Background: A. fumigatus is an opportunistic pathogen responsible for pulmonary invasive aspergillosis.
Results: Multiplexed ABPP revealed significant changes in A. fumigatus metabolism and stress response during culture with human serum over time.
Conclusion: Changes in functional pathways indicate robust adaptation to environmental change.
Significance: A. fumigatus grows under stress by altering metabolism, energy production, and protein biosynthesis, which is relevant for lung colonization.
Keywords: Aging, Aspergillus, Metabolism, Proteomics, Stress Response, ABPP, Aspergillus Fumigatus, Human Serum
Abstract
Environmental adaptability is critical for survival of the fungal human pathogen Aspergillus fumigatus in the immunocompromised host lung. We hypothesized that exposure of the fungal pathogen to human serum would lead to significant alterations to the organism's physiology, including metabolic activity and stress response. Shifts in functional pathway and corresponding enzyme reactivity of A. fumigatus upon exposure to the human host may represent much needed prognostic indicators of fungal infection. To address this, we employed a multiplexed activity-based protein profiling (ABPP) approach coupled to quantitative mass spectrometry-based proteomics to measure broad enzyme reactivity of the fungus cultured with and without human serum. ABPP showed a shift from aerobic respiration to ethanol fermentation and utilization over time in the presence of human serum, which was not observed in serum-free culture. Our approach provides direct insight into this pathogen's ability to survive, adapt, and proliferate. Additionally, our multiplexed ABPP approach captured a broad swath of enzyme reactivity and functional pathways and provides a method for rapid assessment of the A. fumigatus response to external stimuli.
Introduction
Aspergillus fumigatus is a ubiquitous filamentous fungus found in soil and decaying matter that plays an important role in recycling carbon and nitrogen from organic debris. Provided the prevalent nature of these conidia in ambient air, it is estimated that individuals inhale hundreds of airborne conidia per day. Exposure to conidia is inconsequential if the immune system is intact, but this saprophytic fungus is an opportunistic pathogen and the leading cause of the pulmonary infection invasive aspergillosis (IA)2 (1). Immunodeficiency associated with solid organ and allogeneic bone marrow transplantation, prolonged corticosteroid therapy, genetic immunodeficiency, HIV infection, or hematological malignancies such as leukemia increase susceptibility to this devastating disease (2). As the at risk population has increased over the past 2 decades, so too has the incidence of IA (3). Key challenges remain in understanding the biology of A. fumigatus responsible for infection as well as detection and treatment of IA. Techniques for reliable early stage diagnosis (4, 5), identification of IA biomarkers with prognostic value for monitoring fungal load and response to therapeutic intervention, identification of new drug targets (6), and understanding of fungal biology under infection-relevant conditions continue to be active areas of research.
Considerable effort has focused on finding and understanding the pathogenicity and virulence factors of the fungus. This opportunistic pathogen lacks true virulence factors because it evolved to break down organic matter (7). However, many characteristics contribute to its pathogenicity (1, 8), making it highly pathogenic in the immunocompromised host (7). These include its environmental and metabolic adaptability: A. fumigatus grows rapidly at high temperatures and within hypoxic conditions (9, 10); it can obtain carbon and nitrogen from diverse sources (11, 12); and it is highly adaptive in its ability to uptake nutrients from its environment (13). We utilized a chemical biology approach to characterize the fungal functional adaptation to changes in nutrient availability in conditions relevant to IA infection.
Global proteome analysis of A. fumigatus has elucidated protein regulation and pathway responses to environmental stimuli (14). However, global (i.e. untargeted) proteomics provides incomplete proteome coverage and generally fails to provide quantifiable measurement of the functional activity of observed proteins because proteins are dynamic and highly regulated and often exist in inactive forms until proteolytically processed or post-translationally modified. Activity-based protein profiling (ABPP) has emerged to overcome the inherent difficulty of differentiating presence versus activity and to facilitate the measurement of low abundance reactive proteins. ABPP employs specific chemical probes to observe the active protein complements of biological systems (15, 16). Synthetic activity-based probes (ABPs) inhibit enzymes by forming an irreversible covalent bond to the enzyme active site. Enriching and identifying ABP-tagged proteins allows proteomic annotation of function, reduces the complexity of the proteome under analysis, and measures low abundance functional proteins. Coupled with high throughput, high resolution LC-MS/MS analysis utilizing a quantitative accurate mass and time (AMT) tag approach (17), environmental adaptation and metabolic response can be quantified using ABPP.
During the course of IA, A. fumigatus hyphae breach host tissue and interact with serum. A. fumigatus is one of the few pathogenic organisms that readily grows in the presence of serum (18) due to its ability to extract iron from human transferrin in this iron-limited environment (19). Furthermore, A. fumigatus can use serum proteins for biomass generation (20, 21), but the full effect of serum on the cellular processes of A. fumigatus and its relevance to disease is not fully understood. We hypothesize that A. fumigatus can adapt to this nutrient source for metabolic purposes and that fungal enzyme activity within human serum (HS) will be relevant to its metabolism, nutrient sensing, and scavenging response within the immunocompromised host environment. This information can provide valuable insight into how A. fumigatus survives in a host environment on a fundamental biological process level. Herein, we describe a novel ABP and label-free quantitative multiplexed ABPP methodology for delineating the protein activity of A. fumigatus during growth in minimal medium in response to HS as a function of growth duration.
EXPERIMENTAL PROCEDURES
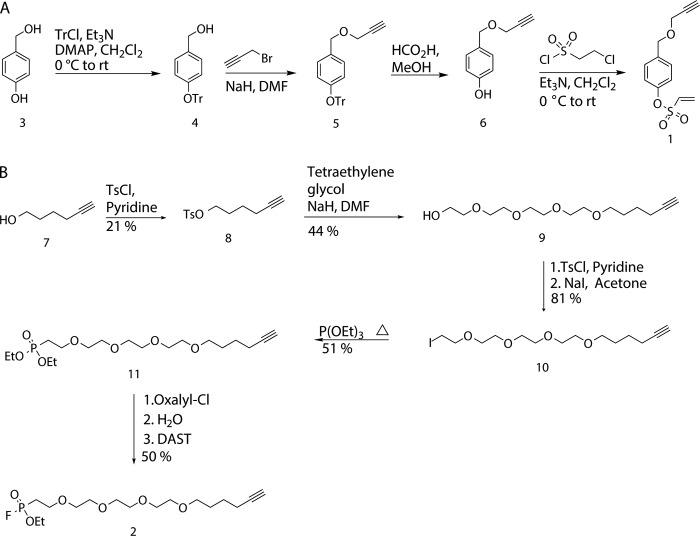
Synthesis of Activity-based Probes 1 and 2
Detailed experimental procedures for the synthesis of 1 and 2 are available in the supplemental material.
Strains, Media, and Culture Conditions
A. fumigatus ATCC® MYA-4609TM (AF 293) obtained from ATCC were stored at −80 °C in individual glycerol. Potato dextrose agar plates were inoculated with conidia from glycerol stocks and harvested by flooding the plate with 0.8% Tween 80 after 4 days. The conidia suspension was filtered through Miracloth (Calbiochem), and the conidia were counted by hemocytometer. For liquid culture, complete Aspergillus minimal medium (AMM; 1% glucose, 2 ml/liter Hunter's trace elements) (22) with or without 10% HS was inoculated to a final concentration of 1 × 106 conidia/ml. Biomass was grown at 37 °C and agitated at 150 rpm on an orbital shaker for 24 or 48 h.
Human Serum
HS (Sigma) was stored at −20 °C and used without further processing.
Growth Curve
AMM and AMM containing 10% HS were inoculated to 1 × 106 conidia/ml with A. fumigatus and cultured as described above. Fungal biomass was harvested in triplicate at each time point by filtering the liquid culture through Miracloth, washing the collected biomass with water, and cryodessication. The mass of the dried fungus was used to generate a growth curve (Fig. 1).
FIGURE 1.
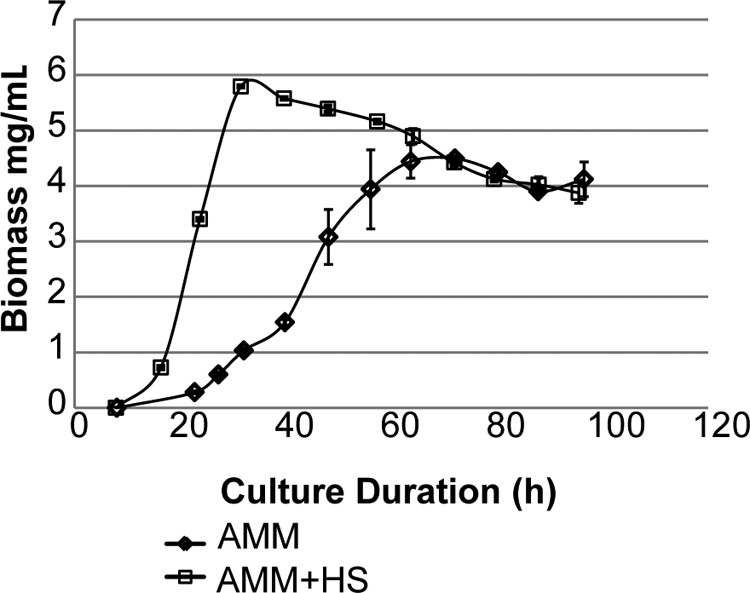
Growth curve generated for AMM and AMM + 10% HS culture. Biomass was harvested, flash-frozen, dried, and then weighed. Data points were collected in triplicate, and error bars represent S.E. of each data point.
Cell Lysis
Biomass was harvested at 24 and 48 h and passed through Miracloth to remove spent medium, and the retained biomass was washed with 15 ml of PBS. Excess medium was squeezed out of the biomass. The biomass was manually cut and then lysed with a Bullet Blender (NextAdvance): 0.55-mm zirconium silicate beads (0.5–1.0 ml) and PBS (1–2 volumes) (1×, pH 7.4: 11.9 mm phosphates, 137 mm NaCl, 2.7 mm KCl) per 5-ml tube. Lysed samples were centrifuged at 3,500 × g for 5 min, pellets were washed with PBS (1 ml) and spun at 3,500 × g for 5 min, and the combined supernatants were spun twice more. Sonication of the final combined supernatant provided global cell lysate (gcl). The gcl was flash frozen in liquid nitrogen and stored at −80 °C until further use. All material was generated in quadruplicate.
Enzyme Assays
Using a BCA assay, the protein concentration of gcl from AMM + HS 24- and 48-h growth was then normalized to 1 mg/ml protein (supplemental Fig. S1A). Alcohol dehydrogenase activity for the gcl was measured as described previously (23). The assay was performed in triplicate, and the slope was calculated using data points from 3–6 min (supplemental Fig. S1C). Citrate synthase activity was measured with a commercial kit (Sigma-Aldrich); assays were run in triplicate (supplemental Fig. S1B).
In Vitro Probe Labeling for Mass Spectrometry Measurement
A. fumigatus mycelia lysate proteomes (1 mg/ml in PBS) were treated with vinyl sulfonate 1 (20 μm) and fluorophosphonate 2 (200 μm) in a 1.5-ml vial and incubated for 1 h at 37 °C. Following probe incubation, proteomes were treated with click chemistry reagents and enriched by the method of Cravatt with the following changes. Peptides were obtained by treating the resin with trypsin (2 μl; trypsin was reconstituted in 40 μl of NH4HCO3 (25 mm, pH 8)) and NH4HCO3 (25 mm, pH 8; 200 μl) and subsequent incubation at 37 °C, 1,200 rpm, for 15 h. Following digestion, peptide supernatant was obtained (6,000 × g), and the pellet was washed with NH4HCO3 (25 mm, pH 8; 150 μl). The combined peptide supernatant was dried by speed vacuum, reconstituted in NH4HCO3 (25 mm, pH 8; 40 μl), and heated for 10 min at 37 °C. The samples were centrifuged at 100,000 × g for 20 min at 4 °C, and 25 μl was collected for MS analysis.
LC-MS Measurement of ABP-labeled Proteins
Digested peptide mixtures were measured on a Thermo Fisher Scientific LTQ Orbitrap Velos MS (San Jose, CA) as described previously (24). The Velos MS data were collected from 400 to 2,000 m/z at a resolution of 100,000 (automatic gain control target: 1 × 106) followed by data-dependent ion trap MS/MS spectra (AGC target: 1 × 104) of the 10 most abundant ions using a collision energy setting of 35–40% and a dynamic exclusion time of 30 or 180 s.
Significant changes were determined at the unique peptide level between growth conditions or culture durations. The resulting MS data were analyzed using the AMT tag pipeline to determine relative protein abundance (17). SEQUEST software (25) was used to search tandem mass spectra against the UniProt A. fumigatus database (April 1, 2011). Peptide identifications filtered with an MS-generating function spectral probability score of ≤1 × 10−9 (26) were assembled into an A. fumigatus-specific AMT tag database. For identification, VIPER software (27) was used to correlate each AMT tag entry with a unique LC-MS feature relying on high mass measurement accuracy (MMAaverage ± 0.683 ppm) and normalized elution time accuracy (NETaverage ± 0.363%) (supplemental Tables S1–S4). Single peptides matching multiple proteins (typically protein isoforms) were removed. Proteins increasing in the presence of HS were manually verified to originate from fungus by looking at MS/MS scans (supplemental Table S5).
For statistical analysis of peptide abundance, DAnTE software (28) was used to perform ANOVA significance tests on culture comparisons; a significant difference was determined as a p value and q value of <0.05. The q value of a test measures the proportion of false positives incurred (i.e. the false discovery rate) for the analysis. Significant peptides found in at least three data sets were rolled up into protein values using Rrollup (28). An absolute -fold change (|fc|) of ≥15, ≥2 peptides/protein, and ≥10% protein coverage were used to ensure high confidence of significantly changing proteins. Unique proteins were obtained by rolling up all unique peptides to protein and using a relative abundance cut-off of ≥18, ≥2 peptides/protein, and 10% protein coverage. Proteins were mapped to function using tools from the Aspergillus Genome Database (29), the Fungifun tools at Omnifung (30), and the KEGG pathway user mapping tool (31).
RESULTS
Growth in the Presence and Absence of Human Serum
Collection of fungal biomass at multiple time points within each culture showed strikingly different growth patterns for A. fumigatus in the presence and absence of 10% HS (Fig. 1). Culture with HS caused a large increase in biomass generation at 24 and 48 h. In the presence of HS, the fungus reaches log phase growth at 16 h, whereas in minimal medium, log phase growth is delayed until 24 h. At 48 h after liquid culture inoculation, A. fumigatus has reached stationary phase in the presence of human serum but is still in log phase growth in minimal medium. In AMM, A. fumigatus reaches stationary phase at 64 h. The decrease in biomass at subsequent time points corresponds to autolysis and death of the mycelia from nutrient deprivation or mechanical shearing forces that break down hyphae as the culture ages (32).
ABP Development and One-dimensional Gel Analysis
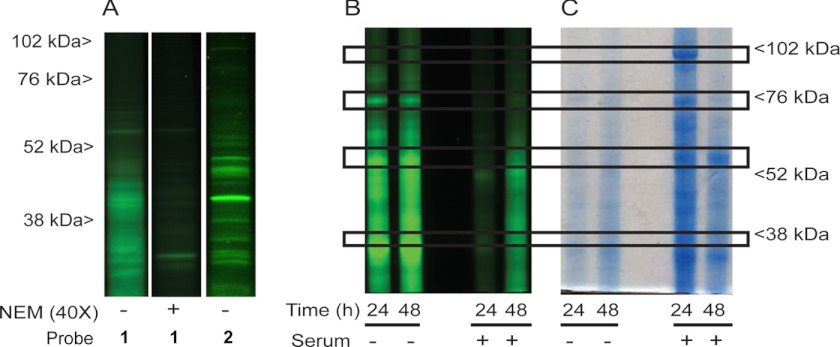
In developing a library of known and novel ABPs that incorporate the click chemistry-compatible alkyne moiety for attachment of azide-derivatized reporter groups, we synthesized vinyl sulfonate ester probe 1 (Fig. 2A). The clickable alkyne probe 1 was synthesized in four steps from commercially available 4-hydroxybenzyl alcohol (33). Optimized labeling conditions of A. fumigatus samples were performed at a final probe concentration of 20 μm at 37 °C for 1 h (supplemental Fig. S2); these labeling conditions were used for all further analysis with 1. Vinyl sulfonate esters are known to irreversibly inhibit cysteine proteases; however, broad labeling of distinct enzyme classes with 1 at 37 °C occurred (supplemental Fig. S2). Furthermore, treatment of purified enzymes and A. fumigatus gcl with 40× cysteine-reactive N-ethylmaleimide followed by probe labeling did not completely eradicate labeling with probe 1, suggesting covalent binding to nucleophilic amino acids other than cysteine in the probe labeling event (Fig. 3A and supplemental Fig. S3), which is akin to prior electrophilic ABP reports (34–36). The generality of 1 allows interrogation of a large system-wide subset of functionally active proteins in A. fumigatus.
FIGURE 2.
Synthesis of vinyl sulfonate ester 1 and fluorophosphonate 2 ABPs used for multiplexed ABPP. A, vinyl sulfonate ester ABP 1 was synthesized in four steps from 4-hydroxybenzyl alcohol. B, fluorophosphonate ABP 2 was synthesized in eight steps from 5-hexyn-1-ol. rt, room temperature. DMF, dimethylformamide; DMAP, 4-dimethylaminopyridine; DAST, diethylaminosulfur trifluoride.
FIGURE 3.
Labeling of gcl of A. fumigatus with 1 and 2 in the presence and absence of HS at 24 and 48 h. A, labeling of AMM + HS 48 h gcl with 1 in the presence and absence of 40× N-ethylmaleimide (NEM) showed incomplete labeling inhibition, suggesting nucleophilic attack of 1 by non-cysteine amino acids. A distinct labeling pattern AMM + HS 48 h gcl is observed when 2 is used. B, one-dimensional SDS-PAGE fluorescent image of A. fumigatus gcl labeled with 1 and 2 showed the highest probe reactivity in AMM at 24 h (lane 1) and 48 h (lane 2). Lower enzyme reactivity was observed in AMM + HS at both time points (lanes 3 and 4). C, Coomassie-stained image of A. fumigatus gcl labeled with probes 1 and 2 showed that labeling was not dependent on protein abundance. The highest protein content corresponded to the lowest fluorescent labeling. Boxes at 102, 76, 60, and 38 kDa show the discrepancy between protein abundance and protein labeling.
To include coverage of serine hydrolases, a serine hydrolase-specific probe (2) was included in the analysis of A. fumigatus (Fig. 3A and supplemental Fig. S5). Probe 2 contains a fluorophosphonate reactive group selective for serine hydrolases (37) and an alkyne for appending a reporter group (Fig. 2B). A probe concentration of 200 μm was optimal for labeling (supplemental Fig. S4). Probes 1 and 2 were used simultaneously to analyze the proteome of A. fumigatus grown with and without HS for 24 and 48 h. Because both probes contain the clickable alkyne tag, it is possible to conjugate both probes with one type of reporter group (i.e. biotin azide or rhodamine azide) using click chemistry. Multiplexing the ABPs permits broader coverage in a single MS measurement.
Protein labeling by ABPs was independent of protein abundance. Equivalent volumes of gcl obtained from A. fumigatus in the presence and absence of HS over time were labeled with 1 and 2 (Fig. 3B). The extent of labeling of the AMM grown samples at both time points was much greater than labeling of the AMM + HS samples (Fig. 3B). In culture with HS, probe labeling was greater at 48 h than at 24 h. Strikingly, Coomassie protein staining of the gel showed higher protein content in AMM + HS samples than in the AMM samples, but probe labeling was lower (Fig. 3C). Three highly abundant proteins by protein staining in AMM + HS 24 h at 60, 80, and 102 kDa were not ABP-labeled. Later time points (4 days) showed a significant overall reduction in enzyme reactivity (supplemental Fig. S4).
Our data clearly indicate that 1 and 2 probe different enzyme reactivity, and labeling is independent of protein abundance. Although probe and protein interaction at a non-catalytic residue within the active site or a ligand binding site with an adjacent nucleophile cannot be ruled out at this time (38, 39), the differences in enzyme reactivity (fluorescent intensity) suggest that there is a different functional state of the tagged enzymes between conditions. If probe labeling was not determined by enzyme functional state, we would expect to see labeling of all proteins within a sample. Therefore, we relate the observed changes in enzyme reactivity with differences in functional activity between the samples.
ABPP by LC-MS and LC-MS/MS
Global cell lysates of A. fumigatus grown in the presence and absence of HS and labeled with 1 and 2 were analyzed by LC-MS/MS, and LC-MS features were identified using quantitative peak matching to an AMT tag database (17). Using the above described criteria, the relative abundance of confident protein identifications were compared over time and growth conditions. When comparing protein abundance between conditions, an absolute fc cut-off of 15 was chosen for manual pathway analysis to ensure high confidence of measured changes and relevance of the reactive proteins to the culture conditions. Multiple enzyme families, including oxidoreductases, transferases, lyases, hydrolases, isomerases, and ligases, were identified in global cell lysates using ABPP.
Comparison of AMM 24 h to AMM + HS 24 h
We compared reactive enzyme abundance in the presence or absence of HS at 24 h (Table 1 and supplemental Table S1). At 24 h, 239 proteins had an fc less than −2 (negative value denotes higher reactivity in AMM 24 h). Eighty-two proteins had a large enzyme reactivity difference (|fc| > 15); only one protein (acetyl-CoA carboxylase; Table 2) had higher enzyme reactivity in the presence of HS, six proteins were observed only in the absence of HS, and 81 proteins had high levels of enzyme reactivity in AMM only (Table 1 and supplemental Table S6). Mapping proteins with absolute value |fc| ≥ 15 onto Functional Catalogues (FunCat) showed enrichment of metabolism, protein synthesis, and energy functional categories (30) (Fig. 4 and supplemental Table S6). KEGG pathway mapping showed enrichment of genetic information processing, metabolism, and cellular processes (supplemental Table S6).
TABLE 1.
Protein trends between conditions
Proteins with high, low, and no fold change between conditions. All proteins counted have >10% peptide coverage and >1 peptide per protein. A p-value >0.05 was used to determine proteins with no significant fold change. The symbol | denotes absolute value of the fold change value. A negative value denotes greater reactivity in condition B, and a positive value denotes greater reactivity in condition A. For unique proteins, a Log2 abundance of >18 was required. For a complete list of proteins and peptides observed in this comparison, see Table S1, S2, S3, S4.
| Condition A | Condition B | Non-changing (A/B) | |fc| > 2 (A/B) | |fc| < 15 (A/B) | fc >15 (A/B) | Unique to A | Unique to B |
|---|---|---|---|---|---|---|---|
| AMM + HS 24 h | AMM 24 h | 196 | 242 | 81 | 1 | 0 | 6 |
| AMM + HS 48 h | AMM 48 h | 166 | 345 | 27 | 0 | 3 | 21 |
| AMM 48 h | AMM 24h | 204 | 342 | 0 | 23 | 6 | 0 |
| AMM + HS 48 h | AMM + HS 24 h | 44 | 320 | 0 | 88 | 10 | 0 |
TABLE 2.
Subset of proteins with metabolic, stress response, secondary metabolism, and cell cycle regulation annotation across all comparisons
Subset of proteins with metabolic, stress response, secondary metabolism, and cell cycle regulation annotation across all comparisons. NA, protein was not detected with confidence in one of the growth conditions and therefore a fold change could not be calculate; NS, no significant change between conditions (p-value > 0.05). For a complete list of proteins and peptides fold changes, see Tables S1–S4.
| Protein description | Locus tag | Enzyme (EC) | -Fold change 24 h (HS/no HS) | -Fold change 48 h (HS/no HS) | -Fold change No HS (48 h/24 h) | -Fold change HS (48 h/24 h) |
|---|---|---|---|---|---|---|
| Histone H3 | AFUA_1G13790 | −9.3 | −17.6 | 4.3 | 2.6 | |
| Acetyl-CoA carboxylase | AFUA_2G08670 | 15.4 | NSa | 13.8 | 3.1 | |
| Sulfur metabolism regulator SkpA | AFUA_5G06060 | −20.9 | −16.4 | NS | 3.3 | |
| Squalene monooxygenase Erg1 | AFUA_5G07780 | NS | NS. | 19.0 | 4.6 | |
| Citrate synthase (Cit1) | AFUA_5G04230 | −13.8 | −3.6 | NS | 11.4 | |
| MAPK MpkA | AFUA_4G13720 | 2.7.11.1 | NS | NS | 18.8 | 15.5 |
| HAD superfamily hydrolase | AFUA_5G08270 | −14.78 | −8.6 | 26.3 | 57.6 | |
| Alcohol dehydrogenase | AFUA_7G01010 | 1.1.1.1 | −145.6 | −8 | 4.5 | 73.7 |
| Aldehyde reductase (AKR1) | AFUA_6G10260 | 1.1.1.2 | NAb | AMM + HS only | NA | 48 only |
| Glutathione oxidoreductase Glr1 | AFUA_1G15960 | 1.8.1.7 | NA | AMM + HS only | NA | 48 only |
| Conserved hypothetical protein | AFUA_2G10170 | NA | AMM + HS only | NA | 48 only | |
| Conserved hypothetical protein | AFUA_8G00430 | AMM only | −2.9 | 19 | NS | |
| Flavin-binding monooxygenase-like protein | AFUA_4G09220 | NS | −2.9 | 16.5 | NS | |
| O-Methyltransferase GliM-like | AFUA_3G12910 | 2.-.-.- | NS | NS | 19.6 | NS |
| Conidial pigment biosynthesis protein Ayg1 | AFUA_2G17550 | NS | NS | 22.5 | NS | |
| Oxidoreductase, 2OG-Fe(II) oxygenase family | AFUA_3G00800 | NS | NS | 24.5 | NS | |
| Conidial pigment biosynthesis 1,3,6,8-tetrahydroxynaphthalene reductase Arp2 | AFUA_2G17560 | NS | NS | 25.2 | NS | |
| Glutathione S-transferase | AFUA_4G14380 | NS | NS | 47.5 | NS | |
| Cytochrome P450 oxidoreductase OrdA-like | AFUA_8G00510 | NS | NS | 50.5 | NS | |
| Glutathione S-transferase Ure2-like | AFUA_4G14530 | NS | NS | 78.9 | NS | |
| Fructosyl amino acid oxidase | AFUA_6G03440 | NS | NS | 48 h only | NS | |
| Catalase Cat | AFUA_2G18030 | 1.11.1.6 | NA | AMM only | 48 h only | NA |
| Cytochrome P450 | AFUA_5G02610 | 1.14.-.- | NA | AMM only | 48 h only | NA |
| Hybrid PKS-NRPS enzyme | AFUA_8G00540 | NS | NS | 28.3 | NA |
a NS, not significant; p-value > 0.05.
b NA, not applicable.
FIGURE 4.
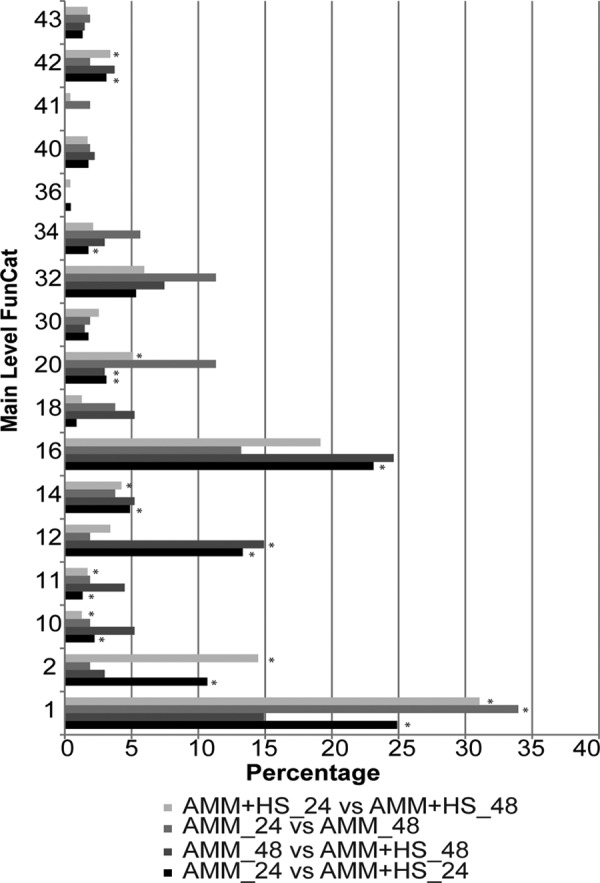
FunCat annotation of |fc| ≥ 15 enzymes in all comparisons. FunCat main categories are on the y axis, and the percentage that each category contributes to total mapping is on the x axis. FunCat categories are as follows: 1, metabolism; 2, energy; 10, cell cycle and DNA processing; 11, transcription; 12, protein synthesis; 14, protein fate; 16, protein with binding function or cofactor requirement; 18, regulation of metabolism and protein function; 30, cellular communication/signal transduction mechanism; 32, cell rescue, defense, and virulence; 34, interaction with the environment; 36, systemic interaction with the environment; 40, cell fate; 41, development; 42, biogenesis of cellular components; 43, cell type differentiation. *, categories found to have a p value of <0.05. For complete annotation mapping, see supplemental Tables S6–S9.
Comparison of AMM 48 h with AMM + HS 48 h
At 48 h, 340 proteins were significantly different between conditions (|fc| > 2), and 51 of these proteins had high enzyme reactivity (|fc| > 15) (supplemental Table S7), 21 were unique to and 27 had higher reactivity in AMM; three were unique to AMM + HS. The enzymes unique to growth in HS were aldehyde reductase akr1 (AFUA_6G10260), glutathione oxidoreductase glr1 (AFUA_1G15960), and a conserved hypothetical protein (AFUA_2G10170) (Table 2). FunCat mapping of all highly reactive proteins in the 48-h growth comparison corresponded to enrichment of protein synthesis, proteins with binding requirements, and cellular transport functions (Fig. 4 and Table S7). Two proteins involved in cell cycle regulation, histone H3 (AUFA_1G13790) and sulfur metabolism regulator SkpA (AFUA_5G06060), had higher ABP probe reactivity in AMM (29).
Comparison of AMM 24 h with AMM 48 h
A time comparison of AMM growth (24 h versus 48 h) showed 204 proteins with no significant change; at 48 h of growth, 342 proteins had a 2-fold increase in enzyme reactivity (Table 1 and supplemental Table S8), 23 proteins had a >15-fold increase, and six proteins were unique. Metabolism was highly enriched, whereas secondary metabolism was the highest enriched subcategory; in addition, detoxification subcategories were also enriched (Fig. 4 and supplemental Table S8). Five proteins with a >15-fold change are annotated with secondary metabolism function (Table 2): catalase fgaCat (AFUA_2G18030), active in fumigaclavine C biosynthesis (40), two melanin conidial pigment biosynthetic proteins Arp2 (AFUA_2G17560) and Ayg1 (AFUA_2G17550) (41), hybrid PKS-NRPS enzyme nrps14 (AFUA_8G00540) of the pseurotin A biosynthesis gene cluster (42), and glutathione S-transferase (AFUA_4G14380) located on chromosome 4 near other secondary metabolite biosynthesis gene clusters (Table 2) (43). Transcription of fgaCat, nrps14, and the glutathione S-transferase changed in the presence of voriconazole (44) and/or hypoxic conditions (45). In addition, melanin is essential for cell wall integrity and protection against neutrophil-mediated phagocytosis (46). Finally, the proteins mpkA (AFUA_4G13720) and squalene monoxygenase erg1 (AFUA_5G07780), both associated with cell wall signaling (47) and membrane synthesis (48), respectively, have altered transcription levels in response to chemical stress (29). ABP-tagged proteins associated with primary metabolic functions were still increasing, albeit with a |fc| of ∼5. In AMM, proteins associated with secondary metabolism and stress response dramatically increased over time (supplemental Table S8).
Comparison of AMM + HS 24 h with AMM + HS 48 h
In the presence of HS, 44 proteins showed no difference between 24 and 48 h, a total of 320 proteins had a 2-fold increase in reactivity, and 10 proteins were unique to and 88 were highly active at 48 h of growth (fc ≥ 15) (Table 1 and supplemental Table S4). The most highly enriched functional categories at 48 h were metabolism (31%) and energy (14%) (Fig. 4 and supplemental Table S9) (30). Furthermore, amino acid and carbohydrate metabolism were the most significant KEGG pathway annotations (supplemental Table S9) (31). The functional categories enriched at 48 h in HS culture (24 h versus 48 h) were also highly enriched at 24 h in minimal medium (AMM 24 h versus AMM + HS 24 h), including transcription, metabolism, energy, protein fate, biogenesis of cellular components, cell cycle, and DNA processing and transport (supplemental Table S9) (Fig. 4).
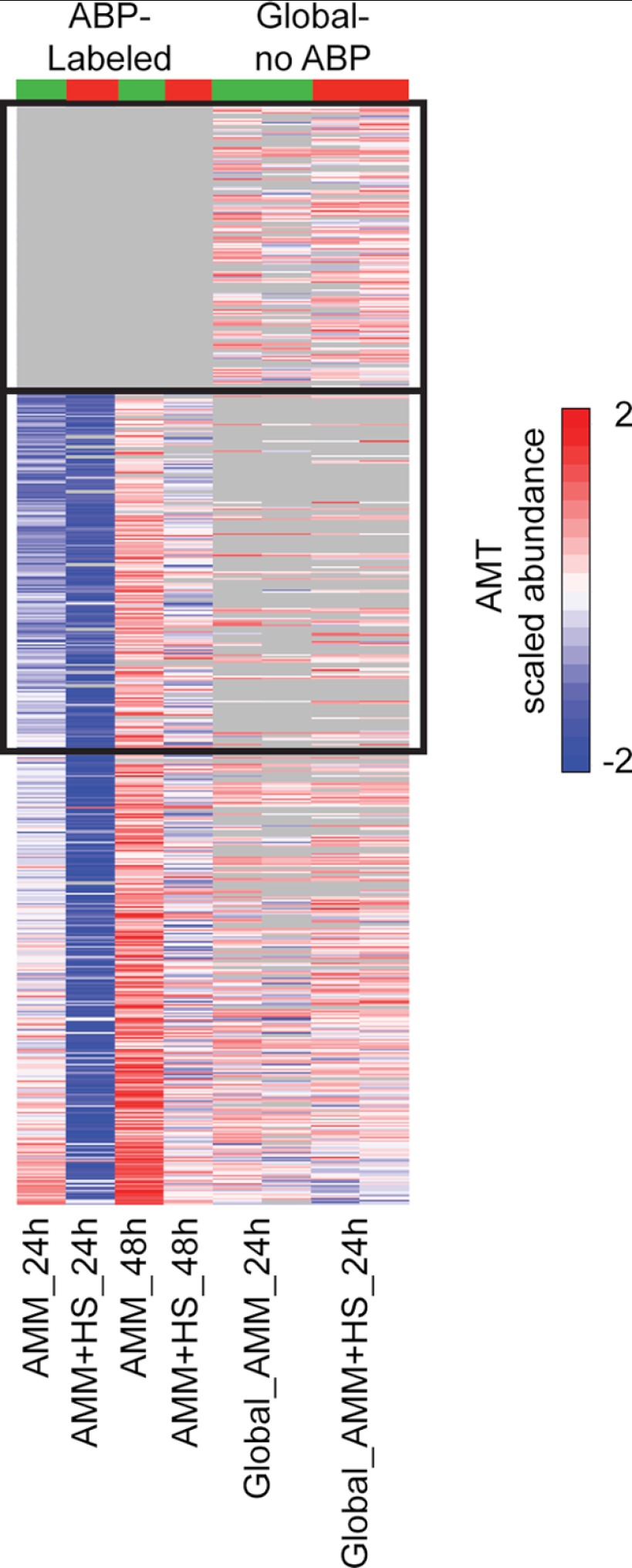
Validation of ABPP by Global Proteomics
To validate ABPP of filamentous fungus, we analyzed gcl obtained from AMM and AMM + HS growth at 24 h without ABP labeling. A total of 647 proteins were confidently identified between the two sample types (Fig. 5 and supplemental Table S10). Of these, 409 and 451 proteins were also detected in the functionally enriched AMM and AMM + HS samples, respectively, but 152 and 82 confident proteins (AMM and AMM + HS, respectively) were not observed in the global analysis. For example, the MAPK MpkA was confidently identified in the enriched AMM and AMM + HS samples (∼15% coverage) but not in the global analysis samples (<10% coverage). The disparity between confident identifications in the global and functionally enriched samples confirms that we are observing a subset of proteins based on enzyme reactivity and not simply protein abundance.
FIGURE 5.
Global ABPP and LC-MS analysis of A. fumigatus from AMM 24 h and AMM + HS 24 h culture. Shown is a comparison of all functionally enriched proteins with the global untargeted proteome analysis of gcl at 24 h. Abundance values are scaled such that a range of −2 to 2 (dark blue to dark red) represents a relative intensity difference of 17 (log2). Each enriched column represents the average of at least three biological replicates. Boxes denote ∼200 proteins observed in global analysis that were not observed in the ABPP analysis and ∼100 proteins observed in ABPP samples that were not observed in global analysis. Differences in reactive protein abundances between ABPP samples correlate to gel-based analysis. See supplemental Table S10 for expanded details.
To further validate the mass spectrometry data, alcohol dehydrogenase and citrate synthase enzyme assays were performed. By MS analysis, a >15-fold change was observed in the alcohol dehydrogenase (AFUA_7G01010, EC 1.1.1.1) reactivity over time in HS culture (supplemental Table S4A). The alcohol dehydrogenase assay showed a 2-fc in alcohol dehydrogenase activity between 24 and 48 h in AMM + 10% HS culture (Table 3). The assay confirmed higher enzyme reactivity in the 48-h culture, but the fc observed by spectrophotometric techniques was much lower than in MS-based analysis. The fc difference is attributed to the higher resolution of MS-based measurements. Citrate synthase (AFUA_5G04230, EC 2.3.3.8, fc = 11.4) activity also increased over time by 2-fold in human serum culture according to the colorimetric enzyme assay (Table 3), which confirms the MS data showing that citrate synthase reactivity is higher in AMM + HS at 48 h (supplemental Table S4A). These enzyme assays independently confirm the ABPP and quantitative AMT tag analysis approach.
TABLE 3.
Enzyme assay results
Enzyme assay results for citrate synthase and alcohol dehydrogenase in the AMM+HS_24 h to AMM+HS_48 h comparison. Units, μmole/ml/min. Time point absorbance was obtained in triplicate, and Student's t-test was applied for each data point. Citrate synthase, p-value < 0.05; alcohol dehydrogenase, p value < 0.1. Units, μmole/ml/min.
| Enzyme | AMM + HS 24 h | AMM + HS 48 h | -Fold change (48 h/24 h) |
|---|---|---|---|
| units | units | ||
| Citrate synthase | 0.04 | 0.06 | 1.5 |
| Alcohol dehydrogenase | 0.008 | 0.019 | 2.4 |
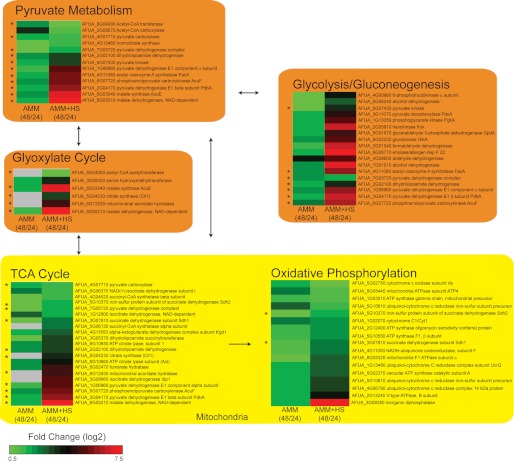
In an effort to visualize changes occurring over time in the presence and absence of HS, confidently identified proteins were mapped onto KEGG pathways. We found 100, 56, 44, 55, and 14% coverage of the TCA, glycolysis/gluconeogenesis, pyruvate metabolism, glyoxylate, and oxidative phosphorylation pathways, respectively (Fig. 6) when all proteins with an |fc| of >2 were used. ABPP revealed that primary metabolism and energy production significantly increase as the fungus ages in HS culture, whereas in minimal medium, the increase of enzyme reactivity within these pathways is much lower. Reactivity is protein dependent, not pathway dependent, as can be seen by negative and positive fc within a particular pathway. This reveals that some steps within a pathway may be more active than others and provides insight into fundamental processes. For example, the alcohol dehydrogenase (AFUA_7G01010) and pyruvate decarboxylase PdcA (AFUA_3G11070), both involved in fermentation within the glycolysis pathway, have higher fc between time points than dihydrolipoamide dehydrogenase (AFUA_2G02100) involved in acetyl-CoA production.
FIGURE 6.
KEGG pathway analysis. All proteins with |fc| > 2 were mapped onto KEGG glycolysis/gluconeogenesis, TCA cycle, pyruvate metabolism, glyoxylate cycle, and oxidative phosphorylation pathways. The yellow color indicates processes occurring in the mitochondria, and the orange represents processes occurring in the cytosol. Arrows between pathways indicate that pathway products are used in other pathways. The heat maps within each pathway represent -fold change of 0.5–7.5 (log2 scale) calculated for enzyme reactivity over time for each condition (i.e. AMM + HS 24 h versus AMM + HS 48 h and AMM 24 h versus AMM 48 h). Gray boxes represent proteins that did not make a 10% protein coverage cut-off. *, protein maps to more than one KEGG pathway.
DISCUSSION
ABPP provides a means for the functional enrichment of a biological system by chemical probe interrogation. Herein, the two new employed ABPs included a general vinyl sulfonate electrophile and a serine hydrolase-selective fluorophosphonate as reactive groups to target functional enzymes and click chemistry-compatible alkynes for downstream fluorescent or LC-MS/MS-based reporting of ABP-labeled enzymes. Probes were used simultaneously in a multiplexed fashion to facilitate efficient use of material and mass spectrometry instrumentation. Multiplexed ABPP of A. fumigatus using 1 and 2 resulted in the isolation of multiple protein families from a complex proteome. Vinyl sulfonate esters react with cysteine residues by Michael addition of the sulfur to the electrophilic vinyl sulfonate (49). Furthermore, an azide derivative of 1 inhibited cysteine-dependent protein phosphatases (33). Usage of the general cysteine-reactive vinyl sulfonate ester and serine hydrolase-specific ABPs allowed enrichment of reactive proteins independent of protein abundance.
ABPP of A. fumigatus over two cross-culture (AMM versus AMM + HS) and temporal comparisons (24 h versus 48 h) showed two key trends; 1) higher enzyme reactivity was always observed in minimal medium, and 2) enzyme reactivity increased over time regardless of growth condition. Within these trends, we observed fluctuations in primary metabolic activity, energy production, and stress response pathways. We hypothesized that growth of the fungus in HS would promote nutrient scavenging and limit de novo biosynthesis due to available proteins and lipids in serum because A. fumigatus can adapt to changing nutrient availability (7). A. fumigatus can use amino acids as both carbon and nitrogen sources, and it can degrade extracellular material by proteinase excretion and uptake of degradation products (13). HS contains 60–80 mg of protein/ml and salts, lipids, amino acids, and sugars, all of which can be utilized by A. fumigatus (50). Indeed, the growth curve generated during this study shows that in liquid culture, the presence of HS induces early logarithmic growth and increases the production of fungal biomass (Fig. 1).
The addition of HS to the culture medium induced a large shift in functional enrichment of A. fumigatus proteins. At both early and late time points, enzyme reactivity was lower in the presence of HS when compared with minimal medium. Two factors may influence the higher enzyme reactivity pattern for AMM culture. First, in minimal medium, the fungus must synthesize all necessary components for growth instead of acquiring them from sera as exemplified by the delay in log phase growth for AMM culture (Fig. 1). Second, Cagas et al. (51) observed high expression of proteins associated with translation and aerobic respiration during early culture when compared with conidia. According to the growth curve, AMM culture is in an earlier phase than HS culture, which may result in higher translation and metabolic activity as the fungus approaches log phase (32).
In HS culture, low reactivity of enzymes associated with aerobic metabolism and protein biosynthesis is consistent with low mRNA levels found in A. fumigatus germlings from murine lung infection (52). Whereas enzyme reactivity associated with metabolic pathways was low at 24 h, enzyme reactivity associated with protein biosynthesis and cell cycle regulation were low at 48 h (Table 2), which is consistent with logarithmic versus stationary growth. During log phase growth, most of the organism biomass is dedicated to hyphae elongation, cellular component generation, and transcription. During stationary phase, when the fungus is no longer actively dividing and has exhausted exogenous nutrients, the fungus can metabolize energy stores, such as starch, glycogen, lipids, and peptides, for cell turnover and maintenance of cell integrity, causing high enrichment of energy and metabolic functional categories (Fig. 6) (32). Cellular components can be broken down and transported from older internal compartments to the newer peripheral hyphae, allowing limited growth, which may account for the enrichment of protein fate, transport, transcription, and protein binding at 48 h of growth. This ultimately leads to autolysis and cell death (32). The lack of the protein biosynthesis category during stationary phase is consistent with the irreversible loss or slowing of Aspergillus niger protein synthesis during fungal aging and autolysis (32).
Only the acetyl-CoA carboxylase, associated with fatty acid biosynthesis, had higher reactivity in the presence of HS at 24 h. Acetyl-CoA carboxylase catalyzes the committed step in fatty acid biosynthesis, and the Saccharomyces cerevisiae (ACC1) and Aspergillus nidulans (accA) orthologs are necessary for growth (53, 54) and have a role in transport of proteins into the nucleus (55). Its high reactivity in AMM + HS 24 h is consistent with log phase growth, where carbohydrates and energy are plentiful and the fungus requires fatty acids for phospholipid biosynthesis (53). The smaller -fold difference between 48 and 24 h in HS culture may correspond with nutrient depletion and no additional accumulation of biomass during stationary phase. Notably, the acetyl-CoA carboxylase activity increases over time in minimal medium culture such that there is no significant change in enzyme reactivity between the presence and absence of HS at 48 h (Table 2). The large fc (∼14) observed in the AMM 24 h and AMM 48 h reflects higher reactivity at mid-log phase growth than at late lag phase/early log phase growth (54). The early high reactivity of the A. fumigatus acetyl-CoA carboxylase in the presence of HS components may provide an opportunity for early detection of fungal load in host tissue (56).
At stationary phase (48 h) in HS culture, two unique proteins were observed (supplemental Table S2C). The ark1 is an aldehyde reductase that converts acetaldehyde to ethanol in the gluconeogenesis/glycolysis and pyruvate metabolism cycles (31). The unique observation and high reactivity of this protein in the AMM + HS at 48 h suggests up-regulation of fermentation possibly due to O2 limitation in aging liquid culture medium (57). Furthermore, the akr1 transcript was up-regulated in the presence of reactive oxygen species (ROS) and neutrophils (58, 59), suggesting a role in response to oxidative stress. The glr1 is a glutathione oxidoreductase whose ortholog helps balance glutathione and glutathione disulfide ratios and redox homeostasis in S. cerevisiae during stationary phase (60). In the phytopathogenic fungus Fusarium decemcellulare, glr activity increased by 5-fold during stationary phase (61). The high enzyme reactivity of these two unique proteins may be associated with a link between metabolism/respiration and ROS generation (62). A connection between up-regulation of glycolysis, ethanol production, and ROS formation due to increased aerobic respiration in an A. fumigatus GTPase RacA mutant has been made previously (63). The observation of these unique enzymes using ABPP may be useful for diagnostic purposes because it is known that conidia and hypha encounter ROS (64), oxygen limitation (65), and alternative carbon sources during infection (66).
As the fungus aged in minimal medium, the functional pathways with high activity were quite different from those during early growth. At 24 h of growth, the log phase is just beginning, and the high level of metabolism, energy production, biosynthesis, and transcription-associated protein activity correlates with initial exit from dormancy (51), germ tube elongation, and hyphal extension (32). During continued log phase, the cell cycle regulators H3 and SkpA had high reactivity because during log phase growth hyphae septation and mitosis are occurring rapidly (32). The S. cerevisiae ortholog of H3 is crucial for activating the spindle assembly checkpoint in response to errors during mitosis (67). Likewise, the S. cerevisiae ortholog Skp1p is necessary for cell cycle progression and may play a regulatory role in activation of a cell cycle checkpoint (68). Finally, as the medium nutrients are depleted and the fungus can no longer maintain exponential growth, the fungus approaches stationary phase, and enzymes associated with stress response and secondary metabolism are highly enriched (Table 2). The nrps14, Glim-Like O-methyltransferase (AFUA_3G11920), Erg1, oxidoreductase 2OG-Fe (II) oxygenase family (AFUA_3G00800), HAD superfamily hydrolase (AFUA_5G08270, down regulated under hypoxia), Ure2-like glutathione Stransferase (AFUA_4G14530), and a conserved hypothetical protein (AFUA_8G00430) transcripts were altered in response to hypoxia. The fructosyl amino acid oxidase (AFUA_6G03440) transcript was up regulated in the presence of the antifungal drug voriconazole. It is intriguing that approaching stationary phase induces an enzyme reactivity response similar to hypoxia and azole responses.
A link between aging fungal culture and oxidative stress has been observed previously (69). In S. cerevisiae, tight control of cellular redox homeostasis, including mitochondrial production of ROS, is necessary for survival as the organism approaches and enters stationary growth phase (60). Six proteins with high reactivity at 48 h in minimal medium, the fgaCat, Arp2, cytochrome P450 (AFUA_5G02610), OrdA-like cytochrome P450 oxidoreductase (AFUA_8G00510), and flavin-binding monooxygenase-like protein (AFUA_4G09220), are annotated with redox regulatory processes, response to oxidative stress, and hydrogen peroxide catabolic processes and may play a role in controlling cellular redox homeostasis in A. fumigatus (Table 2). The high reactivity of the oxidative stress response regulator MpkA under these conditions supports this hypothesis (70). ROS accumulation may trigger an apoptotic response in A. fumigatus as it enters stationary phase, and studies found that a rapid loss of mycelia viability (>95%) occurred soon after stationary phase began (71). As stationary phase begins, the high levels of enzyme reactivity associated with oxidative stress correlates could be due to increasing ROS accumulation.
A. fumigatus can undergo autophagy in response to aging and nutrient depletion stress (72). This process allows limited growth of the fungus at the mycelium periphery to enable foraging of new nutritional sources in the environment and conidiation in response to starvation (73). Conidia are more robust to environmental stressors and can be a means of survival after nutrients cannot sustain growth (74). The two conidial melanin pigment biosynthetic proteins Arp1 and Ayg2 were previously found to be up-regulated in conidia when compared with mycelia and in swollen conidia at 4 h of germination (51, 75). In addition to possible roles in oxidative stress response, the high reactivity of these proteins at 48 h in minimal growth suggests that A. fumigatus is producing conidia under these conditions. In contrast, these pigment proteins were not significantly changing over time in HS culture, and at each time point enzyme reactivity levels were higher in minimal medium. This suggests that HS culture conidia formation is suppressed.
The drastic differences in protein reactivity during the aging process in the presence and absence of HS were visualized by mapping confidently identified proteins onto KEGG pathways. The generality of probe 1 allowed capture of many proteins within primary metabolism and energy pathways (Fig. 6). By using heat maps to express fc, we observed that proteins involved in the glycolysis/gluconeogenesis, glyoxylate and TCA pathways have greater probe reactivity over time in HS culture than in minimal medium. The high reactivity of pyruvate decarboxylase pdc and alcohol dehydrogenase adh1 within these pathways is consistent with a switch from aerobic respiration to fermentation (65). High reactivity of enzymes within the gluconeogenesis pathway and TCA pathway is further evidence that ethanol is being used as a carbon source (75, 76). The apparent switch to fermentation processes at 48 h of growth may be due to reduced dissolved O2 levels in the medium (57) and reduced efficient transfer of O2 through the pellet (32), thereby creating a hypoxic-like environment. Due to the differences in growth state between the culture conditions at 48 h, it is difficult to determine which effects are attributable to the availability of human serum components and which result from stationary phase growth. Instead, the enhanced biomass production, early log phase, and stationary phase are a direct result of HS culture. A comparison of biomass obtained from stationary phase in AMM and AMM + HS culture would show if these proteins and the metabolic change during stationary phase are truly unique to HS culture or growth phase-dependent.
It is apparent that ABPP using 1 and 2 coupled with a label-free quantitative AMT tag proteomics approach showed different active functional states between two growth conditions and two time points and is a powerful tool in analyzing a complex filamentous fungus. As expected, HS induces early log phase growth and an increase in biomass production. Whereas metabolism and energy function are greatly increasing from mid-log phase growth to stationary phase in the presence of HS, in the absence of HS, the fungus appears to be responding to stress as it approaches stationary phase. The low reactivity of stress response and secondary metabolism proteins observed in HS culture when compared with AMM suggests that A. fumigatus is not encountering the same stressors during growth in HS, possibly due to the ability to use nutrients present in HS. The ability of A. fumigatus to adapt well to a host environment is considered one of its virulence factors, and our study clearly shows adaptation to a complex environment. Furthermore, the high reactivity of stress response proteins observed from minimal medium biomass at 48 h provides some insight into functional activity during nutrient deprivation and the aging process. Because exogenous stressors were not applied in our experiment, it would be beneficial to observe enzyme reactivity in the presence of antifungal agents and under hypoxic conditions.
Our results show on a protein reactivity level biological and metabolic pathways and mechanisms of A. fumigatus likely to be operative in opportunistic infections because in the human lung, the fungus encounters stress due to immune response and alternative carbon sources such as human proteins and lipids. Furthermore, the identification of highly reactive unique enzymes, indicating fungal presence, load, and fungal homeostasis, may provide an opportunity for diagnosis of IA. In combination with these methods, cross-species comparisons and incorporation of host responses to Aspergillus infection could yield further insights into the complexity of A. fumigatus pathogenesis during IA.
Supplementary Material
Acknowledgments
We thank the Biological Separations and Mass Spectrometry group and the Chemical and Biological Processes Development Group for helpful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant (NIH), NCRR, Grant 5P41RR018522-10 and NIH, NIGMS, Grant 8 P41 GM103493-10. This work was also supported in part by the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory (PNNL), a multiprogram national laboratory operated by Battelle for the United States Department of Energy under Contract DE-AC05-76RL01830. This work used instrumentation and capabilities developed under support from the National Institutes of Health (Grant P41 GM103493-10) and the United States Department of Energy Office of Biological and Environmental Research (DOE/BER). Work was performed in the Environmental Molecular Sciences Laboratory, a DOE/BER national scientific user facility at PNNL.
The raw LC-MS/MS data and SEQUEST search results for all biological replicates and culture conditions have been deposited in the Biological MS Data and Software Distribution Center data repository under publication number 1057. The data can be accessed at http://omics.pnl.gov/view/publication_1057.html.

This article contains supplemental Tables S1–S10 and Figs. S1–S5.
- IA
- invasive aspergillosis
- ABPP
- activity-based protein profiling
- ABP
- activity-based probe
- AMT
- accurate mass and time
- HS
- human serum
- fc
- fold change
- AMM
- Aspergillus minimal medium
- ROS
- reactive oxygen species
- gcl
- global cell lysate.
REFERENCES
- 1. Latgé J. P. (1999) Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dagenais T. R., Keller N. P. (2009) Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22, 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Michael A., Pfaller P. G., Wingard J. R. (2006) Invasive fungal pathogens. Current epidemiological trends. Clin. Infect. Dis. 43, S3–S14 [Google Scholar]
- 4. Lai C. C., Tan C. K., Huang Y. T., Shao P. L., Hsueh P. R. (2008) Current challenges in the management of invasive fungal infections. J. Infect. Chemother. 14, 77–85 [DOI] [PubMed] [Google Scholar]
- 5. Hummel M., Buchheidt D. (2007) Molecular and serological diagnosis of invasive aspergillosis. New answers to old questions? Mycoses 50, 18–23 [DOI] [PubMed] [Google Scholar]
- 6. Denning D. W., Hope W. W. (2010) Therapy for fungal diseases. Opportunities and priorities. Trends Microbiol. 18, 195–204 [DOI] [PubMed] [Google Scholar]
- 7. Rhodes J. C., Askew D. S. (2010) Aspergillus fumigatus. in Cellular and Molecular Biology of Filamentous Fungus (Borkovich K. A., Ebbole D. J., eds) pp. 697–716, American Society for Microbiology, Washington D. C [Google Scholar]
- 8. Hohl T.M., Feldmesser M. (2007) Aspergillus fumigatus. Principles of pathogenesis and host defense. Eukaryot. Cell 6, 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhabhra R., Askew D.S. (2005) Thermotolerance and virulence of Aspergillus fumigatus. Role of the fungal nucleolus. Med. Mycol. 43, S87–S93 [DOI] [PubMed] [Google Scholar]
- 10. Willger S. D., Grahl N., Cramer R. A. (2009) Aspergillus fumigatus metabolism. Clues to mechanisms of in vivo fungal growth and virulence. Med. Mycol. 47, S72–S79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliver B. G., Panepinto J. C., Askew D. S., Rhodes J. C. (2002) cAMP alteration of growth rate of Aspergillus fumigatus and Aspergillus niger is carbon source-dependent. Microbiology 148, 2627–2633 [DOI] [PubMed] [Google Scholar]
- 12. Fleck C. B., Schöbel F., Brock M. (2011) Nutrient acquisition by pathogenic fungi. Nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 301, 400–407 [DOI] [PubMed] [Google Scholar]
- 13. Schoberle T., May G. (2007) Fungal genomics. A tool to explore central metabolism of Aspergillus fumigatus and its role in virulence. Adv. Genet. 57, 263–283 [DOI] [PubMed] [Google Scholar]
- 14. Kniemeyer O. (2011) Proteomics of eukaryotic microorganisms. The medically and biotechnologically important fungal genus Aspergillus. Proteomics 11, 3232–3243 [DOI] [PubMed] [Google Scholar]
- 15. Lenz T., Fischer J. J., Dreger M. (2011) Probing small molecule-protein interactions. A new perspective for functional proteomics. J. Proteomics 75, 100–115 [DOI] [PubMed] [Google Scholar]
- 16. Cravatt B. F., Wright A. T., Kozarich J. W. (2008) Activity-based protein profiling. From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 17. Zimmer J. S., Monroe M. E., Qian W. J., Smith R. D. (2006) Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 25, 450–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gifford A. H., Klippenstein J. R., Moore M. M. (2002) Serum stimulates growth of and proteinase secretion by Aspergillus fumigatus. Infect. Immun. 70, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hissen A. H., Chow J. M., Pinto L. J., Moore M. M. (2004) Survival of Aspergillus fumigatus in serum involves removal of iron from transferrin. The role of siderophores. Infect. Immun. 72, 1402–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toyotome T., Yamaguchi M., Iwasaki A., Watanabe A., Taguchi H., Qin L., Watanabe H., Kamei K. (2012) Fetuin A, a serum component, promotes growth and biofilm formation by Aspergillus fumigatus. Int. J. Med. Microbiol. 302, 108–116 [DOI] [PubMed] [Google Scholar]
- 21. Rodrigues A. G., Araujo R., Pina-Vaz C. (2005) Human albumin promotes germination, hyphal growth, and antifungal resistance by Aspergillus fumigatus. Med. Mycol. 43, 711–717 [DOI] [PubMed] [Google Scholar]
- 22. Shimizu K., Keller N.P. (2001) Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kägi J. H., Vallee B. L. (1960) The role of zinc in alcohol dehydrogenase. J. Biol. Chem. 235, 3188–3192 [PubMed] [Google Scholar]
- 24. Livesay E. A., Tang K., Taylor B. K., Buschbach M. A., Hopkins D. F., LaMarche B. L., Zhao R., Shen Y., Orton D. J., Moore R. J., Kelly R. T., Udseth H. R., Smith R. D. (2008) Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 80, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 26. Kim S., Gupta N., Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra. A strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monroe M. E., Tolić N., Jaitly N., Shaw J. L., Adkins J. N., Smith R. D. (2007) VIPER. An advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 28. Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G., 2nd, Anderson G. A., Smith R. D. (2008) DAnTE. A statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arnaud M. B., Chibucos M. C., Costanzo M. C., Crabtree J., Inglis D. O., Lotia A., Orvis J., Shah P., Skrzypek M. S., Binkley G., Miyasato S. R., Wortman J. R., Sherlock G. (2010) The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein, and sequence information for the Aspergillus research community. Nucleic Acids Res. 38, D420–D427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Priebe S., Linde J., Albrecht D., Guthke R., Brakhage A. A. (2011) FungiFun. A web-based application for functional categorization of fungal genes and proteins. Fungal Genet. Biol. 48, 353–358 [DOI] [PubMed] [Google Scholar]
- 31. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012) KEGG for integration and interpretation of large scale molecular data sets. Nucleic Acids Res. 40, D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Papagianni M. (2004) Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 22, 189–259 [DOI] [PubMed] [Google Scholar]
- 33. Liu S., Zhou B., Yang H., He Y., Jiang Z. X., Kumar S., Wu L., Zhang Z. Y. (2008) Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein-tyrosine phosphatases. J. Am. Chem. Soc. 130, 8251–8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weerapana E., Simon G. M., Cravatt B. F. (2008) Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 4, 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barglow K. T., Cravatt B. F. (2004) Discovering disease-associated enzymes by proteome reactivity profiling. Chem. Biol. 11, 1523–1531 [DOI] [PubMed] [Google Scholar]
- 36. Böttcher T., Sieber S.A. (2008) β-Lactones as privileged structures for the active-site labeling of versatile bacterial enzyme classes. Angew. Chem. Int. Ed. Engl. 47, 4600–4603 [DOI] [PubMed] [Google Scholar]
- 37. Gillet L. C., Namoto K., Ruchti A., Hoving S., Boesch D., Inverardi B., Mueller D., Coulot M., Schindler P., Schweigler P., Bernardi A., Gil-Parrado S. (2008) In-cell selectivity profiling of serine protease inhibitors by activity-based proteomics. Mol. Cell Proteomics 7, 1241–1253 [DOI] [PubMed] [Google Scholar]
- 38. Schmidinger H., Hermetter A., Birner-Gruenberger R. (2006) Activity-based proteomics. Enzymatic activity profiling in complex proteomes. Amino Acids 30, 333–350 [DOI] [PubMed] [Google Scholar]
- 39. Köster H., Little D. P., Luan P., Muller R., Siddiqi S. M., Marappan S., Yip P. (2007) Capture compound mass spectrometry. A technology for the investigation of small molecule protein interactions. Assay Drug Dev. Technol. 5, 381–390 [DOI] [PubMed] [Google Scholar]
- 40. Wallwey C., Matuschek M., Xie X. L., Li S. M. (2010) Ergot alkaloid biosynthesis in Aspergillus fumigatus. Conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org. Biomol. Chem. 8, 3500–3508 [DOI] [PubMed] [Google Scholar]
- 41. Tsai H. F., Wheeler M. H., Chang Y. C., Kwon-Chung K. J. (1999) A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181, 6469–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maiya S., Grundmann A., Li X., Li S. M., Turner G. (2007) Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. ChemBioChem 8, 1736–1743 [DOI] [PubMed] [Google Scholar]
- 43. Lodeiro S., Xiong Q., Wilson W. K., Ivanova Y., Smith M. L., May G. S., Matsuda S. P. (2009) Protostadienol biosynthesis and metabolism in the pathogenic fungus Aspergillus fumigatus. Org. Lett. 11, 1241–1244 [DOI] [PubMed] [Google Scholar]
- 44. da Silva Ferreira M. E., Malavazi I., Savoldi M., Brakhage A. A., Goldman M. H., Kim H. S., Nierman W. C., Goldman G. H. (2006) Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50, 32–44 [DOI] [PubMed] [Google Scholar]
- 45. Vödisch M., Scherlach K., Winkler R., Hertweck C., Braun H. P., Roth M., Haas H., Werner E. R., Brakhage A. A., Kniemeyer O. (2011) Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J. Proteome Res. 10, 2508–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai H. F., Chang Y. C., Washburn R. G., Wheeler M. H., Kwon-Chung K. J. (1998) The developmentally regulated alb1 gene of Aspergillus fumigatus. Its role in modulation of conidial morphology and virulence. J. Bacteriol. 180, 3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valiante V., Heinekamp T., Jain R., Härtl A., Brakhage A.A. (2008) The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet. Biol. 45, 618–627 [DOI] [PubMed] [Google Scholar]
- 48. Ferreira M. E., Colombo A. L., Paulsen I., Ren Q., Wortman J., Huang J., Goldman M. H., Goldman G. H. (2005) The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus. Med. Mycol. 43, S313–S319 [DOI] [PubMed] [Google Scholar]
- 49. Roush W. R., Gwaltney S. L., Cheng J., Scheidt K. A., McKerrow J. H., Hansell E. (1998) Vinyl sulfonate esters and vinyl sulfonamides. Potent, irreversible inhibitors of cysteine proteases. J. Am. Chem. Soc. 120, 10994–10995 [Google Scholar]
- 50. Krappmann S. (2003) in Aspergillus fumigatus and Aspergillosis (Latgé J.-P., Steinbach W. J., eds) pp. 63–74, ASM Press, Washington DC [Google Scholar]
- 51. Cagas S. E., Jain M. R., Li H., Perlin D. S. (2011) The proteomic signature of Aspergillus fumigatus during early development. Mol. Cell Proteomics 10, M111.010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McDonagh A., Fedorova N.D., Crabtree J., Yu Y., Kim S., Chen D., Loss O., Cairns T., Goldman G., Armstrong-James D., Haynes K., Haas H., Schrettl M., May G., Nierman W. C., Bignell E. (2008) Sub-telomere-directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 4, e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hasslacher M., Ivessa A. S., Paltauf F., Kohlwein S. D. (1993) Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 268, 10946–10952 [PubMed] [Google Scholar]
- 54. Morrice J., MacKenzie D. A., Parr A. J., Archer D. B. (1998) Isolation and characterization of the acetyl-CoA carboxylase gene from Aspergillus nidulans. Curr. Genet. 34, 379–385 [DOI] [PubMed] [Google Scholar]
- 55. Gao H., Sumanaweera N., Bailer S. M., Stochaj U. (2003) Nuclear accumulation of the small GTPase Gsp1p depends on nucleoporins Nup133p, Rat2p/Nup120p, Nup85p, Nic96p, and the acetyl-CoA carboxylase Acc1p. J. Biol. Chem. 278, 25331–25340 [DOI] [PubMed] [Google Scholar]
- 56. Dorr P. K., Parkinson T., Bulawa C. E. (2003) U.S. Patent 6, 514, 726B
- 57. Taubitz A., Bauer B., Heesemann J., Ebel F. (2007) Role of respiration in the germination process of the pathogenic mold Aspergillus fumigatus. Curr. Microbiol. 54, 354–360 [DOI] [PubMed] [Google Scholar]
- 58. Sugui J. A., Kim H. S., Zarember K. A., Chang Y. C., Gallin J. I., Nierman W. C., Kwon-Chung K. J. (2008) Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS ONE 3, e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oosthuizen J. L., Gomez P., Ruan J., Hackett T. L., Moore M. M., Knight D. A., Tebbutt S. J. (2011) Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS ONE 6, e20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drakulic T., Temple M. D., Guido R., Jarolim S., Breitenbach M., Attfield P. V., Dawes I. W. (2005) Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and aging in Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1215–1228 [DOI] [PubMed] [Google Scholar]
- 61. Gessler N., Aver'yanov A., Belozerskaya T. A. (2007) Reactive oxygen species in regulation of fungal development. Biochemistry 72, 1091–1109 [DOI] [PubMed] [Google Scholar]
- 62. Bai Z., Harvey L. M., McNeil B. (2003) Oxidative stress in submerged cultures of fungi. Crit. Rev. Biotechnol. 23, 267–302 [DOI] [PubMed] [Google Scholar]
- 63. Li H., Barker B. M., Grahl N., Puttikamonkul S., Bell J. D., Craven K. D., Cramer R. A. (2011) The small GTPase RacA mediates intracellular reactive oxygen species production, polarized growth, and virulence in the human fungal pathogen Aspergillus fumigatus. Eukaryot. Cell 10, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McCormick A., Loeffler J., Ebel F. (2010) Aspergillus fumigatus. Contours of an opportunistic human pathogen. Cell. Microbiol. 12, 1535–1543 [DOI] [PubMed] [Google Scholar]
- 65. Grahl N., Puttikamonkul S., Macdonald J. M., Gamcsik M. P., Ngo L. Y., Hohl T. M., Cramer R. A. (2011) In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 7, e1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ibrahim-Granet O., Dubourdeau M., Latgé J. P., Ave P., Huerre M., Brakhage A. A., Brock M. (2008) Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell. Microbiol. 10, 134–148 [DOI] [PubMed] [Google Scholar]
- 67. Luo J., Xu X., Hall H., Hyland E. M., Boeke J. D., Hazbun T., Kuo M. H. (2010) Histone H3 exerts a key function in mitotic checkpoint control. Mol. Cell. Biol. 30, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Connelly C., Hieter P. (1996) Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. SáMi L., Emri T., Pócsi I. (2001) Autolysis and aging of Penicillium chrysogenum cultures under carbon starvation. Glutathione metabolism and formation of reactive oxygen species. Mycol. Res. 105, 1246–1250 [Google Scholar]
- 70. Jain R., Valiante V., Remme N., Docimo T., Heinekamp T., Hertweck C., Gershenzon J., Haas H., Brakhage A. A. (2011) The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production, and iron adaptation in Aspergillus fumigatus. Mol. Microbiol. 82, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mousavi S. A., Robson G. D. (2003) Entry into the stationary phase is associated with a rapid loss of viability and an apoptotic-like phenotype in the opportunistic pathogen Aspergillus fumigatus. Fungal Genet. Biol. 39, 221–229 [DOI] [PubMed] [Google Scholar]
- 72. Palmer G. E., Askew D. S., Williamson P. R. (2008) The diverse roles of autophagy in medically important fungi. Autophagy 4, 982–988 [DOI] [PubMed] [Google Scholar]
- 73. Richie D. L., Fuller K. K., Fortwendel J., Miley M. D., McCarthy J. W., Feldmesser M., Rhodes J. C., Askew D. S. (2007) Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell 6, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Adams T. H., Wieser J. K., Yu J. H. (1998) Asexual Sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Teutschbein J., Albrecht D., Pötsch M., Guthke R., Aimanianda V., Clavaud C., Latgé J. P., Brakhage A. A., Kniemeyer O. (2010) Proteome profiling and functional classification of intracellular proteins from conidia of the human-pathogenic mold Aspergillus fumigatus. J. Proteome Res. 9, 3427–3442 [DOI] [PubMed] [Google Scholar]
- 76. Kniemeyer O., Lessing F., Scheibner O., Hertweck C., Brakhage A. (2006) Optimization of a two-dimensional gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr. Genet. 49, 178–189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.