Background: Cholesterol is believed to be sensed via direct binding to the homeostatic machinery rather than by altering membrane properties.
Results: Enantiomeric cholesterol, which exerts membrane effects but not specific interactions, also elicited homeostatic responses.
Conclusion: Cholesterol mediates its own homeostasis through changing membrane properties in addition to specific cholesterol-protein binding.
Significance: This work indicates a significant role of membrane effects in cholesterol homeostasis.
Keywords: Cholesterol, Cholesterol Metabolism, Lipids, Membrane, Sterol, Enantiomer, SREBP, Scap, Squalene Monooxygenase
Abstract
How cholesterol is sensed to maintain homeostasis has been explained by direct binding to a specific protein, Scap, or through altering the physical properties of the membrane. The enantiomer of cholesterol (ent-cholesterol) is a valuable tool in distinguishing between these two models because it shares nonspecific membrane effects with native cholesterol (nat-cholesterol), but not specific binding interactions. This is the first study to compare ent- and nat-cholesterol directly on major molecular parameters of cholesterol homeostasis. We found that ent-cholesterol suppressed activation of the master transcriptional regulator of cholesterol metabolism, SREBP-2, almost as effectively as nat-cholesterol. Importantly, ent-cholesterol induced a conformational change in the cholesterol-sensing protein Scap in isolated membranes in vitro, even when steps were taken to eliminate potential confounding effects from endogenous cholesterol. Ent-cholesterol also accelerated proteasomal degradation of the key cholesterol biosynthetic enzyme, squalene monooxygenase. Together, these findings provide compelling evidence that cholesterol maintains its own homeostasis not only via direct protein interactions, but also by altering membrane properties.
Introduction
Cholesterol is an important lipid in human health and disease. Its unique physicochemical properties allow it to perform diverse cellular roles. Apart from being a precursor for vital classes of molecules (e.g. steroid hormones and bile acids), cholesterol itself has crucial functions within the cell. Cholesterol reinforces membranes, organizes membrane subdomains such as lipid rafts, and is an intimately associated modulator of a growing list of integral membrane proteins (1, 2). Moreover, cholesterol is intricately involved in its own homeostasis.
A major mechanism by which cholesterol feeds back on its own synthesis and uptake is by regulating the activation of the sterol regulatory element-binding protein (SREBP)2 transcription factors (3). SREBP-2 is the major isoform involved in regulating cholesterol homeostasis. When cholesterol levels in the endoplasmic reticulum (ER) exceed a critical threshold (4), the SREBP cleavage-activating protein, Scap, undergoes a conformational change (5), which assists binding of the tethering protein, Insig (6). This traps SREBP-2 in the ER in its inactive precursor form. When cholesterol levels are low, Scap escorts SREBP-2 to the Golgi, where it can be proteolytically activated to induce the expression of a program of genes involved in cholesterol synthesis and uptake.
This paradigm is built on strong genetic and biochemical evidence linking Scap conformation with SREBP-2 processing. Thus, Scap mutants that are sterol-resistant in terms of suppressing SREBP-2 processing are also resistant to cholesterol-induction of conformational change (5, 6). Conversely, Scap mutants that are trapped in the cholesterol-induced conformation cannot mediate SREBP-2 processing in intact cells (7, 8). Furthermore, titration experiments with Insig proteins increase sterol sensitivity to both SREBP-2 processing in cells and cholesterol-induced Scap conformational change in vitro (6, 9). Moreover, the conformational change switches the MELADL sorting signal in Scap to a new location with respect to the ER membrane, preventing COPII protein binding and hence blocking transport of SREBP-2 to the Golgi for activation (10). Together, these studies provide compelling evidence that the Scap conformation is the central determinant that dictates the rate of SREBP-2 processing.
Recently, we have introduced yet another player in cholesterol homeostasis, with cholesterol feeding back on a key cholesterol biosynthetic enzyme, squalene monooxygenase (SM), by accelerating its proteasomal degradation (11). In addition, certain oxysterols and intermediates in cholesterol synthesis also participate in cholesterol homeostasis (12), for example, by inhibiting SREBP activation (13) and by accelerating the Insig-dependent proteasomal degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR), another key cholesterol biosynthetic enzyme (14, 15).
How sterols operate to achieve cholesterol homeostasis has been explained by two alternate models. The first and prevailing model is that the sterols are sensed directly by the homeostatic machinery. Supporting this is evidence that cholesterol directly binds Scap (8, 16, 17) and that oxysterols directly bind Insig (18). The alternative view is that sterols induce a change in the physical properties of the membrane bilayer in the environs of the sensing proteins. Earlier studies on the specificity of the Scap conformational change were also consistent with this latter model, in that sterols that could induce the conformational change (5) most altered membrane fluidity (19), and this effect could be mimicked by particular amphiphiles known to influence membrane properties (6). Moreover, studies of the SREBP system in Drosophila cells indicated that phosphatidylethanolamine, rather than sterols, are sensed by Scap in flies, and it was suggested that cholesterol in mammals and phosphatidylethanolamine in flies may both be sensed by Scap through similar perturbations of the ER membrane (20). Employing an enantiomer of 25-hydroxycholesterol (25-HC), Gale et al. implicated oxysterol-membrane interactions in the acute regulation of sterol homeostatic responses (21). The relative importance of these two modes of sterol action in cholesterol homeostasis requires clarification.
The enantiomer of cholesterol (ent-cholesterol) is the mirror image of native cholesterol (nat-cholesterol, Fig. 1A), with both molecules supporting normal growth of mammalian cells (22) and sharing major physicochemical properties and hence effects on membranes. In contrast, direct protein binding is enantioselective. Therefore, ent-cholesterol is a valuable tool to determine whether the effects of cholesterol on protein function derive from changes in membrane properties or from the direct binding of this sterol to a membrane protein (23, 24). In this report, we have compared the effects of ent-cholesterol with nat-cholesterol on major measures of cholesterol homeostasis.
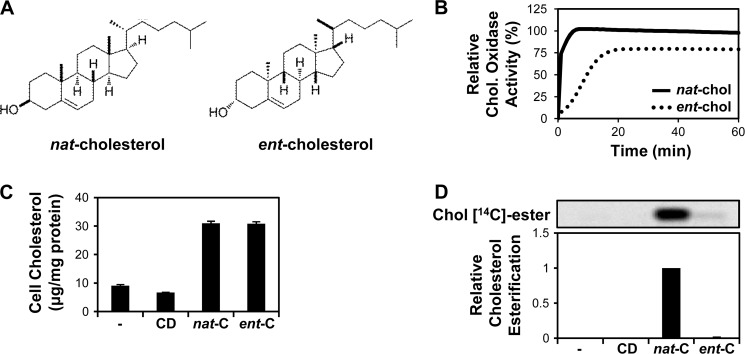
FIGURE 1.
Structures and characterization of nat-cholesterol versus ent-cholesterol. A, chemical structures of native cholesterol (nat-cholesterol) and its enantiomer (ent-cholesterol). B, nat-cholesterol and ent-cholesterol (200 ng) measured using the Amplex red cholesterol assay kit. Fluorescence was monitored during incubation with cholesterol oxidase at 1-min intervals for 60 min at 37 °C. Data are the average of two separate experiments (each performed in triplicate), with the plateau given by nat-cholesterol set to 100% relative cholesterol oxidase activity. C, CHO-7 cells pretreated with statin and then incubated with 20 μg/ml nat-cholesterol (nat-C) or ent-cholesterol (ent-C) for 4 h. Nontreated and CD-treated conditions served as controls. CD was delivered at an amount equivalent to the sterol-CD complex. Lysates were analyzed for cholesterol content by HPLC/UV (performed in triplicate, ±S.E. (error bars)). D, CHO-7 cells pretreated with statin and then incubated with 20 μg/ml nat-cholesterol or ent-cholesterol for 4 h in the presence of 1 μCi of [1-14C]palmitate. Lipids were separated by TLC and detected by phosphorimaging. Cholesteryl [14C]esters were expressed relative to the maximal condition, which was set to 1 (n = 4, ±S.E.).
EXPERIMENTAL PROCEDURES
Materials
Chinese hamster ovary-7 (CHO-7), SRD-1, SRD-13A, and CHO/pGFP-Scap cells were generous gifts of Drs. Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern, Dallas). Dulbecco's modified Eagle's medium/Ham's Nutrient Mixture F-12 (DMEM/F12) and newborn calf serum (NCS) were purchased from Invitrogen. Lipoprotein-deficient serum (LPDS) was prepared from NCS as described previously (25). Nat-cholesterol (Steraloids) and ent-cholesterol (synthesized according to Ref. 26) were complexed to methyl-β-cyclodextrin (CD) as described (5). The molar ratio of sterol to CD was ∼0.1 for nat-cholesterol and ∼0.07 for ent-cholesterol. The lower complexing efficiency for ent-cholesterol is likely due to the altered ability of the enantiomer to interact with the optically active interior of CD (27). Compactin (mevastatin), mevalonate, sodium oleate, Dulbecco's phosphate-buffered saline (PBS), bovine serum albumin (BSA), trypsin, soybean trypsin inhibitor, 25-HC, CD, 2-hydroxypropyl-β-cyclodextrin (HPCD), and U18666A were purchased from Sigma-Aldrich. The SM inhibitor GR144000X (equivalent to NB-598), was a kind gift from GlaxoSmithKline.
Cell Culture
All cells were grown in monolayer at 37 °C in 5% CO2. CHO-7, SRD-1, and CHO/pGFP-Scap cells were cultured in DMEM/F12 supplemented with 5% (v/v) LPDS. SRD-13A cells were maintained in DMEM/F12 (containing 5 μg/ml cholesterol, 1 mm mevalonate, and 20 μm sodium oleate) supplemented with 5% (v/v) NCS. Before treatment, cells were pretreated overnight with statin to deplete sterols. Cells were then treated as indicated in the figure legends. Sterols were delivered to cells as CD complexes. Pretreatment and treatment media were DMEM/F12 containing 5% (v/v) LPDS, 5 μm compactin, and 50 μm mevalonate. “Enantiomer medium” was DMEM/F12 supplemented with 5% (v/v) LPDS containing 3 μg/ml ent-cholesterol (complexed to CD) and 5 μm SM inhibitor to inhibit sterol synthesis.
Transfection
Cells were transfected for 24 h using a ratio of 1 μg of DNA to 4 μl of Lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer's instructions. pTK-SM-V5 encodes full-length SM driven by a TK promoter (11).
Cholesterol Oxidase Assay
The Amplex Red Cholesterol Assay kit (Invitrogen) contains cholesterol oxidase from Streptomyces and was used according to the manufacturer's instructions. The fluorescence signal was measured using a POLARstar Omega microplate spectrofluorometer (excitation λ 544 nm, emission λ 590 nm; BMG Labtech) and was expressed relative to protein levels, measured using the bicinchoninic acid assay (Pierce).
Sterol Analysis by HPLC
Lipids were extracted and prepared for HPLC analysis as described previously (28). Lipids were extracted in a total aqueous volume of 1 ml, containing 2 mm EDTA and 20 μm butylated hydroxytoluene. Methanol (2.5 ml) was added, followed by 5 ml of hexane. Hexane extracts were evaporated and redissolved in the mobile phase, acetonitrile:isopropyl alcohol (30:70, v/v). Both nat-cholesterol and ent-cholesterol were analyzed by HPLC using a Supelco reverse-phase C18 column (0.46 × 25 cm, 2-cm Pelliguard column, 5-μm particle size (Sigma-Aldrich)), run at room temperature with a flow rate of 1 ml/min. UV detection was at 210 nm. All solvents were HPLC grade (Merck).
Cholesterol Esterification
Cells were labeled with 1 μCi of [1-14C]palmitate (specific activity 56.0 mCi/mmol; American Radiolabeled Chemicals) for 4 h during treatment. Lipid extracts were analyzed for cholesteryl [14C]esters by thin layer chromatography (TLC) followed by phosphorimaging, as described previously (29).
[3H]Cholesterol esterification was also determined. [1,2-3H]Cholesterol (specific activity 46.0 Ci/mmol; PerkinElmer Life Sciences) was complexed to HPCD and used as described (30, 31), with minor modifications. [3H]Cholesterol (10 μCi) was dried under nitrogen, redissolved in 2.5 μl of 20% (w/v) HPCD, vortexed for 1 min five times, and incubated for 10 min at 37 °C before adding 17.5 μl of PBS. The complex was then vortexed for 1 min three times and incubated for 3 min at 37 °C. Following statin pretreatment, cells were washed twice in PBS and pulse-labeled in PBS for 10 min at room temperature with the [3H]cholesterol-HPCD complex. After washing three times with 0.5 mg/ml BSA/PBS, cells were treated for 4 h as indicated. Lipid extracts were separated by TLC as described above. [3H]Chol ester and [3H]Chol bands were detected and captured using a specific phosphorimaging plate for tritium detection, BAS-IP TR 2040 (Fujifilm).
Immunoblot Analysis
Immunoblot analysis was performed as described (29) with minor modifications. Cells were lysed in 2% SDS lysis buffer (10 mm Tris-HCl (pH 7.6), 100 mm NaCl, 2% (w/v) SDS) for SREBP-2 or in 10% (w/v) SDS for SM, containing 0.05 volumes of Protease Inhibitor Mixture (Sigma-Aldrich). Lysates were then homogenized using a Kontes motorized micropestle. Samples (40 μg of protein) were subjected to SDS-PAGE and immunoblotted with the following antibodies: anti-V5 (Invitrogen), anti-SQLE (SM) (Proteintech Group), and anti-α-tubulin (Sigma-Aldrich). IgG-7D4, a mouse monoclonal antibody against hamster SREBP-2 (amino acids 32–250) (32) was prepared from a mouse hybridoma cell line (ATCC CRL-2198). IgG-R139, kindly provided by Drs. Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern, Dallas), is a rabbit polyclonal antibody against hamster Scap (33). Bound antibodies were visualized by enhanced chemiluminescence with Immobilon HRP reagents (Millipore) and exposed to Hyperfilm ECL (GE Healthcare). The relative intensities of bands were quantified using ImageJ software 1.45s.
Quantitative Real-time PCR
Cells were harvested for total RNA using TRI Reagent (Sigma-Aldrich), according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed to cDNA with Superscript Reverse Transcriptase III (Invitrogen). Quantitative real-time PCR was performed using the SensiMix SYBR No-ROX kit (Bioline) in a Corbett Rotorgene 3000 and analyzed using Rotor-Gene version 6.0 (Build 27) (Corbett Research). Primers were used to amplify the cDNA of hamster low density lipoprotein receptor (LDLR), HMGCR (34), squalene monooxygenase (SQLE) (11), and the housekeeping control porphobilinogen deaminase (PBGD) (35). Changes in gene expression levels of LDLR, HMGCR, and SQLE were normalized to PBGD for each sample.
Trypsin Cleavage Assay of Scap
The Scap trypsin cleavage assay was performed as described previously (5), with minor modifications. Cells were harvested in buffer A (10 mm Hepes (pH 7.4), 1.5 mm MgCl2, 10 mm KCl, 5 mm EDTA, 5 mm EGTA, and 250 mm sucrose) and homogenized with a 22-gauge needle (40×). Membrane fractions were isolated (5), and aliquots (40 μg of protein) were incubated with sterols as indicated for 20 min at room temperature. Fractions were then pelleted, resuspended in buffer A, and incubated with 1 μg of trypsin for 20 min at 30 °C. Trypsin digestion was stopped by addition of 10 μg of soybean trypsin inhibitor. Samples were subsequently treated with PNGase F (Genesearch) for 2 h at 37 °C, according to the manufacturer's instructions. Samples were mixed with 0.25 volumes of 5× SDS loading buffer and subjected to 16% Tris-Tricine SDS-PAGE and immunoblotted with IgG-R139.
Data Presentation and Statistical Analysis
Unless otherwise stated, quantitative data are presented as averages, and all error bars represent the S.E. from at least three separate experiments. Statistical differences were determined by paired t tests (two-tailed), where a p value of ≤0.05 (*) and ≤0.01 (**) were considered statistically significant.
RESULTS
First, we confirmed that our preparation of ent-cholesterol performed as described in published assays. According to earlier reports, ent-cholesterol can serve as a substrate for bacterial cholesterol oxidase, which is routinely used in the enzymatic determination of cholesterol levels. However, the kinetics for oxidation of ent-cholesterol by this enzyme are reportedly slower and less complete than for nat-cholesterol, yielding 15 and 76% of signal after 5 and 60 min at 37 °C, respectively (36). We found that cholesterol oxidase treatment of ent-cholesterol yielded similar values at these times (22 and 79%, respectively) (Fig. 1B). Therefore, the ent-cholesterol behaves similarly to published data.
Previously, we have shown that cholesterol complexed to CD rapidly and robustly produces cholesterol homeostatic responses in CHO-7 cells, a commonly used model of cellular cholesterol homeostasis (4, 11). Here, we prepared CD complexes for nat- and ent-cholesterol which were then used to treat CHO-7 cells for 4 h. Using HPLC/UV analysis, we found that at equivalent cholesterol concentrations, ent-cholesterol resulted in cellular cholesterol loading comparable with that of nat-cholesterol (Fig. 1C).
Cholesterol esterification by the ER resident enzyme acyl-CoA:cholesterol acyltransferase (ACAT) is enantioselective, in that ent-cholesterol is a very poor substrate for ACAT (37). This was confirmed by co-treating CHO-7 cells with sterol and radiolabeled fatty acid, when ent-cholesterol was unable to be esterified, in contrast to nat-cholesterol (Fig. 1D).
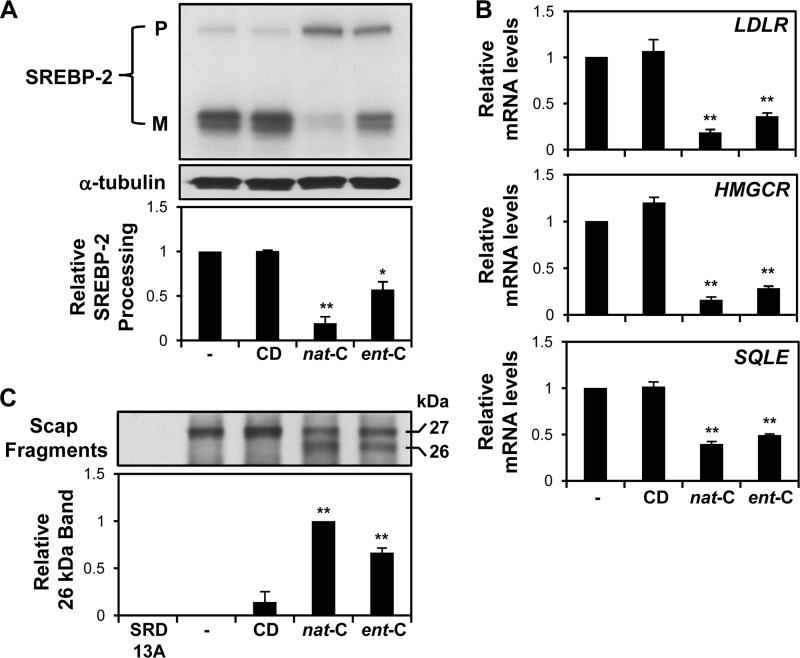
Next, we tested activation of SREBP-2. Nat-cholesterol inhibited SREBP-2 processing, as indicated by increased precursor and decreased mature forms of SREBP-2, relative to the control cells. Importantly, ent-cholesterol also suppressed SREBP-2 processing, although to a lesser extent than nat-cholesterol (Fig. 2A). Ent-cholesterol, like nat-cholesterol, also had a striking ability to suppress the expression of three SREBP-2 target genes: LDLR, HMGCR, and SQLE (Fig. 2B).
FIGURE 2.
Ent-cholesterol suppresses activation of the SREBP-2 pathway. A and B, CHO-7 cells were pretreated with statin and then incubated with 20 μg/ml nat-cholesterol or ent-cholesterol for 4 h. Nontreated and CD-treated conditions served as controls. A, cell lysates were subjected to 7.5% SDS-PAGE and assayed for SREBP-2 by immunoblot analysis with IgG-7D4. P, precursor; M, mature (active) form. Relative SREBP-2 processing was expressed as a ratio of mature to total SREBP (M/P + M) and normalized to the nontreated condition, which was set to 1 (n = 4, ±S.E. (error bars)). *, p ≤ 0.05; and **, p ≤ 0.01 versus nontreated condition. B, relative mRNA levels of three SREBP-2 target genes (LDLR, HMGCR, and SQLE) were determined by quantitative real-time PCR (n = 4, ±S.E., each performed in triplicate). **, p ≤ 0.01 versus nontreated condition, which was set to 1. C, SRD-13A (lacking Scap) and CHO/pGFP-Scap cells were pretreated with statin overnight. After harvesting, microsomes were prepared and subjected to the trypsin cleavage assay of Scap (see “Experimental Procedures”). The densitometric values of the lower (26-kDa) band were normalized relative to nat-cholesterol, which was set to 1 (n = 3, ±S.E.). **, p ≤ 0.01 versus nontreated condition.
The ability of cholesterol to suppress SREBP-2 activation is due to its induction of a conformational change in Scap, which favors its retention in the ER (5, 6). The Scap conformational change can be ascertained by a trypsin cleavage assay, where the addition of cholesterol to Scap-containing microsomes in vitro exposes a previously buried trypsin cleavage site, resulting in a slightly smaller Scap cleavage product (26 versus 27 kDa) (5). Employing this Scap trypsin cleavage assay, we found that, as has been established for nat-cholesterol, ent-cholesterol was also able to induce a conformational change in Scap (Fig. 2C). Together, these findings suggest that the ability of cholesterol to reduce SREBP-2 activation is not necessarily enantiospecific.
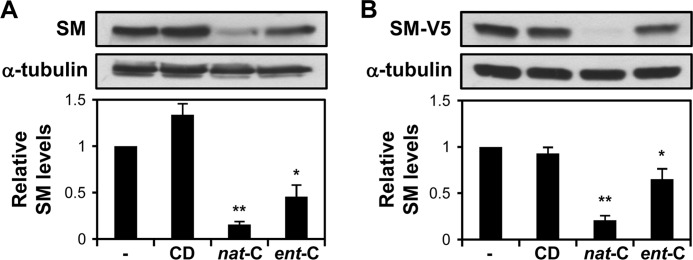
Recently, we proposed that the enzyme SM is an important control point in cholesterol synthesis, beyond HMGCR. We demonstrated that cholesterol feeds back at SM, accelerating its proteasomal degradation (11). Like nat-cholesterol, ent-cholesterol also accelerates the degradation of SM, albeit less potently (Fig. 3). This effect was observed with endogenous SM in SRD-1 cells, which lack sterol regulation (Fig. 3A), and in CHO-7 cells overexpressing full-length SM (Fig. 3B).
FIGURE 3.
Ent-cholesterol causes degradation of squalene monooxygenase. A, SRD-1 cells were pretreated with statin and then incubated with 20 μg/ml nat-cholesterol or ent-cholesterol in medium containing cycloheximide (10 μg/ml) for 8 h. Nontreated and CD-treated conditions served as controls. B, CHO-7 cells were transfected with 1 μg of pTK-SM-V5 for 24 h. Cells were pretreated with statin and then treated as in A. A and B, cell lysates were subjected to 10% SDS-PAGE and immunoblotted with anti-SQLE for endogenous SM (A) or anti-V5 for ectopic SM (B). Densitometric values of SM were expressed relative to the nontreated condition, which was set to 1 (n ≥ 4, ±S.E. (error bars)). *, p ≤ 0.05 for ent-cholesterol versus nontreated condition; **, p ≤ 0.01 for nat-cholesterol versus nontreated condition.
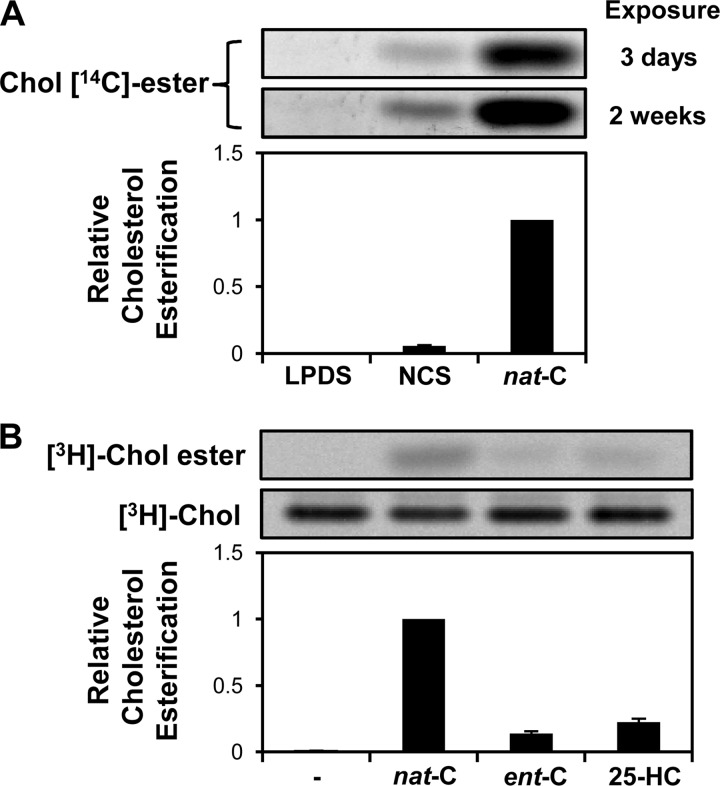
It is possible that endogenous cholesterol might be displaced or “activated” by the addition of ent-cholesterol, thereby potentially confounding the results. Thus, Lange et al. (38) have shown that following cholesterol enrichment with a deuterated cholesterol probe, the endogenous cholesterol is internalized prior to the exogenously loaded cholesterol complexed to cyclodextrin. Therefore, it might be argued that ACAT would first have been presented with endogenous cholesterol and raises the question of why this esterification was not detected with ent-cholesterol in Fig. 1D. However, the CHO-7 cells used in these experiments were selected to grow in cholesterol-deficient medium (39) and were further cholesterol starved by pretreating with a statin. These culturing conditions result in an extremely low cell cholesterol level. Incubating the CHO-7 cells with full serum (NCS) increases the cell cholesterol content by ∼50% (from 7.2 to 11 μg/mg cell protein) and produces a perceptible cholesteryl ester band (Fig. 4A). By contrast, no such band was observed under our basal cholesterol-starved condition (LPDS with statin), even when the phosphorimage was exposed for five times as long (2 weeks versus 3 days). To address this issue further, we took a lead from the work of Lange et al. (30, 38), pulse-labeling cells with [3H]cholesterol complexed to HPCD, and measured [3H]cholesterol esterification after incubation with nat-cholesterol, ent-cholesterol, or the oxysterol 25-HC, which is known to activate endogenous cholesterol (31, 40). Under these conditions, very little of the pulsed [3H]cholesterol is esterified (Fig. 4B). Moreover, very little esterification (∼14%) is observed with the ent-cholesterol compared with nat-cholesterol. These data argue against the effects of ent-cholesterol being attributable to any activation of endogenous cholesterol.
FIGURE 4.
Basal cholesterol esterification in cholesterol-starved CHO-7 cells is undetectable, and loading with ent-cholesterol induces minimal esterification of endogenous cholesterol. A, CHO-7 cells were pretreated overnight with statin (LPDS and nat-C) or with DMEM/F12 supplemented with 5% (v/v) NCS (NCS). They were then incubated in the treatment medium without sterols (LPDS and NCS) or with 20 μg/ml nat-cholesterol (nat-C) for 4 h in the presence of 1 μCi of [1-14C]palmitate. Lipids were separated by TLC and detected by phosphorimaging. This phosphorimage was exposed for 3 days (top panel) or 2 weeks (bottom panel) and is representative of four separate experiments. Cholesteryl [14C]esters were expressed relative to the maximal condition, which was set to 1 (n = 4, ±S.E. (error bars)). B, CHO-7 cells were pretreated with statin overnight and then pulse-labeled for 10 min with 10 μCi of [3H]cholesterol-HPCD complex. After washing three times with 0.5 mg/ml BSA/PBS, cells were treated for 4 h with 20 μg/ml nat-cholesterol or ent-cholesterol, or 1 μg/ml 25-HC. Lipids were separated by TLC and detected by phosphorimaging. This phosphorimage is representative of four separate experiments. [3H]Chol esters were expressed relative to the maximal condition, which was set to 1 (n = 4, ±S.E.).
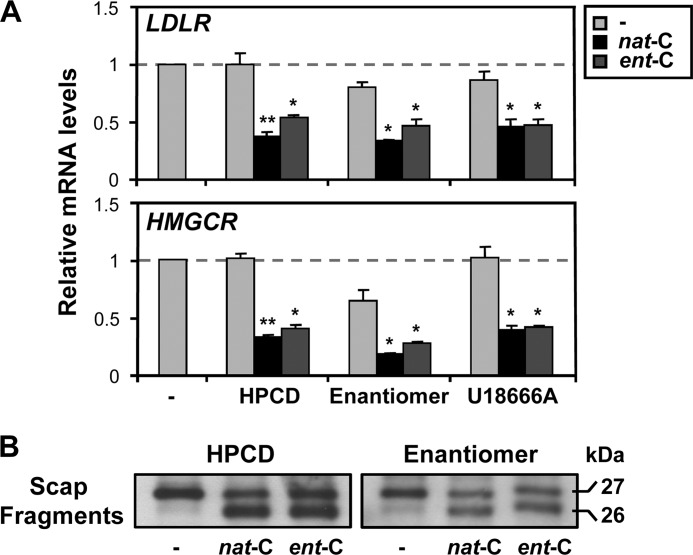
Along similar lines, we performed experiments on SREBP-2 target gene expression under conditions where endogenous cholesterol was severely depleted. Our first approach was to replace nat-cholesterol by prolonged culturing (10 days) with a maintenance concentration of ent-cholesterol (3 μg/ml), while also inhibiting cholesterol synthesis (employing an inhibitor that targets SM). These culturing conditions (designated here as enantiomer medium) have previously been used to demonstrate that ent-cholesterol can substitute for nat-cholesterol in supporting normal cell growth (22). A second approach involved removing residual cholesterol by preincubating cells with 1% (w/v) HPCD for 1 h (5), which decreases total cell cholesterol by ∼30% (data not shown). Under both of these conditions, nat-cholesterol and ent-cholesterol still suppressed the expression of two classical SREBP-2 target genes, HMGCR and LDLR (Fig. 5A). Moreover, this ability of nat-cholesterol and ent-cholesterol to suppress SREBP-2 target gene expression was observed even when transport of endogenous cholesterol was inhibited by treatment with the prototypic class 2 amphiphile U18666A (Fig. 5A). We employed a concentration of U18666A (4.5 μm) that has previously been shown to inhibit 27-hydroxycholesterol formation in the mitochondria and to increase HMGCR activity in the ER (38). It is also noteworthy that reducing cell cholesterol further by cyclodextrin treatment did not increase SREBP-2 target gene expression (as was also seen in the CD control in Fig. 2B), suggesting that ER cholesterol status is already extremely low. Importantly, long term culturing in ent-cholesterol as the sole sterol (with synthesis of nat-cholesterol inhibited) also reduced SREBP-2 target gene expression, reinforcing the concept that ent-cholesterol can elicit cholesterol homeostatic responses.
FIGURE 5.
Ent-cholesterol rather than endogenous cholesterol is able to suppress SREBP-2 target genes and induce Scap conformational change. A, CHO-7 cells were pretreated with statin overnight. They were then preincubated with 1% (w/v) HPCD for 1 h or grown in enantiomer medium for 10 days, or preincubated with 4.5 μm U18666A for 2 h before treatment. Cells were then treated with 20 μg/ml nat-cholesterol or ent-cholesterol for 4 h, with U18666A maintained in the last three conditions during this period. The nontreated condition served as a control in each category. Relative mRNA levels of two SREBP-2 target genes (LDLR and HMGCR) were determined by quantitative real-time PCR (n = 3, ±S.E. (error bars), each performed in triplicate). Values were normalized to the nontreated condition, which was set to 1. *, p ≤ 0.05 and **, p ≤ 0.01 versus nontreated condition for each category. B, CHO/pGFP-Scap cells were pretreated with statin overnight and then incubated with 1% (w/v) HPCD for 1 h (left panel) or grown in enantiomer medium for 10 days (right panel). After harvesting, microsomes were prepared and subjected to the trypsin cleavage assay of Scap (see “Experimental Procedures”). The immunoblots are each representative of three separate experiments.
Similarly, a case could be made that the membranes used in the Scap conformation experiments in Fig. 2C still contains some endogenous cholesterol, and so the conformational change may not be due entirely to ent-cholesterol. To address this, we performed the Scap trypsin cleavage assay using microsomes from cells grown in enantiomer medium for 10 days, or incubated with 1% (w/v) HPCD for 1 h. Under these severely cholesterol-depleted conditions, ent-cholesterol still induced a conformational change in Scap (Fig. 5B), arguing that ent-cholesterol rather than any displaced endogenous cholesterol is able to induce the Scap conformational change.
DISCUSSION
How is cholesterol sensed in the mammalian cell to ensure that its levels are kept in check? Due to the lipophilic nature of cholesterol, it is almost exclusively localized to membranes, and consequently an intricate system has evolved, comprising an assortment of membrane-embedded proteins. The ER is home to many of these. Because the ER normally contains only a low level of cholesterol (4, 41), it is very responsive to small changes in total cell cholesterol levels. Scap is the ER cholesterol sensor for the SREBP transcriptional pathway and binds cholesterol at a saturable, specific receptor-like site, based on studies of cholesterol transfer to recombinant Scap in detergent micelles (17). The cholesterol-binding site was narrowed down to the first loop of Scap, which faces the ER lumen (8), consistent with data indicating that photocholesterol cross-links to this region of Scap (16).
However, these studies do not exclude the possibility that changes in the physical properties of the ER membrane may also contribute to cholesterol homeostasis. For example, the Scap conformational change tends to be induced by membrane-active sterols (5) and by certain amphiphiles known to exert membrane effects (6). Furthermore, processing of SREBP in cultured Drosophila cells is inhibited by a lipid derived from palmitate and ethanolamine, most likely phosphatidylethanolamine, but not by sterols, which may also suggest a role for membrane effects (20). Moreover, treatment with the saturated fatty acid palmitate affected acute sterol homeostatic responses in mammalian cells (21). Thus, there remains the possibility that mammalian Scap also serves as a sensor of the physical properties of the ER membrane.
Because enantiomers have proved powerful tools for discriminating between membrane effects and specific protein interactions (23, 24), here we determined the effects of ent-cholesterol on a number of major parameters of cholesterol homeostasis. In agreement with previous work (36, 37), ent-cholesterol demonstrated selectivity in reacting with bacterial cholesterol oxidase (Fig. 1B) and the mammalian ER cholesterol-esterifying enzyme, ACAT (Fig. 1D). Notably, equivalent cholesterol loading with ent-cholesterol suppressed SREBP-2 processing (Fig. 2A) and gene activation (Fig. 2B), albeit not to the same extent as nat-cholesterol. Moreover, as shown previously for nat-cholesterol (5), ent-cholesterol induced a conformational change in Scap (Fig. 2C). These findings support the contention that in addition to binding cholesterol directly, Scap can also sense cholesterol-induced changes in the physical properties of the ER membrane.
Another key finding was that ent-cholesterol accelerated the proteasomal degradation of the ER resident enzyme SM (Fig. 3), an important control point in cholesterol synthesis beyond HMGCR (11). Previously, we showed that SM degradation requires its membrane-bound N-terminal domain and is restricted to selective sterols most similar in their membrane properties to cholesterol (e.g. desmosterol and ring-oxygenated sterols). Our current study with ent-cholesterol expands these observations by indicating that changes in membrane properties also contribute to the proteasomal degradation of SM.
What are the physical properties of the ER membrane that may be sensed by proteins like Scap and SM? Cholesterol is known to influence membrane fluidity and thickness (42). In addition, cholesterol may form particular structures in membranes like the hexagonal configuration also observed with phosphatidylethanolamine (20). There is also the notion that chemically active cholesterol may escape complexing with phospholipids, to become available for sensing (41, 43). Thus, side chain oxysterols like 25-HC or 27-hydroxycholesterol have been proposed to exert their effects on cholesterol homeostasis by shifting cholesterol from an inactive to active pool (44). However, such a shift is unlikely to explain the ability of ent-cholesterol to elicit homeostatic responses observed in this study. The CHO-7 cells used in the majority of our experiments can grow with minimal cholesterol, and hence their cholesterol status can be dialed down to extremely low levels (∼7 μg/mg cell protein), much lower than cell cholesterol levels previously shown to activate cholesterol esterification (>30 μg/mg cell protein) (40). Moreover, ent-cholesterol itself did not increase cholesterol esterification (Figs. 1D and 4B) but did suppress SREBP-2 target gene expression (Fig. 2B) and alter Scap conformation in isolated membranes in vitro (Fig. 2C), even when steps were taken to eliminate potential confounding effects from endogenous cholesterol (Fig. 5). Our results therefore indicate that ent-cholesterol rather than endogenous cholesterol is able to induce Scap conformational change and suppress SREBP-2-mediated gene transcription. Additional experiments would be required to determine whether ent-cholesterol can bind directly to Scap.
Our collective data indicate, albeit indirectly, that ent-cholesterol reaches the ER to suppress SREBP-2 processing and gene activation, as well as to degrade the ER resident enzyme SM. Although direct analysis of ER cholesterol will complement our data, this was not possible in the present study due to limited stocks of ent-cholesterol.
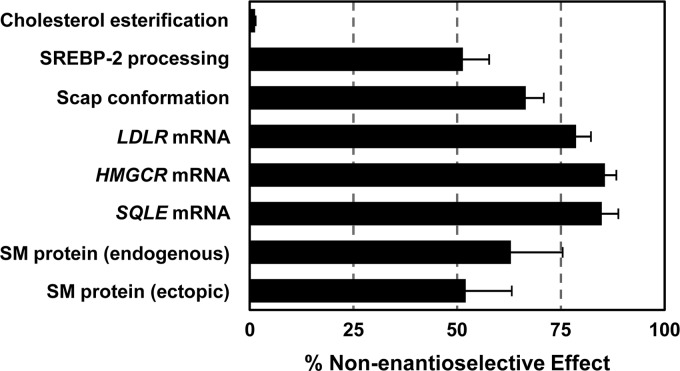
The relative nonenantioselective effects on the various cholesterol homeostatic parameters measured were estimated and are summarized in Fig. 6. Cholesterol esterification was almost exclusively enantioselective, whereas we estimated that the nonenantioselective effects of cholesterol contributed to at least 50% for parameters associated with SREBP-2 activation and SM degradation. However, we cannot rule out the possibility that the apparent lower efficiency of ent-cholesterol compared with nat-cholesterol may be at least in part due to a lower efficiency of delivery to the ER. This outcome is possible if the movement of cholesterol from the plasma membrane to the ER is protein-mediated (i.e. enantioselective). Further work will be required to define more precisely the contribution of altered membrane properties versus specific binding to proteins in cholesterol homeostasis.
FIGURE 6.
Summary of the estimated nonenantioselective effects on various cholesterol homeostatic parameters. Data from Figs. 1D, 2, and 3 (sharing the same culturing conditions) were used to estimate the nonenantioselective effects, calculated as the difference of ent-cholesterol from the nontreated condition, expressed as a percentage of the difference of nat-cholesterol from the nontreated condition (n ≥ 3, ±S.E. (error bars)). It should be noted that these values assume that equivalent amounts of nat-cholesterol and ent-cholesterol reached the ER (not directly measured).
Thus, in summary, we found that ent-cholesterol shares many of the same homeostatic responses as nat-cholesterol. Our data provide strong evidence that there are molecular contributions to cholesterol homeostasis beyond those specific cholesterol-protein interactions currently recognized and are consistent with an additional role of membrane effects in cholesterol homeostasis.
Acknowledgments
We thank members of the Brown Research Laboratory for critical feedback and colleagues for providing cell lines and reagents in this study.
This work was supported by the National Health and Medical Research Council Grant 1008081, the Prostate Cancer Foundation Grant PG2710, and the National Heart Foundation of Australia Grant G11S 5757 (to the Brown Research Laboratory).
- SREBP
- sterol regulatory element-binding protein
- ACAT
- acyl-CoA:cholesterol acyltransferase
- CD
- methyl-β-cyclodextrin
- CHO-7
- Chinese hamster ovary-7
- ent-cholesterol
- enantiomer of cholesterol
- ER
- endoplasmic reticulum
- 25-HC
- 25-hydroxycholesterol
- HMGCR
- 3-hydroxy-3-methylglutaryl-coenzyme A reductase
- HPCD
- 2-hydroxypropyl-β-cyclodextrin
- LPDS
- lipoprotein-deficient serum
- nat-cholesterol
- native cholesterol
- NCS
- newborn calf serum
- Scap
- SREBP cleavage-activating protein
- SM
- squalene monooxygenase
- TK
- thymidine kinase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 2. Maxfield F. R., van Meer G. (2010) Cholesterol, the central lipid of mammalian cells. Curr. Opin. Cell Biol. 22, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown M. S., Goldstein J. L. (2009) Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J. Lipid Res. 50, S15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. (2002) Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10, 237–245 [DOI] [PubMed] [Google Scholar]
- 6. Adams C. M., Goldstein J. L., Brown M. S. (2003) Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proc. Natl. Acad. Sci. U.S.A. 100, 10647–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feramisco J. D., Radhakrishnan A., Ikeda Y., Reitz J., Brown M. S., Goldstein J. L. (2005) Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl. Acad. Sci. U.S.A. 102, 3242–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Motamed M., Zhang Y., Wang M. L., Seemann J., Kwon H. J., Goldstein J. L., Brown M. S. (2011) Identification of luminal loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J. Biol. Chem. 286, 18002–18012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 10. Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill S., Stevenson J., Kristiana I., Brown A. J. (2011) Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13, 260–273 [DOI] [PubMed] [Google Scholar]
- 12. Gill S., Chow R., Brown A. J. (2008) Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog. Lipid Res. 47, 391–404 [DOI] [PubMed] [Google Scholar]
- 13. Wong J., Quinn C. M., Gelissen I. C., Brown A. J. (2008) Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J. Biol. Chem. 283, 700–707 [DOI] [PubMed] [Google Scholar]
- 14. Song B. L., Javitt N. B., DeBose-Boyd R. A. (2005) Insig-mediated degradation of HMG-CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 [DOI] [PubMed] [Google Scholar]
- 15. Lange Y., Ory D. S., Ye J., Lanier M. H., Hsu F. F., Steck T. L. (2008) Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 283, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 16. Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., Goldstein J. L. (2004) Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279, 52772–52780 [DOI] [PubMed] [Google Scholar]
- 17. Radhakrishnan A., Sun L. P., Kwon H. J., Brown M. S., Goldstein J. L. (2004) Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell 15, 259–268 [DOI] [PubMed] [Google Scholar]
- 18. Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gimpl G. (2010) Cholesterol-protein interaction: methods and cholesterol reporter molecules. Subcell. Biochem. 51, 1–45 [DOI] [PubMed] [Google Scholar]
- 20. Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. (2002) Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296, 879–883 [DOI] [PubMed] [Google Scholar]
- 21. Gale S. E., Westover E. J., Dudley N., Krishnan K., Merlin S., Scherrer D. E., Han X., Zhai X., Brockman H. L., Brown R. E., Covey D. F., Schaffer J. E., Schlesinger P., Ory D. S. (2009) Side chain oxygenated cholesterol regulates cellular cholesterol homeostasis through direct sterol-membrane interactions. J. Biol. Chem. 284, 1755–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu F., Rychnovsky S. D., Belani J. D., Hobbs H. H., Cohen J. C., Rawson R. B. (2005) Dual roles for cholesterol in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 102, 14551–14556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mickus D. E., Levitt D. G., Rychnovsky S. D. (1992) Enantiomeric cholesterol as a probe to ion-channel structure. J. Am. Chem. Soc. 114, 359–360 [Google Scholar]
- 24. Westover E. J., Covey D. F. (2004) The enantiomer of cholesterol. J. Membr. Biol. 202, 61–72 [DOI] [PubMed] [Google Scholar]
- 25. Goldstein J. L., Basu S. K., Brown M. S. (1983) Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 26. Belani J. D., Rychnovsky S. D. (2008) A concise synthesis of ent-cholesterol. J. Org. Chem. 73, 2768–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitchell C. R., Armstrong D. W. (2004) Cyclodextrin-based chiral stationary phases for liquid chromatography: a twenty-year overview. Methods Mol. Biol. 243, 61–112 [DOI] [PubMed] [Google Scholar]
- 28. Gelissen I. C., Brown A. J., Mander E. L., Kritharides L., Dean R. T., Jessup W. (1996) Sterol efflux is impaired from macrophage foam cells selectively enriched with 7-ketocholesterol. J. Biol. Chem. 271, 17852–17860 [DOI] [PubMed] [Google Scholar]
- 29. Kristiana I., Sharpe L. J., Catts V. S., Lutze-Mann L. H., Brown A. J. (2010) Antipsychotic drugs up-regulate lipogenic gene expression by disrupting intracellular trafficking of lipoprotein-derived cholesterol. Pharmacogenomics J. 10, 396–407 [DOI] [PubMed] [Google Scholar]
- 30. Lange Y., Ye J., Steck T. L. (2002) Effect of protein kinase C on endoplasmic reticulum cholesterol. Biochem. Biophys. Res. Commun. 290, 488–493 [DOI] [PubMed] [Google Scholar]
- 31. Du X., Pham Y. H., Brown A. J. (2004) Effects of 25-hydroxycholesterol on cholesterol esterification and sterol regulatory element-binding protein processing are dissociable: implications for cholesterol movement to the regulatory pool in the endoplasmic reticulum. J. Biol. Chem. 279, 47010–47016 [DOI] [PubMed] [Google Scholar]
- 32. Yang J., Brown M. S., Ho Y. K., Goldstein J. L. (1995) Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines: role of introns in protein evolution. J. Biol. Chem. 270, 12152–12161 [DOI] [PubMed] [Google Scholar]
- 33. Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. (1997) Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272, 20213–20221 [DOI] [PubMed] [Google Scholar]
- 34. Du X., Kristiana I., Wong J., Brown A. J. (2006) Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol. Biol. Cell 17, 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wong J., Quinn C. M., Brown A. J. (2006) SREBP-2 positively regulates transcription of the cholesterol efflux gene, ABCA1, by generating oxysterol ligands for LXR. Biochem. J. 400, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luker G. D., Pica C. M., Kumar A. S., Covey D. F., Piwnica-Worms D. (2000) Effects of cholesterol and enantiomeric cholesterol on P-glycoprotein localization and function in low-density membrane domains. Biochemistry 39, 7651–7661 [DOI] [PubMed] [Google Scholar]
- 37. Liu J., Chang C. C., Westover E. J., Covey D. F., Chang T. Y. (2005) Investigating the allosterism of acyl-CoA:cholesterol acyltransferase (ACAT) by using various sterols: in vitro and intact cell studies. Biochem. J. 391, 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lange Y., Steck T. L., Ye J., Lanier M. H., Molugu V., Ory D. (2009) Regulation of fibroblast mitochondrial 27-hydroxycholesterol production by active plasma membrane cholesterol. J. Lipid Res. 50, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. (1989) Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 264, 15634–15641 [PubMed] [Google Scholar]
- 40. Lange Y., Ye J., Rigney M., Steck T. L. (1999) Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J. Lipid Res. 40, 2264–2270 [PubMed] [Google Scholar]
- 41. Steck T. L., Lange Y. (2010) Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 20, 680–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaudhuri A., Chattopadhyay A. (2011) Transbilayer organization of membrane cholesterol at low concentrations: implications in health and disease. Biochim. Biophys. Acta 1808, 19–25 [DOI] [PubMed] [Google Scholar]
- 43. Radhakrishnan A., McConnell H. M. (2000) Chemical activity of cholesterol in membranes. Biochemistry 39, 8119–8124 [DOI] [PubMed] [Google Scholar]
- 44. Bielska A. A., Schlesinger P., Covey D. F., Ory D. S. (2012) Oxysterols as nongenomic regulators of cholesterol homeostasis. Trends Endocrinol. Metab. 23, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]