Background: Insulin regulation of human growth hormone has not been studied in non-tumor pituitary cells.
Results: Insulin decreases growth hormone RNA levels via an insulin-induced transcription factor and chromatin remodeling.
Conclusion: The somatotroph and growth hormone gene expression are targets for insulin signaling.
Significance: This may explain the early and rapid suppression of growth hormone levels seen in hyperinsulinemic individuals.
Keywords: Cell Culture, Gene Regulation, Growth Hormone, Hypoxia-inducible Factor (HIF), Insulin, Pituitary Gland, Transgenic Mice
Abstract
Insulin controls growth hormone (GH) production at multiple levels, including via a direct effect on pituitary somatotrophs. There are no data, however, on the regulation of the intact human (h) GH gene (hGH1) by insulin in non-tumor pituitary cells, but the proximal promoter region (nucleotides −496/+1) responds negatively to insulin in transfected pituitary tumor cells. A DNA-protein interaction was also induced by insulin at nucleotides −308/−235. Here, we confirmed the presence of a hypoxia-inducible factor 1 (HIF-1) binding site within these sequences (−264/−259) and investigated whether HIF-1 is associated with insulin regulation of “endogenous” hGH1. In the absence of primary human pituitary cells, transgenic mice expressing the intact hGH locus in a somatotroph-specific manner were generated. A significant and dose-dependent decrease in hGH and mouse GH RNA levels was detected in primary pituitary cell cultures from these mice with insulin treatment. Increasing HIF-1α availability with a hypoxia mimetic significantly decreased hGH RNA levels and was accompanied by recruitment of HIF-1α to the hGH1 promoter in situ as seen with insulin. Both inhibition of HIF-1 DNA binding by echinomycin and RNA interference of HIF-1α synthesis blunted the negative effect of insulin on hGH1 but not mGH. The insulin response is also sensitive to histone deacetylase inhibition/trichostatin A and associated with a decrease in H3/H4 hyperacetylation in the proximal hGH1 promoter region. These data are consistent with HIF-1-dependent down-regulation of hGH1 by insulin via chromatin remodeling specifically in the proximal promoter region.
Introduction
There is evidence to suggest growth hormone (GH)3 production is under insulin control. Excess insulin may act indirectly on the hypothalamus by affecting growth hormone-releasing hormone and somatostatin secretion tone or directly at the pituitary level. More recently, somatotrophs have been implicated as the primary target of hyperinsulinemia in animal models. A reduction in GH synthesis and/or secretion was associated with increased circulating levels of insulin in both high fat diet-induced obesity and the leptin-deficient ob/ob mice (1, 2). The effect of insulin on GH, however, was not accompanied by any alteration in growth hormone-releasing hormone and somatostatin expression in the hypothalamus of the obese mice, supporting a more direct effect of insulin at the pituitary level (1). Certainly, the mouse pituitary gland appears to be an insulin-sensitive tissue as insulin receptors are present at levels comparable to “classical” insulin-sensitive tissues, such as adipose, liver, and skeletal muscle. Furthermore, there is evidence that pituitary cells continue to be sensitive to insulin, whereas the cells from other peripheral tissues become more resistant from chronic hyperinsulinemia in the obese state (1, 3).
In humans, blunted GH secretion in obese individuals has been linked with metabolic derangements (4, 5). Elevated levels of insulin as a component of metabolic syndrome might be a major contributor to the obesity-related reduction in GH secretion (6, 7). Abnormal GH production in obese patients is characterized by impaired spontaneous secretion as well as a decline in pituitary somatotroph responsiveness to all known pharmacological provocative stimuli. Specifically, GH-producing somatotrophs become significantly hyporesponsive to exogenous secretagogues including growth hormone-releasing hormone, ghrelin/growth hormone secretagogue (GHS) and arginine (8–11). The mechanism, however, for this insulin-related suppression of GH synthesis is poorly understood.
There are no available data on the effect of insulin on endogenous human (h) GH gene (hGH1) expression despite evidence for negative control of human and non-human primate GH by excess insulin as well as obesity (5, 12, 13). The proximal region (nucleotides −496/+1) of the hGH1 promoter was shown, however, to respond negatively to insulin treatment after transfection into a rat pituitary tumor cells (13–16). This correlates with a study in which insulin treatment was associated with induction of a DNA-protein interaction at nucleotides −308/−235 in the proximal hGH1 promoter region (13–16). Inspection of these sequences reveals an E-box (Enhancer Box), 5′-CCACGTGACC-3′, which includes a palindromic hexanucleotide sequence (underlined) at position −264 to −259, and potential hypoxia-inducible factor-1 (HIF-1) binding sequence (HBS) (17).
HIF-1α and -β are members of the basic-helix-loop helix transcription factor family. Under normoxic conditions, HIF-1α protein is ubiquitinated and subject to continuous proteasomal degradation because it contains an oxygen-dependent degradation domain targeted by a specific HIF-prolyl hydroxylase (HPH1–3 also referred to as PHD1–3). Hypoxia, however, attenuates hydroxylation, and HIF-1α is stabilized. HIF-1α is then able to heterodimerize with its constitutively present HIF-1β partner, and form a functional complex that regulates genes via a specific hypoxia-responsive element (18, 19). HIF-1α is a critical factor involved in developmental and physiological events (20–22). Deregulation of HIF-1α predominantly in relation to effects on pancreatic gene expression and β cell function has been linked to the development of type 2 diabetes (23). Furthermore, elevated levels of HIF-1α in adipose tissue are postulated as contributing to obesity-related insulin resistance and metabolic dysfunction. As such, HIF-1 is considered an obesity-related transcription factor complex (24–26). Insulin shares the ability with hypoxia to induce the HIF-1α transcription factor complex and is considered to be a downstream molecule of insulin signaling (27–29). HIF-1α has also been detected in the pituitary (30, 31). Thus, these properties make HIF-1 a candidate to mediate insulin regulation of hGH1 expression.
A difficulty in assessing regulation of endogenous hGH production has been the absence of an appropriate non-tumor pituitary culture system expressing hGH1. We generated transgenic (TG) mice containing a single copy of the intact hGH gene locus including its locus control region (LCR), which includes essential hypersensitive site (HS) regions I, II, III, and V (32). This LCR was shown to confer a site of integration-independent pituitary-specific expression (33), and pituitary hGH1 expression was confirmed in two-independent hGH/CS-TG mouse lines (32). More recently we generated primary pituitary cell cultures from these hGH/CS-TG mice and showed that hGH1 is expressed by somatotrophs, as hGH protein co-localized with mouse (m) GH in pituitary tissue and cultures (34). Here we have used these hGH/CS-TG mouse pituitary cell cultures to investigate insulin control of endogenous hGH versus mouse GH gene expression. Our data indicate negative regulation of GH at the RNA level and, in the case of hGH1 but not mGH, a role for HIF-1 and remodeling of the proximal promoter region in this response. These results are discussed in relation to the effects of excess insulin resulting from overeating and obesity on hGH production in vivo.
EXPERIMENTAL PROCEDURES
Primary Pituitary Cell Culture
All procedures involving animals, their tissues, and cells conform to the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication 85–23, revised 1996) and were approved by the animal Protocol Management and Review Committee at the University of Manitoba. Primary pituitary cell cultures were generated from anterior pituitary glands harvested from 10 adult male and female (8–10 weeks) TG mice containing the intact hGH gene and LCR in a 171-kb fragment of human chromosome 17 (32) as reported previously (34). Cells were plated at 1 × 105 cells/well in 24-well plates in 1 ml of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and antibiotics (10 IU/ml penicillin, 10 mg/ml streptomycin) and maintained in this medium for 72 h. For insulin treatment, cells were re-fed DMEM supplemented with 1% double charcoal-stripped FBS for 24 h and then treated with 1–200 nm insulin (#I9278, Sigma) for 24 h. Induction of HIF-1α protein levels was pursued by cobalt (II) chloride hexahydrate (CoCl2) (C2644, Sigma) treatment in which cells were incubated with 150–500 μm CoCl2 for 5 and/or 24 h. To interfere with HIF-1α DNA binding, cultures were preincubated for 60 min with 10 or 20 nm echinomycin (ALX-380–201, Enzo Life Sciences) or vehicle (dimethyl sulfoxide) before insulin treatment. Similarly, inhibition of histone deacetylase (HDAC) class I, II, or III was done using pretreatment with trichostatin A (300 nm, T1952, Sigma) or nicotinamide (15 mm, N0636, Sigma), respectively, for 60 min.
RNA Preparation and Quantitative Real-time Reverse Transcriptase-PCR
Total RNA extraction and quantitative real-time reverse transcriptase (RT)-PCR (qPCR) using specific primers (Table 1) were done as described previously (34). Minus reverse transcriptase controls were performed using the same PCR primers and thermal cycle conditions as a control for the presence of genomic DNA. Specific amplifications were identified by a single peak in the melting curve and a single band in the final PCR product visualized on an agarose gel. The gene expression level in each sample (absolute quantification) was calculated from the standard curve and normalized to mouse β2 microglobulin expression as appropriate. Tests were normally run in duplicate on three independent samples.
TABLE 1.
Primers used for qPCR
| RNA | Primer sequence |
|---|---|
| hGH | Forward, CCTAGAGGAAGGCATCCAAA |
| Reverse, GCAGCCCGTAGTTCTTGAGTAG | |
| mGH | Forward, ACGCGCTGCTCAAAAACTAT |
| Reverse, CACAGGAGAGTGCAGCAGAG | |
| GHS-R | Forward, CTGGACAAAGTCGAGCATCA |
| Reverse, CTGCCCATCTGGCTCTACTC | |
| VEGF | Forward, AGCACAGCAGATGTGAATGC |
| Reverse, AATGCTTTCTCCGCTCTGAA | |
| β2- Microglobulin | Forward, GCTATCCAGAAAACCCCTCAAA |
| Reverse, GCGGGTGGAACTGTGTTACG |
Nuclear Protein Extraction and Detection
Nuclear extraction from cultures was performed using a nuclear extraction kit according to the manufacturer's instructions (Active Motif, Carlsbad, CA, catalog no. 54001) (35). Briefly, after treatment, cells were suspended in hypotonic buffer (20 mm HEPES, pH 7.5, 5 mm sodium fluoride, 10 μm sodium molybdate, and 0.1 mm edta) containing phosphatase inhibitors and incubated on ice for 15 min. Detergent (Nonidet P-40) was added to a final concentration of 0.5%, and incubation was continued for 15 min. Nuclei were separated by centrifugation (14,000 × g for 30 s at 4 °C), and the nuclear pellet was next extracted in a hypertonic lysis buffer for 30 min on ice. After centrifugation (14,000 × g for 10 min at 4 °C), the supernatant containing the pure nuclear fraction was collected. Protein concentration was determined by a Bradford protein assay (Bio-Rad). For detection of HIF-1α, 25 μg (after CoCl2 treatment) or 100 μg (after insulin treatment) of whole cell protein or nuclear-extracted proteins were analyzed by protein immunoblotting as previously described (36). Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and immunoblotted with anti-HIF-1α antibody (NB100–134, Novus Biologicals, Oakville, ON, CA). GAPDH(sc-25778, Santa Cruz), upstream stimulatory factor 1 (USF-1; sc-229, Santa Cruz), and/or histone H1 (C-17, Santa Cruz) were assessed as controls for protein loading for whole cell lysates and nuclear proteins, respectively. The proteins were visualized using horseradish peroxidase-conjugated anti-immunoglobulin G (IgG) secondary antibody and ECL plus immunoblotting detection reagents (Thermo Fisher Scientific Inc., Nepean, ON, Canada).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA and competition with oligonucleotides was performed essentially as previously described (36). Briefly, 500 ng of recombinant HIF-1α and 500 ng of HIF-1β were incubated with EMSA buffer containing 2 μg of poly-dIdC for 5 min. Radiolabeled oligonucleotide probes (1 ng) were then added, and the reactions were incubated for a further 10 min at room temperature. In competition experiments, 2.5-, 5-, and 50-fold molar excesses of unlabeled oligonucleotide duplexes were added during a preincubation period. The DNA-protein complexes were resolved in non-denaturing 5% (w/v) polyacrylamide gels and visualized by autoradiography.
Lentivirus-mediated Short Hairpin RNA (shRNA) Treatment
HIF-1α shRNA (sc-35562-V, Santa Cruz) and control/scrambled shRNA lentiviral particles (sc-108080, Santa Cruz) were used for HIF-1α “knock down” as previously reported (37). The viral infection was performed according to the manufacturer's instructions. Briefly, cultures were incubated with DMEM containing lentiviral particles in the presence of Polybrene (5 μg/ml, Santa Cruz) for 24 h. Lentiviral particles were washed away, and cells were incubated for a further 48 h. Medium was then changed to 1% double charcoal-stripped FBS-DMEM for 24 h before treatment with insulin for 24 h, harvesting, and analysis.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay with cultures treated with or without insulin or CoCl2 were done as previously described (38). In brief, cells were harvested 24 h after insulin or CoCl2 treatments in cold phosphate-buffered saline buffer and cross-linked with 1% formaldehyde before lysis. Chromatin was fragmented by sonication (100 s in 10-s pulses), then insoluble material was removed by centrifugation, and the DNA content was measured by spectrophotometry. Soluble chromatin was precleared for 1 h and then immunoprecipitated with 5 μg of HIF-1α (Novus Biologicals), HIF-1α (Santa Cruz, sc-10790), or histone H3-H4 hyperacetylation antibodies (Catalog # 06-598 and 06-599, Millipore) overnight at 4 °C as well as with normal rabbit IgG (Millipore) as a negative control. Protein A/G plus agarose (Santa Cruz) was added to immunoprecipitation reaction with rotation for 1 h at 4 °C to collect antibody-chromatin complexes. The agarose-antibody complexes were subjected to a series of washes and elution. The eluted antibody complexes were reverse-cross-linked at 65 °C overnight, and DNA was isolated using QIAquick PCR purification kit (Qiagen). Quantitative PCR was performed in a 7500 Fast Real Time CR system (AB Applied Biosystems) under conditions standardized for each primer set (Table 2). Each qPCR reaction was carried out in duplicate in a 20-μl reaction volume by using 5 μl of the 1% of input DNA and 5 μl of pre-amplified (15 cycles) eluted immunoprecipitated DNA and 10 μl of Power SYBR Green Master mix (AB Applied Biosystems). Dissociation curves were analyzed as a mean to ensure the quality of amplicons and to monitor primer dimers. Final PCR products were visualized as a single band in an agarose gel. ChIP enrichment was determined based on a percent input method (39). Briefly, the signals obtained from the immunoprecipitated DNA amplification were divided by the signals obtained from an input sample. Because the starting input fraction was 1%, a dilution factor of 100 or 6.644 cycles (i.e. log 2 of 100) was subtracted from the cycle threshold (CT) value of diluted input. Enrichment was calculated based on the formula, 100 × 2̂(CT-adjusted input − CT-enriched), and data are presented as percent input and relative -fold change compared with the control, which is arbitrarily set to 1. For the ChIP assay of histone acetylation in the hGH locus, hyperacetylation levels are presented as relative -fold changes compared with levels detected at HS V, which is arbitrary set to 1.
TABLE 2.
Primers used for ChIP-qPCR
| Region | Primer sequence |
|---|---|
| HS V | Forward, TCCCTCGGACCAGAACAC |
| Reverse, CCCAGGTAAAAGCAGCATGT | |
| HS III | Forward, CACTGATGAGCTTGGCGTCAC |
| Reverse, CCTGCCACTTCCGCTCTCCA | |
| HS I/II | Forward, CATGGGCCTCAAGCTGACCT |
| Reverse, CGTTCCGGGCAGCCCCAGAT | |
| −2- kb GH | Forward, CTGTGTCCACCTGCAGAGTT |
| Reverse, AGCTTCTTCCATGTTCCTCC | |
| −0.5-kb GHp | Forward, CCCCTTCTCTCCCACTGTTG |
| Reverse, AACCCTCACAACACTGGTGAC | |
| GHp-HBS | Forward, CACAGAGTGTCAGCCAGAGATA |
| Reverse, GGATGTGGTCGGTAGGGGGT | |
| Untr6 | Forward, TCAGGCATGAACCACCATAC |
| Reverse, AACATCCACACGTCCAGTGA |
Statistical Analysis
Statistical analysis was performed using GraphPad Instat® and Prism® software. For single comparisons, paired t tests were applied, and for multiple comparisons one-way analysis of variance was used with the Tukey-Kramer or Dunnett's post-test as appropriate. For multiple comparisons with more than one variable, data were analyzed using two-way analysis of variance with the Bonferroni post-test. A value of p < 0.05 is considered statistically significant and is represented in figures as: * or #, p < 0.05; ** or ##, p < 0.01; *** or ###, p < 0.001.
RESULTS
Endogenous hGH and mGH RNA Levels Are Negatively Regulated by Insulin
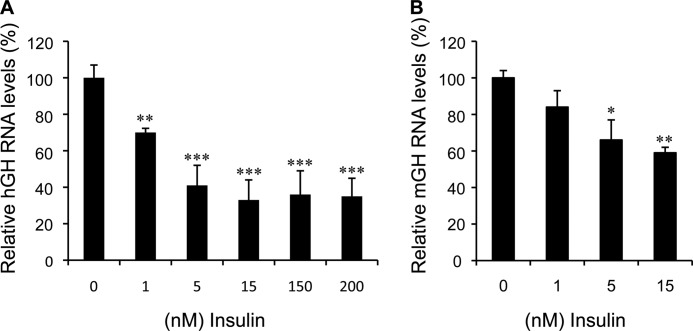
Cultures were generated from hGH/CS-TG mouse pituitaries and treated with insulin to determine whether the endogenous or intact hGH1 is responsive. Human GH RNA levels decreased significantly in response to both physiological (1 nm) and pharmacological (200 nm) doses of insulin (Fig. 1A). This decrease reached a plateau (∼60% decrease) at 5 nm (p < 0.001, n = 12). A similar decrease in hGH RNA levels was observed in cultures from a second independent (141hGH/CS-TG) mouse line (32) with insulin (data not shown). A decrease in mGH transcripts was also seen with 1 nm insulin treatment but, unlike hGH RNA levels, was not significant (Fig. 1B); however, a significant and increasing ∼40% reduction in mGH RNA levels was also observed with 5 and 15 nm insulin (p < 0.01, n = 6).
FIGURE 1.
Endogenous hGH and mGH RNA levels are negatively regulated by insulin. Primary pituitary cells were de-induced in 1% double charcoal-stripped FBS-DMEM for 24 h and treated with 1–200 nm insulin for 24 h. RNA was isolated and assessed by qPCR. A, human GH RNA levels were decreased at physiological (1 nm) and pharmacological (200 nm) concentrations of insulin. B, mouse GH transcripts were also reduced in response to insulin treatment but were only significant at insulin concentrations above 1 nm. RNA levels in each sample (absolute quantification) were calculated from the standard curve and normalized to mouse β2 microglobulin RNA. The results are expressed as relative mean change plus or minus S.E. of the mean compared with the control (0) value, which is arbitrarily set to 100%. Data were assessed by one-way analysis of variance with the Tukey-Kramer post-test. A value of p < 0.05 is considered statistically significant: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
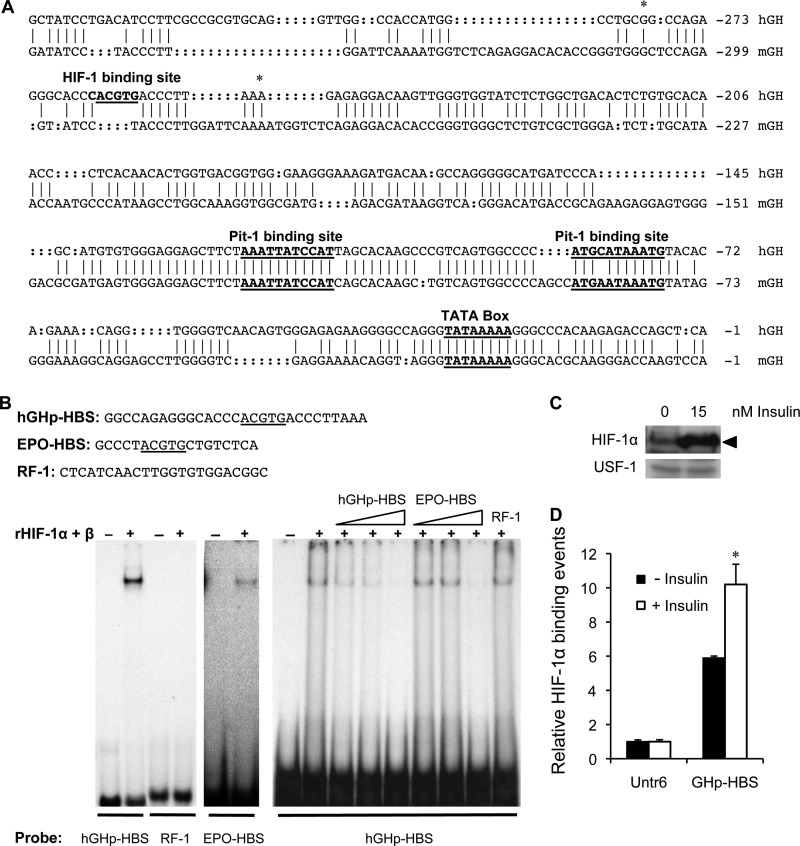
A HIF-1α Binding Site Is Located within Nucleotides −278/−250 of the Proximal hGH1 Promoter Region
To confirm binding of HIF-1 to sequences in the hGH1 proximal promoter region, EMSA was done using combined recombinant HIF-1α and HIF-1β proteins with radiolabeled hGH1 nucleotides −278/−250 (Fig. 2A). A known HBS from the erythropoietin (EPO) gene was used as a positive control (19), and RF-1 sequences (40) were also radiolabeled as a negative control. A single specific complex was observed with both hGH and EPO gene but not RF-1 sequences in the presence of HIF-1 protein (Fig. 2B). The complex was more readily detected with hGH than EPO gene sequences. In addition, the complex formed on nucleotides −278/−250 of hGH1 was competed efficiently by non-labeled hGH1 sequences compared with the EPO HBS, suggesting a higher affinity/specificity for the HBS in −278/−250 hGH1 sequences.
FIGURE 2.
A HIF-1 binding site is located within nucleotides −278/−250 of the proximal hGH1 promoter region. A, alignment and comparison of human (nucleotides −327/−1) and mouse (nucleotides −355/−1) growth hormone gene proximal promoter regions (64) is shown. Conserved TATA box and Pit-1 binding sites are indicated. The hGH but not the mGH promoter region contains a palindromic hexanucleotide (bold) that includes the core HBS at nucleotide position −263/−259 (underlined). Asterisks are used to indicate the boundaries of the −279/−250 hGHp-HBS fragment used for EMSA. B, EMSA was done with a combination of recombinant HIF-1α (rHIF-1α) and HIF-1β protein together with radiolabeled probes: hGHp-HBS, EPO-HBS (positive control), and RF1 (negative control), with sequences as indicated. A single complex was seen in the presence of recombinant protein with both hGHp-HBS and EPO-HBS probes by autoradiography but required prolonged exposure to detect the EPO-HBS-protein complex. Competition of the hGHp-HBS-protein complex with a 2.5-, 5-, and 50-fold molar excess of unlabeled hGHp-HBS and EPO-HBS oligonucleotide as well as a 50-fold molar excess of RF-1 oligonucleotides was also performed to assess affinity/specificity. Competition was detected with hGHp-HBS and EPO-HBS but was more evident with a 5-fold molar excess of unlabeled hGHp-HBS than EPO-HBS; no competition with RF1 was observed. C, protein immunoblotting was done to assess HIF-1α protein in nuclear extracts (100 μg) of primary pituitary cells treated with 15 nm insulin for 24 h. Nuclear proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoreactive proteins were detected by chemiluminescence. The HIF-1α protein band (∼120 kDa) is indicated by black arrowhead. Upstream stimulatory factor 1 (USF-1; 43 kDa) was used as a loading control for nuclear proteins. D, a ChIP assay was performed with an anti-HIF1α antibody on chromatin isolated from primary pituitary cells treated with 15 nm insulin for 24 h. Binding events were calculated based on the signals obtained from the immunoprecipitated/input DNA amplification using specific primers to hGHp-HBS and the control untranscribed region (Untr6) by qPCR. The results are expressed as relative mean change plus or minus S.E. of the mean compared with the control (0 nm insulin) Untr6 value, which is arbitrarily set to 1. Significant differences are indicated by *, p < 0.05.
To assess whether insulin affects HIF-1α levels in primary pituitary cells, nuclear extracts were isolated from cultures treated with or without 15 nm insulin for 24 h and analyzed by protein immunoblotting using antibodies to HIF-1α and another transcription factor, upstream stimulatory factor 1 (USF-1) that is a ubiquitously expressed transcription factor (41). A band of the expected size for HIF-1α was detected (120 kDa), and levels were stimulated reproducibly by insulin (Fig. 2C). Ponceau staining of the membranes was used to confirm equivalent protein loading in each lane (see the supplemental data, Fig. S1).
A ChIP assay was used to assess HIF-1α association with the HBS-containing hGH1 proximal promoter region in the context of pituitary cell chromatin. Anti-HIF-1α antibody was used for independent immunoprecipitations, and normal rabbit IgG was employed as a negative control. PCR primers (Table 2) were designed to specifically amplify the −308/−235 region of hGH1 promoter as well as an untranscribed region on chromosome 6 (Untr6), which served as a measure of “background” or nonspecific association of protein-DNA detected by ChIP assay. HIF-1α associates preferentially with the hGH1 proximal promoter region by ChIP assay, relative to the signal seen with Untr6 background control sequences (Fig. 2D). The level of association increased significantly 1.7-fold after insulin (15 nm) treatment of pituitary cell cultures for 24 h (p < 0.05, n = 4).
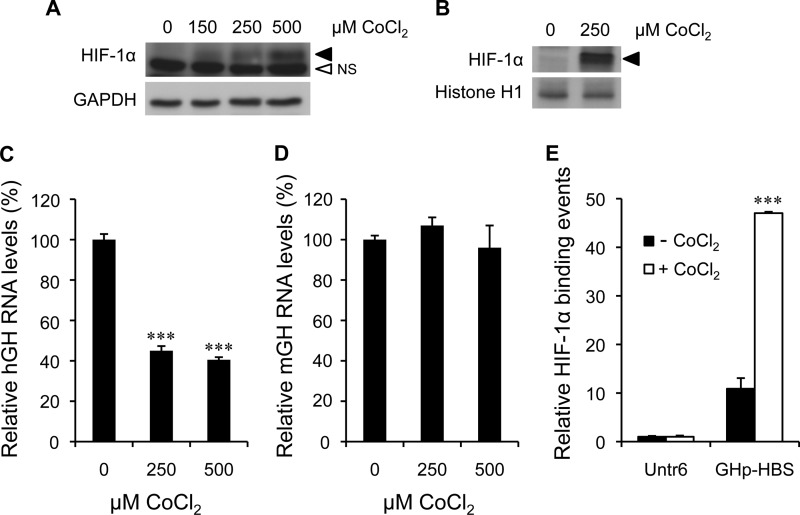
Induction of HIF-1α Protein by CoCl2 Treatment Mimics the Effect of Insulin on hGH1 RNA Levels
The effect of increased HIF-1 availability on hGH RNA levels was examined by treating primary pituitary cells with CoCl2, which can mimic the effects of hypoxia by inhibiting prolyl hydroxylase activity, thereby stabilizing HIF-1α (42). Cultures were treated with or without 150–500 μm CoCl2 for 5 h in serum substitute, and whole cell or nuclear extract (250 μm only) was assessed for HIF-1α by protein immunoblotting (Fig. 3, A and B). A dose-dependent stimulation of a band of the expected size (120 kDa) was observed in the whole cell extract and was also detectable in nuclear protein.
FIGURE 3.
Induction of HIF-1α protein by CoCl2 treatment mimics the effect of insulin on hGH RNA levels. Immunodetection of induced HIF-1α (120 kDa) in whole cell lysate (A) and nuclear extracts (B) of primary pituitary cells treated with 250 and 500 μm CoCl2 for 5 h is shown. GAPDH (37 kDa) and histone H1 (32 kDa) were used as loading controls for whole cell and nuclear proteins, respectively. The HIF-1α protein band (black arrowhead) and the nonspecific protein band (NS), which was only detected in whole cell lysate, are indicated. The effects of 250 and 500 μm CoCl2 treatment on hGH (C) and mGH (D) RNA levels were assessed by qPCR after 24 h. Significant decreases in hGH but not mGH RNAs were observed. E, a ChIP assay was performed with an anti-HIF1α antibody on chromatin isolated from primary pituitary cells treated with 250 μm CoCl2 for 24 h. Binding events were calculated and are expressed as relative mean change, as described in Fig. 2. Significant differences are indicated by ***, p < 0.001.
The effect of 250 and 500 μm CoCl2 on hGH and mGH RNA was also assessed by qPCR after 24 h of treatment. Vascular endothelial growth factor (VEGF) expression was also assessed as positive control. It is up-regulated by hypoxia through direct involvement of HIF-1α (43) and is also produced by hormone-producing as well as capillary endothelial cells of the anterior pituitary gland (44, 45). CoCl2 decreased hGH RNA levels significantly (p < 0.001, n = 6) and by a similar extent (∼60%) to that observed with insulin treatment (Fig. 3C). In contrast, there was no significant effect of CoCl2 treatment on endogenous mGH RNA levels (Fig. 3D); however, a significant 2-fold increase in VEGF RNA levels (p < 0.01, n = 3) was observed (see the supplemental data, Fig. S3B).
Primary pituitary cells were treated with CoCl2 for 24 h and assessed by ChIP assay for HIF-1α association with the HBS-containing hGH1 proximal promoter region (Fig. 3E). The level of association increased significantly 4–5-fold (p < 0.001, n = 4).
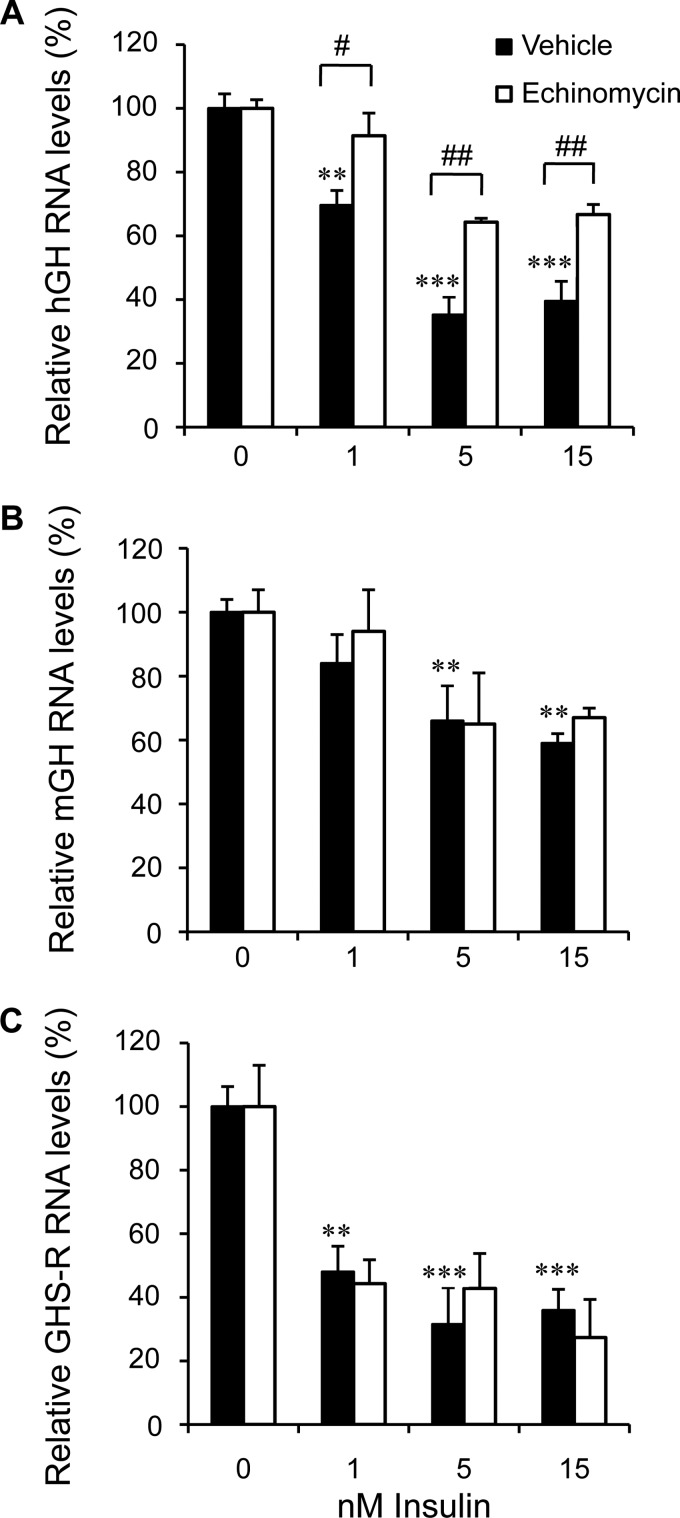
Interference with HIF-1α Binding and Production Reduces the Effect of Insulin on hGH RNA Levels
The effect of inhibiting HIF-1 binding on hGH RNA levels in primary pituitary cell cultures was assessed. Echinomycin is a cyclic peptide, antibiotic, and specific DNA binding factor with a strong affinity for the 5′- ACGTG-3′ sequence, which is at the core of a HBS (46, 47). Cultures were incubated with 10 nm echinomycin 1 h before insulin treatment. Human GH, mGH, and mouse ghrelin receptor (GHS-R) RNAs were assessed by qPCR after 24 h. Pretreatment with echinomycin interfered significantly with the negative effect of 1, 5, and 15 nm insulin on hGH transcript levels (Fig. 4A); this effect was greater than 50% at all doses of insulin. In contrast, the significant decreases in mGH and GHS-R RNA levels were not affected by echinomycin pretreatment (p < 0.01, n = 6) (Fig. 4, B and C). It is also noted that pretreatment with a higher concentration of echinomycin (20 nm) led to 100% inhibition of the negative effect of 5 nm insulin on hGH RNA levels. Again, pretreatment with 20 nm echinomycin had no effect on insulin regulation of mGH transcripts (see the supplemental data, Fig. S2, A and B).
FIGURE 4.
Interference with HIF-1α binding reduces the effect of insulin on hGH RNA levels. Primary pituitary cells were incubated without (solid columns) or with (open columns) 20 nm echinomycin 1 h before insulin (1, 5, and 15 nm) treatment. Human GH (A), mGH (B), and GHS-R RNA (C) levels were assessed by qPCR 24 h post-insulin treatment. Dose-dependent decreases in hGH, mGH, and GHS-R RNA levels were observed; however, the decrease in hGH RNA was the only one affected significantly by echinomycin treatment. Results are expressed as relative mean change plus or minus S.E. of the mean, compared with control values, which are arbitrarily set to 100. Data were analyzed by two-way analysis of variance with the Bonferroni post-test. Significant differences between groups at each concentration of insulin are indicated with brackets; #, p < 0.5; ##, p < 0.01. Significant differences between insulin treatments compared with no insulin group (0) as control are indicated by **, p < 0.01; ***, p < 0.001.
It was also noted that the decrease in hGH RNA and increase in VEGF RNA levels seen in response to CoCl2 (Fig. 3, B and C) were blocked by echinomycin pretreatment (p < 0.01, n = 3) (supplemental data, Fig. S3, A and B).
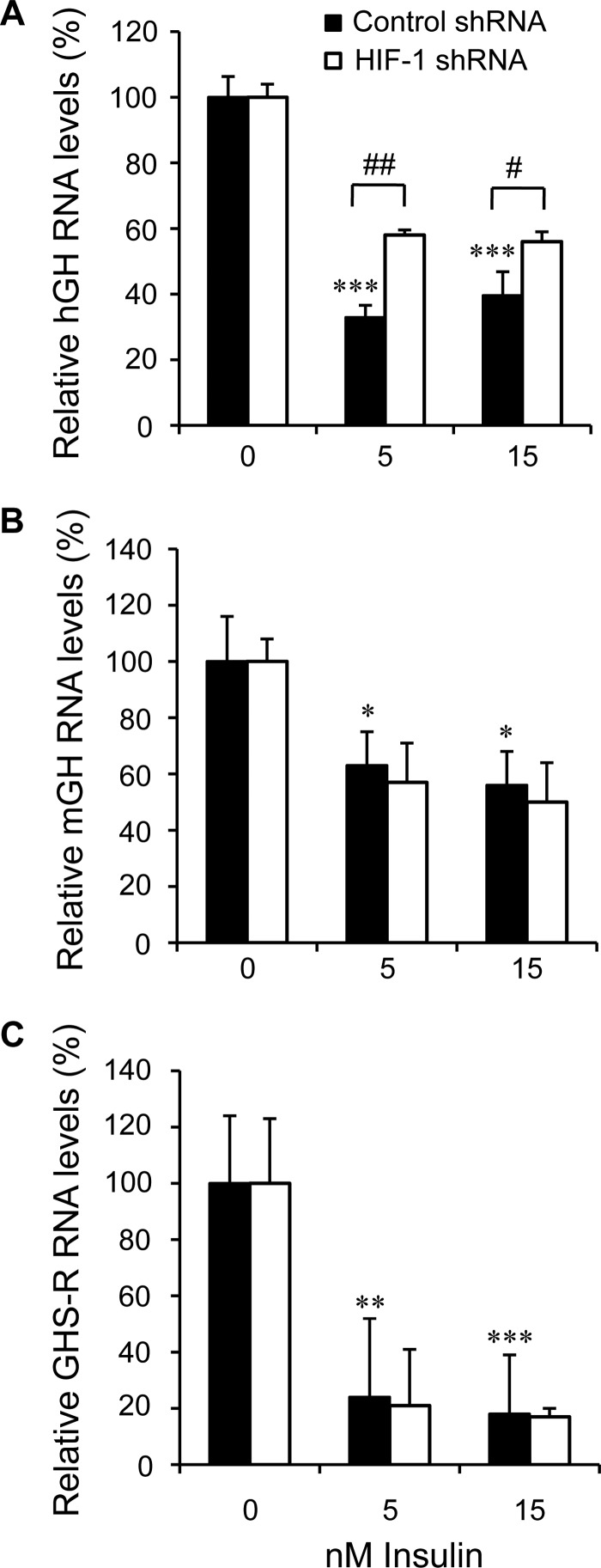
Lentiviral transduction particles of shRNA against HIF-1α were used to limit HIF-1α induction in response to insulin and examine the effect on hGH RNA levels. Pituitary cell cultures were treated with HIF-1α and control/scrambled shRNA for 24 h, maintained for 72 h, and then treated for 24 h with insulin. Human GH, mGH, and mGHS-R RNAs were assessed by qPCR. Partial but significant interference with the negative effect of 5 and 15 nm insulin on hGH RNA levels was observed with HIF-1α shRNA versus control treatment (p < 0.01, n = 5) (Fig. 5A). As with echinomycin (Fig. 4, B and C); however, there was no significant effect on the insulin responsiveness of mGH and mGHS-R transcript levels (Fig. 5, B and C).
FIGURE 5.
Interference with HIF-1α production reduces the effect of insulin on hGH RNA levels. Primary pituitary cells were treated with lentiviral transduction particles of scrambled/control (solid columns) or HIF-1α shRNA (open columns) before insulin (5 and 15 nm) treatment. Human GH (A), mGH (B), and GHS-R RNA (C) levels were assessed by qPCR 24 h post-insulin treatment. Dose-dependent decreases in hGH, mGH, and GHS-R RNA levels were seen. Partial but significant blockage of the inhibitory effect of insulin on hGH RNA levels was observed with HIF-1α but not control shRNA. There was no significant effect of shRNA on mGH GHS-R RNA levels in response to insulin. Results were analyzed and are expressed as relative mean change, as described in Fig. 4.
Histone Hyperacetylation Decreases in the hGH1 Proximal Promoter Region with Insulin Treatment
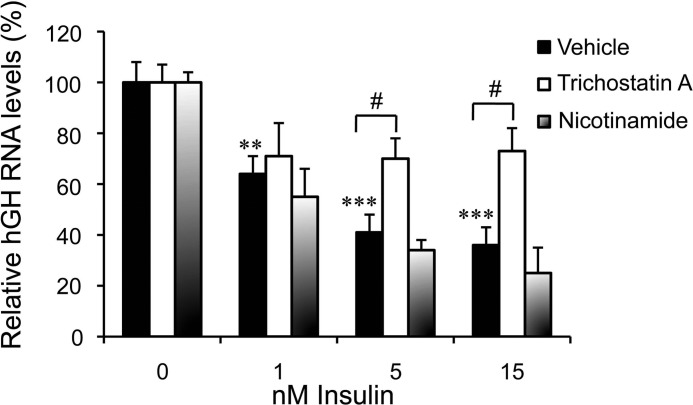
A potential role for histone acetylation status on the hGH1 insulin responsiveness was examined. HIF-1α is reported to interact with members of class I, II, and III HDACs and to recruit them to regulatory elements in target genes (48–53). Thus, pituitary cell cultures were treated with increasing concentrations of insulin in the presence of the class I and II HDAC inhibitor, trichostatin A, and class III HDAC (sirtuins) inhibitor, nicotinamide (Fig. 6). A dose-dependent decrease in hGH RNA was observed with increasing insulin concentration as assessed by qPCR; however, the decreases with 5 and 15 nm insulin were blunted significantly in response to trichostatin A but not nicotinamide treatments (p < 0.05, n = 6) (Fig. 6).
FIGURE 6.
The negative effect of insulin on hGH RNA levels is trichostatin A-sensitive. Primary pituitary cells were treated with HDACs inhibitors, trichostatin A (300 nm), nicotinamide (15 mm), or vehicle 1 h before insulin (1, 5, and 15 nm) treatment. Human GH RNA was assessed by qPCR 24 h post-insulin treatment. Trichostatin A but not nicotinamide muted the insulin effect on hGH RNA levels and was significant with 5 and 15 nm insulin. Results were analyzed and are expressed as relative mean change as described in Fig. 4.
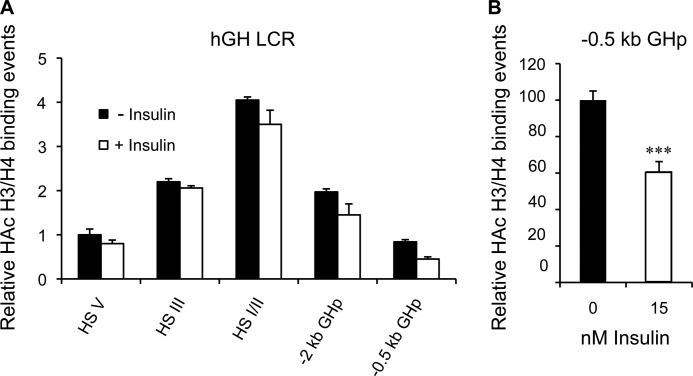
A distinctive histone H3/H4 hyperacetylation pattern has been reported for the hGH LCR in pituitary versus non-pituitary tissue/cells and is associated with an active hGH1 promoter (54); this includes the pattern detected in transgenic mice containing the intact hGH locus and LCR (55). Specifically, H3/H4 hyperacetylation is centered around HS I/II about 14.5 kb upstream of hGH1 and extends outwards, upstream to HS V (−32 kb hGH1) and downstream to include the hGH1 promoter in an “umbrella-like” manner (54, 55). A ChIP assay was done to assess the relative level of H3/H4 hyperacetylation across the hGH LCR and hGH1 promoter in response to 24-hour insulin treatment in chromatin from primary hGH/CS-TG pituitary cells in situ. Specific antibodies to hyperacetylated histones H3/H4 as well as control IgG were used for precipitations, and this was combined with specific PCR primers to analyze potentially bound sequences across the LCR (Table 2). Histone hyperacetylation levels, relative to HS V in untreated cultures, were determined for HS I/II and III as well as proximal (includes the HBS) and slightly more distal hGH1 promoter regions (−2 kb hGH1). As expected, hyperacetylation showed the characteristic pituitary cell pattern with the highest levels centered and extending out from HS I/II (Fig. 7A). This pattern was also seen after insulin treatment; however, there was a significant ∼40% decrease in H3/H4 hyperacetylation at the proximal (not more distal) hGH1 promoter region after insulin treatment (p < 0.001, n = 4) (Fig. 7B).
FIGURE 7.
Histone hyperacetylation decreases in the hGH1 proximal promoter region with insulin treatment. A, ChIP assay was performed with antibodies specific to the hyperacetylated (HAc) histone H3 and H4 (three or more acetylated lysine residues) on chromatin isolated from primary pituitary cells treated without (solid columns) or with 15 nm insulin (open columns) for 24 h. Quantitative PCR was performed on both input and immunoprecipitated (bound) chromatin fractions with primer sets designed to detect upstream HS V, HS III, and HS I/II as well as distal (−2 kb) and proximal (−0.5 kb) hGH1 promoter regions. Results are presented as relative hyperacetylated H3/H4 “binding events” compared with the control value for HS V, which is arbitrarily set to 1. A peak for hyperacetylated H3/H4 is seen at HS I/II in the presence and absence of insulin. There is also the suggestion of a modest decrease in hyperacetylation in the region spanning HS I/II and the hGH1 promoter. B, when re-graphed (control arbitrarily set to 100), a significant ∼40% decrease in hyperacetylated H3/H4 is detected only in the proximal hGH1 promoter region. Significance was assessed by t test; ***, p < 0.001.
DISCUSSION
Using cultures derived from “humanized” transgenic mouse pituitaries containing the intact hGH locus and LCR, we show for the first time that hGH RNA synthesis is negatively regulated by a physiological concentration of insulin in primary non-tumor pituitary cells. This regulation is dependent on the HIF-1 transcription factor. Increasing HIF-1α levels with CoCl2 increased association with the proximal hGH1 promoter region and mimicked the decrease in hGH RNA seen with insulin. Furthermore, interference with both HIF-1 binding and levels using echinomycin and lentiviral delivery of HIF-1α shRNA, both, blunted the negative response of hGH1 to insulin significantly. In addition we characterized a HBS in the proximal promoter region of hGH1, which supports a previous report by others for the presence of an insulin-responsive element in this region (14). Our data also implicate histone modification of the proximal hGH1 promoter region in its response to insulin, as specific inhibition of class I and II (but not III) HDACs, significantly blunted the effect. Furthermore, insulin treatment was associated with a significant and specific reduction in H3/H4 hyperacetylation in the proximal promoter region of hGH1 containing the HBS. These data suggest that HIF-1 binding and chromatin remodeling are required for insulin regulation of endogenous hGH synthesis at the level of the pituitary somatotroph.
Activation of genes and regulation of expression typically involve posttranslational modification of histone proteins, making DNA more or less accessible to the transcription machinery in response to environmental cues. Based on elegant transgenic mouse studies, activation of the hGH locus requires “global” acetylation of a 32-kb chromatin domain centered and extending outward from HS I/II to HS V upstream and the hGH1 promoter downstream in pituitary somatotroph development (56, 57). This leads to generation of a “specificity” loop between HS III/V and HS I/II, linked with appropriate (site of integration independent and somatotroph-specific) synthesis, and an “expression” loop between HS I/II and the hGH1 promoter that is associated with efficient promoter activity (38, 56, 57). The global acetylation pattern spanning the 32-kb hGH LCR is observed in our transgenic mouse pituitary cell cultures, consistent with somatotroph expression of hGH1. This pattern was largely unaffected by insulin treatment except for a significant decrease in the proximal promoter region containing the HBS. Our data indicate that the insulin response requires histone deacetylation via class I or II HDACs and a decrease in H3/H4 hyperacetylation “locally” in this proximal promoter region. Target genes for both insulin and HIF-1 are subject to the histone modification by deacetylation in their regulatory elements via recruitment of class I and II HDACs (48–50, 58). This suggests that a regulatory element(s) in the hGH1 promoter is a target for insulin and HIF-1-mediated histone deacetylation. This would be expected to result in “inactive/repressed” chromatin, thereby decreasing accessibility to factors including RNA polymerase (59) and/or affecting the expression loop and thus lessening hGH1 promoter function in pituitary cells.
GH is a major metabolic homeostatic factor involved in maintaining the balance between excess energy intake and expenditure via its lipolytic activity (60); however, obesity has a significant negative impact on GH production. Thus, GH insufficiency is considered to be a classical feature of obesity, but the underlying mechanisms responsible for this blunted GH production is unclear. Hyperleptinemia, hyperglycemia, and high levels of free fatty acids as well as hyperinsulinemia are among the deregulated metabolic states/factors that have been proposed to contribute to this phenomenon (4, 5). Recent studies, however, strongly suggest a significant role for excess insulin as the GH-suppressive factor. Overeating for only 3 days resulted in profound hyperinsulinemia and contributed to a 77% decrease in plasma hGH levels without any significant weight gain in a group of healthy 24-year-old individuals (61). Thus, our data are consistent with a direct effect of “excess” insulin on endogenous hGH synthesis at the level of the somatotroph, contributing to the observed reduced plasma hGH levels. If “overeating” is seen as a stage of developing “obesity,” then the initial decrease in GH might be interpreted as a positive response by increasing the whole body insulin sensitivity for a limited period of time due to its anti-insulin activity (62, 63). This was suggested based on the observation that adult onset isolated GH deficiency in mice was associated with higher insulin sensitivity (62). If conditions of excess caloric intake were to persist, however, then the decrease in GH is expected to be a primary or secondary cause of obesity by contributing to further weight gain due to loss of lipolytic and anabolic activities of GH as well as impaired insulin output (62, 65–68). If, however, the initial response to excess insulin is a decrease in GH and a compensatory increase in insulin sensitivity, then this would be consistent with HIF-1 involvement. HIF-1 is a significant player in energy metabolism (52, 69–71) and as a signaling molecule induced by elevated levels of insulin (hyperinsulinemia) can offer an explanation for blunted GH levels seen in obese individuals. High concentrations of insulin can provoke the accumulation of HIF-1 via phosphatidylinositol 3 kinase/Akt-dependent and/or mitogen-activated protein kinase signaling pathways (72).
Like hGH, endogenous mGH RNA was also down-regulated in response to low doses of insulin, although this was only significant above 1 nm. Unlike hGH, however, the negative effect of insulin on mGH RNA levels was not responsive to changes in HIF-1α levels or interference with HIF-1 DNA binding. This is similar to the effect of insulin on mGHS-R RNA levels, which was decreased in agreement with previous reports using non-human primate pituitary cultures (12). Insulin regulation of both mGH and mGHS-R appear to be independent of HIF-1. An alignment of mGH with hGH gene promoter regions, including nucleotides −279/−250, revealed no putative HIF-1 binding site in equivalent mGH sequences or in a search of 2 kb of upstream mGH and mGHS-R flanking DNA. Thus, although there appears to be convergence in terms of the overall response of the hGH and mGH genes to insulin, hGH1 and by extension human somatotrophs may be more sensitive to insulin levels, and this may reflect distinct regulatory mechanisms and the involvement of HIF-1 in hGH but not mGH synthesis. Certainly, primate and murine GH are structurally and, therefore, functionally distinct, and differences, as with the presence of a HBS in the proximal promoter region, appear to extend beyond the coding to regulatory sequences (33, 73–78).
In summary, these observations provide the first evidence that human GH synthesis is regulated directly by insulin at the pituitary somatotroph level. The insulin effect is HIF-1 dependent, and our observations are consistent with an important role for HIF-1α in mediating the early and rapid suppression of GH levels seen in obese individuals. Further examination of the roles of hyperinsulinemia, HIF-1, and HIF-1-mediated GH reduction in obesity and diabetes models may provide valuable insight into the physiological regulation and pathophysiological dysregulation of somatotrophs as it applies to the pathogenesis of obesity and insulin resistance.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Grant MT-10853.

This article contains supplemental Figs. S1–S3.
- GH
- growth hormone
- GHS
- growth hormone secretagogue
- GHS-R
- GHS receptor
- HIF-1
- hypoxia-inducible factor-1
- HBS
- HIF-1 binding sequence
- TG
- transgenic
- LCR
- locus control region
- HS
- hypersensitive site
- HDAC
- histone deacetylase
- qPCR
- quantitative PCR
- EPO
- erythropoietin
- Untr6
- untranscribed region on chromosome 6
- CS
- chorionic somatomammotropin.
REFERENCES
- 1. Luque R. M., Kineman R. D. (2006) Impact of obesity on the growth hormone axis. Evidence for a direct inhibitory effect of hyperinsulinemia on pituitary function. Endocrinology 147, 2754–2763 [DOI] [PubMed] [Google Scholar]
- 2. Buettner R., Newgard C. B., Rhodes C. J., O'Doherty R. M. (2000) Correction of diet-induced hyperglycemia, hyperinsulinemia, and skeletal muscle insulin resistance by moderate hyperleptinemia. Am. J. Physiol. Endocrinol Metab. 278, E563–E569 [DOI] [PubMed] [Google Scholar]
- 3. Brothers K. J., Wu S., DiVall S. A., Messmer M. R., Kahn C. R., Miller R. S., Radovick S., Wondisford F. E., Wolfe A. (2010) Rescue of obesity-induced infertility in female mice due to a pituitary-specific knockout of the insulin receptor. Cell Metab. 12, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pataky Z., Bobbioni-Harsch E., Golay A. (2010) Obesity. A complex growing challenge. Exp. Clin. Endocrinol. Diabetes 118, 427–433 [DOI] [PubMed] [Google Scholar]
- 5. De Marinis L., Bianchi A., Mancini A., Gentilella R., Perrelli M., Giampietro A., Porcelli T., Tilaro L., Fusco A., Valle D., Tacchino R. M. (2004) Growth hormone secretion and leptin in morbid obesity before and after biliopancreatic diversion. Relationships with insulin and body composition. J. Clin. Endocrinol. Metab. 89, 174–180 [DOI] [PubMed] [Google Scholar]
- 6. Lanzi R., Luzi L., Caumo A., Andreotti A. C., Manzoni M. F., Malighetti M. E., Sereni L. P., Pontiroli A. E. (1999) Elevated insulin levels contribute to the reduced growth hormone (GH) response to GH-releasing hormone in obese subjects. Metabolism 48, 1152–1156 [DOI] [PubMed] [Google Scholar]
- 7. Lanzi R., Manzoni M. F., Andreotti A. C., Malighetti M. E., Bianchi E., Sereni L. P., Caumo A., Luzi L., Pontiroli A. E. (1997) Evidence for an inhibitory effect of physiological levels of insulin on the growth hormone (GH) response to GH-releasing hormone in healthy subjects. J. Clin. Endocrinol. Metab. 82, 2239–2243 [DOI] [PubMed] [Google Scholar]
- 8. Scacchi M., Orsini F., Cattaneo A., Grasso A., Filippini B., Pecori Giraldi F., Fatti L. M., Moro M., Cavagnini F. (2010) The diagnosis of GH deficiency in obese patients. A reappraisal with GHRH plus arginine testing after pharmacological blockade of lipolysis. Eur. J. Endocrinol. 163, 201–206 [DOI] [PubMed] [Google Scholar]
- 9. Pijl H., Langendonk J. G., Burggraaf J., Frölich M., Cohen A. F., Veldhuis J. D., Meinders A. E. (2001) Altered neuroregulation of GH secretion in viscerally obese premenopausal women. J. Clin. Endocrinol. Metab. 86, 5509–5515 [DOI] [PubMed] [Google Scholar]
- 10. Procopio M., Maccario M., Grottoli S., Oleandri S. E., Boffano G. M., Camanni F., Ghigo E. (1995) Short-term fasting in obesity fails to restore the blunted GH responsiveness to GH-releasing hormone alone or combined with arginine. Clin Endocrinol. (Oxf.) 43, 665–669 [DOI] [PubMed] [Google Scholar]
- 11. Maccario M., Procopio M., Grottoli S., Oleandri S. E., Razzore P., Camanni F., Ghigo E. (1995) In obesity the somatotrope response to either growth hormone-releasing hormone or arginine is inhibited by somatostatin or pirenzepine but not by glucose. J. Clin. Endocrinol. Metab. 80, 3774–3778 [DOI] [PubMed] [Google Scholar]
- 12. Luque R. M., Gahete M. D., Valentine R. J., Kineman R. D. (2006) Examination of the direct effects of metabolic factors on somatotrope function in a non-human primate model, Papio anubis. J. Mol. Endocrinol. 37, 25–38 [DOI] [PubMed] [Google Scholar]
- 13. Prager D., Melmed S. (1988) Insulin regulates expression of the human growth hormone gene in transfected cells. J. Biol. Chem. 263, 16580–16585 [PubMed] [Google Scholar]
- 14. Prager D., Gebremedhin S., Melmed S. (1990) An insulin-induced DNA-binding protein for the human growth hormone gene. J. Clin. Invest. 85, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peritz L. N., Fodor E. J., Silversides D. W., Cattini P. A., Baxter J. D., Eberhardt N. L. (1988) The human growth hormone gene contains both positive and negative control elements. J. Biol. Chem. 263, 5005–5007 [PubMed] [Google Scholar]
- 16. Lefevre C., Imagawa M., Dana S., Grindlay J., Bodner M., Karin M. (1987) Tissue-specific expression of the human growth hormone gene is conferred in part by the binding of a specific transacting factor. EMBO J. 6, 971–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schödel J., Oikonomopoulos S., Ragoussis J., Pugh C. W., Ratcliffe P. J., Mole D. R. (2011) High resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117, e207–e217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Löfstedt T., Jögi A., Sigvardsson M., Gradin K., Poellinger L., Påhlman S., Axelson H. (2004) Induction of ID2 expression by hypoxia-inducible factor-1. A role in dedifferentiation of hypoxic neuroblastoma cells. J. Biol. Chem. 279, 39223–39231 [DOI] [PubMed] [Google Scholar]
- 20. Weidemann A., Johnson R. S. (2008) Biology of HIF-1α. Cell Death Differ. 15, 621–627 [DOI] [PubMed] [Google Scholar]
- 21. Adams J. M., Difazio L. T., Rolandelli R. H., Luján J. J., Haskó G., Csóka B., Selmeczy Z., Németh Z. H. (2009) HIF-1. A key mediator in hypoxia. Acta Physiol. Hung. 96, 19–28 [DOI] [PubMed] [Google Scholar]
- 22. Ratan R. R., Siddiq A., Aminova L., Lange P. S., Langley B., Ayoub I., Gensert J., Chavez J. (2004) Translation of ischemic preconditioning to the patient. Prolyl hydroxylase inhibition and hypoxia-inducible factor-1 as novel targets for stroke therapy. Stroke 35, 2687–2689 [DOI] [PubMed] [Google Scholar]
- 23. Cheng K., Ho K., Stokes R., Scott C., Lau S. M., Hawthorne W. J., O'Connell P. J., Loudovaris T., Kay T. W., Kulkarni R. N., Okada T., Wang X. L., Yim S. H., Shah Y., Grey S. T., Biankin A. V., Kench J. G., Laybutt D. R., Gonzalez F. J., Kahn C. R., Gunton J. E. (2010) Hypoxia-inducible factor-1α regulates β cell function in mouse and human islets. J. Clin. Invest. 120, 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X., Lam K. S., Ye H., Chung S. K., Zhou M., Wang Y., Xu A. (2010) Adipose tissue-specific inhibition of hypoxia-inducible factor 1α induces obesity and glucose intolerance by impeding energy expenditure in mice. J. Biol. Chem. 285, 32869–32877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weng Q., Zhang J., Cao J., Xia Q., Wang D., Hu Y., Sheng R., Wu H., Zhu D., Zhu H., He Q., Yang B. (2010) Q39, a quinoxaline 1,4-Di-N-oxide derivative, inhibits hypoxia-inducible factor-1α expression and the Akt/mTOR/4E-BP1 signaling pathway in human hepatoma cells. Investig. New Drugs 9, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 26. Erman A., Wabitsch M., Goodyer C. G. (2011) Human growth hormone receptor (GHR) expression in obesity. II. Regulation of the human GHR gene by obesity-related factors. Int. J. Obes. (Lond.) 35, 1520–1529 [DOI] [PubMed] [Google Scholar]
- 27. Zelzer E., Levy Y., Kahana C., Shilo B. Z., Rubinstein M., Cohen B. (1998) Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1α/ARNT. EMBO J. 17, 5085–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L., Zhou W., Gou S., Wang T., Liu T., Wang C. (2010) Insulin promotes proliferative vitality and invasive capability of pancreatic cancer cells via hypoxia-inducible factor 1α pathway. J. Huazhong Univ. Sci. Technolog. Med. Sci. 30, 349–353 [DOI] [PubMed] [Google Scholar]
- 29. Kietzmann T., Samoylenko A., Roth U., Jungermann K. (2003) Hypoxia-inducible factor-1 and hypoxia response elements mediate the induction of plasminogen activator inhibitor-1 gene expression by insulin in primary rat hepatocytes. Blood 101, 907–914 [DOI] [PubMed] [Google Scholar]
- 30. Yoshida D., Kim K., Noha M., Teramoto A. (2006) Anti-apoptotic action by hypoxia-inducible factor 1-α in human pituitary adenoma cell line, HP-75 in hypoxic condition. J. Neurooncol. 78, 217–225 [DOI] [PubMed] [Google Scholar]
- 31.Deleted in proof
- 32. Jin Y., Lu S. Y., Fresnoza A., Detillieux K. A., Duckworth M. L., Cattini P. A. (2009) Differential placental hormone gene expression during pregnancy in a transgenic mouse containing the human growth hormone/chorionic somatomammotropin locus. Placenta 30, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones B. K., Monks B. R., Liebhaber S. A., Cooke N. E. (1995) The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15, 7010–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vakili H., Jin Y., Nagy J. I., Cattini P. A. (2011) Transgenic mice expressing the human growth hormone gene provide a model system to study human growth hormone synthesis and secretion in non-tumor-derived pituitary cells. Differential effects of dexamethasone and thyroid hormone. Mol. Cell. Endocrinol. 345, 48–57 [DOI] [PubMed] [Google Scholar]
- 35. Aza-Carmona M., Shears D. J., Yuste-Checa P., Barca-Tierno V., Hisado-Oliva A., Belinchón A., Benito-Sanz S., Rodríguez J. I., Argente J., Campos-Barros A., Scambler P. J., Heath K. E. (2011) SHOX interacts with the chondrogenic transcription factors SOX5 and SOX6 to activate the aggrecan enhancer. Hum. Mol. Genet. 20, 1547–1559 [DOI] [PubMed] [Google Scholar]
- 36. Norquay L. D., Yang X., Jin Y., Detillieux K. A., Cattini P. A. (2006) Hepatocyte nuclear factor-3α binding at P sequences of the human growth hormone locus is associated with pituitary repressor function. Mol. Endocrinol. 20, 598–607 [DOI] [PubMed] [Google Scholar]
- 37. Nam S. Y., Ko Y. S., Jung J., Yoon J., Kim Y. H., Choi Y. J., Park J. W., Chang M. S., Kim W. H., Lee B. L. (2011) A hypoxia-dependent up-regulation of hypoxia-inducible factor-1 by nuclear factor-κB promotes gastric tumor growth and angiogenesis. Br J. Cancer 104, 166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X., Jin Y., Cattini P. A. (2010) Appearance of the pituitary factor Pit-1 increases chromatin remodeling at hypersensitive site III in the human GH locus. J. Mol. Endocrinol. 45, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yochum G. S., McWeeney S., Rajaraman V., Cleland R., Peters S., Goodman R. H. (2007) Serial analysis of chromatin occupancy identifies β-catenin target genes in colorectal carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 104, 3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lytras A., Cattini P.A. (1994) Human chorionic somatomammotropin gene enhancer activity is dependent on the blockade of a repressor mechanism. Mol. Endocrinol. 8, 478–489 [DOI] [PubMed] [Google Scholar]
- 41. Sirito M., Lin Q., Maity T., Sawadogo M. (1994) Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xi L., Taher M., Yin C., Salloum F., Kukreja R. C. (2004) Cobalt chloride induces delayed cardiac preconditioning in mice through selective activation of HIF-1α and AP-1 and iNOS signaling. Am. J. Physiol. Heart Circ. Physiol. 287, H2369–H2375 [DOI] [PubMed] [Google Scholar]
- 43. Fukuda R., Kelly B., Semenza G. L. (2003) Vascular endothelial growth factor gene expression in colon cancer cells exposed to prostaglandin E2 is mediated by hypoxia-inducible factor 1. Cancer Res. 63, 2330–2334 [PubMed] [Google Scholar]
- 44. Vidal S., Lloyd R. V., Moya L., Scheithauer B. W., Kovacs K. (2002) Expression and distribution of vascular endothelial growth factor receptor Flk-1 in the rat pituitary. J. Histochem. Cytochem. 50, 533–540 [DOI] [PubMed] [Google Scholar]
- 45. Lombardero M., Vidal S., Hurta R., Román A., Kovacs K., Lloyd R. V., Scheithauer B. W. (2006) Modulation of VEGF/Flk-1 receptor expression in the rat pituitary GH3 cell line by growth factors. Pituitary 9, 137–143 [DOI] [PubMed] [Google Scholar]
- 46. Formica J. V., Waring M. J. (1983) Effect of phosphate and amino acids on echinomycin biosynthesis by Streptomyces echinatus. Antimicrob. Agents Chemother. 24, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Dyke M. M., Dervan P. B. (1984) Echinomycin binding sites on DNA. Science 225, 1122–1127 [DOI] [PubMed] [Google Scholar]
- 48. Kato H., Tamamizu-Kato S., Shibasaki F. (2004) Histone deacetylase 7 associates with hypoxia-inducible factor 1α and increases transcriptional activity. J. Biol. Chem. 279, 41966–41974 [DOI] [PubMed] [Google Scholar]
- 49. Lee K. J., Lee K. Y., Lee Y. M. (2010) Down-regulation of a tumor suppressor RECK by hypoxia through recruitment of HDAC1 and HIF-1α to reverse HRE site in the promoter. Biochim. Biophys. Acta 1803, 608–616 [DOI] [PubMed] [Google Scholar]
- 50. Qian D. Z., Kachhap S. K., Collis S. J., Verheul H. M., Carducci M. A., Atadja P., Pili R. (2006) Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1α. Cancer Res. 66, 8814–8821 [DOI] [PubMed] [Google Scholar]
- 51. Du J., Chen Y., Li Q., Han X., Cheng C., Wang Z., Danielpour D., Dunwoodie S. L., Bunting K. D., Yang Y. C. (2012) HIF-1α deletion partially rescues defects of hematopoietic stem cell quiescence caused by Cited2 deficiency. Blood 119, 2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He Q., Gao Z., Yin J., Zhang J., Yun Z., Ye J. (2011) Regulation of HIF-1α activity in adipose tissue by obesity-associated factors, adipogenesis, insulin, and hypoxia. Am. J. Physiol. Endocrinol. Metab. 300, E877–E885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rajendran R., Garva R., Krstic-Demonacos M., Demonacos C. (2011) Sirtuins. Molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription.J. Biomed. Biotechnol. 2011, 368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elefant F., Cooke N. E., Liebhaber S. A. (2000) Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 275, 13827–13834 [DOI] [PubMed] [Google Scholar]
- 55. Elefant F., Su Y., Liebhaber S. A., Cooke N. E. (2000) Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19, 6814–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ho Y., Elefant F., Cooke N., Liebhaber S. (2002) A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9, 291–302 [DOI] [PubMed] [Google Scholar]
- 57. Ho Y., Tadevosyan A., Liebhaber S. A., Cooke N. E. (2008) The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep. 9, 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee Y. S., Sohn D. H., Han D., Lee H. W., Seong R. H., Kim J. B. (2007) Chromatin remodeling complex interacts with ADD1/SREBP1c to mediate insulin-dependent regulation of gene expression. Mol. Cell. Biol. 27, 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rönsch K., Jäger M., Schöpflin A., Danciu M., Lassmann S., Hecht A. (2011) Class I and III HDACs and loss of active chromatin features contribute to epigenetic silencing of CDX1 and EPHB tumor suppressor genes in colorectal cancer. Epigenetics 6, 610–622 [DOI] [PubMed] [Google Scholar]
- 60. Yang S., Mulder H., Holm C., Edén S. (2004) Effects of growth hormone on the function of β-adrenoceptor subtypes in rat adipocytes. Obes. Res. 12, 330–339 [DOI] [PubMed] [Google Scholar]
- 61. Cornford A. S., Barkan A. L., Horowitz J. F. (2011) Rapid suppression of growth hormone concentration by overeating. Potential mediation by hyperinsulinemia. J. Clin. Endocrinol. Metab. 96, 824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Luque R. M., Lin Q., Córdoba-Chacón J., Subbaiah P. V., Buch T., Waisman A., Vankelecom H., Kineman R. D. (2011) Metabolic impact of adult-onset, isolated, growth hormone deficiency (AOiGHD) due to destruction of pituitary somatotropes. PLoS One 6, e15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yakar S., Setser J., Zhao H., Stannard B., Haluzik M., Glatt V., Bouxsein M. L., Kopchick J. J., LeRoith D. (2004) Inhibition of growth hormone action improves insulin sensitivity in liver IGF-1-deficient mice. J. Clin. Invest. 113, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krawczak M., Chuzhanova N. A., Cooper D. N. (1999) Evolution of the proximal promoter region of the mammalian growth hormone gene. Gene 237, 143–151 [DOI] [PubMed] [Google Scholar]
- 65. Møller N., Gjedsted J., Gormsen L., Fuglsang J., Djurhuus C. (2003) Effects of growth hormone on lipid metabolism in humans. Growth Horm. IGF Res. 13, S18–S21 [DOI] [PubMed] [Google Scholar]
- 66. Takahashi S., Satozawa N. (2002) The 20-kDa human growth hormone reduces body fat by increasing lipolysis and decreasing lipoprotein lipase activity. Horm. Res. 58, 157–164 [DOI] [PubMed] [Google Scholar]
- 67. Christoforidis A., Maniadaki I., Stanhope R. (2005) Growth hormone/insulin-like growth factor-1 axis during puberty. Pediatr. Endocrinol. Rev. 3, 5–10 [PubMed] [Google Scholar]
- 68. Scacchi M., Pincelli A. I., Cavagnini F. (1999) Growth hormone in obesity. Int. J. Obes. Relat. Metab. Disord. 23, 260–271 [DOI] [PubMed] [Google Scholar]
- 69. Zhang H., Zhang G., Gonzalez F. J., Park S. M., Cai D. (2011) Hypoxia-inducible factor directs POMC gene to mediate hypothalamic glucose sensing and energy balance regulation. PLoS Biol. 9, e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jiang C., Qu A., Matsubara T., Chanturiya T., Jou W., Gavrilova O., Shah Y. M., Gonzalez F. J. (2011) Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high fat diet-fed mice. Diabetes 60, 2484–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Krishnan J., Danzer C., Simka T., Ukropec J., Walter K. M., Kumpf S., Mirtschink P., Ukropcova B., Gasperikova D., Pedrazzini T., Krek W. (2012) Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 26, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Treins C., Giorgetti-Peraldi S., Murdaca J., Semenza G. L., Van Obberghen E. (2002) Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J. Biol. Chem. 277, 27975–27981 [DOI] [PubMed] [Google Scholar]
- 73. Strasburger C. J. (1990) Antigenic epitope mapping of the human growth hormone molecule. A strategy to standardize growth hormone immunoassays. Acta Paediatr. Scand. Suppl. 370, 82–86 [DOI] [PubMed] [Google Scholar]
- 74. Yi S., Bernat B., Pál G., Kossiakoff A., Li W. H. (2002) Functional promiscuity of squirrel monkey growth hormone receptor toward both primate and nonprimate growth hormones. Mol. Biol. Evol. 19, 1083–1092 [DOI] [PubMed] [Google Scholar]
- 75. Nickel B. E., Kardami E., Cattini P. A. (1990) The human placental growth hormone variant is mitogenic for rat lymphoma Nb2 cells. Endocrinology 126, 971–976 [DOI] [PubMed] [Google Scholar]
- 76. Wells J. A., Cunningham B. C., Fuh G., Lowman H. B., Bass S. H., Mulkerrin M. G., Ultsch M., deVos A. M. (1993) The molecular basis for growth hormone-receptor interactions. Recent Prog. Horm. Res. 48, 253–275 [PubMed] [Google Scholar]
- 77. Wells J. A., de Vos A. M. (1993) Structure and function of human growth hormone. Implications for the hematopoietins. Annu. Rev. Biophys. Biomol. Struct. 22, 329–351 [DOI] [PubMed] [Google Scholar]
- 78. Lira S. A., Kalla K. A., Glass C. K., Drolet D. W., Rosenfeld M. G. (1993) Synergistic interactions between Pit-1 and other elements are required for effective somatotroph rat growth hormone gene expression in transgenic mice. Mol. Endocrinol. 7, 694–701 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.