Background: The transcriptional network governing cancer metastasis is largely unexplored.
Results: BACH1 regulates multiple metastasis genes and promotes breast cancer metastasis to bone.
Conclusion: BACH1 is a master regulator of breast cancer bone metastasis and transcriptional network reverse engineering is helpful to identify novel functional genes of metastasis.
Significance: This study provides a systems biology approach to identify master regulators of complicated biological processes.
Keywords: Bone, Breast Cancer, Cancer, Hypoxia, Metastasis, SMAD Transcription Factor, Transcription Factors
Abstract
The application of functional genomic analysis of breast cancer metastasis has led to the identification of a growing number of organ-specific metastasis genes, which often function in concert to facilitate different steps of the metastatic cascade. However, the gene regulatory network that controls the expression of these metastasis genes remains largely unknown. Here, we demonstrate a computational approach for the deconvolution of transcriptional networks to discover master regulators of breast cancer bone metastasis. Several known regulators of breast cancer bone metastasis such as Smad4 and HIF1 were identified in our analysis. Experimental validation of the networks revealed BACH1, a basic leucine zipper transcription factor, as the common regulator of several functional metastasis genes, including MMP1 and CXCR4. Ectopic expression of BACH1 enhanced the malignance of breast cancer cells, and conversely, BACH1 knockdown significantly reduced bone metastasis. The expression of BACH1 and its target genes was linked to the higher risk of breast cancer recurrence in patients. This study established BACH1 as the master regulator of breast cancer bone metastasis and provided a paradigm to identify molecular determinants in complex pathological processes.
Introduction
Breast cancer is the most common cancer and the leading cause of cancer deaths in both developed and developing countries (1). Most breast cancer-related deaths are not caused by the growth of primary tumors but by the metastatic spread of cancer cells to distant organs such as bone, lung, brain, and liver. Better treatments of cancer metastasis rely on the identification of molecular determinants of this deadly process and the understanding of the regulatory networks governing the activities of these functional molecules.
Recently, genomic profiling using microarrays have identified several gene expression signatures associated with breast cancer metastasis phenotypes (2–8). These studies have provided us with molecular markers of diagnostic and prognostic importance. However, the molecular events that activate the metastasis signatures remain unclear, and more importantly, therapeutic application of these metastasis markers is limited by the apparent diversity of the markers identified in different studies. For example, there is only one gene in common between the 70-gene signature identified by van't Veer et al. (5) from the NKI patient cohort and the 76-gene signature identified by Wang et al. (8) from the EMC patient cohort. Furthermore, none of these two poor prognosis signatures display statistically significant overlap with the signatures that determine the metastasis capacity of breast cancer cells to lung and bone (4, 6). The diversity of metastasis markers presumably indicates the complexity of metastasis regulatory networks and the existence of functionally redundant molecular routes that lead to the cellular behavior of metastasis. Therefore, the analysis of the molecular networks governing cancer metastasis will not only help understand the regulatory mechanisms of metastasis genes but also uncover the converging nodes in the network that control multiple signal pathways of cancer metastasis. These master regulators, often signal transducers or transcription factors (TFs),3 may provide us with new targets for more effective therapeutics to prevent or limit the metastatic spread of cancers.
Genome-wide deconvolution of the molecular networks of mammalian cells had been a formidable challenge for computational biology. However, with the recent development of bioinformatic approaches and increasing availability of high-throughput genomic data, a number of studies have demonstrated the feasibility of inference of mammalian transcriptional networks from gene expression profiles. Several methods are reported for such analysis, commonly called network reverse engineering, to construct the network graphs where the nodes and edges represent gene species and interactions between genes, respectively (9–19). Although most of these methods have only been applied to analyze organisms with relatively simple genomes, a few of them, which mainly fall into two categories, have been used for reverse engineering of mammalian transcriptional networks. The first class of methods take a “subgenome” approach to analyze the enriched sequence motifs in the promoters of a particular set of genes such as the top list that are correlated to a cellular phenotype or process to search for TFs regulating these genes (16, 17). This approach is limited by the fact that only a short list of genes are analyzed, and thus, some key factors regulating a collection of genes that individually display only modest expression differences, but work synergistically to drive a biological process can be overlooked. The other approaches, exemplified by the method named ARACNe (algorithm for the reconstruction of accurate cellular networks) developed by Margolin et al. (18, 19), use mutual information on the expression data as measurements of the dependence between genes to look for regulatory targets of TFs. The data transmission theory “data processing inequality” can be applied to enrich for gene pairs with direct dependence and thus filter out indirect targets of TFs. ARACNe has been used for network inference of a number of physiological and pathological conditions of mammalian cells, including human B cell development (20, 21), mouse lung response to oxidative stress (22), and mesenchymal transformation of brain tumors (23). These studies revealed the TFs regulating the cellular processes, and importantly, the bioinformatic analyses were experimentally validated. Furthermore, ARACNe has also been applied to the analysis of metastasis regulatory networks and showed that although metastasis signature genes identified by differential expression analysis from various clinical samples were largely inconsistent, the TFs predicted to be master regulators of metastasis networks displayed a much higher overlap rate (24). This work indicated that the master regulators, rather than the signature genes, were better biomarkers and probably better therapeutic targets for cancer metastasis with desired generality across samples. However, the regulatory roles of those identified TFs on metastasis were not experimentally tested in the report. Therefore, transcriptional network analysis has fallen short of uncovering functionally validated molecular connections that govern metastasis behaviors of cancer cells.
Here, we report the reverse engineering and experimental validation of the molecular networks for breast cancer metastasis to bone. Our bioinformatic analyses identified Smad4 and HIF1, the key TFs in TGFβ and hypoxia pathways that have been previously proven to regulate breast cancer bone metastasis (25–28). Additionally, BACH1, a transcription factor belonging to the basic region leucine zipper (bZip) TF family with a Cap'n'collar (CNC)-type bZip domain and a broad complex, tramtrack, and bric-a-brac (BTB) domain, was shown to transcriptionally regulate a list of genes that are involved in osteolytic metastasis of breast cancer, and more importantly, it promoted the invasiveness and metastasis of breast cancer cells. Therefore, our study demonstrates an approach to infer the molecular interactions for complex biological processes and establishes BACH1 as a master regulator of breast cancer bone metastasis.
EXPERIMENTAL PROCEDURES
Transcriptional Network Analysis
The inference of transcriptional network in metastasis was performed in three steps: biclustering, gene set enrichment analysis (GSEA) (29), and motif analysis. Biclustering of the gene expression microarray data set was performed with a previously reported algorithm (30) with modifications (see supplemental data for details). The expression clusters resulted from biclustering were tested by GSEA for their association with bone metastasis. The clusters that were significantly correlated with bone metastasis (FDR-q < 0.25, p < 0.05) were kept for further motif analysis. To test the enrichment of TF binding sites in the promoters of expression cluster (EC) genes, the regulons of all TFs were first found by motif search. The DNA region of upstream 3000 bp to downstream 500 bp from the transcription start site of each gene was scanned with the Match program and the TF binding site matrices available in the TRANSFAC Professional Database (31). For each binding site matrix, TRANSFAC provided the cutoffs of minimal matrix match score and core match score to minimize false positive matches, which were used in our motif search. The matched sites of each matrix were sorted according to the matrix match scores, and the top 3000 matches (if there are >3000 matches) were defined as the regulon of the corresponding TF. The overlap of a TF regulon and an EC set was analyzed by binomial test, and the p values were corrected for multiple testing. An overlap with Benjamini-Hochberg FDR-q < 0.1 resulted in a transcription module.
BACH1 Overexpression and Knockdown
The BACH1 cDNA clone (BC063307) was ordered from ATCC and cloned into the pTRE2puro plasmid (BD Clontech). The SCP4 cells were engineered to express the tetracycline transactivator with the plasmid pUHD15-1-Neo and then transfected with pTRE2puro-BACH1 for Tet-Off inducible overexpression. Constitutive expression of BACH1 was achieved with the plasmid pCMV5–2xHA (Addgene). Inducible knockdown of BACH1 was achieved by cloning a shRNA construct (target sequence, 5′-GCGTCTTGAAAGCCTAATAT-3′) into the shRNA-expressing plasmid pRSMX-puro, which was modified from the pSuper-Retro-puro (OligoEngine) system by adding a tetO operator into the promoter region. The SCP2 cells were engineered to express TetR with the plasmid pQCXIH-TetR and then transfected with the pRSMX construct for Tet-On inducible knockdown. The overexpression and knockdown effects were validated by Western blot analysis with anti-BACH1 antibody (Atlas Antibodies) and real-time PCR (forward primer, 5′-GGCTGATGGAGAGCTGAACATT-3′ and reverse primer, 5′-AGCAGTGTAGGCAAACTGAATTAAAG-3′).
ChIP
ChIP assays were performed in HeLa and SCP4 cells using the Fast ChIP method (32) with some modifications. Briefly, cells were cross-linked with 1% formaldehyde, and 125 mm glycine was used to quench the formaldehyde. The nuclear extracts were sonicated and incubated with control IgG or anti-HA antibody (Santa Cruz Biotechnology) for immunoprecipitation. The precipitated complexes were eluted and reverse cross-linked. The captured genomic DNA was purified with the silica membrane purification kit (TIANGEN) and used for PCR analysis. 2.5% of the total genomic DNA from the nuclear extract was used as input. An intron region of MMP1 was used as the negative control.
MMP1 Promoter Analysis
For MMP1 promoter analysis, the −4334 bp to +62 bp region of MMP1 (NM002421.1) was cloned to pGL3-basic. The 5′ end of the fragment was further truncated to −2685 bp, −272 bp, and −63 bp, respectively. The mutated sequences for AP1, PEA-3, and BACH1 binding sites located at the −272 bp to −63 bp region were as follows: 5′-GTTTGAAGTACTTCATGACAT-3′ (AP1), 5′-TAATCAAGATTATGTTATAA-3′ (PEA-3), and 5′-ATAAAGCATTCTTCAGACAGC-3′ (BACH1).
Two-chamber Migration and Invasion Assays
105 cancer cells in serum-free medium were seeded into the upper chamber of the insert membranes with a 3-μm pore size (BD Bioscience) with or without Matrigel (BD Bioscience) coating in a 24-well plate. FBS was used in the bottom chamber as the attractant. 12–16 h later, the cells in the upper chamber were removed using a cotton swab, and the invaded cells was stained with crystal violet and counted.
Bone Metastasis Assays in Nude Mice
105 cells were washed in PBS and injected into the left ventricle of female athymic Ncr-nu/nu mice to study the bone metastasis activity as described previously (4). Weekly non-invasive bioluminescence imaging was performed to quantify the metastasis burden at the target organs using the IVIS 200 Imaging System (Caliper Life Sciences) and the NC100 Imaging System (Berthold).
X-ray Radiography and Micro-computed Tomography Imaging
Bone damages were detected by x-ray radiography. Mice were anesthetized, arranged in prone position on single-wrapped films (X-OMAT Kodak) and exposed at 24 kV for 180 s with a Faxitron instrument (Faxitron Bioptics). In vivo micro-computed tomography (micro-CT) images were obtained using a Skyscan-1076 micro-CT scanner (Skyscan) while the animals were anesthetized. The micro-CT scanner was operated at 55 kV, 181 μA, 0.5 mm Al filter, and a scan resolution of 17.4 μm. The cross-sections were reconstructed using the NRecon software (Skyscan).
Gene Expression Microarray Data Analysis
The gene expression microarray profiling data for TGFβ treatment and hypoxic culturing of MDA-MB-231 (MDA231) derivative cells have been described (25, 33). For microarray analysis of BACH1 overexpression, SCP4 cells with BACH1-inducible overexpression were cultured in medium with 1 μg/ml doxycycline. 72 h after doxycycline removal, the cells were harvested, and RNA was isolated with RNeasy mini kit (Qiagen). The quality of purified RNA samples was monitored using a 2100 bioanalyzer (Agilent). Gene expression profiling was performed with the human Affymetrix U133A microarrays as described previously (4). The cells cultured in doxycycline-containing medium were used as a control. The microarray data were processed with GeneSpring (version 7.2) and normalized according to the chip median. The genes with fold changes of more than 2 after BACH1 induction were selected as the BACH1-regulated genes. The microarray data of these genes are available in supplemental Table S4.
BACH1 Clinical Analysis
Fresh tumor specimens were obtained with informed consent from patients who underwent surgical resection of breast cancer at the Department of Breast Surgery of Qilu Hospital of Shandong University. The study was approved by the Institutional Review Board. RNA was extracted from the tumors, and the quality was monitored by OD reading. Finally, a cohort of 73 specimens with high-quality RNA samples was used for BACH1 analysis. The patients were classified to BACH1 high and low expression groups according to the median expression level of BACH1 and were compared for their metastasis-free survival.
To analyze the prognostic power of BACH1-regulated module genes in the EMC and NKI data sets, the gene expression pattern of these genes in each tumor was compared with those in the SCP4 cells with BACH1 overexpression turned on and off. Pearson's correlation coefficient was used as the measurement of expression pattern similarity. The patients was stratified into two groups according to the expression similarity to the cells with BACH1 on and off, and their organ-specific metastasis, overall metastasis, and overall survival were compared by Kaplan-Meir analysis and Cox hazard ratio analysis. Only eight of the 11 BACH1-regulted module genes were found in the Hu25K microarray platform used in the NKI data set and were used for the analysis. To analyze the prognostic power of each module gene, the patients were classified according to the expression level of each gene using the median expression as the cut-off, and the patient prognosis was analyzed by Cox hazard model.
Statistical Analysis
The Kaplan-Meier method was used to estimate survival curves for the patients, and Cox proportional hazard regression was used to compare the survival. A two-sided Wilcoxon rank test was performed to analyze the bioluminescent imaging results in the in vivo studies. A two-sided independent Student's t test without equal variance assumption was performed to analyze the results of in vitro assays.
RESULTS
Reverse Engineering of Transcriptional Networks of Breast Cancer Bone Metastasis
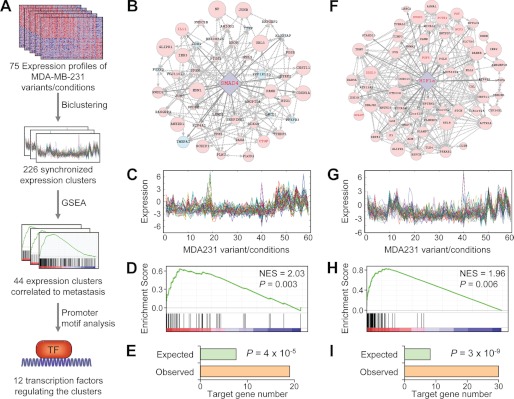
Previously, through single cell cloning and in vivo selection, we and others (4, 6) established 47 derivative sublines from the MDA231 breast cancer cells. These sublines displayed varied metastasis capabilities and specificities to bone and lung when tested in mice (4, 6). Genome-wide gene expression microarray analyses were performed for these cell lines under different culture conditions with or without TGFβ treatment, resulting in a total of 75 expression profiles (4, 6, 33). The expression data set for these cell lines, which were isogenic but with different metastasis behaviors, provided a good starting point for us to reconstruct the molecular networks governing cancer metastasis. Therefore, we designed a stepwise bioinformatic approach to identify metastasis regulatory modules, each consisting of a TF and its target genes (Fig. 1A). The gene set of a metastasis-regulating transcriptional module is expected to show the following three characteristics: 1) they show synchronized expression pattern over a spectrum of cellular conditions; 2) they are coordinately over/underexpressed in metastatic cells; and 3) the promoter regions of these genes are enriched for the binding sites of a certain TF. We used a series of analytical methods to pinpoint the gene sets with such characteristics. First, we performed an unsupervised biclustering analysis on the microarray data set of MDA231 sublines to identify co-expressed gene sets. Biclustering (30, 34) is a technique for simultaneous clustering of both genes and conditions and separates itself from clustering or conventional two-way clustering by that it reveals gene sets that are co-expressed within a subset of conditions and that overlaps among gene sets are allowed. These features were important for our transcriptional network analysis in that co-regulated gene sets may display nonsynchronous expression in some samples because of sporadic genetic or epigenetic alterations and that a single gene could be regulated by more than one TF. We used both the genes whose expression was previously found to be correlated to breast cancer metastasis to bone or lung (4, 6) and the “random” pseudogenes that were identified from a preliminary k-means clustering of the data set as the seeds for biclustering (see supplemental data for details), to identify gene clusters with sizes of at least 20 that were co-expressed in at least 60 (80%) of the cellular conditions. 226 ECs with a median size of 155 genes were identified. Then, we performed GSEA to screen for the ECs that were collectively correlated to breast cancer organ-specific metastasis and discovered 44 clusters for bone tropism with FDR-q < 0.25 and p value <0.05 (21 enriched in highly metastatic cells and 23 enriched in lowly metastatic cells). Another set of 14 clusters were found to be correlated to breast-to-lung metastasis and will be discussed elsewhere. Finally, we performed a promoter motif search to see whether the binding sites of any TFs were enriched in the promoters of these EC genes using TRANSFAC TF binding site matrices (31). Binomial test was used to examine the significance of overlap between the EC set and the predicted target gene sets (regulons) of the 553 TFs available in the TRANSFAC database, yielding 12 TFs with their binding sites significantly enriched in 13 ECs (Table 1) of which nine were up-regulated in bone metastasis and four were down-regulated. Among them, HIF1 was the only TF associated with two ECs.
FIGURE 1.
Transcription network analysis of breast cancer metastasis. A, schematic illustration of the bioinformatic approach. See text for details. B–I, two transcription modules of breast cancer bone metastasis revealed by the network analysis. B, the transcription module regulated by Smad4. The network includes a cluster of genes with synchronized expression (EC) and the hub factor Smad4. Pink and blue filled circles indicate the genes with expression positively and negatively, respectively, correlated to metastasis. The size of filled circles denotes the extent of the correlation. The gene name in red font indicates it is a BMS gene identified previously. Dashed lines indicate tight expression correlation between gene pairs. Solid arrowed lines indicate direct regulation of the genes by the TF. C, the expression pattern of Smad4 EC genes in various MDA231 cell derivatives and culture conditions. Each colored curve denotes an EC gene that was listed in supplemental Table S1. D, GSEA of Smad4 EC gene enrichment in breast cancer bone metastasis. All genes on the U133A microarray were ordered by their expression correlation to metastasis in descending order from left to right and then were examined for their presence in the EC set. When the gene was found in the EC set (vertical black bar), the enrichment score was awarded with a correlation rank-weighted score; otherwise, it was penalized. The significance of the final score was estimated by gene set permutation. See Subramanian et al. (29) for a detail description of the GSEA algorithm. NES, normalized enrichment score. E, putative Smad4 target genes were enriched in the EC set. Shown are observed numbers of EC genes with Smad4 binding sites in their promoter and the gene number expected by random chances. F–I, another transcription module regulated by HIF1.
TABLE 1.
The predicted transcriptional modules of breast cancer bone metastasis
| Cluster ID | Cluster size | Correlation to metastasis | TF | Expected target no. | Observed target no. | p value | FDR-q |
|---|---|---|---|---|---|---|---|
| 4 | 38 | Positive | Bach1 | 3.6 | 18 | 3.70 × 10−10 | 4.32 × 10−4 |
| 23 | 72 | Positive | HIF1 | 10.3 | 30 | 6.27 × 10−9 | 5.59 × 10−4 |
| 24 | 183 | Negative | Egr1 | 18.0 | 38 | 6.08 × 10−6 | 3.21 × 10−3 |
| 11 | 52 | Positive | SMAD4 | 7.6 | 19 | 4.38 × 10−5 | 6.44 × 10−3 |
| 9 | 86 | Positive | E2F | 23.1 | 39 | 1.42 × 10−4 | 5.99 × 10−2 |
| 123 | 44 | Positive | MEF2C | 4.0 | 12 | 1.89 × 10−4 | 6.59 × 10−2 |
| 58 | 409 | Positive | NFIL3 | 35.0 | 57 | 2.19 × 10−4 | 6.99 × 10−2 |
| 146 | 322 | Positive | HIF1 | 46.0 | 70 | 2.22 × 10−4 | 7.99 × 10−2 |
| 87 | 328 | Positive | POU3F2 | 49.0 | 73 | 3.24 × 10−4 | 6.11 × 10−2 |
| 59 | 84 | Negative | AP-1 | 14.2 | 27 | 3.98 × 10−4 | 6.87 × 10−2 |
| 64 | 360 | Negative | Elk-1 | 55.2 | 80 | 4.24 × 10−4 | 7.87 × 10−2 |
| 73 | 86 | Positive | STAT5A | 8.2 | 18 | 7.94 × 10−4 | 9.20 × 10−2 |
| 54 | 253 | Negative | ELSPBP | 23.6 | 40 | 7.65 × 10−4 | 9.22 × 10−2 |
Notably, two TFs in the top four transcriptional modules, Smad4 and HIF1, have been already proven to be critical mediators of bone metastasis in breast cancer (25–28, 33). Smad4 is the essential TF complex component of TGFβ pathway, which has been well documented as a signal cascade that inhibits early tumorigenesis but paradoxically promotes metastasis. Specifically, TGFβ activation and Smad4 activity plays a central role to regulate the metastasis capability of MDA231 derivative cells in mice (33). HIF1 is a heterodimeric basic helix-loop-helix TF complex composed of the hypoxia-responsive subunit HIF1α and the constitutively expressed subunit Arnt (HIF1β). Oxygen-dependent degradation of HIF1α mediates the hypoxic regulation of HIF1 downstream genes and has been recognized to be critical in various aspects of tumor progression including cell proliferation, angiogenesis, and cell survival (35). HIF1α expression is also correlated to cancer metastasis (36). More importantly, we recently documented the evidence to show that HIF1 activity was essential for bone metastasis of breast cancer cells (25). Interestingly, HIF1α, Smad4, and receptor-regulated Smads (Smad2 and Smad3) did not any differential expression in the gene expression microarray data of the MDA231 sublines with different metastasis propensities. They were not in the list of bone metastasis signature (BMS) genes (4). Statistical analysis (Student's t test) of their expression from the microarray data set did not reach any significance (data not shown), indicating the role of post-transcriptional regulation in these signal pathways. The discovery of the regulatory modules of these TFs not only demonstrated the effectiveness of our transcriptional network analysis to identify master regulators of metastasis but also emphasized a feature of our approach that separates it from previously reported methods for transcription network inference, in that it does not rely on the transcription measurements of the TFs and thus is able to reveal the regulation relationship that is hidden in transcription analysis.
EC Genes Are Regulated by the TFs in Transcription Modules
In each transcription module, a cluster of genes of co-expression, an EC, were predicted to be regulated by a common TF. For example, in one module (Fig. 1B and supplemental Table S1), 52 genes displayed a synchronized expression pattern in various cell lines (Fig. 1C), and they were collectively up-regulated in the cells metastatic to bone as revealed by the GSEA analysis (Fig. 1D). These genes were significantly enriched for the predicted direct targets of Smad4, with 19 of them containing the Smad4 binding site in their promoters (Fig. 1E). This gene subset of predicted TF targets will be denoted as the target cluster (TC) hereafter in the text. Notably, several Smad4 TC genes, such as IL11 and CTGF, in the module were bona fide transcriptional targets of Smad4 and also functional drivers of bone metastasis (4, 33). Another module contained 72 EC genes (Fig. 1, F and G, and supplemental Table S2), which were also globally correlated to bone metastasis capability (Fig. 1H), and enriched with HIF1 direct targets (Fig. 1I). A number of the HIF1 TC genes in the cluster, such as DUSP1 and FGF5, were previously shown to be transcriptionally regulated by hypoxia (25). These two and other genes, including PCTK2, FST, NCF2, DDX10, and SPAST, were among the BMS genes predicting the bone tropism of breast cancer cells (4).
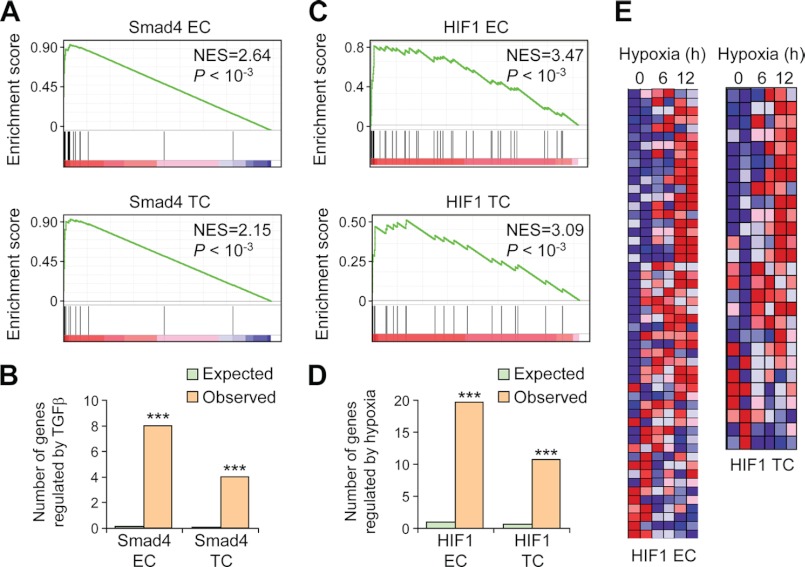
To test objectively whether the genes in these transcriptional modules were regulated by the corresponding TFs, we analyzed the genome-wide expression data of MDA231 cells following TGFβ or hypoxia treatment. The gene expression profiles of 28 MDA231 derivative lines with and without TGFβ treatment, and those of two derivative lines after 6 and 12 h of hypoxic culturing, were analyzed by microarray (4, 25). When we performed the GSEA of the Smad4 EC and TC genes, a global up-regulation of these gene sets was observed after TGFβ activation (Fig. 2A). Indeed, all but two of the 52 genes were activated by TGFβ. Furthermore, eight of these genes were among the top 49 TGFβ-responsive genes, with folds of enrichment of 70.0 for EC and 91.0 for TC (Fig. 2B). Similarly, the HIF1 EC and TC genes were globally up-regulated in hypoxic conditions (Fig. 2C) and enriched for the top HIF1-responsive genes (Fig. 2D, folds of enrichment were 18.4 and 18.3, respectively). The majority of the genes in this module was activated after 6 h of hypoxic challenge and further up-regulated after 12 h (Fig. 2E). These analyses demonstrated that the predicted target genes in the transcription modules were actually regulated by the TFs, and thus, our bioinformatic approach was able to reveal bona fide regulatory relationship between TFs and downstream genes from transcriptional data.
FIGURE 2.
Validation of the transcriptional regulation of Smad4 and HIF1 modules. A, GSEA of the expression of Smad4 EC (top) and TC (bottom) genes after TGFβ treatment. Genes were ordered by the expression fold changes after TGFβ treatment in descending order from left to right. B, the number of genes in Smad4 EC and TC that were up-regulated by TGFβ was compared with the number of genes expected by random chance. C, GSEA of HIF1 EC (top) and TC (bottom) genes after hypoxic culturing. Genes were ordered by the expression fold changes after hypoxia culturing in descending order from left to right. D, the number of genes in HIF1 EC and TC clusters that were up-regulated by hypoxia was compared with the number of genes expected by random chance. E, expression patterns of HIF1 EC and TC genes in hypoxic condition. ***, binomial test, p < 0.001.
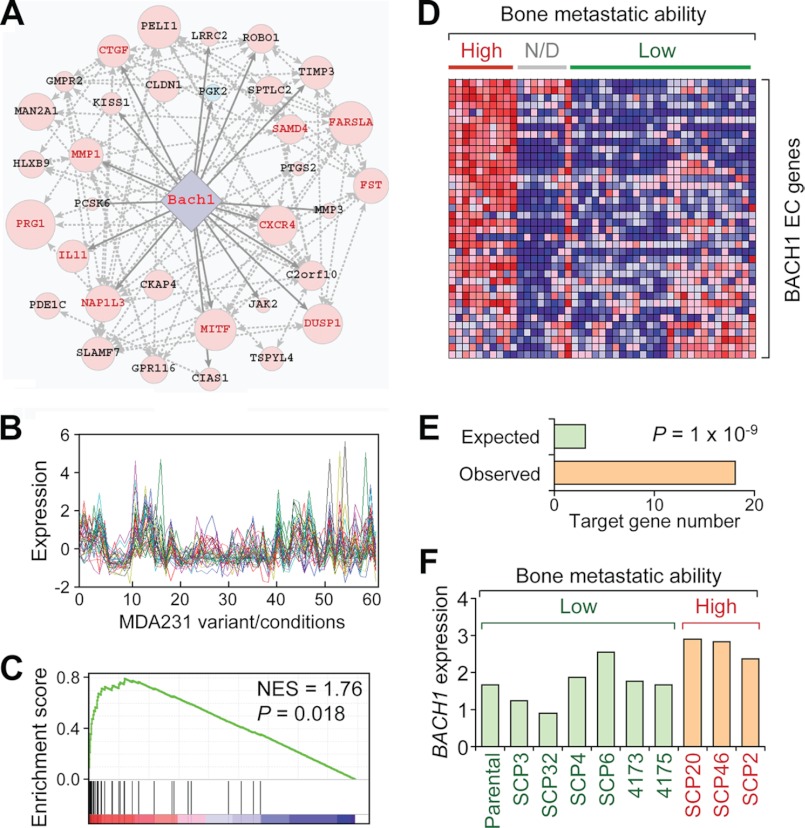
A BACH1 Module Is Linked with Breast Cancer Bone Metastasis
BACH1 in the overall top module (Fig. 3A) is a heme-binding transcription factor. BACH1 was previously found to function mainly in the physiological regulation of oxidative stress, by repressing the transcription of HMOX1, the key enzyme for heme degradation and radical scavenge (37–41), with a few of most recent reports on its roles in cancer progression (42, 43). Our analysis identified a cluster of 38 probes (33 unique genes) with co-expression (Fig. 3B and supplemental Table S3), which were significantly up-regulated in bone-tropic cancer cells (Fig. 3, C and D). 18 genes in the set were predicted to be direct targets of BACH1, an enrichment of 5-fold over the statistical background (Fig. 3E, p = 1.0 × 10−9). Interestingly, BACH1 itself was up-regulated in cells with elevated bone metastasis capability according to the gene expression microarray data (Fig. 3F, fold change, 1.55; p = 0.01 by Student's t test). However, the difference was rather weak, and therefore, it was not identified as a BMS gene previously (4). Although BACH1 was reported to mainly function as a transcription repressor, it could also act as an activator either on different target genes (40) or on the same genes in different cellular contexts (38). Considering the expression pattern of BACH1 and the EC genes in the module, BACH1 seemed to mainly function as a transcriptional activator in the transcription module. Notably, as much as 11 of the 43 BMS genes that were up-regulated in bone-tropic cells were observed in the BACH1 EC set (Fig. 3A, folds of enrichment = 152, Binomial test p < 10−10), indicating a role of BACH1 to regulate breast cancer bone metastasis.
FIGURE 3.
The BACH1 transcription module. A, the network view of BACH1 module. Note that several BMS genes (red font) were included in the module. B, expression of BACH1 EC genes in various cell conditions. C, GSEA of BACH1 EC genes in bone metastasis. D, expression heatmap of BACH1 EC genes in breast cancer cells with varied capabilities of bone metastasis. N/D, not determined. E, the number of BACH1 putative target genes in EC. F, BACH1 expression in highly metastatic and lowly metastatic breast cancer cells. NES, normalized enrichment score.
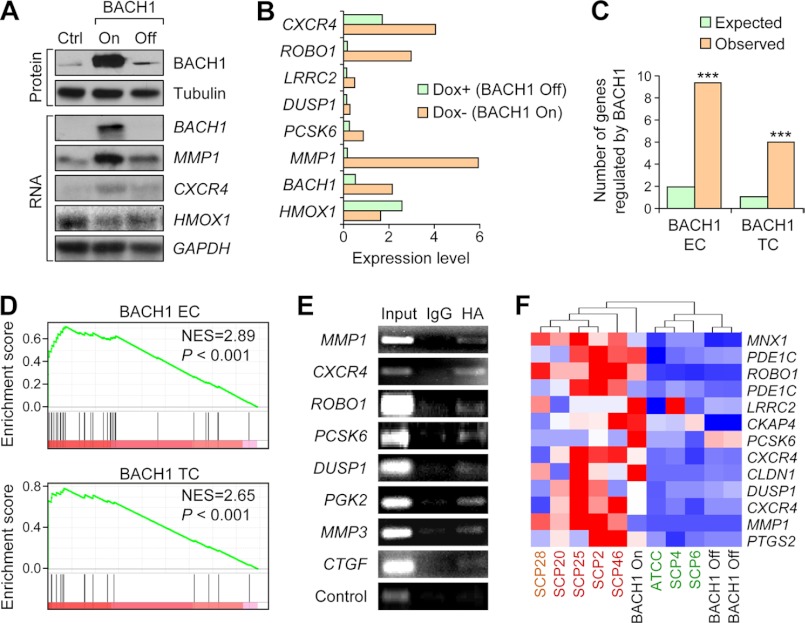
To validate the regulatory role of BACH1 in the transcriptional module and bone metastasis, we overexpressed BACH1 with a Tet-Off inducible system in the weakly metastatic SCP4 cells. Northern and Western blot were performed to confirm the overexpression of BACH1 following induction by doxycycline removal from the culture medium (Fig. 4A). Microarray analysis was performed to compare the gene expression profiles of SCP4 cells before and after doxycycline removal and identified 1350 genes with expression elevation of >2-fold after BACH1 activation (supplemental Table S4). Among the 1350 genes, six genes, namely MMP1, CXCR4, LRRC2, ROBO1, DUSP1, and PCSK6, were found in the BACH1 TC (Fig. 4B, enrichment, p = 0.0001). Another five genes were included in the BACH1 EC set (Fig. 4C, enrichment, p = 4.3 × 10−8). BACH1 also repressed another set of 872 genes at least 2-fold (supplemental Table S4). The known target gene of BACH1, HMOX1, was mildly repressed by BACH1 in SCP4 cells (Fig. 4A). Notably, there was no significant overlap between the BACH1-regulated genes in this study and those previously identified in HEK 293 (40), emphasizing the dependence of BACH1 activity on cellular context.
FIGURE 4.
BACH1 module genes were regulated by BACH1. A, Western blot (top) and Northern blot (bottom) analysis of BACH1 and the module genes in SCP4 parental cells and the cells with BACH1 inducible overexpression. B, the mRNA expression levels of BACH1 and the module genes after BACH1 induction. HMOX1 is a gene known to be repressed by BACH1. C, the number of genes regulated by BACH1 in the EC and TC sets. ***, binomial test, p < 0.001. D, GESA of BACH1 EC (top) and TC (bottom) genes after BACH1 induction. The genes were ordered by the expression fold changes after BACH1 induction in descending order from left to right. E, ChIP analysis of BACH1 module genes in HeLa cells with HA-BACH1 overexpression. F, unsupervised clustering analysis of SCP4 with inducible BACH1 overexpression (black) and cells with strong (red), medium (yellow), and weak (green) metastasis tendencies by BACH1-regulated EC genes. NES, normalized enrichment score; Ctrl, control.
We further analyzed the global expression pattern of BACH1 EC and TC genes after BACH1 activation. GESA showed that both EC and TC genes were globally up-regulated by BACH1 (Fig. 4D). Indeed, nearly 80% of the EC and TC genes were activated by BACH1 (supplemental Fig. S1A), arguing that BACH1 is the common regulator of this transcriptional module. To test whether the genes activated by BACH1 are its direct targets, we performed chromatin immunoprecipitation (ChIP) assays in HeLa and SCP4 cells to analyze the physical binding of BACH1 on the gene promoters. BACH1 was overexpressed with an HA tag in the cells, and DNA fragments bound by BACH1 were immunoprecipitated with the anti-HA antibody. The promoter regions containing the BACH1 binding sites of the six TC genes that were significantly activated by BACH1 and three other TC genes (PGK2, MMP3, and CTGF) that were modestly regulated by BACH1 were analyzed by PCR following ChIP. A nonspecific region without any surrounding BACH1 binding site was used as the negative control. The ChIP analysis showed that the promoter regions of all but one gene (LRRC2) were indeed enriched by BACH1 immunoprecipitation (Fig. 4E and supplemental Fig. S1B). In contrast, no enrichment was found when an empty HA vector was transfected into the cells (data not shown).
Interestingly, when we performed unsupervised hierarchical clustering of the MDA231 sublines with varied metastasis tendencies using the expression pattern of BACH1-regulated module genes, the cancer cells were segregated in a way that clearly reflected the metastasis behaviors of these cells. In addition, when BACH1 was turned on in the weakly metastatic SCP4 cells, the cell line was relocated from the weakly metastatic group to the highly metastatic group by the clustering analysis (Fig. 4F and supplemental Fig. S1C), indicating a regulatory role of BACH1 and its target genes in tuning the metastasis ability of cancer cells.
BACH1 Regulation of the MMP1 Promoter
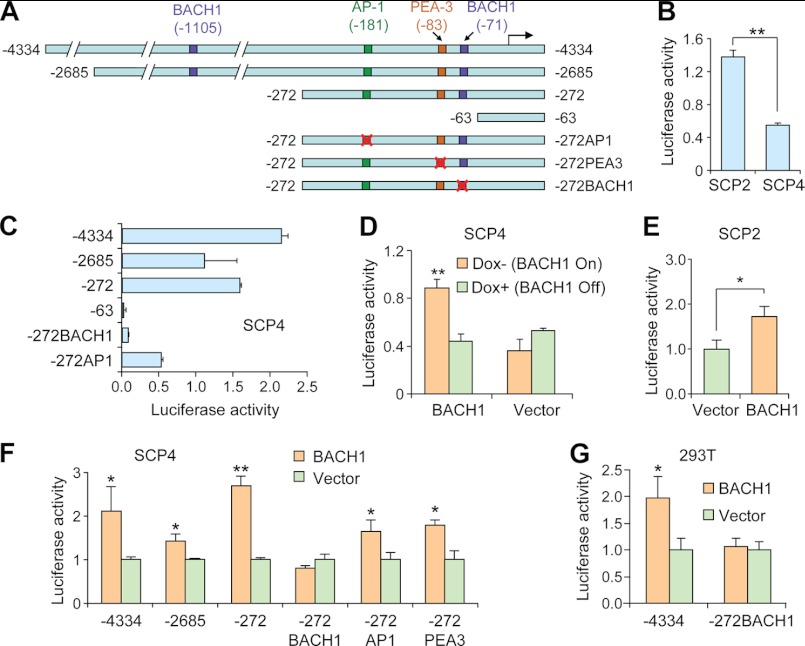
Among the confirmed BACH1 direct targets, MMP1 was well recognized for its role in tissue remodeling, tumor progression, and invasion (44) and specifically, the osteolytic bone metastasis of breast cancer (4, 45). Therefore, we chose MMP1 to further study BACH1 regulation of metastasis genes. MMP1 has been shown to be up-regulated in highly metastatic MDA231 sublines such as SCP2, as compared with the weakly metastatic counterparts including SCP4 (4). We constructed a luciferase reporter plasmid with the MMP1 promoter region spanning from −4334 bp to +65 bp (Fig. 5A) and first tested its activity in MDA231 sublines. Luciferase analysis showed that the MMP1 promoter was more active in SCP2 than in SCP4 (Fig. 5B), indicating that this gene was transcriptionally regulated in these cells. To further study the mechanism of MMP1 regulation, we created a series of MMP1 promoter truncation constructs (Fig. 5A) to map out the functional cis-elements. Deletion of the region upstream of −272 bp from the transcription start site did not significantly affect the promoter activity, whereas removal of the segment from −272 bp to −63 bp completely abolished the transcriptional activity (Fig. 5C), suggesting the existence of cis-elements regulating MMP1 expression in this region. Furthermore, ectopic BACH1 expression in SCP4, SCP2, and 293T cells, via either inducible or constitutive overexpression systems, was able to turn on MMP1 promoters (Fig. 5, D–G). There was a BACH1 binding site at −71 bp and an AP-1 binding site at −181 bp, which contains a core sequence similar to that of BACH1 binding site, in the functional region of the MMP1 promoter. We mutated these two sites and a PEA-3 binding site as a control (Fig. 5B). BACH1 site mutation, but not AP-1 or PEA-3 site mutations, abolished the activating effect of BACH1 on MMP1 promoter activities (Fig. 5, F and G). Furthermore, BACH1 site mutation, rather than the AP-1 site mutation, diminished the transcriptional activity of MMP1 promoter (Fig. 5C). In addition, the DNA region tested in the ChIP assay for MMP1 promoter (Fig. 4E) was also the fragment containing this BACH1 binding site. Together, our data demonstrated that BACH1 regulated MMP1 transcription in the bone-metastatic cells via binding to the −71 bp site.
FIGURE 5.
MMP1 was a transcription target of BACH1. A, the MMP1 promoter and its truncation and site mutation constructs for luciferase reporter analysis. B, MMP1 promoter activities in SCP2 and SCP4. C, the transcriptional activities of MMP1 promoter with serial truncation and binding site mutation in SCP4. D, the transcriptional activities of MMP1 wild type promoter (−4334) in SCP4 transfected with inducible BACH1 or control plasmids. E, MMP1 wild type promoter (−4334) activities in SCP2 with or without constitutive BACH1 overexpression. F, the transcriptional activities of different MMP1 promoters in SCP4 with or without BACH1 constitutive overexpression. G, the transcriptional activities of MMP1 promoters in 293T cells with or without BACH1 overexpression. *, p < 0.05; **, p < 0.01 by Student's t test.
BACH1 Regulates Breast Cancer Cell Invasiveness and Bone Metastasis
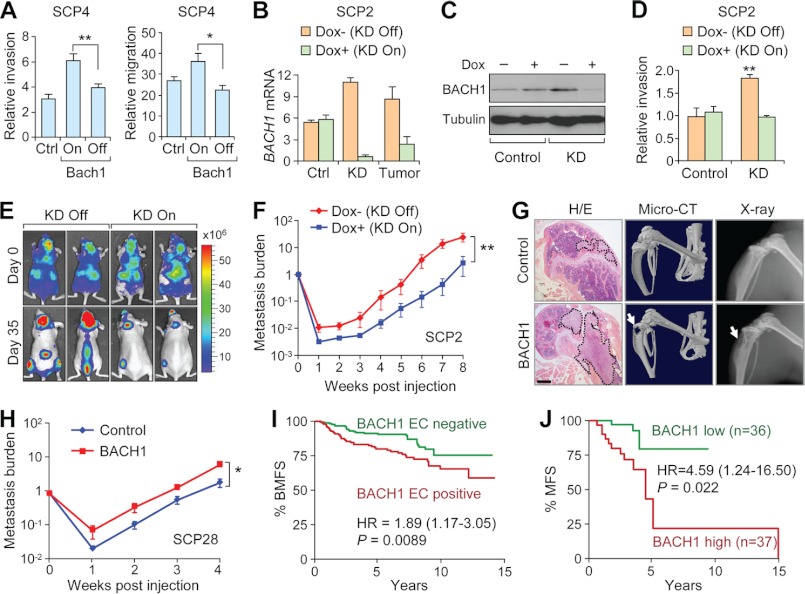
To study whether BACH1 functions to regulate breast cancer metastasis as our bioinformatic analysis indicated, we tested the effects of BACH1 inducible overexpression on cancer cell migration and invasion. When BACH1 expression was suppressed by doxycycline in SCP4, no significant changes were observed on cellular migration or invasion as compared with the parental SCP4 cells. However, when BACH1 was activated by doxycycline removal, the cells appeared more migratory and invasive (Fig. 6A). To further study the functional role of BACH1 in metastasis, we depleted its expression in the highly metastatic cells SCP2 with a Tet-On shRNA system, in which the BACH1-targeting shRNA was expressed only when doxycycline was supplied in the culture medium. Real-time PCR and Western blot assays showed that doxycycline addition significantly suppressed the expression of BACH1 in SCP2 cells when the cells were transfected with the shRNA construct, but doxycycline itself could not reduce BACH1 expression (Fig. 6, B and C). As expected, BACH1 depletion, but not doxycycline addition, reduced the invasion of cancer cells (Fig. 6D).
FIGURE 6.
BACH1 was a regulator of breast cancer metastasis to bone. A, Trans-well invasion and migration analyses of SCP4 cells with and without BACH1 induction. B, BACH1 expression levels in SCP2 cells with inducible BACH1 shRNA or control (Ctrl) vectors and in the metastasis tumors in mice implanted with SCP2 shRNA cells. KD, knockdown. C, BACH1 protein levels in SCP2 cells with inducible BACH1 shRNA or control vectors. D, Trans-well invasion of the SCP2 cells as described in C. E, in vivo analysis of SCP2 metastasis capabilities. Shown are representative mice at day 0 and day 35. F, bioluminescence imaging quantification of SCP2 bone metastasis burden (10 mice per group). G, in vivo analysis of bone metastasis after BACH1 overexpression in SCP28 cells. Shown are the hematoxylin and eosin (H/E) staining, micro-CT, and x-ray analyses of animal hind limbs 5 weeks after cancer cell implantation. Areas circled by dotted lines denote the tumor regions, and arrows indicate the regions of bone damages. Scale bar, 400 μm. H, bioluminescence imaging quantification of SCP28 bone metastasis burden. I, bone metastasis-free survival (BMFS) analysis of the EMC patients stratified with the expression pattern of BACH1-regulated EC genes. HR, hazard ratio. J, metastasis-free survival (MFS) analysis of the Qilu cohort with BACH1 expression. *, p < 0.05; **, p < 0.01 by repeated measures analysis of variance. Dox, doxycyclin.
To assess the in vivo function of BACH1 on bone metastasis, SCP2 cells with inducible BACH1 shRNA were stably labeled with a luciferase-expressing retrovirus and injected into the left ventricle of nude mice for in vivo bone metastasis analysis. The mice were fed with doxycycline-containing water to shut down BACH1 expression and examined every week after injection by bioluminescent imaging to analyze the metastasis of cancer cells to different organs. Using human-specific real-time PCR primers, we confirmed that BACH1 expression in the cancer cells was effectively knocked down by doxycycline feeding of the animals (Fig. 6B). Doxycycline addition was also able to suppress SCP2 metastasis to limbs, spine, and skull (Fig. 6, E and F). At the 5th week post injection, the bone metastasis burden from SCP2 with doxycycline feeding was over 10-fold weaker than the control group (Fig. 6F). We also analyzed the effect of BACH1 in bone metastasis by overexpressing it in a mildly metastatic MDA231 derivative cell line SCP28 (4). BACH1 overexpression led to a significant increment in the metastatic capability of SCP28 (Fig. 6, G and H). Histology staining, as well as micro-CT and x-ray imaging analyses demonstrated obvious bone damages in the animal limbs by the overexpression cells, arguing a regulating role of BACH1 in breast cancer bone metastasis.
Clinical Relevance of the BACH1 Module
To study the clinical significance of BACH1-regulated transcription module, we analyzed the expression pattern of module genes in the published EMC patient cohort for which the patient follow-up information of organ-specific metastasis was available (7, 8). The patients were stratified according the expression pattern of the 11 BACH1-regulated EC genes and were compared for their metastasis-free survival. The patients with the expression pattern similar to that in the BACH1-ovexpressing cells suffered earlier bone metastasis and overall metastasis than other patients (Fig. 6I and supplemental Fig. S2A). Interestingly, survival analysis with the expression levels of any individual gene in this EC set did not significantly segregate the patients with different clinical outcomes. Only the expression pattern of the whole gene set predicted the patient prognosis (supplemental Table S5). The prognosis significance of the EC set was also seen to predict lung metastasis, albeit to a lesser extent (supplemental Fig. S2B). Similar results were observed when the analyses were performed using the BACH1-regulated TC genes (supplemental Fig. S2, C and D, and supplemental Table S5). We further tested the prognosis power of BACH1 module in the NKI breast cancer clinical data set (2, 7). There were only eight genes in the BACH1-regulated EC set matched to the NKI microarray platform. Similar results were observed when the NKI patients were stratified by the expression pattern of these genes; the expression pattern of the whole set, but not the individual genes, was correlated to worse outcomes in bone metastasis, as well as lung metastasis, overall metastasis, and overall survival of the patients (supplemental Table S5 and supplemental Fig. S3). These results not only demonstrated the robustness of BACH1 module for prognosis prediction but also suggested that the prognosis power was from the collective action of this module, but not the individual genes. The mRNA levels of BACH1 itself did not seem to be correlated to metastasis in these microarray data sets, probably due to the complication of post-transcriptional modification in these data sets. Therefore, we analyzed BACH1 expression in another cohort of breast cancer patients with prognosis information from Qilu Hospital of Shandong University (see details under “Experimental Procedures”). Because the BACH1 antibody available to us was not suitable for immunohistochemical analysis, we had to perform real-time PCR to assess the mRNA levels of BACH1 in these patients. In this data set, BACH1 expression was significantly linked to worse overall metastasis-free survival (Fig. 6J). We also analyzed the correlation of BACH1 and its module genes with ER, PR, HER2, and triple negative status in the EMC cohort for which the tumor molecular subtype information is available. The expression of BACH1 and the whole module was not linked to any of these parameters (supplemental Fig. S4). Importantly, the BACH1 module retained its prognosis power in the multivariate Cox analysis together with these parameters (supplemental Table S6). Therefore, our clinical analysis demonstrated that BACH1 and its transcriptional module genes were prognosis factors for breast cancer metastasis.
DISCUSSION
A well recognized caveat of gene expression analysis is that microarray data are only a snapshot of the transcriptome and thus not optimal to probe the regulatory activities of TFs. This is because 1) the activity changes of some TFs might be subtle but significant due to the synergistic effects of multiple downstream genes. However, this subtle changes could be masked by the vast noise of microarray data; 2) TFs are among the proteins that tend to be subject to post-transcriptional modification such as phosphorylation, degradation, and translocation, which are neglected by the microarray survey; 3) the transcriptome snapshot cannot reveal the transient changes of regulatory molecules. Therefore, TFs tend to escape from the screening by gene expression differential analysis (4). Transcriptional network analysis, which reconstructs the dynamic interactions between the TFs and their targets, is able to pinpoint the regulatory molecules that work via the collective action of multiple downstream mediators and therefore is a useful alternative for differential expression screening. Previous efforts, such as the studies with the ARACNe algorithm, have proven the feasibility of this strategy (18–22, 24). However, the transcriptional network analysis to reveal master regulators of metastasis, one of the most complicated biological processes, is yet to produce experimentally validated discovery. Furthermore, ARACNe algorithm is characterized by measuring the dependences between TFs and the target genes with their pair-wise mutual information, which is calculated from the expression levels of TFs and target genes. Therefore, this method still relies on the accuracy of mRNA measurements of TFs and thus will not be able to discover the TFs whose activity changes are not faithfully recapitulated by microarray. Our analysis took a bottom-up approach by first searching for the target genes that were regulated by an unknown TF and thus averted the dependence on TF mRNA quantification. This feature was demonstrated by the three transcriptional modules identified in our study, for which the expression levels of the TFs were either not associated with metastasis at all (HIF1 and Smad4), or rather weakly (BACH1), and therefore could not be found by differential analysis. Thus, our analysis algorithm provided an approach to uncover the elusive regulators not only for cancer metastasis but also for other biological processes that require multigenic synergism.
Two of the top four modules identified by the network reverse engineering analysis, HIF1 and Smad4, have been shown to be master regulators of breast cancer metastasis. Together with BACH1 that was validated here, these transcription modules demonstrated a high level of reliability of the prediction from the bioinformatic analysis. This could be due to both the carefully designed analysis approach and the quality of the microarray data set used in the analysis. In our analysis, we used the expression data set from a large collection of MDA231 isogenic sublines. These cells shared the same genetic background but varied with their metastasis proclivities, allowing us to search for the metastasis-disturbing factors while at the same time minimizing non-relevant alterations. Previously, Califano and colleagues (24) analyzed the NKI and Wang data sets of breast cancer clinical samples to search for metastasis regulators. Although they showed that the TFs identified in their analysis classified breast cancer samples of different prognosis with cross-data set generality, the functional roles of these TFs in metastasis were not validated, which is at least partially due to the complication by the extensive diversities in patient genetic background and the molecular subtypes of breast cancer in the clinical data sets.
At the time of our bioinformatic analysis, BACH1 had not been linked to any processes of cancer progression. Therefore, BACH1 appeared as a novel TF from the analysis and was validated by our experimental assays. BACH1 regulated four BMS genes, MMP1, CXCR4, DUSP1 and FHL1, among which MMP1 and CXCR4 had been proven as functional drivers of metastasis. In addition, five other genes in BMS were also activated by BACH1, only to a lesser extent (supplemental Table S4). Consistent with its transcription activity, BACH1 overexpression promoted the migration and invasion of cancer cells, whereas knockdown significantly suppressed these processes. We also noticed that most recently two studies reported BACH1 in the context of cancer progression (42, 43). Alvarez et al. (42) predicted that BACH1 might be a regulator of the prostate cancer marker ACPP, although this was not experimentally verified. Yun et al. (43) reported that BACH1 is a pro-metastatic gene and a direct target of the tumor-suppressive microRNA Let-7. We showed that BACH1 depletion resulted in significant reduction of experimental metastasis, and more importantly, the expression of BACH1 and its target genes were linked to the metastasis probability of clinical samples. Although individual BACH1 module genes were not prognostic for clinical metastasis, their overall expression pattern consistently predicted metastasis and patient deaths in multiple data sets, which again argues the role of BACH1 as a master regulator of cancer metastasis. Therefore, BACH1 may provide us an important target for breast cancer diagnosis and therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Dr. Etsuro Ito and Dr. Masayuki Yamamoto for help on BACH1 expression and luciferase reporter plasmids and Dr. Andrea Califano for the discussion on transcriptional network analysis.

This article contains supplemental data, Tables S1–S6, Figs. S1–S4, and an additional reference.
- TF
- transcription factor
- GSEA
- gene set enrichment analysis
- EC
- expression cluster
- Tet
- tetracycline
- BMS
- bone metastasis signature
- TC
- target cluster
- micro-CT
- micro-computed tomography
- ECM
- Erasmus Medical Center
- NKI
- Netherlands Cancer Institute.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA-Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E. T., Friend S. H., Bernards R. (2002) A gene expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 3. Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. (2003) A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 [DOI] [PubMed] [Google Scholar]
- 4. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 5. van 't Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., Schreiber G. J., Kerkhoven R. M., Roberts C., Linsley P. S., Bernards R., Friend S. H. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 [DOI] [PubMed] [Google Scholar]
- 6. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bos P. D., Zhang X. H., Nadal C., Shu W., Gomis R. R., Nguyen D. X., Minn A. J., van de Vijver M. J., Gerald W. L., Foekens J. A., Massagué J. (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M., Atkins D., Foekens J. A. (2005) Gene expression profiles to predict distant metastasis of lymph node-negative primary breast cancer. Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 9. Butte A. J., Tamayo P., Slonim D., Golub T. R., Kohane I. S. (2000) Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc. Natl. Acad. Sci. U.S.A. 97, 12182–12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yeung M. K., Tegnér J., Collins J. J. (2002) Reverse engineering gene networks using singular value decomposition and robust regression. Proc. Natl. Acad. Sci. U.S.A. 99, 6163–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gardner T. S., di Bernardo D., Lorenz D., Collins J. J. (2003) Inferring genetic networks and identifying compound mode of action via expression profiling. Science 301, 102–105 [DOI] [PubMed] [Google Scholar]
- 12. Friedman N., Linial M., Nachman I., Pe'er D. (2000) Using Bayesian networks to analyze expression data. J. Comput. Biol. 7, 601–620 [DOI] [PubMed] [Google Scholar]
- 13. Hartemink A. J., Gifford D. K., Jaakkola T. S., Young R. A. (2001) Pacific Symposium on Biocomputing (PSB), Hawaii, January 4–9, 2000, pp. 6422–6433, The National Library of Medicine/National Institutes of Health and the International Society for Computational Biology, Bethesda, MD [Google Scholar]
- 14. Butte A. J., Kohane I. S. (2000) Pacific Symposium on Biocomputing (PSB), Hawaii, January 3–7, 2001, pp. 5418–5429, The National Library of Medicine/National Institutes of Health and the International Society for Computational Biology, Bethesda, MD [Google Scholar]
- 15. Huynh-Thu V. A., Irrthum A., Wehenkel L., Geurts P. (2010) Inferring regulatory networks from expression data using tree-based methods. PloS One 5, e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhodes D. R., Kalyana-Sundaram S., Mahavisno V., Barrette T. R., Ghosh D., Chinnaiyan A. M. (2005) Mining for regulatory programs in the cancer transcriptome. Nat. Genet. 37, 579–583 [DOI] [PubMed] [Google Scholar]
- 17. Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. (2003) Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Margolin A. A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., Dalla Favera R., Califano A. (2006) BMC Bioinformatics 7, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margolin A. A., Wang K., Lim W. K., Kustagi M., Nemenman I., Califano A. (2006) Reverse engineering cellular networks. Nature Protoc. 1, 662–671 [DOI] [PubMed] [Google Scholar]
- 20. Lefebvre C., Rajbhandari P., Alvarez M. J., Bandaru P., Lim W. K., Sato M., Wang K., Sumazin P., Kustagi M., Bisikirska B. C., Basso K., Beltrao P., Krogan N., Gautier J., Dalla-Favera R., Califano A. (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 [DOI] [PubMed] [Google Scholar]
- 22. Taylor R. C., Acquaah-Mensah G., Singhal M., Malhotra D., Biswal S. (2008) Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLoS Comput. Biol. 4, e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carro M. S., Lim W. K., Alvarez M. J., Bollo R. J., Zhao X., Snyder E. Y., Sulman E. P., Anne S. L., Doetsch F., Colman H., Lasorella A., Aldape K., Califano A., Iavarone A. (2010) The transcriptional network for mesenchymal transformation of brain tumors. Nature 463, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim W. K., Lyashenko E., Califano A. (2009) Pacific Symposium on Biocomputing (PSB), Hawaii, January 5–9, 2009, pp. 14504–14515, The National Library of Medicine/National Institutes of Health and the International Society for Computational Biology, Bethesda, MD [Google Scholar]
- 25. Lu X., Yan C. H., Yuan M., Wei Y., Hu G., Kang Y. (2010) In vivo dynamics and distinct functions of hypoxia in primary tumor growth and organotropic metastasis of breast cancer. Cancer Res. 70, 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korpal M., Yan J., Lu X., Xu S., Lerit D. A., Kang Y. (2009) Imaging transforming growth factor-β signaling dynamics and therapeutic response in breast cancer bone metastasis. Nature Med. 15, 960–966 [DOI] [PubMed] [Google Scholar]
- 27. Mohammad K. S., Javelaud D., Fournier P. G., Niewolna M., McKenna C. R., Peng X. H., Duong V., Dunn L. K., Mauviel A., Guise T. A. (2011) TGF-β-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 71, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dunn L. K., Mohammad K. S., Fournier P. G., McKenna C. R., Davis H. W., Niewolna M., Peng X. H., Chirgwin J. M., Guise T. A. (2009) Hypoxia and TGF-β drive breast cancer bone metastases through parallel signaling pathways in tumor cells and the bone microenvironment. PloS One 4, e6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tagkopoulos I., Slavov N., Kung S. Y. (2005) Fifth IEEE Symposium on Bioinformatics and Bioengineering (BIBE'05), Washington, DC, October 19–21, 2005 [Google Scholar]
- 31. Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., Kloos D. U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. (2003) TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson J. D., Denisenko O., Bomsztyk K. (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nature Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 33. Kang Y., He W., Tulley S., Gupta G. P., Serganova I., Chen C. R., Manova-Todorova K., Blasberg R., Gerald W. L., Massagué J. (2005) Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng Y., Church G. M. (2000) Biclustering of expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 8, 93–103 [PubMed] [Google Scholar]
- 35. Harris A. L. (2002) Hypoxia–a key regulatory factor in tumor growth. Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 36. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 37. Hintze K. J., Katoh Y., Igarashi K., Theil E. C. (2007) Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, β-globin, and NADP(H) quinone(oxido) reductase1. J. Biol. Chem. 282, 34365–34371 [DOI] [PubMed] [Google Scholar]
- 38. Oyake T., Itoh K., Motohashi H., Hayashi N., Hoshino H., Nishizawa M., Yamamoto M., Igarashi K. (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J., Brand M., Zenke Y., Tashiro S., Groudine M., Igarashi K. (2004) Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. U.S.A. 101, 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warnatz H. J., Schmidt D., Manke T., Piccini I., Sultan M., Borodina T., Balzereit D., Wruck W., Soldatov A., Vingron M., Lehrach H., Yaspo M. L. (2011) The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem. 286, 23521–23532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toki T., Katsuoka F., Kanezaki R., Xu G., Kurotaki H., Sun J., Kamio T., Watanabe S., Tandai S., Terui K., Yagihashi S., Komatsu N., Igarashi K., Yamamoto M., Ito E. (2005) Transgenic expression of BACH1 transcription factor results in megakaryocytic impairment. Blood 105, 3100–3108 [DOI] [PubMed] [Google Scholar]
- 42. Alvarez A., Woolf P. J. (2011) RegNetB: Predicting relevant regulator-gene relationships in localized prostate tumor samples. BMC Bioinformatics 12, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yun J., Frankenberger C. A., Kuo W. L., Boelens M. C., Eves E. M., Cheng N., Liang H., Li W. H., Ishwaran H., Minn A. J., Rosner M. R. (2011) Signaling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 30, 4500–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 45. Lu X., Wang Q., Hu G., Van Poznak C., Fleisher M., Reiss M., Massagué J., Kang Y. (2009) ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 23, 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.