Background: Insulin signaling regulates luteinizing hormone (LH) production in pituitary gonadotrope cells through unknown mechanisms.
Results: FOXO1 phosphorylation and cellular localization are regulated by insulin signaling, and FOXO1 represses LH β-subunit (Lhb) transcription.
Conclusion: Our study highlights a novel mechanism for regulation of Lhb gene expression.
Significance: FOXO1 may act as a metabolic sensor in controlling LH levels and fertility.
Keywords: Foxo, Neuroendocrinology, Pituitary Gland, Reproduction, Transcription Regulation, LH, Gonadotrope
Abstract
Synthesis of luteinizing hormone (LH) is tightly controlled by a complex network of hormonal signaling pathways that can be modulated by metabolic cues, such as insulin. One group of candidate genes that may be regulated by insulin signaling in pituitary gonadotrope cells is the FOXO subfamily of forkhead transcription factors. In this study we investigated whether FOXO1 is expressed in gonadotropes and if it can modulate LH β-subunit (Lhb) gene expression. We demonstrated that FOXO1 is expressed in murine gonadotrope cells and that insulin signaling increased FOXO1 phosphorylation and cytoplasmic localization in a PI3K-dependent manner. We also showed that FOXO1 repressed basal transcription and gonadotropin-releasing hormone (GnRH) induction of both the murine and human LHB genes in LβT2 cells, suggesting that FOXO1 regulation of LHB transcription may be conserved between rodents and humans. Although we did not detect FOXO1 binding to the proximal Lhb promoter, the FOXO1 DNA binding domain was necessary for the suppression, suggesting that FOXO1 exerts its effect through protein-protein interactions with transcription factors/cofactors required for Lhb gene expression. FOXO1 repression mapped to the proximal Lhb promoter containing steroidogenic factor 1 (SF1), pituitary homeobox 1 (PTX1), and early growth response protein 1 (EGR1) binding elements. Additionally, FOXO1 blocked induction of the Lhb promoter with overexpressed SF1, PTX1, and EGR1, indicating that FOXO1 repression occurs via these transcription factors but not through regulation of their promoters. In summary, we demonstrate that FOXO1 phosphorylation and cellular localization is regulated by insulin signaling in gonadotropes and that FOXO1 inhibits Lhb transcription. Our study also suggests that FOXO1 may play an important role in controlling LH levels in response to metabolic cues.
Introduction
LH2 regulates critical aspects of reproduction, including steroidogenesis and ovulation (1–3). It is produced by gonadotropes within the anterior pituitary and secreted into the blood stream as a result of pulsatile GnRH activation of the GnRH receptor (4–6). LH is a heterodimeric glycoprotein composed of an α subunit and a unique β subunit (7). Transcription of the β subunit is the rate-limiting step for LH production and is essential for reproduction (8, 9). Decreased Lhb mRNA levels result in infertility due to hypogonadotropic hypogonadism, whereas increased Lhb gene expression, such as in the LHβCTP transgenic mouse model, result in infertility due to anovulation (1, 10).
Basal transcription of Lhb occurs upon the binding of specificity protein 1 (SP1), SF1, and PTX1 transcription factors to response elements in the Lhb promoter (for review, see Ref. 11). GnRH signaling via protein kinase C and mitogen-activated protein kinase pathways (12–16) results in increased synthesis of EGR1 (17, 18). EGR1 binds to the promoter and interacts in a synergistic manner with SP1, SF1, and PTX1 to up-regulate Lhb gene expression (19, 20). Activin also induces Lhb synthesis via the binding of SMA/mothers against decapentaplegic (SMAD) transcription factors to the proximal Lhb promoter (21–23).
In addition to GnRH and activin, other peptide hormones and growth factors may regulate LH production. The receptors for insulin, insulin-like growth factor 1, and epidermal growth factor as well as downstream components of their signaling pathways are present in gonadotrope cells (24–27). A recent study demonstrated that pituitary insulin signaling may play an important role in obesity-related infertility (28). Although insulin has been shown to increase Lhb gene expression and LH secretion in immortalized gonadotrope cells and in primary pituitary cultures (24, 29–32), the mechanisms by which insulin modulates LH production at the level of the gonadotrope remain unclear.
One possibility is that insulin regulates Lhb transcription in gonadotropes via the FOXO subfamily of forkhead transcription factors. The activity of FOXOs is tightly controlled by posttranslational modifications including phosphorylation, acetylation, and ubiquitination (33). Insulin/growth factor signaling has been shown to negatively regulate FOXOs through phosphorylation by AKT, resulting in their active nuclear export and inhibition of their transcriptional activities (34). Phosphorylation of FOXOs by other kinases, such as c-Jun N-terminal kinase, in response to stress results in their translocation to the nucleus (35, 36). Studies have also demonstrated that FOXO can be acetylated by cAMP-response element-binding protein (CREB)-binding protein (CBP)/p300 and deacetylation by sirtuins such as SIRT1 (37, 38).
FOXOs are the mammalian orthologs of DAF-16, which regulates longevity, metabolism, and fertility in the nematode Caenorhabditis elegans. The FOXO family is composed of four members including FOXO1, FOXO3, FOXO4, and FOXO6 (39, 40). FOXOs regulate diverse cell functions, such as cell cycle progression, apoptosis, stress resistance, and metabolism (41, 42). Because disruption of the FOXO1 gene in mice results in embryonic lethality, the role of FOXO1 in the regulation of reproduction is unknown (43). Although disruption of FOXO4 has no overt phenotype, FOXO3 null mice have an age-dependent reduction in fertility caused by defective follicular growth in the ovary similar to premature ovarian failure in women (44). In addition to genetic analyses, mechanistic studies have established a role for FOXOs in reproductive tissues, such as the ovary and uterus (45–52), but the function of FOXOs in the neuroendocrine control of reproduction remains to be elucidated.
Given that insulin has been reported to regulate Lhb gene expression and that FOXOs are downstream effectors of insulin signaling, the purpose of this study was to determine whether FOXOs can regulate Lhb transcription. We demonstrate that FOXO1 is expressed in adult mouse gonadotrope cells in vivo and that insulin signaling can regulate FOXO1 phosphorylation and cellular localization in an immortalized gonadotrope-derived cell line. More importantly, we show that overexpression of FOXO1 in LβT2 cells resulted in suppression of basal and GnRH-induced Lhb synthesis.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit anti-rat LHB (anti-rβLH-IC-3), guinea pig anti-rat LHB (anti-rβLH-IC-2) and guinea pig anti-rat thyroid stimulating hormone β-subunit (TSHB) antibodies were provided by Dr. A. F. Parlow from the NIDDK, National Institutes of Health National Hormone and Pituitary Program (NHPP), Harbor-UCLA Medical Center. Rabbit anti-human FOXO1 (H-128; sc-11350) and rabbit anti-human β-Tubulin (H-235; sc-9104) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit anti-human phospho-FOXO1 (Ser-256) (9461) antibody was purchased from Cell Signaling Technology, Inc. (Beverly, MA).
Immunohistochemistry (IHC)
Adult mouse pituitary tissue sections embedded in paraffin (Zyagen, San Diego, CA) were dewaxed with xylene washes, rehydrated through a series of graded ethanol baths, and washed in H2O. The sections were immersed in 10 mm sodium citrate, pH 6.0, heated in a standard microwave twice for 5 min, and allowed to cool for 20 min at room temperature. After washing in water, sections were incubated in 0.3% hydrogen peroxide for 10 min to quench endogenous peroxidase and then washed in 1× phosphate buffered saline (PBS). The sections were blocked in 5% goat serum (Vector Laboratories, Burlingame, CA), 0.3% Triton X-100, PBS for 60 min at room temperature. The primary antibodies used were rabbit anti-rat LHB (1:1000 dilution in 5% goat serum, 0.3% Triton X-100, PBS) and rabbit anti-human FOXO1 (1:100). Biotinylated goat anti-rabbit or anti-rat IgG (Vector Laboratories, 1:300) were used as secondary antibodies, and proteins were visualized using the Vectastain ABC Elite and VIP peroxidase substrate kits (Vector Laboratories). Sections were then counterstained with methyl green (Vector Laboratories), destained in water, immersed in a 0.05% acetic acid, acetone solution, rehydrated with a series of ethanol and xylene baths, and coverslips were mounted using Vectamount (Vector Laboratories). Sections were viewed using a Nikon Eclipse E800 microscope (Nikon, Tokyo, Japan). Digital images were collected using a Nikon Digital Sight camera (DS-Fi1) and analyzed with Version 2.3 NIS-elements image analysis system.
Immunofluorescence (IF)
Adult mouse pituitary tissue sections were rehydrated and processed for antigen retrieval as described above. Dual-fluorescence labeling was performed on the same section with a guinea pig anti-rat LHB (1:200 dilution) or TSHB primary antibody (1:2000 dilution in 10% goat serum, 0.3% Triton X-100) plus a rabbit anti-human FOXO1 primary antibody (1:100) for 48 h at 4 °C. The sections were incubated with goat anti-guinea pig IgG Texas Red (Abcam, ab6906; 1:300) and goat anti-rabbit IgG FITC (Vector Laboratories, FI-1000; 1:300) secondary antibodies for 60 min at room temperature. Coverslips were mounted using Vectashield HardSet mounting media with DAPI (Vector Laboratories). Sections were viewed using a Nikon Eclipse TE 2000-U inverted fluorescence microscope. Digital images were collected using a Sony CoolSNAP EZ cooled CCD camera (Roper Scientific, Trenton, NJ) and analyzed with Version 2.3 NIS-elements image analysis system.
β-Gal Staining of Lhb-Cre x Rosa26 Pituitaries
Heterozygous gonadotrope-specific Lhb-Cre mice (53) were crossed with homozygous Rosa26 mice containing a Cre-responsive β-gal reporter gene (54). Mice were housed in a vivarium animal facility under a 12-h light-dark cycle and provided with food and water ad libitum. All procedures were approved by the University of California, San Diego Institutional Animal Care and Use Committee. Mice were anesthetized with isofluorane and decapitated, and their pituitaries were removed for analysis. Pituitaries were fixed in 4% paraformaldehyde for 1 h, washed with PBS, and incubated overnight at 37 °C with 1 mg/ml X-gal (5-bromo-4-chloro-3-indolyl β-d-galactoside), 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 2 mm MgCl2, PBS. The pituitaries were then washed with PBS, incubated overnight with 70% EtOH at 4 °C, and embedded in paraffin. 7-μm-thick sections were processed for IHC using a rabbit anti-human FOXO1 primary antibody (1:100), as described above.
Tissue Cell Culture
Cell culture was performed using αT1-1, αT3-1, and LβT2 cell lines (55, 56). The αT1-1 cells represent a precursor to the gonadotrope/thyrotrope lineages as they express pituitary glycoprotein hormone α subunit (CGA), whereas the αT3-1 cells represent a precursor to the gonadotrope lineage because they express CGA, the GnRH receptor, and SF1 but not LHB and FSHB. The LβT2 cell line has many characteristics of a mature, differentiated gonadotrope (57, 58). CV-1 cells, a monkey kidney cell line that does not express detectable levels of SF1 or PTX1 (59, 60), were also used. The cells were maintained in 10-cm plates in DMEM (Dulbecco's modification of Eagle's medium) from Mediatech Inc., (Herndon, VA) with 10% fetal bovine serum (Omega Scientific, Inc., Tarzana, CA) and penicillin/streptomycin antibiotics (Invitrogen) at 37 °C and 5% CO2. αT1-1 cells were maintained on 10-cm plates treated with Matrigel (1:30 dilution). 1× trypsin-EDTA (Sigma) was used for cell dissociation.
Western Blot Analysis
Before hormone treatment, LβT2 cells were switched to serum-free DMEM containing 0.1% BSA, 5 mg/liter transferrin, and 50 mm sodium selenite. After overnight starvation in serum-free media, the cells were pretreated for 1 h with vehicle (DMSO) or 50 μm LY294002 (EMD Biosciences, San Diego, CA) then treated for 10 min with vehicle (0.1% BSA), 1–100 nm insulin (Sigma), 50 μm LY294002, or 10 nm insulin and 50 μm LY294002. Cells were harvested by incubating in a lysis buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40 (Nonidet P-40), 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, complete protease inhibitor mixture pellet (Roche Applied Science) and phosphatase inhibitor mixture pellet (Roche Applied Science) for 10 min at 4 °C. The protein concentration was determined with Bradford reagent (Bio-Rad), and an equal amount of protein per sample was loaded on a 10% SDS-PAGE gel. Proteins were resolved by electrophoresis and transferred for 2 h at 100 V onto polyvinylidene difluoride (Millipore, Billerica, MA) in 1× Tris-glycine, 20% methanol. Blots were blocked for 1 h in 5% milk, then probed overnight at 4 °C with rabbit anti-human FOXO1 (1:1000 in blocking buffer), rabbit anti-human phospho-FOXO1 (Ser-256) (1:1000), or anti-human β-Tubulin (1:500). Blots were then incubated with an anti-rabbit horseradish peroxidase-linked secondary antibody (Santa Cruz Biotechnology), and bands were visualized using the Western Lightning Plus enhanced chemiluminescence substrate (PerkinElmer Life Sciences).
Immunofluorescence of Cultured Cells
LβT2 cells were plated onto poly l-lysine coverslips (BD Biosciences, 354085). After 24 h the media were changed to serum-free media for overnight serum starvation. The next morning cells were pretreated for 30 min with vehicle (DMSO) or 50 μm LY294002 then treated for 30 min with DMSO, 10 nm insulin, 50 μm LY294002, or 10 nm insulin and 50 μm LY LY294002. Cells were then washed twice with PBS and fixed with 4% paraformaldehyde for 10 min. Cells were washed in PBS twice and permeabilized with Nonidet P-40 solution (PBS containing 0.2% Nonidet P-40, 20% goat serum, 1% BSA) for 1 h at room temperature. Cells were washed twice in PBS and incubated with FOXO1 primary antibody (1:100) in blocking buffer (PBS containing 20% goat serum, 1% BSA) for 48 h at 4 °C. Cells were washed 3 times with PBS for 5 min and incubated with Alexa594-conjugated goat anti-rabbit secondary antibody (Invitrogen, A-11012) (1:400) in blocking buffer for 1 h at room temperature. Cells were washed 3 times with PBS for 5 min, coverslips were mounted, and the cells were imaged as described above for IF.
Plasmid Constructs
The pcDNA3 FLAG human FOXO1, pcDNA3 FLAG FOXO1-CA, pALTER FOXO1-CA, and pALTER FOXO1-CA + mut DBD (W209G/H215L) expression vectors and 3xIRS-luciferase (luc) were previously described (61). We obtained the pcDNA3 FOXO1-ΔDBD (Δ208–220) from Dr. William Sellers (Addgene plasmid 10694). The pcDNA3 mouse PTX1 expression vector was also described previously (62). The pCMV rat EGR1 and mouse SF1 were kindly provided by Dr. Jacques Drouin and Dr. Bon-chu Chung, respectively. The −1800 rat Lhb-luc reporter in pGL3 was provided by Dr. Mark Lawson. Construction of 5′ truncations of the −1800 Lhb-luc reporter plasmid (−500, −300, −150, −87) was previously described (63). The −1068/+9 human LHB-luc reporter in pA3 was generously provided by Dr. Daniel Bernard.
Transient transfection-LβT2 cells were seeded at 3 × 105 cells/well on 12-well plates and transfected 18 h later using FuGENE 6 reagent (Roche Applied Science) following the manufacturer's instructions. CV-1 cells were seeded at 1.5 × 105 cells/well. For all experiments, the cells were transfected with 400 ng of the indicated luciferase reporter plasmid and 200 ng of a β-gal reporter plasmid driven by the herpes virus thymidine kinase promoter to control for transfection efficiency. LβT2 cells were switched to serum-free DMEM containing 0.1% BSA, 5 mg/liter transferrin, and 50 mm sodium selenite 6 h after transfection. After overnight starvation in serum-free media, the cells were treated with vehicle (0.1% BSA) or 10 nm GnRH (Sigma) for 6 h.
Luciferase and β-Galactosidase Assays
To harvest the cells, they were washed with 1× PBS and lysed with 0.1 m potassium phosphate buffer, pH 7.8, containing 0.2% Triton X-100. Lysed cells were assayed for luc activity using a buffer containing 100 mm Tris-HCl pH 7.8, 15 mm MgSO4, 10 mm ATP, and 65 μm luciferin. β-Gal activity was assayed using the Tropix Galacto-light assay (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Both assays were measured using a Veritas Microplate Luminometer (Promega, Madison, WI).
Adenoviral Infection
Adenoviral vectors containing cDNA of green fluorescent protein (Ad-GFP) and constitutively active FOXO1 (T24A/S256A/S319A) (Ad-FOXO1-CA) were generously provided by Dr. Domenico Accili. LβT2 cells were seeded at 2 × 106 cells/well on 6-well plates. The next morning cells were transduced with a multiplicity of infection of 200 of Ad-GFP or Ad-FOXO1-CA for 6 h, then switched to serum-free DMEM. 24 h after adenoviral infection, cells were treated with vehicle (0.1% BSA) or 10 μm GnRH for 6 h. GFP and FOXO1-CA protein expression was verified by Western blot analysis.
Quantitative RT-PCR
Total RNA was extracted from LβT2 cells with TRIzol reagent (Invitrogen) following the manufacturer's protocol. Contaminating DNA was removed with DNA-free reagent (Invitrogen). 2 μg of RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. Quantitative real-time PCR was performed in an iQ5 iCycler using iQ SYBR Green Supermix (Bio-Rad) and the primers Lhb forward, (CTGTCAACGCAACTCTGG), Lhb reverse (CAGGAGGCAAAGCAGC), Gapdh forward (TGCACCACCAACTGCTTAG), and Gapdh reverse (GGATGCAGGGATGATGTTC) under the following conditions: 95 °C for 5 min followed by 40 cycles at 95 °C for 45 s, 54 °C for 45 s, and 72 °C for 45 s. Each sample was assayed in triplicate, and the experiment was repeated four times. Standard curves with dilutions of a plasmid containing Lhb or Gapdh cDNA were generated with the samples in each run. In each experiment, the amount of Lhb and Gapdh was calculated by comparing the threshold cycle obtained for each sample with the standard curve generated in the same run. Replicates were averaged and divided by the mean value of Gapdh in the same sample. After each run, a melting curve analysis was performed to confirm that a single amplicon was generated in each reaction.
Electrophoretic Mobility Shift Assay (EMSA)
FLAG-FOXO1-CA was transcribed and translated using a TnT Coupled Reticulocyte System (Promega). The oligonucleotides were end-labeled with T4 polynucleotide kinase and [γ-32P]ATP. 4 μl of TnT lysate was incubated with 1 fmol of 32P-labeled oligo at 4 °C for 30 min in a DNA binding buffer (10 mm Hepes, pH 7.8, 50 mm KCl, 5 mm MgCl2, 0.1% Nonidet P-40, 1 mm dithiothreitol, 2 μg of poly(dI-dC), and 10% glycerol). After 30 min, the DNA binding reactions were run on a 5% polyacrylamide gel (30:1 acrylamide:bisacrylamide) containing 2.5% glycerol in a 0.25× Tris borate-EDTA buffer. Mouse anti-FLAG M2 antibody (Sigma) was used to supershift FLAG-FOXO1-CA; mouse IgG was used as a control for nonspecific binding. 100× cold oligo was used as a competitor. The following oligonucleotides were used for EMSA: −150/−115 (5′-CTGGAGCTGGTCCCTGGCTTTTCTGACCTTGTCTG-3′); −122/−87 (5′-TTGTCTGTCTCGCCCCCAAAGAGATTAGTGTCTAG-3′); −95/−60, (5′-GTGTCTAGGTTACCCAAGCCTGTAGCCTCTGCTTA-3′); −65/−30 (5′-GCTTAGTGGCCTTGCCACCCCCACAACCCGCAGGT-3′), the consensus forkhead binding element (FBE) (64) (5′-CTAGATGGTAAACAACTGTGACTAGTAGAACACGG-3′), and the mutated consensus FBE (5′-CTAGATGGTGGGCAACTGTGACTAGTAGAACACGG-3′).
Statistical Analyses
Transient transfections were performed in triplicate, and each experiment was repeated at least three times. The data were normalized for transfection efficiency by expressing luc activity relative to β-gal and relative to the empty reporter plasmid to control for hormone effects on the vector DNA. The data were analyzed by Student's t test for independent samples or one-way analysis of variance (ANOVA) followed by post-hoc comparisons with the Tukey-Kramer honestly significant difference test using the statistical package JMP 9.0 (SAS, Cary, NC). Significant differences were designated as p < 0.05.
RESULTS
FOXO1 Is Expressed in Gonadotrope Cells
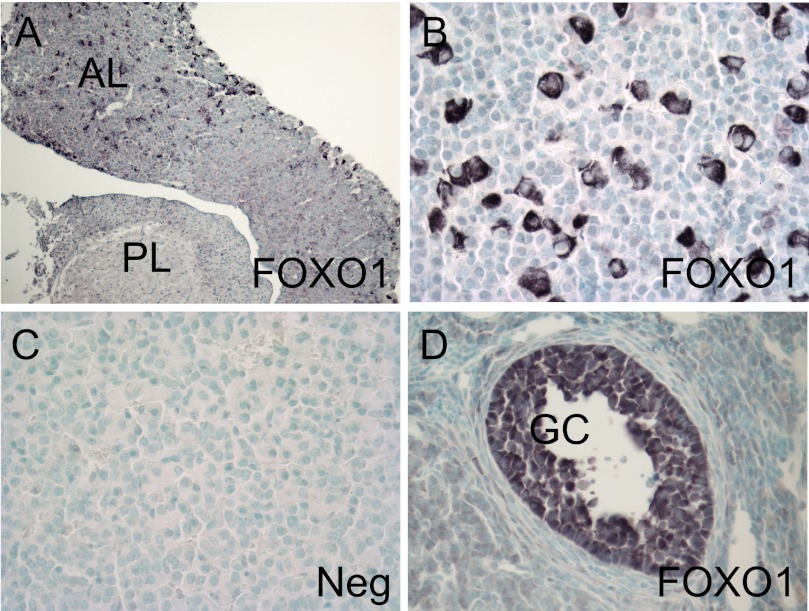
Using IHC with a primary FOXO1 antibody, we demonstrated that FOXO1 is expressed in a discrete population of cells within the anterior lobe (AL) of the pituitary of adult mice (Fig. 1, A and B). FOXO1 was not expressed in the posterior lobe (PL). The negative control, lacking primary antibody, showed no staining (Fig. 1C). We also performed IHC on sections of mouse ovaries using the FOXO1 antibody as a positive control (Fig. 1D). FOXO1 expression was specifically detected in ovarian granulosa cells (GC), as previously reported (50).
FIGURE 1.
FOXO1 transcription factor is expressed in adult mouse anterior pituitary. IHC was performed with a FOXO1 primary antibody on paraffin-embedded sections of adult mouse pituitary with the anterior lobe (AL) and posterior lobe (PL) as indicated (A and B) or ovary (D). FOXO1 protein was visualized with peroxidase. The negative control (Neg) lacked primary antibody (C). The positive control shows FOXO1 expression in granulosa cells (GC) in the ovary (D). Representative images were obtained using a Nikon Eclipse E800 microscope at 10× (A) or 40× (B–D) magnification.
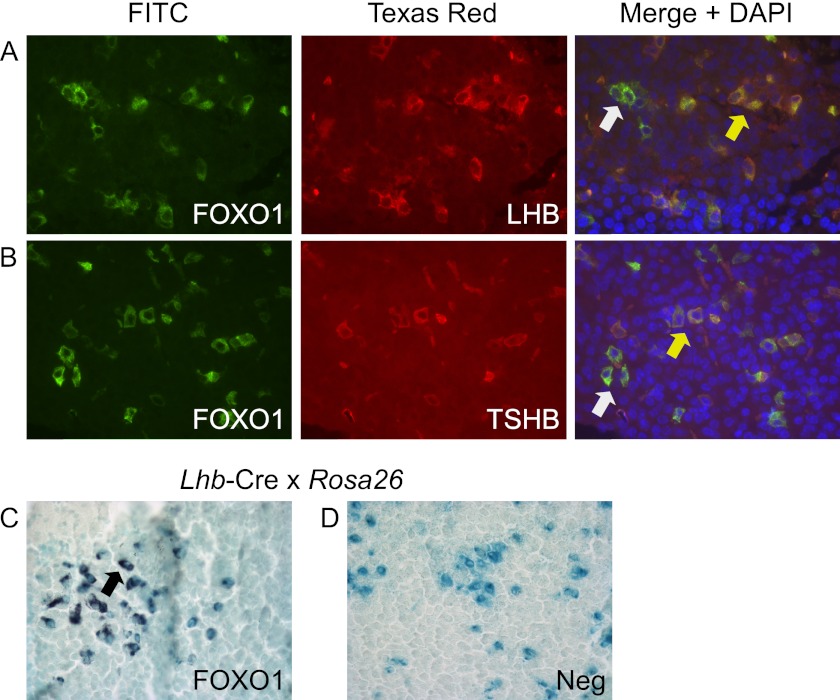
We then ascertained whether FOXO1 co-localized with LHB-expressing gonadotrope cells. First, we showed that FOXO1 co-localized with LHB- and TSHB-expressing cells in adult mice using dual label IF. The yellow arrow indicates co-localization between FOXO1 and LHB (Fig. 2A, merge panel) or FOXO1 and TSHB (Fig. 2B, merge panel), whereas the white arrow indicates cells that express FOXO1 and do not co-localize. Second, we demonstrated that FOXO1 co-localized with β-gal produced in LHB-expressing gonadotrope cells in heterozygous Lhb-Cre mice (53) crossed with Rosa26 mice (54) as indicated by the black arrow (Fig. 2C). The negative control, lacking primary antibody, did not have any peroxidase staining (Fig. 2D).
FIGURE 2.
FOXO1 is expressed in adult mouse gonadotrope cells. A and B, dual label IF was performed on paraffin-embedded sections of adult mouse pituitary with FOXO1 plus LHB or TSHB primary antibodies. Proteins were visualized with secondary antibodies coupled to FITC or Texas Red. Representative images were obtained using a Nikon Eclipse TE 2000-U inverted fluorescence microscope at 40× magnification. Yellow arrows indicate colocalization, whereas white arrows show lack of colocalization. C and D, β-gal staining was performed on paraffin-embedded sections of adult mouse pituitary from Lhb-Cre x Rosa26 mice. IHC was then performed with a FOXO1 primary antibody (C). A black arrow indicates colocalization. The negative control (Neg) lacked primary antibody (D). Representative images were obtained using a Nikon Eclipse E800 microscope at 40× magnification.
FOXO1 Phosphorylation and Cellular Localization Is Regulated by Insulin Signaling in Immortalized Gonadotrope Cells
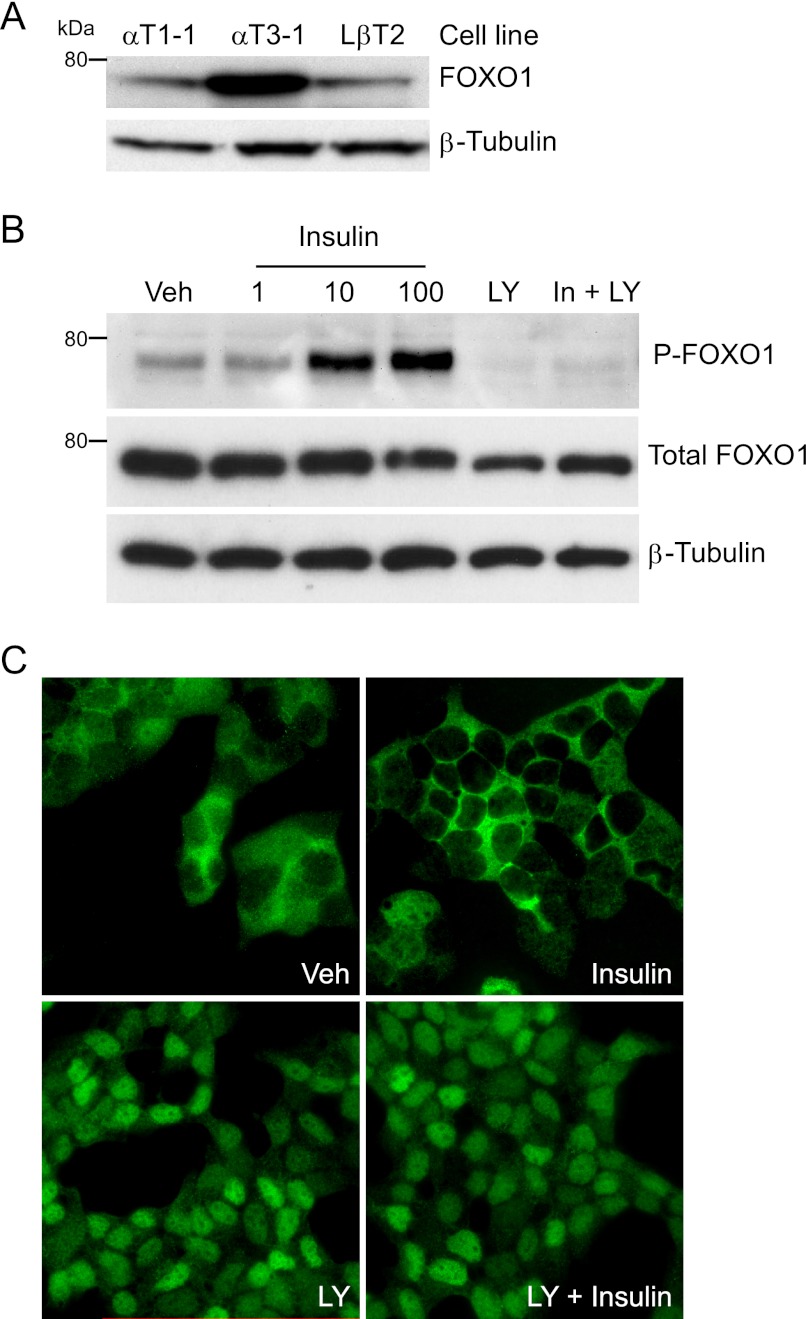
After demonstrating in vivo FOXO1 expression in pituitary gonadotrope cells, we determined whether FOXO1 is expressed in immortalized gonadotrope cells. We performed Western blot analysis of whole cell extracts from αT1-1, αT3-1, and LβT2 cells with a FOXO1 antibody. As described under “Experimental Procedures,” these immortalized cell lines represent a precursor to the gonadotrope/thyrotrope lineages, an immature gonadotrope cell and a mature gonadotrope cell, respectively (55, 56). FOXO1 was expressed in all three cell lines (Fig. 3A). Western blot analysis was also used to determine whether insulin signaling results in FOXO1 phosphorylation in LβT2 cells. Fig. 3B demonstrates that 10 min of 10 and 100 nm insulin treatment resulted in increased phosphorylation of FOXO1 serine 256 compared with vehicle control. In contrast, total FOXO1 or β-tubulin protein levels were unaffected by insulin treatment. We also demonstrated that FOXO1 serine 256 phosphorylation induced by insulin was dependent on PI3K signaling using LY294002, an inhibitor of PI3K activity (Fig. 3B). We then determined whether insulin treatment resulted in a change in endogenous FOXO1 cellular localization in LβT2 cells using IF. Vehicle treatment of LβT2 cells after an overnight incubation in serum-free media resulted in FOXO1 localization distributed between the nucleus and cytoplasm (Fig. 3C). Insulin treatment resulted in predominantly cytoplasmic FOXO1 localization, whereas treatment with LY294002 led to nuclear localization. Co-treatment of LY294002 and insulin prevented FOXO1 export to the cytoplasm and resulted in nuclear localization.
FIGURE 3.
FOXO1 phosphorylation and cellular localization is regulated by insulin signaling in immortalized gonadotrope cells. A, Western blot analysis was performed on whole cell extracts from the indicated cell lines using a FOXO1 primary antibody and a horseradish peroxidase-linked secondary antibody. β-Tubulin was used as a loading control. B, LβT2 cells were incubated overnight in serum-free media, pretreated for 1 h with vehicle (Veh) or 50 μm LY294002 (LY), and then treated for 10 min with vehicle, 1–100 nm insulin, 50 μm LY, or 10 nm insulin and 50 μm LY, as indicated. Western blot analysis was performed using FOXO1 serine 256 (P-FOXO1), total FOXO1, and β-Tubulin antibodies. C, LβT2 cells were incubated overnight in serum-free media, pretreated for 30 min with vehicle (Veh), or 50 μm LY294002 (LY), and then treated for 30 min with vehicle, 10 nm insulin, 50 μm LY294002, or 10 nm insulin and 50 μm LY294002, as indicated. IF was performed with a FOXO1 primary antibody and an Alexa594-conjugated goat anti-rabbit secondary antibody. Representative images were obtained using a Nikon Eclipse TE 2000-U inverted fluorescence microscope at 60× magnification.
FOXO1 Overexpression Suppresses Basal and GnRH-induced Lhb Transcription
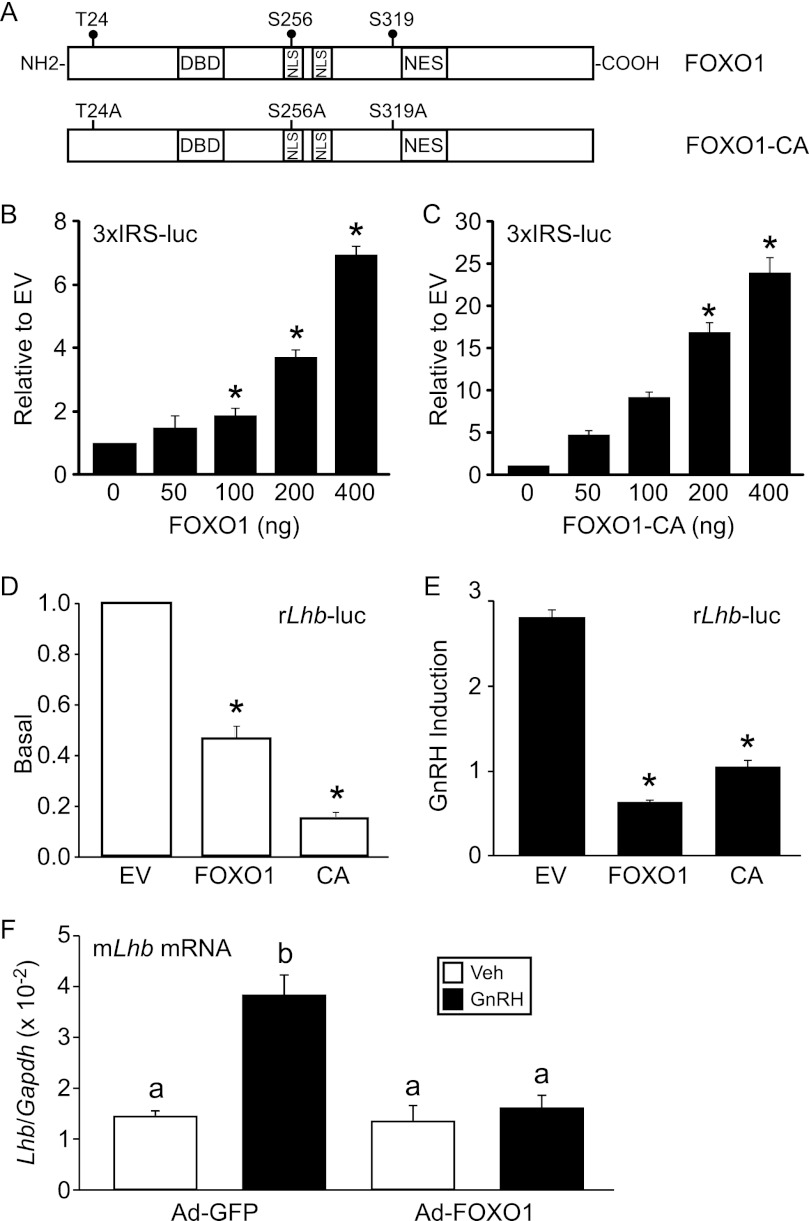
Overexpression of FOXO1 and constitutively active FOXO1-CA (T24A/S256A/S319A) (Fig. 4A) in LβT2 cells enhanced transcription of a 3xIRS-luc plasmid (61) containing three copies of a FBE from the IGFBP-1 gene linked to a luc reporter gene (Fig. 4, B and C). In these experiments, transfection of increasing concentrations of FOXO1 and FOXO1-CA resulted in higher levels of reporter gene expression, indicating that FOXO1 levels did not reach saturation and that the effects were not likely due to supraphysiological levels of FOXO1 in the cells. Intriguingly, when 200 ng of FOXO1 or FOXO1-CA was transiently transfected with the −1800 rat Lhb promoter linked to a luc reporter gene (rLhb-luc), we observed repression of basal and GnRH-induced Lhb synthesis (Fig. 4, D and E).
FIGURE 4.
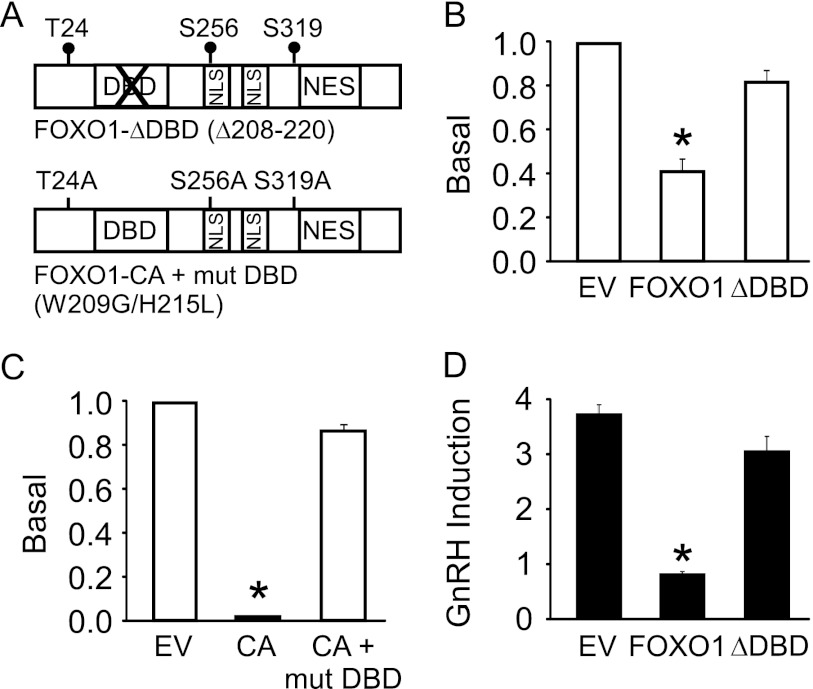
FOXO1 suppresses basal and GnRH induction of Lhb synthesis in LβT2 cells. A, shown is a schematic illustrating wild-type FOXO1 and constitutively active FOXO1-CA (T24A/S256A/S319A). NES, nuclear export signal; NLS, nuclear localization signal. B and C, the 3xIRS-luc was transiently transfected into LβT2 cells along with increasing amounts of FOXO1 or FOXO1-CA expression vectors as indicated. Total amount of expression vector was balanced using the empty pcDNA3 vector. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented relative to empty vector (EV). * indicates that the induction by FOXO1 or FOXO1-CA is significantly different from the empty vector using one-way ANOVA followed by Tukey's post-hoc test. D and E, the −1800 rat Lhb-luc reporter was transfected into LβT2 cells along with 200 ng of pcDNA3 empty vector, FOXO1, or FOXO1-CA (CA) as indicated. After overnight starvation in serum-free media, cells were treated for 6 h with 0.1% BSA vehicle (Veh) or 10 nm GnRH. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as basal transcription relative to EV (D) or-fold GnRH induction relative to the vehicle control (E). * indicates that Lhb-luc transcription is significantly repressed by FOXO1 or FOXO1-CA compared with EV using one-way ANOVA followed by Tukey's post-hoc test. F, Ad-expressing GFP or FOXO1-CA was transduced into LβT2 cells as indicated. After overnight starvation in serum-free media, cells were treated for 6 h with 0.1% BSA vehicle (Veh) or 10 nm GnRH. The results represent the mean ± S.E. of four experiments performed in triplicate and are presented as Lhb mRNA relative to Gapdh. The letter b indicates that the GnRH induction in the presence of Ad-FOXO1-CA is significantly different using one-way ANOVA followed by Tukey's post-hoc test.
To ascertain whether FOXO1 can suppress endogenous Lhb transcription, we transduced LβT2 cells with adenovirus (Ad) expressing FOXO1-CA or GFP and measured Lhb mRNA levels using quantitative RT-PCR. Although basal transcription of Lhb was not significantly altered, we demonstrated that GnRH-induced Lhb gene expression was significantly decreased in cells infected with Ad-FOXO1-CA versus Ad-GFP (Fig. 4F). These results indicate that FOXO1 can suppress Lhb synthesis in the context of the native chromatin.
The DNA Binding Domain of FOXO1 Is Required for Suppression of Lhb Synthesis
To determine whether FOXO1 repression occurs at the level of the Lhb promoter, we tested whether DNA binding-deficient FOXO1 could still elicit a suppressive response using a deletion in the FOXO1-DNA binding domain (DBD) (Δ208–220) and a FOXO1-CA with a mutated DBD (Fig. 5A). We demonstrated that although overexpression of FOXO1 and FOXO1-CA reduced Lhb gene expression, transcription was not suppressed in the presence of the mutated transcription factors. FOXO1-ΔDBD and FOXO1-CA + mut DBD did not alter basal Lhb synthesis (Fig. 5, B and C), whereas the FOXO1-ΔDBD mutant did not suppress GnRH-induced Lhb gene expression (Fig. 5D). These results indicate that the DBD of FOXO1 is necessary for FOXO1 repression of Lhb basal transcription and GnRH induction on the Lhb promoter and suggest that FOXO1 exerts an effect by either binding to the Lhb promoter or indirectly through FOXO1 DBD interaction with other transcription factors/cofactors.
FIGURE 5.
DNA binding domain of FOXO1 is required to suppress basal and GnRH-induced Lhb gene expression. A, shown is a schematic illustrating FOXO1-ΔDBD (Δ208–220) and FOXO1-CA + mut DBD (W209G/H215L). NES, nuclear export signal; NLS, nuclear localization signal. B–D, the −1800 rat Lhb-luc reporter was transfected into LβT2 cells along with 200 ng of pcDNA3 empty vector (EV), FOXO1, or FOXO1-CA (CA) as indicated. After overnight starvation in serum-free media, cells were treated for 6 h with 0.1% BSA vehicle (Veh) or 10 nm GnRH. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as basal transcription relative to empty vector (B and C) or -fold GnRH induction relative to the vehicle control (D). * indicates that Lhb-luc transcription is significantly repressed by FOXO1 or FOXO1-CA compared with empty vector using one-way ANOVA followed by Tukey's post-hoc test.
FOXO1 Suppression Maps to the Proximal Promoter of Lhb
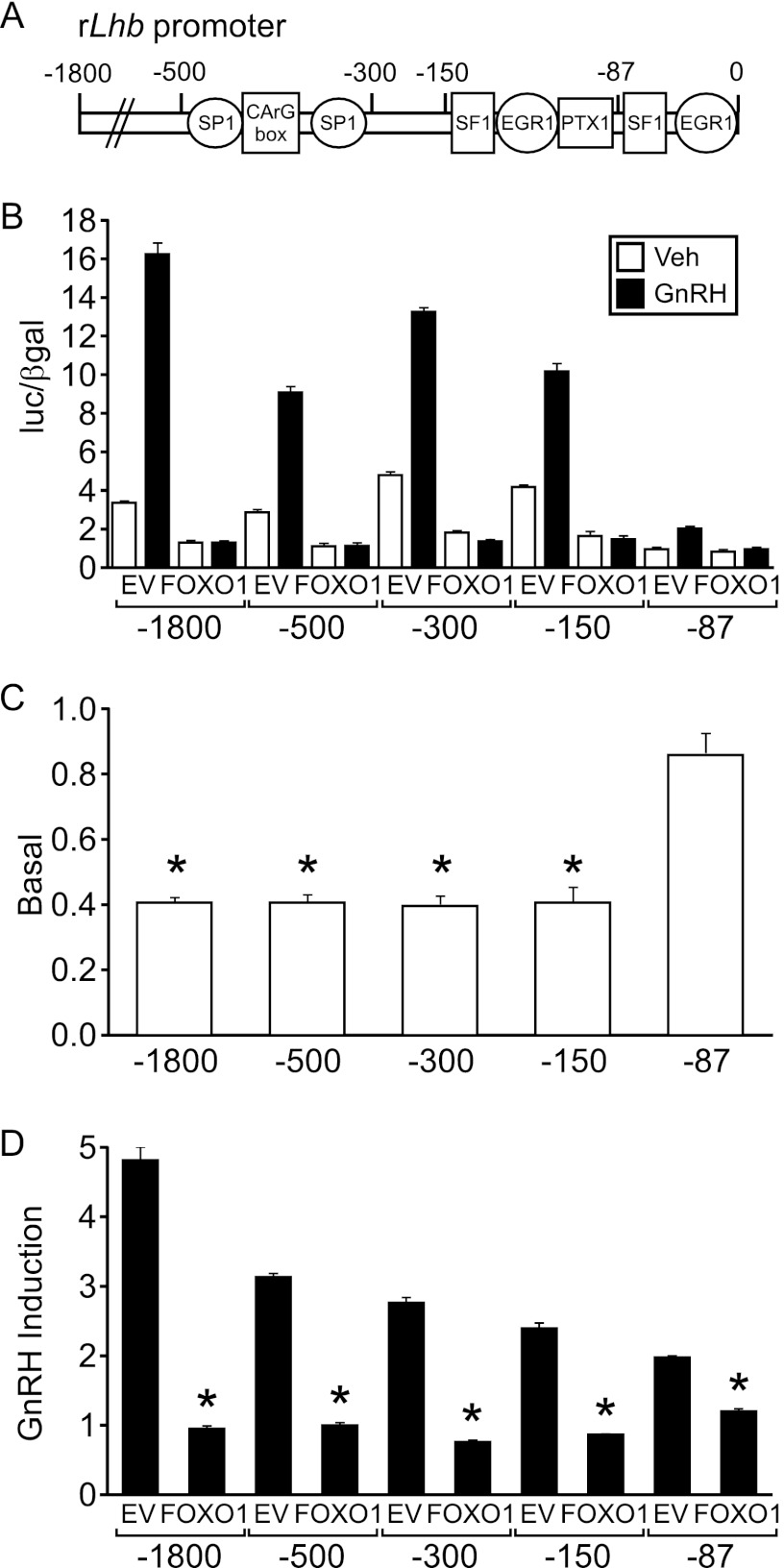
We determined which regions of the Lhb promoter were required for FOXO1 suppression using 5′ truncation analysis of the promoter. Basal transcription and GnRH induction were measured using −1800, −500, −300, −150, and −87 Lhb-luc reporter plasmids (Fig. 6A). The suppressive effect of FOXO1 mapped to the proximal region on the Lhb promoter (Fig. 6B), which contains binding elements for SF1, PTX1, and EGR1 (11). In particular, FOXO1 suppression of basal Lhb gene expression mapped to a region between −150 and −87 of the promoter as repression was lost with the −87 Lhb-luc reporter plasmid (Fig. 6C). In contrast, FOXO1 suppression of GnRH induction was reduced but not abolished with the −87 Lhb-luc reporter (Fig. 6D), suggesting that elements present in the 87 base pairs upstream of the transcription start site are important for FOXO1 repression of GnRH-induced Lhb transcription.
FIGURE 6.
FOXO1 suppression maps to the proximal Lhb promoter. A, a schematic illustrates the location of binding elements for SP1, SF1, EGR1, and PTX1 transcription factors. B–D, the −1800, −500, −300, −150, and −87 Lhb-luc reporters were transiently transfected into LβT2 cells along with pcDNA3 empty vector (EV) or FOXO1 as indicated. After overnight starvation in serum-free media, cells were treated for 6 h with 0.1% BSA vehicle (Veh) or 10 nm GnRH. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as luc/β-gal (B), basal transcription relative to empty vector (C), or -fold GnRH induction relative to the vehicle control (D). * indicates that Lhb-luc transcription is significantly repressed by FOXO1 compared with EV using one-way ANOVA followed by Tukey's post-hoc test.
FOXO1 Does Not Bind Directly to the Lhb Promoter
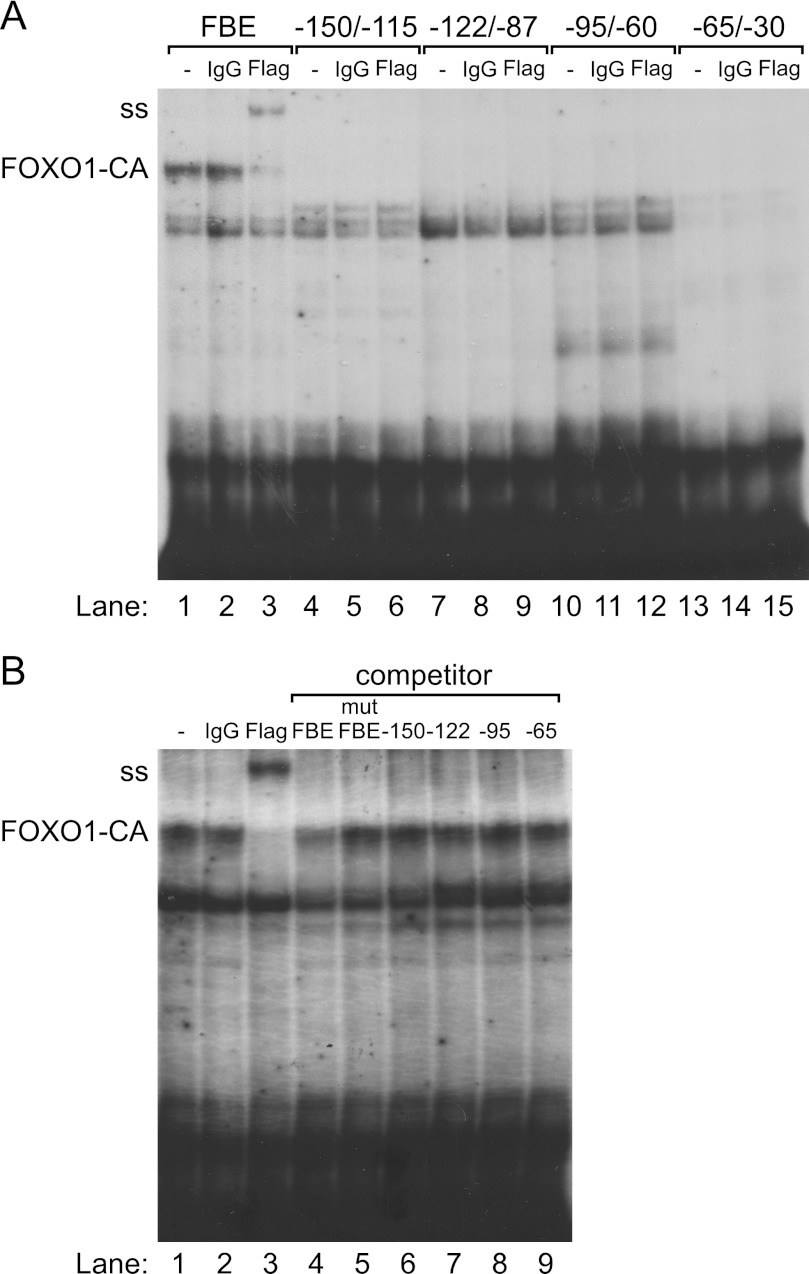
EMSA was performed to assess whether FOXO1 can bind to the proximal Lhb promoter in vitro. Four 35-mer oligonucleotide probes were designed to span the −150/−30 region. FLAG-FOXO1-CA, synthesized with TnT rabbit reticulocyte lysate, bound to an oligonucleotide probe containing a consensus FBE (Fig. 7, A and B, lanes 1) but not to probes encompassing the −150/−30 region of the Lhb promoter (Fig. 7A, lanes 4, 7, 10, and 13). TnT FLAG-FOXO1 also bound to a consensus FBE (data not shown). To identify which complex contained the FLAG-FOXO1-CA bound to the FBE, we supershifted the complex with an anti-FLAG antibody (Fig. 7, A and B, lanes 3) but not with IgG (Fig. 7, A and B, lanes 2). This complex was not present when rabbit reticulocyte lysate containing the pcDNA3 empty vector was used (data not shown). This complex also showed evidence of self-competition (Fig. 7B, lane 4) but did not compete with a mutated consensus FBE (Fig. 7B, lane 5). Incubation with oligos encompassing the −150/−30 region of the Lhb promoter did not result in competition (Fig. 7B, lanes 6–9), indicating that FOXO1 does not bind directly to the proximal Lhb promoter.
FIGURE 7.
FOXO1 does not bind to the proximal Lhb promoter. TnT FLAG-FOXO1-CA was incubated with a consensus FBE, −150/−115, −122/−87, −95/−60, or −65/30 Lhb probes and tested for complex formation in EMSA. A, FOXO1-CA-DNA complex on the FBE is shown in lane 1, whereas anti-FLAG supershift is shown in lane 3, and IgG control is shown in lane 2. The FOXO1-CA-DNA complex and antibody supershift (ss) are indicated on the left of the gel. B, self-competition with excess cold FBE is shown in lane 4, and lack of competition with mutant FBE, −150/−115, −122/−87, −95/−60, or −65/−30 Lhb oligos is shown in lanes 5–9.
FOXO1 Suppression Involves SF1, PTX1, and EGR1
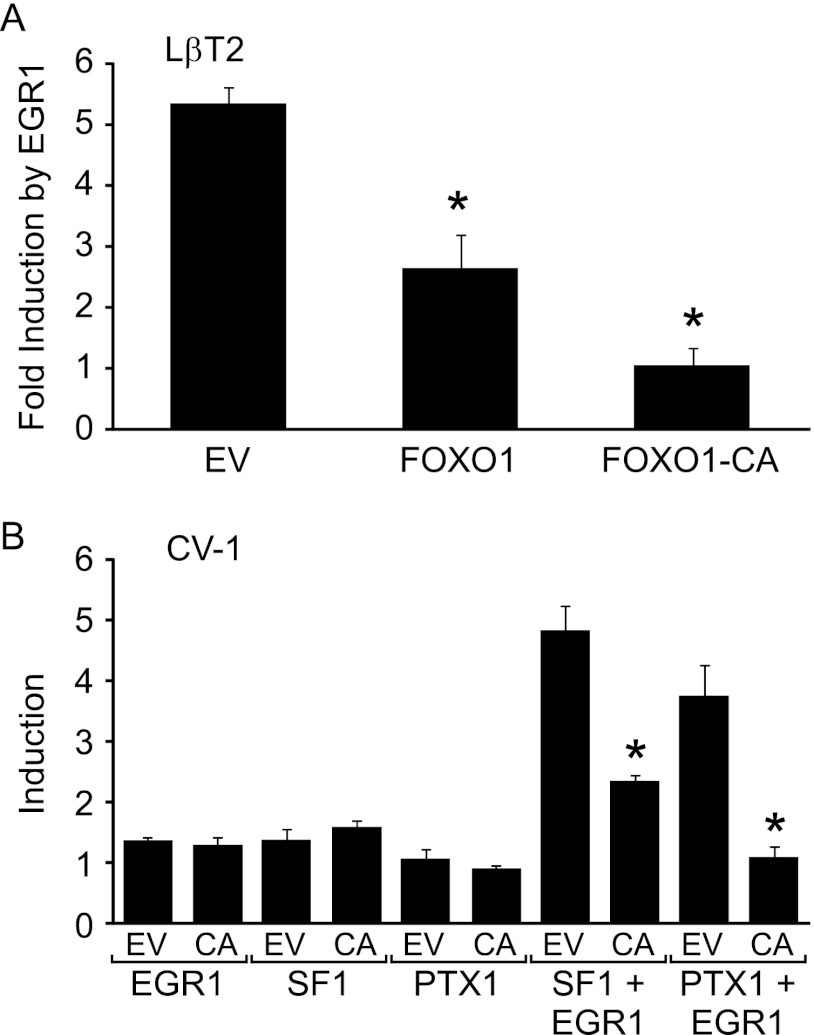
We examined the role of EGR1 in FOXO1 repression of GnRH-induced Lhb synthesis by determining whether Lhb transcription due to EGR1 overexpression in LβT2 cells was reduced by FOXO1. Overexpression of EGR1 in LβT2 cells resulted in a 5.3-fold induction of the Lhb promoter that was suppressed 51 or 81% by overexpression of FOXO1 or FOXO1-CA, respectively (Fig. 8A). Because LβT2 cells contain abundant amounts of SF1 and PTX1, we also performed transient transfection experiments in CV-1 cells lacking these transcription factors (59, 60). Overexpression of SF1, PTX1, or EGR1 by themselves did not result in a significant induction of Lhb gene expression (Fig. 8B). However, coexpression of EGR1 with either SF1 or PTX1 increased Lhb synthesis in CV-1 cells by 4.8- and 3.7-fold, respectively, whereas FOXO1-CA reduced this induction by 52 and 71% (Fig. 8B).
FIGURE 8.
FOXO1 suppresses induction of Lhb synthesis by SF1, PTX1, and EGR1. A, the −1800 rat Lhb-luc reporter was transfected into LβT2 cells along with 50 ng EGR1 as well as 200 ng of pcDNA3 empty vector (EV), FOXO1, or FOXO1-CA (CA) as indicated. Cells were harvested 36 h after transfection. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as -fold induction by EGR1 relative to the EV control. * indicates that Lhb-luc transcription is significantly repressed by FOXO1 or FOXO1-CA compared with EV using one-way ANOVA followed by Tukey's post-hoc test. B, the −1800 Lhb-luc reporter was transfected into CV-1 cells along with 100 ng of EGR1, 100 ng of SF-1, and 50 ng of PTX1 as well as 200 ng of pcDNA3 EV or FOXO1-CA (CA) as indicated. Total amount of expression vectors was balanced using empty pcDNA3 and pCMV vectors. Cells were harvested 36 h after transfection. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as -fold induction relative to the empty vector control. * indicates that Lhb-luc transcription is significantly repressed by FOXO1-CA compared with empty vector using Student's t test.
FOXO1 Suppresses Human LHB Synthesis
Given that the SF1, PTX1, and EGR1 elements in the proximal Lhb promoter are highly conserved between mice and humans (20), we hypothesized that FOXO1 would have a similar repressive effect on the human LHB promoter as the murine promoter. When 200 ng of FOXO1 was transiently transfected with the −1068/+9 human LHB promoter linked to a luciferase reporter gene (hLHB-luc), we observed repression of basal and GnRH-induced Lhb synthesis (Fig. 9, A and B). Basal Lhb transcription was reduced 79% by FOXO1, whereas GnRH-induced LHB gene expression was reduced by 87%.
FIGURE 9.
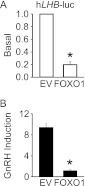
FOXO1 suppresses basal and GnRH induction of the human LHB promoter. A and B, the −1068/+9 human LHB-luc reporter was transfected into LβT2 cells along with pcDNA3 empty vector (EV) or FOXO1 as indicated. After overnight starvation in serum-free media, cells were treated for 6 h with 0.1% BSA vehicle (Veh) or 10 nm GnRH. The results represent the mean ± S.E. of at least three experiments performed in triplicate and are presented as basal transcription relative to empty vector (A) or -fold GnRH induction relative to the vehicle control (B). * indicates that LHB-luc transcription is significantly repressed by FOXO1 compared with empty vector using Student's t test.
DISCUSSION
Although FOXO1 has been shown to be expressed in reproductive tissues such as the ovary and uterus (65), this is the first report that FOXO1 is expressed in adult murine anterior pituitary. Co-localization of FOXO1 with LHB- and TSHB-expressing cells demonstrated that FOXO1 expression occurred in gonadotropes and thyrotropes. Co-localization of FOXO1 and CGA (present only in gonadotrope and thyrotrope cells within the pituitary) provides supporting evidence that FOXO1 expression is restricted to these two pituitary cell types.3 FOXO1 is considered to be a relatively ubiquitously expressed protein (66, 67), although there are many instances where FOXO1 expression is restricted to a specific cell type. For example, FOXO1 is highly expressed in granulosa cells of proliferating follicles but not in adjacent cells within the ovary (48, 50). FOXO1 is also expressed at high levels in POMC and NPY/AgRP neurons in the arcuate nucleus of the hypothalamus (68). Our results indicate that future studies investigating the function of FOXO1 in thyrotrope cells as well as the pituitary cell type-specific regulation of FOXO1 gene expression may be informative.
After demonstrating that FOXO1 is expressed in gonadotrope cells in vivo, we also showed that the FOXO1 protein can be detected in immortalized gonadotrope-derived cell lines, in agreement with a previous study (26). We then investigated whether FOXO1 is a target of insulin signaling in gonadotropes, as previously reported for IRS1/2 and AKT (24, 27). Our results showed that insulin treatment of LβT2 cells increased FOXO1 serine 256 phosphorylation and cytoplasmic localization. These data are in agreement with a previous study that showed IGF-1 signaling increased FOXO1 phosphorylation in αT3-1 cells (26). We also used the PI3K inhibitor LY294002 to demonstrate that regulation of FOXO1 phosphorylation and cellular localization by insulin signaling in LβT2 cells was dependent on PI3K, which is consistent with the insulin effect on FOXO1 in other tissues (33).
Our studies in LβT2 cells also demonstrated that FOXO1 may regulate fertility through modulation of Lhb transcription, the rate-limiting step in the production of the mature hormone (8, 9). Overexpression of wild-type and constitutively active FOXO1 reduced basal and GnRH-induced Lhb-luc expression, whereas overexpression of FOXO1-CA decreased endogenous GnRH-induced Lhb mRNA levels. Our results also indicate that FOXO1 regulation of LH levels may be conserved among mammals as FOXO1 suppressed transcription of both the murine and human LHB promoters. The modulation of Lhb by FOXO1 in LβT2 cells does not occur because of a general repressive mechanism as the 3xIRS-luc reporter plasmid is not repressed by FOXO1. Rather, FOXO1 suppression of Lhb appears to be due to specific elements in the Lhb promoter lacking in the 3xIRS. Although FOXO1 is often portrayed as an activator of transcription such as on the IGFBP-1, phosphoenolpyruvate carboxykinase, and glucose-6-phosphatase catalytic subunit promoters (69, 70), it can also act as a repressor. For example, when FOXO1, -3, and -4 were knocked out in liver endothelial cells and the mRNA profiles were compared with wild-type cells using microarray analysis, transcription of many genes increased, indicating that FOXOs act as repressors (71). Many of these genes were confirmed as FOXO target genes using quantitative RT-PCR and chromatin immunoprecipitation. FOXO1 has also been reported to repress transcription of the peroxisome proliferator-activated receptor γ (72).
To test whether FOXO1 binding to the Lhb promoter is necessary for FOXO1 suppression, we used FOXO1 with a mutated or deleted DBD and showed that repression of Lhb required the FOXO1 DBD. We also tested whether FOXO1 binds directly to the Lhb promoter. Using EMSA, we showed that in vitro transcribed and translated FOXO1 and FOXO1-CA do not bind to the proximal −150/−30 Lhb promoter, although they can bind to an oligo containing a consensus FBE. This is in agreement with in silico analysis of the proximal Lhb promoter, which did not show any putative FBEs. Together, our results indicate that FOXO1 does not bind directly to the Lhb promoter but rather that FOXO1 modulates Lhb gene expression indirectly through interactions between the FOXO1 DBD and other transcription factors/cofactors.
To further our understanding of FOXO1 regulation of Lhb synthesis, we ascertained which regions of the Lhb promoter were necessary for FOXO1 suppression. The SP1/CArG box is apparently not required to elicit FOXO1 repression of basal and GnRH-induced Lhb gene expression because the suppression maps to the proximal promoter, which lacks these elements. That FOXO1 suppression of basal Lhb transcription was lost with the −87 5′ truncation suggests FOXO1 requires elements located between −150/−87 such as the 5′ SF1 or PTX1 binding elements. In contrast, the FOXO1 suppression of GnRH-induced Lhb synthesis was reduced with the −87 5′ truncation but not abolished, suggesting that FOXO1 affects GnRH induction through elements located in the region downstream of −87 as well as between −150/−87. These regions include SF1, PTX1, and EGR1 binding elements.
In addition to mapping the FOXO1 repression to the proximal Lhb promoter, we also determined whether FOXO1 can elicit a repressive effect though SF1, PTX, and/or EGR1. Our studies demonstrated that FOXO1 could suppress Lhb transcription induced by EGR1 overexpression in LβT2 cells. Moreover, FOXO1 suppressed Lhb gene expression induced by SF1 and EGR1 or PTX1 and EGR1 overexpression in CV-1 cells. Because we did not observe induction of Lhb when exogenous SF1 or PTX1 was expressed in the CV-1 cells by themselves, we were unable to determine whether these transcription factors are essential for FOXO1 suppression. However, our results do indicate that FOXO1 suppression of Lhb synthesis can occur in the context of either SF1 or PTX1 combined with EGR1. It is interesting to speculate that this may be due to FOXO1 interactions with SF1, PTX1, and/or EGR1, although it is not known whether such interactions can occur. Alternatively, FOXO1 could interact with cofactors that are recruited to the proximal Lhb promoter. For example, β-catenin has been reported to act as a coactivator on the Lhb promoter through interactions with SF1 (73) and potentially PTX1 (74). Because FOXO1 can interact with β-catenin (75, 76), FOXO1 repression of Lhb may be mediated through FOXO1 interaction with β-catenin.
It is also possible that FOXO1 exerts a repressive effect on Lhb mRNA levels through transcriptional regulation of transcription factors/cofactors necessary for Lhb gene expression. For instance, FOXO1 has been reported to decrease EGR1 expression in other cells (77). However, our results indicate that FOXO1 does not require the endogenous SF1, PTX1, or EGR1 promoters to exert its repressive effect as SF1, PTX1, and EGR1 were overexpressed using a heterologous promoter and support the idea that FOXO1 repression is likely due to effects of FOXO1 on the proximal Lhb promoter. Additional studies are needed to determine how interactions among FOXO1, SF1, PTX1, EGR1, and cofactors such as β-catenin contribute to the suppression of Lhb gene expression.
Obese women and women with polycystic ovarian syndrome have reduced fertility and increased LH levels (78–81). They also have a high prevalence of insulin resistance and hyperinsulinemia (82). Female mice with diet-induced obesity are also subfertile with elevated LH production and insulin resistance (83). Interestingly, obese mice with the insulin receptor knocked out in gonadotrope cells displayed a substantial improvement in reproductive function, indicating that pituitary insulin signaling may play an important role in the etiology of obesity-related infertility (28). If, as our data strongly suggest, FOXO1 is regulated by insulin signaling in gonadotropes and represses Lhb transcription, then it may prove to be an important factor in the regulation of LH production in situations of caloric insufficiency or excess.
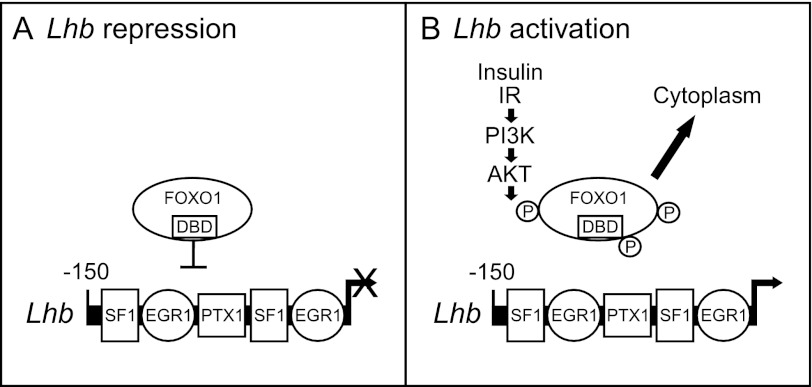
In summary, we show that FOXO1 is expressed in vivo in pituitary gonadotropes and that FOXO1 phosphorylation and cellular localization is regulated by insulin signaling in a PI3K-dependent manner in LβT2 cells. We also demonstrate that FOXO1 suppresses basal transcription and GnRH induction of the Lhb gene, likely through protein-protein interactions between the FOXO1 DBD and transcription factors/cofactors recruited to the proximal Lhb promoter (see model of Lhb repression in Fig. 10). Future in vivo studies using mouse knock-out and transgenic models will help elucidate the potential role of FOXO1 as a pituitary metabolic sensor that regulates reproduction. Further research on the function of specific FOXO1 posttranslational modifications in gonadotrope cells is also needed to understand how FOXO1 integrates input from multiple cellular signaling pathways to regulate reproduction under favorable or adverse environmental conditions.
FIGURE 10.
Model of FOXO1 suppression of Lhb transcription in pituitary gonadotrope cells. A, Lhb repression is shown. In the absence of insulin signaling, FOXO1 is located in the nucleus of the cell where it blocks Lhb gene expression. Although FOXO1 does not bind to the proximal Lhb promoter containing EGR1, PTX1, and SF1 binding elements, the FOXO1 DBD is necessary for the suppression, suggesting that FOXO1 exerts its effect through protein-protein interactions with transcription factors/cofactors required for Lhb transcription. B, Lhb activation is shown. Activated IR/PI3K/AKT signaling leads to phosphorylation and nuclear export of FOXO1. Removal of FOXO1 repression results in increased Lhb synthesis.
Acknowledgments
We thank Djurdjica Coss, Kellie Breen, and Pamela Mellon for helpful discussions and comments. We also thank Scott Kelley for suggestions and critical reading of the manuscript. We acknowledge using the UCSD Cancer Center DNA Sequencing Shared Resource funded in part by the NCI Cancer Support Grant P30 CA023100 for DNA sequencing.
This work was supported, in whole or in part, by National Institutes of Health Grants K01 DK080467 and R01 HD067448 (to V. G. T.) as well as T32 HD007203 (to D. V. S.). This work was also supported by a pilot and feasibility grant from the UCSD/UCLA Diabetes and Endocrinology Research Center (P30 DK063491) by NICHD, National Institutes of Health through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and by a UCSD Academic Senate Health Sciences Research Grant.
D. V. Skarra and V. G. Thackray, unpublished data.
- LH
- luteinizing hormone
- GnRH
- gonadotropin-releasing hormone
- SP1
- specificity protein 1
- SF1
- steroidogenic factor 1
- PTX1
- pituitary homeobox 1
- EGR1
- early growth response protein 1
- IHC
- immunohistochemistry
- IF
- immunofluorescence
- FBE
- forkhead binding element
- Ad
- adenovirus
- DBD
- DNA binding domain
- ANOVA
- analysis of variance
- TSH
- thyroid stimulating hormone
- luc
- luciferase.
REFERENCES
- 1. Ma X., Dong Y., Matzuk M. M., Kumar T. R. (2004) Targeted disruption of luteinizing hormone β-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility. Proc. Natl. Acad. Sci. U.S.A. 101, 17294–17299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huhtaniemi I. (2006) Mutations along the pituitary-gonadal axis affecting sexual maturation. Novel information from transgenic and knockout mice. Mol. Cell. Endocrinol. 254, 84–90 [DOI] [PubMed] [Google Scholar]
- 3. Huhtaniemi I., Ahtiainen P., Pakarainen T., Rulli S. B., Zhang F. P., Poutanen M. (2006) Genetically modified mouse models in studies of luteinizing hormone action. Mol. Cell. Endocrinol. 252, 126–135 [DOI] [PubMed] [Google Scholar]
- 4. Stojilkovic S. S., Reinhart J., Catt K. J. (1994) Gonadotropin-releasing hormone receptors. Structure and signal transduction pathways. Endocr. Rev. 15, 462–499 [DOI] [PubMed] [Google Scholar]
- 5. Kaiser U. B., Conn P. M., Chin W. W. (1997) Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocr. Rev. 18, 46–70 [DOI] [PubMed] [Google Scholar]
- 6. Sealfon S. C., Weinstein H., Millar R. P. (1997) Molecular mechanisms of ligand interaction with the gonadotropin-releasing hormone receptor. Endocr. Rev. 18, 180–205 [DOI] [PubMed] [Google Scholar]
- 7. Pierce J. G., Parsons T. F. (1981) Glycoprotein hormones. Structure and function. Ann. Rev. Biochem. 50, 465–495 [DOI] [PubMed] [Google Scholar]
- 8. Kaiser U. B., Jakubowiak A., Steinberger A., Chin W. W. (1997) Differential effects of gonadotropin-releasing hormone (GnRH) pulse frequency on gonadotropin subunit and GnRH receptor messenger ribonucleic acid levels in vitro. Endocrinology 138, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 9. Papavasiliou S. S., Zmeili S., Khoury S., Landefeld T. D., Chin W. W., Marshall J. C. (1986) Gonadotropin-releasing hormone differentially regulates expression of the genes for luteinizing hormone α and β subunits in male rats. Proc. Natl. Acad. Sci. U.S.A. 83, 4026–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mann R. J., Keri R. A., Nilson J. H. (2003) Consequences of elevated luteinizing hormone on diverse physiological systems. Use of the LHβCTP transgenic mouse as a model of ovarian hyperstimulation-induced pathophysiology. Recent Prog. Horm. Res. 58, 343–375 [DOI] [PubMed] [Google Scholar]
- 11. Jorgensen J. S., Quirk C. C., Nilson J. H. (2004) Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr. Rev. 25, 521–542 [DOI] [PubMed] [Google Scholar]
- 12. Sundaresan S., Colin I. M., Pestell R. G., Jameson J. L. (1996) Stimulation of mitogen-activated protein kinase by gonadotropin-releasing hormone. Evidence for the involvement of protein kinase C. Endocrinology 137, 304–311 [DOI] [PubMed] [Google Scholar]
- 13. Vasilyev V. V., Lawson M. A., Dipaolo D., Webster N. J., Mellon P. L. (2002) Different signaling pathways control acute induction versus long term repression of LHβ transcription by GnRH. Endocrinology 143, 3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris D., Bonfil D., Chuderland D., Kraus S., Seger R., Naor Z. (2002) Activation of MAPK cascades by GnRH. ERK and Jun N-terminal kinase are involved in basal and GnRH-stimulated activity of the glycoprotein hormone LHβ-subunit promoter. Endocrinology 143, 1018–1025 [DOI] [PubMed] [Google Scholar]
- 15. Liu F., Austin D. A., Mellon P. L., Olefsky J. M., Webster N. J. (2002) GnRH activates ERK1/2 leading to the induction of c-fos and LHβ protein expression in LβT2 cells. Mol. Endocrinol. 16, 419–434 [DOI] [PubMed] [Google Scholar]
- 16. Naor Z. (2009) Signaling by G-protein-coupled receptor (GPCR). Studies on the GnRH receptor. Front. Neuroendocrinol. 30, 10–29 [DOI] [PubMed] [Google Scholar]
- 17. Kaiser U. B., Halvorson L. M., Chen M. T. (2000) Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-β gene promoter. An integral role for SF-1. Mol. Endocrinol. 14, 1235–1245 [DOI] [PubMed] [Google Scholar]
- 18. Weck J., Anderson A. C., Jenkins S., Fallest P. C., Shupnik M. A. (2000) Divergent and composite gonadotropin-releasing hormone-responsive elements in the rat-luteinizing hormone subunit genes. Mol. Endocrinol. 14, 472–485 [DOI] [PubMed] [Google Scholar]
- 19. Dorn C., Ou Q., Svaren J., Crawford P. A., Sadovsky Y. (1999) Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J. Biol. Chem. 274, 13870–13876 [DOI] [PubMed] [Google Scholar]
- 20. Tremblay J. J., Drouin J. (1999) Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol. Cell. Biol. 19, 2567–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Attardi B., Miklos J. (1990) Rapid stimulatory effect of activin-A on messenger RNA encoding the follicle-stimulating hormone β-subunit in rat pituitary cell cultures. Mol. Endocrinol. 4, 721–726 [DOI] [PubMed] [Google Scholar]
- 22. Burger L. L., Dalkin A. C., Aylor K. W., Haisenleder D. J., Marshall J. C. (2002) GnRH pulse frequency modulation of gonadotropin subunit gene transcription in normal gonadotrope assessment by primary transcript assay provides evidence for roles of GnRH and follistatin. Endocrinology 143, 3243–3249 [DOI] [PubMed] [Google Scholar]
- 23. Coss D., Thackray V. G., Deng C. X., Mellon P. L. (2005) Activin regulates luteinizing hormone β-subunit gene expression through Smad binding and homeobox elements. Mol. Endocrinol. 19, 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buggs C., Weinberg F., Kim E., Wolfe A., Radovick S., Wondisford F. (2006) Insulin augments GnRH-stimulated LHβ gene expression by Egr-1. Mol. Cell. Endocrinol. 249, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armstrong J., Childs G. V. (1997) Changes in expression of epidermal growth factor receptors by anterior pituitary cells during the estrous cycle. Cyclic expression by gonadotropes. Endocrinology 138, 1903–1908 [DOI] [PubMed] [Google Scholar]
- 26. Rose A., Froment P., Perrot V., Quon M. J., LeRoith D., Dupont J. (2004) The luteinizing hormone-releasing hormone inhibits the anti-apoptotic activity of insulin-like growth factor-1 in pituitary αT3 cells by protein kinase Cα-mediated negative regulation of Akt. J. Biol. Chem. 279, 52500–52516 [DOI] [PubMed] [Google Scholar]
- 27. Navratil A. M., Song H., Hernandez J. B., Cherrington B. D., Santos S. J., Low J. M., Do M. H., Lawson M. A. (2009) Insulin augments gonadotropin-releasing hormone induction of translation in LβT2 cells. Mol. Cell. Endocrinol. 311, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brothers K. J., Wu S., DiVall S. A., Messmer M. R., Kahn C. R., Miller R. S., Radovick S., Wondisford F. E., Wolfe A. (2010) Rescue of obesity-induced infertility in female mice due to a pituitary-specific knockout of the insulin receptor. Cell Metab. 12, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soldani R., Cagnacci A., Yen S. S. (1994) Insulin, insulin-like growth factor I (IGF-I), and IGF-II enhance basal and gonadotrophin-releasing hormone-stimulated luteinizing hormone release from rat anterior pituitary cells in vitro. Eur. J. Endocrinol. 131, 641–645 [DOI] [PubMed] [Google Scholar]
- 30. Xia Y. X., Weiss J. M., Polack S., Diedrich K., Ortmann O. (2001) Interactions of insulin-like growth factor-I, insulin, and estradiol with GnRH-stimulated luteinizing hormone release from female rat gonadotrophs. Eur. J. Endocrinol. 144, 73–79 [DOI] [PubMed] [Google Scholar]
- 31. Leblanc P., L'Héritier A., Kordon C. (1997) Cryptic gonadotropin-releasing hormone receptors of rat pituitary cells in culture are unmasked by epidermal growth factor. Endocrinology 138, 574–579 [DOI] [PubMed] [Google Scholar]
- 32. Dorn C., Mouillet J. F., Yan X., Ou Q., Sadovsky Y. (2004) Insulin enhances the transcription of luteinizing hormone-β gene. Am. J. Obstet. Gynecol. 191, 132–137 [DOI] [PubMed] [Google Scholar]
- 33. Calnan D. R., Brunet A. (2008) The FoxO code. Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
- 34. Van Der Heide L. P., Hoekman M. F., Smidt M. P. (2004) The ins and outs of FoxO shuttling. Mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 380, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 36. Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 38. van der Horst A., Tertoolen L. G., de Vries-Smits L. M., Frye R. A., Medema R. H., Burgering B. M. (2004) FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 279, 28873–28879 [DOI] [PubMed] [Google Scholar]
- 39. Jacobs F. M., van der Heide L. P., Wijchers P. J., Burbach J. P., Hoekman M. F., Smidt M. P. (2003) FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278, 35959–35967 [DOI] [PubMed] [Google Scholar]
- 40. Burgering B. M. (2008) A brief introduction to FOXOlogy. Oncogene 27, 2258–2262 [DOI] [PubMed] [Google Scholar]
- 41. Greer E. L., Brunet A. (2005) FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]
- 42. Accili D., Arden K. C. (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 43. Hosaka T., Biggs W. H., 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., Arden K. C. (2004) Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U.S.A. 101, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castrillon D. H., Miao L., Kollipara R., Horner J. W., DePinho R. A. (2003) Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301, 215–218 [DOI] [PubMed] [Google Scholar]
- 45. Buzzio O. L., Lu Z., Miller C. D., Unterman T. G., Kim J. J. (2006) FOXO1A differentially regulates genes of decidualization. Endocrinology 147, 3870–3876 [DOI] [PubMed] [Google Scholar]
- 46. Labied S., Kajihara T., Madureira P. A., Fusi L., Jones M. C., Higham J. M., Varshochi R., Francis J. M., Zoumpoulidou G., Essafi A., Fernandez de Mattos S., Lam E. W., Brosens J. J. (2006) Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol. Endocrinol. 20, 35–44 [DOI] [PubMed] [Google Scholar]
- 47. Kajihara T., Jones M., Fusi L., Takano M., Feroze-Zaidi F., Pirianov G., Mehmet H., Ishihara O., Higham J. M., Lam E. W., Brosens J. J. (2006) Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol. Endocrinol. 20, 2444–2455 [DOI] [PubMed] [Google Scholar]
- 48. Park Y., Maizels E. T., Feiger Z. J., Alam H., Peters C. A., Woodruff T. K., Unterman T. G., Lee E. J., Jameson J. L., Hunzicker-Dunn M. (2005) Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J. Biol. Chem. 280, 9135–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cunningham M. A., Zhu Q., Unterman T. G., Hammond J. M. (2003) Follicle-stimulating hormone promotes nuclear exclusion of the forkhead transcription factor FoxO1a via phosphatidylinositol 3-kinase in porcine granulosa cells. Endocrinology 144, 5585–5594 [DOI] [PubMed] [Google Scholar]
- 50. Richards J. S., Sharma S. C., Falender A. E., Lo Y. H. (2002) Expression of FKHR, FKHRL1, and AFX genes in the rodent ovary. Evidence for regulation by IGF-I, estrogen, and the gonadotropins. Mol. Endocrinol. 16, 580–599 [DOI] [PubMed] [Google Scholar]
- 51. John G. B., Gallardo T. D., Shirley L. J., Castrillon D. H. (2008) Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev. Biol. 321, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. John G. B., Shidler M. J., Besmer P., Castrillon D. H. (2009) Kit signaling via PI3K promotes ovarian follicle maturation but is dispensable for primordial follicle activation. Dev. Biol. 331, 292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Charles M. A., Mortensen A. H., Potok M. A., Camper S. A. (2008) Pitx2 deletion in pituitary gonadotropes is compatible with gonadal development, puberty, and fertility. Genesis 46, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 55. Alarid E. T., Windle J. J., Whyte D. B., Mellon P. L. (1996) Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 122, 3319–3329 [DOI] [PubMed] [Google Scholar]
- 56. Alarid E. T., Holley S., Hayakawa M., Mellon P. L. (1998) Discrete stages of anterior pituitary differentiation recapitulated in immortalized cell lines. Mol. Cell. Endocrinol. 140, 25–30 [DOI] [PubMed] [Google Scholar]
- 57. Graham K. E., Nusser K. D., Low M. J. (1999) LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to active A. J. Endocrinol. 162, R1–R5 [DOI] [PubMed] [Google Scholar]
- 58. Pernasetti F., Vasilyev V. V., Rosenberg S. B., Bailey J. S., Huang H. J., Miller W. L., Mellon P. L. (2001) Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology 142, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 59. Tremblay J. J., Lanctôt C., Drouin J. (1998) The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol. Endocrinol. 12, 428–441 [DOI] [PubMed] [Google Scholar]
- 60. Jiang Q., Jeong K. H., Horton C. D., Halvorson L. M. (2005) Pituitary homeobox 1 (Pitx1) stimulates rat LHβ gene expression via two functional DNA-regulatory regions. J. Mol. Endocrinol. 35, 145–158 [DOI] [PubMed] [Google Scholar]
- 61. Tang E. D., Nuñez G., Barr F. G., Guan K. L. (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274, 16741–16746 [DOI] [PubMed] [Google Scholar]
- 62. Rosenberg S. B., Mellon P. L. (2002) An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol. Endocrinol. 16, 1280–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thackray V. G., Hunnicutt J. L., Memon A. K., Ghochani Y., Mellon P. L. (2009) Progesterone Inhibits basal and gonadotropin-releasing hormone induction of luteinizing hormone β-subunit gene expression. Endocrinology 150, 2395–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang X., Gan L., Pan H., Guo S., He X., Olson S. T., Mesecar A., Adam S., Unterman T. G. (2002) Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J. Biol. Chem. 277, 45276–45284 [DOI] [PubMed] [Google Scholar]
- 65. Brosens J. J., Wilson M. S., Lam E. W. (2009) FOXO transcription factors. From cell fate decisions to regulation of human female reproduction. Adv. Exp. Med. Biol. 665, 227–241 [DOI] [PubMed] [Google Scholar]
- 66. Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Biggs W. H., 3rd, Cavenee W. K., Arden K. C. (2001) Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome 12, 416–425 [DOI] [PubMed] [Google Scholar]
- 68. Kitamura T., Feng Y., Kitamura Y. I., Chua S. C., Jr., Xu A. W., Barsh G. S., Rossetti L., Accili D. (2006) Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat. Med. 12, 534–540 [DOI] [PubMed] [Google Scholar]
- 69. Schweizer-Groyer G., Fallot G., Cadepond F., Girard C., Groyer A. (2006) The cAMP-responsive unit of the human insulin-like growth factor-binding protein-1 coinstitutes a functional insulin-response element. Ann. N.Y. Acad. Sci. 1091, 296–309 [DOI] [PubMed] [Google Scholar]
- 70. Mounier C., Posner B. I. (2006) Transcriptional regulation by insulin. From the receptor to the gene. Can. J. Physiol. Pharmacol. 84, 713–724 [DOI] [PubMed] [Google Scholar]
- 71. Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Armoni M., Harel C., Karni S., Chen H., Bar-Yoseph F., Ver M. R., Quon M. J., Karnieli E. (2006) FOXO1 represses peroxisome proliferator-activated receptor-γ1 and -γ2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J. Biol. Chem. 281, 19881–19891 [DOI] [PubMed] [Google Scholar]
- 73. Salisbury T. B., Binder A. K., Grammer J. C., Nilson J. H. (2007) Maximal activity of the luteinizing hormone β-subunit gene requires β-catenin. Mol. Endocrinol. 21, 963–971 [DOI] [PubMed] [Google Scholar]
- 74. Olson L. E., Tollkuhn J., Scafoglio C., Krones A., Zhang J., Ohgi K. A., Wu W., Taketo M. M., Kemler R., Grosschedl R., Rose D., Li X., Rosenfeld M. G. (2006) Homeodomain-mediated β-catenin-dependent switching events dictate cell lineage determination. Cell 125, 593–605 [DOI] [PubMed] [Google Scholar]
- 75. Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 76. Hoogeboom D., Essers M. A., Polderman P. E., Voets E., Smits L. M., Burgering B. M. (2008) Interaction of FOXO with β-catenin inhibits β-catenin/T cell factor activity. J. Biol. Chem. 283, 9224–9230 [DOI] [PubMed] [Google Scholar]
- 77. Cabodi S., Morello V., Masi A., Cicchi R., Broggio C., Distefano P., Brunelli E., Silengo L., Pavone F., Arcangeli A., Turco E., Tarone G., Moro L., Defilippi P. (2009) Convergence of integrins and EGF receptor signaling via PI3K/Akt/FoxO pathway in early gene Egr-1 expression. J. Cell. Physiol. 218, 294–303 [DOI] [PubMed] [Google Scholar]
- 78. Yoo R. Y., Dewan A., Basu R., Newfield R., Gottschalk M., Chang R. J. (2006) Increased luteinizing hormone pulse frequency in obese oligomenorrheic girls with no evidence of hyperandrogenism. Fertil. Steril. 85, 1049–1056 [DOI] [PubMed] [Google Scholar]
- 79. Morales A. J., Laughlin G. A., Bützow T., Maheshwari H., Baumann G., Yen S. S. (1996) Insulin, somatotropic, and luteinizing hormone axes in lean and obese women with polycystic ovary syndrome. Common and distinct features. J. Clin. Endocrinol. Metab. 81, 2854–2864 [DOI] [PubMed] [Google Scholar]
- 80. Hall J. E., Taylor A. E., Hayes F. J., Crowley W. F., Jr. (1998) Insights into hypothalamic pituitary dysfunction in polycystic ovary syndrome. J. Endocrinol. Invest. 21, 602–6011 [DOI] [PubMed] [Google Scholar]
- 81. Chang R. J. (2007) The reproductive phenotype in polycystic ovary syndrome. Nat. Clin. Pract. Endocrinol. Metab. 3, 688–695 [DOI] [PubMed] [Google Scholar]
- 82. Dunaif A., Thomas A. (2001) Current concepts in the polycystic ovary syndrome. Annu. Rev. Med. 52, 401–419 [DOI] [PubMed] [Google Scholar]
- 83. Tortoriello D. V., McMinn J., Chua S. C. (2004) Dietary-induced obesity and hypothalamic infertility in female DBA/2J mice. Endocrinology 145, 1238–1247 [DOI] [PubMed] [Google Scholar]