Background: Chromatin-HP1 (heterochromatin protein 1) interaction is crucial for heterochromatin assembly.
Results: hHP1β uses alternative interfaces to bind nucleosomes depending on histone 3 methylation within a highly dynamic complex.
Conclusion: hHP1β explores chromatin for sites of methyl-mark enrichment where it can bind histone 3 tails from adjacent nucleosomes.
Significance: We provide a conceptual framework to understand the molecular basis of dynamic interactions regulated by histone modification.
Keywords: Chromatin Regulation, Epigenetics, Heterochromatin, Histone Methylation, NMR, Structural Biology, Dynamics
Abstract
Binding of heterochromatin protein 1 (HP1) to the histone H3 lysine 9 trimethylation (H3K9me3) mark is a hallmark of establishment and maintenance of heterochromatin. Although genetic and cell biological aspects have been elucidated, the molecular details of HP1 binding to H3K9me3 nucleosomes are unknown. Using a combination of NMR spectroscopy and biophysical measurements on fully defined recombinant experimental systems, we demonstrate that H3K9me3 works as an on/off switch regulating distinct binding modes of hHP1β to the nucleosome. The methyl-mark determines a highly flexible and very dynamic interaction of the chromodomain of hHP1β with the H3-tail. There are no other constraints of interaction or additional multimerization interfaces. In contrast, in the absence of methylation, the hinge region and the N-terminal tail form weak nucleosome contacts mainly with DNA. In agreement with the high flexibility within the hHP1β-H3K9me3 nucleosome complex, the chromoshadow domain does not provide a direct binding interface. Our results report the first detailed structural analysis of a dynamic protein-nucleosome complex directed by a histone modification and provide a conceptual framework for understanding similar interactions in the context of chromatin.
Introduction
A large number of diverse post-translational modifications on histones modulate chromatin structure and dynamics. Methylation of lysine 9 within the H3 N terminus is a crucial determinant of heterochromatin formation (1). The trimethylated form (H3K9me3) of this modification is found at pericentric heterochromatin in virtually all higher eukaryotes and is viewed as a hallmark of silenced chromatin (2). H3K9me3 provides a binding site for heterochromatin protein 1 (HP1), a family of non-histone chromatin proteins found in different isoforms from Schizosaccharomyces pombe to human. HP1 proteins induce formation of heterochromatin when artificially targeted to heterologous sites within the genome (3), an effect that is concomitant with induction of local H3K9me3 (4). Given the important roles of heterochromatin in gene regulation, it is surprising that mammalian HP1β is the only HP1 protein essential for viability (5).
All HP1 proteins share the same architecture consisting of two structured and conserved domains, a chromodomain (CD)7 and a chromoshadow domain (CSD), connected by a less conserved hinge region (6). Using histone peptides, it was shown that a conserved aromatic cage within the CD binds methylated H3K9 with low (micromolar) affinity but high sequence specificity and a preference for the trimethylated states (7–9). However, biochemical work on individual HP1 domains, HP1 proteins of different species and isoforms, has suggested that the CD might not be sufficient for HP1 nucleosome binding but that the hinge region or CSD make major contacts (10). These findings are supported by the varying dependence on the different HP1 domains for heterochromatin localization (5, 11). In particular, the CSD, which mediates dimerization of all HP1 proteins (12, 13), has been shown to interact with a plethora of nuclear proteins containing a short PXVXL consensus sequence (14, 15). Other studies imply a chromatin interaction largely independent of the histone tails. Interactions between hHP1s with the core region of H3 (12, 16, 17), of hHP1α with H2A.Z (18), of hHP1α with linker histone H1 (12, 19, 20), and of human or Xenopus HP1α with DNA (19, 21, 22) or RNA (23) have been reported. In addition, it has been questioned whether DmHP1 makes stable contacts with chromatin in the absence of accessory factors (24). Overall, the importance and the functional consequence of the HP1-H3K9me3 interaction in HP1 chromatin targeting are still unclear (25).
Understanding the molecular determinants of HP1-chromatin interaction requires biochemical analysis that goes beyond work on isolated HP1 domains or usage of nucleosomes and oligonucleosomes isolated from cells and carrying heterogeneous histone modification patterns. Recently, work on Swi6 using a fully defined experimental system has suggested that specific interaction with H3K9me3 chromatin requires multimerization of the protein on the template (26). However, it is unclear whether HP1 proteins of higher eukaryotes work via similar mechanisms and form fixed complexes on chromatin, because no structural insights into the HP1-nucleosome interaction could be obtained so far.
Despite the key role of histone post-translational modifications in chromatin biology, no structural analysis of any protein-nucleosome complex dependent on covalent histone modifications has been reported. An NMR-based docking model of the nucleosome with HMG2 (27) and crystal structures of nucleosome in complex with KSHV LANA peptide (28), with Drosophila RCC1 (29), and with yeast Sir3 BAH (30) revealed interactions that involve the acidic patch of the H2A-H2B dimer and that are independent of histone post-translational modifications. By using a combination of NMR, biochemical, and biophysical measurements, we reveal here detailed molecular insights into the hHP1β-nucleosome complex in dependence of H3K9me3.
EXPERIMENTAL PROCEDURES
Sample Preparation
An extensive description of materials and methods for nucleosome and hHP1β sample preparation is given in supplemental Methods.
Nucleosome Pulldown Experiments
1 μg of biotinylated nucleosomes (reconstituted with DNA ligated via an EcoRI site to a biotinylated oligonucleotide) were preincubated with 50 μg of streptavidin-coated magnetic beads (Promega) in binding buffer (10 mm triethanolamine-HCl, 150 mm NaCl, 0.1% v/v Triton X-100, 5% v/v glycerol, 0.1 mm EDTA, pH 7.5) for 4 h at 4 °C. Unbound peptides or nucleosomes were removed by two washes with binding buffer. Immobilized nucleosomes were incubated with recombinant HP1 protein for 1 h at 4 °C in binding buffer. Beads were then washed three times with washing buffer (10 mm triethanolamine-HCl, 300 mm NaCl, 0.1% v/v Triton X-100, 5% v/v glycerol, 0.1 mm EDTA, pH 7.5), and bound protein was eluted by boiling the beads in sample loading buffer. Samples were run on SDS-polyacrylamide gels either stained with Coomassie or transferred to nitrocellulose membranes for Western blotting. For Western analysis, primary antibodies were used as follows: anti-H3 1:40,000 (Abcam) and anti-HP1β 1:2000 (Millipore).
Fluorescence Polarization Measurements
Fluorescence polarization assays were carried out and analyzed as described previously (31). Experiments were performed at 293 K in 10 mm triethanolamine-HCl, 150 mm NaCl, 0.1 mm EDTA, 2 mm DTT, pH 7.4. Titration series of a 10-μl volume in 384-well plates were read multiple times on a Plate Chameleon II plate reader (HIDEX Oy). Multiple readings and independent titration series were averaged after data normalization.
Isothermal Titration Calorimetry
ITC measurements were performed on an iTC200 calorimeter (Microcal) at 293 K in 10 mm triethanolamine, 150 mm NaCl, 0.1 mm EDTA, 2 mm DTT, pH 7.4. Reaction heats were recorded by sequences of 37 injections of 0.6, 1.8, or 3 mm hHP1β, spaced at 120-s intervals, into 250 μl of 20, 60, or 150 μm H3 peptides or nucleosomes under constant stirring at 1000 rpm (injections 1–10, 0.5 μl each; injections 11–32, 1 μl each; and injections 33–37, 2 μl each). Data analysis was performed with Origin® software, as described in supplemental Methods.
Surface Plasmon Resonance
SPR measurements were performed on a Biacore 2000 instrument (GE Healthcare) at 298 K in 10 mm triethanolamine, 150 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 0.005% Tween 20, pH 7.4. Varying amounts of biotinylated ligands were immobilized on streptavidin-coated sensor chips SA (GE Healthcare) using different contact times and concentrations. hHP1β proteins were injected at 24 nm to 50 μm (serial 2-fold dilutions) for 2 min (30 μl/min). Dissociation was recorded for up to 5 min. Data evaluation was performed using BIAevaluation 4.1 and Prism 5.04 (GraphPad) software, as described in supplemental Methods.
NMR Experiments
NMR spectra were recorded on Bruker spectrometers operating at 1H frequencies of 600, 700, 800, and 900 MHz equipped with cryogenic probes. Unless otherwise specified, the temperature was 303 K. Data were processed with NMRPipe (32) and analyzed using the software Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Mathematical and graphical analysis was performed using Origin® and Wolfram Mathematica®. Three-dimensional structures were visualized using PyMOL.
Assignment of backbone chemical shifts for WT hHP1β was achieved by standard TROSY-based three-dimensional (33) and APSY five-, six-, and seven-dimensional experiments (34, 35). Because of the high correspondence of 1H-15N HSQC spectra, the assignment was transferred to the shorter mutants ΔNΔC and CSD and verified by additional multidimensional experiments where required. Backbone assignment of hHP1βI161A was completed with three-dimensional HNCA and three-dimensional CBCA(CO)NH experiments. Methyl resonances were assigned with three-dimensional CC(CO)NH, three-dimensional H(CC)(CO)NH, and three-dimensional HCCH-TOCSY experiments.
Titration of hHP1β with H3 peptides or nucleosomes was followed by TROSY 1H-15N HSQC and 1H-13C HMQC experiments. 1H-13C HMQC spectra were recorded at 800 MHz, 310 K, and in a 100% D2O-based buffer when the [U-2H]hHP1β with selective methyl [13CH3,12CD3]Val, Leu labeling was used (36).
Cross-saturation experiments were recorded at 700 MHz, 282 K using 0.1 mm U-2H,15N-labeled hHP1β and unlabeled 0.01 mm H3KC9me3 nucleosome. Saturation of nucleosome protons was achieved by a WURST-2 decoupling scheme followed by TROSY 1H-15N HSQC (37). The irradiation frequency was set to 0.9 ppm for selective saturation of aliphatic protons and to 30 ppm for the reference. The irradiation time was 2 s, and the recycling delay was 4 s. Additional cross-saturation experiments were performed at different temperatures (289, 296, 303, and 310 K) and using different irradiation frequencies (2.9 ppm) and irradiation times (1 s). hHP1β in the absence of nucleosome, measured under the same experimental condition, was used as reference for data normalization.
To measure the rates of cross-correlation between 15N chemical shift anisotropy and 1H-15N dipole-dipole interaction (ηxy) of U-2H,15N-labeled hHP1β, we used the TROSY-based pulse sequence developed for proteins larger than 50 kDa (38). The experiments were recorded at 800 MHz, 303 K with a relaxation delay 2τ = 1/JNH = 10.8 ms.
The hydrodynamic radius of CD(19–79) was determined at two different concentrations from diffusion coefficients measured by pulsed field gradient NMR. Stimulated echo-diffusion experiments were performed at 303 K.
RESULTS
Specific Binding of hHP1β to the H3K9me3 Nucleosome Is Noncooperative
To dissect the molecular mechanisms of the hHP1β-nucleosome interaction, we employed a fully recombinant nucleosome system. Histone H3 was uniformly modified introducing H3K9me3 by native chemical ligation as described in supplemental Methods and supplemental Fig. 1.
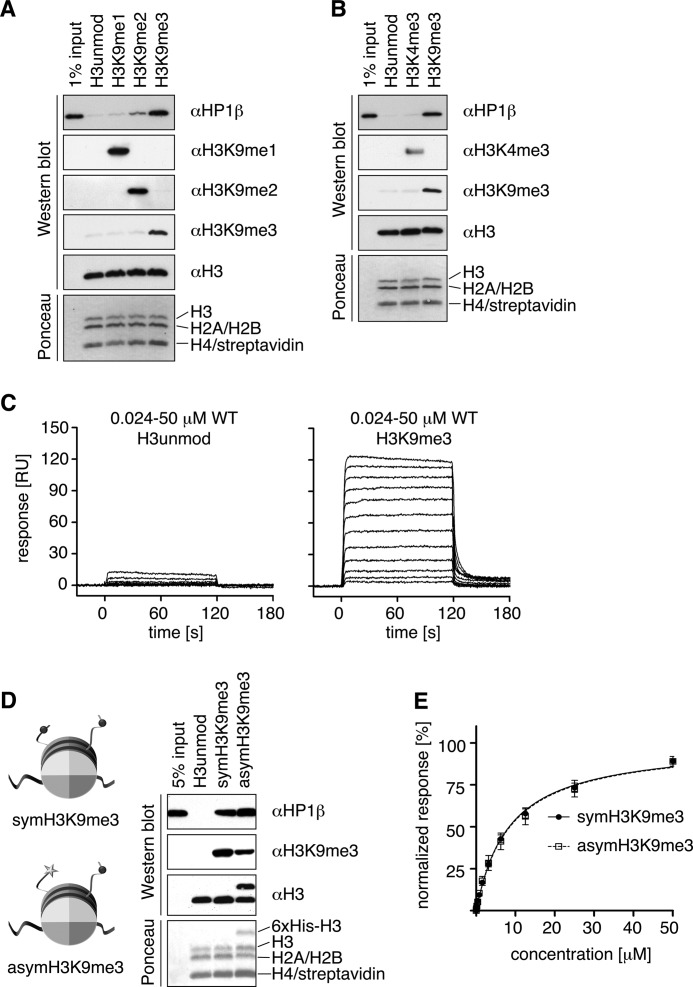
Pulldown experiments showed specific interaction of hHP1β with H3K9me templates, with a clear preference for the trimethylated state (me3), over the di- (me2) and mono (me1)-H9-methylated nucleosomes (Fig. 1A). In contrast, almost no interaction was observed with unmodified or H3K4me3 nucleosomes (Fig. 1, A and B).
FIGURE 1.
hHP1β specifically interacts with H3K9me3 nucleosomes. A and B, pulldown experiments using synthetic nucleosomes uniformly containing the indicated histone H3 modification status immobilized via biotinylated DNA on streptavidin-coated magnetic beads and soluble recombinant hHP1β. Recovered material was analyzed by Western blotting using the indicated antibodies and by Ponceau staining. Note that streptavidin stripped from the beads runs with H4 on the SDS-polyacrylamide gel. C, SPR analysis of titration series of soluble hHP1β WT interacting with unmodified (immobilization of 940 response units (RU)) and H3K9me3 (immobilization of 950 RU) nucleosomes immobilized via biotinylated DNA on a sensor chip. D, pulldown experiments using symmetrically or asymmetrically H3KC9me3-modified nucleosomes. E, comparative steady state evaluation of hHP1β binding to symmetric or asymmetric H3KC9me3 nucleosomes. SPR binding responses shortly before the end of the association phase were plotted against the analyte concentration and fitted by nonlinear regression assuming a 1:1 interaction model. Data of two independent measurements were normalized to Rmax = 100% and averaged.
To obtain quantitative interaction data at thermodynamic equilibrium, we performed SPR measurements (Fig. 1C). In titration experiments using different immobilization levels of H3K9me3 nucleosomes, we could reach close to saturation of hHP1β binding. With exemption of very high immobilization levels of H3K9me3 nucleosomes where some binding that could not be washed off by buffer occurred (supplemental Fig. 1N), all binding curves could be fitted well using a one-site binding model. In agreement, analysis by Hill plotting did not reveal any cooperativity (supplemental Fig. 1O). We deduced the dissociation constant for H3K9me3 nucleosomes using SPR at 2.4 ± 1.9 μm, a value similar to the binding affinity to a histone H3 (residues 1–15) K9me3 peptide (0.9 ± 0.3 μm) determined by the same method (see Table 1). Retention on unmodified nucleosomes, in contrast, was far lower implying high specificity of interaction. Although no faithful binding constant could be deduced due to the limited number of titration points that showed significant retention (supplemental Fig. 1N), we note that binding to unmodified nucleosomes occurred and was increased compared with the isolated unmodified H3-tail.
TABLE 1.
Apparent dissociation constants of hHP1β as determined by different experimental methods
The following abbreviations are used: FP, fluorescence polarization; ITC, isothermal titration calorimetry; and SPR, surface plasmon resonance. Kd values represent averages and standard deviation of multiple independent measurements (FP), including different concentrations of hHP1β and peptides or nucleosomes (ITC) or varying immobilization levels of peptides or nucleosomes (SPR).
| Kd (μm), hHP1β | Kd (μm), CSD | |
|---|---|---|
| H3K9me3 nucleosome | 2.4 ± 1.9 at SPR | |
| H3KC9me3 nucleosome | 8.0 ± 0.3 at SPR | NBa at ITC |
| 22 ± 17 at ITC | ||
| Unmodified nucleosome | NDb at SPR | |
| K9me3 H3(1–15) peptide | 1.9 ± 0.6 at FP | NBa at FP |
| 0.9 ± 0.3 at SPR | ||
| 1.4 ± 0.6 at ITC | ||
| KC9me3 H3(1–15) peptide | 7.5 ± 2.6 at FP | |
| 11 ± 4 at ITC | ||
| Unmodified H3(1–15) peptide | >300c at FP | NBa at FP |
| >300c at SPR | ||
| >500c at ITC |
a NB means not binding; titration curves could not be analyzed due to lack of binding signals.
b ND means not determined; although interaction was clearly observed, no binding constants could be deduced faithfully due to the limited number of data points (see supplemental Fig. 1N for details).
c In the case where titration curves did not reach the inflection points, minimal values for Kd are given.
To further understand the molecular details of hHP1β nucleosome interaction, we used a structural approach. Because stable association of hHP1β with nucleosomes could not be found in analytical ultracentrifugation experiments (supplemental Fig. 1L), we took advantage of nuclear magnetic resonance spectroscopy. NMR is particularly suited for the study of dynamic multicomponent systems as it allows insights at atomic levels into both structural effects and dynamic behavior for each domain while working with the full-length complex.
NMR Analysis of hHP1β Bound to the H3KC9me3 Nucleosome
To overcome limitations in the amounts of H3K9me3 nucleosomes, a methyl lysine analog (H3KC9me3) was used for the NMR experiments (see supplemental Methods and supplemental Fig. 1 for details on reconstitution). H3KC9me3 provided a reasonable mimic of the methyl-lysine in nucleosome recognition by hHP1β as determined by ITC and SPR experiments (Table 1 and supplemental Fig. 1M) (39, 40).
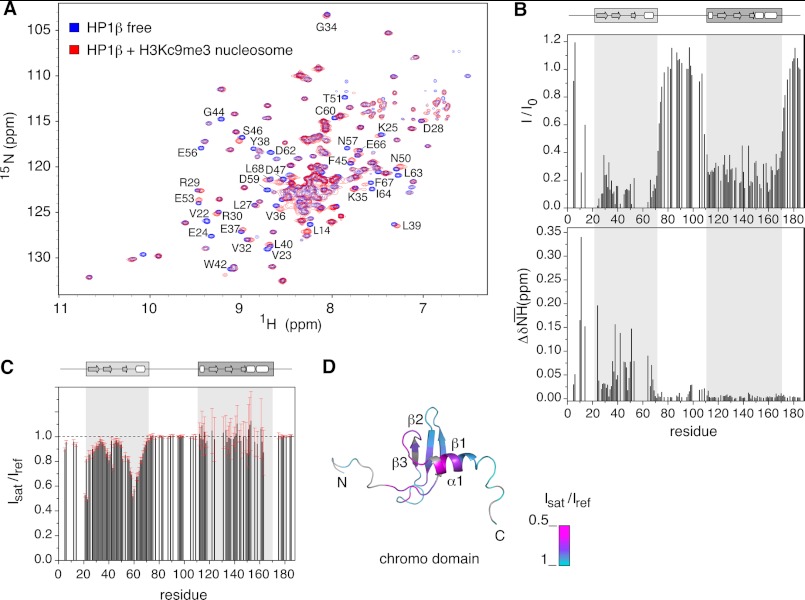
We performed a series of titrations using two-dimensional relaxation-optimized 1H,15N-heteronuclear correlation NMR experiments (33). Addition of unlabeled H3KC9me3 nucleosome to U-2H,12C- and 15N-labeled hHP1β resulted in gradual loss of signal intensity for many backbone resonances concomitant with small shifts occurring on the fast to intermediate NMR time scale (Fig. 2A). As shown by residual signal intensity and chemical shift difference plots at a molar ratio of 1:2 of hHP1β:H3KC9me3 nucleosome, residues within the CD were most affected (Fig. 2B). In addition, signals of the CSD were strongly broadened but without any chemical shift perturbation.
FIGURE 2.
Molecular determinants of hHP1β binding to H3KC9me3 nucleosome defined by NMR spectroscopy. A, two-dimensional TROSY-1H,15N HSQC spectra of hHP1β alone (blue) and with H3KC9me3 nucleosome at a molar ratio of 1:2 (red). Residues experiencing large changes are annotated. B, intensity loss (I/I0) and weighted chemical shift difference (ΔδNH) values from the spectra in A are plotted as a function of hHP1β primary sequence. A cluster of signals (Val-22 to Val-23, Lys-43 to Gly-44, and Glu-55 to Asp-62) disappeared already at low molar ratio. Other missing values correspond to proline residues or residues with severe signal overlap. The domain organization of hHP1β is schematically shown at the top. C, nucleosome-to-hHP1β cross-saturation transfer experiments: intensity ratio of hHP1β signals recorded with (Isat) or without (Iref) selective saturation of nucleosome aliphatic protons. Error bars were calculated on the basis of the signal-to-noise-ratios in the two spectra. Missing signals are due to severe overlap, and some residues (i.e. 164–173) broadened beyond detection below 290 K (supplemental Fig. 2). D, intensity ratio values representing the success of cross-saturation transfer via direct binding are mapped onto the three-dimensional structure of the chromodomain (Protein Data Bank code 1AP0 (6)). Residues without experimental values (proline or overlapped) are in gray.
In the hinge region, a bell-shaped intensity profile with the maximum around residue Asp-90 was observed (Fig. 2B). The intensity values were highly similar to those observed for the protein in its free state, indicating that the central part of the linker retained similar dynamics as in the absence of nucleosome but gradually lost flexibility at the joints with the structured CD and CSD. In addition, residues in the vicinity of Lys-99 experienced small but reproducible changes in NMR signal position (Fig. 2B, lower panel, and supplemental Fig. 2A). As this effect was absent in the hHP1β-H3KC9me3 peptide complex (supplemental Fig. 2, A and B), it points to a possible interaction between the hinge region and the core of the nucleosome particle. Instead, chemical shift changes in the unstructured N-terminal tail of hHP1β were observed both in the hHP1β-H3KC9me3 nucleosome and the hHP1β-H3KC9me3 peptide complex (Fig. 2B and supplemental Fig. 2B), suggesting that they are linked to the recognition of the H3K9me3-tail by the CD.
CD Is the Only Region of hHP1β That Stably Contacts the H3KC9me3 Nucleosome
To clearly define the contact interfaces of hHP1β in binding to the H3KC9me3 nucleosome, we performed NMR cross-saturation experiments (37). We selectively saturated the nucleosome aliphatic resonances and then allowed transfer of the magnetization to the amide protons of hHP1β. The signal attenuation profile of Fig. 2C, which represents the level of success of saturation transfer over the protein sequence, unambiguously demonstrates that the only region of hHP1β that stably contacts the nucleosome is the CD (Fig. 2, C and D). Consistent with the observed changes in chemical shifts (Fig. 2B), the N-terminal tail experienced a small but considerable saturation effect suggesting a weak interaction with the nucleosome. However, the N-terminal tail does not contribute to the structural changes observed for the CD upon histone-tail binding, as addition of H3K9me3 peptide induced highly similar chemical shift changes in the CD of wild-type (WT) and mutant hHP1β lacking the N- and C-terminal tails (hHP1βΔNΔC) (supplemental Fig. 2C). In addition, no changes were seen for the hinge region, the CSD, and the C-terminal tail at a wide range of temperatures (282–310 K) and different saturation times and saturation schemes (Fig. 2C and supplemental Fig. 2). The results imply that these regions do not bind directly or may contact the nucleosome only weakly, such as the hinge region (see below), with a kinetic modality that prevents detection by NMR cross-saturation.
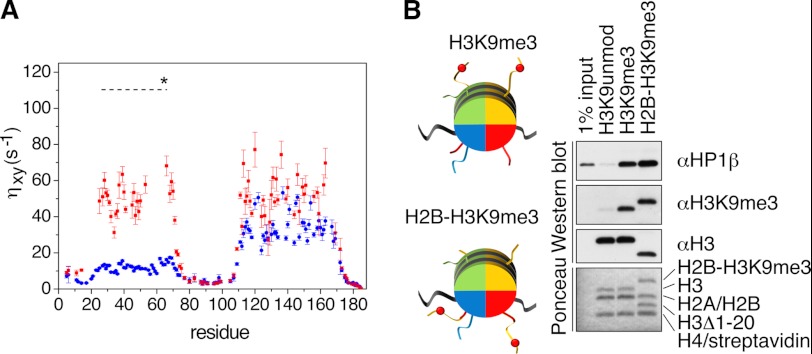
In contrast to the N-terminal tail and the hinge region, no NMR chemical shift changes were observed in the CSD (Fig. 2B). Moreover, when using the isolated CSD (residues 107–176), we did not detect any interaction with the H3K9me3 peptide or the H3KC9me3 nucleosome (Table 1). However, addition of H3KC9me3 nucleosomes induced a strong reduction in the NMR intensities of the CSD in full-length hHP1β (Fig. 2B). The reason for the observed signal broadening (in absence of chemical shift changes) most likely lies in hydrodynamic coupling (41, 42). Because of the spatial proximity of the CD nucleosome complex, the rotational diffusion of the CSD will be affected, even in the absence of a direct contact of the CSD with the nucleosome. Indeed, 15N spin-relaxation measurements (ηxy) (38) of hHP1β in the free state compared with those of hHP1β bound to the H3KC9me3 nucleosome showed enhanced relaxation in the context of the complex for both CD and CSD (Fig. 4A). As the ηxy rate does not contain contributions from chemical exchange, its elevated value following binding to nucleosome indicates a decreased rotational diffusion of both domains. Moreover, we observed a remarkable decrease in signal broadening of the CSD when we used a hHP1βI161A mutant, in which dimerization of the CSD is impaired but the overall fold is retained (supplemental Fig. 3), in line with a partial relief of the hydrodynamic coupling. Signal broadening on CSD peaks due to hydrodynamic coupling was further abolished when using the isolated CSD(107–176) (supplemental Fig. 3, A–C). Taken together, the results demonstrate that the CSD of hHP1β does not provide any additional contact interface to the nucleosome.
FIGURE 4.
Dynamics of the hHP1β-nucleosome complex. A, 15N cross-correlation rates (ηxy) measured for U-2H,15N- labeled hHP1β in the free state (blue) or with H3KC9me3 nucleosome at a molar ratio of 1:2 (red). The dashed line at 110 s−1 marks the expected ηxy value for the CD if it would tumble together with the core of the nucleosome particle as a single rigid body. In the calculation (see supplemental Methods), the population-weighted average of the rates of free and bound forms was considered. B, “tail-transplantation” pulldown experiments of nucleosomes uniformly containing unmodified H3, H3K9me3, or a truncated version of H3(Δ1–20) together with a H2B hybrid where the first 20 amino acids of H3 containing K9me3 were fused to the N terminus (see schematic representation on the left). Recovered material was analyzed by Western blotting using the indicated antibodies and by Ponceau staining.
Binding of the CD of hHP1β to the H3KC9me3 Nucleosome Is Structurally Equivalent to Interaction with a Free H3K9me3 Histone Tail Peptide
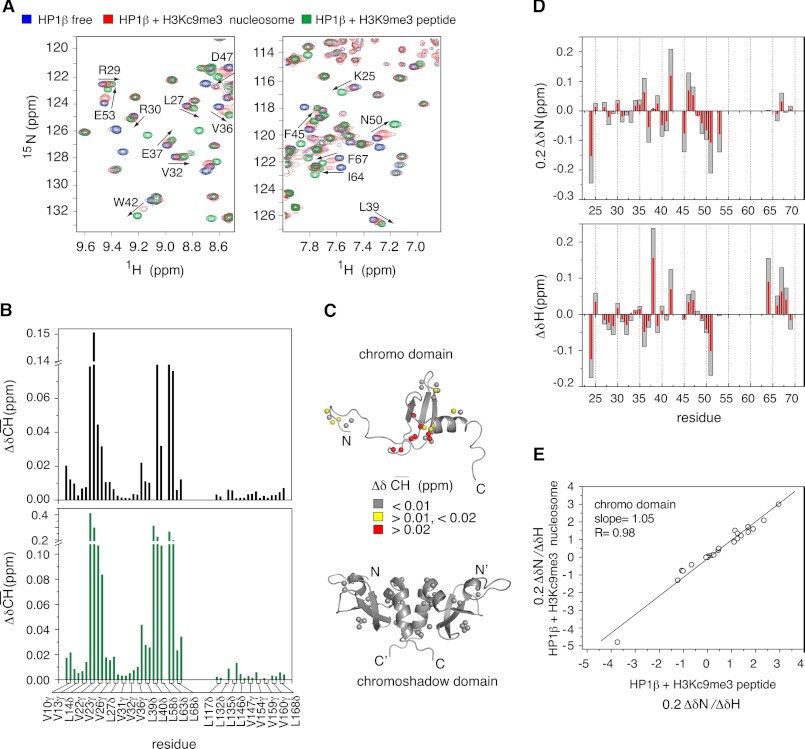
To reveal the molecular basis of the dominant role of the CD in H3KC9me3 nucleosome binding, we directly compared chemical shift changes induced in the CD upon binding the H3-tail within the H3KC9me3 nucleosome and within the free H3K9me3 peptide (Fig. 3). The NMR analysis clearly demonstrated that the same cluster of residues of the CD comprising the aromatic cage was perturbed (Figs. 2B and 3A and supplemental Fig. 2B). Moreover, changes in chemical shifts of the methyl groups of valine and leucine residues in the CD were highly similar in the two complex structures (Fig. 3B and supplemental Fig. 4). In contrast, no changes were observed in the CSD (Fig. 3, B and C). However, at identical molar ratios, the absolute values of chemical shift deviation were higher in the case of the peptide (Fig. 3A), consistent with the higher affinity of hHP1β for the H3K9me3 peptide compared with the H3KC9me3 nucleosome complex (Table 1).
FIGURE 3.
Effect of H3KC9me3 nucleosome and H3K9me3/H3KC9me3 peptide binding on the structure of hHP1β CD. A, selected regions from TROSY 1H,15N HSQC spectra of hHP1β alone (blue), with unlabeled H3KC9me3 nucleosome at a 1:2 molar ratio (red), or with H3K9me3 peptide at a 1:2 molar ratio (green). Black arrows highlight some of the CD peaks shifting in the same direction in the two binding events. B, chemical shift differences of methyl resonances (ΔδCH) in U-2H,12C, selective methyl Val and Leu 13CH3,12CD3-labeled hHP1β, upon binding to H3KC9me3 nucleosome (black; hHP1β/nucleosome molar ratio of 1:2) and to H3K9me3 peptide (green; hHP1β/peptide molar ratio of 1:2) relative to the free state. C, 1H,13C chemical shift changes of methyl groups of hHP1β upon binding to H3KC9me3 nucleosome are mapped onto the three-dimensional structures of CD (Protein Data Bank code 1AP0, top panel) and CSD (Protein Data Bank code 1DZ1 (13), bottom panel). Methyl groups are shown as spheres. D, comparison of 15N (upper panel) and 1H (lower panel) chemical shift changes in the CD upon binding of hHP1β to H3KC9me3 peptide (gray) and H3KC9me3 nucleosome (red). E, correlation plot of relative changes of 15N and 1H chemical shifts (i.e. 0.2ΔδN/ΔδH) between hHP1β bound to H3KC9me3 peptide (x axis) and H3KC9me3 nucleosome (y axis) with respect to the free form. Only CD residues with chemical exchange that is fast on the NMR time scale were considered.
For detailed comparison of the chemical shift changes of the CD in the two complexes, we used the H3KC9me3 peptide as reference. We observed strong correlation between the direction of 15N/1H chemical shift changes upon binding to H3KC9me3 nucleosome (red) and those upon binding to H3KC9me3 peptide (gray) (Fig. 3D). In addition, the affinity-independent values of Δδ15N/Δδ1H were almost identical (Fig. 3E). As chemical shifts are influenced by even minute alterations in the chemical environment, the high correspondence of the shift trajectories indicates that the structure of the CD in the peptide and nucleosome-bound state is the same. The chemical shifts of the hHP1β-H3KC9me3 peptide complex at saturation, which was already reached at a molar ratio of 1:4, served as good approximation for the chemical shifts of the hHP1β-bound state interacting with the nucleosome. Therefore, these allowed us to estimate the amount of the nucleosome-bound hHP1β under the exact conditions of temperature, concentration, and buffer of the NMR experiment. Based on the chemical shift changes of hHP1β residues that are in the fast-exchange binding regime, ∼54% of hHP1β is bound to the H3KC9me3 nucleosome at a hHP1β/H3KC9me3 nucleosome molar ratio of 1:2. The high correspondence of the chemical shift changes observed for hHP1β in complex with the H3K9me3 peptide, the H3KC9me3 peptide, and the H3KC9me3 nucleosome (Fig. 3 and supplemental Fig. 2D) strongly suggests that there are no additional interactions of the CD, such as CD-CD dimerization in the context of the nucleosome.
hHP1β-H3KC9me3 Nucleosome Complex Is Highly Flexible
The NMR analysis of the hHP1β-H3KC9me3 nucleosome interaction suggested that the H3-tails, from the viewpoint of binding to hHP1β, behave independently from the core of the nucleosome. This is further supported by a “tail-transplantation” experiment (Fig. 4B) where nucleosomes reconstituted with a chimeric H2B-H3K9me3 (see supplemental Methods for details) bound hHP1β as efficiently as the wild-type H3K9me3 material. The result further confirms interaction with the nucleosome that is solely directed via CD-H3K9me3 H3-tail binding of hHP1β independent of other contacts.
Beyond the dynamic nature of the H3-tail, 15N spin-relaxation rates showed that all hHP1β domains are very flexible within the complex with the H3KC9me3 nucleosome (Fig. 4A). Assuming that the CD tumbles together with the core of the nucleosome as a rigid unit (see supplemental Methods for details), we estimated a 15N cross-correlation rate of ∼110 s−1 on the basis that 54% of hHP1β is nucleosome-bound at the conditions of the NMR experiment (see above). Experimentally, however, we found a 15N cross-correlation rate of ∼50 s−1, demonstrating that the CD remains highly mobile when bound to the H3KC9me3 nucleosome. The NMR relaxation data also showed that the CSD moves independently from the core of the nucleosome, whereas the dynamic properties of the N and C terminus as well as the hinge region were highly similar to those observed in the free state. Taken together, the results reveal that HP1β-H3KC9me3 nucleosome complex is a highly flexible assembly.
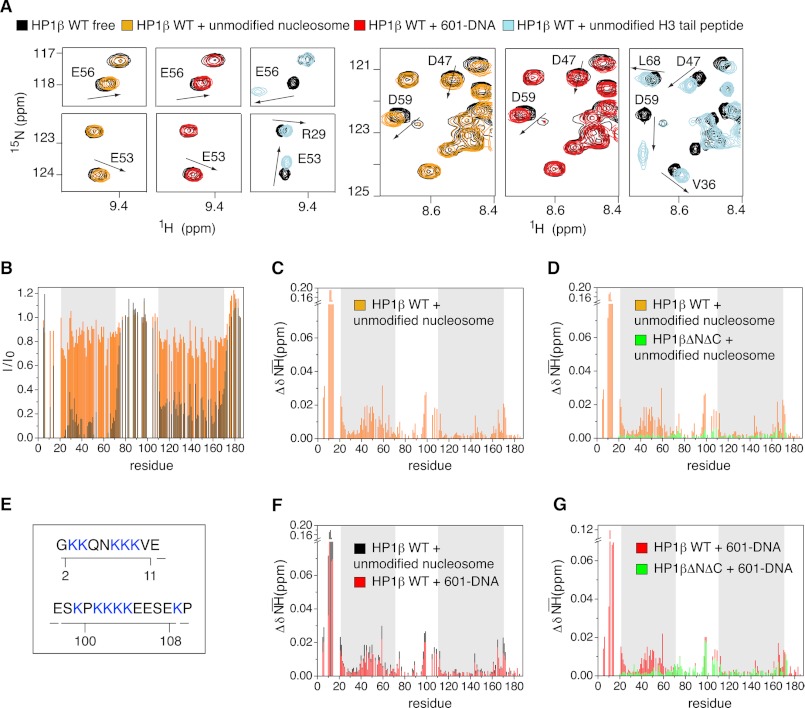
Binding Modes of hHP1β to Unmodified and H3KC9me3 Nucleosome Are Different
Analysis of hHP1β-nucleosome interaction using pulldown, ITC, and SPR experiments had shown clear differences in binding strength to unmodified and H3K9me3 nucleosomes. However, the nature of hHP1β binding to the unmodified template remained unclear. We therefore analyzed the interaction by NMR. Little signal broadening and small chemical shift changes point to a relatively weak interaction that is in fast exchange on the NMR time scale (Fig. 5, A and B). Although the CD was affected, the largest chemical shift changes were observed in the N-terminal tail of hHP1β (Fig. 5C). Importantly, chemical shift changes induced in the CD did not match those observed upon binding to the H3KC9me3 nucleosome (Fig. 2B) or free unmodified H3-tail peptide (Fig. 5A and supplemental Fig. 5). The observed perturbations not only involved different residues but also had different direction and magnitude (Fig. 5A). Indeed, no changes in the chemical shifts of methyl resonances of the CD were detected (supplemental Fig. 5), suggesting that the CD does not directly contact the unmodified nucleosome. Consistent with this hypothesis, titration of the tail-less mutant protein of hHP1β (hHP1βΔNΔC) with unmodified nucleosome did not affect the CD (Fig. 5D). We conclude that hHP1β interacts with the unmodified and H3KC9me3 nucleosome by distinct mechanisms.
FIGURE 5.
Interaction interfaces of hHP1β with unmodified and H3KC9me3 nucleosome are different. A, selected regions of the TROSY 1H,15N HSQC spectrum of free hHP1β (black) and of hHP1β in the presence of unmodified nucleosome (orange), free 601-DNA (red), or unmodified H3-tail peptide (light blue). In all three cases, the hHP1β/ligand molar ratio was 1:1. Arrows highlight the direction of peak shifts for some residues in the CD. B, hHP1β-1H,15N resonance intensity loss upon binding to unmodified nucleosome (orange) and H3KC9me3 nucleosome (black) as a function of residue number. The hHP1β/ligand molar ratio was 1:2 in both cases. C, chemical shift changes (ΔδNH) upon hHP1β binding to unmodified nucleosome. D, comparison of chemical shift changes (ΔδNH) in WT hHP1β (orange) and hHP1βΔNΔC (green) in the presence of unmodified nucleosome, both at 1:1 molar ratio. E, lysine patches in the hHP1β N-terminal and hinge region (see also supplemental Fig. 5F). F, comparison of chemical shift changes induced in WT hHP1β upon binding to free 601-DNA (red) and unmodified nucleosome (black), both at 1:1 molar ratio. G, 1H,15N chemical shift changes in WT hHP1β (red) and hHP1βΔNΔC (green) in the presence of 601-DNA at molar ratios of 1:1.
Which parts of the unmodified nucleosome are involved in the binding to hHP1β? Residues 97–109 in the hinge region of hHP1β showed highly similar chemical shift changes in the presence of H3KC9me3 nucleosome and unmodified nucleosome (Figs. 2B and 5C and supplemental Fig. 2A). In contrast, no changes were observed in the hinge region for hHP1β bound to the H3KC9me3 peptide (supplemental Fig. 2, A and B). Thus, an additional component in the nucleosome core that is not present in the peptide interacts with the hinge region. Based on the quantitative binding measurements and the inability to detect the contact in NMR cross-saturation experiments (Fig. 2C), this interaction must be weak.
Previous studies have shown that the positively charged hinge region of human or Xenopus HP1α can bind DNA (19, 21, 22) or RNA (23). The very N terminus of hHP1β also contains patches of charged residues (Fig. 5E and supplemental Fig. 5F). To further elucidate the nature of hHP1β binding to the unmodified nucleosome, we probed the interaction of free 601-DNA, which was used for nucleosome reconstitution, with WT as well as ΔNΔC hHP1β. Strikingly, the chemical shift changes induced by free 601-DNA in hHP1β WT were highly similar to those seen in the presence of unmodified nucleosome (Fig. 5F). In addition, for hHP1βΔNΔC changes were restricted to the hinge region (Fig. 5G). The high correspondence of NMR signal perturbations indicated that the very N terminus and the hinge region of hHP1β transiently associate with DNA of the unmodified nucleosome. The most affected part inside the hinge region has been identified within residues 97–109, which includes the lysine patch (residues 101–104) that previous mutagenesis studies have shown to be essential for the interaction between hHP1α and DNA (19). The results indicate that binding of hHP1β to unmodified and H3K9me3 nucleosome involves completely different contact interfaces.
hHP1β Can Bridge Adjacent H3KC9me3 Nucleosomes
In light of the high flexibility of hHP1β protein in its free form and in complex with nucleosome, where CD-H3K9me3 represents the only anchoring point of interaction, we tested the ability of the protein to engage immobilized H3K9me3 nucleosomes in bridging interactions. To this end, we designed a pulldown experiment with immobilized asymmetrically H3KC9me3-modified nucleosome, i.e. nucleosomes with only one of the two H3 tails carrying K9me3 and the other one being unmodified (see supplemental Fig. 1R for scheme of reconstitution). Binding analysis of hHP1β to this species showed only minor differences in interaction compared with the symmetrically modified templates (Fig. 1D). These results were further confirmed by SPR measurements, for which the protein concentration dependences of hHP1β binding to asymmetrical or symmetrically methylated nucleosomes (Fig. 1E) was comparable, a behavior that is not possible unless the unbound CD of hHP1β dimer finds its partner in a nearby nucleosome. Thus, the data indicate that simultaneous contacts of both CDs of an hHP1β dimer to two H3K9me3 marks of a symmetrically modified nucleosome are not a prerequisite for binding. More importantly, bridging interactions of hHP1β to nucleosomes deposited on a chromatin mimicking surface occur, facilitated by the high flexibility of the system.
DISCUSSION
Although HP1 proteins are implicated in establishing and maintaining heterochromatin in many experimental systems, mammalian HP1β is the only HP1 protein essential for viability (5). Previous work on component systems using isolated histone peptides and/or individual protein domains, as well as analysis of HP1 of different species and isoforms, has determined parameters of HP1-chromatin binding implying various interaction interfaces (5, 11, 43). On the basis of the many different findings, it is not possible to deduce a coherent picture of general HP1-chromatin interaction and function.
Our study using a fully defined system of uniformly modified nucleosomes unambiguously defines the binding mode of hHP1β to unmodified and H3K9me3 nucleosomes. We provide quantitative and single-residue resolution details of alternative binding interfaces in dependence of H3K9 methylation. The hHP1β-H3K9me3 nucleosome complex is a highly dynamic assembly (Figs. 2 and 4). Methylation at lysine 9 in histone 3 is the major determinant of the interaction. Serving as an on/off switch it generates a binding site for the CD. There are no other constraints, and even the CD bound to the methyl-mark, which represents the only stable anchoring point of the complex, remains highly mobile (Fig. 4).
Previous studies reported that the CSD of hHPIα and hHPIγ bind a portion of the globular domain of H3, named the “Shadock” region (16). However, because those results came from biochemical assays with isolated histones, it is possible that the interaction does not occur in the integral nucleosome assembly, where the “Shadock” region is partially buried (44). Indeed, our NMR (Figs. 2 and 3), SPR, and ITC data (Table 1) indicate that the CSD is not involved in direct binding to the nucleosome. Further studies using different assembly states of the binding partners are required to resolve whether CSD-Shadock or other interactions with histone globular cores (12) occur when chromatin gets unfolded and exposes epitopes otherwise occluded such as proposed in the S-phase (17).
In the absence of H3K9 methylation, the CD is not involved in binding to the nucleosome, and different interfaces as compared with the CD-H3K9me3 interaction are used. The N-terminal tail and the hinge region of hHP1β contact nucleosomal DNA (Fig. 5). This is in clear contrast to the binding properties to the isolated H3-tail peptide that binds to the CD even in the absence of the methyl-mark (Fig. 5A and supplemental Fig. 5). In peptide studies, the methyl-mark primarily regulates the affinity to hHP1β but not the binding interface.
What is the role of the weak DNA binding in the context of chromatin? We hypothesize that in the absence of H3K9me3, the interaction of hHP1β with DNA may result in the diffusion of hHP1β along the surface of chromatin. Such explorative binding has been postulated for transcription factors that search for their high affinity target sites while having unspecific binding to general DNA sequences (45, 46). At regions of low-to-medium local concentration of H3K9me3, this diffusion will be slowed down, as the hHP1β CD will engage in interaction directed by the H3 tail modification. We envision that only at areas of the genome that contain high density of H3K9me3, both CDs of the hHP1β dimer can simultaneously bind their cognate target leading to stable, i.e. prolonged, binding to chromatin.
The only other study of an HP1 protein, Swi6 of S. pombe, in the context of fully defined chromatin systems recently provided a model for heterochromatin spreading that involves CD-mediated oligomerization (26). Our findings on hHP1β using the same (SPR, analytical ultracentrifugation, and size exclusion chromatography-multiangle laser light scattering (SEC-MALLS) as well as other (ITC, pulldown, and NMR) analysis tools indicate remarkable differences in binding of the two HP1 factors to chromatin templates. Swi6 showed, besides CSD dimerization, homo-interaction via the CD that was suggested to increase the specificity for H3KC9me3-chromatin in a cooperative manner. We and others (6, 13, 47) did not find a similar multimerization behavior of hHP1β (supplemental Fig. 3, D and E). Indeed, NMR diffusion measurements indicated a hydrodynamic radius of the isolated CD of 1.55 ± 0.05 nm for concentrations up to 1 mm in full agreement with monomeric behavior. Moreover, the stoichiometry of the hHP1β-H3K9me3 nucleosome interaction is low, which is indicative of binding of individual hHP1β dimers (Fig. 1). Detailed Hill analysis of the quantitative binding data revealed no cooperativity (supplemental Fig. 1), a finding that is in agreement with the very high flexibility of the complex. Indeed, in comparing the NMR chemical shifts within the hHP1β-H3K9me3 peptide, hHP1β-H3KC9me3 peptide, and hHP1β-H3KC9me3 nucleosome complexes, we found no evidence for CD-CD interaction induced by immobilization on the nucleosome (Fig. 3 and supplemental Fig. 2). In addition, binding strength of hHP1β to H3K9Cme3 nucleosomes was at least 1 order of magnitude lower as that reported for Swi6. Furthermore, discrimination of binding to unmodified and H3K9me3 nucleosomes is high for hHP1β and similar to what is seen on isolated peptides (Table 1), whereas substantial interaction with unmodified nucleosomes had been reported for Swi6, both in gel shift and SPR experiments (26).
Our studies point to putatively distinct biochemistry and divergent chromatin binding modes of HP1 proteins. Interestingly, earlier studies have found that the CD domain of hHP1β but not hHP1α is essential for heterochromatin targeting (48, 49). Human and Xenopus HP1α, but not Xenopus Hp1γ, show strong interaction with DNA (22) and nucleosomes or nucleosomal arrays independent of H3K9me3 (19, 21). Also, Swi6 binding strength to free DNA and H3K9me3 chromatin templates is comparable (26). In this context, CD-mediated multimerization of some HP1 proteins but not others might be required to regulate DNA-mediated unspecific versus H3K9me3-specific binding to nucleosomes. Discrete impact of additional parameters on HP1 biochemistry also needs to be considered as follows. (i) binding of distinct HP1 proteins to other factors, e.g. via the CSD and/or hinge region, might modulate interaction differentially (5, 43). (ii) Post-translational modification of the different domains of HP1 proteins could specifically tune chromatin-binding events (43, 50). (iii) The status of the chromatin template itself might be an important regulator, e.g. linker histones of the H1 type could preclude or enhance HP1 binding either directly (12, 19, 20) or indirectly by modulating chromatin structure (51). (iv) Timing of interaction within the cell cycle could allow or prevent certain types of binding (12, 17).
We suggest that the highly flexible nature of hHP1β in its free and bound forms plays an important role in its fundamental biological activity to connect different nuclear processes, as it enables the second hHP1β binding platform, i.e. the chromoshadow domain, to be adapted within the dense chromatin architecture to recruit additional proteins. Moreover, it allows sampling of a relatively large structural space for binding of chromodomain to H3K9me3 tails of nearby nucleosomes. The ability of the protein to engage immobilized nucleosomes in bridging interactions, confirmed by our SPR and pulldown experiments (Fig. 1, D and E), provides an effective structural linker of distinct chromatin layers (18, 25).
Overall, we provide detailed structural insights into a dynamic protein-nucleosome complex directed by histone modification and offer a conceptual framework to understand the molecular basis of similar dynamic and flexible interactions regulated by histone post-translational modification in the context of chromatin and epigenetics.
Supplementary Material
Acknowledgments
We thank the mass spectrometry facility of Henning Urlaub for performing MS analysis and Winfried Lendeckel for excellent technical assistance.
This work was supported in part by the Max Planck Society (to P. J. W., W. F., and M. Z.), the European Union Marie Curie reintegration fellowship and NET fellowship of the NOE Epigenome (to W. F.), the Fonds der Chemischen Industrie (to P. J. W.), the Deutsche Forschungsgemeinschaft (to P. J. W.), Heisenberg scholarship (to M. Z.), and Deutsche Forschungsgemeinschaft Grants ZW 71/2-2 and 3-2, SFB 860 (to M. Z.).

This article contains supplemental Methods, Figs. 1–5, and additional references.
- CD
- chromodomain
- CSD
- chromoshadow domain
- ITC
- isothermal titration calorimetry
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Ebert A., Lein S., Schotta G., Reuter G. (2006) Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14, 377–392 [DOI] [PubMed] [Google Scholar]
- 2. Grewal S. I., Elgin S. C. (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447, 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K., Rauscher F. J., 3rd (2003) Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing. A mammalian cell culture model of gene variegation. Genes Dev. 17, 1855–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verschure P. J., van der Kraan I., de Leeuw W., van der Vlag J., Carpenter A. E., Belmont A. S., van Driel R. (2005) In vivo HP1 targeting causes large scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell. Biol. 25, 4552–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh P. B. (2010) HP1 proteins. What is the essential interaction? Genetika 46, 1424–1429 [PubMed] [Google Scholar]
- 6. Ball L. J., Murzina N. V., Broadhurst R. W., Raine A. R., Archer S. J., Stott F. J., Murzin A. G., Singh P. B., Domaille P. J., Laue E. D. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 16, 2473–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacobs S. A., Khorasanizadeh S. (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 8. Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., Duan S., Wasney G. A., Li Z., Vedadi M., Schapira M., Min J., Arrowsmith C. H. (2011) Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., Kouzarides T., Murzin A. G., Murzina N. V., Laue E. D. (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 [DOI] [PubMed] [Google Scholar]
- 10. Dawson M. A., Bannister A. J., Göttgens B., Foster S. D., Bartke T., Green A. R., Kouzarides T. (2009) JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hediger F., Gasser S. M. (2006) Heterochromatin protein 1. Don't judge the book by its cover! Curr. Opin. Genet. Dev. 16, 143–150 [DOI] [PubMed] [Google Scholar]
- 12. Nielsen A. L., Oulad-Abdelghani M., Ortiz J. A., Remboutsika E., Chambon P., Losson R. (2001) Heterochromatin formation in mammalian cells. Interaction between histones and HP1 proteins. Mol. Cell 7, 729–739 [DOI] [PubMed] [Google Scholar]
- 13. Brasher S. V., Smith B. O., Fogh R. H., Nietlispach D., Thiru A., Nielsen P. R., Broadhurst R. W., Ball L. J., Murzina N. V., Laue E. D. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromodomain dimer. EMBO J. 19, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwon S. H., Workman J. L. (2008) The heterochromatin protein 1 (HP1) family. Put away a bias toward HP1. Mol. Cells 26, 217–227 [PubMed] [Google Scholar]
- 15. Smothers J. F., Henikoff S. (2000) The HP1 chromoshadow domain binds a consensus peptide pentamer. Curr. Biol. 10, 27–30 [DOI] [PubMed] [Google Scholar]
- 16. Lavigne M., Eskeland R., Azebi S., Saint-André V., Jang S. M., Batsché E., Fan H. Y., Kingston R. E., Imhof A., Muchardt C. (2009) Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 5, e1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dialynas G. K., Makatsori D., Kourmouli N., Theodoropoulos P. A., McLean K., Terjung S., Singh P. B., Georgatos S. D. (2006) Methylation-independent binding to histone H3 and cell cycle-dependent incorporation of HP1β into heterochromatin. J. Biol. Chem. 281, 14350–14360 [DOI] [PubMed] [Google Scholar]
- 18. Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 19. Meehan R. R., Kao C. F., Pennings S. (2003) HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22, 3164–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R. (2005) HP1 binds specifically to Lys-26-methylated histone H1.4, whereas simultaneous Ser-27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280, 38090–38095 [DOI] [PubMed] [Google Scholar]
- 21. Zhao T., Heyduk T., Allis C. D., Eissenberg J. C. (2000) Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 275, 28332–28338 [DOI] [PubMed] [Google Scholar]
- 22. Sugimoto K., Yamada T., Muro Y., Himeno M. (1996) Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein that possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C). J. Biochem. 120, 153–159 [DOI] [PubMed] [Google Scholar]
- 23. Muchardt C., Guilleme M., Seeler J. S., Trouche D., Dejean A., Yaniv M. (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3, 975–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eskeland R., Eberharter A., Imhof A. (2007) HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 27, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henikoff S., Shilatifard A. (2011) Histone modification. Cause or cog? Trends Genet. 27, 389–396 [DOI] [PubMed] [Google Scholar]
- 26. Canzio D., Chang E. Y., Shankar S., Kuchenbecker K. M., Simon M. D., Madhani H. D., Narlikar G. J., Al-Sady B. (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato H., van Ingen H., Zhou B. R., Feng H., Bustin M., Kay L. E., Bai Y. (2011) Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 108, 12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barbera A. J., Chodaparambil J. V., Kelley-Clarke B., Joukov V., Walter J. C., Luger K., Kaye K. M. (2006) The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 [DOI] [PubMed] [Google Scholar]
- 29. Makde R. D., England J. R., Yennawar H. P., Tan S. (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E. (2011) Structural basis of silencing. Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334, 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacobs S. A., Fischle W., Khorasanizadeh S. (2004) Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 376, 131–148 [DOI] [PubMed] [Google Scholar]
- 32. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 33. Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narayanan R. L., Durr U. H., Bibow S., Biernat J., Mandelkow E., Zweckstetter M. (2010) Automatic assignment of the intrinsically disordered protein Tau with 441 residues. J. Am. Chem. Soc. 132, 11906–11907 [DOI] [PubMed] [Google Scholar]
- 35. Hiller S., Fiorito F., Wüthrich K., Wider G. (2005) Automated projection spectroscopy (APSY). Proc. Natl. Acad. Sci. U.S.A. 102, 10876–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruschak A. M., Kay L. E. (2010) Methyl groups as probes of supra-molecular structure, dynamics, and function. J. Biomol. NMR 46, 75–87 [DOI] [PubMed] [Google Scholar]
- 37. Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I. (2000) A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 7, 220–223 [DOI] [PubMed] [Google Scholar]
- 38. Wang C., Rance M., Palmer A. G., 3rd (2003) Mapping chemical exchange in proteins with MW >50 kDa. J. Am. Chem. Soc. 125, 8968–8969 [DOI] [PubMed] [Google Scholar]
- 39. Seeliger D., Soeroes S., Klingberg R., Schwarzer D., Grubmuller H., Fischle W. (2012) Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem. Biol. 7, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simon M. D., Chu F., Racki L. R., de la Cruz C. C., Burlingame A. L., Panning B., Narlikar G. J., Shokat K. M. (2007) The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walsh J. D., Meier K., Ishima R., Gronenborn A. M. (2010) NMR studies on domain diffusion and alignment in modular GB1 repeats. Biophys. J. 99, 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halle B. (2009) The physical basis of model-free analysis of NMR relaxation data from proteins and complex fluids. J. Chem. Phys. 131, 224507. [DOI] [PubMed] [Google Scholar]
- 43. Lomberk G., Wallrath L., Urrutia R. (2006) The heterochromatin protein 1 family. Genome Biol. 7, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richart A. N., Brunner C. I., Stott K., Murzina N. V., Thomas J. O. (2012) Characterization of chromoshadow domain-mediated binding of heterochromatin protein 1α (HP1α) to histone H3. J. Biol. Chem. 287, 18730–18737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Hippel P. H., Berg O. G. (1989) Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 [PubMed] [Google Scholar]
- 46. Slutsky M., Mirny L. A. (2004) Kinetics of protein-DNA interaction. Facilitated target location in sequence-dependent potential. Biophys. J. 87, 4021–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ye Q., Callebaut I., Pezhman A., Courvalin J. C., Worman H. J. (1997) Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272, 14983–14989 [DOI] [PubMed] [Google Scholar]
- 48. Yamada T., Fukuda R., Himeno M., Sugimoto K. (1999) Functional domain structure of human heterochromatin protein HP1(Hsα). Involvement of internal DNA-binding and C-terminal self-association domains in the formation of discrete dots in interphase nuclei. J. Biochem. 125, 832–837 [DOI] [PubMed] [Google Scholar]
- 49. Thiru A., Nietlispach D., Mott H. R., Okuwaki M., Lyon D., Nielsen P. R., Hirshberg M., Verreault A., Murzina N. V., Laue E. D. (2004) Structural basis of HP1/PXVXL motif peptide interactions and HP1 localization to heterochromatin. EMBO J. 23, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. LeRoy G., Weston J. T., Zee B. M., Young N. L., Plazas-Mayorca M. D., Garcia B. A. (2009) Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell. Proteomics 8, 2432–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McBryant S. J., Adams V. H., Hansen J. C. (2006) Chromatin architectural proteins. Chromosome Res. 14, 39–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.