Background: How the root of stereocilia tip link is designed to withstand mechanical stretching is poorly understood.
Results: Cadherin23 tail and harmonin form polymeric protein assemblies via nonsynergistic, multisite interactions.
Conclusion: The cadherin23 tail/harmonin/actin filament polymer can account for the mechanic strength required by the root of upper tip link.
Significance: The findings of this study suggest a mechanism for constructing a strong root for the stereocilia tip link.
Keywords: Cilia, Hair Cell, Hearing, Protein Structure, Protein-Protein Interactions, Usher Syndrome, Cadherin23, Harmonin, Tip Link
Abstract
Stereocilia tip links of inner ear hair cells are subjected to constant stretching during hair-bundle deflection, and accordingly are well designed to prevent from being broken by mechanical tensions. The roots of tip links, which couple tip links with the cytoskeleton, supposedly play important roles in withstanding large forces under stimulated conditions. The upper root of the tip link is mainly formed by the cytoplasmic tail of cadherin23 and its actin-anchoring protein harmonin. However, the detailed organization mode of the two proteins that gives rise to a strong upper root remains unclear. Here we show that the exon68-encoded peptide of cadherin23 can either interact with the N-terminal domain (NTD) of harmonin or form a homodimer. We demonstrate that the three harmonin binding sites of cadherin23, namely the NTD-binding motif, the exon68 peptide, and the C-terminal PDZ binding motif, do not synergize with each other in binding to harmonin, instead they facilitate formation of polymeric cadherin23/harmonin complexes. The exon68 peptide can promote the cadherin23/harmonin polymer formation via either binding to harmonin NTD or self-dimerization. We propose that the polymeric cadherin23/harmonin complex formed beneath the upper tip link membranes may serve as part of the stable rootlet structure for anchoring the tip links of stereocilia.
Introduction
Stereocilia bundle of the inner ear hair cell is mechanosensing organelle that converts mechanical forces evoked by sound waves and head movements into electric signals to generate the sense of hearing and balance. The mechanoelectrical transduction channel is present at the tips of all but the tallest stereocilia (1), and gated by a filamentous ultrastructure, called tip link, to control stereocilia depolarization and electric signal propagation (2, 3). Tip link connects the tip of one stereocilium to the side of the nearby taller stereocilium (4), and is formed by cadherin23 homodimer in trans association with protocadherin15 homodimer (5–8). Both cadherin23 and protocadherin15 are usher syndrome I, single-transmembrane adhesion proteins with a long extracellular cadherin repeats followed by a short cytoplasmic tail. Protocadherin15 forms the lower part of tip link, and has been proposed to be coupled with a mechanoelectrical transduction channel to participate in its gating. Cadherin23 assembles the upper part of tip link, and its cytoplasmic tail is anchored to the actin filaments of stereocilia via binding to the actin-binding protein harmonin (9). Deflection of stereocilia bundles induces stretching force on tip links, and the tensile stresses force them to pull open the coupled mechanoelectrical transduction channels and trigger the electrical signal transduction (3).
Although no experimental data are available so far to precisely determine the strength of mechanical forces applied onto a tip link during hair-bundle deflection, biophysical estimates of its stiffness (∼1 mN m−1) indicate that it should withstand forces of >100 pN (10). There is a report showing a pulling force of ∼150 pN is sufficient to pull out Band 3, an eight-transmembrane anion exchange channel associated with actin filaments, from the plasma membrane of red blood cells (11). Tip link is made of single-transmembrane proteins, and subjected to constant stretching forces ranging likely from 0 to >100 pN (3, 12). How tip link could sustain such strong and frequent stretching forces and avoid being uprooted from plasma membrane is an important question to understand the working mechanism of inner ear hair cells.
Harmonin has three isoforms: a, b, and c (Fig. 1A) (13). Harmonin-b plays the most important role in cadherin23 anchorage (14, 15). It contains a NTD and three PDZ domains together with two coiled-coil regions and a proline-serine-threonine-rich (PST) region situated between PDZ2 and PDZ3. On the one hand, harmonin binds to a N-terminal domain (NTD)4 binding motif (NBM) and PDZ binding motif (PBM) of cadherin23 cytoplasmic tail via its NTD and PDZ2, respectively (Fig. 1A) (16). On the other hand, harmonin can directly attach to actin filaments through its PST region (9), or indirectly via forming a complex with sans-myosin VIIa complex (17). Thus, a popular working model proposes that the upper root of the tip link is composed of a dimer of the cadherin23/harmonin complex attached to actin filaments (16).
FIGURE 1.
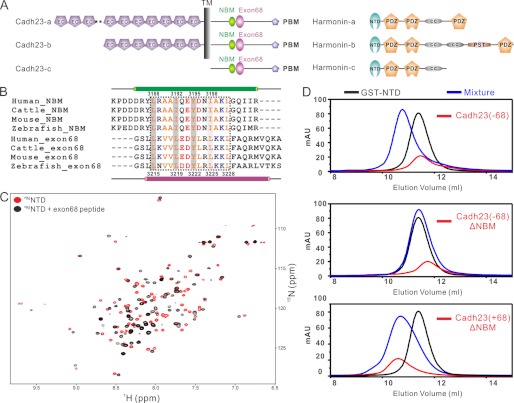
The exon68 peptide specifically binds to harmonin NTD. A, schematic diagram depicting the domain organizations of different isoforms of cadherin23 and harmonin. Exon68 can be further spliced out of all three cadherin23 isoforms. B, amino acid sequence alignment of cadherin23 NBM and the exon68-encoded peptide. The highly similar region of NBM and exon68 is highlighted within a black dotted line. The crucial residues involved in NTD binding are highlighted by gray background. C, overlay plot of the 1H,15N-HSQC spectra of the free NTD and the protein saturated with the exon68 peptide. D, analytical gel filtration analysis showing that the exon68-containing cadherin23 cytoplasmic tail specifically binds to harmonin NTD.
The major isoform of cadherin23 expressed in stereocilia contains a 35-amino acid sequence encoded by an alternatively spliced exon, exon68 (Fig. 1A) (18–22). Although cadherin23 is widely distributed in many tissues including brain, kidney, heart, and spleen, exon68 is exclusively expressed in the inner ear and retina (18, 19), implying its yet unknown but important functions in these two tissues. Previous studies of cadherin23/harmonin interaction either did not include the exon68 encoding residues (16), or did not account for its potential role in the interaction (19, 23, 24). In this study, we found that the exon68-encoded peptide can either bind to harmonin NTD or self-dimerize. Furthermore, the three harmonin binding sites of cadherin23, the exon68 peptide, NBM, and PBM, do not synergize with each other in binding to harmonin. Instead these three regions facilitate formation of concentration-dependent, polymeric cadherin23/harmonin complexes. Interestingly, exon68, either via binding to NTD or self-dimerization, can effectively promote the polymeric cadherin23/harmonin complex formation. Considering the special role of tip link as the mechanoelectrical transduction channel gating spring, as well as the specific expression of exon68 in the stereocilia bundle (20), we propose that exon68-facilitated cadherin23/harmonin polymer formation may provide a mechanistic explanation for the formation of the sturdy upper tip link root organized by the cdherin23/harmonin/actin filaments network.
EXPERIMENTAL PROCEDURES
Protein Preparation
The coding sequences of the full-length (FL) harmonin-a and NTD (residues 1–97) were PCR-amplified from human Ush1C (NCBI accession number NP_005700.2) and cloned into pET32a or pGEX4T-1 vector. The coding sequence of the cytoplasmic tail of cadherin23(±68) was PCR amplified from human Ush1D and cloned into pET32a vector. Different cadherin23 mutants and fragments were generated using standard PCR-based methods. The GB1-exon68 was generated by fusing the GB1 tag with the exon68-encoded sequence (amino acid sequence: GSLLKVVLEDYLRLKKLFAQRMVQKASSC—HSSISE). For generating the cadherin23(GCN4) chimera, the coding sequence of the GCN4 dimer (amino acid sequence: ARMKQLEDKIEELLSKIYHLENEIARLKKLIGER) is inserted into cadherin23 to replace the exon68 sequence. For the cadherin23(NBM) chimera, the coding sequence of part of the exon68 peptide (amino acid sequence: LKVVLEDYLRLKKL) is replaced with NBM (amino acid sequence: LRAAIQEYDNIAKL). All recombinant proteins were expressed in BL21(DE3) Escherichia coli cells and purified by using either Ni2+-nitrilotriacetic acid-agarose or GSH-Sepharose affinity chromatography, followed by size exclusion chromatography.
NMR Spectroscopy
The protein samples for NMR studies were concentrated to ∼0.2 mm for titration experiments and ∼0.6 mm for structural determinations in 100 mm potassium phosphate at pH 6.5. NMR spectra were acquired at 30 °C on Varian Inova 500 or 750 MHz spectrometers. Backbone and side chain resonance assignments were achieved by a combination of the standard heteronuclear correlation experiments and two-dimensional 1H NOESY experiments (25). Approximate interproton distance restraints were derived from two-dimensional 1H-NOESY, three-dimensional 15N-separated NOESY, and 13C-separated NOESY spectra. Structures were calculated with the program CNS (26). The figures were prepared with the programs MOLSCRIPT (27), PyMOL, and MOLMOL (28).
Analytical Gel Filtration Chromatography
Analytical gel filtration chromatography was carried out on an AKTA FPLC system (GE Healthcare). Proteins were loaded onto a Superose 12 10/300 GL column 20 (GE Healthcare) equilibrated with a buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm DTT, and 1 mm EDTA.
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed on a Beckman XL-I analytical ultracentrifuge equipped with an eight-cell rotor at 25 °C. The partial specific volume of different protein samples and the buffer density were calculated using the program SEDNTERP (29). The final sedimentation velocity data were analyzed and fitted to a continuous sedimentation coefficient distribution model using the program SEDFIT (29). The fitting results are further output to the Origin 7.0 (MicroCal) software.
For sedimentation equilibrium experiments, ∼110 μl of cadherin23(+68) at three different concentrations (A280 absorption about 0.3, 0.6, and 0.9, respectively) were loaded into a six-chamber analytical ultracentrifuge cuvette. As a blank, ∼120 μl of protein buffer (50 mm Tris-HCl, 100 mm NaCl, and 1 mm EDTA, pH 7.5) was also loaded into the adjacent chamber. The experiment was performed on a Beckman XL-I analytical ultracentrifuge equipped with an eight cell rotor under 13,000 rpm at 25 °C. The final sedimentation equilibrium data were analyzed and fitted to a monomer-dimer self-association model to obtain the dimerization Kd constant using the SEDPHAT software (29).
Isothermal Titration Calorimetry Assay
ITC measurements were carried out on a VP-ITC calorimeter (Microcal, North Hampton, MA) at 25 °C. All protein samples were in 50 mm Tris-HCl buffer at pH 7.5 plus 500 mm NaCl and 1 mm EDTA. The titration processes were performed by injecting 10-μl aliquots of the His-tagged cadherin23 cytoplasmic tail (450 μm) into His-tagged full-length harmonin-a (45 μm) at time intervals of 3 min to ensure that the titration curve returned to baseline. The titration data were analyzed with Origin7.0 from Microcal and fitted with a one-site binding model.
Large Complex Formation Assay
For the high-speed centrifugation assay, the full-length harmonin-a and cytoplasmic tail of cadherin23 were freshly purified and mixed at 1:1 molar ratio at the final concentration of 45 μm in a buffer composed of 50 mm Tris-HCl, 500 mm NaCl, 1 mm EDTA, and 1 mm DTT at pH 7.5 and incubated at 4 °C for 10 min. The samples were centrifuged by an Optima Max-XP ultracentrifuge (Beckman Coulter) with a TLA-55 rotor at 4 °C, 98,000 × g for 30 min. The supernatant and pellet were collected and analyzed by SDS-PAGE.
RESULTS
Exon68 Binds to the N-terminal Domain of Harmonin
Previously we found that the α-helix encoded by cadherin23 NBM binds to the solvent-exposed pocket of harmonin NTD through hydrophobic interactions involving Leu-3188, Ile-3192, and Tyr-3195 of NBM (16). Surprisingly, deletion of this NBM did not abolish the specific interaction between the cytoplasmic tail of cadherin23(+68) and harmonin NTD, indicating the existence of an additional NTD binding site in the cadherin23 tail (data not shown). Further NMR-based titration studies narrowed this additional NTD binding sequence down to the exon68-encoded region (data not shown). Interestingly, sequence alignment analysis revealed that, for each cadherin23 NBM residue essential for harmonin NTD binding, an identical or highly similar amino acid can be found in the corresponding positions of the exon68-encoded peptide (referred to as the exon68 peptide hereafter, Fig. 1B). Additionally, this exon68 peptide is also predicted to form an α-helix. The above analysis implies that the exon68 peptide may function as a NTD binder. To test this hypothesis, we synthesized the exon68 peptide (SLLKVVLEDYLRLKKLFAQR) and investigated its possible binding to harmonin NTD by titrating 15N-labeled NTD with the peptide. The obvious chemical shift changes of NTD after titrating with the exon68 peptide indicated the specific interaction between exon68 and NTD (Fig. 1C). To investigate whether the exon68 peptide in the intact cadherin23 tail can bind to NTD, we purified the recombinant, entire cytoplasmic tail of cadherin23. Our analytical gel filtration data demonstrated that deletion of NBM from cadherin23(−68) abrogated its interaction with harmonin NTD. In contrast, deletion of NBM from cadherin23(+68) did not disrupt its interaction with harmonin NTD (Fig. 1D), indicating that the exon68 segment of cadherin23 functions as a harmonin NTD binding sequence. Additionally, individual point mutations of the conserved hydrophobic residues within the exon68 region (e.g. L3215Q, L3219Q, and Y3222E) either disrupted or weakened the binding of the exon68 peptide to NTD, further supporting the specific interaction between the exon68 peptide and harmonin NTD (supplemental Fig. S1, A–C). As the control, replacements of Leu-3223, Leu-3225, and Leu-3228, which are aligned with noncrucial residues in NBM (Fig. 1A), individually with Gln, had no obvious effect on its binding to the NTD (supplemental Fig. S1, D–F).
The Exon68/NTD Complex Structure
To better understand the exon68 peptide/NTD interaction, we intended to solve the structure of the exon68 peptide/NTD complex using NMR spectrometry. First, we fused the exon68 peptide to the C terminus of NTD to get a stable 1:1 complex. However, as it occurred to the NTD/NBM complex (16), the solubility of the exon68 peptide/NTD complex was too low (<0.1 mm) for structure determination by NMR. Following what we did for the NBM/NTD complex (16), we substituted Val-3218 with Glu to increase the solubility of the exon68 peptide/NTD complex. And this mutation successfully improved the solubility of the complex to up to 0.5 mm without affecting the overall binding mode of the exon68 peptide and NTD as evidenced by the good overlay of the 1H,15N-HSQC spectra of the NTD complexes formed with the WT peptide and the mutant peptide, respectively (data not shown).
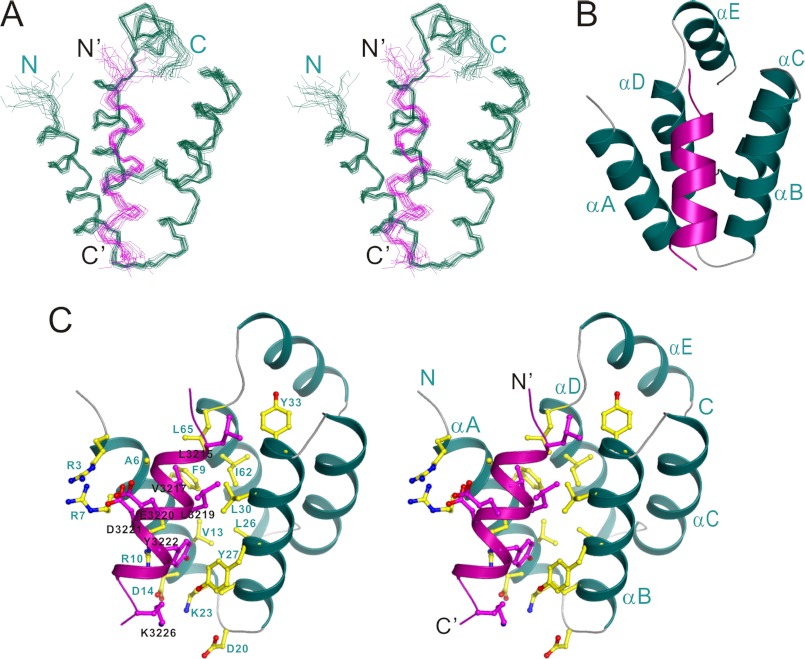
We determined the high resolution structure of the exon68/NTD complex by NMR spectroscopy (Table 1). Not surprisingly, in the complex structure the exon68 peptide adopts an α-helical conformation and occupies the same pocket of NTD as NBM does (Fig. 2, A and B). The interaction between the amphipathic exon68 α-helix and NTD buries a total of ∼760 Å2 surface area and is mainly mediated by hydrophobic interactions. Specifically, the hydrophobic side chains of Leu-3215, Val-3217, and Leu-3219 of the exon68 peptide extend into the hydrophobic pocket of the NTD formed by the side chains of Ala-6, Phe-9, Leu-30, Tyr-33, Ile-62, and Leu-65. Notably, the side chain of Val-3217 is deeply embedded in the hydrophobic pocket and completely occluded from the solvent. In addition, the aromatic side chain of exon68 Tyr-3222 inserts into another pocket located at the base of the αA/αB-helix hairpin formed by the hydrophobic side chains of Val-13, Leu-26, and Tyr-27 as well as the aliphatic side chain of Lys-23. The side chain of Tyr-3222 is further stabilized by Arg-10 through a cation-π interaction (Fig. 2C). Additional charge-charge interactions (e.g. between Asp-3221 of exon68 and Arg-7 of NTD) further contribute to the overall complex formation (Fig. 2C).
TABLE 1.
Structural statistics for the family of 20 NMR structures of harmonin NTD in complex with the exon68 peptide
| Statistics | |
|---|---|
| Distance restraints | |
| Intraresidue (i-j = 0) | 670 |
| Sequential (|i-j| = 1) | 386 |
| Medium range (2 ≤ |i-j| ≤ 4) | 362 |
| Long range(|i-j| ≥ 5) | 277 |
| Hydrogen bonds | 88 |
| Total | 1783 |
| Dihedral angle restraints | |
| Φ | 69 |
| Ψ | 69 |
| Total | 138 |
| Mean root mean square deviations from the experimental restraints | |
| Distance (Å) | 0.010 ± 0.000 |
| Dihedral angle (Å) | 0.166 ± 0.021 |
| Mean root mean square deviations from idealized covalent geometry | |
| Bond (Å) | 0.002 ± 0.0000 |
| Angle (°) | 0.355 ± 0.004 |
| Improper (°) | 0.192 ± 0.005 |
| Mean energies (kcal mol−1)a | |
| ENOEb | 13.23 ± 0.43 |
| Ecdihb | 0.23 ± 0.06 |
| EL-J | −356.77 ± 15.98 |
| Ramachandran plotc | |
| % Residues in the most favorable regions | 93.4 |
| Additional allowed regions | 5.7 |
| Generously allowed regions | 0.3 |
| Atomic rroot mean square differences (Å)d | |
| Backbone heavy atoms (N, Cα, and C′) | 0.42 |
| Heavy atoms | 0.97 |
a None of the structures exhibits distance violations greater than 0.3 Å or dihedral angle violations greater than 4°.
b The final values of the square-well NOE and dihedral angle potentials were calculated with force constants of 50 kcal mol−1Å−1 and 200 kcal mol−1rad−1.
c The PROCHECK-NMR program (www.ebi.ac.uk/thornton-srv/software/PROCHECK/) was used to assess the overall quality of the structures.
d Residues 3–78 of NTD plus the residues 3214–3226 of the cadherin23 exon68 peptide.
FIGURE 2.
The structure of the NTD/exon68 peptide complex. A, stereo view showing the backbones of 20 superimposed NMR structures of harmonin NTD in complex with the exon68 peptide. The exon68 peptides are drawn in purple. B, ribbon diagram of a representative NMR structure of the NTD/exon68 peptide complex. C, stereo view showing the detailed interactions of the exon68 peptide with the residues from harmonin NTD.
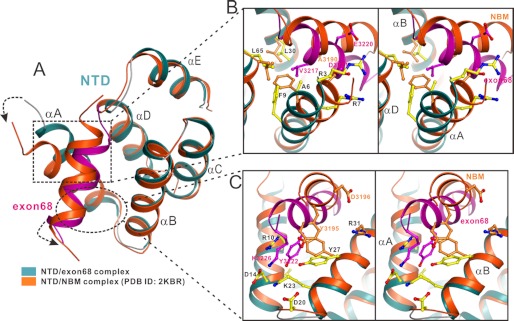
Because the exon68 peptide and NBM occupy the same NTD pocket, we are curious about which site has a stronger binding affinity. Unfortunately, the synthetic exon68 peptide is highly hydrophobic and not soluble in aqueous buffer for measuring its binding affinity to NTD using various direct binding assays. Fortunately, it is soluble when in complex with NTD. When the exon68 peptide was titrated into the 15N-labeled NTD, the peaks of NTD showed a slow exchange pattern in the NMR spectrum (supplemental Fig. S2, A and B). In contrast, titration of the NBM peptide showed a fast exchange pattern (supplemental Fig. S2C). Based on these NMR results, we conclude that the exon68 peptide has a stronger NTD binding affinity than NBM. Superimposing the two complex structures showed that although the overall conformations of the two complex structures are very similar, the relative orientations of bounded NBM and the exon68 peptide are noticeably different (Fig. 3A). Consistent with a stronger NTD binding affinity observed in the NMR study, the exon68 helix fits deeper into the cleft between the V-shaped αA/αB helix-hairpin by “pushing” the αA helix of NTD a bit more outwards (Fig. 3A). Although both Ala-3190 of NBM and Val-3217 of exon68 are embedded in the NTD hydrophobic pocket, the bulkier hydrophobic side chain of Val-3217 is much more favorable for binding to NTD compared with Ala-3190 in NBM (Fig. 3B). Additionally, the side chain of Tyr-3222 forms more extensive interactions in the exon68/NTD complex as described above (Fig. 2C), whereas the corresponding aromatic side chain of Tyr-3195 in the NBM/NTD complex stacks only with the side chain of Tyr-27 from NTD (Fig. 3C). Finally, the NBM/NTD complex further lacks charge-charge interactions observed in the exon68/NTD complex (Figs. 2A and 3, B and C). Taken together, these structural differences clearly explain why the exon68 peptide has a stronger NTD binding affinity than NBM.
FIGURE 3.
Structural comparison of NTD/exon68 peptide complex and NTD/NBM complex. A, superimposed schematic diagram showing the comparison of the NTD/exon68 peptide complex with the NTD/NBM complex (PDB access code 2KBR). The overall conformation of NTD is highly similar except the αA helix region in the two structures. The bounded cadherin23 NBM and exon68 peptide (purple) both adopt α-helical conformations but have different orientations in the two complexes. B and C, stereo views showing the detailed different orientations of NTD αA helix and the bounded cadherin23 α-helix in the two complexes. In particular, Val-3217 of the exon68 peptide makes much more extensive hydrophobic contacts with residues from NTD compared with Ala-3190 in NBM (B), and Tyr-3222 of exon68 peptide inserts deeper into another hydrophobic pocket in harmonin NTD than Tyr-3195 of NBM does (C). The weak charge-charge interactions formed between NTD and the exon68 peptide are also indicated in B and C.
Exon68 Dimerizes the Cytoplasmic Tail of Cadherin23
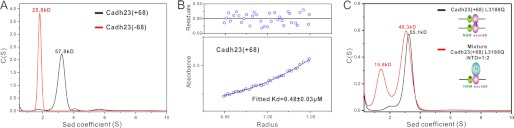
To our surprise, besides its NTD binding, the exon68 encoding sequence can induce dimerization of the cadherin23 cytoplasmic tail. Analytical ultracentrifugation analysis showed that the fitted molecular mass of the cadherin23(−68) tail is about ∼25.8 kDa, close to its theoretical monomer mass. In contrast, the cadherin23(+68) tail has a molecular mass of ∼57.8 kDa, which is about twice of the monomer mass, indicating that exon68 can induce dimer formation of the cadherin23 cytoplasmic tail (Fig. 4A). The dissociation constant (Kd) of the cadherin23(+68) tail dimer is 0.48 μm based on the sedimentation equilibrium analysis (Fig. 4B).
FIGURE 4.
Dimerization and NTD binding of exon68 are mutually exclusive. A, overlay plot of the sedimentation velocity analytical ultracentrifugation profiles of cadherin23(+68) and cadherin23(−68). B, equilibrium ultracentrifugation-based measurement of the dissociation constant for cadherin23(+68) dimer. C, molecular weight of the L3188Q-cadherin23(+68)/NTD complex determined by the sedimentation velocity analysis indicates that self-dimerization and NTD binding are mutually exclusive for the exon68 peptide.
We further mapped the minimal dimerization region of the cadherin23(+68) cytoplasmic tail using analytical ultracentrifugation. By fusing the exon68 sequence with the GB1 tag, we can get a soluble GB1-exon68 fusion protein. The GB1-exon68 protein forms nonspecific aggregates, indicating that exon68 alone is not sufficient for forming a stable dimer (supplemental Fig. S3, A and I). Its upstream fragments 2 and 3, and downstream fragment 6 help to form a stable dimer, and deletion of each of them individually weakens the stability of the dimer as shown by a broader dimer peak in each obtained sedimentation velocity profile (supplemental Fig. S3, A, E, H, and J). Fragments 1 and 7 are not required for dimerization, as deletion of each fragment individually did not affect the dimer stability (supplemental Fig. S3, A, D, and F). Interestingly, despite its high similarity to the exon68 peptide, NBM is not involved in dimer formation of the cadherin23(+68) tail (supplemental Fig. S3, A and G).
NTD Binding and Dimerization Are Mutually Exclusive for Exon68 Sequence
We next asked whether the exon68 sequence can simultaneously function as a harmonin NTD binder and a self-dimerization motif. To address this question, we substituted Leu-3188 with Gln to eliminate complications by disrupting the NBM/NTD interaction (16). The L3188Q-cadherin23(+68) mutant still forms a dimer (Fig. 4C). The molecular weight of the complex formed by NTD and L3188Q-cadherin23(+68) is ∼49.3 kDa (Fig. 4C), which is roughly equal to the theoretical mass of the 1:1 complex. This analysis clearly demonstrates that binding to NTD and self-dimerization are two mutually exclusive properties of the exon68-containing cadherin23 cytoplasmic tail. In agreement with this conclusion, mutations of the amino acids that directly participate in NTD binding (L3215Q, L3219Q, and Y3222E) also impaired dimer formation of the cadherin23(+68) cytoplasmic tail (supplemental Fig. S4).
Large Cadherin23(+68)/Harmonin Complex Formation Mediated by Multiple Nonsynergistic Binding Sites
We have demonstrated that the cadherin23(+68) cytoplasmic tail contains three harmonin binding sites: the harmonin NTD binding NBM and the exon68 sequence, and the PDZ2 binding PBM (Fig. 5A). Therefore, the interaction between cadherin23(+68) and harmonin are much more complicated than we have initially anticipated.
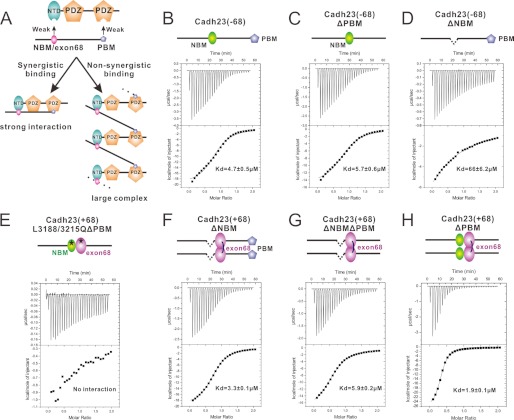
FIGURE 5.
NTD and PDZ2 do not synergize in binding to cadherin23. A, two models illustrating multiple site-mediated complex formation based on whether NTD and PDZ2 synergize with each other or not in binding to cadherin23. B-H, binding affinities of different forms of cadherin23 to the full-length harmonin-a determined by ITC. The L3188Q and L3215Q mutations on the cadherin23(+68) tail that abrogate its NTD binding as well as the exon68 peptide dimerization are indicated by black stars.
To determine what kind of complex can form between cadherin23 and the full-length harmonin, we first asked whether NTD and PDZ2 of harmonin can synergize with each other in binding to NBM/exon68 and PBM of cadherin23, respectively. If the bindings were synergistic, harmonin and the cadherin23 tail would form a 1:1 complex with an affinity much higher than those of the individual interaction sites. If the bindings were not synergistic, harmonin and the cadherin23 tail would form a concentration-dependent polymer with a binding affinity comparable with (or rather a simple sum of) the individual sites (see Fig. 5A for schematic diagrams of the two models). To address which model fits the cadherin23/harmonin interaction, we purified the cadherin23(±68) cytoplasmic tail with different deletions or mutations and the full-length harmonin-a protein, and measured binding affinities of their interactions using isothermal titration calorimetry. We did not use harmonin-b in our assay, as we were not able to obtain purified protein from the bacterial expression system. Because harmonin-a and harmonin-b share the same cadherin23 interaction motifs, we believe that the findings described in our study using harmonin-a can likely be extended to harmonin-b.
First, deletion of PBM (ΔPBM) and mutation of NBM and exon68 (L3188/3215Q) completely abolished the cadherin23/harmonin interaction (Fig. 5E), confirming that there are no additional harmonin binding sites in cadherin23 other than the three studied here. The dissociation constant for cadherin23(−68)ΔPBM/full-length harmonin-a is ∼5.7 μm (Fig. 5C). For the cadherin23(−68)ΔNBM/full-length harmonin-a interaction, the dissociation constant is ∼66 μm (Fig. 5D). If NTD and PDZ2 could synergize, the dissociation constant of the cadherin23(−68)/full-length harmonin-a complex should be much stronger than any single binding site, such as the characterized myosin VIIa MyTH4-FERM/sans, harmonin/sans interaction β-Catenin/E-Cadherin, Grb2 SH3 tandem/Sos, liprin-α/CASK, and Myosin VI CBD/Dab2 interactions (30–35). However, if they are nonsynergistic interactions, the dissociation constant of the cadherin23(−68)/full-length harmonin-a complex should be an additive value of each individual binding site, such as the interactions between two RNA binding domains of heterogeneous nuclear ribonucleoprotein A1 and a single-stranded RNA, and the Dap160/dynamin interaction (36, 37). The affinity of the cadherin23(−68)/full-length harmonin-a complex is ∼4.7 μm (Fig. 5B), roughly equal to the additive effect of NBM and PBM binding, suggesting that the NTD and PDZ2 do not synergize with each other in binding to cadherin23(−68). Similarly, the dissociation constant of the exon68 dimer (cadherin23(+68)ΔNBMΔPBM)/full-length harmonin-a complex is ∼5.9 μm (Fig. 5G). The cadherin23(+68)ΔNBM/full-length harmonin-a complex has a dissociation constant ∼3.3 μm (Fig. 5F), suggesting that the cadehrin23 exon68 peptide and PBM do not synergize with each other in binding to harmonin-a either. NBM and exon68 peptide cannot synergize with each other as they compete for harmonin NTD. Therefore, the multiple interaction sites between the cadherin23 tail and harmonin do not synergize with each other. Instead, the two proteins are predicted to form concentration-dependent polymers as depicted in the nonsynergistic binding model in Fig. 5A.
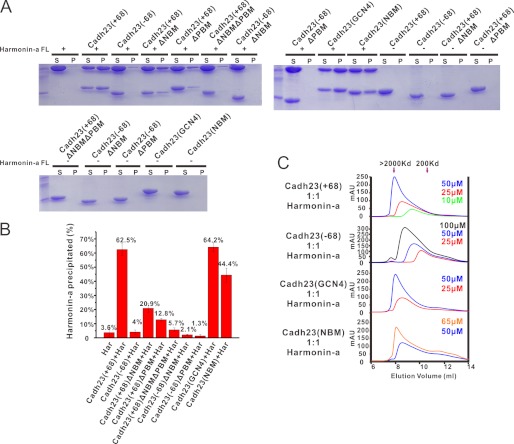
To obtain experimental evidence for the nonsynergistic binding model, analytical size exclusion gel filtration and ultracentrifugation assays were used to determine the size of the cadherin23/harmonin complex at different concentrations. When mixed at a 1:1 stoichiometric ratio, the cadherin23/harmonin complex always showed a concentration-dependent size increase, regardless of whether cadherin23(−68) or cadherin23(+68) were used (Fig. 6C). For cadherin23(+68), at a concentration of 50 μm, the complex already reached the detection limit of the gel filtration column (i.e. molecular mass >2000 kDa). Because the protein size detection by gel filtration can be influenced by the protein conformation, we further confirmed the large cadherin23/harmonin complex formation using ultracentrifugation. Cadherin23 was mixed with the full-length harmonin-a protein at a concentration of 50 μm at a 1:1 molar ratio, and the mixtures were centrifuged at 98,000 × g for 30 min. As expected, isolated harmonin or cadherin cannot be pelleted under this centrifugation force. However, ∼62.5% of the cadherin23(+68)/harmonin complex was spun down into the pellet presumably due to the formation of large molecular polymers (Fig. 6, A and B). It is noted that cadherin23(+68) and harmonin remain at ∼1:1 ratio both in supernatant and pellet fractions, suggesting that the pelleted complex is not due to the nonspecific protein aggregations. Together with the gel filtration data, we clearly demonstrated that cadherin23(+68) and the full-length harmonin-a can form a concentration-dependent large protein complex.
FIGURE 6.
Both self-dimerization and NTD binding of exon68 can promote formation of the polymeric cadherin23/harmonin complex. A, ultracentrifugation-based analysis of the formation of the high molecular weight protein complexes of the full-length harmonin-a and various forms of cadherin23. The proteins and their mixtures recovered from the supernatant and pellet for each sample are analyzed by SDS-PAGE with Coomassie Blue staining. B, quantification of results of panel A showing the fraction of harmonin recovered from the pellet. Data are obtained from three independent ultracentrifugation-based experiments. C, analytical gel filtration profiles of the mixtures of harmonin-a and various forms of cadherin23 showing the concentration-dependent large complex formation. The protein size markers are labeled by arrows at the top of the panel.
To better understand this protein clustering process, we deleted one harmonin binding site of cadherin23(+68) at a time and measured their individual contribution to the cadherin23/harmonin polymer formation. Both NBM and PBM are important in this process, as individual deletion of them from cadherin23(+68) dramatically decrease the pellet fraction from 62.5 to 20.9 and 12.8%, respectively. Deletion of both reduced the pellet fraction to the basal level (Fig. 6, A and B).
Consistent with the proposed model in Fig. 5A, cadherin23(−68) also underwent concentration-dependent large protein complex formation (Fig. 6C), albeit the complex is not as large as that formed by cadherin23(+68) (Fig. 6, A–C). Even at the concentration of 100 μm, the complex formed by cadherin23(−68) and harmonin-a is still much smaller than that formed by cadherin23(+68) at 25 μm (Fig. 6C). Taking all of the above data together, we conclude that each site of cadherin23 fulfills their harmonin binding function independently, and together they facilitate formation of concentration-dependent cadherin23/harmonin polymers. The function of the exon68 peptide is to further promote the cadherin23/harmonin polymer formation.
Either NTD Binding or Self-dimerization of Exon68 Peptide Could Facilitate Large Complex Formation
In theory, the exon68 peptide can facilitate large cadherin23/harmonin complex formation by either directly binding to harmonin NTD or self-dimerization. We dissected the potential roles of the two distinct properties of the exon68 sequence in assembling the cadherin/harmonin complex by replacing exon68 of the cadherin23 tail either with the dimer-forming GCN4 leucine zipper, or with the monomeric harmonin NTD-binding NBM motif. The cadherin23(GCN4) chimera formed a stable dimer in solution (supplemental Fig. S3L), and only contains two harmonin binding sites (a NTD binding site and a PDZ2 binding motif). Cadherin23(NBM) instead contains three harmonin binding sites (two NTD binding sites and a PDZ2 binding motif), but has a much weaker dimerization capacity compared with cadherin23(+68) (supplemental Fig. S3, B and M). The formation of large complexes with harmonin-a by cadherin23(GCN4) is as efficient as that by cadherin23(+68) (Fig. 6, A–C), suggesting that exon68-mediated dimerization is necessary for forming large cadherin23(+68)/harmonin complexes. The cadherin23(NBM) is also effective, albeit a little weaker than cadherin23(+68) does, in forming large complexes with harmonin-a (Fig. 6, A–C). The formations of large complexes with harmonin by cadherin23(GCN4) and cadherin23(NBM) are also concentration-dependent. Therefore, via either binding to harmonin NTD or forming homodimer, harmonin exon68 can effectively promote the formation of the cadherin23/harmonin polymer.
DISCUSSION
Tip link molecules take chief responsibilities to maintain the structural integrity of the stereocilia bundle, as well as to control the mechanotransduction channel gating during sound perception. Although several tip link-associated proteins including cadherin23, protocadherin15, and harmonin have been identified, little is known about how these proteins act together to construct sturdy tip links that can endure ceaseless mechanic stretching.
In the present study, we made several discoveries on the interaction between harmonin and the cytoplasmic tail of cadherin23. First, apart from our previously reported NTD/NBM and PDZ2/PBM interactions (16), we discovered that the exon68 peptide functions as a new NTD binding sequence of cadherin23. The structure of the NTD/exon68 peptide complex reveals that the exon68 peptide adopts a similar conformation as NBM and occupies the same pocket of NTD as NBM does. The NMR-based binding experiment suggests the exon68 peptide has a stronger NTD binding affinity than NBM. Second, the exon68 peptide-containing cadherin23 tail can form a stable homodimer. The self-dimerization and harmonin NTD binding are mutually exclusive properties of cadherin23 exon68. We note that the ITC-based assay and the NMR titration assay give different binding affinities for the exon68 peptide and NBM (Fig. 5, C and G, and supplemental Fig. S2). These two sets of data actually do not contradict each other. ITC-based assay was performed using the whole cytoplasmic domain of cadherin23(+68), in which exon68 forms a stable dimer with its upstream and downstream residues (supplemental Fig. S3, A–K), whereas the NMR titration assay used the isolated synthetic exon68 peptide. Because NTD binding and self-dimerization of exon68 are mutually exclusive and require the participation of the same amino acids in the exon68 peptide (Leu-3215, Leu-3219, Tyr-3222) (Fig. 4C, supplemental Figs. S1 and S4, A–C), dimerization of the cadherin(+68) tail would decrease its apparent binding affinity to harmonin NTD when compared with binding of the isolated exon68 peptide to harmonin NTD. Third and perhaps most importantly, NTD and PDZ2 of harmonin do not synergize with each other in binding to the multiple harmonin-binding sites of cadherin23 (Fig. 5, B–H). This finding lead to our discovery that multiple nonsynergistic binding sites between harmonin and the cytoplasmic tail of cadherin23(±68) result in the formation of a concentration-dependent, inter-linked, very large cadherin23/harmonin complex (Fig. 6, A–C). Compared with cadherin23(−68), the existence of exon68 in cadherin23(+68) further promotes formation of the cadherin23/harmonin polymer (Fig. 6, A–C).
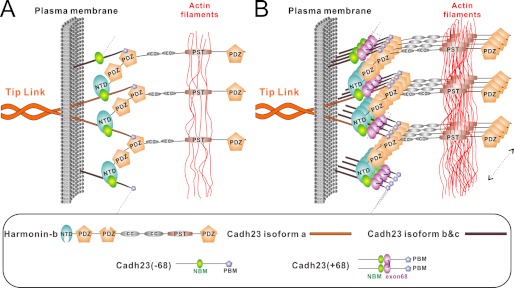
Combining published data and the findings in the present study, we propose a new model depicting the upper tip link root formation for hair cell stereocilia (Fig. 7). Each upper tip link is expected to have very limited amounts of cadherin23 homodimers, considering that the diameter of each tip link is about 8–10 nm (4), and that the first and second cadherin repeats of cadherin23 are about 2.5 nm in width (6, 38). We also assume here that cadherin23 in hair cell stereoclilia contains exon68 (18–22). The cytoplasmic tails of cadherin23(+68) at each upper tip link can bind to harmonin through nonsynergistic PBM/PDZ2 and NBM/NTD interactions, thus forming an inter-linked multimer. The self-dimerization capacity of the exon68 sequence of cadherin23(+68) allows the cytoplasmic tail of cadherin23 to recruit additional cadherin23 molecules (e.g. cadherin23-b and/or -c) (Fig. 7B). The exon68 sequence can also allow cadherin23 to recruit additional harmonin via binding to NTD (not drawn here). Thus, a polymerized, inter-linked cadherin23/harmonin web-like structure can form below the plasma membranes of upper tip link, as long as sufficient amounts of harmonin and various isoforms of cadherin23 are available (see below). This massive cadherin23/harmonin complex is further anchored to actin filaments through the PST sequence of harmonin and/or via binding to the sans/myosin VIIa complex (not shown here). It is envisioned that this actin filament-anchored, highly dense cadherin23/harmonin protein web is highly stable to root the upper tip link of stereocilia. In agreement with this model, EM and fluorescence imaging studies have shown that the base of each upper tip link contains densely packed proteins including harmonin, sans, and myosin VIIa (4, 14, 17, 39).
FIGURE 7.
Cadherin23/harmonin interaction model at the root of the upper tip link. A, cadherin23(−68) binds to NTD and PDZ2 of harmonin-b via its NBM and PBM, and forms a one-layered polymeric complex that anchors cadherin23 onto actin filaments via the PST region of the harmonin. B, the one-layered complex in panel A can be further extended into multiple layers by the exon68 encoding sequence either via its self-dimerization (as shown) or by binding to harmonin NTD (not shown here). This conceptual model is only for demonstrating that exon68 can facilitate formation of a strong root. The real role of exon68 in the upper tip link root construction may depend on the local molar ratio of harmonin to cadherin23.
Although exon68 can promote multimer formation either by self-dimerization or by binding to NTD, its real role in upper tip link root construction may depend on the local concentrations of harmonin to cadherin23. Our ITC data shows that NBM and dimerized exon68 have comparable NTD binding affinity (5.7 and 5.9 μm, respectively) (Fig. 5, C and G). Therefore, harmonin cannot differentiate the two sites. If the molar ratio of harmonin to cadherin23 is higher than 2:1, all exon68 would be occupied by NTD and hence no dimer would form. If the molar ratio is lower than 2:1, some exon68 would be free from NTD binding and form dimer. Under both scenarios, a large interlinked protein polymer would effectively form with the presence of the exon68 peptide sequence. For simplicity purpose, the exon68 peptides are drawn as dimers in the schematic model shown in Fig. 7B.
Our biochemistry data and the proposed model provide a plausible explanation as why exon68 is specifically expressed in hair cells (9, 18, 20). The presence of exon68 allows cadherin23(+68) to form a more stable tip link root organized by the cadherin/harmonin polymer. Compared with cadherin23(+68), the corresponding cadherin23(−68)/harmonin complex can only form a thin, one-layered structure (Fig. 7A) and thus is not as stable as the multiple layered complex formed by cadherin(+68) and harmonin (Fig. 7B).
The validity of the model shown in Fig. 7B critically depends on the amounts of cadherin23 and harmonin available at the base of each upper tip link. As stated before, each upper tip link is expected to have only a few pairs of cadherin23 isoform a homodimers at most. Such a small amount of cadherin23 is not sufficient for large complex formation. This apparent contradiction can be reconciled by taking into account the existence of cadherin23 isoforms b and c in stereocilia. Cadherin23 have three major isoforms, a, b, and c, all of which have the same cytoplasmic tail (±exon68). They mainly differ in their extracellular region. Isoform a is the longest one with 27 cadherin repeats and is responsible to form tip link. Isoform b contains 7 cadherin repeats and isoform c contains no extracellular and transmembrane regions (Fig. 1A) (3). All three cadherin23 isoforms are known to express in cochlea (18, 21), and the majority contain exon68 (18, 21). Because only stereocilia was positively stained when cochlea was probed by an exon68-specific antibody (20), it is assumed that expressed cadherin23-c is also present in stereocilia. Although the functions of cadherin 23 b and c are unknown, we propose that they may function to provide a sufficient amount of cadherin23 for formation of a stable upper tip link root by forming harmonin/cadherin23 polymer (Fig. 7B). Further experiments are required to test this hypothesis directly.
Polymerized protein complexes are widely present in a variety of biological processes. For example, postsynaptic proteins Homer and Shank form polymerized large complex to maintain the structural framework of a highly dense protein web-like structure called post-synaptic density (40); nephrin, Nck, and N-WASP are known to form protein aggregates to stimulate rapid nucleation of actin filaments (41); and the harmonin/cadherin23 polymer found in this study may function to construct a strong root for the upper part of the tip link. Although the detailed interaction mechanisms are different, formation of high-order polymerized protein complexes via multiple nonsynergistic bindings are a shared feature in these systems. It has been proposed that such multisite, nonsynergistic protein-protein interactions are highly effective and efficient in building macromolecular assemblies for both structural and signaling roles in diverse biological processes (41).
Supplementary Material
This work was supported by Grants 664009, 660709, 663610, 663811, HKUST6/CRF/10, and SEG_HKUST06 from the Research Grants Council of Hong Kong (to M. Z.).
The atomic coordinates and structure factors (code 2LSR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

This article contains supplemental Figs. S1–S4.
- NTD
- N terminal domain
- NBM
- NTD binding motif
- PBM
- PDZ binding motif
- ITC
- isothermal titration calorimetry
- HSQC
- heteronuclear single quantum coherence
- N
- newton.
REFERENCES
- 1. Beurg M., Fettiplace R., Nam J. H., Ricci A. J. (2009) Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat. Neurosci. 12, 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Markin V. S., Hudspeth A. J. (1995) Gating-spring models of mechanoelectrical transduction by hair cells of the internal ear. Annu. Rev. Biophys. Biomol. Struct. 24, 59–83 [DOI] [PubMed] [Google Scholar]
- 3. Richardson G. P., de Monvel J. B., Petit C. (2011) How the genetics of deafness illuminates auditory physiology. Annu. Rev. Physiol. 73, 311–334 [DOI] [PubMed] [Google Scholar]
- 4. Kachar B., Parakkal M., Kurc M., Zhao Y., Gillespie P. G. (2000) High-resolution structure of hair-cell tip links. Proc. Natl. Acad. Sci. U.S.A. 97, 13336–13341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E. M., Milligan R. A., Müller U., Kachar B. (2007) Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91 [DOI] [PubMed] [Google Scholar]
- 6. Elledge H. M., Kazmierczak P., Clark P., Joseph J. S., Kolatkar A., Kuhn P., Müller U. (2010) Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members. Proc. Natl. Acad. Sci. U.S.A. 107, 10708–10712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Söllner C., Rauch G. J., Siemens J., Geisler R., Schuster S. C., Müller U., Nicolson T., and Tübinger 2000 Screen Consortium (2004) Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature 428, 955–959 [DOI] [PubMed] [Google Scholar]
- 8. Ahmed Z. M., Goodyear R., Riazuddin S., Lagziel A., Legan P. K., Behra M., Burgess S. M., Lilley K. S., Wilcox E. R., Riazuddin S., Griffith A. J., Frolenkov G. I., Belyantseva I. A., Richardson G. P., Friedman T. B. (2006) The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26, 7022–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boëda B., El-Amraoui A., Bahloul A., Goodyear R., Daviet L., Blanchard S., Perfettini I., Fath K. R., Shorte S., Reiners J., Houdusse A., Legrain P., Wolfrum U., Richardson G., Petit C. (2002) Myosin VIIa, harmonin, and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 21, 6689–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaramillo F., Hudspeth A. J. (1992) Displacement-clamp measurement of forces exerted by hair bundles during adaptation. FASEB J. 6, A516–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Afrin R., Ikai A. (2006) Force profiles of protein pulling with or without cytoskeletal links studied by AFM. Biochem. Biophys. Res. Commun. 348, 238–244 [DOI] [PubMed] [Google Scholar]
- 12. Pickles J. O., Comis S. D., Osborne M. P. (1984) Cross-links between stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hear Res. 15, 103–112 [DOI] [PubMed] [Google Scholar]
- 13. Verpy E., Leibovici M., Zwaenepoel I., Liu X. Z., Gal A., Salem N., Mansour A., Blanchard S., Kobayashi I., Keats B. J., Slim R., Petit C. (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat. Genet. 26, 51–55 [DOI] [PubMed] [Google Scholar]
- 14. Grillet N., Xiong W., Reynolds A., Kazmierczak P., Sato T., Lillo C., Dumont R. A., Hintermann E., Sczaniecka A., Schwander M., Williams D., Kachar B., Gillespie P. G., Müller U. (2009) Harmonin mutations cause mechanotransduction defects in cochlear hair cells. Neuron 62, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michalski N., Michel V., Caberlotto E., Lefèvre G. M., van Aken A. F., Tinevez J. Y., Bizard E., Houbron C., Weil D., Hardelin J. P., Richardson G. P., Kros C. J., Martin P., Petit C. (2009) Harmonin-b, an actin-binding scaffold protein, is involved in the adaptation of mechanoelectrical transduction by sensory hair cells. Pflugers Arch 459, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan L., Yan J., Wu L., Zhang M. (2009) Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23. Proc. Natl. Acad. Sci. U.S.A. 106, 5575–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grati M., Kachar B. (2011) Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. U.S.A. 108, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lagziel A., Ahmed Z. M., Schultz J. M., Morell R. J., Belyantseva I. A., Friedman T. B. (2005) Spatiotemporal pattern and isoforms of cadherin 23 in wild type and waltzer mice during inner ear hair cell development. Dev. Biol. 280, 295–306 [DOI] [PubMed] [Google Scholar]
- 19. Siemens J., Kazmierczak P., Reynolds A., Sticker M., Littlewood-Evans A., Müller U. (2002) The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl. Acad. Sci. U.S.A. 99, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siemens J., Lillo C., Dumont R. A., Reynolds A., Williams D. S., Gillespie P. G., Müller U. (2004) Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428, 950–955 [DOI] [PubMed] [Google Scholar]
- 21. Michel V., Goodyear R. J., Weil D., Marcotti W., Perfettini I., Wolfrum U., Kros C. J., Richardson G. P., Petit C. (2005) Cadherin 23 is a component of the transient lateral links in the developing hair bundles of cochlear sensory cells. Dev. Biol. 280, 281–294 [DOI] [PubMed] [Google Scholar]
- 22. Di Palma F., Pellegrino R., Noben-Trauth K. (2001) Genomic structure, alternative splice forms and normal and mutant alleles of cadherin 23 (Cdh23). Gene 281, 31–41 [DOI] [PubMed] [Google Scholar]
- 23. Bahloul A., Michel V., Hardelin J. P., Nouaille S., Hoos S., Houdusse A., England P., Petit C. (2010) Cadherin-23, myosin VIIa, and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum. Mol. Genet. 19, 3557–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng L., Zheng J., Whitlon D. S., García-Añoveros J., Bartles J. R. (2010) Targeting of the hair cell proteins cadherin 23, harmonin, myosin XVa, espin, and prestin in an epithelial cell model. J. Neurosci. 30, 7187–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bax A., Grzesiek S. (1993) Methodological advances in protein NMR. Acct. Chem. Res. 26, 131–138 [Google Scholar]
- 26. Daniels D. L., Cohen A. R., Anderson J. M., Brünger A. T. (1998) Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat. Struct. Biol. 5, 317–325 [DOI] [PubMed] [Google Scholar]
- 27. Kraulis P. J. (1991) Molscript, a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 28. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL, A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 29. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu C., Feng W., Wei Z., Miyanoiri Y., Wen W., Zhao Y., Zhang M. (2009) Myosin VI undergoes cargo-mediated dimerization. Cell 138, 537–548 [DOI] [PubMed] [Google Scholar]
- 31. Wei Z., Zheng S., Spangler S. A., Yu C., Hoogenraad C. C., Zhang M. (2011) Liprin-mediated large signaling complex organization revealed by the liprin-α/CASK and liprin-α/liprin-β complex structures. Mol Cell 43, 586–598 [DOI] [PubMed] [Google Scholar]
- 32. Wu L., Pan L., Wei Z., Zhang M. (2011) Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science 331, 757–760 [DOI] [PubMed] [Google Scholar]
- 33. Yan J., Pan L., Chen X., Wu L., Zhang M. (2010) The structure of the harmonin/sans complex reveals an unexpected interaction mode of the two Usher syndrome proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huber A. H., Weis W. I. (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 35. Simon J. A., Schreiber S. L. (1995) Grb2 SH3 binding to peptides from Sos. Evaluation of a general model for SH3-ligand interactions. Chem. Biol. 2, 53–60 [DOI] [PubMed] [Google Scholar]
- 36. Roos J., Kelly R. B. (1998) Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J. Biol. Chem. 273, 19108–19119 [DOI] [PubMed] [Google Scholar]
- 37. Shamoo Y., Abdul-Manan N., Patten A. M., Crawford J. K., Pellegrini M. C., Williams K. R. (1994) Both RNA-binding domains in heterogenous nuclear ribonucleoprotein A1 contribute toward single-stranded RNA binding. Biochemistry 33, 8272–8281 [DOI] [PubMed] [Google Scholar]
- 38. Sotomayor M., Weihofen W. A., Gaudet R., Corey D. P. (2010) Structural determinants of cadherin-23 function in hearing and deafness. Neuron 66, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Furness D. N., Hackney C. M. (1985) Cross-links between stereocilia in the guinea pig cochlea. Hear. Res. 18, 177–188 [DOI] [PubMed] [Google Scholar]
- 40. Hayashi M. K., Tang C., Verpelli C., Narayanan R., Stearns M. H., Xu R. M., Li H., Sala C., Hayashi Y. (2009) The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 137, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.