Background: Pks13 is involved in the final biosynthesis step of mycolic acids.
Results: We report the full characterization of a 52-kDa fragment containing the acyltransferase domain of Pks13.
Conclusion: Pks13 is able to load unusually long chain acyl-CoAs through an unprecedented hydrophobic channel.
Significance: This study could constitute a key step toward the development of new antibiotics against mycobacterial infections.
Keywords: Mycobacterium tuberculosis, Polyketides, Structural Biology, Tuberculosis, X-ray Crystallography, Crystal Structure, Mycolic Acids, Polyketide Synthase
Abstract
Pks13 is a type I polyketide synthase involved in the final biosynthesis step of mycolic acids, virulence factors, and essential components of the Mycobacterium tuberculosis envelope. Here, we report the biochemical and structural characterization of a 52-kDa fragment containing the acyltransferase domain of Pks13. This fragment retains the ability to load atypical extender units, unusually long chain acyl-CoA with a predilection for carboxylated substrates. High resolution crystal structures were determined for the apo, palmitoylated, and carboxypalmitoylated forms. Structural conservation with type I polyketide synthases and related fatty-acid synthases also extends to the interdomain connections. Subtle changes could be identified both in the active site and in the upstream and downstream linkers in line with the organization displayed by this singular polyketide synthase. More importantly, the crystallographic analysis illustrated for the first time how a long saturated chain can fit in the core structure of an acyltransferase domain through a dedicated channel. The structures also revealed the unexpected binding of a 12-mer peptide that might provide insight into domain-domain interaction.
Introduction
Type I polyketide synthases (PKSs)3 are large multifunctional enzymes responsible for the biosynthesis of a wide array of natural compounds, the so-called polyketides. Polyketide biosynthesis is very similar to that of fatty acids. Indeed, a carbon skeleton is formed through successive decarboxylative condensations between the growing polyketide chain and short chain acyl-CoA extender units. Different PKS domains are involved in the catalysis of the various steps leading to polyketide synthesis (1). Three domains are absolutely required for the elongation of the starter unit and form the core of all PKSs: an acyltransferase (AT) domain that plays a major role in the selection and transfer of the extender unit, an acyl carrier protein (ACP) domain activated by a phosphopantetheine arm on which the extender unit is transferred, and a β-ketoacylsynthase (KS) domain that is involved in the condensation of the starter and extender units, thus leading to the formation of the β-ketoacyl intermediate. In addition, PKSs may contain other domains that will for instance help modify the β-carbon position. In type I PKSs, all domains are on a single polypeptide chain.
The structure elucidation of type I PKSs is of both fundamental and applied interest, and no structure of such full-length enzyme has been solved so far presumably because of their intrinsic flexibility. Nevertheless, the x-ray structures of entire type I FAS enzymes (2–9) have been resolved. The current knowledge about structure-function relationships of type I PKSs is presently deduced from these FAS-I structures, from the structure of the KS-AT didomains of modules 3 and 5 from 6-deoxyerythronolide-B synthase (DEBS) (10–13), and from structural homologues of the individual domains (14). Thus, there is a clear need for information related to structural details as well as the exact interplay between the various catalytic domains of type I PKSs to establish the molecular and structural bases responsible for their molecular mechanism and programming.
Polyketides form a structurally diverse family of natural compounds that mostly exhibit very interesting biological activities and have widely used pharmacological properties. The polyketide family also comprises compounds involved in important biological processes, such as cell wall biogenesis, and/or processes that are essential for the virulence of various bacteria. This is especially the case of human mycobacterial pathogens, namely Mycobacterium tuberculosis, Mycobacterium leprae, and Mycobacterium ulcerans (15–18). The genome sequence of M. tuberculosis contains more than 20 pks genes that mostly encode type I PKSs (19). Among the type I PKSs produced by M. tuberculosis, Pks13 is involved in the final assembly of mycolic acids, key structural components of the mycobacterial cell envelope, and is essential for the viability of mycobacteria (20). Pks13 catalyzes the decarboxylative condensation of two long chain fatty acid derivatives, a very long (C40–C60) meromycoloyl-AMP and a shorter (C24–C26) 2-carboxyacyl-CoA (21). Thus, Pks13 is an intriguing PKS in the sense that it performs only one cycle of condensation between two unusually long substrates. Moreover, because of its essentiality and the highly specific nature of its substrates, Pks13 has emerged as an interesting pharmacological target in the context of an urgent need for new antituberculosis compounds (22). Pks13 from M. tuberculosis includes 1733 amino acid residues for a molecular mass of 186,446 Da. It comprises the three mandatory PKS domains described above plus an additional ACP domain at the N terminus and a thioesterase (TE) domain at the C terminus with the organization ACP-KS-AT-ACP-TE. All five domains are separated by linker regions ranging from about 30 to 200 residues. The catalytic mechanism of Pks13 has been described previously (21). Briefly, the central AT domain is involved in loading the carboxyacyl-CoA extender unit, which is subsequently transferred onto the C-terminal ACP domain (see Fig. 1A). The meromycoloyl chain, activated and loaded by the FadD32 enzyme onto the N-terminal ACP domain (23), is transferred onto the KS domain. The KS domain is involved in catalyzing the Claisen-type condensation between the meromycoloyl and the carboxyacyl chains to produce an α-alkyl β-ketothioester linked to the C-terminal ACP domain. The thioesterase domain may then be involved in the release of the product. Here, we report the functional and structural characterization of a 52-kDa fragment of Pks13 from M. tuberculosis containing the AT domain and part of the upstream and downstream linkers. We show that this protein preferentially binds long carboxyacylated chains. The high resolution crystallographic characterization of the protein in the apo form and in palmitoylated and carboxypalmitoylated states reveals for the first time how a long fatty acyl chain is accommodated inside the core structure of an AT domain.
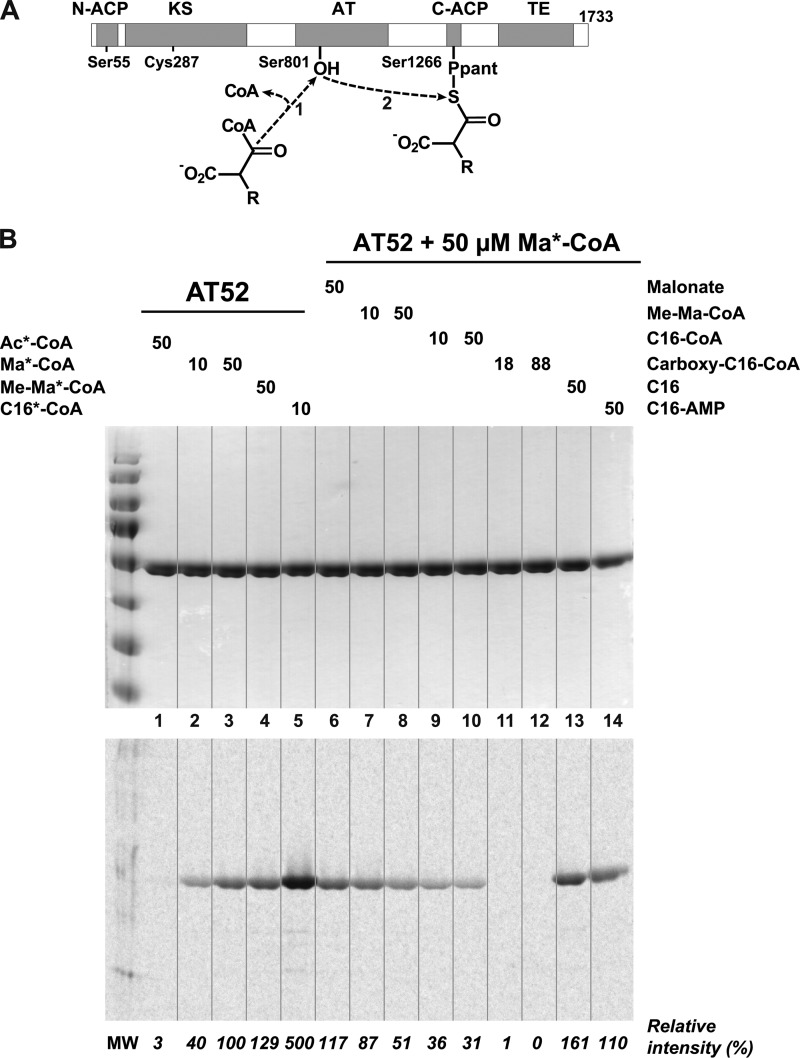
FIGURE 1.
Characterization of the AT52 fragment from Pks13. A, scheme of the reaction performed by the AT domain. The acyl chain of long carboxyacyl-CoA substrates (R = C22–C24) are covalently attached to the active site of the AT domain (1) and are subsequently transferred onto the phosphopantetheinylated C-terminal ACP domain (2). B, loading and competitive binding assays. Top, Coomassie Blue staining; bottom, phosphorimaging. Lane 1, 50 μm [1-14C]acetyl-CoA (Ac*-CoA); lanes 2 and 3, 10 and 50 μm [1-14C]malonyl-CoA (Ma*-CoA), respectively; lane 4, 50 μm [1-14C]methylmalonyl-CoA (Me-Ma*-CoA); lane 5, 50 μm [1-14C]palmitoyl-CoA (C16*-CoA). Lanes 6–14, 50 μm [1-14C]malonyl-CoA (plus 50 μm malonate in lane 6); lanes 7 and 8, 10 and 50 μm methylmalonyl-CoA (Me-Ma-CoA), respectively; lanes 9 and 10, 10 and 50 μm palmitoyl-CoA (C16-CoA), respectively; lanes 11 and 12, 18 and 88 μm carboxypalmitoyl-CoA (carboxy-C16-CoA), respectively; lane 13, 50 μm palmitic acid (C16); lane 14, 50 μm palmitoyl-AMP (C16-AMP). Relative intensities were calculated versus the band intensity obtained in the presence of 50 μm [1-14C]malonyl-CoA alone (lane 3).
EXPERIMENTAL PROCEDURES
Identification of the AT52 Fragment from Pks13
The overexpression and purification of the full-length Pks13 protein from M. tuberculosis H37Rv will be described elsewhere. The protein was then subjected to limited proteolysis using α-chymotrypsin (Nα-p-tosyl-l-lysine chloromethyl ketone-treated; Fluka) with a protease/protein ratio of 1:100 (w/w) in 50 mm Tris, 300 mm NaCl, 10 mm CaCl2, pH 8.0 at 12 °C for 10 h. Digestion was stopped with 2 mm phenylmethylsulfonyl fluoride (PMSF). Purification of the different proteolytic fragments prior to mass determination by mass spectroscopy was achieved by gel filtration. Purified fragments were desalted using a Zip-Tip (C4) with a final elution at 80% acetonitrile, 1% formic acid, and mass determination was performed using a QSTAR XL (Applied Biosciences) mass spectrometer. A potential of 1–2 kV was applied to the precoated nanoelectrospray needles (Picotips and Econotips, New Objective) in the ion source. Instrument operation, data acquisition, and analysis were carried out using Analyst® QS 1.0 software and BioAnalystTM extensions. Proteolytic fragments were also separated by SDS-PAGE on an acrylamide gel (12%) and visualized using Coomassie Blue staining. The gel lane corresponding to the polypeptide at 52 kDa was cut out to perform in-gel tryptic digestion. For this, after several washing steps to eliminate the stain, the corresponding protein was reduced and alkylated by successive incubation in solutions of 10 mm dithiothreitol (DTT) in 100 mm NH4HCO3 for 45 min at 56 °C and 55 mm iodoacetamide in 100 mm NH4HCO3 for 30 min at room temperature, respectively. In-gel tryptic digestion was then performed by incubating the gel slice in a sufficient covering volume of trypsin solution (12.5 ng/μl modified sequencing grade trypsin (Promega) in 12.5 mm NH4HCO3) overnight at 37 °C with shaking. MALDI-TOF mass spectroscopy analyses were carried out on a MALDI-TOF/TOF instrument (4700 Proteomics Analyzer, Applied Biosystems). A total of 0.5 μl of trypsin digest was loaded onto the MALDI target plate and air-dried. Then, 0.3 μl of matrix solution (α-cyano-4-hydroxycinnamic acid; 5 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid) was added. Mass spectra were acquired in automated positive reflector mode from m/z 700 to 4000. Trypsin autolytic peptides (m/z 842.5100 and 2211.1046) were used to internally calibrate each spectrum to a mass accuracy within 50 ppm. Peak lists from peptide mass mapping spectra were compared manually with the theoretical molecular masses of the trypsin peptides of Pks13.
Cloning, Overexpression, and Purification of AT52
The DNA encoding the AT52 fragment was amplified by PCR from M. tuberculosis H37Rv genomic DNA using primers F2A (5′-TTCATTAGCGGTTCGACGAGTTCGGC-3′) and F2B (5′-TTAAGCTTGAACCGGGTCGGCGGAAT-3′). The PCR product was digested with NdeI and HindIII and cloned between the NdeI and HindIII sites of pET28aII, a derivative of the pET28a expression vector (Novagen), to yield pWM71. The pET28aII vector was constructed by inserting a DNA linker harboring a stop codon that was prepared by annealing primers 5′-AGCTTTGACAGGTACCATC-3′ and 5′-TCGAGATGGTACCTGTCAA-3′ between the XhoI and HindIII site of pET28a. pWM71 was then transferred in the Escherichia coli BL21 (DE3) pLysS strain (Novagen), and the resulting strain was grown at 37 °C in terrific broth medium (Invitrogen) supplemented with chloramphenicol (25 μg/ml) and kanamycin (35 μg/ml) until reaching an A600 nm of 0.6. The culture was then cooled to 30 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and grown for 4 h at 30 °C. Cells were harvested by centrifugation (4,000 × g for 20 min), resuspended in lysis/wash buffer (50 mm Tris/HCl, 300 mm NaCl, 10 mm imidazole, pH 8.0) supplemented with 0.1% Triton X-100, and lysed by sonication with a Vibra Cell apparatus (Bioblock Scientific) for 3 × 30 s (microtip 4, 50% duty cycle) in ice. After centrifugation at 30,000 × g for 30 min, cellular debris was removed, and the supernatant was loaded into a column (10 ml) containing Nickel Chelating Fast Flow (Amersham Biosciences) connected to an ÄKTA purifier (GE Healthcare) and equilibrated with lysis buffer. The column was extensively washed with the wash buffer supplemented with 60 mm imidazole and then eluted with an increasing linear gradient of imidazole (up to 300 mm) in 50 mm Tris/HCl, 300 mm NaCl, pH 8.0 in 10 column volumes. Fractions containing high concentrations of pure protein were identified by SDS-PAGE, pooled, and concentrated by ultrafiltration (Vivaspin, 10 kDa) to 5–8 mg/ml in the digestion buffer (10 mm Tris/HCl, 150 mm NaCl, 2.5 mm CaCl2, pH 8.4). The His6 tag was cleaved by trypsin digestion (1 unit of enzyme for 4 mg of protein; Novagen) at 20 °C for 12 h, and the reaction was stopped with 2 mm PMSF. The remaining tagged protein was removed by a second nickel affinity column equilibrated in wash buffer. The untagged protein was concentrated to 5–8 mg/ml and applied to HiLoad 16/60 Superdex 75 (Amersham Biosciences) connected to an ÄKTA purifier using 20 mm Tris/HCl, 300 mm NaCl, 2 mm DTT, pH 8.0 as elution buffer. The purified protein was finally buffer-exchanged into 20 mm Tris/HCl, 300 mm NaCl, 0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 1 mm tris(2-carboxyethyl)phosphine, 2 mm EDTA, pH 8.0 and concentrated to 3–10 mg/ml. For 250 ml of culture, 12 mg of purified protein were obtained. Protein concentration was determined by measuring A280 and using an extinction coefficient of 40,125 m−1 cm−1.
Ligand Loading and Competition Binding Assays
The protocols used for the synthesis of carboxypalmitoyl-CoA and ligand loading were as described previously (21). For ligand loading, the assays (total volume, 15 μl) were performed in 50 mm Tris, pH 8.0. [1-14C]Malonyl-CoA (PerkinElmer Life Sciences; 51.5 Ci/mol), [1-14C]acetyl-CoA (Amersham Biosciences; 51 Ci/mol), [1-14C]methylmalonyl-CoA (PerkinElmer Life Sciences; 54.3 Ci/mol), and [1-14C]palmitoyl-CoA (American Radiolabeled Chemicals, Inc.; 55 Ci/mol) were used at 50 μm (except for malonyl-CoA, which was also used at 10 μm), and the protein was diluted to 3 μm. After 1-h incubation at 30 °C, the samples were denatured at 100 °C for 10 min, separated by SDS-PAGE (12% acrylamide), and analyzed by phosphorimaging (Variable Mode Imager Typhoon Trio, Amersham Biosciences) and Coomassie Blue staining. The competition binding assays were performed with the same experimental procedures using [1-14C]malonyl-CoA (50 μm) as the radiolabeled ligand and various cold ligands (malonate (50 μm), methylmalonyl-CoA (10 and 50 μm), palmitoyl-CoA (10 and 50 μm), carboxypalmitoyl-CoA (18 and 88 μm), palmitate (50 μm), and palmitoyl-AMP (50 μm)) as competitors.
Crystallization, Soaking, and Data Collection
Small fragile plates of AT52 were obtained at 20 °C by using the hanging drop vapor diffusion method with a 1:1 (v/v) ratio of protein (5 mg/ml in 20 mm Tris/HCl, 50 mm NaCl, pH 8) to precipitation solution (100 mm KH2PO4/K2HPO4, 1.3 m (NH4)2SO4, 10 mm malonate, pH 6.25). These crystals were cryoprotected in precipitation solution supplemented by 20% ethylene glycol and frozen in a stream of nitrogen gas, giving anisotropic diffraction patterns upon x-ray exposure. They belong to space group P21212 with one molecule per asymmetric unit. Large and robust bipyramidal crystals could be grown at 12 °C by using the hanging drop vapor diffusion method and mixing an equal volume of protein (3–7 mg/ml) and precipitation solution containing 0.1 m HEPES, 1.7 m (NH4)2SO4, 15% glycerol, 1.7% PEG 400, pH 7.2–7.7. These crystals diffract up to 2.2-Å resolution and belong to space group P41212 with two molecules per asymmetric unit. Complexes with carboxypalmitoyl-CoA were prepared by incubating tetragonal crystals with 6 mm ligand at 20 °C for 24 h. Soaking of crystals with methylmalonate (10 mm; 3 min at 12 °C), malonyl-CoA and methylmalonyl-CoA (25 mm; 26 h at 12 °C), palmitoyl-CoA (40 mm; 16 h at 12 °C), and tetracosanoyl-CoA (144 μm; 24 h at 12 °C) as well as co-crystallization of the protein with excess (20×) malonyl-CoA and palmitoyl-CoA at 12 °C were also performed. All ligands were solubilized in the crystallization solution. All tetragonal crystals were directly flash cooled in a stream of nitrogen gas.
Diffraction data were collected at 100 K on the ID14–1, ID14–2, and ID14–4 beamlines at the European Synchrotron Radiation Facility (Grenoble, France). Diffracted intensities were integrated using MOSFLM (24) and scaled with SCALA (25) from the CCP4 software package (26). Crystal parameters, space groups, and data collection statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set |
||||
|---|---|---|---|---|
| Apo orthorhombic | Apo tetragonal | Palmitoylated | Carboxypalmitoylated | |
| Data collection | ||||
| Beam line | ID14-4 | ID14-1 | ID14-4 | ID14-2 |
| Space group | P21212 | P41212 | P41212 | P41212 |
| Unit cell dimensions a, b, c (Å) | 108.66, 119.81, 50.16 | 106.56, 106.56, 258.61 | 105.89, 105.89, 259.62 | 106.33, 106.33, 261.69 |
| Resolution limits (Å) | 45.0-2.6 (2.7-2.6)a | 41.7-2.3 (2.4-2.3) | 52.9-2.2 (2.3-2.2) | 49.4-2.5 (2.6-2.5) |
| No. of measured reflections | 79,508 | 274,785 | 481,764 | 191,134 |
| No. of unique reflections | 20,741 | 65,097 | 69,116 | 49,363 |
| Completeness (%) | 99.6 (99.9) | 96.9 (83.4) | 91.5 (89.5) | 92.8 (91.5) |
| Rsym | 10.6 (36.8) | 6.7 (32.9) | 7.0 (31.6) | 7.2 (47.6) |
| I/σI | 3.9 (1.6) | 8.6 (2.3) | 7.0 (2.4) | 8.5 (1.6) |
| BWilson (Å2) | 43.0 | 41.5 | 42.2 | 63.3 |
| Refinement statistics | ||||
| Resolution range (Å) | 42.6–2.6 | 41.1–2.3 | 50.0–2.2 | 47.0–2.5 |
| No. of reflections work/test | 19,692/1,016 | 61,727/3,291 | 65,571/3,439 | 46,742/2,524 |
| No. of molecules/A.U. | 1 | 2 | 2 | 2 |
| Rwork | 0.187 | 0.200 | 0.223 | 0.228 |
| Rfree | 0.264 | 0.260 | 0.265 | 0.278 |
| No. of atoms | ||||
| Protein atoms | 3,466 | 6,968 | 6,959 | 6,888 |
| Peptide atoms | 72 | 161 | 148 | 73 |
| Ligand atoms | —b | — | 34 | 47 |
| Water/other solvent heteroatoms | 145/50 | 351/44 | 234/52 | 98/60 |
| r.m.s.d.c bond lengths (Å) | 0.024 | 0.019 | 0.011 | 0.007 |
| r.m.s.d. bond angles (°) | 2.147 | 2.068 | 1.475 | 1.327 |
| Mean temperature factor (Å2) | ||||
| Protein main chain | 26.6 | 38.7/35.7d | 44.8/41.9 | 44.3/51.6 |
| Protein side chain | 27.9 | 40.4/37.4 | 46.9/44.2 | 45.6/52.5 |
| Peptide overall | 55.8 | 88.0/54.4 | 84.4/56.4 | —/72.2 |
| Ligand | — | — | 58.6/55.5 | 40.4/66.2 |
| Water/other solvent entities | 26.6/55.7 | 39.2/67.1 | 45.7/67.1 | 43.1/76.8 |
a Values in parentheses are for highest resolution shell.
b —, not applicable.
c r.m.s.d., root mean square deviation.
d Mean temperature factor values related to each of the two molecules in the asymmetric unit (A.U.) have been indicated separately.
Structure Determination
An initial model was obtained using the molecular replacement program PHASER (27) with the crystal structure of the KS-AT didomain of module 5 from DEBS (13) (Protein Data Bank code 2HG4; residues 467–892 encompassing the AT domain and the KS to AT linker) as a model and the data set of the tetragonal form. Improved quality maps were generated by performing histogram matching, solvent flattening, and 2-fold non-crystallographic symmetry averaging by using the program DM (26). Refinement was carried out with REFMAC5 (28), whereas the model was built manually using Coot (29). Water molecules were added via the Coot/findwaters subprogram and inspected manually. The structure was refined to 2.3-Å resolution with final Rwork and Rfree values of 0.200 and 0.260, respectively. The final model is composed of two AT52 molecules (chain A (residues 595–1059) and chain B (residues 596–1062)), two supplementary peptides of 12 residues, 351 water molecules, three glycerol molecules, and two sulfate ions. This structure was in turn used to solve the structure of AT52 in the orthorhombic form and the structures of protein-ligand complexes. The 2.6-Å resolution refined orthorhombic structure contained one AT52 molecule (residues 596–1062) and the supplementary peptide of 12 residues, 145 water molecules, six sulfate ions, and six ethylene glycol molecules leading to Rwork and Rfree values of 0.187 and 0.264, respectively. Refinement statistics are detailed in Table 1. All structures were validated using PROCHECK (30) and Coot validation tools. Atomic coordinates and restraints of ligands used for the structures of the different complexes were generated with the PRODRG2 (31) server and JLigand (59). Composite simulated annealing omit maps were calculated in PHENIX (32) using Protein Data Bank coordinates missing the palmitoyl and carboxypalmitoyl groups. Electrostatic potential of protein surfaces was calculated via the PDB2PQR (33, 34) server and using the program APBS. All structures and the electrostatic potential of protein surfaces were visualized with PyMOL (35). Structure alignment analyses were performed using ESCET (36) and PROFIT. Rendering of sequence alignment was performed using ESPript (37). Atomic coordinates have been deposited in the Protein Data Bank under the following accession codes: 3TZW (orthorhombic apo form), 3TZX (tetragonal apo form), 3TZY (palmitoylated form), and 3TZZ (carboxypalmitoylated form).
RESULTS
Production of a Functionally Active Acyltransferase Fragment of Pks13
The purified full-length Pks13 polyketide synthase from M. tuberculosis H37Rv was subjected to limited proteolysis using α-chymotrypsin. Two stable fragments (with molecular masses of approximately 32 and 52 kDa) persisted after 10 h of incubation. Purification of the highest molecular weight fragment by gel filtration experiment followed by electrospray ionization MS analysis gave an experimental mass of 52,612 ± 10 Da. The band at 52 kDa obtained by SDS-PAGE was also analyzed by tandem mass spectrometry. The MS/MS experiments revealed that this 52-kDa fragment comprises Pks13 residues 576–1062 (80% sequence coverage), which correspond to the AT domain plus partial interdomain linkers between the upstream KS domain and the downstream ACP domain. The theoretical mass (52,615 Da) deduced from the sequence is in agreement with the experimental mass. The DNA sequence corresponding to the Pks13(576–1062) fragment, hereafter referred to as AT52, was then subcloned into a pET expression vector with an N-terminal His6 tag, and the resulting construct was used to transform an E. coli strain. The recombinant protein was overexpressed and purified to homogeneity in two chromatographic steps, and the histidine tag was removed.
Previous in vitro experiments on full-length Pks13 revealed the predilection of the AT domain for long carboxyacyl-CoA substrates in accordance with the role it played for the selection of mycolic acid C24–C26 alkyl chain (21). These experiments also demonstrated the position of the acylation on the catalytic serine (Ser801). A similar study on the loading of functional ligands and a competition binding assay were undertaken to finely characterize the AT52 substrate specificity (Fig. 1B). First, the AT52 fragment was incubated with various radioactive ligands, and the relative intensity of each lane was measured. Acetyl-CoA (lane 1) did not exhibit any detectable affinity to AT52. The presence of a carboxylic acid group (malonyl-CoA; lanes 2 and 3) and the introduction of a methyl group (methylmalonyl-CoA; lane 4) significantly increased ligand loading, which was also dependent on the ligand concentration. Finally, the presence of a long fatty acyl chain (palmitoyl-CoA; lane 5) dramatically raised the affinity of the protein in agreement with previous data (21). These results demonstrated that the AT52 fragment retains ligand loading ability as found for intact Pks13.
A competition binding assay was then carried out to evaluate the relative affinity of non-radioactive substrates in comparison with [1-14C]malonyl-CoA (50 μm). In these conditions, malonate, which harbors two free carboxylic acid groups, could not be selected by the AT52 fragment (lane 6). Increasing amounts of methylmalonyl-CoA (lanes 7 (10 μm) and 8 (50 μm)) reduced the quantity of radiolabeled protein by up to 50%, thus confirming that malonyl-CoA and racemic methylmalonyl-CoA share comparable affinities for the AT fragment of Pks13. On the other hand, it is noteworthy that the presence of a methyl substituent enhances the affinity of the substrate for the full-length protein (21). Long chain (C16) fatty acyl-CoA derivatives with and without a carboxylic acid group on the β-carbon were then challenged. Palmitoyl-CoA at either 10 or 50 μm (lanes 9 and 10, respectively) significantly reduced the signal on the autoradiogram, whereas complete suppression of the radiolabeled intensity was achieved by using carboxypalmitoyl-CoA even at 18 μm (lanes 11 and 12), demonstrating that the carboxylated ligand has the highest affinity. Finally, neither palmitic acid (lane 13) nor palmitoyl-AMP (lane 14) could decrease the amount of radiolabeled protein. Thus, the presence of long fatty acyl chains is not sufficient for ligands to be selected by AT52. These results are in agreement with a gatekeeper role of the acyltransferase domain and demonstrate that AT52 predominantly selects atypical substrates that are long carboxylated fatty acid chains esterified by coenzyme A.
Structure Determination
Several structures of the AT52 fragment were elucidated using x-ray crystallography. Two crystal forms of apoAT52 were obtained: one in space group P21212 with one molecule per asymmetric unit and the other in space group P41212 with two molecules per asymmetric unit. Because of their higher diffraction limit, tetragonal crystals were later used to better understand the selection and loading of substrates into the acyltransferase reaction chamber. Several soaking and co-crystallization experiments with available commercial molecules, such as malonyl-CoA, racemic (2R,2S)-methylmalonate and (2R,2S)-methylmalonyl-CoA, and palmitoyl-CoA, as well as non-commercial racemic (2R,2S)-carboxypalmitoyl-CoA and tetracosanoyl-CoA were performed. For every experiment with the exception of soaking with methylmalonate where crystals dramatically suffered, diffraction data were collected, and the corresponding structures were solved. Among all the structures solved, only those of the two apo forms and of the palmitoylated and carboxypalmitoylated enzymes will be described in detail here (Table 1). All structures have >99% of their residues in the favored or allowed regions of the Ramachandran plot as defined by PROCHECK.
A Conserved Molecular Organization with Intrinsic Plasticity
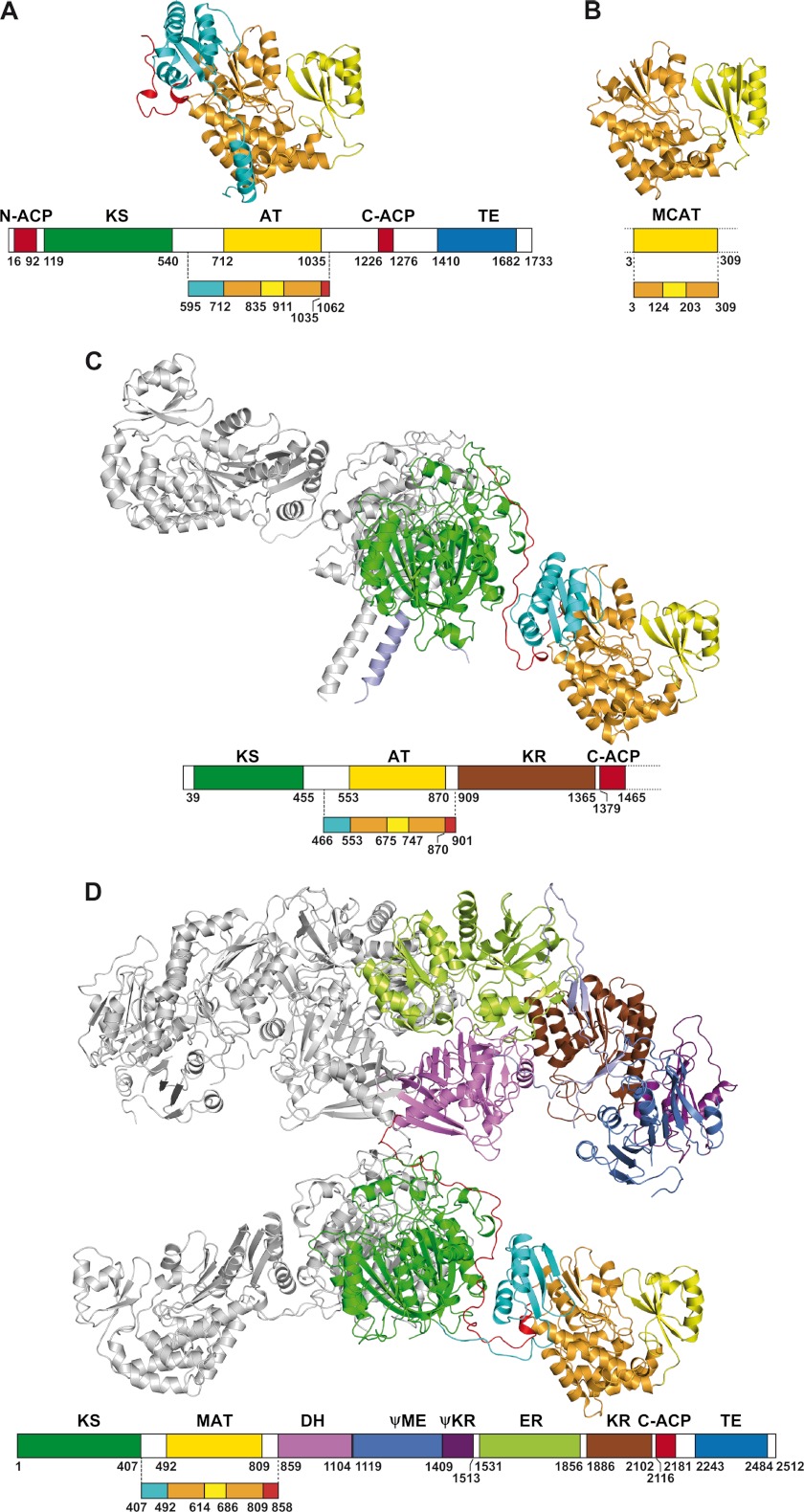
The structure of the AT52 fragment comprises the Pks13 AT domain and parts of the upstream KS-AT and downstream AT-ACP linkers (Fig. 2). The AT domain consists of a large α/β-hydrolase-like subdomain delineated by residues 712–835 and 911–1035 and a smaller ferredoxin-like subdomain, which comprises residues 836–910. Thus, the AT domain of the AT52 fragment adopts the canonical fold found in various individual AT enzymes, such as malonyl-CoA:ACP transferases (MCATs) (38–41); in the AT domain associated with DEBS (7, 13, 41, 42); and in malonyl-acetyltransferase (MAT) domains associated with FAS enzymes (7, 13, 41, 42) (Fig. 3). Pairwise superposition of all protomers found in the four AT52 structures described here led to root mean square deviation values in the range of 0.2–1.5 Å. Further structural comparison revealed that the AT52 protomers can be divided into two distinct groups that differ by the respective positioning of two conformationally invariant regions (supplemental Fig. 1A). The first region is delineated by the α/β-hydrolase subdomain plus the KS-AT and AT-ACP linkers. The second region includes the ferredoxin-like subdomain. Thus, our data show that no flexibility is observed in AT52 between the linkers and the α/β-hydrolase-like subdomain, whereas the ferredoxin-like subdomain, helped by the two anchoring loops, can be considered as a flexible element of the protein. A similar observation has been reported in the x-ray structures of fungal, yeast, and mammalian FASs (4, 6, 7) and of both the KS-MAT didomain (42) and isolated MAT domain of human FAS (41). Pairwise superposition of Pks13 AT52, E. coli MCAT, KS-AT of module 5 from DEBS, and FAS structures also illustrates this flexibility (supplemental Fig. 1B).
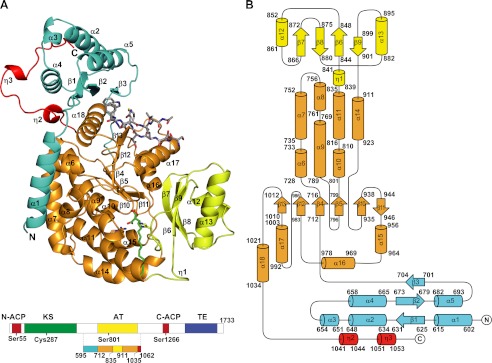
FIGURE 2.
Overall structure of AT52. A, ribbon representation. B, topology diagram. The α/β-hydrolase subdomain is in orange, the ferredoxin-like subdomain is in yellow, the KS-AT linker is in cyan, and the AT-ACP linker is in red. Helices and β-strands are numbered. The carboxypalmitoyl chain and the 12-mer peptide are shown as sticks with nitrogen and oxygen atoms in blue and red, respectively. Carbon atoms of the carboxypalmitoyl chain and of the peptide are in green and light gray, respectively. The side chains of active site residues Ser801, Arg826, and His909 are also depicted. The same color scheme has been applied throughout all the figures.
FIGURE 3.
Comparison of known structures comprising AT, MCAT, and MAT domains. A, AT52 of Pks13 from M. tuberculosis. B, E. coli MCAT (Protein Data Bank code 2G2Z). C, KS-AT didomain of module 5 from DEBS (Protein Data Bank code 2HG4). D, mammalian FAS-I (Protein Data Bank code 2VZ8). The structures are shown such that the AT domains are in the same orientation. The domains and linkers are color-coded as follows: α/β-hydrolase and ferredoxin-like subdomains, orange and yellow, respectively; KS-AT linkers, cyan; post-AT linkers, red; KS, green; dehydratase (DH), violet; pseudo-methyltransferase (ΨME), blue; pseudo-ketoreductase (ΨKR), magenta; enoyl reductase (ER), lime green; ketoreductase (KR), brown. The second chains of the KS-AT didomain of module 5 from DEBS and mammalian FAS dimers are in light gray.
Upstream of the AT domain, residues 625–704 of the Pks13 KS-AT linker form a compact domain made of an antiparallel three-stranded β-sheet (β1, β2, and β3), which faces the AT domain, covered on the opposite side by three moderately long (10–16 residues) α-helices (α2, α4, and α5) themselves capped by a helical turn (Fig. 2A). The folded linker domain of Pks13 is strikingly similar to that found in the structures of the KS-AT didomain of DEBS modules 3 and 5 (12, 13) with a root mean square deviation value of 0.9 Å for 76 aligned positions sharing 38% sequence identity. The same type of fold was also reported in the available structures of FAS although with a lower structural similarity (7, 42). As in DEBS and FAS, a short peptide segment connects the KS-AT adaptor and the AT domain in Pks13 (Fig. 4).
FIGURE 4.
Structure-based multiple sequence alignment of AT52 from Pks13 and other MCAT/MAT/AT-containing enzymes. The sequence numbering is for AT52. Sequence homology is highlighted in red, whereas sequence identity is shown as white letters on a red background. Secondary structure elements (arrows for β-strands and coils for α-helices) of AT52 are indicated at the top and colored according to the different subdomains depicted in Fig. 2. Residues of AT52 that are disordered in the crystal structure have been underlined in black. Residues located at the C-terminal end of the adjacent KS domain have been underlined in green. The catalytic residues are indicated by triangles, the arginine residue involved in selecting carboxylated substrates is indicated by a star, and other important residues are indicated by ellipses. MYCTU, M. tuberculosis; SACER, Saccharopolyspora erythraea; ECOLI, E. coli.
In contrast, the structure-based sequence alignment revealed that in Pks13 an 84-residue-long peptide stretch separates the first β-strand of the folded KS-AT linker domain (residue 625) from the last β-strand of the KS domain (which presumably ends at residue 540), whereas this connection comprises about 15 residues in DEBS and FAS (Fig. 4). This long insertion is not found in other mycobacterial PKSs and is thus specific to Pks13. The AT52 construct starts at residue 576, but residues 576–594 could not be seen in the electron density maps and are thus absent from the crystal structure due to intrinsic flexibility. Indeed, we verified by MS of dissolved crystals that the protein did not undergo any proteolytic degradation during the crystallization process. Residues 595–624 comprise an α-helix (α1; Glu602–Ala617) that runs opposite to the Pks13 KS domain, assuming a similar KS and AT arrangement as in FAS and DEBS (Fig. 3), and wraps around the AT domain (Fig. 2A). In this configuration, residue 595 is about 60 Å from the expected position for residue 540, a distance that could be easily spanned by the 54 residues found between residues 540 and 595. The N terminus of AT52 is associated to the catalytic domain through a set of apolar interactions involving residues belonging to helix α1 on the one side and to helices α6, α7, and α8 on the other side, providing an interaction area of 800 Å2.
The structure of AT52 also comprises part of the ∼200-residue-long post-AT linker that connects the AT and C-terminal ACP domains of Pks13. All 27 residues downstream of the AT domain of the AT52 construct, i.e. residues 1036–1062, could be traced in the electron density. They form a long V-shaped loop containing two helical turns (η2 and η3) that wraps around the KS-AT linker (Fig. 2A) as observed previously in the structures of KS-AT didomains and full-length mammalian FAS (7, 12, 13, 42) (Fig. 3). Compared with the N terminus of AT52, residues of the post-AT linker make extensive polar and apolar contacts with the remaining part of the structure, mostly with helix α4, leading to an interaction area of 1100 Å2.
Description of the Active Site
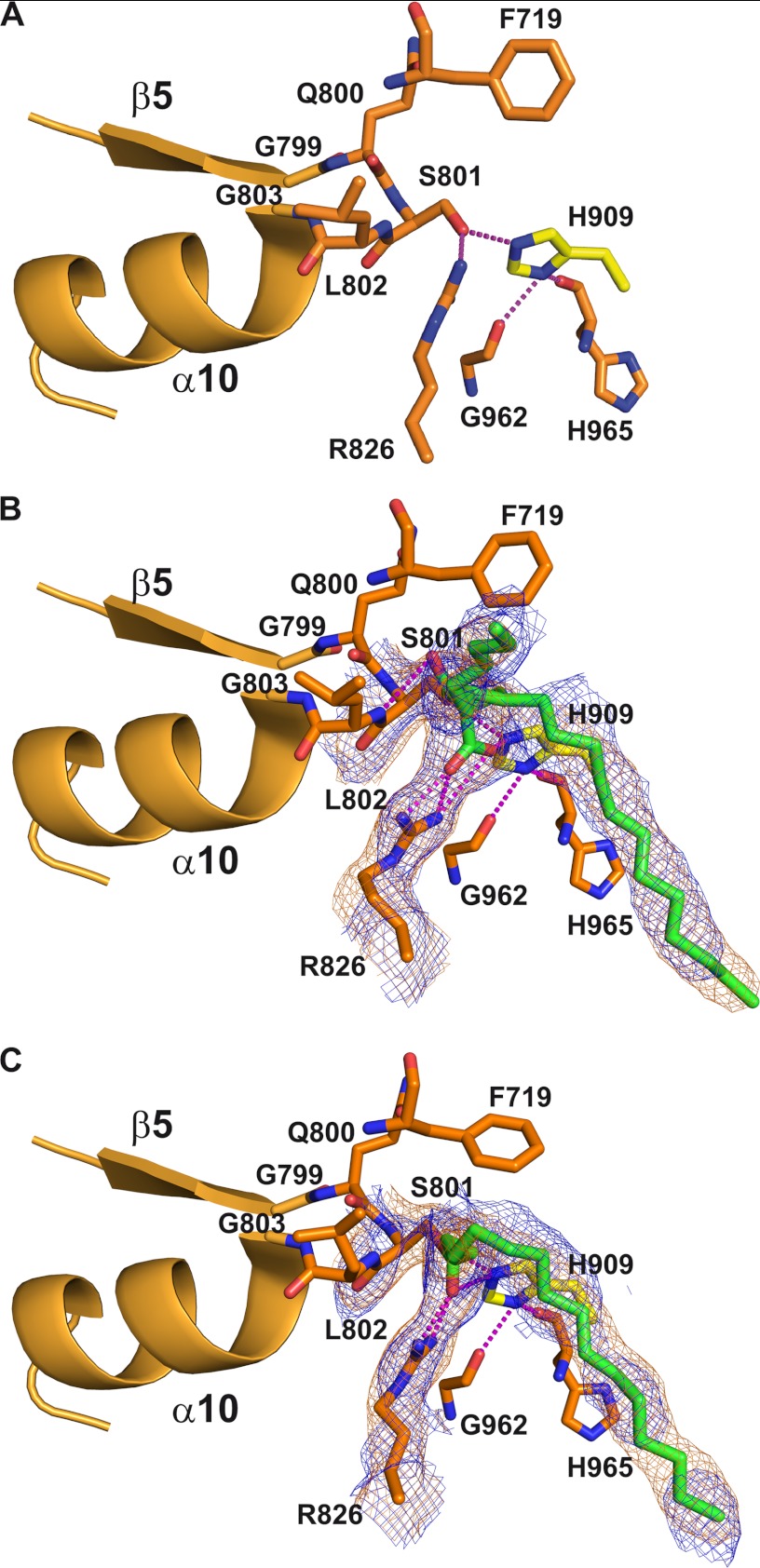
Sequence alignment between AT52 and various acyltransferase catalytic domains revealed that Ser801 and His909 could play the role of catalytic residues (Fig. 4). This prediction was further confirmed by analysis of the AT52 structure (see below) and is in agreement with MS/MS fragmentation data in the case of the catalytic serine (21). The active site of the AT domain of Pks13 is located in a gorge at the interface between the two subdomains (Fig. 2A). Ser801 is found in a nucleophilic elbow between strand β5 and helix α10 of the large subdomain and is part of the highly conserved consensus sequence Gly-X-Ser-X-Gly (38) (Figs. 4 and 5A). The nucleophilic power and reactivity of the catalytic serine is enhanced by the macrodipole effect of helix α10. Ser801 is also constrained into an energetically unfavorable conformation characteristic of the nucleophilic residue in α/β-hydrolase proteins (43, 44). This geometry contributes to the formation of an oxyanion hole that should help stabilize the negatively charged reaction intermediate during the catalytic process. Based on previous studies (39, 45), one can anticipate that the oxyanion hole of AT52 likely involves the backbone amides of residues Leu802 and Phe719. The Phe719 amide is not in an optimal orientation to interact with the carbonyl group of the substrate and requires rearrangement of the backbone dihedral angles to contribute to an active oxyanion hole (39). His909, the other catalytic residue, is located in a loop at the C-terminal end of the ferredoxin-like subdomain and faces Ser801, thereby increasing its reactivity (Fig. 5A). Indeed, the δ-nitrogen atom of His909 is at hydrogen bond distance from the carbonyl groups of Gly962 and His965. The two hydrogen bonds enable an optimal orientation of the side chain of His909 where the ϵ-nitrogen atom establishes a hydrogen bond with the hydroxyl group of the catalytic serine. A highly conserved arginine residue within the family of AT and MCAT proteins is also found within the active site of AT52 (Arg826). Different studies, including the structure of E. coli MCAT esterified by a malonyl residue (40) and bioengineering of the MCAT from Streptomyces coelicolor (46), have demonstrated that this arginine is involved in the recognition and selection of carboxylated substrates. In AT52, Arg826 is located in helix α11 at the base of the active site. In the tetragonal apo form, both NH side chain atoms of Arg826 make hydrogen bonds with the hydroxyl group of the catalytic serine. The conformation of Arg826 is also held in position through interaction with the side chain of Gln773. It is noteworthy that Gln773 is conserved in MCAT from E. coli and in DEBS (residues 63 and 614, respectively) but not in mammalian and fungal FASs where a phenylalanine is found (Phe553 and Phe250, respectively).
FIGURE 5.
Detailed description of the active site of AT52. A, apo form. B, carboxypalmitoylated state with the R stereoisomer. C, palmitoylated state. The nucleophilic elbow comprising strand β5 and helix α10 is shown in ribbon representation. Important residues defining the active site are shown and labeled. Hydrogen bonds are represented by magenta dotted lines. The final 2mFo − DFc and 2Fo − Fc simulated annealing omit electron density maps contoured at 1.0σ around the carboxypalmitoyl or palmitoyl moiety and selected residues are shown as orange and blue mesh, respectively. For sake of clarity, the side chain of Gln773, which interacts with the side chain of Arg826, has been omitted.
Structural Study of AT52 in Complex with Lipidic Ligand Analogues of Pks13
Although many structures of AT enzymes have been solved, it is noteworthy that only the structure of E. coli MCAT has been determined in the acyl enzyme state (40) undoubtedly because of rapid hydrolysis of acyl enzyme intermediates (46, 47).
In our work, no sufficiently defined electron density could be observed in the maps obtained from crystals soaked with malonyl-CoA and methylmalonyl-CoA, resulting in ambiguous positions and orientations of the corresponding short acyl chains even in the presence of a large excess of substrates (>200) and with long incubation periods (26 h). On the contrary, well defined extra electron density was identified in the active site when crystals were soaked with palmitoyl-CoA or carboxypalmitoyl-CoA. In both cases, this allowed positioning of esterified fatty chains in the two molecules of the asymmetric unit of the tetragonal form (Table 1). This is in agreement with previous results obtained for full-length Pks13 where stable acyl enzyme intermediates could be analyzed by mass spectrometry, demonstrating the formation of a covalent link between the AT domain and a carboxy-C16 chain (21). It might be possible that the crystallization conditions and crystal packing environment could even slow the deacylation process in the case of AT52. A structural comparison of the organization of the active site in the apo, acylated, and carboxyacylated forms is given in Fig. 5. Esterification did not induce any significant rearrangement of the active site geometry and residue conformation. The only major conformational rearrangement occurred upon carboxyacylation and concerns the side chain of Arg826. Indeed, this residue plays a role in selecting the carboxylated substrate as expected by establishing a bidentate salt bridge between its guanidinium group and the carboxylate group of the ligand. To establish such a planar interaction, the side chain of the arginine must move back, and these positional and conformational changes of Arg826 are only observed in the case of the carboxypalmitoyl chain (Fig. 5B). In the case of the palmitoyl chain, the movement of Arg826 also occurred but to a lesser extent, and the carbonyl group of the ester has flipped to establish hydrogen bonding interactions with the Arg826 guanidinium group (Fig. 5C). It is noteworthy that the interaction between Arg826 and Gln773 is maintained upon acylation.
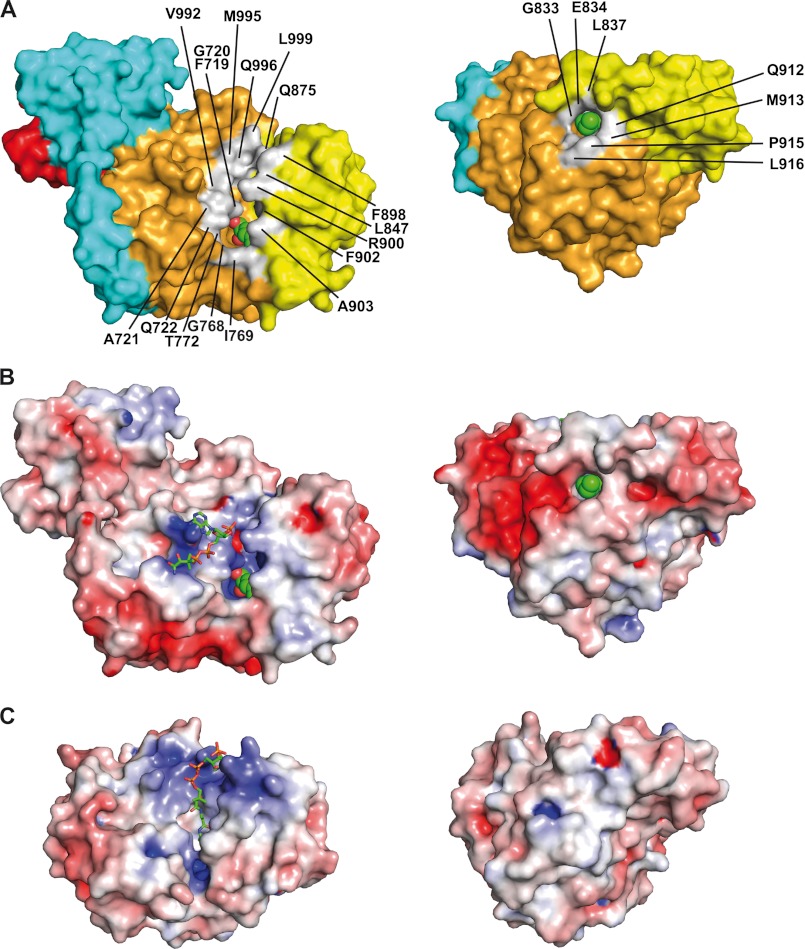
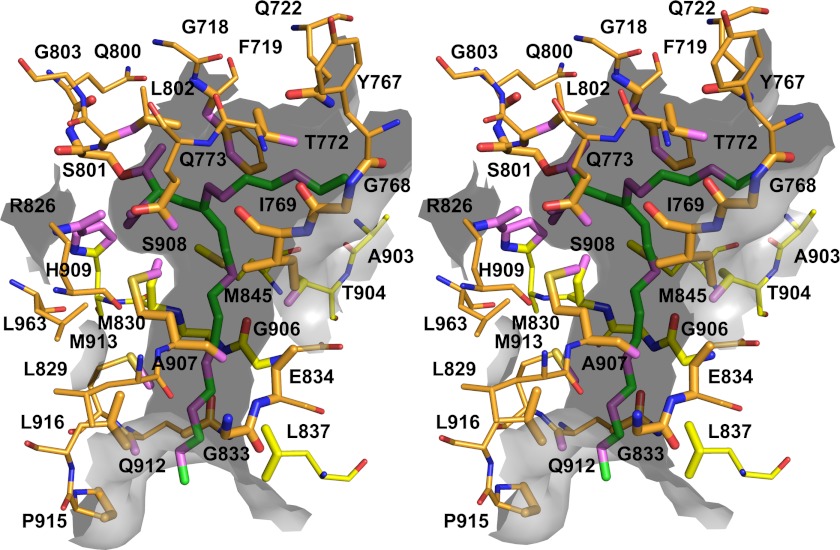
Analysis of the topography of AT52 revealed an open groove at the apex of the active site followed by a long open channel that exits the catalytic chamber and runs parallel to the major axis of the protein (Fig. 6A). The groove lies at the border between the α/β-hydrolase and the ferredoxin-like subdomains and comprises 17 residues scattered at the surface of the enzyme (Phe719, Gly720, Ala721, Gln722, Gly768, Ile769, Thr772, Leu847, Gln875, Phe898, Arg900, Phe902, Ala903, Val992, Met995, Gln996, and Leu999), defining a rather hydrophobic crown (Fig. 6A). Direct access to the catalytic site is partially impeded by residues Arg900, Phe902, and Phe719 from top to bottom and in the tetragonal crystal structures of both the apo and acylated forms by a molecule of glycerol provided by the crystallization medium. Upon acylation, the first five carbon atoms of the acyl chain sit at the base of the catalytic site in the same plane and are accessible from the groove opening (Figs. 5, B and C, and 6A). The acyl chain then becomes kinked to enter and fit inside the long channel (delineated by residues Met830, Gly833, Glu834, Leu837, Met845, Thr904, Gly906, Ala907, Ser908, Gln912, Met913, Pro915, and Leu916), thereby burying the nine following carbon atoms whereas the last two carbons remain accessible to solvent (Fig. 7). Electrostatics calculations revealed the presence of an electropositive area corresponding to the floor of the active site cavity due to the presence of Arg826 and His909 and an electronegative area close to the channel exit (Fig. 6B). They also confirmed the hydrophobic character of the channel. A second conformation was observed for one of the two molecules in the asymmetric unit of the carboxyacylated form. This conformation arises from rotation around the C3-C4 bond of the acyl chain. Only 11 of the 16 carbon atoms could be traced in the electron density maps, thus illustrating its high flexibility (Fig. 5B). The stabilized part of the acyl chain is found in a deep, solvent-exposed cleft delineated by atoms belonging to residues Phe719, Gln722, Tyr767, Gly768, Ile769, Thr772, Gln773, Leu802, and Ala903 (Fig. 7).
FIGURE 6.
Topography of the active site and hydrophobic channel of AT52. A, molecular surface. B, electrostatic potential surface. Two perpendicular views are shown. Residues delineating the groove, at the apex of the active site, and the exit of the channel are colored in light gray and labeled. The catalytic serine and the carboxypalmitoyl chain are shown as spheres. C, electrostatic potential surface of E. coli MCAT (Protein Data Bank code 2G2Z). The CoA-SH moieties are shown as balls and sticks with green carbon atoms (nitrogen, blue; oxygen, red; phosphorus, orange; sulfur, yellow). Electrostatic potential is color-coded from red (−6 kBT/e) to blue (+6 kBT/e); white is neutral.
FIGURE 7.
Stereoview of the carboxypalmitoyl group in its dedicated tunnel in AT52. Residues delineating the tunnel are represented as sticks and labeled. Atoms found within 5 Å of the ligand are shown as enlarged sticks and were used to depict the tunnel surface, shown as the gray semitransparent surface. Protein (respectively ligand) atoms found within 3.5 Å distances of ligand (respectively protein) atoms are colored violet. The second incomplete conformation of the carboxypalmitoyl group and its chemical environment are also depicted using the same representation scheme.
Peptide Binding at the Surface of the Protein
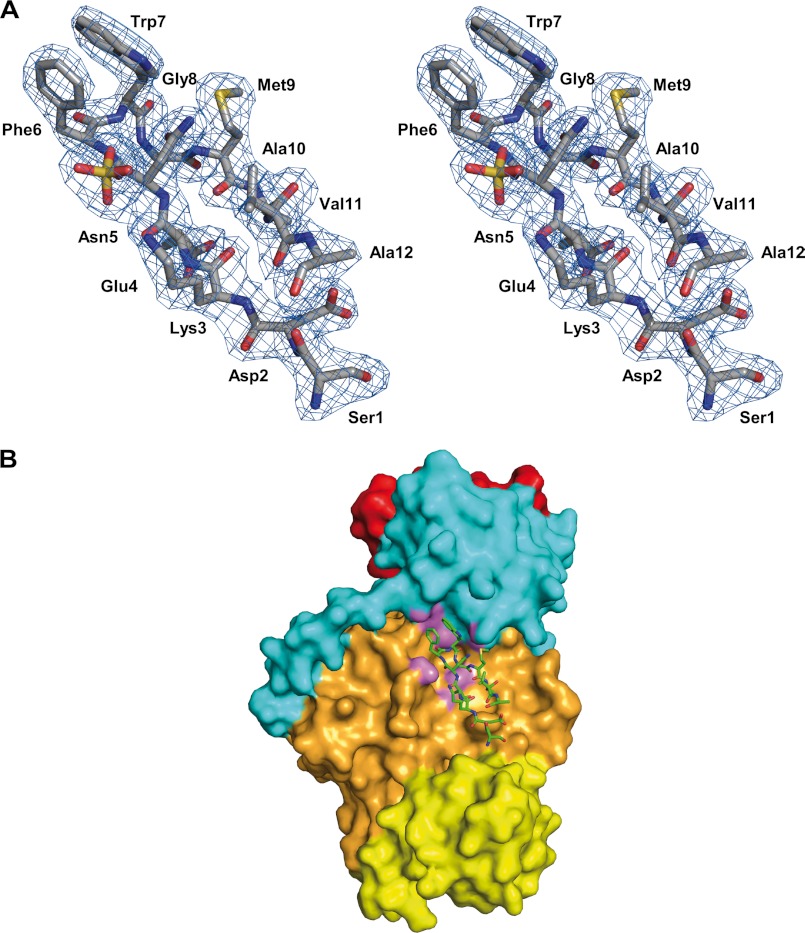
The crystallographic analysis of AT52 revealed an intriguing feature. Additional electron density was found in all resolved structures that corresponds to a co-purified and crystallized peptide (Fig. 8A). However, in the tetragonal form, the electron density map was better defined for one of the two protein molecules in the asymmetric unit, suggesting that the binding site is not fully occupied and/or the peptide displays some flexibility. Although ambiguities remained concerning the exact sequence and length of the peptide (>12 residues), the following putative sequence could be assigned for the best defined electron density map: Ser25-Asp/Asn75-Lys80-Glu/Gln65-Asp/Asn75-Phe85-Trp100-Gly/Ala50-Met70-Ala10-Thr/Val50-Ala10 (where subscript numbers indicate the confidence level based on electron density and most probable side chain rotamers).
FIGURE 8.
Peptide binding at the surface of AT52. A, putative sequence derived from the 2mFo − DFc electron density map (shown here at 1.0σ contour level). B, map of the interaction of the peptide at the molecular surface of the protein. Atoms of the protein within 3.5 Å of any peptide atom are shown in magenta.
We verified that this peptide did not arise from proteolysis of AT52, and important efforts were undertaken to determine its exact sequence using Edman sequencing and tandem mass spectrometry. Experiments carried out with protein from several purification batches or dissolved crystals were unsuccessful. Further experiments based on the separation of the peptide from the purified protein by Tris-Tricine SDS-PAGE (48) before analysis by mass spectrometry and Edman sequencing suggested the presence of several low molecular weight, difficult to ionize species in the protein solution. They also suggested that the co-crystallized peptide could be modified or cyclized, which impeded sequence assignment. Unfortunately, a database sequence similarity search using the crystallographically derived sequence of the peptide did not result in any hits.
The peptide-binding site is defined by a hydrophobic cavity at the interface between the KS-AT linker and the AT α/β-hydrolase subdomain (Fig. 8B) and is delineated by the β2-α5-β3 and β13-α18 structural segments, respectively. The peptide adopts β-hairpin geometry with residues Asp/Asn-Phe-Trp-Gly/Ala forming a type I turn and the two aromatic side chains in a stacking interaction. Protein-peptide interactions are mostly hydrophobic, and there are few polar interactions, giving a total buried surface area of 2070 Å2.
DISCUSSION
M. tuberculosis and other mycobacteria have developed the remarkable ability to synthesize a large variety of lipids. These unique lipids primarily play a structural role and therefore contribute to the low permeability of the cell envelope to many hydrophilic molecules. Being positioned at the bacterial surface, they also intervene in the interplay between host and pathogen, and their direct role in pathogenicity and virulence has been clearly established (49). The strong capacity of M. tuberculosis to synthesize such a unique lipid repertoire, including mycolic acids, relies on FAS and PKS systems as well as activating enzymes. Pks13, an essential type I PKS containing the ACP-KS-AT-ACP-TE domains, catalyzes the condensation of two fatty acids chains to form a β-keto ester, the ultimate intermediate of the mycolic motif (20). Whereas the catalytic mechanism of Pks13 has been well characterized (21), nothing is known about its three-dimensional structure. This prompted us to set up a global structural characterization, which included a limited proteolysis approach to identify isolated functional domains of this complex enzyme, such as the 52-kDa fragment containing the AT catalytic domain (Pks13(576–1062) or AT52) described in this study. This fragment could be purified to homogeneity and was subjected to biochemical and crystallographic analyses where several x-ray structures in the apo form and in complex with substrate analogs have been solved.
The structure of AT52 provides another example of the remarkable structural conservation among type I FAS and PKS enzymes. Indeed, besides the canonical catalytic domain per se, Pks13 also displays folded KS-AT and AT-ACP linkers similar in conformation and position to those found in other known structures. In contrast, the longer upstream connection between the KS and AT domains of Pks13 that results in an unprecedented α-helix may exemplify the singularity displayed by these multifunctional, multidomain enzymes: specific structural elements and fine tuning of the relative position between linkers and catalytic domains may be related to the distinct specificities of the various PKSs. In line with this idea, it has been shown recently for DEBS that the KS-AT and post-AT linkers play an important role for both acylation and transacylation of AT and ACP domains, respectively (50).
Acyltransferase domains function as the primary “gatekeepers” for selecting the extender units that become incorporated into polyketide or fatty acid chains. These building blocks are most often in the form of (methyl)malonyl-CoA with also acetyl-CoA, propionyl-CoA, and other derivatives, and several functional, structural, and computational studies have been conducted to understand key features involved in substrate specificity and selectivity (39, 40, 45, 46, 51). In contrast, the AT domain of the mycobacterial Pks13 enzyme exhibits a very unusual substrate specificity (C24–C26 carboxyacyl-CoAs in M. tuberculosis) in comparison with other PKS enzymes of known function. A binding and competition assay with various ligands confirmed that the isolated AT domain of Pks13 keeps the ability to load long chain carboxyacyl-CoA molecules. In accordance with the functional study, the crystallographic analysis revealed that no or only moderate binding could be observed in the presence of malonyl-CoA or methylmalonyl-CoA compared with palmitoyl-CoA or carboxypalmitoyl-CoA. Although racemic (2R,2S)-carboxypalmitoyl-CoA was used for this study, neither the electron density nor the chemical environment could help discriminate between stereoisomers, a somewhat surprising result considering that AT domains select α-substituted substrates usually in S configuration (51). Previous results with S. coelicolor MCAT revealed that two residues (Met126 and Phe200) could form a selectivity filter against α substituents due to steric clashes (39). Met126 and Phe200 are replaced by Met830 and Ser908, respectively, in AT52. Because the methionine residue is conserved, the sole presence of a small side chain at position 908 of AT52 could indeed explain the accommodation of an α-substituted substrate but does not seem to play a role for R/S selectivity. The apparent non-discrimination of substrate stereoisomers in the case of AT52 is in contrast with what is currently known for FAS and other PKS enzymes. In all systems studied so far, a single configuration of the substrate could be selected by the AT domain. Because the asymmetric α-carbon of mycolic acids is in the R configuration, this configuration should then be determined upstream by the acyl-CoA carboxylase complex or downstream by the Pks13 KS domain, which performs the Claisen condensation reaction between the activated meromycolic and carboxyacyl chains. To our knowledge, it is not known whether the acyl-CoA carboxylation reaction is stereospecific. In contrast, in type I FASs from animals and yeast, KS-mediated condensation occurs with inversion of configuration at the C2 position. In type I PKSs, the situation is more complex because the KS-mediated condensation reaction can occur with inversion or retention of stereochemistry (52). Finally, although the crystallographic study could not help determine any preference of AT52 for one of the two substrate stereoisomers, we cannot exclude that such discrimination (if any) is kinetically controlled.
Complex formation experiments with palmitoyl-CoA and carboxypalmitoyl-CoA revealed the cleavage of the substrates and for the first time how a C16 fatty acid chain is accommodated inside a dedicated hydrophobic channel. However, the AT52 structures did not allow us to visualize where the remaining carbon atoms would be located in the case of a longer than C16 fatty acyl chain. Indeed, crystallographic analysis after soaking with tetracosanoyl-CoA (C24-CoA) revealed that only the 16 proximal carbon atoms of the ligand are visible in the corresponding electron density map and that binding occurs in a way similar to that observed for the palmitoylated form (data not shown). In addition, the volume of the active site cavity is not big enough so that a C24–C26 fatty chain could be pleated inside. Therefore, at this stage, we cannot conclude whether C24–C26 carboxyacyl-CoAs will still protrude outside the full-length enzyme and consequently retain the flexibility observed in the case of AT52. If this is the case, substrate selectivity could then rely on the mycobacterial acyl-CoA carboxylase (53). The atypical hydrophobic channel found in AT52 differs from the hydrophobic cleft identified in the malonyl/palmitoyl transferase domain from fungal and yeast FAS-I (4, 6) that is notably responsible for the back-transfer of C16 or C18 fatty acids to CoA to terminate the catalytic cycle. The AT52 dedicated hydrophobic channel is due to changes of the position of helix α11 and of the two loops connecting the ferredoxin-like and α/β-hydrolase subdomains, including the insertion of the segment formed by residues 838–841. It also arises from differences in the primary sequences at specific positions. Subtle changes to design long hydrophobic channels on a canonical fold have also been reported for the mycobacterial type III polyketide synthase Pks18, which adopts a thiolase fold and uses C6–C20 acyl-CoA starter units to synthesize tri- and tetraketide pyrones (54). A long hydrophobic channel allowing accommodation of the α-branch of mycolic acids has also been identified from the crystal structure of antigen 85C. Antigen 85C is structured as a α/β-hydrolase fold and catalyzes the formation of trehalose dimycolate (cord factor) through the transfer of a mycolic acid from one molecule of trehalose monomycolate to a second molecule of trehalose monomycolate (55). Although the hydrophobic channel is preformed in the structure of apoAT52, binding and dissociation of a fatty acid chain could imply further opening of the active site, which in turn might rely on the observed flexibility between the α/β-hydrolase and ferredoxin-like subdomains.
In contrast to E. coli MCAT where the crystallographic analysis of the complex formed with malonyl-CoA revealed the position of the CoA-SH moiety, this part of the processed substrates could hardly be assigned in the electron density maps corresponding to the structures of the AT52 complexes. In fact, residual densities were found in nearly all refined structures obtained in the presence of CoA derivatives, but only in the case of soaking with a high concentration of ligands was the electron density sufficiently defined to partially build the CoA-SH moiety, i.e. where the β-alanine and cysteamine groups were missing. The putative CoA-SH-binding site of AT52 is delineated by an electropositive area comprising four basic residues (Arg724, Lys725, Arg900, and Arg1017) located close to the active site main entrance (Fig. 6B). In the case of E. coli MCAT, the CoA-SH is well anchored in the active site gorge above the β-sheet of the small domain with the pantothenic and thiol groups plunging toward the catalytic serine (Fig. 6C).
Little is known about the interaction of MCAT/MAT/AT domains with ACP. ACP plays a critical role in type I FAS and PKS systems by translocating covalently bound intermediates from an active site to another, and it has been shown that productive interactions between ACP and other catalytic domains deeply rely on a high degree of structural flexibility (56). Although partly experimentally supported, physical interaction between ACP and MCAT/MAT/AT has been studied by molecular docking, leading to different modes of interaction (39, 45, 50, 57). ACP domains or individual proteins exhibit a conserved and rather dynamic three-dimensional structure with a common fold consisting of three major α-helices (58). It has been suggested that a loop region upstream of the second ACP helix recognizes a large hydrophobic pocket formed by the helical flap of MCAT (39). In contrast, other studies indicated that the second helix of ACP mostly makes electrostatic interactions with the ferredoxin-like subdomain of MCAT (57). In the case of DEBS, a type I PKS, it has been demonstrated recently that the KS-AT and post-AT linkers play an important role for acylation of the AT domain and for the transacylation to the cognate ACP, and a docking model between the AT and ACP domains of DEBS module 3 revealed that these linkers, but not the ferredoxin-like subdomain, are physically involved in the interaction (50). Interestingly, the peptide that was identified in the crystal structure of the AT52 fragment from Pks13 binds in the same region as that identified for ACP in the docked AT-ACP model of DEBS module 3 (Fig. 8B).
This first characterization of a mycobacterial type I PKS strengthens our knowledge on structure-function relationships of the AT domains. Given the important role of the AT domain as a gatekeeper, it might help the rational design of engineered enzymes for the combinatorial biosynthesis of new compounds. Because Pks13 is an essential enzyme for mycobacterial growth, this study should constitute a breakthrough for the structure-based rational design of inhibitors, a key step toward the development of new antibiotics against the mycobacterial infections tuberculosis and leprosy, which represent major threats to public health worldwide.
Supplementary Material
Acknowledgments
We thank the staff of synchrotron beamlines ID14-1, ID14-2, and ID14-4 at the European Synchrotron Radiation Facility (Grenoble, France); Dr. Sandrine Uttenweiler (Toulouse) for help during mass spectrometry characterization; the Mass Spectrometry Facility at IPBS (Toulouse) for constant support; Dr. Emilie Huc (Toulouse) for help and advice in peptide analysis; and Françoise Viala for help in preparing the figures.
This work was supported by European Commission Grant LSHP-CT-2006-037217, “Vaincre la Mucoviscidose” (France) Grant IC0716, French “Agence Nationale de la Recherche” Grant 09-BLAN-0298-01, “Région Midi-Pyrénées” Grant 09005193, and European Regional Development Fund Grant 34249.

This article contains supplemental Fig. 1.
The atomic coordinates and structure factors (codes 3TZW, 3TZX, 3TZY, and 3TZZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PKS
- polyketide synthase
- AT
- acyltransferase
- ACP
- acyl carrier protein
- KS
- β-ketoacylsynthase
- FAS
- fatty-acid synthase
- DEBS
- 6-deoxyerythronolide-B synthase
- TE
- thioesterase
- MCAT
- malonyl-CoA:ACP transferase
- MAT
- malonyl-acetyltransferase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Liou G. F., Khosla C. (2003) Building-block selectivity of polyketide synthases. Curr. Opin. Chem. Biol. 7, 279–284 [DOI] [PubMed] [Google Scholar]
- 2. Jenni S., Leibundgut M., Maier T., Ban N. (2006) Architecture of a fungal fatty acid synthase at 5 A resolution. Science 311, 1263–1267 [DOI] [PubMed] [Google Scholar]
- 3. Maier T., Jenni S., Ban N. (2006) Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science 311, 1258–1262 [DOI] [PubMed] [Google Scholar]
- 4. Jenni S., Leibundgut M., Boehringer D., Frick C., Mikolásek B., Ban N. (2007) Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316, 254–261 [DOI] [PubMed] [Google Scholar]
- 5. Leibundgut M., Jenni S., Frick C., Ban N. (2007) Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290 [DOI] [PubMed] [Google Scholar]
- 6. Lomakin I. B., Xiong Y., Steitz T. A. (2007) The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319–332 [DOI] [PubMed] [Google Scholar]
- 7. Maier T., Leibundgut M., Ban N. (2008) The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 8. Leibundgut M., Maier T., Jenni S., Ban N. (2008) The multienzyme architecture of eukaryotic fatty acid synthases. Curr. Opin. Struct. Biol. 18, 714–725 [DOI] [PubMed] [Google Scholar]
- 9. Maier T., Leibundgut M., Boehringer D., Ban N. (2010) Structure and function of eukaryotic fatty acid synthases. Q. Rev. Biophys. 43, 373–422 [DOI] [PubMed] [Google Scholar]
- 10. Khosla C. (2009) Structures and mechanisms of polyketide synthases. J. Org. Chem. 74, 6416–6420 [DOI] [PubMed] [Google Scholar]
- 11. Khosla C., Tang Y., Chen A. Y., Schnarr N. A., Cane D. E. (2007) Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 76, 195–221 [DOI] [PubMed] [Google Scholar]
- 12. Tang Y., Chen A. Y., Kim C. Y., Cane D. E., Khosla C. (2007) Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol. 14, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Y., Kim C. Y., Mathews I. I., Cane D. E., Khosla C. (2006) The 2.7-angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U.S.A. 103, 11124–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. White S. W., Zheng J., Zhang Y. M., Rock. (2005) The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 [DOI] [PubMed] [Google Scholar]
- 15. Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. (1999) Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267 [DOI] [PubMed] [Google Scholar]
- 16. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 17. George K. M., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., Small P. L. (1999) Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283, 854–857 [DOI] [PubMed] [Google Scholar]
- 18. Ng V., Zanazzi G., Timpl R., Talts J. F., Salzer J. L., Brennan P. J., Rambukkana A. (2000) Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell 103, 511–524 [DOI] [PubMed] [Google Scholar]
- 19. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 20. Portevin D., De Sousa-D'Auria C., Houssin C., Grimaldi C., Chami M., Daffé M., Guilhot C. (2004) A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. U.S.A. 101, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gavalda S., Léger M., van der Rest B., Stella A., Bardou F., Montrozier H., Chalut C., Burlet-Schiltz O., Marrakchi H., Daffé M., Quémard A. (2009) The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 284, 19255–19264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sacchettini J. C., Rubin E. J., Freundlich J. S. (2008) Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat. Rev. Microbiol. 6, 41–52 [DOI] [PubMed] [Google Scholar]
- 23. Léger M., Gavalda S., Guillet V., van der Rest B., Slama N., Montrozier H., Mourey L., Quémard A., Daffé M., Marrakchi H. (2009) The dual function of the Mycobacterium tuberculosis FadD32 required for mycolic acid biosynthesis. Chem. Biol. 16, 510–519 [DOI] [PubMed] [Google Scholar]
- 24. Leslie A. G. W., Powell H. R. (2007) in Evolving Methods for Macromolecular Crystallography (Read R. J., Sussman J. L., eds) Vol. 245, pp. 41–51, Springer, Dordrecht, The Netherlands [Google Scholar]
- 25. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 36. Schneider T. R. (2004) Domain identification by iterative analysis of error-scaled difference distance matrices. Acta Crystallogr. D Biol. Crystallogr. 60, 2269–2275 [DOI] [PubMed] [Google Scholar]
- 37. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 38. Serre L., Verbree E. C., Dauter Z., Stuitje A. R., Derewenda Z. S. (1995) The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5-Å resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270, 12961–12964 [DOI] [PubMed] [Google Scholar]
- 39. Keatinge-Clay A. T., Shelat A. A., Savage D. F., Tsai S. C., Miercke L. J., O'Connell J. D., 3rd, Khosla C., Stroud R. M. (2003) Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure 11, 147–154 [DOI] [PubMed] [Google Scholar]
- 40. Oefner C., Schulz H., D'Arcy A., Dale G. E. (2006) Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 62, 613–618 [DOI] [PubMed] [Google Scholar]
- 41. Bunkoczi G., Misquitta S., Wu X., Lee W. H., Rojkova A., Kochan G., Kavanagh K. L., Oppermann U., Smith S. (2009) Structural basis for different specificities of acyltransferases associated with the human cytosolic and mitochondrial fatty acid synthases. Chem. Biol. 16, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pappenberger G., Benz J., Gsell B., Hennig M., Ruf A., Stihle M., Thoma R., Rudolph M. G. (2010) Structure of the human fatty acid synthase KS-MAT didomain as a framework for inhibitor design. J. Mol. Biol. 397, 508–519 [DOI] [PubMed] [Google Scholar]
- 43. Derewenda Z. S., Derewenda U. (1991) Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem. Cell Biol. 69, 842–851 [DOI] [PubMed] [Google Scholar]
- 44. Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. (1992) The α/β hydrolase fold. Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 45. Zhang L., Liu W., Xiao J., Hu T., Chen J., Chen K., Jiang H., Shen X. (2007) Malonyl-CoA:acyl carrier protein transacylase from Helicobacter pylori: crystal structure and its interaction with acyl carrier protein. Protein Sci. 16, 1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koppisch A. T., Khosla C. (2003) Structure-based mutagenesis of the malonyl-CoA:acyl carrier protein transacylase from Streptomyces coelicolor. Biochemistry 42, 11057–11064 [DOI] [PubMed] [Google Scholar]
- 47. Ruch F. E., Vagelos P. R. (1973) Characterization of a malonyl-enzyme intermediate and identification of the malonyl binding site in malonyl coenzyme A-acyl carrier protein transacylase of Escherichia coli. J. Biol. Chem. 248, 8095–8106 [PubMed] [Google Scholar]
- 48. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 49. Neyrolles O., Guilhot C. (2011) Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91, 187–195 [DOI] [PubMed] [Google Scholar]
- 50. Wong F. T., Chen A. Y., Cane D. E., Khosla C. (2010) Protein-protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry 49, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yadav G., Gokhale R. S., Mohanty D. (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 [DOI] [PubMed] [Google Scholar]
- 52. Kwan D. H., Schulz F. (2011) The stereochemistry of complex polyketide biosynthesis by modular polyketide synthases. Molecules 16, 6092–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oh T. J., Daniel J., Kim H. J., Sirakova T. D., Kolattukudy P. E. (2006) Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J. Biol. Chem. 281, 3899–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sankaranarayanan R., Saxena P., Marathe U. B., Gokhale R. S., Shanmugam V. M., Rukmini R. (2004) A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 11, 894–900 [DOI] [PubMed] [Google Scholar]
- 55. Ronning D. R., Klabunde T., Besra G. S., Vissa V. D., Belisle J. T., Sacchettini J. C. (2000) Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7, 141–146 [DOI] [PubMed] [Google Scholar]
- 56. Brignole E. J., Smith S., Asturias F. J. (2009) Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat. Struct. Mol. Biol. 16, 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arthur C. J., Williams C., Pottage K., Płoskoń E., Findlow S. C., Burston S. G., Simpson T. J., Crump M. P., Crosby J. (2009) Structure and malonyl CoA-ACP transacylase binding of Streptomyces coelicolor fatty acid synthase acyl carrier protein. ACS Chem. Biol. 4, 625–636 [DOI] [PubMed] [Google Scholar]
- 58. Byers D. M., Gong H. (2007) Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 85, 649–662 [DOI] [PubMed] [Google Scholar]
- 59. Lebedev A. A., Young P., Isupov M. N., Moroz. O. V., Vagin A. A., Murshudov G. N. (2012) JLigand: a graphical tool for the CCP4 template-restraint library. Acta Cryst. D 68, 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.