Background: C1q opsonizes apoptotic cells to facilitate clearance.
Results: Annexin A2 and A5 as well as DNA and histones serve as ligands for C1q on apoptotic cells.
Conclusion: Disturbances in the interaction between C1q and these ligands might exacerbate autoimmune diseases.
Significance: The knowledge of new C1q ligands helps to further the understanding of autoimmune diseases.
Keywords: Annexin, Apoptosis, Complement System, DNA, Histones, Systemic Lupus Erythematosus
Abstract
C1q is the initiator of the classical complement pathway and opsonizes apoptotic cells to facilitate phagocytosis. Deficiency of C1q is the strongest known risk factor for development of systemic lupus erythematosus (SLE), which appears to be related to ensuing impaired clearance of apoptotic material. The objective of the current study was to investigate new ligands for C1q on the surface of apoptotic cells. We revealed that the two phospholipid-binding proteins annexin A2 and A5 are, beside DNA, significant C1q ligands. We furthermore, demonstrated that C1q binds directly to histones exposed on the surface of dying cells but we did not detect significant interaction with phosphatidylserine. The complement inhibitors C4b-binding protein and factor H also interact with dying cells, most likely to decrease complement activation beyond the level of C3 to allow noninflammatory clearance. Despite the fact that C4b-binding protein, factor H, and C1q share some ligands on dying cells, we showed that these three proteins did not compete with one another for binding to apoptotic cells. We additionally demonstrated that the way in which apoptosis is induced influenced both the degree of apoptosis and the binding of C1q. The knowledge, that annexin A2 and A5 act as ligands for C1q on apoptotic cells, sheds new light on the pathophysiology of autoimmune diseases.
Introduction
The complement system constitutes an integral part of innate immunity and serves as a first line of defense against many pathogens. Besides this, it contributes significantly to clearance of immune complexes and dying cells (1). The classical complement pathway is initiated when the C1-complex binds its target, e.g. apoptotic cells, which triggers a cascade of proteolytic cleavages of downstream complement proteins (2). The globular head domains of the C1q subunit comprise the recognition units of the C1 complex. There are six head domains to each C1q molecule and simultaneous binding of several ligands is required for activation of C1. Some C1q ligands are known to bind the N-terminal collagen-like stalk region but these usually do not trigger the classical pathway. C1q binds to surface blebs of apoptotic cells (3) and the interaction is mediated by the globular head region of C1q causing complement activation and deposition of C3b on dying cells (4–7). The aim of the current study was to investigate new ligands for C1q on the surface of apoptotic cells. So far the only widely accepted C1q ligand on dying cells is DNA, which becomes accessible already very early on apoptotic cells, even before phosphatidylserine (PS)3 (8). However, the precise region of C1q involved in DNA binding is a matter of controversy, because both the collagen-like stalk region and the globular head region have been implicated (7, 9–12). C1q has also been proposed to bind PS (13). C1q appears to bind relatively late apoptotic cells and necrotic cells.
Two complement inhibitors, C4b-binding protein (C4BP) and factor H (FH), have also been shown to interact with apoptotic and necrotic cells (14–16). The binding of C4BP, which circulates mainly in complex with protein S, to dying cells is mediated by interaction of protein S with PS (14) and to a much lesser extent via an interaction of the C4BP α-chains with DNA (16). In comparison, we showed recently that FH binds to annexin A2, DNA, and histones on the surface of apoptotic cells (17). These two complement inhibitors interfere with the cascade at the C3 level to minimize proinflammatory and lytic effects of full-blown complement activation. They also compensate for the loss of membrane-bound complement inhibitors such as membrane cofactor protein (MCP, CD46), which in turn, when down-regulated during apoptosis, act as an “eat-me” signal for effective clearance (8). Efficient and noninflammatory clearance of dying cells is crucial to avoid autoimmune reactions. Failure to do so, for example, in the case of genetic C1q deficiency, is proposed to be one of the underlying mechanisms in systemic lupus erythematosus (SLE). In SLE, autoantibodies directed against antigens present on dying cells are frequently found, which indicates less efficient or proinflammatory clearance of effete cells. Reasons for induced inflammatory clearance range from genetic or acquired deficiencies of C1q to potential alterations in ligands for proteins that prevent inflammatory clearance, such as the fluid phase complement inhibitors. Autoantibodies directed against apoptotic cells might further promote an FcγR-mediated clearance, which is proinflammatory. Autoantibodies might also block binding sites for opsonins or complement inhibitors. Autoantibodies directed against annexins have been described in SLE (18).
Annexins are unique proteins that interact with membrane phospholipids in a Ca2+-dependent manner, providing a link between Ca2+ signaling and membrane functions. The human annexin protein family encompasses over 10 members, which are involved in intracellular transport and function as bridging molecules to phospholipid membranes (19). It has been suggested that they play a role in many types of diseases including autoimmune diseases such as SLE (20–22). Annexin A5 is also widely used as an apoptotic marker because it recognizes PS on the surface of apoptotic cells.
In the current study, we further characterized the proposed binding partners for C1q and C4BP on apoptotic cells, and identified annexin A2 and A5 as new ligands for C1q. This knowledge helps to further the understanding of the pathophysiology of autoimmune diseases such as SLE.
EXPERIMENTAL PROCEDURES
Cells and Induction of Cell Death
Jurkat T-cells (ATCC) were grown in RPMI containing glutamine and 10% heat-inactivated fetal calf serum (all from Invitrogen). Apoptosis was generally induced using 0.75 μm staurosporine (Sigma) for 16 h in RPMI without fetal calf serum at 37 °C and 5% CO2. Apoptosis was additionally induced for 4 h using: 1 μm staurosporine, 10 μm camptothecin, 100 μm etoposide, 10 μm actinomycin D, or 100 μm cyclohexamide, all from Calbiochem or anti-fas and protein G (each 1 μg/ml), from Calbiochem and Sigma, respectively.
Isolation of Primary T-lymphocytes
Peripheral blood leukocytes were isolated from leukocyte filters within 5 h of blood donation. Cells were washed from the filter using PBS with 2 mm EDTA (PBS/EDTA), and peripheral blood mononuclear cells were isolated by density media centrifugation using Lymphoprep (Axis Shield) according to the supplied instructions. Isolated peripheral blood mononuclear cells were washed twice in PBS/EDTA and total CD4+ T cells were purified by positive selection using anti-CD4 coupled magnetic beads (Miltenyi Biotech) according to supplied instructions, with an average purity of over 95%. Isolated CD4+ T-cells were resuspended in RPMI for further use.
Proteins, Antibodies, and Sera
The C4BP-protein S complex (23), FH (24), C1q (25), protein S (26), and α1-anti trypsin (27) were purified from human plasma as described. The preparation of stalk and head portions of C1q was performed according to published protocols (28), where pepsin (Worthington) and collagenase (from Clostridium histolyticum, Worthington), respectively, were used for partial digestion of C1q. Histones (H2A/H2B + H1 and H3/H4) were purified from Jurkat T-cells (17), human IgG were from Immuno, annexin A2 (437622) was from Calbiochem, and annexin A5 (5165871A) from BD Biosciences. All isolated proteins were at least 95% pure, as judged by Coomassie staining of proteins separated by SDS-PAGE. Antibodies used were: anti-dsDNA (21227771, Immunotools), anti-C4BP PK9008 (home made), anti-FH (A312, Quidel), anti-C1q (A0136, Dako), anti-C4c (Q0369, Dako), anti-annexin A2 (ab41803, Abcam), and anti-annexin A5 (ab14196, Abcam). Secondary antibodies horseradish peroxidase-conjugated were anti-rabbit (P0399, Dako) and anti-goat (P0449, Dako), and fluorescently labeled were anti-rabbit Alexa Fluor 647 (A21246, Invitrogen), anti-mouse Alexa Fluor 488 (A11059, Invitrogen), and anti-goat Alexa Fluor 647 (A21446, Invitrogen). Normal human serum (NHS) and heat-inactivated serum (Hi-NHS) were prepared as described (26) according to a permit granted by the Ethics Committee of Lund University. C1q-deficient sera (A112) and normal control sera (A509) were purchased from Quidel.
Labeling of Proteins with Fluorescent Dyes
C1q, C4BP, and FH were either labeled with Alexa Fluor 488 (Invitrogen) or DyLight 488 (Pierce), respectively, following the manufacturer's instructions. C1q was easily negatively affected by the labeling and therefore sometimes was replaced with unlabeled protein followed by detection with antibodies.
Complement Deposition and Binding Assay
For deposition assays, 2.5 μg/ml, and for binding assays, 4 μg/ml of annexin A2, annexin A5, H2A/H2B + H1, and H3/H4 were coated onto a Maxisorp (Nunc) microtiter plate in 75 mm sodium carbonate buffer, pH 9.6, at 4 °C overnight. Aggregated IgG (2.5 μg/ml for deposition and 2 μg/ml for binding assays) was used as positive control, 1% BSA or 4 μg/ml of α1-antitrypsin were used as negative controls. The unbound proteins were washed with 50 mm Tris-HCl, 150 mm NaCl, and 0.1% Tween 20, pH 7.5 (immunowash), blocked in 1% BSA (binding assays) or immunowash containing 3% fish gelatin (deposition assays), washed, and incubated with either NHS or Hi-NHS in 5 mm veronal buffer with 0.1% gelatin, 1 mm MgCl2, and 0.15 mm CaCl2 (GVB++) or purified proteins in 50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 50 μg/ml of BSA for 2 h at room temperature (binding assays) or 25 min at 37 °C (deposition assays). For competition studies, the purified proteins were preincubated with 5 to 25 m excess of additional proteins for 45 min at room temperature, in 50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 50 μg/ml of BSA, washed, incubated with antibodies specific for C1q, C4c, or FH, washed and incubated with appropriate horseradish peroxidase-conjugated secondary antibody, and developed using OPD tablets (Dako) according to the manufacturer's instructions. Absorbance was measured at 490 nm in a Cary50 Bio UV spectrophotometer connected to a 50MPR microplate reader (Varian). For assessment of ionic strength dependence, the same basic assay was used. However, 10 μg/ml of C1q was incubated in binding buffer with NaCl concentrations ranging from 0 to 1000 mm. The binding assay assessing calcium/magnesium dependence was performed using binding buffer supplemented with 5 or 20 mm EDTA.
Binding of Radiolabeled Proteins to C1q Heads and Tails
4 μg/ml of purified C1q, C1q globular heads, and collagenous tails were coated as above. 1% BSA or 4 μg/ml of α1-antitrypsin were used as negative controls. The unbound proteins were washed with immunowash, blocked in 1% BSA, and incubated with 125I-radiolabeled histones H2A/H2B + H1 and H3/H3 as well as annexin A2 and A5 in 50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 50 μg/ml of BSA overnight (for histones) or 5 h (for annexins) at room temperature. After washing, the radiolabeled bound proteins were measured using a 1277 GammaMaster (LKB Wallac).
Flow Cytometry
For binding or expression analysis by flow cytometry, apoptosis was induced for the indicated time points as above. For liposomes, liposome-coated Dynabeads were prepared as described below. For binding experiments, cells or liposomes were washed in 10 mm HEPES, 150 mm NaCl, 5 mm KCl, 1 mm MgCl2, and 2 mm CaCl2 (binding buffer) and incubated with unlabeled or labeled proteins. For competition experiments, proteins were preincubated for 30 min at room temperature with competing protein in 5 to 25 m excess. Proteins in binding buffer were subsequently incubated with cells or liposomes for 30 min at 37 °C. For experiments with unlabeled proteins or expression analysis, samples were washed in PBS supplemented with 1% BSA and 30 mm NaN3, incubated with primary antibody for 30 min, washed, incubated with fluorochrome-labeled secondary antibody for 30 min, washed, and taken up in binding buffer supplemented with 30 mm NaN3 and analyzed in a CyFlow Space (Partec). For discrimination between live, apoptotic, or necrotic cells, samples were stained with Annexin A5-PE (BD Bioscience) and Via-Probe (BD Bioscience) in binding buffer after antibody staining, incubated 20 min, and analyzed as above.
Confocal Microscopy
Apoptosis was directly induced on SuperFrostPlus object slides (Menzel). Therefore 80,000 cells diluted in RPMI without fetal calf serum containing 1 μm staurosporine were pipetted within a Dako Pen inked circle and incubated for the indicated time at 37 °C in 5% CO2. After washing twice with 10 mm HEPES, 150 mm NaCl, 5 mm KCl, 1 mm MgCl2, and 2 mm CaCl2 (binding buffer), the proteins were added to the cells in a final volume of 30 μl and incubated for 30 min at 37 °C in a humidified chamber. After washing 3 times for 5 min in PBS, the cells were fixed with 3.7% paraformaldehyde for 15 min, washed again, and centrifuged for 1 min at 500 × g in a Cellspin (Tharmac). After blocking with PBS + 1% BSA + 30 mm NaN3 for 30 min at 37 °C, the primary antibody was incubated either overnight at 4 °C or for 1 h at room temperature. After washing, the cells were incubated with the secondary antibody for 1 h at room temperature followed by washing and addition of 100 ng/ml of propidium iodide or 50 μl of Alexa Fluor-labeled annexin A5 diluted 1:2–1:6 in 10 mm HEPES, 140 mm NaCl, 2.5 mm CaCl2, pH 7.4, for 15 min. For annexin A5 staining, the cells where preincubated for 30 min and washed with the corresponding Ca2+-containing buffer. After the final wash, the slides were briefly dipped in 99% ethanol, dried, and mounted with DakoCytomation fluorescent mounting medium. All slides were analyzed in a LSM 510 Meta confocal microscope (Zeiss); co-localization was calculated using CoLocalizer Express (CoLocalization Research Software).
Liposome Preparation
All used phospholipids are listed in Table 1. They were purchased from Avanti Polar Lipids and kept in chloroform with 10% methanol at −20 °C. For surface plasmon resonance experiments, lipids were mixed in the desired molar ratios and the chloroform was removed. Liposomes were generated by extrusion. Lipids were resuspended in 10 mm HEPES, pH 7.4, and 150 mm NaCl, frozen, and thawed 5 times using dry ice/acetone and a 40 °C water bath, extruded 19 times through two polycarbonate membranes with 100-nm pores (Avestin) using a LiposoFast-Basic (Avestin), and stored at 4 °C for use within 7 days. For flow cytometric assays, 1 μmol of lipids in the desired molar ratio were mixed with 10 nmol of N-(biotinyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanol-amine, triethylammonium (Invitrogen), and 6.4 nmol of 2-(3-(diphenylhexatrienyl)propanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (Invitrogen), chloroform was evaporated and lipids were resuspended in 25 mm HEPES and 150 mm NaCl, pH 7.7 (HBS), sonicated, and mixed with M-280 streptavidin-coated Dynabeads (Invitrogen) as previously described.
TABLE 1.
Phospholipids used for surface plasmon resonance and flow cytometric analyses
C1q did not interact with any of the tested phospholipids.
| Lipid abbreviation | Lipid (underlined) | Lipid source | Mol % | Treatments |
|---|---|---|---|---|
| PC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Synthetic | 100 | |
| PS/PC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine | Synthetic | 10/90 | |
| PE/PC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Synthetic | 10/90 | |
| LysoPC/PC | 1-Oleoyl-2-hydroxy-sn-glycero-3-phosphocholine | Synthetic | 3/97 | |
| LysoPC/PC | 10/90 | |||
| LysoPC/PC | 60/40 | |||
| LysoPS/PC | 1-Oleoyl-2-hydroxy-sn-glycero-3-phospho-l-serine | Synthetic | 60/40 | |
| LysoPE/PC | 1-Oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | Synthetic | 60/40 | |
| LysoPS/PC | 10/90 | |||
| PC | 100 | Oxidized | ||
| LysoPC/PC | 40/60 | Oxidized | ||
| LysoPC/PC | 40/60 | |||
| PS/PC | 10/90 | Oxidized | ||
| LysoPS/PC | 10/90 | Oxidized | ||
| Natural PC | l-α-Phosphatidylcholine | Egg, chicken | 100 | |
| Natural PS/natural PC | l-α-Phosphatidylserine | Brain, porcine | 10/90 | |
| Natural LysoPC/natural PC | l-α-Lysophosphatidylcholine | Egg, chicken | 20/80 |
Surface Plasmon Resonance
To measure the kinetics of the DNA-C1q interaction, biotinylated DNA was coupled to a streptavidin-coated SA chip (GE Healthcare). Double-stranded 25-mer oligonucleotides (G-C)25 and (A-T)25 were produced using equimolar amounts of single-stranded 5′-biotinylated 25-mer oligonucleotides (MWG) as described (16). Briefly, equal concentrations of G-C and A-T were coupled to the streptavidin sensor chip surface to a level of 300 response units (Biacore 2000, GE Healthcare). All experiments were performed at a continuous flow rate of 30 μl/min using running buffer (150 mm NaCl, 10 mm HEPES, 2.5 mm CaCl2, 0.002% Tween 20, pH 7.4). C1q was run over the chip in a concentration gradient from 2 to 200 nm. The chip was regenerated using pulse injection of 3 m guanidium chloride followed by 1 m NaCl. The obtained sensograms were analyzed using Bio-evaluation software 3.0 (GE Healthcare).
To measure kinetics of lipid-protein interactions (29) the lipophilic L1 sensor chip was washed for 1 min at a flow of 20 μl/min with 40 mm n-octyl-β-d-glucopyranoside (Calbiochem). Phospholipid liposomes (500 μm), generated by extrusion, were injected for 17 min at a 3 μl/min flow rate in HBS running buffer. The average immobilization level for all liposomes was 1000 response units. The chip was then washed five times with 10 mm EDTA, pH 8.0, injections (2 min at 20 μl/min), after which the membrane surface was stable. For protein binding experiments, running buffer was changed to HBC (HBS with 10 mg/ml of BSA and 5 mm CaCl2) and flow cells were equilibrated until the baseline stabilized. Purified C1q and C4BP were diluted in HBC and injected over the liposome surfaces at several serial dilutions ranging from 400 to 6 μg/ml. The flow was set at 20 μl/min and the proteins were allowed to associate for 240 s followed by a 200-s dissociation. The immobilized liposome surface could be regenerated with an injection of 10 mm EDTA, pH 8.0, which returned the baseline to the value prior to introducing protein. To calculate the dissociation constant (KD) of interaction between C4BP and liposomes containing 90% PC, 10% PS, the response obtained at equilibrium was plotted against each concentration used and the one-site binding hyperbola was fitted using Prism (GraphPad).
RESULTS
C1q Binds to Late Apoptotic Cells and Interacts with DNA and Histones
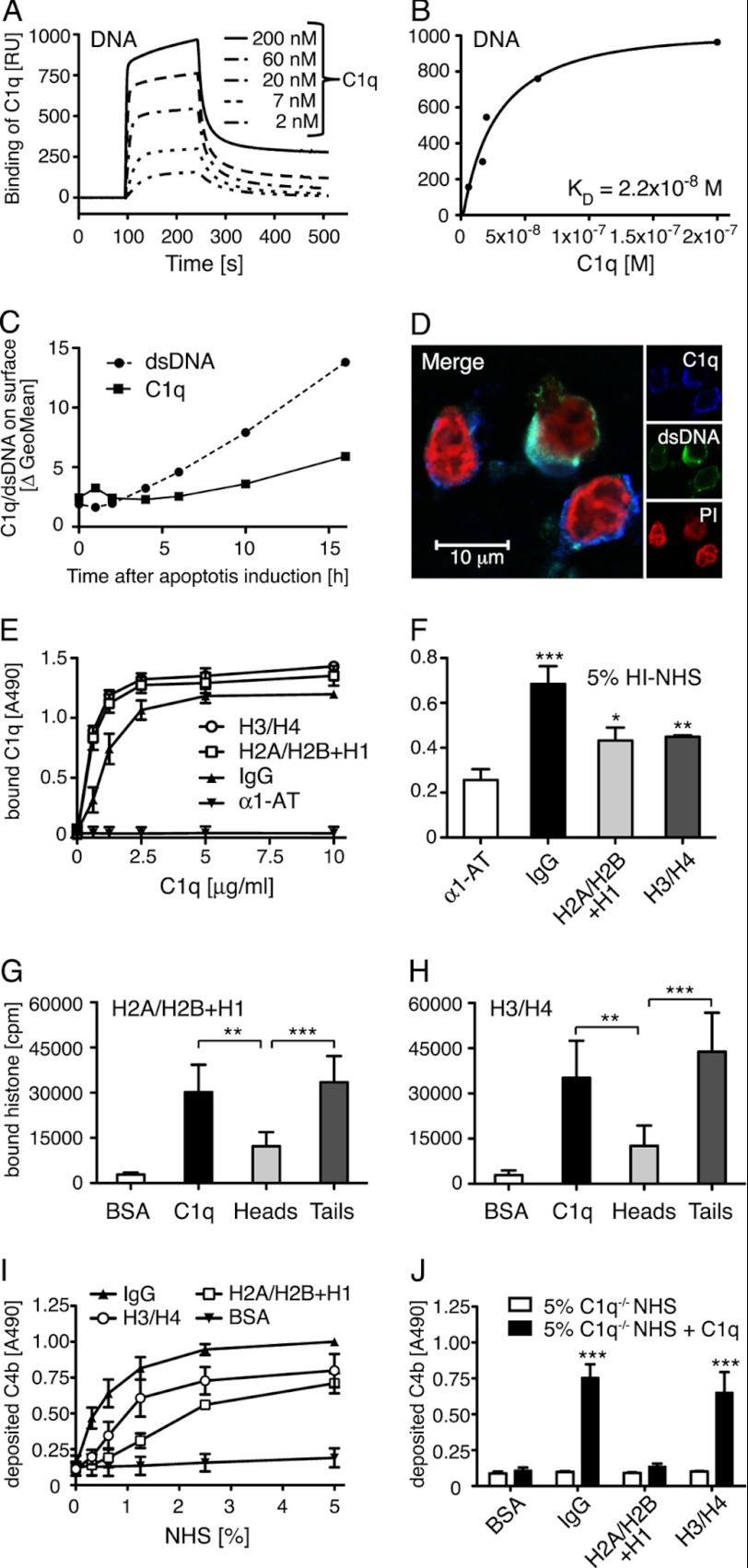
DNA has been previously suggested to act as a C1q ligand on apoptotic cells (8). To further investigate the relevance of this interaction, surface plasmon resonance was used to analyze the binding affinity between C1q and DNA (Fig. 1, A and B). Biotinylated DNA was immobilized onto a streptavidin chip and C1q was injected at several concentrations. KD derived from the plot of response at equilibrium versus C1q concentration was estimated to 2.2 × 10−8 m and indicates very strong binding between the two molecules.
FIGURE 1.
C1q binds DNA and histones. A, increasing concentrations of C1q were flown over immobilized biotinylated DNA on a plasmon surface resonance (Biacore) streptavidin chip. Dose-dependent binding of C1q to DNA was observed and is presented in arbitrary response units (RU). B, the response obtained at equilibrium was plotted against C1q concentration and KD value of the C1q DNA interaction was determined to 2.2 × 10−8 m. C, by flow cytometry, double-stranded DNA expression and C1q binding (50 μg/ml) on apoptotic cells were investigated after the indicated time of apoptosis induction. An increase of expression of dsDNA as well as binding of C1q was seen during the course of apoptosis although the increase was more pronounced for dsDNA than the C1q binding. D, localization of C1q (blue), dsDNA (green), and propidium iodide (PI) (red) on apoptotic cells was analyzed using confocal microscopy. E, binding of C1q to immobilized histones, IgG (positive control), and α1-AT (negative control) was analyzed in an ELISA setup. Clear concentration dependent binding was seen for C1q to both histone complexes. F, binding of C1q from Hi-NHS to immobilized histones or IgG was detected and compared with α1-AT as a control. G and H, radiolabeled histone H2A/H2B + H1 and H3/H4 were incubated with immobilized C1q, C1q globular heads, C1q collagenous tails, and BSA (negative control). After washing, the amount of bound protein was determined in a γ-counter. Binding occurs mainly to the collagenous tail region. I and J, C4b deposition on immobilized histones or controls was measured from NHS (I), C1q-deficient sera or C1q-deficient sera reconstituted with 70 μg/ml of C1q (J). Deposition of C4b was detected on all histone complexes from NHS and it decreased in C1q-deficient sera but could only be restored upon addition of C1q to the H3/H4 histone complex. For C, representative values from two independent experiments are shown. For D, one representative image of two independent experiments is shown. E-H, values are shown as mean of duplicates, n = 4 (G and H) and n = 3 (E and F, I and J) ± S.D. Significance of differences was calculated using one-way analysis of variance followed by Tukey's (G and H) or Dunnett's (F and J) multiple comparison post-test and is displayed as: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To validate the interaction of C1q and DNA on apoptotic cells, C1q was incubated with apoptotic Jurkat T-cells and the amount of bound C1q as well as available DNA were assessed using an antibody against dsDNA. C1q did not bind to live Jurkat cells but did bind to apoptotic cells that were induced with 0.75 μm staurosporine for at least 6 h (Fig. 1C). Very weak or no binding occurred to apoptotic cells, which were induced for shorter time, whereas the binding increased with longer induction times (up to 16 h). C1q bound to both the A5+/Via-Probe− (early apoptotic) and A5+/Via-Probe+ (late apoptotic/necrotic) populations of cells induced for at least 6 h, whereby it bound stronger to the double positive cells. After 16 h of incubation ∼45% of the cells were A5+/Via-Probe− and 45% were A5+/Via-Probe+. To exclude that serum deprivation alone led to induction of C1q binding, we tested binding to cells that have been incubated for 16 h in RPMI without addition of staurosporine. Almost no cells were A5+ or Via-Probe+ indicating that C1q binding is induced by apoptosis (data not shown).
With the apoptosis induction procedure used in this study, an elevated surface level of DNA was detected 4 h after apoptosis induction and increased strongly with longer induction times (Fig. 1C). This indicates that DNA is present on the surface before C1q starts to bind. Together with the observation that the elevated surface level of DNA rises more strongly than C1q binding, we assumed that DNA cannot be the only C1q ligand on dying cells or that the cell surface must be reorganized before DNA is available for interaction with C1q.
To visualize and confirm the interaction of C1q and DNA on the surface of apoptotic cells, confocal microscopy was performed (Fig. 1D). Co-localization was calculated for different areas on two microscope slides from separate experiments by using the overlap coefficient (R) according to Manders (30). C1q and DNA displayed a mean coefficient of 0.74 ± 0.04, which clearly indicates co-localization.
DNA can be present as nucleosomes on the surface of apoptotic cells (31) and therefore we tested whether C1q could interact directly with histones. The interaction was evaluated using purified histones from Jurkat T-cells in an ELISA setup. Clear binding of purified C1q to both H2A/H2B + H1 and H3/H4 histones was observed (Fig. 1E). This interaction was also detected from heat inactivated serum, Hi-NHS (Fig. 1F). To verify whether histones bind to the globular head or to the collagenous tail region of C1q, proteins were immobilized and incubated with radiolabeled histones. Both histone fractions bound mainly to the collagenous tail region of C1q but binding to the globular head region was also significant (Fig. 1, G and H). Interaction of histones with C1q could result in both inhibition and activation of complement depending on the C1q region involved in the interaction. Therefore immobilized histones were incubated with NHS and deposition of C4b was detected using specific antibodies. Both histone pairs induced strong complement activation (Fig. 1I). To evaluate if C1q was responsible for the observed activation, histones were incubated with C1q-deficient NHS and C1q-deficient NHS reconstituted with 70 μg/ml of purified C1q. Although no deposition was seen in the absence of C1q, surprisingly only histones H3/H4 were able to induce C4b deposition upon reconstitution (Fig. 1J). This indicates the presence of other activating factors in NHS that are lacking in the C1q-deficient serum.
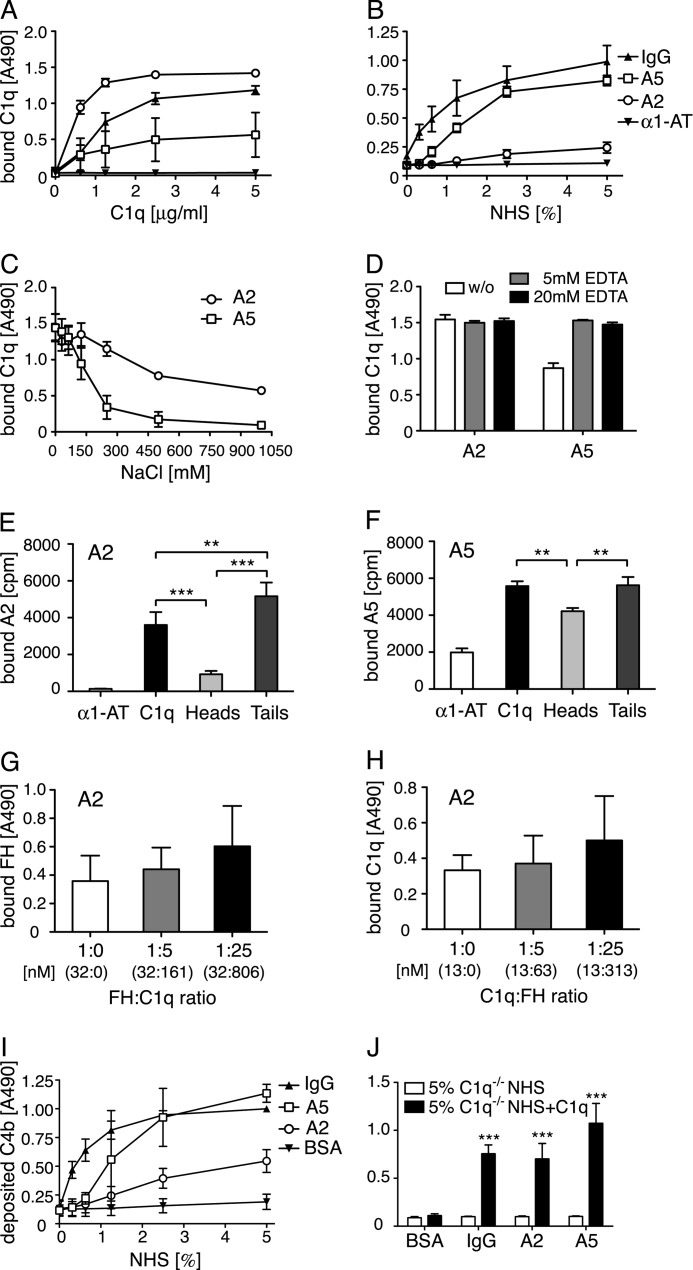
Purified Annexins A2 and A5 Bind C1q and Activate the Classical Pathway of Complement
A direct binding assay was used to assess interactions of purified C1q with annexins A2 and A5 immobilized in microtiter plates. C1q bound specifically in a dose-dependent and saturable manner (Fig. 2A), whereby the binding to annexin A2 was stronger than to annexin A5. Aggregated IgG was used as a positive control and α1-antitrypsin as a negative control for C1q binding. To confirm that C1q from serum binds to annexin A2 and A5, the binding experiment was repeated using NHS as C1q source. Binding to both proteins was confirmed in NHS. The binding of C1q from serum to annexin A5 was comparable with the binding obtained with purified C1q. However, the binding to A2 from serum was much weaker than that of purified C1q (Fig. 2B). This indicates that either the serine proteases C1r and C1s, which are present in the C1 complex or some other serum proteins attenuate the interaction between C1q and annexin A2.
FIGURE 2.
Annexin A2 and A5 bind C1q and activate complement. A and B, increasing concentrations of C1q were incubated with immobilized annexin A2, annexin A5, IgG (positive control), or α1-AT (negative control). Clear binding to both annexins was seen both by purified protein (A) and NHS (B). C and D, C1q (10 μg/ml) diluted in buffers with increasing ionic strength or containing 5 or 20 mm EDTA was added to immobilized annexins. The interaction of C1q with both annexin A2 and annexin A5 decreased with increasing ionic strength (C) but was not dependent on bivalent cations (D). E and F, radiolabeled annexin A2 and A5 were incubated with immobilized C1q, C1q globular heads, C1q collagenous tails, and α1-AT (negative control). After washing, the amount of bound protein was determined in a γ-counter. The annexins bound well to the collagenous tail region of C1q and annexin A5 also bound strongly to the globular head domain of C1q. G and H, binding of C1q (6.25 μg/ml) to annexin A2 was competed with FH (G) and reverse (5 μg/ml and FH, H). The actual concentration of each protein is depicted in nanomolar in parentheses. No competition was observed in either setting. I and J, C4b deposition was measured on annexins or controls from NHS (I), C1q-deficient sera or C1q-deficient sera reconstituted with 70 μg/ml of C1q (J). Clear deposition was seen from NHS, which was completely abolished in C1q-deficient sera and could be restored upon addition of C1q. Values are shown as mean of duplicates, n = 3 (E, n = 4) ± S.D. E–H, significance of differences was calculated using one-way analysis of variance followed by Tukey's multiple comparisons post-test and is displayed as: *, p < 0.05; **, p < 0.01; ***, p < 0.001. For J, the significance of differences was calculated using two-way analysis of variance with a Bonferroni post-test and is displayed as: ***, p < 0.001.
To evaluate the nature of the C1q-annexin A2 and A5 binding, C1q was incubated with immobilized annexins in buffers of varying ionic strength. Both interactions were of ionic nature and the interaction of C1q to A5 was more sensitive to the present ionic strength (Fig. 2C). Furthermore, neither the binding of C1q to annexin A2 nor to annexin A5 was dependent on the presence of calcium and magnesium ions (Fig. 2D). Both radiolabeled annexin A2 and A5 bound weaker to C1q heads in comparison to the collagenous tail region (Fig. 2, E and F). This difference was more pronounced for annexin A2 compared with annexin A5.
We showed recently that annexin A2 is one of the ligands localizing FH to the surface of apoptotic cells (17). Therefore competition experiments were performed whereby annexin A2 was immobilized in microtiter plates and binding of C1q and FH, respectively, was evaluated in the presence of no, 5-, or 25-fold m excess of the other protein. Protein and competitor were preincubated at room temperature. Neither FH (Fig. 2G) nor C1q (Fig. 2H) competed with the other protein for the binding to immobilized annexin A2.
To assay the complement activating potential of the annexins, immobilized annexin A2 and A5 were incubated with NHS as a source of complement and deposition of C4b was detected using specific antibodies. Both annexin A2 and A5 induced complement activation, where activation by annexin A5 was comparable with the strongly activating aggregated IgG (Fig. 2I). The requirement of C1q for complement activation to occur was confirmed using C1q-deficient NHS as well as reconstituted C1q-deficient NHS. No deposition of C4b was observed for IgG or the two annexins from C1q-deficient NHS, but could be fully restored upon addition of C1q (Fig. 2J).
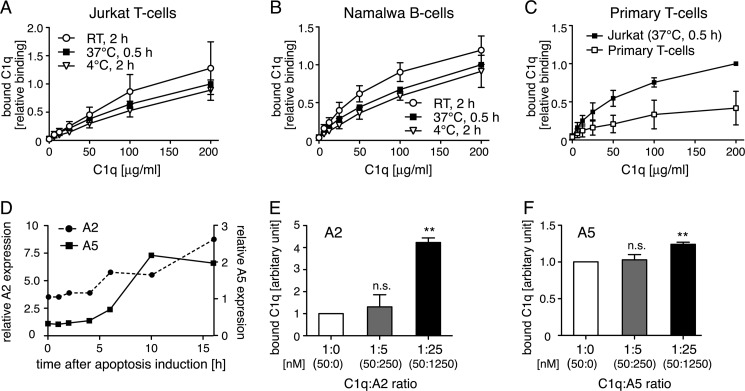
C1q Binds to Apoptotic T- and B-cells and Interacts with Annexin A2 and A5 Exposed on the Cell Surface
C1q bound dose-dependently to apoptotic Jurkat T-cells and Namalwa B-cells (Fig. 3, A and B). No significant differences were found when binding of C1q to apoptotic cells was evaluated at room temperature for 2 h compared with incubations at 4 °C for 2 h or at 37 °C for 30 min. The incubation at 37 °C for 30 min was chosen for all following experiments. Using this setup, C1q bound dose-dependently to apoptotic primary T-lymphocytes, although the binding reached lower levels compared with Jurkat T-cells (Fig. 3C). Both annexin A2 and A5 were only present in a negligible amount on the cell surface of live Jurkat T-cells but both proteins became exposed during the course of apoptosis induction (Fig. 3D). The same course of expression was detected on Namalwa B-cells (data not shown). C1q bound to both annexins when purified proteins were used (Fig. 2A) and thus we investigated whether externally added annexin A2 or A5 could affect the binding of C1q to apoptotic cells. Both proteins were able to increase the binding of C1q to the cell surface, confirming interaction in a setting of the whole cell membrane (Fig. 3, E and F). Due to the fact that both annexins bind to negatively charged phospholipids, which become exposed on the cell surface during apoptosis, it was expected that these proteins, when preincubated with C1q, localize even more C1q to the cell membrane.
FIGURE 3.
C1q binds to apoptotic T- and B-cells and interacts with annexin A2 and A5 exposed on the cell surface. A and B, binding curves of C1q to apoptotic Jurkat T-cells and apoptotic Namalwa B-cells were obtained at different temperatures. C1q bound dose-dependently to both cell lines. C, binding of C1q to apoptotic Jurkat T-cells and apoptotic primary T-lymphocytes. C1q bound dose-dependently, but reached lower levels for primary T-lymphocytes compared with Jurkat T-cells. D, change in annexin A5 and A2 expression on the surface of apoptotic Jurkat T-cells expressed relative to antibody controls. Both annexin A2 and A5 expressions increased during the course of apoptosis induction. E and F, preincubation of annexin A2 (E) and annexin A5 (F) increased binding of C1q (25 μg/ml) to apoptotic Jurkat T-cells. The ΔGeoMean for the binding of C1q in the absence of competitor was defined as 1 and the binding in the presence of competitor has been calculated in relationship. The actual concentration of each protein is depicted in nanomolar in parentheses. A–C, values are shown as mean of three independent experiments, n = 3 ± S.D. Values were normalized to binding for 200 μg/ml of C1q at 37 °C for 30 min. D, representative values from two independent experiments are shown. For E and F, values are shown as mean of three independent experiments, n = 3 ± S.D. Significance of differences was calculated using one-way analysis of variance followed by Dunnett's multiple comparison post-test and is displayed as: n.s., not significant; **, p < 0.01.
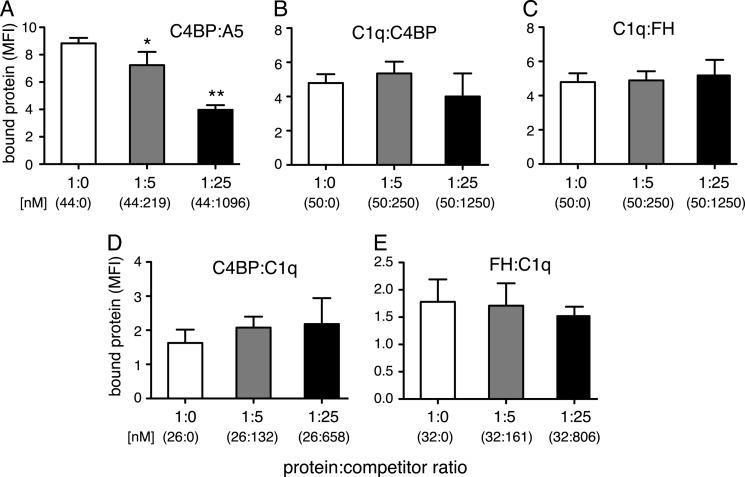
C1q, C4BP, and FH Do Not Compete for One Another's Binding to Apoptotic Cells
C1q, C4BP, and FH have all been proven to bind to apoptotic cells (3, 14, 15). To clarify whether they influence one another's binding, competition assays were performed. Jurkat T-cells were rendered apoptotic and incubated for 30 min with 25 μg/ml of C1q, 15 μg/ml of C4BP-Alexa Fluor 488, or 5 μg/ml of FH-Alexa Fluor 488 either alone or in the presence of 5- or 25-fold m excess of each of the other proteins separately. The protein and potential competitor were preincubated for 30 min at room temperature before the addition to the cells. Neither C4BP (Fig. 4B) nor FH (Fig. 4C) competed with C1q binding and neither did C1q compete with C4BP (Fig. 4D) or FH binding (Fig. 4E). To verify that the experimental settings were correct, C4BP (25 μg/ml) was also incubated with 5- and 25-fold m excess of annexin A5. Both C4BP and annexin A5 bind strongly to negatively charged phospholipids, and annexin A5 was indeed able to compete with C4BP in a statistically significant manner for the binding of apoptotic Jurkat cells at a 5-fold molar excess (Fig. 4A).
FIGURE 4.
C1q, C4BP, and FH do not bind the same surface ligand on apoptotic cells. A–E, C4BP, C1q, and FH were allowed to compete with one another for binding to apoptotic cells in 0, 5, and 25 m ratios, whereby the protein and potential competitor had been preincubated for 30 min at room temperature before the addition to the cells. Binding of C4BP (25 μg/ml) was competed out by annexin A5 (A), whereas no competition was seen for any of the other proteins: C1q binding (25 μg/ml) by C4BP (B), C1q binding (25 μg/ml) by FH (C), C4BP binding (15 μg/ml) by C1q (D), or FH (5 μg/ml) binding by C1q (E). Data are presented as mean of three independent experiments, n = 3 ± S.D. Significance of differences was calculated using one-way analysis of variance followed by Dunnett's multiple comparison post-test and is displayed as: *, p < 0.05; **, p < 0.01.
C1q Does Not Interact with Phospholipids
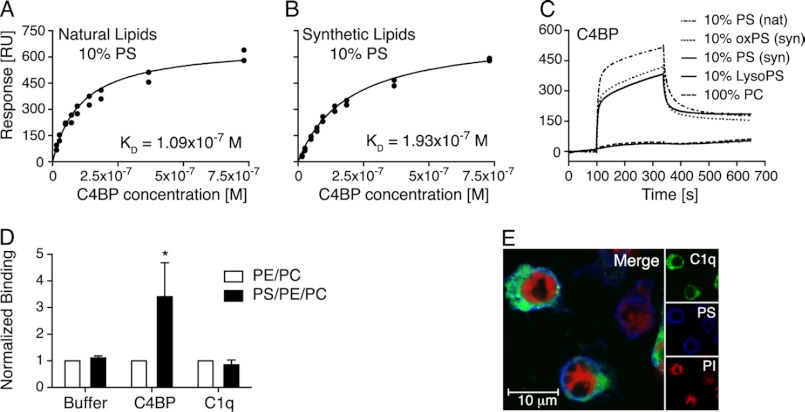
To clarify whether C1q interacts with phospholipids that might be present on the surface of apoptotic cells, surface plasmon resonance was performed using different compositions of extruded liposomes immobilized on an L1 chip. C1q or C4BP, used as a positive control, were injected at different concentrations. During the course of apoptosis, endogenous phospholipases are activated that generate lysophospholipids on the apoptotic cell surface (32). Lipid oxidation also occurs during the onset of programmed cell death (33). Therefore C1q binding to different compositions of lysophospholipids and oxidized phospholipids was also investigated. All tested liposome compositions are listed in Table 1. We found that purified C1q bound to none of the tested liposomes (data not shown). C4BP on the other hand, as expected, bound strongly to liposomes containing 10% PS, whereby it bound slightly better to natural PS (Fig. 5A) than to synthetic PS (Fig. 5B). The binding constants were determined as 1.09 × 10−7 and 1.93 × 10−7 m to natural and synthetic PS containing liposomes, respectively. It bound equally strongly to oxidized and nonoxidized synthetic PS (Fig. 5C). Phosphatidylcholine (PC) alone (100%) was used as a negative control. C4BP did not interact with lyso-PS, which has not been previously reported.
FIGURE 5.
C4BP binds both synthetic and natural PS incorporated in liposomes and on apoptotic cells. A and B, binding of C4BP to extruded PS-PC liposomes generated with natural (A) and synthetic (B) lipids was assayed using surface plasmon resonance. The response obtained at equilibrium was plotted against C4BP concentrations and KD value of the interaction was determined to 1.09 × 10−7 m for natural lipids and 1.93 × 10−7 m for synthetic lipids. C, binding of C4BP to PC liposomes containing PS, oxidized PS as well as lyso-PS was assayed (nat, natural; syn, synthetic). D, binding of Alexa Fluor 488-labeled C4BP (50 μg/ml) and Alexa Fluor 488-labeled C1q (150 μg/ml) to sonicated liposomes containing PC and PE with or without PS was assayed using flow cytometry. E, localizations of Alexa Fluor 488-labeled C1q (green), PS (blue, detected with Alexa Fluor 647-labeled annexin A5), and propidium iodide (PI) (red) on apoptotic cells were studied using confocal microscopy and co-localization was calculated according to Manders et al. (30). D, values are shown as mean of three independent experiments, n = 3 ± S.D. Significance of differences was calculated using Welch's t test assuming equal S.D. and is displayed as: *, p < 0.05. E, one representative image of two independent experiments is shown. RU, response units.
To confirm the results with a different method, liposome-coated Dynabeads were used for flow cytometry analysis. Binding of C1q-Alexa Fluor 488 (150 μg/ml) and C4BP-Alexa Fluor 488 (50 μg/ml) to PE/PC (20/80 mol %) and PS/PE/PC (10/20/70 mol %) liposome-coated Dynabeads was determined. As expected, C4BP bound strongly to the PS-containing liposomes, whereas C1q, even when applied in a three times higher concentration, did not bind to these liposomes (Fig. 5D). The signal obtained from the PE/PC liposomes was defined as background binding and set to 1.
Confocal microscopy was performed to visualize C1q and PS on the surface of apoptotic cells, whereby Alexa Fluor 647-labeled A5 was used to stain PS, and propidium iodide to counterstain nucleic DNA (Fig. 5E). Potential co-localization was calculated for different areas on two microscope slides from separate experiments and the mean overlap coefficient was determined to 0.51 ± 0.06, indicating no significant co-localization for C1q and PS. A time course experiment, using the same time points as displayed in Fig. 1, was performed to determine the overlap coefficient for the whole course of apoptosis induction. The overlap coefficient for live cells was 0.14 ± 0.04 and increased with time up to 0.51 ± 0.06 for the 16-h induced cells, indicating perhaps that C1q localized close to PS with the increasing apoptosis but that the two ligands never co-localized strongly.
Comparison of Different Apoptosis Inducers and Their Impact on C4BP, FH, and C1q Binding
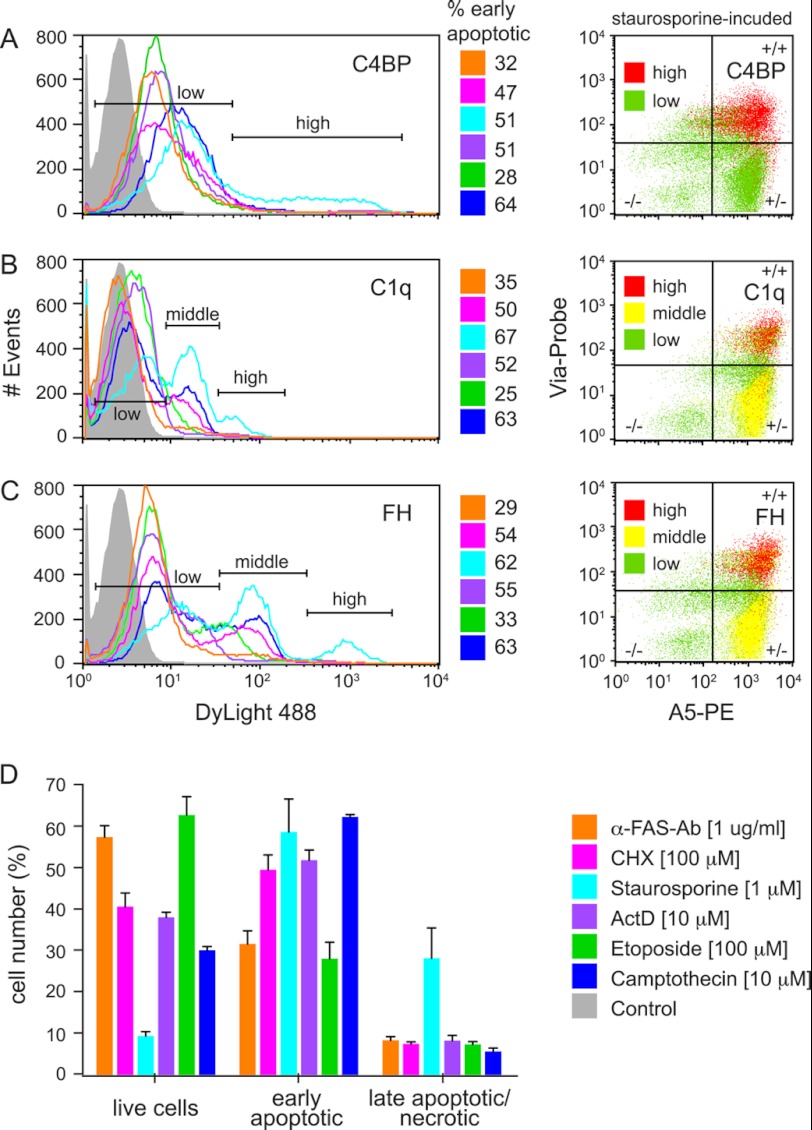
To see whether the mode of apoptosis induction influences the binding of C4BP, FH, and C1q to apoptotic cells, Jurkat T-cells were stimulated with commonly used concentrations of different inducers of the intrinsic (staurosporine, etoposide and camptothecin) or the extrinsic (anti-fas Ab, cycloheximide, and actinomycin D) apoptosis induction pathway. The histograms in Fig. 6 display the binding of C4BP (A), C1q (B), and FH (C) to the whole populations of cells induced to become apoptotic with various agents. The numbers next to the histograms indicate the percentage of early apoptotic cells in the whole cell population. C4BP bound well to all apoptotic cells irrespectively of inducing agent. However, the binding to α-fas Ab, cycloheximide, actinomycin D, and etoposide-induced cells was weaker than to camptothecin and staurosporine-induced cells. Apart from the staurosporine-treated cells, for which two peaks appeared in the histogram, only one peak corresponding to bound C4BP was observed for all other inducers (Fig. 6A). C1q bound generally weaker than C4BP and almost no binding could be detected to the anti-fas Ab and very little binding to the actinomycin D and etoposide-induced cells (Fig. 6B). Only one peak appeared in the histogram for those cells, whereas two peaks were present for the camptothecin and cycloheximide-induced cells and even three peaks for the staurosporine-induced cells. These may be corresponding to two or three distinct cell populations that vary in their ability to bind C1q. FH bound well to all apoptotic cells irrespectively of the induction method (Fig. 6C), whereby the anti-fas Ab-induced cells appeared as one homogenous population (one peak in the histogram). The camptothecin-, cycloheximide-, and etoposide-induced cells were divided into two distinct populations (two peaks) and the staurosporine-induced cells into three distinct populations (three peaks). In general all three proteins bound strongest to staurosporine-induced cells and weakest to anti-fas Ab-induced cells. However, no specific difference in binding strength could be observed between extrinsic or intrinsic apoptosis inducers. None of the three studied proteins bound to live Jurkat cells (data not shown). To verify whether observed different peaks correspond to different stages of apoptosis, the peaks were classified as low, middle, or high intensity binding. Staining with annexin A5 and Via-Probe allowed the differentiation into live (A5-/Via-Probe-), early apoptotic (A5+/Via-Probe−), and late apoptotic (A5+/Via-Probe+) populations, which are depicted in the dot plots (only for the staurosporine-induced cells). The cells that bound with high intensity (red) to C4BP, C1q and FH were, as expected, mainly the late apoptotic population. The cells that bound with middle intensity (yellow) to C1q and FH were mainly the early apoptotic population and the cells that bound with low intensity (green) were heterogeneously averaged through all three populations. The percentage of live, early, and late apoptotic cells for all inducers are indicated in Fig. 6D. After a 4-h stimulation with the different inducers, between 30 and 60% early apoptotic and around 10% late apoptotic cells were detected. Staurosporine was the only exception here yielding ∼35% late apoptotic cells and thus less than 10% live cells.
FIGURE 6.
C4BP, C1q, and FH bind to apoptotic cells generated by different apoptosis inducers. A–C, Jurkat T-cells were rendered apoptotic with different extrinsic and intrinsic apoptosis inducers and the binding of 50 μg/ml of DyLight 488-labeled C4BP (A), C1q (B), and FH (C) was investigated. All proteins bound to the cells were induced to become apoptotic with various agents, although the binding intensity differed for the individual inducers. Generally, all three proteins bound strongest to the staurosporine-induced cells. The percentage of early apoptotic cells generated in the experiments is indicated beside the flow cytometric histograms. The dot plots display the live (A5-/Via-Probe−), early (A5+/Via-Probe−), and late apoptotic (A5+/Via-Probe+) population of the cells that bound with high (red), middle (yellow), or low (green) intensity to the staurosporine-induced cells. D, percentage of live, early, and late apoptotic cells for all different apoptosis inducers. All inducers, except for staurosporine, generated populations that contained 30–60% live cells and 10% late apoptotic cells. Staurosporine-generated apoptotic population contained only 10% live but 40% late apoptotic cells. All inducers generated 30–60% early apoptotic cells. A–C, representative histograms and dot plots of two independent experiments performed in duplicates in which the binding was once detected with labeled proteins (shown data) and once with FITC-labeled antibodies (data not shown) are presented. D, the mean ± S.D. for the cells treated with DyLight 488-labeled C4BP, C1q, and FH are shown.
DISCUSSION
Proper clearance of apoptotic cells is crucial to secure homeostasis. For this purpose, a complex and multifaceted system has evolved containing a vast range of recognition mechanisms of the apoptotic cells from down-regulation of “do not eat me” signals to up-regulation of “find me” as well as “eat me” signals. Deficiencies of key components of this complex network such as C1q as well as phagocytic receptors decrease the efficiency of clearance, causing apoptotic cells to lose their integrity before being properly cleared. This in turn leads to a more inflammatory environment, which could induce the formation of autoantibodies paving the path for development of autoimmunity.
C1q has been shown to interact with apoptotic cells both directly (7), which is the subject of this study, and indirectly via C-reactive protein (34), serum amyloid P, pentraxin 3 (35), and natural IgM (36). DNA has long been known as a ligand for C1q and here we further characterized the interaction by determining the KD value using surface plasmon resonance, confirming a very strong interaction. However, due to the fact that C1q binds at a later stage to apoptotic cells than DNA becomes present on their surface and with different kinetics over induction time, we hypothesized that there must be other ligands. Histones are present on apoptotic cells in the form of nucleosomes. The pentraxins C-reactive protein and serum amyloid P have been shown to bind to histones and activate the classical complement pathway (37). Here we showed that C1q also binds directly to histones, which leads to complement activation. C4b deposition on all histone complexes was abolished in C1q-deficient sera but surprisingly it could not be restored upon addition of C1q for the histone complex H2A/H2B + H1, even though deposition was restored for both IgG and H3/H4. This indicates that there is a factor additional to C1q lacking in the C1q-deficient sera, which is required for complement activation by C1q on the H2A/H2B + H1 complex, which is not needed for complement activation on H3/H4. We further assayed MBL, factor D, and COLEC11 as well as ficolin-L and ficolin-H activity in the different sera, but all tested proteins were functional in both C1q-deficient sera and in the control sera (not shown). The importance of the interaction between C1q and nucleosomes (DNA and histones) on apoptotic cells is underscored by the observation that patients with low serum levels of immune complex-containing DNA at the onset of a flare developed a more severe flare compared with patients for whom the level remained stable. This coincided with low C1q serum levels and high disease activity and was not seen among patients with normal levels of C1q at flare (38).
C1q bound dose-dependently to Jurkat T- and Namalwa B-cells with the binding not becoming completely saturated at the concentrations used. We revealed that there are at least four different and abundant ligands for C1q on the surface of apoptotic cells. The surface expression of annexins A2 and A5, DNA, and histones increases during the course of apoptosis induction. Hence the overall concentration of possible ligands for C1q is very high on the 16-h induced cells and it is likely that the binding cannot be fully saturated unless very high concentrations are used. The binding of C1q to primary T-lymphocytes was also dose-dependent, almost saturated, and reaching lower levels than for Jurkat T-cells. Primary T-lymphocytes are much smaller than Jurkat T-cells and thus exhibit a lesser total amount of ligands for C1q. As a consequence, the molar ratio of C1q to C1q ligands is larger and the binding saturates at lower concentrations.
Annexin A2 and A5 are both present in serum (39, 40) and can bind negatively charged phospholipids. Additionally, their surface expression on lymphocytes increases during apoptosis. C1q preincubated with annexins therefore bound stronger to the apoptotic cell surface and this might be relevant also in an in vivo situation. Depending on the apoptosis stage, annexins A2 and A5 co-localized with C1q on some cells, most likely the later apoptotic ones. No co-localization was found on another population of cells, which comprises most likely the early apoptotic ones (data not shown).
PS has been reported to function as a C1q ligand (13), but we could not find experimental evidence for this hypothesis in our assays. Using annexin A5, we found PS already exposed on the cell surface at an early time point (∼1 h) after apoptosis induction and detected a further increase with induction time up to 4 h. After this time point, almost no further elevation was detected (data not shown). C1q binding occurred significantly later and increased further over the whole induction time. Additionally, we did not detect any interaction of C1q with PS included in liposomes that were immobilized either on L1 chips in Biacore or on beads and analyzed by flow cytometry. The positive control, C4BP-protein S complex bound as expected to PS. Finally, we did not detect significant co-localization of C1q and PS on apoptotic cells at various time points of apoptotic cell induction from early to late apoptotic cells. If C1q bound PS on apoptotic cells we would expect that C1q could compete with C4BP (or the reverse) for binding to apoptotic cells, as A5 does. However, no such competition could be observed. In the same set up, we did not see any competition for C1q with FH, which is in concordance with a recent report (41). The discrepancies with the study by Paidassi et al. (13) might arise from technical differences because that study employed HeLa cells, an epithelial carcinoma cell line, induced to become apoptotic using UV irradiation. HeLa cells were trypsinized for the experiments and it must be kept in mind that trypsinzation might have removed protein ligands for C1q on the surface. In addition, liposomes containing 100% PS, PE, or PC were used to study binding of C1q by surface plasmon resonance. This approach is less accurate than our employed mixtures of PC with PS and PE, because even though PS appears in patches on the surface of apoptotic cells, there are always other lipids present. In addition, whereas the confocal pictures presenting co-localization between C1q and PS looked very convincing, no appropriate software was used to objectively quantify the degree of co-localization. Our software analysis of images found no significant co-localization. An inhibitory effect of annexin A5 on the binding of C1q was also determined, after cells were preincubated with annexin A5 and then C1q was applied. We on the other hand preincubated the two proteins together and then added the mixture to the cells and thus detected increased C1q binding, possibly due to a bridging function of annexin A5 in guiding bound C1q to negatively charged phospholipids.
Furthermore, we found no interaction of C1q with oxidized liposomes, which are normally found on apoptotic cells (33) or myeloperoxidase-treated apoptotic cells. Inclusion of lyso-PS in liposomes did not generate C1q binding but abolished binding of the C4BP-protein S, which binds with high affinity to PS. Interestingly, the interaction of C4BP-protein S to PS was not affected by oxidation of the lipids, which is in contrast to what has been observed for free, recombinant protein S (42).
Association of inherited C1q deficiency and SLE is very strong, but it is also very rare and thus not responsible for the majority of SLE cases. It is therefore possible that there are other factors preventing interaction of C1q with apoptotic cells in SLE patients, such as autoantibodies to C1q ligands. Interestingly, autoantibodies to both annexin A2 and annexin A5 (21, 22), as well as to other C1q ligands and C1q itself (43, 44), have been found in patients suffering from autoimmune diseases such as SLE. These antibodies could either decrease direct binding of C1q to apoptotic cells or recruit additional C1q. Both scenarios could lead to an imbalance in complement activation, which could result in either insufficient removal of the cells or excessive complement activation resulting in a proinflammatory environment. Both are commonly seen in SLE.
The fact that C1q deficiency is a much stronger genetic risk factor for development of SLE than deficiency of C4 or C2 (45) suggests that C1q has a unique function in addition to initiation of complement leading to the opsonization with C3b. To support this, evidence has been presented supporting the role of C1q in development of tolerance (46), clearance of immune complexes (47), and cytokine regulation (48). It differentially modulates the phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells as well as of apoptotic neurons by microglia (49, 50). Clearly, C1q has many functions crucial for homeostasis and prevention of SLE. Accordingly, low serum levels of C1q correlates with severe active SLE (51).
Apoptosis can be induced by two distinct pathways, the intrinsic and the extrinsic. It has been shown that the percentage of generated early and late apoptotic cells depends on the inducing agent. This on the other hand influences among others the amount of released DNA by Jurkat T-cells and monocytic U937 cells (52). Lesional late-stage scleroderma fibroblasts have been shown to be relatively resistant to Fas-induced apoptosis, but sensitive to intrinsic induction (53). In the current study we found that the method of inducing apoptosis did influence the binding of C1q and the complement inhibitors, C4BP and FH, to apoptotic Jurkat T-cells. All three proteins bound to the majority of apoptotic cells independently of the route of apoptosis induction although the intensity of binding differed strongly. Furthermore, no general difference was observed between intrinsic or extrinsic inducers and the order of binding strength with which all three proteins bound to the differently induced cells was the same, even though all three proteins bind at least partially to different ligands and did not compete for one another's binding. Jurkat T-cells are transformed cells and the results obtained might be different for primary cells. Some apoptosis inducers generated cells that bound C1q with high, middle, and/or low intensity. As expected, the late apoptotic cells mainly bound C1q with high intensity and a quite homogenous population of early apoptotic cells bound with middle intensity. Interestingly, the low intensity binding cells were averaged heterogeneously through the live, early, and late apoptotic populations. This indicates that it is not sufficient for a cell to become apoptotic (as determined by annexin A5 and Via-Probe staining) to bind C1q and that, other surface changes also must occur to induce C1q binding. Such changes may involve increased surface expression and spatial organization of endogenous annexin A2, A5, DNA, or histones or of yet unknown additional ligands.
In conclusion, the current study further characterizes the interactions of complement and apoptotic cells and reveals two members of the annexin family, annexin A2 and A5, as new ligands for C1q on dying cells. This phenomenon gives a further explanation for the observations of the presence of pathologic annexin autoantibodies in SLE. This study hence sheds new light on the interactions underlying complement-mediated clearance of apoptotic cells and helps to further the understanding of the pathophysiology of autoimmune diseases such as SLE.
Acknowledgments
We thank Dr. L. A. Trouw for help with the Biacore experiment assessing affinity of C1q-DNA interaction and Dr. Ben King for language revision of the manuscript.
This work was supported in part by the Söderberg Foundation, Swedish Research Council Grant K2009-68X-14928-06-3, the Swedish Foundation for Strategic Research, the Swedish Rheumatism Association, the National Board of Health and Welfare and Skåne University Hospital, and the Österlund, Greta and Johan Kock, King Gustaf V's 80th Birthday, Knut and Alice Wallenberg, and Inga-Britt and Arne Lundberg Foundations.
- PS
- phosphatidylserine
- A2
- annexin A2
- A5
- annexin A5
- α1-AT
- α1-antitrypsin
- C4BP
- C4b-binding protein
- FH
- factor H
- Hi-NHS
- heat inactivated normal human serum
- NHS
- normal human serum
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- SLE
- systemic lupus erythematosus.
REFERENCES
- 1. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010) Complement, a key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wallis R., Mitchell D. A., Schmid R., Schwaeble W. J., Keeble A. H. (2010) Paths reunited. Initiation of the classical and lectin pathways of complement activation. Immunobiology 215, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korb L. C., Ahearn J. M. (1997) C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes. Complement deficiency and systemic lupus erythematosus revisited. J. Immunol. 158, 4525–4528 [PubMed] [Google Scholar]
- 4. Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., Henson P. M. (2001) C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Navratil J. S., Watkins S. C., Wisnieski J. J., Ahearn J. M. (2001) The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J. Immunol. 166, 3231–3239 [DOI] [PubMed] [Google Scholar]
- 6. Vandivier R. W., Ogden C. A., Fadok V. A., Hoffmann P. R., Brown K. K., Botto M., Walport M. J., Fisher J. H., Henson P. M., Greene K. E. (2002) Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro. Calreticulin and CD91 as a common collectin receptor complex. J. Immunol. 169, 3978–3986 [DOI] [PubMed] [Google Scholar]
- 7. Nauta A. J., Trouw L. A., Daha M. R., Tijsma O., Nieuwland R., Schwaeble W. J., Gingras A. R., Mantovani A., Hack E. C., Roos A. (2002) Direct binding of C1q to apoptotic cells and cell blebs induces complement activation. Eur. J. Immunol. 32, 1726–1736 [DOI] [PubMed] [Google Scholar]
- 8. Elward K., Griffiths M., Mizuno M., Harris C. L., Neal J. W., Morgan B. P., Gasque P. (2005) CD46 plays a key role in tailoring innate immune recognition of apoptotic and necrotic cells. J. Biol. Chem. 280, 36342–36354 [DOI] [PubMed] [Google Scholar]
- 9. Jiang H., Cooper B., Robey F. A., Gewurz H. (1992) DNA binds and activates complement via residues 14–26 of the human C1q A chain. J. Biol. Chem. 267, 25597–25601 [PubMed] [Google Scholar]
- 10. Kishore U., Gupta S. K., Perdikoulis M. V., Kojouharova M. S., Urban B. C., Reid K. B. (2003) Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J. Immunol. 171, 812–820 [DOI] [PubMed] [Google Scholar]
- 11. Palaniyar N., Nadesalingam J., Clark H., Shih M. J., Dodds A. W., Reid K. B. (2004) Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose-binding lectin. J. Biol. Chem. 279, 32728–32736 [DOI] [PubMed] [Google Scholar]
- 12. Tissot B., Daniel R., Place C. (2003) Interaction of the C1 complex of complement with sulfated polysaccharide and DNA probed by single molecule fluorescence microscopy. Eur. J. Biochem. 270, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 13. Païdassi H., Tacnet-Delorme P., Garlatti V., Darnault C., Ghebrehiwet B., Gaboriaud C., Arlaud G. J., Frachet P. (2008) C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Webb J. H., Blom A. M., Dahlbäck B. (2002) Vitamin K-dependent protein S localizing complement regulator C4b-binding protein to the surface of apoptotic cells. J. Immunol. 169, 2580–2586 [DOI] [PubMed] [Google Scholar]
- 15. Trouw L. A., Bengtsson A. A., Gelderman K. A., Dahlbäck B., Sturfelt G., Blom A. M. (2007) C4b-binding protein and factor H compensate for the loss of membrane-bound complement inhibitors to protect apoptotic cells against excessive complement attack. J. Biol. Chem. 282, 28540–28548 [DOI] [PubMed] [Google Scholar]
- 16. Trouw L. A., Nilsson S. C., Gonçalves I., Landberg G., Blom A. M. (2005) C4b-binding protein binds to necrotic cells and DNA, limiting DNA release and inhibiting complement activation. J. Exp. Med. 201, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leffler J., Herbert A. P., Norström E., Schmidt C. Q., Barlow P. N., Blom A. M., Martin M. (2010) Annexin-II, DNA, and histones serve as factor H ligands on the surface of apoptotic cells. J. Biol. Chem. 285, 3766–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iaccarino L., Ghirardello A., Canova M., Zen M., Bettio S., Nalotto L., Punzi L., Doria A. (2011) Anti-annexins autoantibodies. Their role as biomarkers of autoimmune diseases. Autoimmun. Rev. 10, 553–558 [DOI] [PubMed] [Google Scholar]
- 19. Gerke V., Creutz C. E., Moss S. E. (2005) Annexins, linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell. Biol. 6, 449–461 [DOI] [PubMed] [Google Scholar]
- 20. Hayes M. J., Moss S. E. (2004) Annexins and disease. Biochem. Biophys. Res. Commun. 322, 1166–1170 [DOI] [PubMed] [Google Scholar]
- 21. Ao W., Zheng H., Chen X. W., Shen Y., Yang C. D. (2011) Anti-annexin II antibody is associated with thrombosis and/or pregnancy morbidity in antiphospholipid syndrome and systemic lupus erythematosus with thrombosis. Rheumatol. Int. 31, 865–869 [DOI] [PubMed] [Google Scholar]
- 22. Hrycek A., Cieślik P. (2012) Annexin A5 and anti-annexin antibodies in patients with systemic lupus erythematosus. Rheumatol. Int. 32, 1335–1342 [DOI] [PubMed] [Google Scholar]
- 23. Dahlbäck B. (1983) Purification of human C4b-binding protein and formation of its complex with vitamin K-dependent protein S. Biochem. J. 209, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blom A. M., Kask L., Dahlbäck B. (2003) CCP1–4 of the C4b-binding protein α-chain are required for factor I mediated cleavage of complement factor C3b. Mol. Immunol. 39, 547–556 [DOI] [PubMed] [Google Scholar]
- 25. Tenner A. J., Lesavre P. H., Cooper N. R. (1981) Purification and radiolabeling of human C1q. J. Immunol. 127, 648–653 [PubMed] [Google Scholar]
- 26. Kask L., Trouw L. A., Dahlbäck B., Blom A. M. (2004) The C4b-binding protein-protein S complex inhibits the phagocytosis of apoptotic cells. J. Biol. Chem. 279, 23869–23873 [DOI] [PubMed] [Google Scholar]
- 27. Laurell C. B., Dahlqvist I., Persson U. (1983) The use of thiol-disulfide exchange chromatography for the automated isolation of α1-antitrypsin and other plasma proteins with reactive thiol groups. J. Chromatogr. 278, 53–61 [DOI] [PubMed] [Google Scholar]
- 28. Pâques E. P., Huber R., Priess H., Wright J. K. (1979) Isolation of the globular region of the subcomponent q of the C1 component of complement. Hoppe-Seyler's Z. Physiol. Chem. 360, 177–183 [DOI] [PubMed] [Google Scholar]
- 29. Krisinger M. J., Guo L. J., Salvagno G. L., Guidi G. C., Lippi G., Dahlbäck B. (2009) Mouse recombinant protein C variants with enhanced membrane affinity and hyper-anticoagulant activity in mouse plasma. FEBS J. 276, 6586–6602 [DOI] [PubMed] [Google Scholar]
- 30. Manders E. M., Verbeek F. J., Aten J. A. (1993) Measurement of colocalization of objects in dual-color confocal images. J. Microsc. 169, 375–382 [DOI] [PubMed] [Google Scholar]
- 31. Radic M., Marion T., Monestier M. (2004) Nucleosomes are exposed at the cell surface in apoptosis. J. Immunol. 172, 6692–6700 [DOI] [PubMed] [Google Scholar]
- 32. Taketo M. M., Sonoshita M. (2002) Phospholipase A2 and apoptosis. Biochim. Biophys. Acta 1585, 72–76 [DOI] [PubMed] [Google Scholar]
- 33. Buttke T. M., Sandstrom P. A. (1994) Oxidative stress as a mediator of apoptosis. Immunol. Today 15, 7–10 [DOI] [PubMed] [Google Scholar]
- 34. Mihlan M., Stippa S., Józsi M., Zipfel P. F. (2009) Monomeric CRP contributes to complement control in fluid phase and on cellular surfaces and increases phagocytosis by recruiting factor H. Cell Death Differ. 16, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 35. Nauta A. J., Bottazzi B., Mantovani A., Salvatori G., Kishore U., Schwaeble W. J., Gingras A. R., Tzima S., Vivanco F., Egido J., Tijsma O., Hack E. C., Daha M. R., Roos A. (2003) Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur. J. Immunol. 33, 465–473 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y., Park Y. B., Patel E., Silverman G. J. (2009) IgM antibodies to apoptosis-associated determinants recruit C1q and enhance dendritic cell phagocytosis of apoptotic cells. J. Immunol. 182, 6031–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hicks P. S., Saunero-Nava L., Du Clos T. W., Mold C. (1992) Serum amyloid P component binds to histones and activates the classical complement pathway. J. Immunol. 149, 3689–3694 [PubMed] [Google Scholar]
- 38. Bengtsson A., Nezlin R., Shoenfeld Y., Sturfelt G. (1999) DNA levels in circulating immune complexes decrease at severe SLE flares. Correlation with complement component C1q. J. Autoimmun. 13, 111–119 [DOI] [PubMed] [Google Scholar]
- 39. Ji N. Y., Park M. Y., Kang Y. H., Lee C. I., Kim D. G., Yeom Y. I., Jang Y. J., Myung P. K., Kim J. W., Lee H. G., Lee K., Song E. Y. (2009) Evaluation of annexin II as a potential serum marker for hepatocellular carcinoma using a developed sandwich ELISA method. Int. J. Mol. Med. 24, 765–771 [DOI] [PubMed] [Google Scholar]
- 40. Matsuda R., Kaneko N., Kikuchi M., Chiwaki F., Toda M., Ieiri T., Horikawa Y., Shimizu M., Shimamoto K. (2003) Clinical significance of measurement of plasma annexin V concentration of patients in the emergency room. Resuscitation 57, 171–177 [DOI] [PubMed] [Google Scholar]
- 41. Kang Y. H., Urban B. C., Sim R. B., Kishore U. (2012) Human complement factor H modulates C1q-mediated phagocytosis of apoptotic cells. Immunobiology 217, 455–464 [DOI] [PubMed] [Google Scholar]
- 42. Baroni M., Pavani G., Marescotti D., Kaabache T., Borgel D., Gandrille S., Marchetti G., Legnani C., D'Angelo A., Pinotti M., Bernardi F. (2010) Membrane binding and anticoagulant properties of protein S natural variants. Thromb. Res. 125, e33–39 [DOI] [PubMed] [Google Scholar]
- 43. Trouw L. A., Daha M. R. (2005) Role of anti-C1q autoantibodies in the pathogenesis of lupus nephritis. Expert Opin. Biol. Ther. 5, 243–251 [DOI] [PubMed] [Google Scholar]
- 44. Shoenfeld Y., Szyper-Kravitz M., Witte T., Doria A., Tsutsumi A., Tatsuya A., Dayer J. M., Roux-Lombard P., Fontao L., Kallenberg C. G., Bijl M., Matthias T., Fraser A., Zandman-Goddard G., Blank M., Gilburd B., Meroni P. L. (2007) Autoantibodies against protective molecules, C1q, C-reactive protein, serum amyloid P, mannose-binding lectin, and apolipoprotein A1. Prevalence in systemic lupus erythematosus. Ann. N.Y. Acad. Sci. 1108, 227–239 [DOI] [PubMed] [Google Scholar]
- 45. Truedsson L., Bengtsson A. A., Sturfelt G. (2007) Complement deficiencies and systemic lupus erythematosus. Autoimmunity 40, 560–566 [DOI] [PubMed] [Google Scholar]
- 46. Baruah P., Simpson E., Dumitriu I. E., Derbyshire K., Coe D., Addey C., Dyson J., Chai J. G., Cook T., Scott D., Botto M. (2010) Mice lacking C1q or C3 show accelerated rejection of minor H disparate skin grafts and resistance to induction of tolerance. Eur. J. Immunol. 40, 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davies K. A., Hird V., Stewart S., Sivolapenko G. B., Jose P., Epenetos A. A., Walport M. J. (1990) A study of in vivo immune complex formation and clearance in man. J. Immunol. 144, 4613–4620 [PubMed] [Google Scholar]
- 48. Lood C., Gullstrand B., Truedsson L., Olin A. I., Alm G. V., Rönnblom L., Sturfelt G., Eloranta M. L., Bengtsson A. A. (2009) C1q inhibits immune complex-induced interferon-α production in plasmacytoid dendritic cells. A novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 60, 3081–3090 [DOI] [PubMed] [Google Scholar]
- 49. Fraser D. A., Laust A. K., Nelson E. L., Tenner A. J. (2009) C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J. Immunol. 183, 6175–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fraser D. A., Pisalyaput K., Tenner A. J. (2010) C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J. Neurochem. 112, 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sturfelt G., Johnson U., Sjöholm A. G. (1985) Sequential studies of complement activation in systemic lupus erythematosus. Scand. J. Rheumatol. 14, 184–196 [DOI] [PubMed] [Google Scholar]
- 52. Choi J. J., Reich C. F., 3rd, Pisetsky D. S. (2004) Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand. J. Immunol. 60, 159–166 [DOI] [PubMed] [Google Scholar]
- 53. Chabaud S., Corriveau M. P., Grodzicky T., Senécal J. L., Chartier S., Raymond Y., Moulin V. J. (2011) Decreased secretion of MMP by nonlesional late-stage scleroderma fibroblasts after selection via activation of the apoptotic Fas-pathway. J. Cell. Physiol. 226, 1907–1914 [DOI] [PubMed] [Google Scholar]