. (2010) The autophagic tumor stroma model of cancer. Role of oxidative stress and ketone production in fueling tumor cell metabolism. Cell Cycle 9, 3485–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montgomery C., Miziorko H. M., Pei Z., Watkins P. A. (2011) Discovery of a human extramitochondrial hydroxymethylglutaryl-CoA lyase. FASEB J. 25, 933 [Google Scholar]
- 11. Lennon G., Auffray C., Polymeropoulos M., Soares M. B. (1996) The I.M.A.G.E. Consortium. An integrated molecular analysis of genomes and their expression. Genomics 33, 151–152 [DOI] [PubMed] [Google Scholar]
- 12. Romanos M. A., Scorer C. A., Clare J. J. (1992) Foreign gene expression in yeast. A review. Yeast 8, 423–488 [DOI] [PubMed] [Google Scholar]
- 13. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 14. Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. (1989) Fluorescence microscopy methods for yeast. Methods Cell Biol. 31, 357–435 [DOI] [PubMed] [Google Scholar]
- 15. Linder S., Schliwa M., Kube-Granderath E. (1996) Direct PCR screening of Pichia pastoris clones. BioTechniques 20, 980–982 [DOI] [PubMed] [Google Scholar]
- 16. Kramer P. R., Miziorko H. M. (1980) Purification and characterization of avian liver 3-hydroxy-3-methylglutaryl coenzyme A lyase. J. Biol. Chem. 255, 11023–11028 [PubMed] [Google Scholar]
- 17. Goldfarb S., Pitot H. C. (1971) Improved assay of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Lipid Res. 12, 512–515 [PubMed] [Google Scholar]
- 18. Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. (1988) Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263, 1784–1790 [PubMed] [Google Scholar]
- 19. Montgomery C., Miziorko H. M. (2011) Influence of multiple cysteines on human 3-hydroxy-3-methylglutaryl-CoA lyase activity and formation of inter-subunit adducts. Arch. Biochem. Biophys. 511, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roberts J. R., Narasimhan C., Hruz P. W., Mitchell G. A., Miziorko H. M. (1994) 3-Hydroxy-3-methylglutaryl-CoA lyase. Expression and isolation of the recombinant human enzyme and investigation of a mechanism for regulation of enzyme activity. J. Biol. Chem. 269, 17841–17846 [PubMed] [Google Scholar]
- 21. Hruz P. W., Miziorko H. M. (1992) Avian 3-hydroxy-3-methylglutaryl-CoA lyase. Sensitivity of enzyme activity to thiol/disulfide exchange and identification of proximal reactive cysteines. Protein Sci. 1, 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bachhawat B., Robinson W. G., Coon M. J. (1955) The enzymatic cleavage of β-hydroxy-β-methylglutaryl coenzyme A to acetoacetate and acetyl coenzyme A. J. Biol. Chem. 216, 727–736 [PubMed] [Google Scholar]
- 23. Guo K., Lukacik P., Papagrigoriou E., Meier M., Lee W. H., Adamski J., Oppermann U. (2006) Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme. A novel, cytosolic type 2R-β-hydroxybutyrate dehydrogenase. J. Biol. Chem. 281, 10291–10297 [DOI] [PubMed] [Google Scholar]
- 24. Green D., Marks A. R., Fleischer S., McIntyre J. O. (1996) Wild-type and mutant human heart (R)-3-hydroxybutyrate dehydrogenase expressed in insect cells. Biochemistry 35, 8158–8165 [DOI] [PubMed] [Google Scholar]
- 25. Pei Z., Oey N. A., Zuidervaart M. M., Jia Z., Li Y., Steinberg S. J., Smith K. D., Watkins P. A. (2003) The acyl-CoA synthetase “bubblegum” (lipidosin). Further characterization and role in neuronal fatty acid β-oxidation. J. Biol. Chem. 278, 47070–47078 [DOI] [PubMed] [Google Scholar]