Background: Copper uptake is tightly regulated to prevent deficiency while avoiding toxicity.
Results: AtCOPT6 localizes to the plasma membrane, is regulated by copper availability, interacts with itself and AtCOPT1, and regulates response to copper limitation and excess.
Conclusion: AtCOPT6 is a novel SPL7 target that functions in copper homeostasis in Arabidopsis.
Significance: Identification and characterization of copper transporters are crucial for understanding of copper homeostasis.
Keywords: Copper Transport, Metals, Plant Molecular Biology, Plant Physiology, Plasma Membrane, Arabidopsis, COPT6, CTR/COPT, SPL7
Abstract
Among the mechanisms controlling copper homeostasis in plants is the regulation of its uptake and tissue partitioning. Here we characterized a newly identified member of the conserved CTR/COPT family of copper transporters in Arabidopsis thaliana, COPT6. We showed that COPT6 resides at the plasma membrane and mediates copper accumulation when expressed in the Saccharomyces cerevisiae copper uptake mutant. Although the primary sequence of COPT6 contains the family conserved domains, including methionine-rich motifs in the extracellular N-terminal domain and a second transmembrane helix (TM2), it is different from the founding family member, S. cerevisiae Ctr1p. This conclusion was based on the finding that although the positionally conserved Met106 residue in the TM2 of COPT6 is functionally essential, the conserved Met27 in the N-terminal domain is not. Structure-function studies revealed that the N-terminal domain is dispensable for COPT6 function in copper-replete conditions but is important under copper-limiting conditions. In addition, COPT6 interacts with itself and with its homolog, COPT1, unlike Ctr1p, which interacts only with itself. Analyses of the expression pattern showed that although COPT6 is expressed in different cell types of different plant organs, the bulk of its expression is located in the vasculature. We also show that COPT6 expression is regulated by copper availability that, in part, is controlled by a master regulator of copper homeostasis, SPL7. Finally, studies using the A. thaliana copt6-1 mutant and plants overexpressing COPT6 revealed its essential role during copper limitation and excess.
Introduction
Copper is a redox-active transition element that serves as an essential micronutrient for all living organisms (1–3). It acts as a cofactor for enzymes involved in electron transfer reactions and, thus, participates in important biological processes such as respiration, photosynthesis, and scavenging of oxidative stress. In addition, copper is involved in the perception of ethylene, nitrogen metabolism, molybdenum cofactor synthesis, cell wall remodeling, and response to pathogens in plants (1, 4, 5). However, free copper ions are toxic to cells in excess due to their ability to promote the formation of free radicals through the Fenton reaction and increased malfunction of important proteins, either through thiol capping or displacement of metal co-factors of metalloenzymes (1, 6–8). Therefore, organisms have evolved sophisticated mechanisms for maintaining copper homeostasis to prevent deficiency while avoiding toxicity.
The response to copper deficiency in plants includes reallocation of intracellular copper and induction of the expression of copper uptake systems. In Arabidopsis thaliana, this regulation is largely attributable to the activity of the transcription factor SPL7 (SQUAMOSA promoter-binding protein-like 7) and its downstream targets, microRNAs (9–13). When copper is limited, SPL7 up-regulates expression of miRNA398, which in turn down-regulates expression of two main isoforms of the major copper enzymes, the cytosol Cu/Zn-superoxide dismutase (SOD),4 CSD1, and the chloroplast stroma SOD, CSD2 (9–13). As CSD1 and CSD2 transcripts and the activity of the encoded proteins decrease, their superoxide-scavenging functions are replaced by an increase in gene expression and total enzyme activity of a plastid-localized Fe-SOD, FSD1 (11, 14). The miRNA-mediated copper economy model has been proposed, where energy-related electron transport functions receive priority in copper delivery over the major copper enzymes (1, 11, 15, 16).
Simultaneously, plants regulate copper homeostasis by controlling its uptake into cells. Copper uptake in plants, the green alga Chlamydomonas reinhardtii, yeast, Drosophila, and humans is maintained mainly through the tight regulation of the expression and stability of copper transporters of the CTR/COPT family (17–25). The founding members, Ctr1p, Ctr2p, and Ctr3p, were identified in Saccharomyces cerevisiae (17–19, 25). Ctr1p and Ctr3p localize to the plasma membrane, are functionally redundant, and mediate high affinity copper uptake from the external medium during copper deficiency (17, 18, 25). Expression of the CTR1 gene is subject to copper metalloregulation; it is transcriptionally induced under copper-deficient conditions via a transcription factor, Mac1, and is post-transcriptionally degraded under copper-replete conditions (26–28). Ctr2p is located on the vacuolar membrane and mobilizes copper from the vacuole during copper deficiency (19). CTR/COPT family members have conserved structural features that include three putative transmembrane (TM) helices, with the N and C termini located toward the extracellular space and cytosol, respectively; methionine-rich N-terminal motifs (Mets motifs); and MXXXM and GXXXXG motifs located in TM2 and TM3, respectively (29–32). Two methionine residues, Met127 in the Mets motif of the N terminus and Met260 in the MXXXM motif of TM2 in S. cerevisiae Ctr1p, are conserved among the majority of CTR/COPT family members and, based on studies of yeast and human Ctr1p, are essential for copper transport (31). The C terminus may contain cysteine-rich CXC motifs, which are suggested to bind copper ions for transfer to cytosolic copper chaperones or to down-regulate Ctr1p activity in response to toxic copper levels (31, 33, 34). CTR/COPT proteins homotrimerize to form a pore within the membrane to transport copper across the lipid bilayer but can also form heterocomplexes with other CTR/COPT family members and/or other proteins (5, 29, 30, 35, 36).
In plants, the CTR/COPT family is best characterized in A. thaliana and Oryza sativa and is represented by six and seven members, respectively (32, 36). A. thaliana COPT1 and COPT2 fully suppress the copper deficiency-associated respiratory defect of the S. cerevisiae ctr1Δctr3Δ mutant, whereas COPT3 and COPT5 partially complement this phenotype (22–24). Subsequent studies showed that COPT1 localizes to the plasma membrane, mediates copper influx, and, similar to CTR proteins from other organisms, is highly specific for copper(I) (20, 23). Studies in A. thaliana showed that COPT1 plays a predominant role in copper acquisition from the soil via root tips (20, 22). In contrast, COPT5 localizes to the tonoplast and prevacuolar compartment and functions by remobilizing copper from these organelles during copper deficiency (37, 38). Expression of COPT1 and COPT2 mRNA is up-regulated in copper-deficient conditions (22), and these responses are controlled by the transcription factor, SPL7 (12). COPT6 is a newly identified member of the CTR/COPT family of A. thaliana transporters (32), and its function in copper homeostasis has not yet been elucidated.
We show that COPT6 is a plasma membrane copper uptake transporter that is distinct from its S. cerevisiae counterpart, Ctr1p, and plays an important role in A. thaliana during copper deficiency and excess.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
All plant lines used in the study were in the A. thaliana Columbia (Col-0) background. Seeds of the copt6-1 (SALK 083438) T-DNA insertion allele were obtained from the Arabidopsis biological Resource Center (39). Seeds of the spl7-1 mutant were obtained from Dr. Shikanai (Kyoto University), and this mutant has been described (12). Before growing different A. thaliana lines on solid medium, seeds were surface-sterilized with 75% (v/v) ethanol before soaking in a solution containing 1.8% bleach (made up by diluting household Clorox) and 0.1% Tween 20. Sterilized seeds were rinsed with sterile water and sowed on half-strength Murashige and Skoog (½ MS) medium (pH 5.7) with 0.1% (w/v) sucrose and 0.7% (w/v) agar (Sigma A1296) supplemented with or without 45 μm CuCl2 or the indicated concentrations of the specific copper chelator, bathocuproine disulfonate (BCS). After stratification at 4 °C for 2 days in darkness, seeds were germinated, and seedlings were grown vertically for 10 days (at 22 °C; 12-h light/12-h dark photoperiod at photosynthetic photon flux density of 120 μmol m−2 s−1) before subsequent analyses.
RNA Extraction and cDNA Synthesis
Root and shoot tissues from 10-day-old seedlings grown under the indicated conditions were separated and flash-frozen in liquid nitrogen before homogenization in liquid nitrogen using a mortar and pestle. Total RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Genomic DNA in total RNA samples was digested with DNase I (Roche Applied Science) prior to first strand cDNA synthesis using the QPCR cDNA synthesis kit (Bio-Rad).
Plasmid Construction
The vectors YES3 (40) and SAT6-N1-EGFP (41) were modified into Gateway destination vectors with the Gateway® vector conversion system (Invitrogen) and designated accordingly as YES3-Gate and SAT6-N1-EGFP-Gate.
To generate the YES3-EGFP vector, the EGFP DNA sequence was amplified from the SAT6-N1-EGFP vector using primer pairs that generated SmaI restriction enzyme recognition sites at 5′ and 3′ ends of the PCR product (supplemental Table S1). The PCR product was then subcloned into SmaI/SmaI restriction enzyme sites of the YES3 vector. The resulting YES3-EGFP was converted into YES3-EGFP-Gate with the Gateway® vector conversion system (Invitrogen).
The COPT6 cDNA with or without the stop codon was amplified by RT-PCR from RNA isolated from A. thaliana leaves. The primers (supplemental Table S1) added attB sites on resulting PCR products, which were then introduced into corresponding vectors by recombination cloning (Invitrogen).
Site-directed mutagenesis of COPT6 was performed directly on a YES3-Gate-COPT6 vector with QuikChange® II site-directed mutagenesis kit (Stratagene). The mutagenic oligonucleotides (supplemental Table 1) were designed to substitute conserved methionines to alanine codons in the Mets motif of the predicted extracellular domain or TM2 of COPT6 or to delete 81 bp corresponding to 27 amino acids of the extracellular N-terminal domain (supplemental Fig. 1). Mutagenesis was confirmed by sequencing the cDNA regions encompassing each mutation.
The cDNA encoding the full-length Ctr1p, and Ctr1p with M127L and M260A point mutations were amplified by PCR from the 416TEF vector (courtesy of Dr. Dennis Thiele, Duke University) using primer pairs (supplemental Table 1). All inserts were introduced into the DONR222 entry vector before recombination with the YES3-Gate destination vector mentioned above.
Functional Complementation of the S. cerevisiae Copper Uptake-deficient ctr1Δctr2Δctr3Δ Mutant Strain
S. cerevisiae SEY6210 (MATa ura3-52 leu2-3,-112 his3Δ200 trp1Δ901 lys2-801 suc2Δ9) wild-type and ctr1Δctr2Δctr3Δ triple mutant (MATa ura3-52 his3Δ200 trp1-901 ctr1::ura3::Knr ctr2::HIS3 ctr3::TRP1) strains that were used for functional complementation assays were the generous gift of Dr. Dennis Thiele (Duke University). Yeast cells were transformed with the YES3-Gate-COPT6 vector or YES3-Gate lacking the cDNA insert using the Frozen-EZ Yeast Transformation II kit (Zymo Research). Transformants were selected for uracil prototrophy on YNB medium containing 6.7% (w/v) yeast nitrogen base without amino acids (Difco), 0.77% (w/v) CSM-URA, 0.5% (w/v) NaCl, 2% glucose, 2% (w/v) agar.
Respiration competence was evaluated by testing the ability of transformants to grow on the non-fermentable carbon sources, glycerol and ethanol (17, 31). Transformants were grown in liquid YNB-URA to an A600 nm = 1.0–1.1, serially 10-fold diluted, and spotted onto YPEG medium containing 1% (w/v) yeast extract, 2% (w/v) bacto-peptone, 3% (v/v) glycerol, 2% (v/v) ethanol, and 2% (w/v) agar and the indicated concentrations of CuSO4. Plates were incubated for 3 days at 30 °C.
Quantitative Real-time (qRT)-PCR and Data Analysis
Prior to qRT-PCR analysis, primer and cDNA concentrations were optimized to reach the target and normalizing gene amplification efficiency of 100 ± 10%. One microliter of 15-fold diluted cDNA was used as a template for qRT-PCR in a total volume of 10 μl containing a 500 nm concentration of each PCR primer, 50 mm KCl, 20 mm Tris-HCl, pH 8.4, 0.2 mm each dNTP, and 1.25 units of iTaq DNA polymerase in iQ SYBR Green Supermix (Bio-Rad). PCR was carried out using the CFX96 real-time PCR system (Bio-Rad). The thermal cycling parameters were as follows: denaturation at 95 °C for 3 min, followed by 39 cycles of 95 °C for 10 s and 55 °C for 30 s. Amplicon dissociation curves (i.e. melting curves) were recorded after cycle 39 by heating from 60 °C to 95 °C with 0.5 °C increments and an average ramp speed of 3.3 °C s−1. Real-time PCR experiments were conducted using three independent biological samples, each consisting of three technical replicates (42), unless indicated otherwise. Data were normalized to the expression of ACTIN 2. The -fold difference (2−ΔΔCt) was calculated using the CFX Manager Software, version 1.5 (Bio-Rad). Statistical analysis was performed using the Relative Expression Software Tool (REST; Qiagen) (43).
Subcellular Localization and Fluorescent Microscopy
For studies of subcellular localization in S. cerevisiae, COPT6 was fused at the C terminus with the EGFP in the YES3-EGFP-Gate vector and expressed under the control of the constitutive phosphoglycerate kinase (PGK) gene promoter. The resulting COPT6-EGFP construct and the empty YES3-EGFP-Gate vector were transformed into S. cerevisiae ctr1Δctr2Δctr3Δ triple mutants using the Frozen-EZ Yeast Transformation II kit (Zymo Research). Plasma membranes were stained with FM 4-64 dye, an endocytic marker that is also used to visualize the plasma membrane after short term staining at 0 °C (44–46). Three milliliters of overnight culture were pelleted by centrifugation, and concentrated cells were resuspended in 1 ml of ice-cold liquid YPD medium containing 20% peptone (w/v), 10% yeast extract (w/v), and 2% glucose (v/v), pH 6.0. The sample was cooled on ice for 5–10 min before adding 3 μl of DMSO and FM 4-64 at a final concentration of 7.5 μm. The sample was incubated at 4 °C before dispensing on precooled slides topped with 1% agarose for visualization. Epifluorescence images were collected using an Axio Imager M2 microscope (Zeiss).
For studies of the subcellular localization of COPT6 in Arabidopsis protoplasts, the full-length COPT6 cDNA without the stop codon was fused at the C terminus with the modified green fluorescent protein (EGFP) using the SAT6-N1-EGFP-Gate vector and expressed under the control of the cauliflower mosaic virus 35S promoter. The resulting 35Spro-COPT6-EGFP construct or SAT6-N1-Gate, lacking the cDNA insert, was transfected into A. thaliana protoplasts isolated from leaf mesophyll tissue using previously established procedures (47–49). Plasma membranes were stained with 50 μm FM 4-64 as described (45). EGFP- and FM 4-64-mediated fluorescence and chlorophyll autofluorescence were visualized using FITC (for EGFP) or rhodamine (FM 4-64 and chlorophyll) filter sets of the Axio Imager M2 microscope equipped with the motorized Z-drive (Zeiss). z stack (1.3-μm-thick) images were collected with the high resolution AxioCam MR Camera and then three-dimensionally deconvoluted using an inverse filter algorithm of the Zeiss AxioVision 4.8 software. Images were processed using the Adobe Photoshop software package, version 12.0.
Bimolecular Fluorescence Complementation (BiFC) Assays and Confocal Microscopy
The vectors used in the BiFC assay, UC-SPYNE and UC-SPYCE, contained the N- or C-terminal fragments of yellow fluorescent protein, YFPN or YFPC, respectively, allowing expression of fused proteins under the control of the 35S promoter (50). To generate COPT6-YFPN and COPT6-YFPC fusion constructs, the C terminus of the COPT6 coding sequence was fused with YFPN or YFPC in the UC-SPYNE or UC-SPYCE vectors, respectively. The coding regions of COPT1 and COPT5 were amplified without stop codons and used to generate COPT1-YFPN and COPT5-YFPN. The resulting constructs and UC-SPYNE and UC-SPYCE vectors lacking cDNA inserts were co-transfected in the indicated combinations into A. thaliana leaf protoplasts as described (47–49). Protoplasts were analyzed using a Zeiss 710 confocal microscope. The YFP fluorophore was excited with a 514-nm laser, and the emission was recorded in the range of 524–595 nm and 650–715 nm for YFP and chlorophyll, respectively. To allow comparison of the relative brightness between different BiFC experiments, the zoom, pinhole, detector gain, amplifier offset, frame size, scan speed, scan average, and laser power were kept consistent between samples. Images were processed using the Zen 2009 LE software of the Zeiss 710 confocal microscope and the Adobe Photoshop software package, version 12.0.
Split Ubiquitin Membrane Yeast Two-hybrid System (MYTH)
Vectors, S. cerevisiae strains THY.AP4 (MATa leu2-3,112 ura3-52 trp1-289 lexA::HIS3 lexA::ADE2 lexA::lacZ) and THY.AP5 (MATα URA3 leu2-3,112 trp1-289 his3-Δ1 ade2Δ::loxP) for MYTH were obtained from the Frommer laboratory (Stanford University) depository at the Arabidopsis Biological Resource Center (ABRC). The full-length COPT6 cDNA was introduced into MetYCgate and XNgate21-3HA vectors by in vivo cloning in yeast as described (51, 52) to generate bait COPT6-CubPLV and prey COPT6-NubG constructs in THY.AP4 and THY.AP5 strains, respectively. In both cases, C- and N-terminal fragments of ubiquitin were placed at the C terminus of COPT6. Protein interactions were selected in diploid cells after 2 days of growth on SC medium lacking adenine and histidine. Interactions were verified using β-galactosidase assays, as detailed (51, 53).
Histochemical Analysis
To examine the pattern of COPT6 expression in planta, a 1,083-bp fragment of the genomic sequence upstream of the COPT6 start codon was amplified by PCR using primer pairs listed in supplemental Table 1. The resulting PCR product was cloned by recombination into the GUS1-Gate vector upstream of uidA, encoding the β-glucuronidase (GUS) reporter gene. The COPT6pro-GUS construct was transformed into wild-type A. thaliana (54). Histochemical staining was performed with 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described (55) with a 20-min (for seedlings) or 3-h (for mature plants) incubation period at 37 °C. Staining patterns were analyzed using the Zeiss 2000 stereomicroscope. Images were collected using a Canon PowerShot S3 IS digital camera and a CS3IS camera adapter. Images were processed using the Adobe Photoshop software package, version 12.0.
Generation of Transgenic Lines
The full-length COPT6 cDNA was introduced by recombination cloning into EarleyGate201 destination vector (56), where COPT6 was fused at the N termini to the human influenza hemagglutinin (HA) epitope tag under the control of the 35S promoter. The resulting 35Spro-COPT6-HA construct or the EarleyGate201 vector lacking the cDNA insert was transformed into wild-type A. thaliana via the floral dip method (54). Based on results of qRT-PCR analyses of T2 transgenic lines, two homozygous lines overexpressing COPT6-HA, 35Spro-COPT6-1-HA and 35Spro-COPT6-2-HA, and one line expressing the empty vector, 35Spro, were selected for subsequent experiments.
SDS-PAGE and Western Blot Analysis
Two grams of leaf tissue were taken from 35-day-old transgenic plants grown on soil and ground in liquid nitrogen using a mortar and pestle before the addition of extraction buffer containing 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 150 mm NaCl, 10% glycerol, 2% polyvinylpolypyrrolidone, 0.25% Triton X-100, 5 mm DTT, 1 mm PMSF, 0.2 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 μg/ml pepstatin. Total protein extracts were cleared from cellular debris by centrifugation at 4,000 × g for 10 min at 4 °C. The supernatant was centrifuged at 100,000 × g for 1 h at 4 °C, and pellet containing total membrane proteins was reconstituted in extraction buffer and stored at −80 °C. Aliquots of proteins (20 μg/lane) were subjected to SDS-PAGE on 12% (w/v) gels and electrotransferred to nitrocellulose filters in Towbin buffer containing 0.05% SDS (57). For detection of the influenza hemagglutinin-HA epitope, nitrocellulose filters were probed with a primary polyclonal anti-HA antibody (1:2,000 dilution; Sigma) and a secondary HRP-conjugated IgG antibody (1:10,000 dilution; GE Healthcare). In both cases, immunoreactive bands were visualized with ECL using the LumiGLO system (KPL).
Analyses of Copper Content
S. cerevisiae wild-type SEY6210 and ctr1Δctr2Δctr3Δ mutant cells transformed with YES3-Gate vector and the ctr1Δctr2Δctr3Δ mutant transformed with YES3-Gate-COPT6 were grown overnight at 30 °C to an A600 nm = 1.2 in liquid YNB medium supplemented with CSM-URA and 2% glucose. One hundred-microliter aliquots of the overnight culture were diluted into 20 ml of the same medium supplemented with the indicated concentrations of CuSO4 and grown for 18 h at 30 °C. Cells were then harvested by centrifugation and washed with deionized water before copper in cell walls was desorbed in a buffer containing 1 mm EDTA and 100 μm BCS, pH 8.0. Cells were then washed two more times with deionized water, dried, digested by heating with a combination of purified concentrated nitric and perchloric acids, and finally dissolved in 10 ml of 5% nitric acid. Copper content in processed yeast and plant samples was analyzed by inductively coupled plasma mass spectroscopy (ICP-MS; Agilent 7500) (58). Copper at mass 63 was measured using helium as the collision gas at 5.0 ml/min to reduce potential polyatomic interferences from sodium-argon ions, which also has a mass at 63.
For analyses of copper content in A. thaliana, 10-day-old seedlings of different plant lines grown as described above were transferred to Arabidopsis hydroponic solution as described (59) and grown at 22 °C and 12-h light/12-h dark photoperiod at photosynthetic photon flux density of 120 μmol m−2 s−1. Roots and shoots of 30-day-old plants were harvested, and roots were desorbed by washing with 10 mm EDTA followed by washing in a solution of 0.3 mm BCS and 5.7 mm sodium dithionite before rinsing with deionized water (60). Shoots were rinsed with deionized water. Root and shoot tissues were dried, processed, and analyzed for copper content as described above.
RESULTS
The COPT6 Polypeptide Contains the Key Features of the CTR/COPT Family
The A. thaliana genome possesses six genes encoding putative CTR/COPT transporters that are designated COPT1 to 6. Of the six family members, COPT6 has been identified in the A. thaliana genome only recently (32), thus making COPT6 the least studied member of the A. thaliana CTR/COPT family. Analysis of the amino acid sequence identity and similarity of COPT6 to other CTR/COPT family members of A. thaliana disclosed that COPT2 and COPT1 are the closest COPT6 homologs, sharing 75%/79% sequence identity/similarity and 71%/75% identity/similarity, respectively (supplemental Fig. 1). In addition, COPT6 clusters in one clade with COPT1 and COPT2 when their amino acid sequences are subjected to phylogenetic analysis (Fig. 1A), suggesting that COPT6 may have similar functions to COPT1 and COPT2 in copper homeostasis.
FIGURE 1.
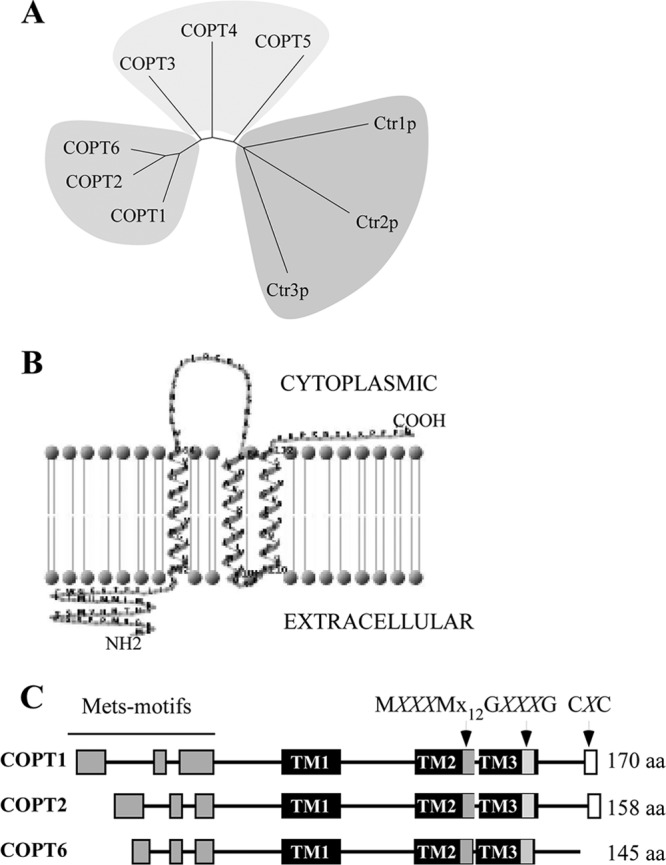
In silico analysis of the putative A. thaliana copper transporter, COPT6. A, phylogenetic tree of A. thaliana COPT proteins and S. cerevisiae Ctr1p, Ctr2p, and Ctr3p. B, schematic representation of the membrane topology of the full-length COPT6 polypeptide. Based on the predicted topology (TMHMM software, version 2.0, and TMRPres2D), the N terminus is located outside (Extracellular), whereas the C terminus is inside the cell (Cytoplasmic). C, schematic diagram showing conserved motifs in the primary sequence of COPT1, COPT2, and COPT6. The conserved features include three transmembrane domains (TM1, TM2, and TM3; black bars); methionine-rich motifs in the predicted extracellular domain (Mets motifs; gray bars); and MXXXM, GXXXG, and CXC motifs (MXXXMX12GXXXG (gray bars) and CXC (white bars)) in TM2, TM3, and the C terminus hydrophilic domain, respectively. Note that the CXC motif is absent in the COPT6 polypeptide. M, G, and C, methionine, glycine, cysteine amino acid residues, respectively; X, any amino acid residue (aa).
Computer algorithm-assisted analysis of the COPT6 polypeptide membrane topology and conserved motif organization revealed an extracellular N-terminal domain; a membrane domain encompassing three TM helices, where TM2 and TM3 are separated by 3 amino acids; and an intracellular C-terminal domain (Fig. 1B). The COPT6 polypeptide includes the MXXXMX12GXXXG signature of CTR/COPT proteins; its N terminus and TM2 contain Mets and MXXXM motifs, respectively, whereas TM3 harbors the GXXXG motif (Fig. 1C). These features are conserved in the majority of the CTR/COPT proteins and are also present in COPT1 and COPT2. However, unlike COPT1 and COPT2, COPT6 does not possess the cysteine-rich CXC motif at the C terminus (Fig. 1C).
Heterologously Expressed COPT6 Complements the Growth Defect of the Triple ctr1Δctr2Δctr3Δ S. cerevisiae Mutant on Non-fermentable Growth Medium
Despite the presence of key motifs conserved in the CTR/COPT family, COPT6 lacks the CXC motif at the C terminus (Fig. 1C), which was shown to play an important role in copper homeostasis in Ctr1p (31, 33, 34). This motif is also present in A. thaliana homologues COPT1 and COPT2, prompting us to test whether the lack of CXC motif in COPT6 still results in a protein capable of transporting copper. In this regard, S. cerevisiae strains harboring mutations in copper uptake genes have been used as a versatile and well defined model system for functional analysis of putative copper transporters from various species. Yeast lacking copper uptake systems cannot grow on non-fermentable carbon sources, such as glycerol and ethanol (YPEG medium), due to low levels of intracellular copper. This deficiency results in the inability of cytochrome c oxidase to obtain its copper cofactor, which, in turn, causes a defect in the mitochondrial respiratory chain unless copper is exogenously supplied to the medium (17, 61).
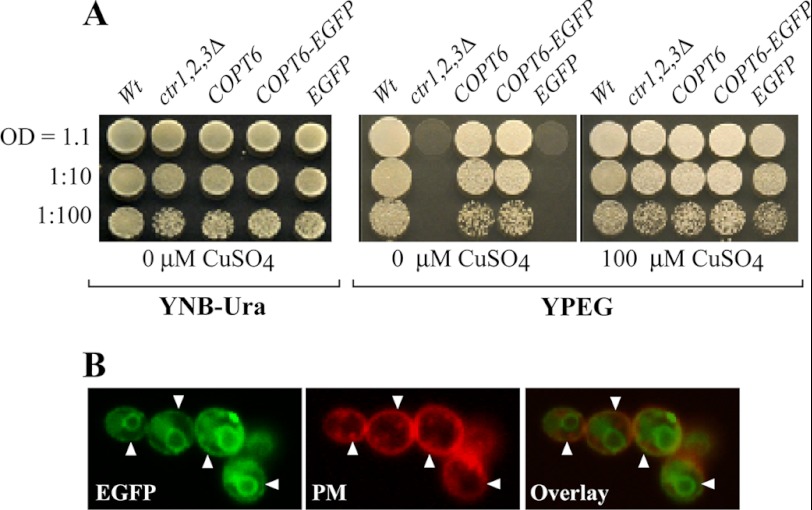
The function of COPT6 in copper transport was tested through complementation studies in the S. cerevisiae ctr1Δctr2Δctr3Δ triple mutant strain, which lacks high affinity plasma membrane transporters Ctr1p and Ctr3p, and a vacuolar membrane transporter, Ctr2p (19). Growth of ctr1Δctr2Δctr3Δ mutant and wild-type strains expressing an empty YES3-Gate vector and the ctr1Δctr2Δctr3Δ mutant transformed with YES3-Gate harboring the COPT6 cDNA insert was compared on medium with glucose (YNB-URA) or ethanol/glycerol (YPEG) as a carbon source. All yeast alleles were able to grow on YNB-URA (Fig. 2A). As shown previously (19), ctr1Δctr2Δctr3Δ cells did not grow on YPEG medium unless the medium was supplemented with exogenous copper (Fig. 2A). In contrast, ctr1Δctr2Δctr3Δ cells expressing COPT6 grew on YPEG regardless of copper supplementation, and their growth was comparable with that of wild-type cells transformed with the empty vector (Fig. 2A).
FIGURE 2.
COPT6 rescues growth defect of the ctr1Δctr2Δctr3Δ S. cerevisiae triple mutant on ethanol/glycerol medium (YPEG). A, the wild-type SEY6210 strain, transformed with the empty vector (Wt), and the ctr1Δctr2Δctr3Δ mutant transformed with the empty vector (ctr1,2,3Δ) or the vector containing COPT6 (COPT6), COPT6-EGFP (COPT6-EGFP), or EGFP (EGFP) cDNA inserts were grown overnight in liquid medium to an A600 nm = 1.1. Cells were then serially 10-fold diluted (indicated on the left) and spotted either onto solid YNB-URA medium (YNB-URA) or YPEG (YPEG) with the indicated concentrations of CuSO4. B, microphotographs of EGFP- and FM 4-64-mediated fluorescence (EGFP and PM, respectively) derived from S. cerevisiae expressing COPT6-EGFP construct and stained with FM 4-64 dye. Superimposed EGFP and FM 4-64 fluorescent images (Overlay) show that COPT6-EGFP localizes at the plasma membrane. A fraction of COPT6-EGFP is also present at ER and inside the vacuole (see the explanation under “Results”).
To determine the subcellular localization of COPT6 in the heterologous system, we fused it at the C terminus to EGFP of the YES3-EGFP-Gate vector and transformed this construct or the empty vector expressing EGFP only into the ctr1Δctr2Δctr3Δ mutant. The COPT6-EGFP construct was functional because, unlike the empty vector, it rescued growth of the ctr1Δctr2Δctr3Δ mutant on YPEG medium (Fig. 2A). The COPT6-EGFP-mediated fluorescence was observed at the cell periphery (Fig. 2B), where it overlapped with FM 4-64 dye, which stained the plasma membrane after short term labeling conducted in low temperatures (44–46). These data suggested that COPT6-EGFP localizes to the plasma membrane in the heterologous system. A fraction of COPT6-EGFP degraded, resulting in EGFP accumulation inside the vacuole and in endoplasmic reticulum (Fig. 2B), as frequently observed for overexpressed plasma membrane proteins (62, 63). Nevertheless, the plasma membrane-localized COPT6-EGFP was sufficient to complement the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium (Fig. 2A). The ability of heterologously expressed COPT6 to rescue the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium, along with its plasma membrane localization, suggest that COPT6 is involved in copper uptake.
Heterologously Expressed COPT6 Facilitates Copper Accumulation in ctr1Δctr2Δctr3Δ S. cerevisiae Cells
To ascertain that COPT6 rescues the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium by transporting copper, we analyzed the copper content of the ctr1Δctr2Δctr3Δ mutant expressing the YES3-Gate vector with or without the COPT6 cDNA and wild-type cells expressing the YES3-Gate vector. Yeast cells were grown on YNB-URA medium supplemented with the indicated concentrations of CuSO4, and copper content was analyzed by ICP-MS. We found that empty vector-expressing wild-type cells accumulated 20.7 ± 0.5 ppb of copper when grown on YNB-URA. In contrast, the ctr1Δctr2Δctr3Δ mutant accumulated 6-fold less copper than the empty vector-expressing wild type (Fig. 3). These data are consistent with previous findings showing that CTR proteins are important for copper transport (17, 25, 31). Expression of COPT6 in the ctr1Δctr2Δctr3Δ mutant increased its ability to accumulate copper to the level of the wild-type strain (Fig. 3). Supplementing YNB-URA with the indicated concentrations of copper increased copper accumulation in all yeast alleles (Fig. 3). Nevertheless, ctr1Δctr2Δctr3Δ cells expressing COPT6 accumulated 2- and 3-fold more copper when grown at 10 and 20 μm CuSO4, respectively, than mutant cells expressing the empty vector and 3- and 4-fold more copper than the empty vector-expressing wild type (Fig. 3). These data indicate unambiguously that A. thaliana COPT6 is a copper uptake transporter.
FIGURE 3.
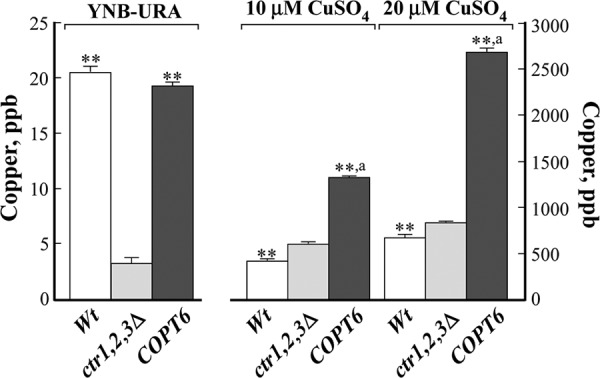
Copper content of the vector-transformed S. cerevisiae wild-type (Wt) and ctr1Δctr2Δctr3Δ mutant cells (ctr1,2,3Δ) and ctr1Δctr2Δctr3Δ mutant cells transformed with the vector containing the COPT6 cDNA insert (COPT6). Yeast cells were cultured in liquid YNB-URA medium that originally contained 0.25 μm CuSO4 (YNB-URA) or on YNB-URA supplemented with additional concentrations of CuSO4, as indicated. Copper content was analyzed by ICP-MS. Statistical significance of differences was determined using analysis of variance single factor analysis. **, statistically significant differences of the mean values from the vector-transformed ctr1Δctr2Δctr3Δ mutant (p ≤ 0.001; n = 5). a, statistically significant differences (p ≤ 0.001) of the mean values from the vector-expressing wild-type cells. Error bars, S.E.
A Positionally Conserved Methionine Residue in TM2 (Met106), but Not a Positionally Conserved N-terminal Methionine Residue (Met27), Is Essential for COPT6 Function
Elegant genetic and biochemical studies have shown that positionally conserved methionine residues within the hydrophilic N-terminal extracellular domain of S. cerevisiae Ctr1p, Met127, and in TM2, Met260, are important for extracellular copper binding and uptake, respectively (31). To test whether the corresponding residues in COPT6 (Met27 in the N terminus and Met106 in TM2; supplemental Fig. 1) are also essential for function, we converted Met27 and Met106 in the full-length COPT6 wild-type protein to alanine, M27A and M106A, respectively, and transformed these mutant alleles into the ctr1Δctr2Δctr3Δ mutant. Concurrently, the ctr1Δctr2Δctr3Δ mutant was transformed with full-length Ctr1p or the previously described point mutants with Met127 and Met260 substituted to leucine or alanine (M127L or M260A, respectively) as controls (31). We then compared the ability of wild-type COPT6, wild-type Ctr1p, and their mutant alleles to suppress the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium.
We found that all yeast alleles grew well on YPEG medium supplemented with 100 μm CuSO4 (Fig. 4). As expected, expression of the full-length wild-type COPT6 and Ctr1p complemented the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium; however, expression of M106A-substituted COPT6 did not (Fig. 4). Consistent with previous findings (31), the Ctr1p mutant allele with a point mutation in the corresponding Met260 residue (M260A) also did not complement the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG (Fig. 4). We also tested whether the adjacent conserved Met102 residue in TM2 of COPT6 is essential for COPT6 function. However, substituting Met102 to alanine did not appear to alter COPT6 function because the ctr1Δctr2Δctr3Δ mutant expressing the M102A allele grew on YPEG medium as well as mutant cells expressing wild-type COPT6 or Ctr1p (Fig. 4). These results show that the function of one of two methionine residues within the TM2 is conserved throughout the CTR/COPT proteins in yeast and plants.
FIGURE 4.
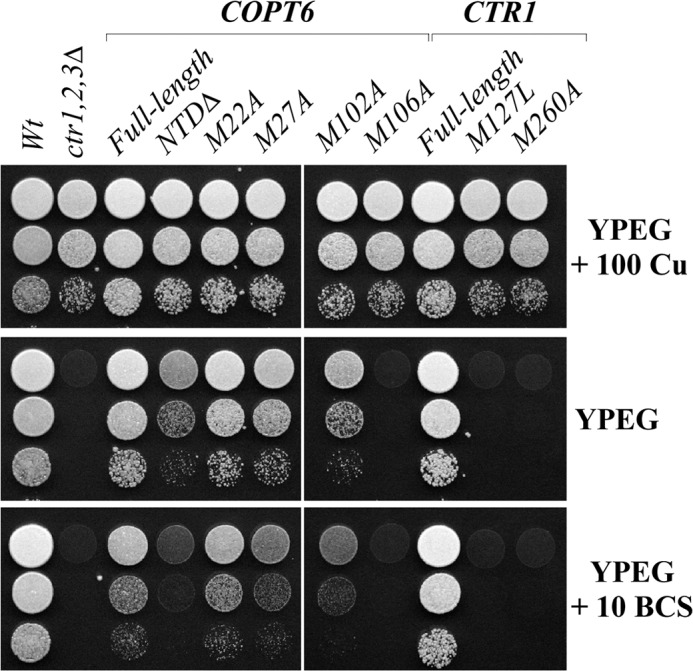
The conserved Met106 residue within the TM2 is essential for COPT6 function. S. cerevisiae wild-type (Wt) and ctr1Δctr2Δctr3Δ mutant cells (ctr1,2,3Δ) were transformed with the empty vector. ctr1Δctr2Δctr3Δ mutant cells were also transformed with the vector containing the full-length COPT6, S. cerevisiae CTR1 cDNA inserts (COPT6 and CTR1, respectively), COPT6 lacking the extracellular N-terminal domain (NTDΔ), Met-to-Ala-substituted COPT6 alleles (M22A, M27A, M102A, and M106A), or Met-to-Leu and Met-to-Ala-substituted CTR1 alleles (M127L and M260A). Yeast alleles were grown, diluted as indicated in the legend to Fig. 3, and spotted on YPEG medium with or without 100 μm CuSO4 (100 Cu) or 10 μm BCS (10 BCS), as indicated on the right.
Consistent with previous findings of the essential role of a positionally conserved Met127 residue in the N terminus of Ctr1p (31), expression of the M127L mutant allele of Ctr1p did not complement the respiratory deficiency of the ctr1Δctr2Δctr3Δ mutant on the YPEG medium (Fig. 4). Surprisingly, substitution of the corresponding Met27 (M27A) residue in COPT6 complemented the ctr1Δctr2Δctr3Δ mutant growth defect on YPEG medium, suggesting that this residue in the N terminus of COPT6 is not required for COPT6 function. In addition, mutation of the semiconserved, adjacent Met22 (M22A) in the N terminus (supplemental Fig. 1) also complemented the growth defect of the ctr1Δctr2Δctr3Δ mutant on YPEG medium. These data suggest that, unlike Ctr1p, the tested conserved Met residues of the N terminus in COPT6 are not essential for its function.
The Methionine-rich N Terminus Is Required for COPT6 Function under Conditions of Copper Limitation
Previous studies suggested that Met127 within the methionine-rich regions (Mets motifs) of the N terminus of Ctr1p is involved in extracellular copper binding prior to its transport into the cell (31). Our data indicate that the corresponding positionally conserved Met27 or adjacent semiconserved Met22 are dispensable for COPT6 function. To determine the function of the N terminus of COPT6, we decided to generate a COPT6 mutant allele without its extracellular N-terminal domain (NTDΔ). We then tested the respiratory competence of the ctr1Δctr2Δctr3Δ mutant expressing NTDΔ-COPT6 compared with the mutant expressing full-length COPT6 or Ctr1p proteins. Surprisingly, cells expressing NTDΔ-COPT6 were still able to grow on YPEG medium (Fig. 4), suggesting that the N-terminal region of COPT6 is not absolutely essential for the function of the transporter. However, we noticed that the cell density of NTDΔ-COPT6-expressing cells on YPEG medium was lower than that of cells expressing the full-length COPT6 or Ctr1p (Fig. 4). We then tested if limiting bioavailable copper through the addition of a specific copper chelator, BCS, to the YPEG medium would alter the growth of cells expressing NTDΔ-COPT6. We found that the growth of cells expressing NTDΔ-COPT6 was severely impacted when extracellular bioavailable copper was limited by the addition of BCS (Fig. 4, YPEG + 10 BCS). These results suggest that the extracellular N-terminal domain of COPT6 plays an important role in copper homeostasis under copper-limiting conditions. Future studies will reveal the specific amino acid residue(s) involved in copper binding.
COPT6 Localizes to the Plasma Membrane in A. thaliana Protoplasts
Members of the CTR/COPT family in different species associate with different endomembranes, including the plasma membrane or endosomal vesicles/vacuoles, and contribute to copper homeostasis either by copper uptake from the external medium or copper release from internal compartments during deficiency (19, 20, 64, 65). Our studies of the subcellular localization of COPT6 in S. cerevisiae showed that in this heterologous system, it localizes to the plasma membrane. Subsequently, we sought to determine its localization in A. thaliana. The coding sequence of COPT6 was fused at the C terminus to EGFP in the SAT6-EGFP-N1-Gate vector and transiently expressed under the control of the constitutive cauliflower mosaic virus 35S promoter in A. thaliana protoplasts. As a control, protoplasts were transfected with the empty SAT6-EGFP-N1-Gate vector.
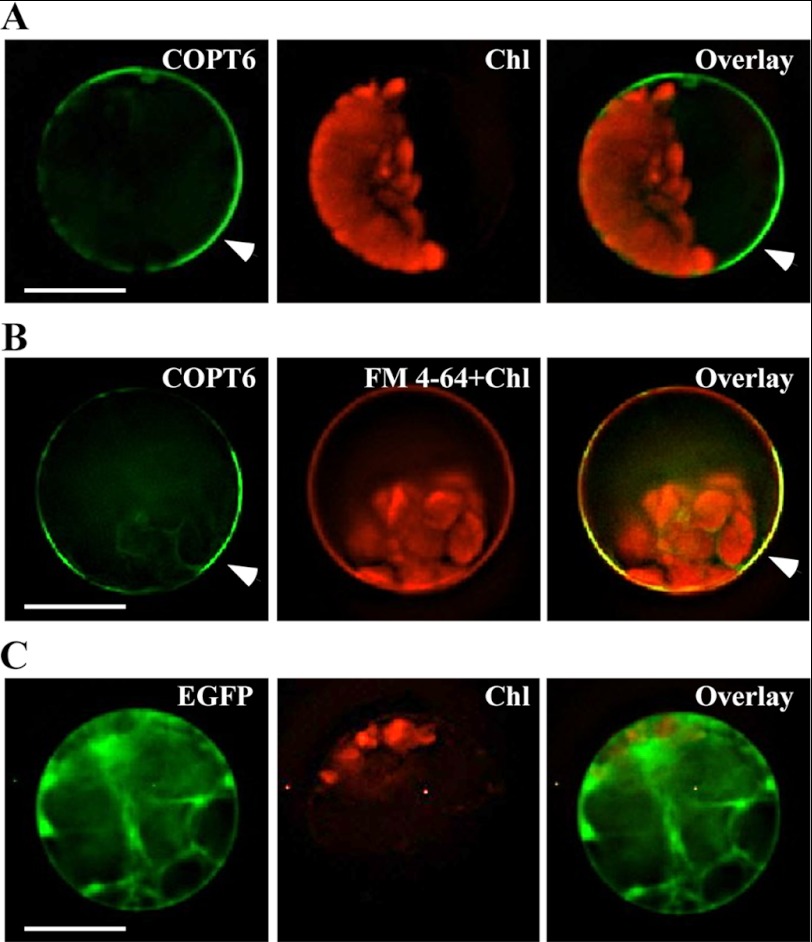
EGFP-mediated fluorescence was present at the periphery of COPT6-EGFP-transfected protoplasts and did not overlap with chlorophyll autofluorescence (Fig. 5A). To ascertain the plasma membrane localization of COPT6-EGFP, transfected protoplasts were co-stained with FM 4-64. After the short term labeling at 4 °C, FM 4-64 stained the plasma membrane (Fig. 5B, FM 4-64+Chl), and FM 4-64-mediated fluorescence overlapped with COPT6-EGFP-mediated fluorescence but not with chlorophyll-mediated fluorescence (Fig. 5B). In protoplasts transfected with the empty vector, EGFP was present as a soluble protein in the cytosol, and its fluorescence did not overlap with chlorophyll autofluorescence (Fig. 5C). These results are consistent with the plasma membrane localization of COPT6.
FIGURE 5.
Subcellular localization of COPT6 in A. thaliana protoplasts. A. thaliana leaf protoplasts were transfected with the vector expressing the COPT6-EGFP fusion (A and B) or vector expressing EGFP without the COPT6 cDNA insert (C). To visualize the plasma membrane, COPT6-EGFP-transfected protoplasts were co-stained with FM 4-64 (B). EGFP-mediated fluorescence, derived from COPT6-EGFP (COPT6) or EGFP (EGFP) and FM 4-64-mediated fluorescence and chlorophyll autofluorescence (FM 4-64 + Chl and Chl, respectively) were visualized using FITC or rhodamine filter sets. Superimposed images of COPT6-EGFP- and FM 4-64-mediated fluorescence and chlorophyll autofluorescence (Overlay) were created to demonstrate that green fluorescence derived from COPT6-EGFP co-localizes with FM 4-64. Scale bar, 10 μm.
We also co-transfected protoplasts with the SAT6-EGFP-N1-Gate vector containing a COPT6-EGFP fusion and BIN20 vector containing a plasma membrane marker, PIP2A, fused to mCherry (66). Fluorescence signals originated from COPT6-EGFP and PIP2A-mCherry co-localized at the periphery of transfected protoplasts (supplemental Fig. 2), suggesting that COPT6 similar to PIP2A localizes to the plasma membrane.
Analyses of the copt6-1 Mutant and Transgenic Plants Ectopically Expressing COPT6
Our studies in S. cerevisiae showed that COPT6 is involved in copper uptake (Figs. 2A and 3). To study the role of COPT6 in planta, we used a T-DNA insertion allele, copt6-1 (SALK 083438) and two transgenic lines, 35Spro-COPT6-1 and 35Spro-COPT6-2. Wild-type plants and a transgenic line expressing the EarlyGate201 vector without the COPT6 cDNA insert (35Spro) were used as controls for the copt6-1 mutant- or COPT6-overexpressing lines, respectively.
Sequencing of the copt6-1 genomic DNA fragment revealed a T-DNA insertion 110 bp upstream of the COPT6 start codon. qRT-PCR analysis disclosed a 10- and 8-fold decrease of COPT6 mRNA in roots and shoots, respectively, in the copt6-1 allele (Fig. 6A). In contrast, transcript abundance of COPT6 was 1,000- and 30-fold higher in roots and shoots of 35Spro-HA-COPT6-1 transgenic plants compared with plants expressing the empty vector (Fig. 6B). Expression of COPT6 in the 35Spro-HA-COPT6-2 transgenic line was 7,000 and 170 times higher in roots and shoots, respectively (Fig. 6B).
FIGURE 6.
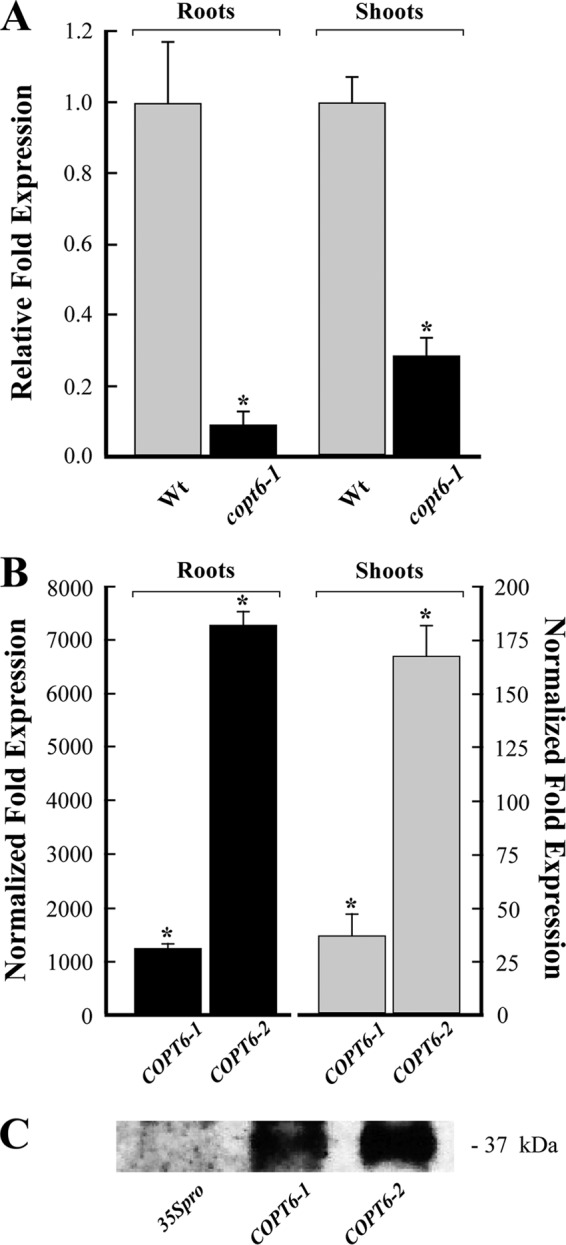
Characterization of the copt6-1 mutant allele and transgenic lines ectopically expressing 35Spro-HA-COPT6. A, qRT-PCR analysis of 10-day-old copt6-1 seedlings (copt6-1) indicates a 10- and 8-fold decrease of the abundance of the COPT6 transcript in roots and shoots, respectively, compared with the wild-type (Wt) seedlings. B, abundance of the COPT6 transcript in roots and shoots of two transgenic lines, 35Spro-HA-COPT6-1 (COPT6-1) and 35Spro-HA-COPT6 (COPT6-2). Results are presented relative to the expression of COPT6 in plants transformed with the empty vector, designated as 1. Error bars, S.E. (n = 9). *, statistically significant differences (p ≤ 0.05) of the mean values. C, immunoblot analysis of the COPT6-HA polypeptide in transgenic plants expressing EarleyGate201 vector lacking the cDNA insert (35Spro) and two lines (COPT6-1 and COPT6-2) expressing 35Spro-HA-COPT6. The apparent molecular weight of COPT6-HA is indicated on the right.
SDS-PAGE and immunoblot analyses of total membrane proteins isolated from leaves of transgenics plants revealed the anti-HA antibody reactive protein with an apparent molecular mass of ∼37 kDa in extracts from 35Spro-HA-COPT6-1 and 35Spro-HA-COPT6-2 transgenic lines but not from the empty vector-expressing plants (Fig. 6C). Consistent with the qRT-PCR results (Fig. 6B), the level of anti-HA immunoreactive polypeptide was higher in the 35Spro-HA-COPT6-2 line than in the 35Spro-HA-COPT6-1 line (Fig. 6C). It is noteworthy that the molecular mass of COPT6-HA is considerably larger than its predicted size of 16.9 kDa. It is possible that this aberrant migration is a consequence of posttranslational modifications (i.e. O-glycosylation) that were shown for the COPT6 homolog from S. cerevisiae, Ctr1p (31).
COPT6 Plays an Important Role in Copper Homeostasis in A. thaliana
Wild type, the copt6-1 mutant, and transgenic plants were germinated and grown on solid ½ MS medium (copper-sufficient conditions; Fig. 7A). To limit copper availability, ½ MS was supplemented with the indicated concentration of BCS (Fig. 7C). To impose copper toxicity, ½ MS was supplemented with 45 μm CuCl2 (Fig. 7D). These concentrations of CuCl2 and BCS were selected based on morphological and molecular phenotypes associated with copper excess or deficiency (supplemental Fig. 3). Copper toxicity and deficiency inhibits root growth of A. thaliana (1, 67, 68) and alters expression of the miRNA398 and its targets, cytosol- and plastid-localized Cu/ZnSODs, CSD1 and CSD2, and iron-containing SOD, FSD1, in shoots of A. thaliana. Specifically, the abundance of miRNA398 mRNA increases during copper deficiency, whereas mRNA abundance of its targets, CSD1 and CSD2, decreases (9–11). The superoxide-scavenging functions of CSD1 and CSD2 during copper deficiency are replaced by an increase in gene expression of FSD1 (11, 16). These morphological and molecular phenotypes of copper excess or deficiency were observed in A. thaliana seedlings grown in the presence of 45 μm CuCl2 or 750 μm BCS (supplemental Fig. 3, A and B).
FIGURE 7.
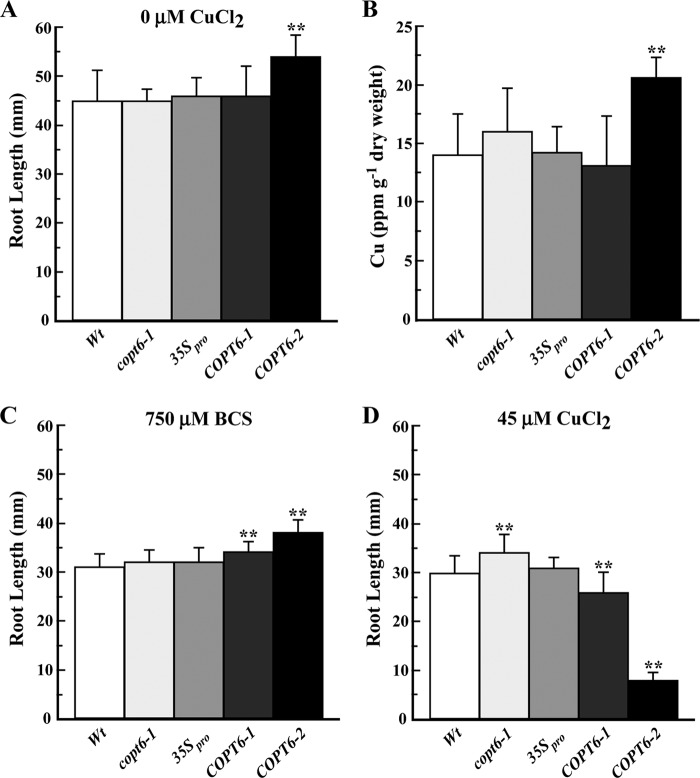
Altered expression of COPT6 in the copt6-1 mutant and COPT6 transgenic plants affects their response to copper limitation and excess. Shown are the root length of wild type (Wt), the copt6-1 mutant (copt6-1), two transgenic lines overexpressing 35Spro-HA-COPT6 (COPT6-1 and COPT6-2), and plants transformed with the empty vector (35Spro) grown in ½ MS (0 μm CuCl2; A) or in ½ MS supplemented with 750 μm BCS (C) or 45 μm CuCl2 (D). B, copper content in shoots of different plant lines grown hydroponically. Plant lines are designated as in A. **, statistically significant differences (p ≤ 0.001) of the mean values (analysis of variance). Error bars, S.E.
Root length of 10-day-old seedlings was used as a measure of tolerance or sensitivity to these growth conditions. The average lengths of primary roots of the copt6-1 mutant and the 35Spro-COPT6-1 transgenic line cultured at ½ MS were the same as of their corresponding controls, the wild type- and the empty vector-expressing plants, respectively (Fig. 7A). We also did not find statistically significant differences in copper accumulation in roots (not shown) or shoots (Fig. 7B) of these plant lines. In contrast, roots of 35Spro-COPT6-2 transgenic line, which manifested the highest level of the COPT6 transcript and polypeptide (Fig. 6, B and C), were 1.2-fold longer then empty vector-expressing plants (Fig. 7A). Although internal copper content of roots of this plant line did not differ from the other plant lines (not shown), its shoots accumulated 1.5-fold more copper then the empty vector-expressing plants (Fig. 7B). These data suggest that significantly increased expression of COPT6 in these plants was beneficial for plants even at copper-sufficient conditions.
We then tested the response of different plant lines to copper limitation or excess. As would be expected for copper-limiting conditions, primary roots were shorter in all plant lines grown on ½ MS supplemented with BCS than on ½ MS without supplements (Fig. 7, A and C). However, there was no difference between the root length of the copt6-1 mutant and wild-type plants in copper-limiting conditions (Fig. 7C). This result was not very surprising because of the possible functional redundancy of COPT6 with other COPT family members. In contrast, roots of 35Spro-COPT6-1 and 35Spro-COPT6-2 transgenic plants grown in copper-limiting conditions were 1.3- and 1.4-fold longer, respectively, compared with the empty vector-expressing plants (Fig. 7C), suggesting that overexpression of COPT6 was beneficial for plant growth also when copper was scarce.
Supplementation of ½ MS medium with a toxic concentration of CuCl2 inhibited root growth of all plant lines in comparison with ½ MS without supplementation (Fig. 7, A and D). COPT6-overexpressing transgenics and the copt6-1 mutant showed opposite responses to toxic copper; roots of plants overexpressing COPT6 were significantly shorter, whereas roots of the copt6-1 mutant were longer when compared with corresponding controls (Fig. 7D). It is important to note that the sensitivity of COPT6-overexpressing plants to copper excess correlated with the level of COPT6 overexpression; the 35Spro-COPT6-2 line was more markedly sensitive and overexpressed COPT6 to a higher degree compared with 35Spro-COPT6-1.
The improved growth of COPT6 transgenics in copper-limiting conditions and their increased sensitivity to copper excess and the decreased sensitivity of the copt6-1 mutant to copper excess not only show that COPT6 functions in copper homeostasis in planta but also further support the notion that tight control of the expression of copper transporters is needed for preventing copper deficiency while avoiding toxicity.
Expression of COPT6 Is Regulated by Copper Status
Regulation of the mRNA expression of CTR/COPT family members by copper status is among the major mechanisms of the control of copper homeostasis in the cell. Therefore, we sought to determine whether the abundance of COPT6 mRNA in A. thaliana would be altered in response to changes in copper supply as well. Wild-type A. thaliana (Col-0) seedlings were grown on ½ MS medium alone (control) or with 45 μm CuCl2 (copper excess) or 500 μm BCS (copper deficiency).
Analyses of COPT6 transcript abundance in A. thaliana cultured on control ½ MS medium showed that COPT6 is mainly expressed in leaves (Fig. 8A). The transcript abundance of COPT6 increased in both roots and shoots of BCS-treated A. thaliana wild-type plants (Fig. 8B). In contrast, copper excess affected the transcript abundance of COPT6 only in shoots and not in roots (Fig. 8B). Based on these results, we concluded that COPT6 expression is differentially regulated in roots and leaves of A. thaliana in response to alterations in copper availability.
FIGURE 8.
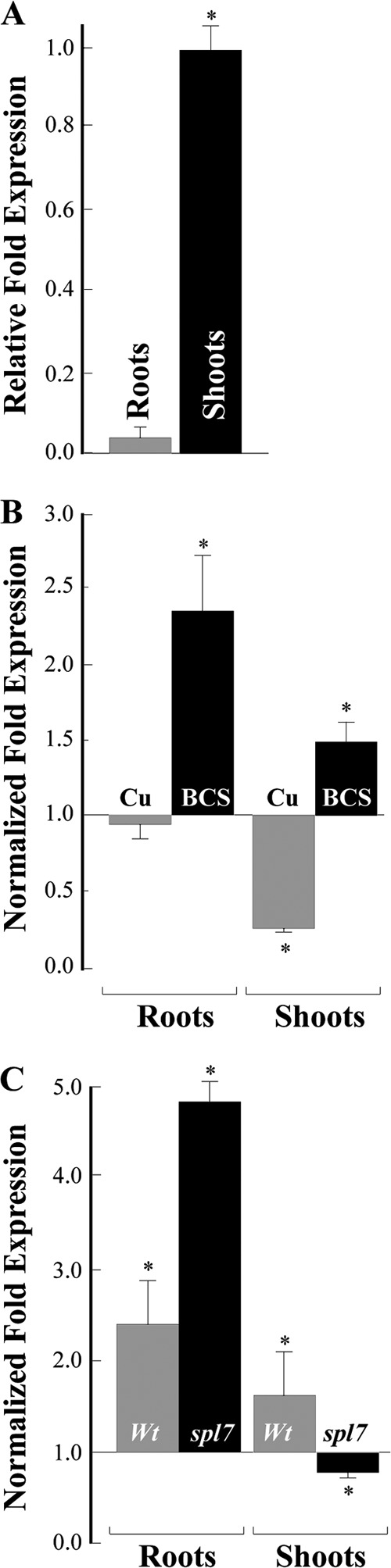
qRT-PCR analysis of COPT6 transcript abundance in 10-day-old A. thaliana seedlings. A, relative transcript abundance of COPT6 in roots (Roots) and shoots (Shoots) of seedlings grown on ½ MS agar plates. B, analysis of the COPT6 mRNA expression in response to copper status in wild-type plants. Roots and shoots were collected from seedlings germinated and grown on ½ MS agar plates without or with the addition of 45 μm CuCl2 (Cu) or 500 μm bathocuproine disulfonate (BCS). Results are presented relative to the expression of COPT6 in the wild type grown without added copper or BCS, designated as 1. C, analysis of the COPT6 expression in the spl7-1 mutant in response to copper limitation. Roots and shoots were collected from wild-type (Wt) and spl7-1 mutant (spl7) seedlings germinated and grown on ½ MS agar plates without or with the addition of 500 μm BCS. Transcript abundance of COPT6 in tissues of the wild type and the spl7-1 mutant grown in the presence of BCS is presented relative to its expression in corresponding plant lines grown without BCS, which is designated as 1. Error bars, S.E. (n = 9–18). *, statistically significant differences (p ≤ 0.05) of the mean values from corresponding control samples.
Expression of COPT6 Depends, in Part, on SPL7
A recent microarray study revealed that up-regulation of the expression of many copper deficiency-responsive genes in A. thaliana, including COPT1 and COPT2, is attributable to the activity of the transcription factor, SPL7 (12). SPL7 activates the transcription of its targets via binding to the GTAC motif (also known as copper-responsive elements) in the transcription regulation regions of copper-responsive genes (12, 69). Because COPT6 is not represented on the Agilent Arabidopsis 3 Oligo Microarray used previously (12), we sought to determine whether the transcriptional response of COPT6 to low copper availability (Fig. 8B) is controlled by SPL7 as well.
In this regard, we identified two GTAC motifs in a region of <200 bp upstream of the COPT6 start codon using the PLACE prediction software, suggesting that COPT6 might be an SPL7 target. We then used the spl7-1 mutant allele of A. thaliana (12) and tested whether decreasing copper availability by supplementing the growth medium with BCS would up-regulate COPT6 expression in this mutant background as it did in the wild type (Fig. 8B). It was found that the transcript abundance of COPT6 was still significantly increased in roots of the BCS-treated spl7-1 mutant (Fig. 8C). In contrast, expression of COPT6 mRNA was not up-regulated in shoots of the BCS-treated spl7-1 mutant (Fig. 8C). Based on these findings, we concluded that SPL7 is essential for the transcriptional response of COPT6 to copper limitation in shoots, whereas other transcription regulators might be involved in its response in roots of A. thaliana.
The Expression Pattern of COPT6 in A. thaliana
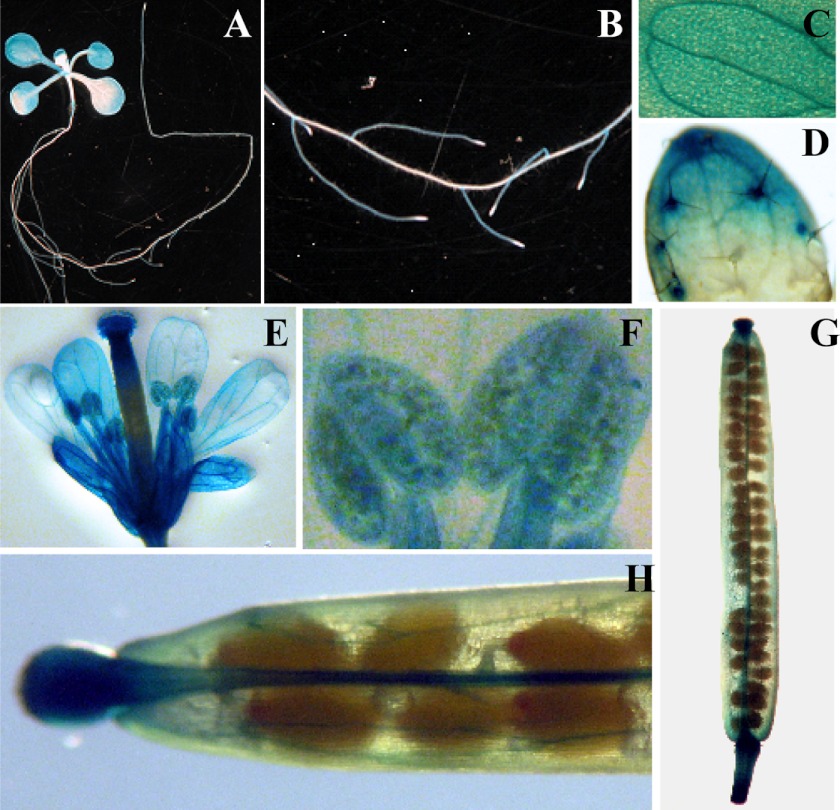
To investigate the sites of COPT6 action in A. thaliana, the COPT6 promoter sequence (COPT6pro) was fused to the uidA reporter gene encoding β-glucuronidase (GUS), and this COPT6pro-GUS construct was transformed into wild-type A. thaliana. From three transgenic lines exhibiting the same pattern of GUS activity, we selected one representative line for subsequent studies. Consistent with results of qRT-PCR analysis (Fig. 8A), the majority of GUS activity was observed in leaves of young seedlings (Fig. 9A). Although the GUS staining was observed throughout different cell types of lateral roots, it was absent in the primary root and at the root tips of lateral roots (Fig. 9B). We also observed strong GUS staining in the vasculature in leaves, in leaf trichomes and trichome basal cells (Fig. 9, C and D). The histochemical analysis of mature plants revealed that COPT6 is also expressed in sepals and petals of inflorescence (Fig. 9E). Analysis of the COPT6pro activity in reproductive organs disclosed COPT6pro activity in stigma and ovary, pollen grains, and filaments of stamens (Fig. 9, E and F). GUS activity in young siliques was less abundant and was primarily located in a gynophore, vasculature, a central replum, funiculus, style, and stigma surfaces (Fig. 9, G and H). Because the bulk of COPT6pro activity is concentrated in the shoots and reproductive organs, it is tempting to suggest that COPT6 may play a primary role in copper delivery and/or redistribution in above ground tissues.
FIGURE 9.
Histochemical analysis of the expression pattern of COPT6 in A. thaliana transformed with the COPT6pro-GUS construct. A, representative expression patterns of COPT6pro-GUS in a 10-day-old seedling. Note the strong GUS activity in lateral roots and vascular tissues of cotyledons (close-up images B and C, respectively), trichomes, and trichome basal cells (shown in a close-up image of a first true leaf (D)). E, staining pattern in a flower; F, close-up of an anther and a filament of a stamen; G, a young silique; H, close-up of a silique.
COPT6 Interacts with Itself and with COPT1 at the Plasma Membrane
CTR/COPT proteins exist as homo- and/or heterocomplexes on the cellular membranes (5, 29, 30, 35, 36). Here we tested whether COPT6 would interact with itself by using split ubiquitin-based MYTH and BiFC approaches (51, 70, 71).
In the MYTH approach, modified ubiquitin is split into C- and N-terminal halves (Cub and NubG) and is fused to membrane-bound bait or prey proteins, respectively. Interactions of bait and prey proteins cause Cub and NubG to reconstitute ubiquitin. The presence of reconstituted ubiquitin is recognized by ubiquitin-specific proteases (UBPs) that release an artificial transcription factor, PLV (protein A-LexA-VP16), which is fused to the C terminus of ubiquitin (CubPVL). Interactions are monitored by the PLV-induced expression of lexA-driven reporter genes, ADE2, HIS3, and lacZ (51, 52).
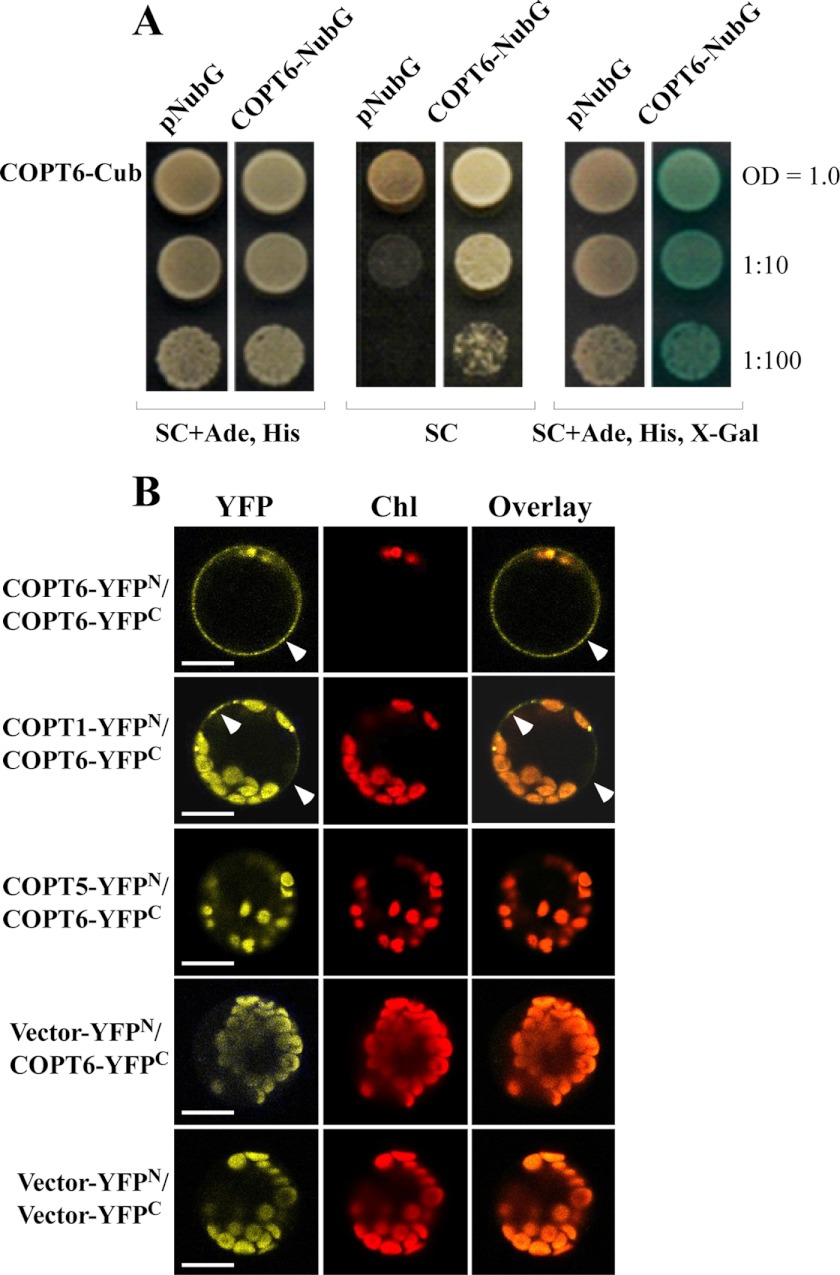
Interactions using the MYTH approach can only be detected when CubPLV and Nub fusions are located in the cytosol (51). Based on computer algorithm analysis of the predicted membrane topology, the C terminus of COPT6 is in the cytosol, whereas the N terminus is located extracellularly (Fig. 1B). Therefore, in order to detect COPT6-COPT6 interactions, NubG and CubPVL were fused to the C terminus of the full-length COPT6, generating COPT6-NubG and COPT6-CubPVL, and co-expressed in S. cerevisiae. To detect false positives due to self-activation, COPT6-CubPLV was co-expressed with pNubG lacking the COPT6 cDNA insert. Our data show that regardless of whether interactions were monitored as colony formation on selective medium (SC) or by β-galactosidase activity, interactions occurred only in cells that co-expressed COPT6-CubPVL and COPT6-NubG constructs (Fig. 10A). These interactions were not caused by self-activation because they did not occur in cells that co-expressed COPT6-CubPLV and pNubG (Fig. 10A).
FIGURE 10.
COPT6-COPT6 protein-protein interactions. A, COPT6 interacts with itself in the MYTH system. Shown are yeast cells co-expressing COPT6-CubPVL construct (COPT6-Cub) with NubG lacking the COPT6 cDNA insert (pNubG) or with COPT6 fused to NubG (COPT6-NubG). Growth was monitored for 2 days under conditions indicated below each panel; concentrations of yeast cells are indicated on the right. Shown are representative results of at least three biological replicates. SC, synthetic complete medium; Ade, adenine; His, histidine; X-gal, bromo-chloro-indolyl-galactopyranoside. B, epifluorescence images of YFP fluorescence complementation. Protoplasts were co-transfected with plasmids containing COPT6-YFPN and COPT6-YFPC (COPT6-YFPN/COPT6-YFPC), COPT1-YFPN and COPT6-YFPC (COPT1-YFPN/COPT6-YFPC), or COPT5-YFPN and COPT6-YFPC (COPT5-YFPN/COPT6-YFPC) or with empty vector-YFPN and COPT6-YFPC (Vector-YFPN/COPT6-YFPC) or empty vector-YFPN and vector-YFPC (Vector-YFPN/Vector-YFPC). Chlorophyll autofluorescence, visible in yellow (YFP) and red filter sets (Chl) does not overlap with yellow fluorescence of reconstituted YFP in superimposed images (Overlay), indicating COPT6-COPT6 and COPT1-COPT6 protein-protein interactions due to assembly of split YFP. Scale bar, 10 μm.
The BiFC method is based on the observation that N- and C-terminal halves of enhanced yellow fluorescent protein, YFPN and YFPC, respectively, reconstitute a functional fluorophore when brought into proximity by two interacting proteins fused to the YFP fragments (70, 71). For BiFC assays, the full-length COPT6 was fused at the C terminus with the N- or C-terminal halves of YFP to generate COPT6-YFPN or COPT6-YFPC, and both constructs were co-transfected into A. thaliana protoplasts. For negative controls, protoplasts were co-transfected with YFPN and YFPC vectors without the COPT6 insert (Vector-YFPN/Vector-YFPC) or with COPT5-YFPN and COPT6-YFPC (COPT5-YFPN/COPT6-YFPC). COPT5 has been shown to localize to the vacuolar membrane (37, 38) and should not interact with the plasma membrane-localized COPT6 protein. Interactions were visualized by following the fluorescence pattern of the reconstituted YFP fluorophore. Strong yellow fluorescence was observed at the plasma membrane only in COPT6-YFPN and COPT6-YFPC co-transfected protoplasts and not in protoplasts co-transfected with Vector-YFPN/Vector-YFPC or COPT6-YFPC/Vector-YFPN (Fig. 10B). Chlorophyll autofluorescence was also detected using the YFP filter set, but it did not overlap with yellow fluorescence associated with the plasma membrane in COPT6-YFPN/COPT6-YFPC-co-expressing protoplasts. These results suggest that COPT6 interacts with itself at the plasma membrane in BiFC assays.
We also tested if COPT6 will form heterologous interactions with COPT1, which is located at the plasma membrane and has a well established role in copper homeostasis (22, 23). Protoplasts co-transfected with COPT1-YFPN and COPT6-YFPC (COPT1-YFPN/COPT6-YFPC) showed a faint yellow fluorescence pattern at the plasma membrane, which did not overlap with chlorophyll autofluorescence (Fig. 10B). This suggests that COPT6 interacts with COPT1. Collectively, these results show that COPT6 at a minimum interacts with itself and COPT1 at the plasma membrane.
DISCUSSION
The essential, yet potentially toxic, nature of copper exemplifies the careful balance required to prevent deficiency while avoiding toxicity in most organisms. Among the central mechanisms in controlling copper homeostasis is the regulation of copper uptake. In this work, we characterized COPT6, a newly identified member of the CTR/COPT family in A. thaliana. The primary sequence of COPT6 contains the family-conserved methionine-rich motifs of which residues corresponding to Met27 and Met106 in COPT6 are important in S. cerevisiae Ctr1p for copper binding and transport functions, respectively. In addition, the C terminus of Ctr1p and two closest COPT6 homologs, COPT1 and COPT2, contain the CXC motif (Fig. 1C). This motif in Ctr1p is also involved in copper binding and transfer to cytosolic copper chaperones and in protein degradation during copper excess (31, 33, 34). In contrast, the primary sequence of COPT6 lacks the CXC motif (Fig. 1C). Nevertheless, COPT6 suppresses the copper-deficient phenotype of the ctr1Δctr2Δctr3Δ mutant and confers copper accumulation (Figs. 2 and 3), suggesting that the CXC motif is not important for its transport activity. Whether it plays a regulatory role under copper excess as was proposed for S. cerevisiae Ctr1p (31, 33, 34) is yet to be determined.
Consistent with the role of the positionally conserved methionine residue in TM2 of the CTR/COPT proteins (31), the positionally conserved Met106 in TM2 is required for COPT6 function (Fig. 4). However, it was surprising to find that the conserved Met27 in the N-terminal extracellular domain as well as the extracellular domain itself were dispensable for COPT6 function when copper was still available in the medium. We found, however, that the extracellular domain is required for COPT6 function when availability of external copper was depleted by the addition of BCS (Fig. 4), suggesting that this domain is important for COPT6 function during high affinity copper uptake under copper limitation. Because the positionally conserved Met27 and adjacent semiconserved Met22 were dispensable for COPT6 function in the presence of BCS in the medium (Fig. 4), we suggest that other residues within the extracellular domains may be involved in copper coordination. In this regard, the N terminus of COPT6 contains five methionine and three histidine residues in addition to Met22 and Met27 (supplemental Fig. 1), which can also coordinate copper ions prior their transport via COPT6.
The role of COPT6 in copper homeostasis in planta was tested by using an A. thaliana knockdown allele, copt6-1, and two transgenic lines ectopically expressing COPT6, 35Spro-HA-COPT6-1 and 35Spro-HA-COPT6-2. Knocking down or overexpressing COPT6 had opposite effects on plant growth during copper limitation or excess; copt6-1 plants were more sensitive to copper limitation while more tolerant to copper toxicity (Fig. 7, C and D). Furthermore, as expected, 35Spro-HA-COPT6-1 and 35Spro-HA-COPT6-2 plants were more tolerant of copper limitation but more sensitive to copper excess (Fig. 7, C and D). Given that our studies in yeast show that COPT6 is an uptake transporter, it is likely that the observed responses of the mutant and transgenic plants to copper availability result from impaired copper uptake and/or partitioning in the mutant and increased copper transport and/or tissue partitioning in transgenic lines.
The transcript abundance of COPT6 is significantly higher in leaves than in roots (Fig. 8A), suggesting its primary role in maintaining copper homeostasis of above ground tissues. We also found that the transcript abundance of COPT6 decreases in leaves but not in roots when copper concentration in external medium reaches its toxic limits (Fig. 8B), suggesting that COPT6 expression must be tightly controlled in leaves to protect the photosynthetic apparatus from copper overload. We hypothesize that COPT6 expression in roots is not altered by high copper because it is already relatively low under control conditions, and thus COPT6 contribution to copper uptake would be minimal under copper-replete or -excess conditions. In contrast, because of the low expression in roots under control conditions, COPT6 expression is up-regulated under copper-limiting conditions to provide an adequate copper supply to plants (Fig. 8, A and B).
The histochemical analysis of the spatial distribution of the COPT6 promoter activity in transgenic plants expressing the COPT6pro-GUS construct shows that although COPT6 is expressed in different cell types, its expression is concentrated in the vasculature (Fig. 9), where the majority of plant copper is stored to ensure rapid remobilization in response to increased copper demand (37). Therefore, it is tempting to speculate that COPT6 is involved in copper partitioning between different plant organs for its delivery to copper-requiring functions. Because COPT6 is also expressed in different cell types of lateral roots except for root tips (Fig. 9), its function in copper uptake from external solution cannot be excluded. In addition, the finding that COPT6 transcript abundance increases in roots and leaves of A. thaliana under copper-deficient conditions (Fig. 8B) further support the suggestion that COPT6-mediated copper influx might be important when copper availability is limited.
Similar to S. cerevisiae Ctr1p, COPT6 interacts with itself on the plasma membrane (Fig. 10). However, unlike Ctr1p, which does not form heterocomplexes with its closest homolog, a plasma membrane transporter, Ctr3p (31), COPT6 interacts with COPT1 (Fig. 10B). However, because COPT6 functions in copper uptake without COPT1 in S. cerevisiae (Figs. 2 and 3), the biological significance of COPT6-COPT1 interactions is yet to be determined. It is possible that COPT1-COPT6 interactions are important for copper uptake in specific A. thaliana cell types (e.g. in trichomes and/or pollen that co-express COPT6 and COPT1) (Fig. 9) (22).
The transcriptional response of some of the CTR/COPT family members is under tight transcriptional control exerted by Mac1 or SPL7 transcriptional factors in S. cerevisiae and A. thaliana, respectively (12, 27, 28). Microarray analysis revealed that A. thaliana COPT1 and COPT2 are among the SPL7 targets (12). Because COPT6 was not represented on the Agilent Arabidopsis 3 Oligo Microarray used previously (12) but is subjected to transcriptional regulation by copper status (Fig. 8B), we tested whether COPT6 is among the SPL7 targets as well. Our studies using the spl7-1 mutant of A. thaliana revealed that the transcript abundance of COPT6 increases independently of SPL7 in roots of A. thaliana (Fig. 8C), suggesting involvement of other transcription factors in this organ. In contrast, the COPT6 transcriptional response to copper limitations in the shoot absolutely depends on SPL7 (Fig. 8C). Although the transcriptional regulation region of COPT6 contains two copper-responsive elements essential for SPL7 binding, whether COPT6 is a direct SPL7 target is not known. It is noteworthy that SPL7 is expressed mainly in roots, where it suggested to sense copper availability (12). Nevertheless, many SPL7 targets (12), including COPT6, are induced by copper deficiency in shoots, suggesting a complex mechanism of SPL7-dependent regulation of the transcriptional copper deficiency response.
COPT6 has been annotated as a vacuolar membrane protein based on studies of the A. thaliana vacuole proteome (72). However, our data showing that 1) COPT6-EGFP, heterologously expressed in S. cerevisiae and transiently expressed in A. thaliana protoplasts is associated with the plasma membrane, 2) COPT6 interacts with the plasma membrane-localized COPT1 but not with vacuolar membrane-localized COPT5 in BiFC assays, 3) heterologously expressed COPT6 complements the S. cerevisiae copper uptake mutant, and 4) transgenic plants that overexpress COPT6 are more sensitive to copper excess are consistent with the suggestion that COPT6 localizes to the plasma membrane and is involved in copper uptake rather than in vacuolar sequestration. To reconcile our experimental data with the results of Carter et al. (72), we suggest that the detection of COPT6 in the vacuolar proteome may reflect a vacuole-mediated degradation pathway of COPT6, as was shown for its S. cerevisiae counterpart Ctr1p (73). To conclude, COPT6 is a plasma membrane copper uptake transporter and is a novel SPL7 target that is essential for maintaining plant growth during extreme copper conditions, acting, possibly, by controlling copper uptake and partitioning to copper requiring functions.
Acknowledgments
We thank Dr. Shikanai (Kyoto University) for A. thaliana spl7-1 seeds, Dr. Thiele (Duke University) for S. cerevisiae strains and Ctr1 constructs, Dr. Jian Hua (Cornell University) for BiFC and SAT6-N1-EGFP plasmids, and Zhiyang Zhai and Sungjin Kim (The Vatamaniuk Laboratory, Cornell University) for modifying vectors into Gateway destination vectors.
This work was supported by the Cornell University Agricultural Experiment Station USDA-CSREES NYC-FFF-Hatch 125433, USDA-CSREES NYC-MRF S1041 125853, and a Cornell start-up grant (to O. K. V).

This article contains supplemental Table 1 and Figs. 1–3.
- SOD
- superoxide dismutase
- BCS
- bathocuproine disulfonate
- MYTH
- split ubiquitin membrane yeast two-hybrid system
- BiFC
- bimolecular fluorescence complementation
- ICP-MS
- inductively coupled plasma mass spectroscopy
- Mets motif
- methionine-rich N-terminal motif
- qRT-PCR
- quantitative RT-PCR
- EGFP
- enhanced green fluorescent protein
- TM
- transmembrane
- ½ MS
- half-strength Murashige and Skoog
- UBP
- ubiquitin-specific protease.
REFERENCES
- 1. Burkhead J. L., Reynolds K. A., Abdel-Ghany S. E., Cohu C. M., Pilon M. (2009) Copper homeostasis. New Phytol. 182, 799–816 [DOI] [PubMed] [Google Scholar]
- 2. Prohaska J. (2000) Long term functional consequences of malnutrition during brain development. Copper. Nutrition 16, 502–504 [DOI] [PubMed] [Google Scholar]
- 3. Puig S., Andrés-Colás N., García-Molina A., Peñarrubia L. (2007) Copper and iron homeostasis in Arabidopsis. Responses to metal deficiencies, interactions, and biotechnological applications. Plant Cell Environ. 30, 271–290 [DOI] [PubMed] [Google Scholar]
- 4. Gavnholt B., Larsen K. (2002) Molecular biology of plant laccase in relation to lignin formation. Physiol. Plant. 116, 273–280 [Google Scholar]
- 5. Yuan M., Chu Z., Li X., Xu C., Wang S. (2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22, 3164–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stadtman E. R. (1990) Metal ion-catalyzed oxidation of proteins. Biochemical mechanism and biological consequences. Free Radic. Biol. Med. 9, 315–325 [DOI] [PubMed] [Google Scholar]
- 7. Gaetke L. M., Chow C. K. (2003) Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189, 147–163 [DOI] [PubMed] [Google Scholar]
- 8. Merchant S. S. (2010) The elements of plant micronutrients. Plant Physiol. 154, 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdel-Ghany S. E., Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sunkar R., Kapoor A., Zhu J. K. (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down-regulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamasaki H., Abdel-Ghany S. E., Cohu C. M., Kobayashi Y., Shikanai T., Pilon M. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282, 16369–16378 [DOI] [PubMed] [Google Scholar]
- 12. Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009) SQUAMOSA Promoter Binding Protein-like 7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 21, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kliebenstein D. J., Monde R. A., Last R. L. (1998) Superoxide dismutase in Arabidopsis. An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohu C. M., Pilon M. (2007) Regulation of superoxide dismutase expression by copper availability. Physiol. Plant. 129, 747–755 [Google Scholar]
- 15. Ravet K., Danford F. L., Dihle A., Pittarello M., Pilon M. (2011) Spatiotemporal analysis of copper homeostasis in Populus trichocarpa reveals an integrated molecular remodeling for a preferential allocation of copper to plastocyanin in the chloroplasts of developing leaves. Plant Physiol. 157, 1300–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abdel-Ghany S. E., Müller-Moule P., Niyogi K. K., Pilon M., Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dancis A., Haile D., Yuan D. S., Klausner R. D. (1994) The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269, 25660–25667 [PubMed] [Google Scholar]
- 18. Pena M. M., Puig S., Thiele D. J. (2000) Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275, 33244–33251 [DOI] [PubMed] [Google Scholar]
- 19. Rees E. M., Lee J., Thiele D. J. (2004) Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229 [DOI] [PubMed] [Google Scholar]
- 20. Andrés-Colás N., Perea-García A., Puig S., Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol. 153, 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Page M. D., Kropat J., Hamel P. P., Merchant S. S. (2009) Two Chlamydomonas CTR copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21, 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sancenón V., Puig S., Mateu-Andrés I., Dorcey E., Thiele D. J., Peñarrubia L. (2004) The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 279, 15348–15355 [DOI] [PubMed] [Google Scholar]
- 23. Sancenón V., Puig S., Mira H., Thiele D. J., Peñarrubia L. (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51, 577–587 [DOI] [PubMed] [Google Scholar]
- 24. Kampfenkel K., Kushnir S., Babiychuk E., Inzé D., Van Montagu M. (1995) Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J. Biol. Chem. 270, 28479–28486 [DOI] [PubMed] [Google Scholar]
- 25. Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., Klausner R. D. (1994) Molecular characterization of a copper transport protein in S. cerevisiae. An unexpected role for copper in iron transport. Cell 76, 393–402 [DOI] [PubMed] [Google Scholar]
- 26. Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S., Klausner R. D. (1996) Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15, 3515–3523 [PMC free article] [PubMed] [Google Scholar]
- 27. Yonkovich J., McKenndry R., Shi X., Zhu Z. (2002) Copper ion-sensing transcription factor Mac1p post-translationally controls the degradation of its target gene product Ctr1p. J. Biol. Chem. 277, 23981–23984 [DOI] [PubMed] [Google Scholar]
- 28. Jungmann J., Reins H. A., Lee J., Romeo A., Hassett R., Kosman D., Jentsch S. (1993) MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12, 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Feo C. J., Aller S. G., Unger V. M. (2007) A structural perspective on copper uptake in eukaryotes. Biometals 20, 705–716 [DOI] [PubMed] [Google Scholar]
- 30. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puig S., Lee J., Lau M., Thiele D. (2002) Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277, 26021–26030 [DOI] [PubMed] [Google Scholar]
- 32. Peñarrubia L., Andrés-Colás N., Moreno J., Puig S. (2010) Regulation of copper transport in Arabidopsis thaliana. A biochemical oscillator? J. Biol. Inorg. Chem. 15, 29–36 [DOI] [PubMed] [Google Scholar]
- 33. Wu X., Sinani D., Kim H., Lee J. (2009) Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J. Biol. Chem. 284, 4112–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao Z., Wedd A. G. (2002) A C-terminal domain of the membrane copper pump Ctr1 exchanges copper(I) with the copper chaperone Atx1. Chem. Commun. Camb. 6, 588–589 [DOI] [PubMed] [Google Scholar]
- 35. Lee J., Peña M. M., Nose Y., Thiele D. J. (2002) Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 277, 4380–4387 [DOI] [PubMed] [Google Scholar]
- 36. Yuan M., Li X., Xiao J., Wang S. (2011) Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. BMC Plant Biol. 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garcia-Molina A., Andrés-Colás N., Perea-García A., Del Valle-Tascón S., Peñarrubia L., Puig S. (2011) The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J. 65, 848–860 [DOI] [PubMed] [Google Scholar]
- 38. Klaumann S., Nickolaus S. D., Fürst S. H., Starck S., Schneider S., Ekkehard Neuhaus H., Trentmann O. (2011) The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 192, 393–404 [DOI] [PubMed] [Google Scholar]
- 39. Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 40. Lu Y. P., Li Z. S., Rea P. A. (1997) AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump. Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Natl. Acad. Sci. U.S.A. 94, 8243–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tzfira T., Tian G. W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005) pSAT vectors. A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57, 503–516 [DOI] [PubMed] [Google Scholar]
- 42. Udvardi M. K., Czechowski T., Scheible W. R. (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bolte S., Talbot C., Boutte Y., Catrice O., Read N. D., Satiat-Jeunemaitre B. (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214, 159–173 [DOI] [PubMed] [Google Scholar]
- 47. Zhai Z., Jung H. I., Vatamaniuk O. K. (August 17, 2009) Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 10.3791/1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhai Z., Sooksa-nguan T., Vatamaniuk O. K. (2009) Establishing RNA interference as a reverse genetic approach for gene functional analysis in protoplasts. Plant Physiol. 149, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung H. I., Zhai Z., Vatamaniuk O. K. (2011) Direct transfer of synthetic double-stranded RNA into protoplasts of Arabidopsis thaliana. Methods Mol. Biol. 744, 109–127 [DOI] [PubMed] [Google Scholar]
- 50. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 51. Kittanakom S., Chuk M., Wong V., Snyder J., Edmonds D., Lydakis A., Zhang Z., Auerbach D., Stagljar I. (2009) Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol. Biol. 548, 247–271 [DOI] [PubMed] [Google Scholar]
- 52. Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., Ellerbrok H., Kamuzinzi R., Ledent V., Blaudez D., Sanders D., Revuelta J. L., Boles E., André B., Frommer W. B. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim S., Selote D. S., Vatamaniuk O. K. (2010) The N-terminal extension domain of the C. elegans half-molecule ABC transporter, HMT-1, is required for protein-protein interactions and function. PLoS One 5, e12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 55. Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) GUS fusions. β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 57. Towbin H., Staehelin T., Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lahner B., Gong J., Mahmoudian M., Smith E. L., Abid K. B., Rogers E. E., Guerinot M. L., Harper J. F., Ward J. M., McIntyre L., Schroeder J. I., Salt D. E. (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 59. Arteca R. N., Arteca J. M. (2000) A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plant. 108, 188–193 [Google Scholar]
- 60. Cailliatte R., Schikora A., Briat J. F., Mari S., Curie C. (2010) High affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22, 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Glerum D. M., Shtanko A., Tzagoloff A. (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271, 14504–14509 [DOI] [PubMed] [Google Scholar]
- 62. Baekgaard L., Mikkelsen M. D., Sørensen D. M., Hegelund J. N., Persson D. P., Mills R. F., Yang Z., Husted S., Andersen J. P., Buch-Pedersen M. J., Schjoerring J. K., Williams L. E., Palmgren M. G. (2010) A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J. Biol. Chem. 285, 31243–31252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jahn T. P., Schulz A., Taipalensuu J., Palmgren M. G. (2002) Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J. Biol. Chem. 277, 6353–6358 [DOI] [PubMed] [Google Scholar]
- 64. van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., Klomp L. W. (2007) Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bellemare D. R., Shaner L., Morano K. A., Beaudoin J., Langlois R., Labbe S. (2002) Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277, 46676–46686 [DOI] [PubMed] [Google Scholar]
- 66. Nelson B. K., Cai X., Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 67. Murphy A., Taiz L. (1995) A new vertical mesh transfer technique for metal tolerance studies in Arabidopsis (ecotypic variation and copper-sensitive mutants). Plant Physiol. 108, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lequeux H., Hermans C., Lutts S., Verbruggen N. (2010) Response to copper excess in Arabidopsis thaliana. Impact on the root system architecture, hormone distribution, lignin accumulation, and mineral profile. Plant Physiol. Biochem. 48, 673–682 [DOI] [PubMed] [Google Scholar]
- 69. Quinn J. M., Merchant S. (1995) Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7, 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schütze K., Harter K., Chaban C. (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol. Biol. 479, 189–202 [DOI] [PubMed] [Google Scholar]
- 71. Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427 [DOI] [PubMed] [Google Scholar]
- 72. Carter C., Pan S., Zouhar J., Avila E. L., Girke T., Raikhel N. V. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16, 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu J., Sitaram A., Burd C. G. (2007) Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8, 1375–1384 [DOI] [PubMed] [Google Scholar]