Background: The cannabinoid receptor CB1 has been refractory to purification and structural analysis, thus limiting mechanistic information about its activation and attenuation.
Results: Fluorescence probes on purified, functional CB1 detect a key agonist-induced structural change that is blocked by a novel allosteric ligand.
Conclusion: Some allosteric GPCR ligands may capture structural intermediates.
Significance: GPCR intermediate structures may be optimal templates for allosteric drug design.
Keywords: 7-Helix Receptor, Allosteric Regulation, Fluorescence, G protein-coupled Receptors (GPCR), Pharmacology, Protein Dynamics, Site-directed Fluorescence Labeling
Abstract
Allosteric ligands that modulate how G protein-coupled receptors respond to traditional orthosteric drugs are an exciting and rapidly expanding field of pharmacology. An allosteric ligand for the cannabinoid receptor CB1, Org 27569, exhibits an intriguing effect; it increases agonist binding, yet blocks agonist-induced CB1 signaling. Here we explored the mechanism behind this behavior, using a site-directed fluorescence labeling approach. Our results show that Org 27569 blocks conformational changes in CB1 that accompany G protein binding and/or activation, and thus inhibit formation of a fully active CB1 structure. The underlying mechanism behind this behavior is that simultaneous binding of Org 27569 produces a unique agonist-bound conformation, one that may resemble an intermediate structure formed on the pathway to full receptor activation.
Introduction
G protein-coupled receptors (GPCRs)2 comprise ∼3% of the protein-coding human genome (1). Due to their involvement in a vast number of signaling systems, these membrane receptors are targeted by numerous therapeutic agents. An exciting field of GPCR research has emerged with the discovery that allosteric ligands can bind to some GPCRs and modulate their activity (2). Allosteric ligands bind to a different site than traditional competitive agonists and antagonists, and thus, they may affect receptor signaling (efficacy) through new mechanisms. Knowing how allosteric GPCR ligands induce their effect is of great therapeutic interest as they can complement endogenous ligands, have less potential for overdose, and specifically target receptor subtypes due to greater evolutionary divergence for allosteric binding sites (3). Clearly, these novel ligands enrich the pharmacological dimensions of GPCR signaling and provide additional ways to further “dial in” GPCR responses.
One of the highest expressed GPCRs in the central nervous system (CNS) is the human neuronal cannabinoid receptor, CB1 (4). Although initial interest in CB1 was linked to its role as the target for psychotropic agents in marijuana (5), CB1 has subsequently been implicated in a wide array of clinically relevant conditions, including Parkinson disease, Alzheimer disease, depression, inflammation, neuropathic pain, and obesity. However, despite its ubiquitous presence in the CNS and its therapeutically exploitable nature, structural and biophysical information about CB1 is limited. The lipophilic nature of cannabinoid ligands has made ligand binding assays technically challenging. Moreover, the CB1 receptor has proven refractory to purification of significant quantities in a functional form (6–11).
In this study, we show that it is possible to purify significant amounts of CB1 in a functional form and investigate how an allosteric ligand interacts with the purified CB1. This ligand, Org 27569, exhibits an interesting behavior; it increases agonist binding to CB1, yet in contrast, inhibits CB1 signaling (i.e. it is a positive allosteric modulator of agonist affinity yet a negative allosteric modulator of agonist signaling efficacy) (12). One possibility is that Org 27569 places the receptor in a distinct, agonist-bound, nonsignaling conformational state or (because the previous studies of Org 27569 were all carried out using unpurified cell membranes) acts indirectly through unidentified component(s) of the CB1 signaling pathway.
We set out to experimentally test both possibilities by determining whether Org 27569 acts directly on CB1 and testing whether it evokes these opposing effects by inducing a distinct structural state in the CB1 receptor. To do this, we first established conditions under which we could obtain a functional, purified CB1 receptor. We then studied this purified CB1 using a site-directed fluorescent labeling (SDFL) approach, in which we placed a fluorescent label on the cytoplasmic end of transmembrane helix six (TM6), a helix shown to move during activation in other GPCRs by SDFL (13–18). We then monitored this probe to determine whether Org 27569 altered conformational changes in or around TM6 when agonists bound to the receptor.
Our results clearly show that agonist binding induces some kind of movement in the cytoplasmic end of TM6 of CB1, whereas antagonist binding does not. We also confirm that Org 27569 stimulates agonist binding, both in membranes and for purified CB1 in detergent. Our SDFL studies of agonist-bound CB1 show that Org 27569 blocks the agonist-induced conformational change at TM6 described above. Together, these results explain how Org 27569 can elicit differential effects on CB1 agonist affinity and efficacy; Org 27569 traps the receptor in a distinct agonist-bound, but nonsignaling conformational state.
EXPERIMENTAL PROCEDURES
Buffers
The buffers used are defined as: PBSSC (137 mm NaCl, 2.7 mm KCL, 1.5 mm KH2PO4, 8 mm Na2HPO4 (pH 7.2)); Hypotonic Buffer (5 mm Tris and 2 mm EDTA (pH 7.5)); TME (20 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 1 mm EDTA); Binding Buffer (TME with 5 mg/ml BSA); Wash Buffer (TME with 1 mg/ml BSA); and Purification Buffer (50 mm Tris (pH 7.5), 200 mm NaCl, 5 mm MgCl2, 20% glycerol, 0.12% CHAPS, 0.02% n-Dodecyl-β-d-Maltopyranoside (DM), and 0.02% Cholesteryl Hemisuccinate (CHS).
Construction of shCB1 Mutants
The site-directed mutants and truncation constructs were made using overlap extension PCR to generate the mutants in the shCB1 (synthetic human CB1) gene (19). The nonreactive mutant, θ, contains only two of the original 13 cysteines (Cys-257 and Cys-264), which appear to be required for a functional receptor (20). We previously established that θ is insensitive to sulfhydryl-modifying reagents when assessed by ligand binding (20). To facilitate purification, we further modified θ by deleting the N and C termini and then introducing the last 9 amino acids of rhodopsin (1D4 epitope: TETSQVAPA) to the C terminus to enable immunoaffinity purification.
For the site-specific fluorescence labeling studies, we then introduced a unique reactive Cys on TM6 at residue A342C (6.34) into the θ background using a two-step PCR procedure. All mutations were verified using restriction enzyme analysis and the dideoxynucleotide sequencing method.
Transfection
The mutant shCB1 genes were expressed in transiently transfected monkey kidney cells (COS1) in 15-cm plates. Samples were incubated for ∼65 h at 5% CO2, 75% relative humidity, and 37 °C. The cells were then harvested in PBSSC, and the pellets were snap-frozen in liquid nitrogen and stored at −80 °C.
SDS-PAGE and Immunoblot Analysis of Cannabinoid Receptor Mutants
SDS-PAGE and immunoblot analysis were performed according to previously published procedures (20). PDT-bimane labeling of the samples was visualized by measuring the in-gel fluorescence using an Alpha Innotech gel documentation system. Subsequently, Coomassie Brilliant Blue R-250 protein staining was carried out using the Imperial protein stain (Thermo Scientific) as described in the manufacturer's protocol.
Purification of Cannabinoid Receptor Mutants
COS1 cell membranes containing mutant CB1 receptor protein were suspended in detergent buffer supplemented with protease inhibitor tablet (Roche Applied Science), as well as 5 μg/ml leupeptin, 10 mm benzamidine, 0.5 mm PMSF, and 1 μm SR141716A and gently nutated for 2–3 h at 4 °C. Samples were then centrifuged for 1 h at 100,000 × g in a Beckman Optima LE-80K ultracentrifuge with a TI60 rotor. The supernatant was removed and then added to an appropriate volume of 1D4 antibody-Sepharose beads (binding capacity ∼1 μg of rhodopsin/μg of resin) and allowed to bind via gentle agitation at 4 °C for 4–5 h. Next, the receptor-bound beads were washed, first with ∼5 ml of buffer containing protease inhibitor and antagonist SR141716A and then two times with 1-ml washes of buffer. Alternatively, for fluorescence labeling of mutant of CB1 receptors, the CB1 bound to 1D4 beads was incubated with 50 μm PDT-bimane overnight followed by extensive washes to remove nonreactive free bimane label. The samples were then eluted from the 1D4 antibody-Sepharose beads with purification buffer containing 200 μm nonapeptide.
Solution Radioligand Binding Measurements
The ability of the detergent-solubilized receptors to bind [3H]CP55940 or [3H]SR141716A was measured using mini size-exclusion chromatography columns, as follows; 50–150 nm of soluble receptors were incubated with ∼25–75 nm 3H-ligand in the presence of increasing amounts of agonist or antagonist for 1 h at 30 °C in a total volume of 100 μl of buffer. Separation of bound from free ligand was achieved by gel filtration and then analyzed by liquid scintillation counting to determine the amount of bound ligand. The one-site competition binding model in SigmaPlot was fit to our data. The Kd and Bmax values were estimated using previously described methods (21). Data were globally fit, and error estimates for the parameters were derived from least square fits.
Additionally, an allosteric ternary complex model, described previously (22), was used to fit our data
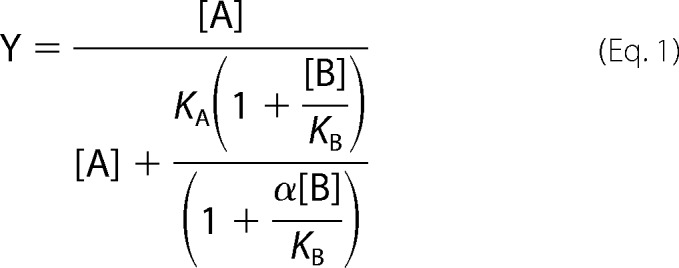 |
where Y denotes the specific bound orthosteric ligand divided by the total concentration of orthosteric ligand [A]. [B] denotes the total concentration of allosteric ligand. KA and KB are the dissociation constants for the orthosteric and allosteric ligand, respectively, and α is the binding cooperativity factor between the orthosteric and allosteric ligands. The [A] was the average radioactive orthosteric ligand concentration employed in the binding assays, and KA was estimated from the fraction bound and [A]. Values of α and KB were determined from least squares fitting of Equation 1.
Binding Measurements in COS1 Membranes
The ligand binding properties of the unpurified CB1 receptor mutants in cell membranes were measured using a previously described competitive inhibition binding assay (20). Briefly, this involved incubating 50 μg of membranes (total membrane protein) at 30 °C for an hour in 500 μl of binding buffer with ∼1 nm tritiated ligands and increasing amounts of agonist or antagonist. The binding reactions were then filtered over 0.2% (w/v) polyethyleneimine-treated Whatman GF/B filters using a Brandel 24- or 48-well filtration apparatus and then washed three times with 5-ml washes of wash buffer. Radioactivity was detected and quantified by liquid scintillation. Data were fit as described above.
Preparation of the Gαiβγ Heterotrimer
Purification of rat Gαi was performed essentially as described previously (23). The transducin βγ subunit was purified from rod outer segments essentially as described (24). In brief, after transducin extraction, subunits were collected contemporaneously on a HiTrap Blue (for the Gα) and a HiTrap Q (for the βγ) columns. The βγ subunits collected on the HiTrap Q were eluted using NaCl gradient. The elution was then subjected to dialysis and further concentrated. The Gαiβγ heterotrimer was generated by overnight incubation at 4 °C on ice; Gαi and βγ were combined at a 1:1 molar ratio (∼2 μm of each) with 1.5 mm dithiothreitol and 75 μm GDP. Gαiβγ heterotrimers were then aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C.
Cannabinoid Functional Efficacy Assessed by Reconstitution with Gαiβγ Heterotrimer
Gαi assays were done in a similar manner to rhodopsin transducin assays (25). The final reaction mixture contained 200–300 nm purified, labeled CB1 in detergent and appropriate buffer, 1 μm G protein heterotrimer, and 2 μm GTPγS. The samples were assayed using [35S]GTPγS that was added to the receptor:G protein mixture and immediately transferred into tubes containing various ligands to be tested. 10-μl aliquots were removed after 30 min and spotted onto prewetted Millipore MF 0.45-μm HA membrane filters using a modified Brandel M-24 cell harvester. Spotted filters were washed three times with 4 ml of wash buffer (10 mm Tris, 100 mm NaCl, 5 mm MgCl2 and 0.1 mm EDTA, pH 7.5), filters were removed, and radioactivity on each filter was measured by liquid scintillation spectroscopy.
Fluorescence Assays
Steady-state fluorescence measurements were performed using a Photon Technology International fluorescence spectrometer at room temperature. The excitation wavelength was 380 nm (2-nm slit settings), and the emission was collected from 400–650 nm (with 12-nm slit settings). All measurements were carried out with the CB1 receptor at a final concentration of 200 nm in Purification Buffer. The CB1 receptor concentrations were estimated from absorbance value at 280 nm (corrected for the contribution of bimane at this wavelength), using an extinction coefficient of 42,525 liters mol−1 cm−1 estimated from the protein sequence (ExPASy ProtParam tool). All ligands were diluted, such that the final solvent concentration was less than 1%. The fluorescence spectra were buffer subtracted and corrected for dilution.
The variable slope sigmoidal dose-response function was fit globally to our bimane response (change in bimane fluorescence) with respect to Org 27569 concentration. The error estimates for the parameters were derived from least square fits.
An operational model of allosterism, as described previously by Price et al. (12), that assumes the allosteric modulator does not process any intrinsic efficacy was also fit to our data (Equation 2).
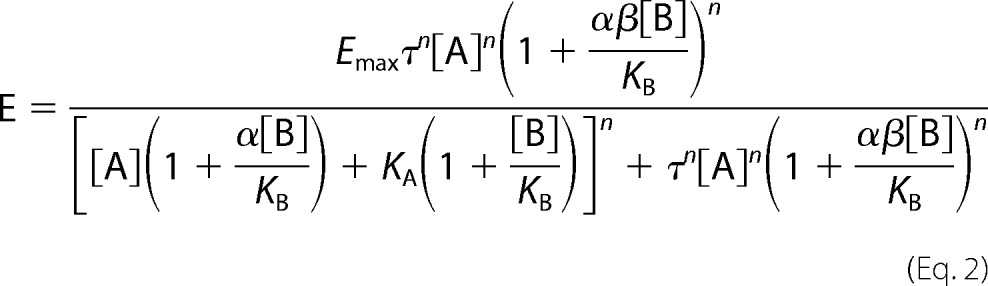 |
[A], KA, KB, and α are as defined above in Equation 1. E represents the bimane effect, n is a logistic slope factor, τ is a measure of orthosteric ligand efficacy, and β is the empirical proportionality constant describing the modulation of an allosteric ligand on agonist-mediated efficacy. When β is less than 1, there is an inhibition of signaling efficacy imparted on the receptor by the allosteric modulator. The fitting used 10 μm for [A] and used values obtained from Equation 1 from our solution binding assay for KA, KB, and α; set Emax to the mean of our empirically derived value from our data sets; and restricted β to be greater than 0.
TCA Precipitation Method to Determine the Extent of Free Label Contamination
To assess whether free (unattached, nonreacted) bimane label was present in the samples, we used a slightly modified version of our previous procedure (15, 26). Briefly, this involves determining whether any bimane fluorescence is present in a sample after TCA precipitation of the protein. To do this, the total bimane fluorescence of a sample containing PDT-bimane-labeled CB1 was measured immediately after adding 10% TCA. The protein was then precipitated by placing the sample on ice for 20 min and then subjected to centrifugation at 14,000 rpm at 4 °C for 20 min using a bench-top Eppendorf centrifuge. The supernatant was then collected, and fluorescence emission was measured. This approach exploits the fact that the 10% TCA precipitates essentially all protein, whereas free bimane is not precipitated. Thus, any fluorescence remaining in the supernatant must be due to (free) bimane that is not attached to the protein. Comparison of these two emission maximum values gave a relative amount of free label in the sample. In all cases, the measured free label concentration was essentially negligible (<1%).
Fluorescence Quenching Experiments
Measurements of the accessibility of the bimane probe were carried out by Stern-Volmer quenching studies to determine the bimolecular quenching coefficient (see Fig. 2). Briefly, the bimane-labeled CB1 samples were incubated in 20 μm CP55940 or SR141716A for 30 min prior to measurements. For the KI quenching assays, the added total salt concentration was kept at ∼40 mm by the addition of a corresponding amount of KCl, and 0.1 nm Na2SO4 was present to inhibit formation of I3 (15, 17). Fluorescence lifetime measurements were carried out using a PicoQuant Fluo Time 200 equipped with a Hamamatsu R3809U-5X series microchannel plate photomultiplier. The excitation was from a 405 nm diode laser, and emission was monitored at 490 nm with 2-nm slits. The average fluorescent lifetime 〈τ〉 and slopes from the KI quenching assay (Ksv) were used to calculate kq (kq = Ksv/〈τ〉) (27). The bimolecular quenching coefficient (kq) is a direct measure of the efficiency of quenching (m−1s−1).
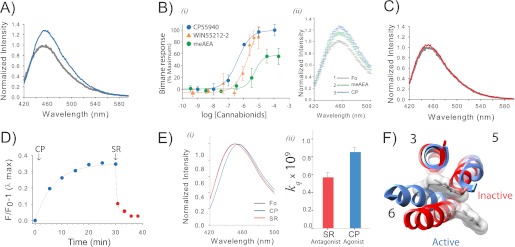
FIGURE 2.
Agonist binding to CB1 induces a conformational change that is detected by a probe at site 6.34 (or 342) on TM6. A, the addition of agonist CP55940 (CP) causes an ∼35% increase in fluorescence intensity for PDT-bimane-labeled mutant sample A342C/θ. The spectra, normalized to the apo state (gray) in Purification Buffer, show before and after a 30-min incubation with 10 μm CP55940 (blue). B, i, the dose-response plot of the agonists (CP55940, WIN55212-2, and meAEA) report the stimulated increase in fluorescence (data normalized to the maximum increase in fluorescence for CP55940). The apparent EC50 are 430 ± 86 nm for CP55940, 3 ± 0.4 μm for WIN, and 6.6 ± 4.0 μm for meAEA. The bimane dose-response plots are the means of at least three independent experiments fit with a sigmoidal dose response function. ii, the partial increase in fluorescence induced by meAEA addition (50 μm, green) is further increased by subsequent addition of CP55940 (CP, 35 μm, blue). Each data point in the spectra shows the range of the S.E. from three independent experiments. Fo, ligand-free receptor. C, in contrast to agonists, adding antagonist (5 μm SR141716A, red) has essentially no effect on the fluorescence when compared with the ligand-free receptor (gray). D, the agonist-induced increase in fluorescence (10 μm CP55940, (CP) blue) occurs slowly, whereas subsequent addition of antagonist (5 μm SR141716A (SR), red) causes a rapid reversal. E, agonist binding induces the probe to move into a more polar, solvent-accessible environment, as indicated by the shift in the λmax of the normalized emission spectra (blue, 10 μm CP55940; red, 5 μm SR141716A; gray, absence of ligands) (i) and a comparison of the bimolecular quenching constants (kq) determined from the Stern-Volmer quenching experiments (ii). Error estimates come from the least squares fitting. F, a movement of the probe on A342C into a more polar environment is consistent with the presumed location of the probe in CB1 models based on rhodopsin in the inactive state (red, Protein Data Bank (PDB): 1GZM) and active state (blue, PDB: 3DQB). For clarity, the figure only shows the probe and TM3, TM5, and TM6. See “Experimental Procedures” for more details. Error bars indicate S.E. unless otherwise noted.
RESULTS
Expression, Purification, and Site-specific Labeling of CB1
Before introducing unique cysteines into CB1 for labeling with a fluorophore, we first had to establish a mutant that showed no background cysteine labeling. To do this, we used our gene construct, called θ, which contains only two cysteines, Cys-257 and Cys-264 (20). These two cysteines are required to produce a functional receptor (20, 28), and we have previously shown that all other cysteines can be mutated to alanine while still retaining a functional CB1 receptor. Together, these results strongly suggest (but do not definitively prove) that Cys-257 and Cys-264 form a disulfide bond (20, 28).
To obtain a unique site for attaching the fluorescent label, we then introduced a cysteine at the cytoplasmic end of TM6, in the θ construct, at residue 342 (or 6.34 via the Weinstein and Ballestros nomenclature). We hereafter refer to this cysteine mutant as A342C/θ.
The θ and A342C/θ gene constructs were expressed in COS cells. Subsequently, the membranes containing mutant CB1 receptors were solubilized in detergent, the samples were clarified by centrifugation, and the supernatant was then applied to a 1D4 immunoaffinity antibody column. The bound CB1 receptors were then incubated with an ∼20-fold excess of PDT-bimane for ∼16 h, and the excess fluorescent label was then washed from the receptor-bound immunoaffinity antibody column. The purified receptors were then eluted from the immunoaffinity antibody column using an excess of nonapeptide corresponding to the 1D4 binding epitope. The yield from this process is ∼15 μg of purified, bimane-labeled receptor per 15-cm plate of transfected COS1 cells.
The Purified CB1 Is Specifically Labeled at TM6
SDS-PAGE analysis shows that the eluted proteins are pure (Fig. 1, C and D). Moreover, the lack of fluorescence in θ when this gel was irradiated with UV light (prior to Coomassie Blue staining) indicates that the background receptor is not reactive to the bimane label (Fig. 1, C and D). Notably, treating θ with a reducing agent prior to labeling resulted in label incorporation (Fig. 1C), providing further direct evidence that cysteines Cys-257/Cys-264 form a disulfide bond in CB1.
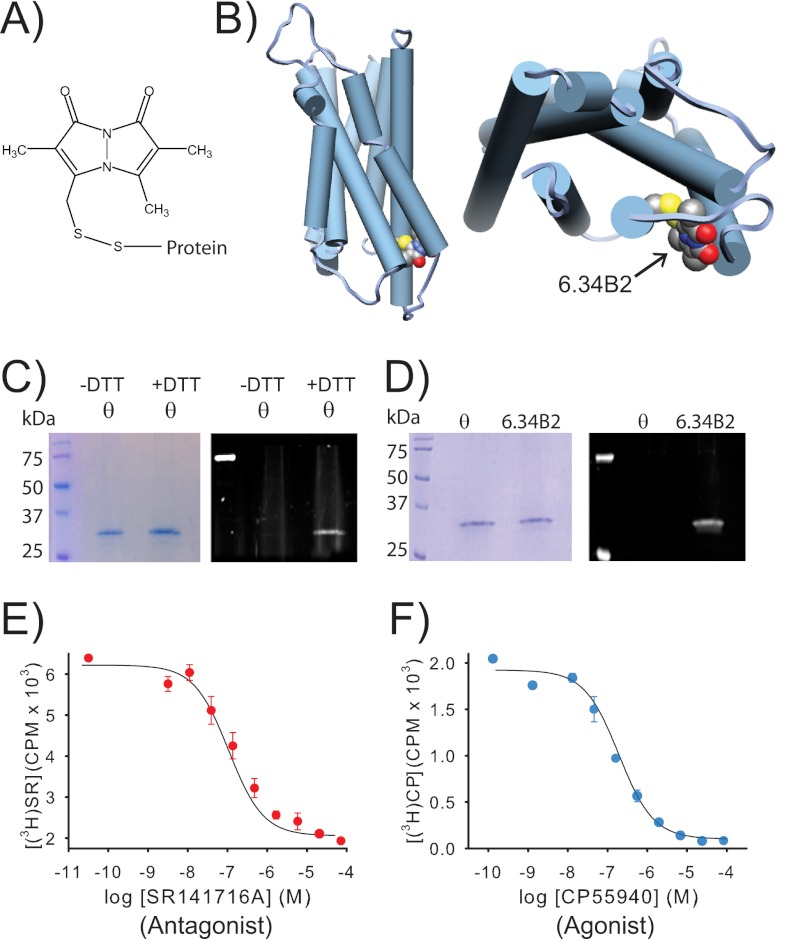
FIGURE 1.
A purified CB1 receptor, specifically labeled with a bimane fluorophore at site 6.34 on TM6, can still bind agonist and antagonist. A, the structure of PDT-bimane. B, a model of CB1 showing the probe covalently attached at A342C (C6.34) on the cytoplasmic face of TM6. C, Coomassie Blue-stained SDS-PAGE gel (left) of purified minimal-cysteine construct mutant θ (which contains only Cys-257 and Cys-264). Ultraviolet irradiation of the same gel (right), before staining, shows that θ does not react with PDT-bimane unless it is first reduced with DTT, prior to labeling (note the bimane fluorescence in the DTT treated sample). This result provides direct chemical evidence that Cys-257 and Cys-264 are in a disulfide bond in CB1. D, left, a Coomassie Blue-stained SDS-PAGE gel showing that the immunopurified CB1 mutants θ and A342C/θ can be purified to homogeneity. Right, in-gel fluorescence of the same gel before Coomassie Blue staining shows only that the A342C/θ mutant exhibits fluorescence, indicating that the bimane is uniquely and specifically covalently attached at A342C in TM6. E and F, the same purified, detergent-solubilized, bimane-labeled A342C/θ from D is functional, as indicated by its ability to bind antagonist, SR141716A (SR) (E), and agonist, CP55940 (CP) (F), in solution. Further details are provided under “Experimental Procedures.” Error bars indicate range.
In contrast to θ, mutant A342C/θ showed robust labeling with the PDT-bimane (Fig. 1D). This result indicates that the fluorophore is specifically attached to the cysteine at site 342. The labeling efficiency was ∼60–80% based on comparison of the ratio of 280 nm (protein)/390 nm (bimane) absorbance. The samples were free of nonreacted label, as determined by TCA precipitation analysis.
The Purified, Bimane-labeled CB1 Retains Its Functional Affinity and Efficacy for Cannabinoid Ligands
Fig. 1, D and E, show that the purified, bimane-labeled A342C/θ mutant is functional in respect to its pharmacological properties. It can bind both agonist and antagonist in a solution binding assay, exhibiting Kd values of 187 ± 27 and 47 ± 23 nm for agonist and antagonist, respectively, or 398 ± 58 and 52 ± 37 nm, respectively, when fit using Swillens approximation to account for possible ligand depletion (20). These values are ∼50–100-fold higher than what we (and others) typically observe in membranes (19, 20). This shift may be partially due to the absence of G proteins in our purified samples, as well as nonspecific effects of the detergent on the receptor. To test whether our bimane-labeled, purified receptor retains functional efficacy, we measured its ability to stimulate [35S]GTPγS binding when reconstituted with G protein (Gαi) and agonist. The results confirm an agonist-induced stimulation of G protein activation and [35S]GTPγS binding when compared with the basal or antagonist-bound states (see Fig. 3B). It is unclear why SR141716A did not affect basal G protein activity. Possibly the intrinsic activity of our G protein preparation could mask this effect and/or our purified samples lack endocannabinoids that may be present in previous in vivo assays that demonstrate basal activity.
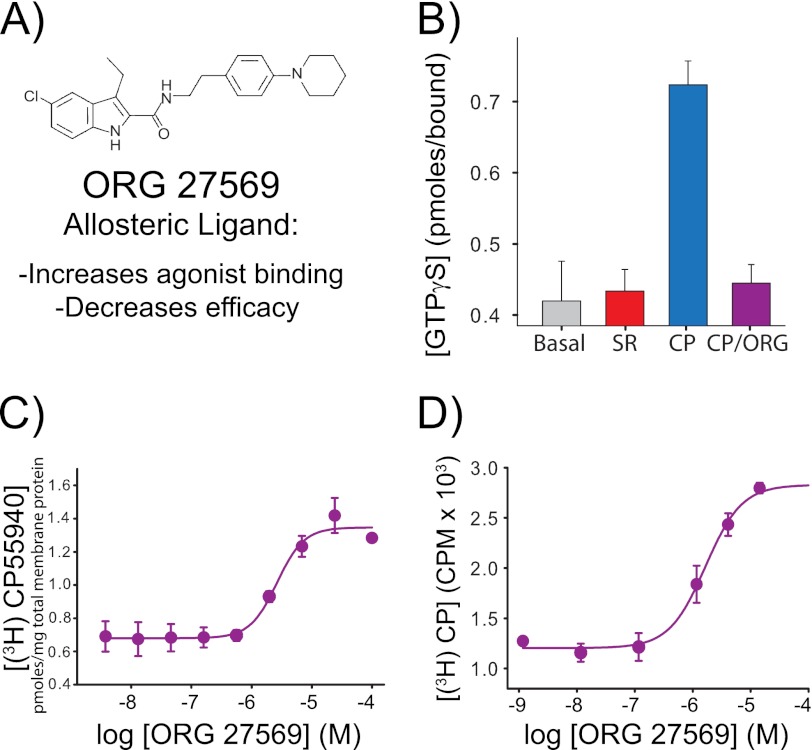
FIGURE 3.
The allosteric CB1 modulator Org 27569 enhances agonist (CP55940) binding yet inhibits agonist-induced G protein activation. A, molecular structure of the allosteric ligand Org 27569. B, the purified, detergent-solubilized bimane-labeled CB1 mutant A342C/θ is functionally active; it stimulates G protein activation upon the addition of agonist (10 μm CP55940 (CP), blue) as measured by [35S]GTPγS binding to purified Gαiβγ. In contrast, no agonist ligands (gray bar) or antagonist ligands (10 μm SR141716A (SR), red bar) show less GTPγS binding. G protein activation is blocked when Org 27569 is added along with agonist (10 μm Org 27569 + 10 μm CP55940 (CP/Org), purple). Note that Org 27569 does not block agonist binding; instead, it actually increases agonist binding ([3H]CP55940) to CB1. C and D, we observed this phenomenon for CB1 mutant A342C/θ in membranes (EC50 for CP55940 binding enhancement = 2.7 ± 0.7 μm) (C) and in a bimane-labeled, detergent-solubilized, purified form (EC50 for CP55940 binding enhancement = 1.9 ± 0.6 μm) (D). All radioactive binding studies are representative of two independent experiments performed in duplicate, shown as mean ± S.E. The specific equilibrium binding of [3H]CP55940 in C and D were determined in the presence of various concentrations of Org 27569 when compared with saturating amounts of cold CP55940. The EC50 values were determined by fitting a variable slope sigmoidal dose-response function to the combined respective data sets, and errors were determined from least squares fitting. See “Experimental Procedures” for more details.
Binding of Agonist to CB1 Induces a Conformational Change in the Cytoplasmic End of TM6, as Detected by Changes in the Fluorescence of the Bimane Probe
The addition of agonist, CP55940, causes an ∼35% increase in the fluorescence intensity of the attached bimane label (Fig. 2A). This fluorescence increase is clearly due to agonist-induced structural changes in CB1 altering the environment around the probe. CP55940 itself is nonfluorescent at the excitation and emission wavelengths used (data not shown). The agonist-induced fluorescence increase is dose-dependent, exhibiting an EC50 of 430 ± 86 nm (Fig. 2Bi). No further increase is seen at ligand concentrations greater than ∼10 μm. A fluorescence increase occurs upon the addition of the endocannabinoid analog meAEA or the CB1 agonist WIN55212-2. Interestingly, the three cannabinoids we tested have the same rank order of potency (CP55940 > WIN55212-2 > AEA) for their ability to induce the bimane fluorescence response in CB1 as they are observed in more traditional pharmacological assays (Fig. 2Bi). We also found that the partial agonist AEA appears to cause less of a fluorescent change, which can be overcome by the addition of more CP55940 (Fig. 2Bii). However, we found meAEA and WIN55212-2 more difficult to work with than CP55940 due to solubility issues and their lower affinities resulting in substantially noisier data. Thus, we did not further explore their behavior in more detail and focused instead on CP55940.
In contrast to agonists, adding antagonist (SR141716A) caused no significant fluorescence change in the sample (Fig. 2C). The antagonist could also reverse the agonist-induced fluorescence increase (Fig. 2D), and it did so much more rapidly (t½ ∼1.5 min) than the slow rate of agonist-induced fluorescence increase (t½ ∼4.7 min).
The Bimane Label on TM6 Moves to a More Polar Environment upon Addition of the Agonist, CP55940
Along with the fluorescence increase, the addition of agonist also induced an ∼6-nm red shift in the bimane fluorescence when compared with the SR141716A form (Fig. 2Ei). We have previously shown that for a soluble protein, the bimane fluorescence emission λmax reflects the solvent accessibility at the site of attachment (26, 29).
However, CB1 is a membrane protein; thus, the λmax shifts could also be affected by interaction of bimane with detergent. Thus, to assess the exposure of the probe to solvent, we carried out fluorescence quenching studies using the aqueous quenching agent, KI. The results show that the probe collides more frequently with I− (i.e. it has a larger bimolecular quenching constant) in the agonist-bound form (Fig. 2Eii). Together, these data confirm that agonist binding causes the probe to relocate to a more solvent-exposed environment, as is expected if CB1 activation involves a conformational change in TM6 (modeled in Fig. 2F), as is observed in other GPCRs (rhodopsin, β2 adrenergic receptor, and adenosine A2A receptor).
The Allosteric Ligand Org 27569 Promotes Agonist Binding to CB1, yet Blocks the Agonist-induced Conformational Changes in TM6
Previous studies have shown that Org 27569 (Fig. 3A) inhibits the ability of CB1 to elicit agonist-induced downstream signaling (12). To test whether this effect occurred at the level of the G protein interaction with the receptor, we measured agonist-stimulated guanine nucleotide exchange for the labeled, purified receptor reconstituted with Gαi. The results show that agonist-stimulated GTPγS binding is completely inhibited in the presence of Org 27569 (Fig. 3B).
We next confirmed previous reports (12) that the allosteric ligand Org 27569 enhances CP55940 binding for CB1 in membranes (Fig. 3C). We then confirmed that Org 27569 also enhances agonist binding to the detergent-solubilized, purified, bimane-labeled CB1 (Fig. 3D). Importantly, these data clearly establish that Org 27569 can enhance specific CP55940 binding independent of the G protein-coupling state of the receptor as our purified, detergent-solubilized CB1 samples are devoid of G protein (Fig. 3D). Together, these results confirm that Org 27569 is not a competitive inhibitor for the orthosteric binding site. Moreover, panel (Fig. 3D) proves that Org 27569: (i) binds to the purified bimane-labeled CB1 receptor and (ii) acts directly on the CB1 receptor.
Additionally, when an allosteric ternary complex (Equation 1) is fit to our data, the allosteric cooperativity factor is 2.74 ± 0.41 and 2.75 ± 0.23 for membrane and solution binding, respectively (Table 1). Both of these values are nearly the same and are greater than one, inicating positive cooperativity.
TABLE 1.
Allosteric ternary complex model (ATCM) and allosteric operational model parameter values for Org 27569. ATCM best fit parameter values for crude membranes expressing A342C/θ, as well as for purified bimane-labeled A342C/θ. KB is the equilibrium dissociation constant for Org 27569, and α is the allosteric cooperativity factor. A value of α > 1 indicates positive cooperativity and governs the magnitude that the allosteric modulator enhances agonist binding. The reported parameter values represent the mean ± S.E. determined from least-squares fitting of Equation 1 from two experiments performed in duplicate. An operational model of allostery was used to fit the data in Fig. 4F. A value of β < 1 indicates attenuation of the orthosteric-induced observable, and it governs the magnitude of this event (in this case the bimane response). Values not shown in the table are the calculated intrinsic efficacy of the orthosteric ligand (τ = 2.21 ± 0.95) and the calculated “fitting” factor (n = 2.03 ± 1.30). The reported parameter values represent the mean ± S.E. determined from least-squares fitting of Equation 2 from two independent experiments
Interestingly, the affinities of the orthosteric ligands are significantly lower in our detergent-purified samples than in membranes, yet the Org 27569 enhancement of agonist binding is essentially unchanged (Fig. 3, C and D, Table 1). We are not sure why this is; it possible that the allosteric site is insensitive to the G protein-coupling state of the receptor (in contrast to the orthosteric ligands) and/or there is a differential “detergent effect” on the samples.
After establishing that Org 27569 does not block but rather increases agonist binding, we next tested the effect of Org 27569 on the agonist-induced conformational changes in TM6 detected by the fluorescence from the bimane probe. Interestingly, the data show that Org 27569 blocks the agonist-induced fluorescence increase of the bimane probe on TM6 (Fig. 4A). Org 27569 can also rapidly reverse the fluorescence increase induced by agonist binding (Fig. 4, B and C). Org 27569 also reversed the fluorescence increase that occurs upon the addition of the endocannabinoid analog meAEA or the CB1 agonist WIN55212-2, although again, the use of these compounds resulted in substantially noisier data (Fig. 4, D and E).
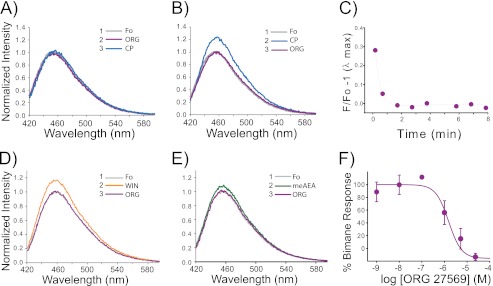
FIGURE 4.
The allosteric modulator Org 27569 inhibits agonist-induced TM6 movement in CB1 detected by a fluorescent probe on site 342. The order in which indicated drugs were added is denoted by the number and compound (inset). Fo represents the ligand-free or apo state (gray) in Purification Buffer. A, Org 27569 (ORG) impairs TM6 movements in CB1. When Org 27569 is preincubated with the bimane-labeled CB1 mutant A342C/θ (10 μm, purple) for 30 min before adding agonist (10 μm CP55940 (CP), blue), the agonist-induced fluorescence change for the bimane probe on TM6 (blue) is no longer observed. B, Org 27569 can also reverse agonist-induced TM6 movements. Adding Org 27569 (10 μm, purple) after agonist (10 μm CP55940) reverses the fluorescence increase in the bimane-labeled CB1 A342C/θ mutant (blue). C, the Org 27569 induced reversal seen in B is rapid, with a t½ < 1 min. Org 267569 (5 μm) also reverses the fluorescence increase caused by CB1 agonists (the figure is an example of data collected from one sample in real time). D and E, WIN55212-2 (WIN, 10 μm, 30 min) (D) and meAEA (38 μm, 30 min) (E). F, the dose-response plot for Org 27569 inhibition of agonist (CP55940)-induced TM6 movement (stimulated increase in fluorescence, EC50 = 2.2 ± 1.2 μm) can be compared with the dose response for Org 27569 enhancement of agonist CP55940 binding (shown in Fig. 3D). The bimane dose-response plot represents the mean of two independent experiments ± range. The EC50 values were determined by fitting a variable slope sigmoidal dose-response function to the combined respective data sets, and errors were determined from least squares fitting. All spectra are background-subtracted from buffer and ligands and are normalized to the background-subtracted emission for the bimane-labeled mutant CB1 in the apo form (Fo, gray). For comparison, the data for the Org 27569 enhancement of fluorescence was normalized to the maximum increase in fluorescence. Further details are provided under “Experimental Procedures.”
Importantly, the inhibition of agonist-induced fluorescence occurs in a dose-dependent manner that closely parallels radioligand CP55940 binding enhancement (compare Figs. 3D and 4F). When fit to an operational model of allostery (Equation 2), we find the β value (magnitude of the allosteric modulation of agonist efficacy) to be less than one and in fact approaches zero (Table 1). This indicates an insurmountable allosteric antagonism of the observable, the bimane response, which we interpret to be transition of the receptor into the active state. The implications of these results are discussed below.
DISCUSSION
In this study, we set out to determine how the allosteric CB1 ligand Org 27569 can enhance agonist binding, yet at the same time inhibit receptor function, a phenomenon first reported by Price et al. (12). GPCRs are inherently under allosteric regulation by G proteins; a bound G protein induces a high agonist affinity binding site in the receptor that is lost when the G protein is activated and released (30). Recently, the cause of this effect has been localized to specifically involve binding of the G protein C terminus to a site exposed by TM6 movement in the receptor (31, 32). Thus, we hypothesized that Org 27569 binding might affect key conformational changes in the cytoplasmic face of the receptor that typically accompany agonist binding and receptor activation/G protein coupling.
To test this hypothesis, we employed an SDFL approach. We introduced a unique and reactive cysteine residue into CB1 and then labeled it with an environmentally sensitive fluorophore, PDT-bimane. We put this probe on the cytoplasmic end of TM6 because this helix has been shown to move during activation in a number of GPCRs (15, 17, 32–41). Thus, we anticipated that activation would cause a change in the fluorescence of the bimane probe.
Our results clearly show activation-induced changes in fluorescence caused by increased solvent exposure for the bimane probe on TM6 upon agonist binding (Fig. 2). Although these results do not delineate precisely how TM6 moves (or the extent) in CB1, they are consistent with an outward TM6 movement observed in other GPCRs (Fig. 2F) (15, 17, 31–33, 35–38, 40–42).
Significantly, the agonist concentration that yields half-maximal bimane fluorescence response (EC50) essentially matches the agonist affinity determined from radioligand binding (compare Fig. 2B with Fig. 1D). The fact that antagonist binding causes no dramatic fluorescence change (Fig. 2C), and can even rapidly reverse the slower agonist-induced changes (Fig. 2D), indicates that the fluorescence increase is specifically linked to agonist activation.
However, it is not clear why the agonist-induced fluorescence change is so slow. The Kobilka laboratory (43) observed a similar slow change in their SDFL studies of the β2 adrenergic receptor, which they determined is due to a multistep binding phenomenon of the ligand to the receptor (44). Thus, the slow change we see for CB1 may represent an analogous multistep binding phenomenon.
Alternatively, the slow fluorescence change we see in CB1 may be caused by interactions of the hydrophobic cannabinoid ligands with the detergent micelles used in our experiments. Interaction of the ligand with empty micelles could slow the amount of agonist cannabinoid ligand available to a receptor/micelle complex. Thus, the more hydrophobic CP55940 would have a slower apparent rate of repartitioning from an empty micelle to a micelle containing a receptor, and this could thus contribute to the slower observed fluorescence change/conformational change in the labeled CB1. Similarly, the faster rate of change observed for the antagonist SR141716A might be partially due to its greater aqueous solubility (lower octanol/water partition coefficient when compared with agonist -1 × 105 versus 1.6 × 106, respectively) (45, 46). Notably, multistep binding models have previously been proposed for cannabinoid ligands to account for their interaction with membranes (47, 48).
How can we reconcile our Org 27569 data with an activation model of CB1? We propose that the binding of the allosteric modulator Org 27569 induces or stabilizes a new ligand-specific conformation, a state that has an agonist bound, but lacks conformational changes in TM6. These results are in agreement with predictions of the allosteric two-state model where the allosteric ligand has positive cooperativity with agonist binding but negative cooperativity with receptor activation (49).
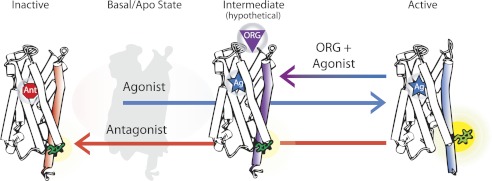
Fig. 5 demonstrates how a multistate model can explain our data. The model shows that agonist binding accompanies a movement of TM6 (right), whereas the antagonist-bound state does not (left). Org 27569 binds with the agonist to CB1, but at least partially inhibits and/or reverses the agonist-induced TM6 movements (middle). Previous experimental studies as well as models have also suggested that multiple GPCR conformations are possible (50–52). The model in Fig. 5 is consistent with our data, which show that Org 27569 puts the CB1 receptor in a distinct conformational state, one in which the binding pocket is occupied by an agonist (Fig. 3, B and C), yet lacks conformational change(s) in TM6 (Fig. 4).
FIGURE 5.
Graphic model proposing that discrete CB1 receptor structures are induced by a bound agonist, antagonist, or agonist plus allosteric ligand. The model suggests that occupation of the traditional (orthosteric) binding site by an agonist alone (Ag, right) accompanies a conformational change in TM6 (blue), detected as an increase in fluorescence from an attached bimane probe (green). In contrast, antagonist alone binding (Ant, left) causes no change in TM6 (red). When the allosteric ligand Org 27569 (ORG) binds to its (currently unknown) site on an agonist-bound CB1, the conformational change in TM6 (purple) is either blocked or reversed. This model proposes that ORG binding traps a distinct and different agonist-bound CB1 structure, which may be a structural intermediate on the pathway to full receptor activation. The basal (ligand-free) CB1 state is depicted in light gray.
Lack of full TM6 movement explains the observed negative allosteric effect Org 27569 has on CB1 signaling efficacy (12). Inhibiting structural changes in the cytoplasmic face of CB1 should impact receptor signaling because movement in this region is associated with the ability of a GPCR to bind and activate its cognate G protein (15, 17, 32, 33, 37, 38).
It is tempting to speculate that the CB1 agonist Org 27569 complex represents not a new conformational state, but rather, the stabilization of an already existing intermediate structure, one that is on the pathway that flows from agonist binding to full receptor activation. There is ample precedence for this possibility. Rhodopsin, the GPCR involved in vision, clearly undergoes several spectrally distinct conformational changes during the conversion of the inactive state to fully activated receptor (53, 54). Structures for many of these inactive intermediates have been solved, and they show that although the agonist (all-trans-retinal) is in the binding pocket of the receptor, only limited changes have propagated throughout the protein to the cytoplasmic face, especially regarding TM6 movements (42, 55–57). Similarly, a structure of a “low-affinity” β2 adrenergic receptor containing an irreversibly bound agonist shows diminished TM6 movement (58). These examples demonstrate the difficulty of trapping a fully active GPCR conformation, even one that contains a covalently attached agonists.
One way that Org 27569 could trap such an early activation intermediate would be by exploiting or lowering the energy of an early agonist-bound intermediate state and/or increasing the energy barrier required for the receptor to take on the active conformation. Interestingly, a similar concept was recently used to determine the structure of an energetic intermediate of the adenosine A2A and β1 adrenergic receptors by extensive mutagenesis designed to produce a more thermally stable receptor. The resulting structures show an intermediate conformation between the inactive and active state, with TM6 partially occluded (59–61). Interestingly, one would expect that the inhibition of full TM6 movement by Org 27569 and the trapping of CB1 in the intermediate state on the pathway to full activation would also increase the dwell time of agonist in the binding pocket. This should enhance the apparent amount of agonist bound to CB1 in the presence of Org 27569, exactly as we observed in our data (Fig. 3).
Understanding how allosteric ligands exert their effects is an exciting and crucial new field of GPCR study (62). Our results here provide insight into one way an allosteric ligand can alter the signaling of its cognate GPCR by either inducing or capturing a previously unidentified and unique receptor conformation or trapping an existing intermediate state formed on the way to receptor activation. These findings also suggest that GPCR intermediate structures may prove to be better templates for designing and screening new allosteric GPCR drugs than either the fully active or the fully inactive state structures.
Acknowledgments
We thank Prof. R. Goodman, O. Davulcu, and members of the Farrens laboratory (A. Jones, E. Lorenzen, A. Sinha, and C. Schafer) for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01DA018169 (to D. L. F.) from the National Institute on Drug Abuse (NIDA), NIDA Training Grant T32 DA007262 (to J. F. F.), and NEI Grant 2R01EY015436-05 (to D. L. F.).
- GPCR
- G protein-coupled receptor
- CB1
- cannabinoid type-1 receptor
- CP55940
- (−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol
- meAEA
- (R)-N-(2-hydroxy-1-methylethyl)-5Z,8Z,11Z,14Z-eicosatetraenamide
- Org 27569
- 5-chloro-3-ethyl-1H-indole-2-carboxylic acid [2-(4-piperidin-1-yl-phenyl)ethyl]amide
- SR141716A
- 5-(4-chloro-phenyl)-1-(2,4-dichloro-phenyl)-4-methyl-1H-pyrazole-3-carboxylic acid piperidin-1-ylamide hydrochloride
- WIN55212-2
- (R)-(+)-[2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenylmethanone mesylate
- PDT-bimane
- 2,3,6-trimethyl-5-[(2-pyridinyldithio)methyl]-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione
- SDFL
- site-directed fluorescent labeling
- shCB1
- synthetic human CB1 receptor
- TM6
- transmembrane helix 6
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Lagerström M. C., Schiöth H. B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 2. Keov P., Sexton P. M., Christopoulos A. (2011) Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology 60, 24–35 [DOI] [PubMed] [Google Scholar]
- 3. May L. T., Leach K., Sexton P. M., Christopoulos A. (2007) Allosteric modulation of G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 47, 1–51 [DOI] [PubMed] [Google Scholar]
- 4. Piomelli D. (2003) The molecular logic of endocannabinoid signaling. Nat. Rev. Neurosci. 4, 873–884 [DOI] [PubMed] [Google Scholar]
- 5. Devane W. A., Dysarz F. A., 3rd, Johnson M. R., Melvin L. S., Howlett A. C. (1988) Determination and characterization of a cannabinoid receptor in rat brain. Mol. Pharmacol. 34, 605–613 [PubMed] [Google Scholar]
- 6. Calandra B., Tucker J., Shire D., Grisshammer R. (1997) Expression in Escherichia coli and characterization of the human central CB1 and peripheral CB2 cannabinoid receptors. Biotechnol. Lett. 19, 425–428 [Google Scholar]
- 7. Chillakuri C. R., Reinhart C., Michel H. (2007) C-terminal truncated cannabinoid receptor 1 coexpressed with G protein trimer in Sf9 cells exists in a precoupled state and shows constitutive activity. FEBS J. 274, 6106–6115 [DOI] [PubMed] [Google Scholar]
- 8. Kim T. K., Zhang R., Feng W., Cai J., Pierce W., Song Z. H. (2005) Expression and characterization of human CB1 cannabinoid receptor in methylotrophic yeast Pichia pastoris. Protein Expr. Purif. 40, 60–70 [DOI] [PubMed] [Google Scholar]
- 9. Link A. J., Skretas G., Strauch E. M., Chari N. S., Georgiou G. (2008) Efficient production of membrane-integrated and detergent-soluble G protein-coupled receptors in Escherichia coli. Protein Sci. 17, 1857–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michalke K., Huyghe C., Lichière J., Gravière M. E., Siponen M., Sciara G., Lepaul I., Wagner R., Magg C., Rudolph R., Cambillau C., Desmyter A. (2010) Mammalian G protein-coupled receptor expression in Escherichia coli: II. Refolding and biophysical characterization of mouse cannabinoid receptor 1 and human parathyroid hormone receptor 1. Anal. Biochem. 401, 74–80 [DOI] [PubMed] [Google Scholar]
- 11. Xu W., Filppula S. A., Mercier R., Yaddanapudi S., Pavlopoulos S., Cai J., Pierce W. M., Makriyannis A. (2005) Purification and mass spectroscopic analysis of human CB1 cannabinoid receptor functionally expressed using the baculovirus system. J. Pept. Res. 66, 138–150 [DOI] [PubMed] [Google Scholar]
- 12. Price M. R., Baillie G. L., Thomas A., Stevenson L. A., Easson M., Goodwin R., McLean A., McIntosh L., Goodwin G., Walker G., Westwood P., Marrs J., Thomson F., Cowley P., Christopoulos A., Pertwee R. G., Ross R. A. (2005) Allosteric modulation of the cannabinoid CB1 receptor. Mol. Pharmacol. 68, 1484–1495 [DOI] [PubMed] [Google Scholar]
- 13. Dunham T. D., Farrens D. L. (1999) Conformational changes in rhodopsin: Movement of helix f detected by site-specific chemical labeling and fluorescence spectroscopy. J. Biol. Chem. 274, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 14. Ghanouni P., Steenhuis J. J., Farrens D. L., Kobilka B. K. (2001) Agonist-induced conformational changes in the G protein-coupling domain of the β2 adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 98, 5997–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janz J. M., Farrens D. L. (2004) Rhodopsin activation exposes a key hydrophobic binding site for the transducin α-subunit C terminus. J. Biol. Chem. 279, 29767–29773 [DOI] [PubMed] [Google Scholar]
- 16. Yao X., Parnot C., Deupi X., Ratnala V. R., Swaminath G., Farrens D., Kobilka B. (2006) Coupling ligand structure to specific conformational switches in the β2 adrenoceptor. Nat. Chem. Biol. 2, 417–422 [DOI] [PubMed] [Google Scholar]
- 17. Tsukamoto H., Farrens D. L., Koyanagi M., Terakita A. (2009) The magnitude of the light-induced conformational change in different rhodopsins correlates with their ability to activate G proteins. J. Biol. Chem. 284, 20676–20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farrens D. L. (2010) What site-directed labeling studies tell us about the mechanism of rhodopsin activation and G protein binding. Photochem. Photobiol. Sci. 9, 1466–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farrens D. L., Dunham T. D., Fay J. F., Dews I. C., Caldwell J., Nauert B. (2002) Design, expression, and characterization of a synthetic human cannabinoid receptor and cannabinoid receptor/ G protein fusion protein. J. Pept Res. 60, 336–347 [DOI] [PubMed] [Google Scholar]
- 20. Fay J. F., Dunham T. D., Farrens D. L. (2005) Cysteine residues in the human cannabinoid receptor: only Cys-257 and Cys-264 are required for a functional receptor, and steric bulk at Cys-386 impairs antagonist SR141716A binding. Biochemistry 44, 8757–8769 [DOI] [PubMed] [Google Scholar]
- 21. DeBlasi A., O'Reilly K., Motulsky H. J. (1989) Calculating receptor number from binding experiments using same compound as radioligand and competitor. Trends Pharmacol. Sci. 10, 227–229 [DOI] [PubMed] [Google Scholar]
- 22. Ehlert F. J. (1988) Estimation of the affinities of allosteric ligands using radioligand binding and pharmacological null methods. Mol. Pharmacol. 33, 187–194 [PubMed] [Google Scholar]
- 23. Skiba N. P., Bae H., Hamm H. E. (1996) Mapping of effector binding sites of transducin α-subunit using Gαt/Gαi1 chimeras. J. Biol. Chem. 271, 413–424 [DOI] [PubMed] [Google Scholar]
- 24. Matsuda T., Fukada Y. (2000) Functional analysis of farnesylation and methylation of transducin. Methods Enzymol. 316, 465–481 [DOI] [PubMed] [Google Scholar]
- 25. Mansoor S. E., Palczewski K., Farrens D. L. (2006) Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. U.S.A. 103, 3060–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mansoor S. E., Farrens D. L. (2004) High-throughput protein structural analysis using site-directed fluorescence labeling and the bimane derivative (2-pyridyl)dithiobimane. Biochemistry 43, 9426–9438 [DOI] [PubMed] [Google Scholar]
- 27. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, p. 279, Springer-Verlag New York Inc., New York [Google Scholar]
- 28. Shire D., Calandra B., Delpech M., Dumont X., Kaghad M., Le Fur G., Caput D., Ferrara P. (1996) Structural features of the central cannabinoid CB1 receptor involved in the binding of the specific CB1 antagonist SR 141716A. J. Biol. Chem. 271, 6941–6946 [DOI] [PubMed] [Google Scholar]
- 29. Mansoor S. E., McHaourab H. S., Farrens D. L. (1999) Determination of protein secondary structure and solvent accessibility using site-directed fluorescence labeling: studies of T4 lysozyme using the fluorescent probe monobromobimane. Biochemistry 38, 16383–16393 [DOI] [PubMed] [Google Scholar]
- 30. De Lean A., Stadel J. M., Lefkowitz R. J. (1980) A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled β-adrenergic receptor. J. Biol. Chem. 255, 7108–7117 [PubMed] [Google Scholar]
- 31. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 34. Sheikh S. P., Zvyaga T. A., Lichtarge O., Sakmar T. P., Bourne H. R. (1996) Rhodopsin activation blocked by metal ion binding sites linking transmembrane helices C and F. Nature 383, 347–350 [DOI] [PubMed] [Google Scholar]
- 35. Xu F., Wu H., Katritch V., Han G. W., Jacobson K. A., Gao Z. G., Cherezov V., Stevens R. C. (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Crystal structure of opsin in its G protein-interacting conformation. Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 37. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Crystal structure of the ligand-free G protein-coupled receptor opsin. Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 38. Yao X. J., Vélez Ruiz G., Whorton M. R., Rasmussen S. G., DeVree B. T., Deupi X., Sunahara R. K., Kobilka B. (2009) The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U.S.A. 106, 9501–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsukamoto H., Terakita A., Shichida Y. (2010) A pivot between helices V and VI near the retinal binding site is necessary for activation in rhodopsins. J. Biol. Chem. 285, 7351–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 41. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye S., Zaitseva E., Caltabiano G., Schertler G. F., Sakmar T. P., Deupi X., Vogel R. (2010) Tracking G protein-coupled receptor activation using genetically encoded infrared probes. Nature 464, 1386–1389 [DOI] [PubMed] [Google Scholar]
- 43. Gether U., Lin S., Kobilka B. K. (1995) Fluorescent labeling of purified β2 adrenergic receptor: evidence for ligand-specific conformational changes. J. Biol. Chem. 270, 28268–28275 [DOI] [PubMed] [Google Scholar]
- 44. Swaminath G., Deupi X., Lee T. W., Zhu W., Thian F. S., Kobilka T. S., Kobilka B. (2005) Probing the β2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J. Biol. Chem. 280, 22165–22171 [DOI] [PubMed] [Google Scholar]
- 45. Katoch-Rouse R., Pavlova O. A., Caulder T., Hoffman A. F., Mukhin A. G., Horti A. G. (2003) Synthesis, structure-activity relationship, and evaluation of SR141716 analogues: development of central cannabinoid receptor ligands with lower lipophilicity. J. Med. Chem. 46, 642–645 [DOI] [PubMed] [Google Scholar]
- 46. Kimura T., Cheng K., Rice K. C., Gawrisch K. (2009) Location, structure, and dynamics of the synthetic cannabinoid ligand CP55940 in lipid bilayers. Biophys. J. 96, 4916–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hurst D. P., Grossfield A., Lynch D. L., Feller S., Romo T. D., Gawrisch K., Pitman M. C., Reggio P. H. (2010) A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J. Biol. Chem. 285, 17954–17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makriyannis A. (1995) Cannabinoid Receptors, p. 105, Academic Press, Orlando, FL [Google Scholar]
- 49. Hall D. A. (2000) Modeling the functional effects of allosteric modulators at pharmacological receptors: an extension of the two-state model of receptor activation. Mol. Pharmacol. 58, 1412–1423 [DOI] [PubMed] [Google Scholar]
- 50. Ghanouni P., Gryczynski Z., Steenhuis J. J., Lee T. W., Farrens D. L., Lakowicz J. R., Kobilka B. K. (2001) Functionally different agonists induce distinct conformations in the G protein-coupling domain of the β2 adrenergic receptor. J. Biol. Chem. 276, 24433–24436 [DOI] [PubMed] [Google Scholar]
- 51. Kenakin T. (2004) Principles: receptor theory in pharmacology. Trends Pharmacol Sci. 25, 186–192 [DOI] [PubMed] [Google Scholar]
- 52. Christopoulos A., Kenakin T. (2002) G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54, 323–374 [DOI] [PubMed] [Google Scholar]
- 53. Okada T., Ernst O. P., Palczewski K., Hofmann K. P. (2001) Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem. Sci 26, 318–324 [DOI] [PubMed] [Google Scholar]
- 54. Tsukamoto H., Szundi I., Lewis J. W., Farrens D. L., Kliger D. S. (2011) Rhodopsin in nanodiscs has native membrane-like photointermediates. Biochemistry 50, 5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakamichi H., Okada T. (2006) Local peptide movement in the photoreaction intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 12729–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruprecht J. J., Mielke T., Vogel R., Villa C., Schertler G. F. (2004) Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salom D., Lodowski D. T., Stenkamp R. E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J. A., Palczewski K. (2006) Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rosenbaum D. M., Zhang C., Lyons J. A., Holl R., Aragao D., Arlow D. H., Rasmussen S. G., Choi H. J., Devree B. T., Sunahara R. K., Chae P. S., Gellman S. H., Dror R. O., Shaw D. E., Weis W. I., Caffrey M., Gmeiner P., Kobilka B. K. (2011) Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature 469, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lebon G., Bennett K., Jazayeri A., Tate C. G. (2011) Thermostabilization of an agonist-bound conformation of the human adenosine A2A receptor. J. Mol. Biol. 409, 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lebon G., Warne T., Edwards P. C., Bennett K., Langmead C. J., Leslie A. G., Tate C. G. (2011) Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474, 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warne T., Moukhametzianov R., Baker J. G., Nehmé R., Edwards P. C., Leslie A. G., Schertler G. F., Tate C. G. (2011) The structural basis for agonist and partial agonist action on a β1 adrenergic receptor. Nature 469, 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Unal H., Karnik S. S. (2012) Domain coupling in GPCRs: the engine for induced conformational changes. Trends Pharmacol. Sci. 33, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]