Abstract
Introduction
Genetic variability affects clinical outcome in pediatric acute lymphocytic leukemia (ALL) patients. Evaluating gene polymorphisms in ABC transporters could help identify relapse risk and predict outcome.
Material and methods
The SNaPshot SNP technique was used to analyze single-nucleotide polymorphisms (SNPs) in the multidrug transporter 1 (MDR1), multidrug resistance associated proteins (MRP1, MRP2) and breast cancer resistance protein (BCRP) genes of 82 pediatric ALL patients. The association between the SNPs with risk of all events and death as well as with survival was evaluated by the univariate Cox proportional hazard model.
Results
The BCRP G34A SNP was the only SNP significantly associated with ALL. Risk factors included pre-treatment WBC counts and post-treatment peripheral and bone marrow leukemic cell counts. We found no association between MDR1 SNPs with these factors. The BCRP C421A C/A and C/C genotypes were significantly associated with low pre-treatment WBC counts while MRP2 G1249A G/G was significantly associated with low levels of post-treatment peripheral and bone marrow leukemic cells. A combination of C1236T, G1249A and/or G34A SNPs was significantly associated with lower EFS and OS.
Conclusions
Polymorphisms associated with risk of ALL and clinical outcome may be potential biomarkers to predict clinical outcome and improve prognosis in childhood ALL.
Keywords: acute lymphoblastic leukemia, single nucleotide polymorphism, survival, outcome
Introduction
Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy worldwide [1, 2]. The ALL is a heterogeneous group of disorders that arise from the clonal proliferation of malignant, lymphoid cells in the bone marrow, blood and other organs [3]. Children between 1 and 10 years of age are most likely to develop ALL and have an overall cure rate of about 80%. Cases in adults are more likely to result from chromosomal abnormalities that make treatment more difficult. The overall survival rates for adult ALL are significantly lower and the frequency of relapses higher compared to childhood ALL [2]. There are a number of variations in clinical characteristics and outcomes seen in pediatric ALL patients, which are thought to be the result of diversity in the genetic backgrounds, mutations and chromosomal changes in leukemic blasts [4]. Response to therapy is evaluated by determining the status of minimal residual disease (MRD), which is important in risk stratification of ALL patients [5]. The most important prognostic factors in ALL are pre-treatment white blood cell (WBC) counts and the early response to treatment [6, 7].
Treatment regimens for ALL are complex and consist of induction chemotherapy (to reduce lymphoblast numbers in the bone marrow and to restore hematopoietic function), consolidation therapy (to eliminate residual disease) and maintenance chemotherapy (to prevent relapse) [8]. Current therapeutic options include the use of 1) thiopurine antimetabolites such as mercaptopurine (MP) and thioguanine (TG); 2) high dose methotrexate (HDMTX) given as consolidation therapy and low dose methotrexate (LDMTX) given as continuation therapy; 3) synthetic glucocorticoids such as dexamethasone and prednisolone, which induce G1 cell cycle arrest and apoptosis, and 4) L-asparaginase [9]. Pediatric ALL patients are also subjected to total body irradiation prior to bone marrow transplant [10, 11].
Single nucleotide polymorphisms (SNPs) are the most frequently inherited sequence variations in a particular gene and occur every 100-300 bp [9]. Such genetic polymorphisms in genes mediating drug transport and drug metabolism have been suggested to play an important role in the variability in survival after cancer therapy [12]. A recent genome-wide analysis showed an association between 102 SNPs and MRD in childhood ALL, suggesting that host genetic variability plays an important role in dictating treatment response [5]. Specific inherited polymorphisms were also shown to be correlated with drug toxicity in leukemias [13].
ATP-binding cassette (ABC) transporters are transmembrane proteins that are involved in ATP-dependent transport of specific substrates across lipid membranes [14]. The ABC transporters play a role in transport of drugs across membranes, excretion of substances out of the cell and in lipid transport. Mutations in these transporters have been shown to be associated with human disease [15]. The three major proteins comprising the ABC family of transporters are 1) MDR1 or ABCB1 (encoding the P-glycoprotein), 2) multidrug resistance associated protein 1 (MRP1) and 3) breast cancer resistance protein (BCRP) [16]. MDR1 expression has been shown to be upregulated in a number of chemoresistant tumors such as renal cell, adrenocortical, hepatocellular and colon cancers and in over 50% of relapsed acute myeloblastic leukemia (AML) patients, suggesting that it is a marker of poor prognosis [17]. P-glycoprotein (P-gp), the product of the MDR1 gene, plays an important role in steroid metabolism and in the export of metabolites, carcinogens and cytotoxic drugs such as anthracyclines, taxanes, vinca alkaloids and epipodophyllotoxins [18–20]. A number of SNPs in the MDR1 gene were recently shown to result in altered expression and functioning of the P-gp protein [18]. Analysis of the C3435T polymorphism of the MDR1 gene showed that the TT genotype was associated with development of childhood ALL, while the CC genotype was associated with poor prognosis [21, 22]. Similarly, patients with high levels of MRP were shown to have a poor prognosis, independent of age [23]. Upregulation of MDR1 in AML and MRP1 in ALL were also associated with response to induction chemotherapy [24].
Although MRP1 and MRP2 have not been specifically associated with prognosis of childhood ALL, the MRP genes have been reported to play a role in extruding substrates such as doxorubicin, etoposide, vincristine and 6-mercaptopurine, which are used to treat ALL [23]. Upregulation of BCRP expression has been suggested to confer drug resistance in ALL [25]. However, it is not clear if BCRP expression levels are associated with prognosis [26, 27]. Nevertheless, these data collectively suggest that SNP analyses of genes such as the ABC transporters could be an important tool in optimizing the efficacy of chemotherapy in ALL patients. Understanding the effect of specific SNPs on the clinical outcome and identifying disease-associated SNPs would help in early identification of ALL patients at risk for relapse.
In this study, we analyzed a total of 9 SNPs in the MDR1, MRP1, MRP2 and BCRP genes and determined the correlation between the SNPs, risk of ALL and clinical outcome in pediatric patients with ALL.
Material and methods
Patients
This study recruited 138 children with ALL who presented for the first time at the Affiliated Children's Hospital of Fudan University between May 2003 and March 2008. The patients were recruited consecutively. ALL diagnosis was based on the criteria developed by WHO and conformed to the FAB (French-American-British) classification system. The study also recruited 93 healthy children to serve as controls. All study participants were Han Chinese. Of the 138 patients who were initially recruited, 56 patients were excluded due to loss of follow-up and only 82 patients completed the study protocol. Complete information on the treatment and follow-up regimens was available for all 82 patients who completed the study. The study protocol was approved by the Ethics Committee of the hospital and informed consent was obtained from the families of all study participants, since the study participants were children. The blood samples obtained for this study were for the sole use of this study and families were immediately informed in the case of a significant, positive result.
Treatment regimen
Children with ALL were classified into low risk, moderate risk or high risk groups and treated based on the modified BFM-2002 regimen [28, 29]. Age, pathological type, immunophenotype, peripheral white blood cell count at disease onset, presence of peripheral leukemic cells at 7 days after prednisone test (day 8), and the proportion of bone marrow leukemic cells at 15 and 33 days after treatment (days 15 or 33) were all used as prognostic indicators (Table I).
Table I.
Patients’ characteristics
| Variable | N = 82 | |
|---|---|---|
| Baseline characteristics | ||
| Age [year] | 4.6 (3.3, 7.0), 1.0-13.2 | |
| Age category | 1-6 years > 6 years |
54 (65.9%) 28 (34.1%) |
| Gender | Male Female |
53 (64.6%) 29 (35.4%) |
| Risk group | Standard Intermedia High |
28 (34.1%) 39 (47.6%) 15 (18.3%) |
| Immunology | T cell Common B cell Pre-B cell Early pre-B cell |
6 (7.3%) 59 (72.0%) 8 (9.8%) 9 (11.0%) |
| Pathologic types | L1 L1/L2 L2 L3 |
52 (63.4%) 16 (19.5%) 13 (15.9%) 1 (1.2%) |
| CNS involvement | – + |
78 (95.1%) 4 (4.9%) |
| Testicular involvement | – + |
79 (96.3%) 3 (3.7%) |
| TEL/AML1 | – + |
70 (85.4%) 12 (14.6%) |
| E2A/PBX1 | – + |
81 (98.8%) 1 (1.2%) |
| BCR/ABL | – + |
81 (98.8%) 1 (1.2%) |
| WBC (109/l) | < 20 20-100 > 100 |
54 (65.9%) 22 (26.8%) 6 (7.3%) |
| Post-treatment characteristics | ||
| D8 peripheral leukemic cells [/μl] | ≤ 1000 > 1000 |
78 (95.1%) 4 (4.9%) |
| D15 bone marrow leukemic cells [%] | None < 5 5-25 > 25 |
41 (50.0%) 23 (28.0%) 10 (12.2%) 8 (9.8%) |
| D33 bone marrow leukemic cells [%] | None < 5 5-25 > 25 |
53 (64.6%) 27 (32.9%) 1 (1.2%) 1 (1.2%) |
Data are presented as count and percentage except for age, which is presented as median, inter-quartile range, and full range
Chemotherapy was performed regularly during the treatment regimen. Intrathecal injection was administered at designated time points and routine blood tests were performed every 1-2 weeks during maintenance therapy. After therapy discontinuation, follow-up was performed every three months in the first year, every 6 months in the second and third year, and thereafter every year for 3 years. Routine blood tests were performed at every follow-up visit and bone marrow aspirates were examined to determine if the patients were in remission. Liver and kidney function tests, echocardiography and ultrasonography of abdomen were also performed during follow-up visits.
Multiplex PCR and SNP analysis
Lymphocytes were isolated from peripheral blood of 175 children including 82 children with ALL and 93 controls. DNA was extracted from blood using Qiagen DNA Blood Kit (Qiagen, Hilden, Germany). A total of 9 SNPs in the MDR1, MRP1, MRP2 and BCRP genes were analyzed in a total of 175 samples using the SNaPshot SNP technique [1, 2]. Briefly, a total of 9 segments (111-330 bp in length) were amplified using 9 pairs of primers (Sangon, Shanghai, China). All primers, as listed in Table II, were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi).
Table II.
Primer sequences (5’-3’) used for PCR
| Mutation | PCR primers | Exon | |
|---|---|---|---|
| MDR1 C3435T | F gagcccatcctgtttgactgc R tgtatgttggcctcctttgctg |
26 | rs1045642 |
| MDR1 G2677T/A | F cccatcattgcaatagcaggagt R gcatgaaaaagattgctttgagga |
21 | rs2032582 |
| MDR1 C1236T | F tcagttcctatatcctgtgtctgtgaa R ccacagccactgtttccaacc |
12 | rs1128503 |
| MRP1 T825C | F gtggtagggggctgcatctct R aagcctccacctcctcattcg |
8 | rs246221 |
| MRP2 C24T | F ccagcatgattcctggactgc R cgattaaatggttgggatgaaagg |
1 | rs717620 |
| MRP2 G1249A | F tggctttgtccatgggtccta R gggcatccacagacatcaggt |
10 | rs2273697 |
| MRP2 C3972T | F cactccacctaccttctccatgc R ccagtttaacaactaccaagtgcggta |
28 | rs3740066 |
| BCRP C421A | F gttgtgatgggcactctgacg R tgaccctgttaatccgttcgttt |
5 | rs2231142 |
| BCRP G34A | F ccagatgtcttccagtaatgtcgaa R cgacaaggtagaaagccactcttca |
2 | rs2231137 |
Multiplex PCR was carried out using HotStarTaq (Qiagen, Dusseldorf, Germany) according to the manufacturer's instructions. The reaction was performed in a total volume of 20 μl containing 1× HotStarTaq buffer, 3.0 mM Mg2 +, 0.3 mM dNTP, 1 U HotStarTaq polymerase (Qiagen), 0.1 μM primers and 1 μl of DNA. Cycling conditions were as follows: 95°C for 15 min; 12 cycles of (94°C for 20 s, 65 ±0.5°C/cycle for 40 s, 72°C for 1 min 40 s); 23 cycles of (94°C for 20 s, 59°C for 30 s, 72°C for 1.5 min); 72°C for 2 min. PCR products were purified using shrimp alkaline enzyme (SAP; Promega, Wisconsin, USA) and exonuclease I (EXO I; EpiCentre, Palmerston North, New Zealand) according to the manufacturer's instructions.
Purified PCR products from 2 panels were mixed and used as a template for extension. Extension was performed in a total volume of 10 μl comprising 5 μl of SNaPshot Multiplex reaction mix (ABI, California, USA), 2 μl of purified PCR products, 1 μl of primers for extension (0.8 μM rs1045642SR, rs2231142SR, rs1128503SR, rs246221SR, rs2273697SR, rs3740066SR, rs2032582SR, rs2231137SF, rs717620SR) and 2 μl of ultra-pure water. Extension was performed using the SNaPshot Multiplex kit (ABI, USA) under the following conditions: 96°C 1 min; 28 cycles of (96°C for 10 s, 50°C for 5 s, 60°C for 30 s); 60°C for 1 min; hold at 4°C.
Extension products were purified using SAP (Promega, California, USA) and loaded onto an ABI 3130XL sequencer (ABI, California, USA) for sequencing. The raw data from the ABI 3130XL sequencer were subjected to analysis with GeneMapper 4.0 (Applied Biosystems, California, USA).
Statistical analysis
The continuous variable age was expressed by median with inter-quartile range and full range. Other category data were expressed as counts with percentages, and the associations between the category variables were tested with Fisher's exact test. We used the χ2 fitness test to test the Hardy-Weinberg equilibrium in the experimental and control groups for the SNPs. The univariate Cox proportional hazard model was used to evaluate the risk factors of all events and death. Since our sample size was small, we did not find a statistical significance between any two variables using the multivariate Cox proportional hazard model and therefore did not include the multivariate Cox proportional hazard model in the results. Kaplan-Meier survival curves described the event-free survival and overall survival rates, and the log-rank test was performed to compare the Kaplan-Meier survival curves. All statistical hypothesis tests were two-sided using a significance level of 0.05. The statistical analyses were performed using the SPSS 15.0 software (SPSS Institute Inc., Chicago, IL).
Results
Association of MDR1, MRP1, MRP2 and BCRP SNPs with ALL
This study recruited a total of 138 children, newly diagnosed with ALL during 2003-2008. Of these, 25 children discontinued treatment or moved to a different hospital before start of treatment. Thirteen and eighteen children were lost to follow-up during the treatment and after treatment, respectively. The remaining 82 children with ALL (53 males and 29 females) were included in the statistical analysis. The study also recruited 93 healthy children as controls.
The ages of the 82 children included in the analyses ranged from 1.0 to 13.2 years (mean age; 5.5 years) and 65.9% of children were younger than 6 years old. The clinical characteristics of the patients are summarized in Table I.
The ALL and healthy control groups were analyzed using the Hardy-Weinberg equilibrium model. Only one SNP (G34A) in the BCRP gene was significantly associated with ALL. Our data showed that 11.8% and 49.5% of the children in the healthy control group had the A/A and G/A genotypes respectively, while only 4.9% and 39.0% in the ALL group had these genotypes (Table III).
Table III.
Frequency of SNPs in the patient and control groups
| Variable | Healthy control (n= 93) | ALL (n= 82) | Value of p | |
|---|---|---|---|---|
| T825C exon 8a Value of p for HWE |
C/C | 21 (24.4%) | 24 (32.0%) | 0.409 |
| C/T | 47 (54.7%) | 33 (44.0%) | ||
| T/T | 18 (20.9%) | 18 (24.0%) | ||
| 0.382 | 0.322 | |||
| C3435T exon 26 Value of p for HWE |
C/C | 40 (43.0%) | 39 (47.6%) | 0.775 |
| C/T | 40 (43.0%) | 34 (41.5%) | ||
| T/T | 13 (14.0%) | 9 (11.0%) | ||
| 0.559 | 0.700 | |||
| G2677T exon 21a Value of p for HWE |
A/A | 7 (7.5%) | 5 (6.2%) | 0.630 |
| G/A | 13 (14.0%) | 13 (16.0%) | ||
| G/G | 18 (19.4%) | 14 (17.3%) | ||
| G/T | 28 (30.1%) | 25 (30.9%) | ||
| T/A | 7 (7.5%) | 12 (14.8%) | ||
| T/T | 20 (21.5%) | 12 (14.8%) | ||
| 0.016 | 0.883 | |||
| C1236T exon 12 Value of p for HWE |
C/C | 18 (19.4%) | 16 (19.5%) | 0.289 |
| C/T | 32 (34.4%) | 37 (45.1%) | ||
| T/T | 43 (46.2%) | 29 (35.4%) | ||
| 0.013 | 0.501 | |||
| C421A exon 5 Value of p for HWE |
A/A | 8 (8.6%) | 8 (9.8%) | 0.733 |
| C/A | 42 (45.2%) | 41 (50.0%) | ||
| C/C | 43 (46.2%) | 33 (40.2%) | ||
| 0.614 | 0.353 | |||
| G34A exon 2 Value of p for HWE |
A/A | 11 (11.8%) | 4 (4.9%) | 0.043* |
| G/A | 46 (49.5%) | 32 (39.0%) | ||
| G/G | 36 (38.7%) | 46 (56.1%) | ||
| 0.523 | 0.599 | |||
| C24T exon 1 Value of p for HWE |
T/T | 6 (6.5%) | 3 (3.7%) | 0.418 |
| C/T | 27 (29.0%) | 31 (37.8%) | ||
| C/C | 60 (64.5%) | 48 (58.5%) | ||
| 0.232 | 0.458 | |||
| G1249A exon 10 Value of p for HWE |
A/A | 1 (1.1%) | 2 (2.4%) | 0.823 |
| G/A | 17 (18.3%) | 15 (18.3%) | ||
| G/G | 75 (80.6%) | 65 (79.3%) | ||
| 0.973 | 0.332 | |||
| C3972T exon 28 Value of p for HWE |
T/T | 6 (6.5%) | 3 (3.7%) | 0.141 |
| C/T | 29 (31.2%) | 37 (45.1%) | ||
| C/C | 58 (62.4%) | 42 (51.2%) | ||
| 0.371 | 0.132 |
There were 14 values missing (7 for each group) in T825C exon 8, and one value missing in G2677T exon 21 in the patient group.
Indicates that a significant association with group was observed for G34A exon 2
Association of MDR1, MRP1, MRP2 and BCRP SNPs with risk of all events and risk of death
Among the baseline characteristics analyzed, children with a pre-treatment WBC count > 100 × 109/l had a significantly higher risk of all events and a significantly higher risk of death (hazard ratios of 10.21, p < 0.001 and 10.64, p = 0.001, respectively). Of the post-treatment characteristics analyzed, children with a peripheral blood leukemic cell count greater than 1000 per μl on the 8th day were more likely to die than those with peripheral leukemic cells less than 1000 per μl (hazard ratio of 5.12, p = 0.036). Children with > 25% bone marrow leukemic cells on the 15th day had a significantly higher risk of all events and death (hazard ratios of 3.57, p = 0.032 and 5.20, p = 0.009, respectively). Similarly, children with bone marrow leukemic cells in the range of 5-25% on the 33rd day had a significantly higher risk of all events and death (hazard ratios of 27.17, p = 0.004 and 41.55, p = 0.002, respectively) (Table IV).
Table IV.
Univariate Cox regression models by patients’ characteristics and the 9 SNPs
| Variable | Risk of all events | Risk of death | |||
|---|---|---|---|---|---|
| HR (95% CI) | Value of p | HR (95% CI) | Value of p | ||
| Baseline characteristics | |||||
| Age | 1-6 years | – | – | ||
| > 6 years | 2.24 (0.86, 5.81) | 0.099 | 2.13 (0.72, 6.37) | 0.174 | |
| Gender | Female | – | – | ||
| Male | 0.97 (0.36, 2.62) | 0.952 | 1.19 (0.37, 3.86) | 0.774 | |
| Risk group | Standard | – | – | ||
| Intermedia | 1.88 (0.50, 7.10) | 0.350 | 1.80 (0.35, 9.30) | 0.481 | |
| High | 3.29 (0.81, 13.35) | 0.096 | 4.70 (0.94, 23.50) | 0.060 | |
| Immunology | T cell | – | 0.158 | – | 0.077 |
| Early pre-B cell | 1.47 (0.27, 8.08) | 0.655 | 1.41 (0.26, 7.74) | 0.691 | |
| Pre-B cell | 0.30 (0.03, 3.28) | 0.321 | NA | ||
| Common B cell | 0.43 (0.09, 1.98) | 0.279 | 0.29 (0.06, 1.43) | 0.129 | |
| Pathologic types | L1 | – | – | ||
| L1/L2 | 0.48 (0.11, 2.13) | 0.336 | 0.27 (0.04, 2.13) | 0.216 | |
| L2 | 0.69 (0.15, 3.09) | 0.629 | 0.40 (0.05, 3.07) | 0.375 | |
| L3 | NA | NA | |||
| CNS involvement | – | – | – | ||
| + | 2.84 (0.62, 12.93) | 0.178 | 3.88 (0.82, 18.23) | 0.086 | |
| Testicular involvement | – | – | – | ||
| + | NA | NA | |||
| TELAML1 | – | – | – | ||
| + | 0.71 (0.16, 3.12) | 0.650 | 0.42 (0.05, 3.22) | 0.401 | |
| E2APBX1 | – | – | – | ||
| + | NA | NA | |||
| BCRABL | – | – | – | ||
| + | 4.40 (0.57, 33.74) | 0.154 | 5.30 (0.68, 41.47) | 0.112 | |
| WBC (109/l) | < 20 | – | – | ||
| 20-100 | 0.92 (0.28, 3.00) | 0.891 | 0.91 (0.23, 3.53) | 0.891 | |
| > 100 | 10.21 (2.82, 36.98) | < 0.001* | 10.64 (2.50, 45.30) | 0.001* | |
| Genotypes | |||||
| T825C exon 8 | T/T | – | – | ||
| C/T | 0.46 (0.13, 1.60) | 0.223 | 0.46 (0.11, 1.83) | 0.269 | |
| C/C | 0.94 (0.30, 2.98) | 0.918 | 0.75 (0.20, 2.81) | 0.672 | |
| C3435T exon 26 | T/T | – | – | ||
| C/T | 1.08 (0.12, 9.68) | 0.948 | 0.74 (0.08, 7.20) | 0.797 | |
| C/C | 2.84 (0.37, 22.00) | 0.317 | 1.93 (0.24, 15.41) | 0.533 | |
| G2677T exon 21 | G/G | – | – | ||
| G/A | 0.80 (0.19, 3.37) | 0.761 | 1.38 (0.28, 6.87) | 0.696 | |
| G/T | 0.24 (0.05, 1.24) | 0.088 | 0.43 (0.07, 2.60) | 0.358 | |
| A/A | 2.33 (0.55, 9.95) | 0.252 | 2.60 (0.43, 15.66) | 0.297 | |
| T/T | 0.22 (0.03, 1.87) | 0.164 | 0.40 (0.04, 3.87) | 0.428 | |
| T/A | 0.72 (0.17, 2.99) | 0.647 | 0.77 (0.13, 4.61) | 0.775 | |
| G2677T/A exon21 | G/G + G/A + A/A | 3.15 (1.16, 8.57) | 0.025* | 2.62 (0.85, 8.07) | 0.093 |
| G/T + T/T +T/A | – | – | |||
| C1236T exon 12 | T/T | – | – | ||
| C/T | 3.04 (0.81, 11.35) | 0.099 | 1.81 (0.45, 7.29) | 0.406 | |
| C/C | 4.63 (1.08, 19.80) | 0.039* | 3.47 (0.76, 15.76) | 0.108 | |
| C421A exon 5 | C/C | – | – | ||
| C/A | 0.68 (0.25, 1.89) | 0.460 | 0.97 (0.30, 3.20) | 0.963 | |
| A/A | 1.05 (0.22, 4.96) | 0.949 | 1.71 (0.33, 8.85) | 0.520 | |
| G34A exon 2 | G/G | – | – | ||
| G/A | 0.86 (0.31, 2.44) | 0.781 | 1.10 (0.33, 3.63) | 0.879 | |
| A/A | 4.19 (0.89, 19.63) | 0.069 | 4.80 (0.96, 23.99) | 0.056 | |
| C24T exon 1 | C/C | – | – | ||
| C/T | 1.30 (0.50, 3.36) | 0.592 | 0.61 (0.19, 2.00) | 0.419 | |
| T/T | NA | NA | |||
| G1249A exon 10 | G/G | – | – | ||
| G/A | NA | NA | |||
| A/A | 8.27 (1.82, 37.59) | 0.006* | 13.07 (2.68, 63.64) | 0.001* | |
| C3972T exon 28 | C/C | – | – | ||
| C/T | 0.94 (0.36, 2.43) | 0.894 | 0.45 (0.14, 1.47) | 0.188 | |
| T/T | NA | NA | |||
| Post-treatment characteristics | |||||
| D8 peripheral leukemic cells [/l] | ≤ 1000 | – | – | ||
| > 1000 | 3.30 (0.75, 14.54) | 0.115 | 5.12 (1.11, 23.49) | 0.036* | |
| D15 bone marrow leukemic cells [%] | < 5 | – | – | ||
| 5-25 | 1.76 (0.48, 6.44) | 0.390 | 1.76 (0.36, 8.52) | 0.482 | |
| > 25 | 3.57 (1.12, 11.38) | 0.032* | 5.20 (1.52, 17.82) | 0.009* | |
| D33 bone marrow leukemic cells [%] | < 5 | – | – | ||
| 5-25 | 27.17 (2.82, 261.57) | 0.004* | 41.55 (3.76, 459.28) | 0.002* | |
| > 25 | 3.03 (0.38, 23.90) | 0.293 | 4.11 (0.51, 33.27) | 0.185 | |
NA – the corresponding odds ratio was not applicable due to zero or small count.
p < 0.05 indicates that a significant impact on the risk of all events, recurrence, or death was observed in the corresponding variable
SNP analyses demonstrated that patients with the G2677T/A polymorphism (G/G, G/A or A/A genotypes) in the MDR1 gene had a significantly higher risk of all events compared to those with G2677T/A polymorphism (G/T or T/T or T/A genotypes) (hazard ratio of 3.15, p = 0.025). Similarly, patients with the C1236T polymorphism of the C/C genotype in the MDR1 gene had a significantly higher risk of all events compared to those with the T/T genotype (hazard ratio of 4.63, p = 0.039). Patients with the MRP2 G1249A polymorphism of the A/A genotype had a significantly higher risk of all events and a significantly higher risk of death compared to those with the G/G genotype (hazard ratios of 8.27, p = 0.006, and 13.07, p = 0.001, respectively) (Table IV).
Effect of SNP combinations on EFS and OS
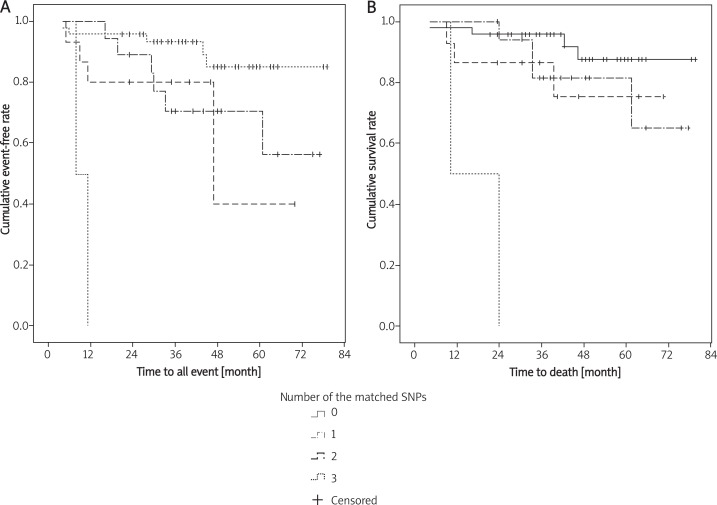
Based on our data showing a significantly higher risk of all events and death in 1) children with MDR1 G2677 T/A (G/G, G/A or A/A genotypes); 2) children with MDR1 C1236T of the C/C genotype; 3) children with MRP-2 G1249A of the A/A genotype; and 4) the association of BCRP G34A of the A/A genotype with ALL, we evaluated the effect of different combinations of these 4 polymorphisms on event-free survival (EFS) and overall survival (OS). We found significantly lower event-free survival in children with three matched SNPs when compared to children who had no matched SNPs (p < 0.001), one matched SNP (p < 0.001), or two matched SNPs (p = 0.014). Children with three matched SNP also had significantly lower overall survival compared to children with no matched SNPs (p < 0.001), one matched SNP (p < 0.001), or two matched SNPs (p = 0.006) (Figure 1).
Figure 1.
Survival analysis for EFS (A ) and OS (B ) using the combinations of the C1236, G1249 and the G34A
Association of MDR1, MRP1, MRP2 and BCRP SNPs with risk factors
Since we found that pretreatment WBC counts and post-treatment leukemic cell counts were significantly associated with risk of all events and risk of death (Table IV), we evaluated the association between the 9 SNPs with the pretreatment WBC, or the post-treatment leukemic cell counts. Our data showed no significant associations between the 3 MDR1 SNPs (C1236T, C3435T, and G2677T/A) and pretreatment WBC counts, post-treatment day 8 peripheral leukemic cell counts or day 15 and day 33 bone marrow leukemic cell counts (Table V). However, the BCRP C421A exon 5 SNP was significantly associated with pretreatment WBC counts. We found that 63.6% and 75.6% of children with the C/C and C/A genotypes, but only 25% of children with the A/A genotype, had pretreatment WBC counts less than 20 × 109/l (p = 0.025).
Table V.
The associations between MDR1 SNPs: C1236T, C3435T, G2677T/A and pre-treatment WBC counts in day 8 peripheral leukemic cells, and in day 15 and day 33 bone marrow leukemic cells
| Variable | G2677T/A | Value of p | C3435T exon 26 | Value of p | C1236T exon 12 | Value of p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T/T + T/G + T/A | A/A + A/G + G/G | C/C | C/T | T/T | C/C | C/T | T/T | |||||
| Pretreatment WBC [109/l] | < 20 | 33 (67.3%) | 20 (62.5%) | 0.437 | 24 (61.5%) | 25 (73.5%) | 5 (55.6%) | 0.564 | 11 (68.8%) | 25 (67.6%) | 18 (62.1%) | 0.270 |
| 20-100 | 14 (28.6%) | 8 (25.0%) | 11 (28.2%) | 7 (20.6%) | 4 (44.4%) | 2 (12.5%) | 10 (27.0%) | 10 (34.5%) | ||||
| > 100 | 2 (4.1%) | 4 (12.5%) | 4 (10.3%) | 2 (5.9%) | 0 (0.0%) | 3 (18.8%) | 2 (5.4%) | 1 (3.4%) | ||||
| Post-treatment | ||||||||||||
| D8 peripheral leukemic cells [/μl] | ≤1000 | 46 (93.9%) | 31 (96.9%) | 1.000 | 38 (97.4%) | 33 (97.1%) | 7 (77.8%) | 0.084 | 15 (93.8%) | 37 (100.0%) | 26 (89.7%) | 0.097 |
| > 1000 | 3 (6.1%) | 1 (3.1%) | 1 (2.6%) | 1 (2.9%) | 2 (22.2%) | 1 (6.3%) | 0 (0.0%) | 3 (10.3%) | ||||
| D15 bone marrow leukemic cells [%] | < 5 | 38 (77.6%) | 25 (78.1%) | 0.672 | 30 (76.9%) | 28 (82.4%) | 6 (66.7%) | 0.798 | 12 (75.0%) | 31 (83.8%) | 21 (72.4%) | 0.765 |
| 5-25 | 7 (14.3%) | 3 (9.4%) | 5 (12.8%) | 3 (8.8%) | 2 (22.2%) | 2 (12.5%) | 3 (8.1%) | 5 (17.2%) | ||||
| > 25 | 4 (8.2%) | 4 (12.5%) | 4 (10.3%) | 3 (8.8%) | 1 (11.1%) | 2 (12.5%) | 3 (8.1%) | 3 (10.3%) | ||||
| D33 bone marrow leukemic cells [%] | < 5 | 49 (100.0%) | 30 (93.8%) | 0.153 | 37 (94.9%) | 34 (100.0%) | 9 (100.0%) | 1.000 | 15 (93.8%) | 37 (100.0%) | 28 (96.6%) | 0.176 |
| 5-25 | 0 (0.0%) | 1 (3.1%) | 1 (2.6%) | 0 (0.0%) | 0 (0.0%) | 1 (6.3%) | 0 (0.0%) | 0 (0.0%) | ||||
| > 25 | 0 (0.0%) | 1 (3.1%) | 1 (2.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (3.4%) | ||||
The MRP2 G1249A SNP was significantly associated with post-treatment peripheral leukemic cell counts and bone marrow leukemic cell counts. Of the 17 children with the A/A or G/A genotypes, 4 had peripheral leukemic cell counts over 1000 per μl on day 8 after treatment, while none of the 65 children with the G/G genotype had peripheral leukemic cell counts over 1000 per μl (p < 0.001). Both children with G1249A in A/A had over 25% bone marrow leukemic cells on day 15 after treatment. Four of the 15 children (26.7%) with G1249A in G/A had over 5% bone marrow leukemic cells on day 15 after treatment, while only 12 of the 65 (18.5%) children with the G/G genotype had over 5% bone marrow leukemic cells on day 15 after treatment (p = 0.023). Fifteen children with the G/A genotype (100%) and 64 children with the G/G genotype (98.5%) had less than 5% bone marrow leukemic cells on day 33 after treatment, while one of the two children with the A/A genotype had 5-25% bone marrow leukemic cells on day 33 after treatment (p = 0.048) (Tables V-VII).
Table VII.
The associations between MRP2 SNPs: C24T exon 1, G1249A exon 10, and C3972T exon 28 and pre-treatment WBC counts in day 8 peripheral leukemic cells and in day 15 and day 33 bone marrow leukemic cells
| Variable | C24T exon 1 | Value of p | G1249A exon 10 | Value of p | C3972T exon 28 | Value of p | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T/T | C/T | C/C | A/A | G/A | G/G | T/T | C/T | C/C | |||||
| Pretreatment WBC [109/l] | < 20 | 1 (33.3%) | 20 (64.5%) | 33 (68.8%) | 0.265 | 1 (50.0%) | 13 (86.7%) | 40 (61.5%) | 0.109 | 1 (33.3%) | 24 (64.9%) | 29 (69.0%) | 0.386 |
| 20-100 | 1 (33.3%) | 10 (32.3%) | 11 (22.9%) | 0 (0.0%) | 2 (13.3%) | 20 (30.8%) | 1 (33.3%) | 11 (29.7%) | 10 (23.8%) | ||||
| > 100 | 1 (33.3%) | 1 (3.2%) | 4 (8.3%) | 1 (50.0%) | 0 (0.0%) | 5 (7.7%) | 1 (33.3%) | 2 (5.4%) | 3 (7.1%) | ||||
| Post-treatment | |||||||||||||
| D8 peripheral leukemic cells [/μl] | ≤ 1000 | 3 (100.0%) | 31 (100.0%) | 44 (91.7%) | 0.270 | 0 (0.0%) | 13 (86.7%) | 65 (100.0%) | < 0.001 | 3 (100.0%) | 37 (100.0%) | 38 (90.5%) | 0.243 |
| > 1000 | 0 (0.0%) | 0 (0.0%) | 4 (8.3%) | 2 (100.0%) | 2 (13.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 4 (9.5%) | ||||
| D15 bone marrow leukemic cells [%] | < 5 | 3 (100.0%) | 24 (77.4%) | 37 (77.1%) | 0.845 | 0 (0.0%) | 11 (73.3%) | 53 (81.5%) | 0.023 | 3 (100.0%) | 30 (81.1%) | 31 (73.8%) | 0.782 |
| 5-25 | 0 (0.0%) | 3 (9.7%) | 7 (14.6%) | 0 (0.0%) | 3 (20.0%) | 7 (10.8%) | 0 (0.0%) | 3 (8.1%) | 7 (16.7%) | ||||
| > 25 | 0 (0.0%) | 4 (12.9%) | 4 (8.3%) | 2 (100.0%) | 1 (6.7%) | 5 (7.7%) | 0 (0.0%) | 4 (10.8%) | 4 (9.5%) | ||||
| D33 bone marrow leukemic cells [%] | < 5 | 3 (100.0%) | 30 (96.8%) | 47 (97.9%) | 0.660 | 1 (50.0%) | 15 (100.0%) | 64 (98.5%) | 0.048 | 3 (100.0%) | 36 (97.3%) | 41 (97.6%) | 0.741 |
| 5-25 | 0 (0.0%) | 0 (0.0%) | 1 (2.1%) | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.4%) | ||||
| > 25 | 0 (0.0%) | 1 (3.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (1.5%) | 0 (0.0%) | 1 (2.7%) | 0 (0.0%) | ||||
Table VI.
The associations between MRP1 SNP: T825C exon 8, BCRP SNP: C421A exon 5, and BCRP SNP: G34A exon 2 and pre-treatment WBC counts in day 8 peripheral leukemic cells, and in day 15 and day 33 bone marrow leukemic cells
| Variable | T825C exon 8 | Value of p | C421A exon 5 | Value of p | G34A exon 2 | Value of p | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C/C | C/T | T/T | A/A | C/A | C/C | A/A | G/A | G/G | |||||
| Pretreatment WBC [109/l] | < 20 | 15 (62.5%) | 24 (72.7%) | 9 (50.0%) | 0.409 | 2 (25.0%) | 31 (75.6%) | 21 (63.6%) | 0.025* | 3 (75.0%) | 24 (75.0%) | 27 (58.7%) | 0.577 |
| 20-100 | 8 (33.3%) | 6 (18.2%) | 7 (38.9%) | 4 (50.0%) | 7 (17.1%) | 11 (33.3%) | 1 (25.0%) | 7 (21.9%) | 14 (30.4%) | ||||
| > 100 | 1 (4.2%) | 3 (9.1%) | 2 (11.1%) | 2 (25.0%) | 3 (7.3%) | 1 (3.0%) | 0 (0.0%) | 1 (3.1%) | 5 (10.9%) | ||||
| Post-treatment | |||||||||||||
| D8 peripheral leukemic [/μl] | ≤ 1000 | 23 (95.8%) | 32 (97.0%) | 16 (88.9%) | 0.450 | 7 (87.5%) | 40 (97.6%) | 31 (93.9%) | 0.300 | 4 (100.0%) | 30 (93.8%) | 44 (95.7%) | 1.000 |
| > 1000 | 1 (4.2%) | 1 (3.0%) | 2 (11.1%) | 1 (12.5%) | 1 (2.4%) | 2 (6.1%) | 0 (0.0%) | 2 (6.3%) | 2 (4.3%) | ||||
| D15 bone marrow leukemic cells [%] | < 5 | 21 (87.5%) | 28 (84.8%) | 12 (66.7%) | 0.258 | 6 (75.0%) | 34 (82.9%) | 24 (72.7%) | 0.636 | 4 (100.0%) | 25 (78.1%) | 35 (76.1%) | 0.902 |
| 5-25 | 1 (4.2%) | 4 (12.1%) | 3 (16.7%) | 1 (12.5%) | 3 (7.3%) | 6 (18.2%) | 0 (0.0%) | 3 (9.4%) | 7 (15.2%) | ||||
| > 25 | 2 (8.3%) | 1 (3.0%) | 3 (16.7%) | 1 (12.5%) | 4 (9.8%) | 3 (9.1%) | 0 (0.0%) | 4 (12.5%) | 4 (8.7%) | ||||
| D33 bone marrow leukemic cells [%] | < 5 | 23 (95.8%) | 33 (100.0%) | 17 (94.4%) | 0.211 | 7 (87.5%) | 40 (97.6%) | 33 (100.0%) | 0.187 | 4 (100.0%) | 31 (96.9%) | 45 (97.8%) | 0.688 |
| 5-25 | 0 (0.0%) | 0 (0.0%) | 1 (5.6%) | 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.2%) | ||||
| > 25 | 1 (4.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.4%) | 0 (0.0%) | 0 (0.0%) | 1 (3.1%) | 0 (0.0%) | ||||
Discussion
Although the cure rate for childhood ALL is as high as 80%, almost 25% of these children will relapse [30]. Multidrug resistance (MDR), mediated by a number of proteins in the ABC transporter family, remains an important challenge during treatment of acute leukemia. In this study, we analyzed the associations between 9 different SNPs in the MDR1, MRP1, MRP2 and BCRP genes, with risk of ALL or clinical outcome in pediatric patients with ALL. We also evaluated the effect of different combinations of the 9 SNPs on event-free survival (EFS) and overall survival (OS) among our study participants. We found that the BCRP G34A SNP was the only SNP significantly associated with ALL. Important risk factors included 1) pre-treatment WBC counts of > 100 × 109/l; 2) post-treatment peripheral leukemic cell counts of > 1000 on day 8 and 3) bone marrow leukemic cells > 25% on day 15 and 5-25% on day 33. Our data suggest that the 3 MDR1 SNPs that we analyzed were not associated with these risk factors. However, the BCRP C421A C/A and C/C genotypes were significantly associated with low pre-treatment WBC counts (< 20 × 109/l), while the MRP2 G1249A G/G genotype was significantly associated with low levels of post-treatment day 8 peripheral leukemic cells and day 15 and day 33 bone marrow leukemic cells. The MDR1 G2677 T/A polymorphism (G/G, G/A or A/A genotypes) was associated with a significantly higher risk of all events compared to the G2677 T/A polymorphism (G/T or T/T or T/A genotypes). Importantly, our data also suggest that a combination of MDR1 C1236T, MRP2 G1249A and/or BCRP G34A SNPs was significantly associated with lower EFS and OS.
Gene expression profiles have been shown to correlate with morphology, immunophenotype and response to therapy and drug resistance in childhood ALL patients [5, 31].The genetic polymorphisms in MDR1 are thought to influence the expression levels of the P-gp protein, thereby influencing the development of ALL [32]. Additionally, the C3435T polymorphism in exon 26 of the MDR1 gene has been suggested to result in an abnormal conformation of the P-gp protein and inhibition of its substrate-binding properties [33, 34]. The C3435T and the G2677G/A but not the C1236T SNPs in the MDR1 gene have been shown to be associated with pharmacokinetics of P-gp substrates such as cyclosporine, digoxin, and fexofenadine, but not vincristine, suggesting a role for these SNPs in drug metabolism in ALL patients [35]. Additionally, it was reported that the wild type MDR1 haplotypes 1236C-2677G-3435C conferred protection against the leukemogenic effect of pesticides [36]. MRP2 was shown to facilitate the transport of a number of anticancer agents such as cisplatin, vinblastine and camptothecin derivatives [37].
We evaluated the association between the MDR1, MRP2 and BCRP SNPs and ALL. Although the MDR1 C3435 polymorphism was previously shown to be associated with the risk of development as well as with clinical outcome of childhood ALL and adult ALL [20, 21], our present study showed that the BCRP G34A SNP was the only SNP that was significantly associated with ALL. The frequency of the T/T genotype of the MDR1 C3435T polymorphism was previously reported to be significantly higher in children with ALL when compared to controls, while the MDR1 2677G/G, 3435C/C and the 2677G-3435C polymorphisms were shown to be significantly associated with poor prognosis in pediatric Taiwanese ALL patients who received the TPOG-ALL-93-SR treatment regimen [38].
The MDR1 C3435T and G2677T/A polymorphisms were reported to be associated with decreased P-glycoprotein levels in the human placenta [39]. Our data showed no significant association between the MDR1 C3435T SNP and risk of death, although the G2677 T/A polymorphism (G/G, G/A or A/A genotypes) was associated with a significantly higher risk of all events compared to the G2677 T/A polymorphism (G/T or T/T or T/A genotypes). Patients with the MDR1 C1236T SNP (C/C genotype) also had a significantly higher risk of all events compared to those with C1236T polymorphism (T/T genotype). In contrast, the MRP2 G1249 SNP (A/A genotype) conferred a significantly higher risk of all events as well as a significantly higher risk of death compared to the G/G genotype.
BCRP expression was upregulated in ventricular samples from cardiomyopathic hearts and was shown to play an important role in multidrug resistance to a number of cytostatic drugs [37]. The BCRP C421A polymorphism was previously shown to be significantly associated with decreased drug resistance when compared to the wild type and the CA/AA genotype was shown to be associated with a higher risk of death when compared to the CC genotype in acute myeloid leukemia [40]. In contrast, our data showed that the BCRP C421A C/A and C/C genotypes were significantly associated with low levels of pretreatment WBC counts. Since low pretreatment WBC counts are a positive prognostic indicator, it is important to expand our understanding of the functional significance of the BCRP 421A C/A and C/C genotypes.
We determined the correlation of the MDR1, MRP2 and BCRP SNPs with survival in pediatric ALL patients. The MDR1 C3435T C/C genotype was previously shown to be associated with a significantly lower EFS and OS in a population of pediatric ALL patients [20]. High expression of all MRP genes, except MRP4, was associated with lower EFS [19, 23] and polymorphisms in the MRP4 gene were also shown to be associated with outcome in childhood ALL [41]. The BCRP G34A SNP AA/AG genotypes were shown to correlate with significantly improved survival compared to the GG genotype [40]. To the best of our knowledge, we are the first to analyze combinations of SNPs and to show that a combination of MDR1 (C1236T), MRP2 (G1249A) and/or the BRCP (G34A) SNPs was associated with a significantly lower EFS and OS in pediatric ALL patients.
Differences between data reported by the various studies have been attributed to a number of reasons such as differences in study design, variation in environmental exposure which could mask the effects of the SNPs or possibly the fact that many ABC transporter proteins bind to the same substrate [42]. It will also be interesting to investigate whether these differences between the data from different studies are due to genetic variation between different populations of ALL patients.
The most important limitation of this study was the small sample size. Factors contributing to the large number of patients lost to follow-up in this study included: 1) lack of national health insurance along with the high cost of medical care in China and 2) initiation of Chinese traditional treatments in preference over western medicine. Our present study is a preliminary, exploratory study, which serves as a useful pilot study based upon which we will design further studies. We plan to validate our data on a larger patient population. Another limitation of this study was that we did not analyze the correlation between the various SNPs and drug resistance in our ALL patients. It has been suggested that ALL patients with the 3435 CC genotype of the MDR1 gene could more efficiently eliminate P-gp substrates such as anthracyclines, daunorubicin, vincristine and mitoxantrone, resulting in poor prognosis [22]. We would like to evaluate the association between the MDR1 C1236T, MRP2 G1249A and/or BCRP G34A SNPs and response to specific chemotherapy regimens in our pediatric ALL patient population. Data from such studies could have important implications in designing therapy for pediatric ALL to overcome the negative prognostic impact conferred by specific SNPs. It will also be an important goal to explore the association of these SNPs with the risk of ALL and with clinical outcome in different ethnic populations and in the context of different drug regimens. We would also investigate the mechanisms underlying the differences in clinical outcome associated with specific SNPs by evaluating the relevant mRNA and protein levels in each SNP population. It will also be interesting to follow the children in the control group until adulthood to evaluate if any of them develop ALL.
In conclusion, we evaluated a population of pediatric ALL patients for 9 SNPs in the MDR1, MRP2 and BCRP genes. Based on our data, evaluation of MDR, MRP and BCRP SNPs at the time of presentation is an important tool to screen for high risk patients and for clinical management of childhood ALL. Polymorphisms associated with risk of ALL and clinical outcome are potential biomarkers which can be used to predict clinical outcome and improve prognosis in childhood ALL.
References
- 1.Howlader N, Noone AM, Krapcho M. SEER Cancer Statistics Review, 1975-2008. National Cancer Institute. SEER Website. http://seer.cancer.gov/csr/1975_2008/. Accessed November 10, 2011.
- 2.Pui CH, Evans WE. Acute lymphoblastic leukemia. N Engl J Med. 1998;339:605–15. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 3.Apostolidou E, Swords R, Alvarado Y, Giles FJ. Treatment of acute lymphoblastic leukaemia: a new era. Drugs. 2007;67:2153–71. doi: 10.2165/00003495-200767150-00004. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–78. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 5.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Angulo G, Yuen C, Palla SL, Anderson PM, Zweidler-McKay PA. Absolute lymphocyte count is a novel prognostic indicator in ALL and AML: implications for risk stratification and future studies. Cancer. 2008;112:407–15. doi: 10.1002/cncr.23168. [DOI] [PubMed] [Google Scholar]
- 7.Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360:2730–41. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock W. Adolescents and young adults with acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2010;2010:21–9. doi: 10.1182/asheducation-2010.1.21. [DOI] [PubMed] [Google Scholar]
- 9.Cheok MH, Pottier N, Kager L, Evans WE. Pharmacogenetics in acute lymphoblastic leukemia. Semin Hematol. 2009;46:39–51. doi: 10.1053/j.seminhematol.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattar E, Hammad L, Al-Mohammed H. Measurement and comparison of skin dose using OneDose MOSFET and mobile MOSFET for patients with acute lymphoblasic leukemia. Med Sci Monit. 2011;17:MT51–55. doi: 10.12659/MSM.881833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Mohammed H, Mahyoub F, Moftah B. Comparative study on skin dose measurement using MOSFET and TLD for pediatric patients with acute lymphoblastic leukemia. Med Sci Monit. 2010;16:CR325–9. [PubMed] [Google Scholar]
- 12.Ekhart C, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. An overview of the relations between polymorphisms in drug metabolising enzymes and drug transporters and survival after cancer drug treatment. Cancer Treat Rev. 2009;35:18–31. doi: 10.1016/j.ctrv.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Kishi S, Cheng C, French D, et al. Ancestry and pharmacogenetics of antileukemic drug toxicity. Blood. 2007;109:4151–7. doi: 10.1182/blood-2006-10-054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarr PT, Tarling EJ, Bojanic DD, Edwards PA, Baldán A. Emerging new paradigms for ABCG transporters. Biochim Biophys Acta. 2009;1791:584–93. doi: 10.1016/j.bbalip.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 16.Lebedeva IV, Pande P, Patton WF. Sensitive and specific fluorescent probes for functional analysis of the three major types of mammalian ABC transporters. PLoS One. 2011;6:e22429. doi: 10.1371/journal.pone.0022429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8:411–24. doi: 10.1634/theoncologist.8-5-411. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh HJ, Park CJ, Jang S, et al. Prognostic significance of multidrug resistance gene 1 (MDR1), multidrug resistance-related protein (MRP) and lung resistance protein (LRP) mRNA expression in acute leukemia. J Korean Med Sci. 2006;21:253–8. doi: 10.3346/jkms.2006.21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamroziak K, Młynarski W, Balcerczak E, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72:314–21. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 21.Jamroziak K, Balcerczak E, Cebula B, et al. Multi-drug transporter MDR1 gene polymorphism and prognosis in adult acute lymphoblastic leukemia. Pharmacol Rep. 2005;57:882–8. [PubMed] [Google Scholar]
- 22.Rao DN, Anuradha C, Vishnupriya S, et al. Association of an MDR1 gene (C3435T) polymorphism with acute leukemia in India. Asian Pac J Cancer Prev. 2010;11:1063–6. [PubMed] [Google Scholar]
- 23.Plasschaert SL, de Bont ES, Boezen M, et al. Expression of multidrug resistance-associated proteins predicts prognosis in childhood and adult acute lymphoblastic leukemia. Clin Cancer Res. 2005;11:8661–8. doi: 10.1158/1078-0432.CCR-05-1096. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan PS, Bhushan B, Singh LC, et al. Expression of genes related to multiple drug resistance and apoptosis in acute leukemia: response to induction chemotherapy. Exp Mol Pathol. 2011;92:44–9. doi: 10.1016/j.yexmp.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Jiang XJ, Wang JS, Fang Q. Gene expression of breast cancer resistance protein in adult acute lymphocytic leukemia and its clinical significance [Chinese] J Exp Hematol. 2008;16:31–34. [PubMed] [Google Scholar]
- 26.Imanishi H, Okamura N, Yagi M, et al. Genetic polymorphisms associated with adverse events and elimination of methotrexate in childhood acute lymphoblastic leukemia and malignant lymphoma. J Hum Genet. 2007;52:166–71. doi: 10.1007/s10038-006-0096-z. [DOI] [PubMed] [Google Scholar]
- 27.Plasschaert SL, van der Kolk DM, de Bont ES, et al. The role of breast cancer resistance protein in acute lymphoblastic leukemia. Clin Cancer Res. 2003;9:5171–7. [PubMed] [Google Scholar]
- 28.Fronkova E, Mejstrikova E, Avigad S, et al. Minimal residual disease (MRD) analysis in the non-MRD-based ALL IC-BFM 2002 protocol for childhood ALL: is it possible to avoid MRD testing? Leukemia. 2008;22:989–97. doi: 10.1038/leu.2008.22. [DOI] [PubMed] [Google Scholar]
- 29.Möricke A, Zimmermann M, Reiter A, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–84. doi: 10.1038/leu.2009.257. [DOI] [PubMed] [Google Scholar]
- 30.Valera ET, Scrideli CA, Queiroz RG, Mori BM, Tone LG. Multiple drug resistance protein (MDR-1), multidrug resistance-related protein (MRP) and lung resistance protein (LRP) gene expression in childhood acute lymphoblastic leukemia. Sao Paulo Med J. 2004;122:166–71. doi: 10.1590/S1516-31802004000400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunger SP, Winick NJ, Sather HN, Carroll WL. Therapy of low-risk subsets of childhood acute lymphoblastic leukemia: when do we say enough? Pediatr Blood Cancer. 2005;45:876–80. doi: 10.1002/pbc.20501. [DOI] [PubMed] [Google Scholar]
- 32.Hattori H, Suminoe A, Wada M, et al. Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia. Leuk Res. 2007;31:1633–40. doi: 10.1016/j.leukres.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 34.Komar AA. Silent SNPs: impact on gene function and phenotype. Pharmacogenomics. 2007;8:1075–80. doi: 10.2217/14622416.8.8.1075. [DOI] [PubMed] [Google Scholar]
- 35.Plasschaert SL, Groninger E, Boezen M, et al. Influence of functional polymorphisms of the MDR1 gene on vincristine pharmacokinetics in childhood acute lymphoblastic leukemia. Clin Pharmacol Ther. 2004;76:220–9. doi: 10.1016/j.clpt.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Urayama KY, Wiencke JK, Buffler PA, Chokkalingam AP, Metayer C, Wiemels JL. MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev. 2007;16:1172–1177. doi: 10.1158/1055-9965.EPI-07-0007. [DOI] [PubMed] [Google Scholar]
- 37.Cascorbi I. Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol Ther. 2006;112:457–73. doi: 10.1016/j.pharmthera.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Yang YL, Lin DT, Chang SK, et al. Pharmacogenomic variations in treatment protocols for childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2010;54:206–11. doi: 10.1002/pbc.22292. [DOI] [PubMed] [Google Scholar]
- 39.Hitzl M, Schaeffeler E, Hocher B, et al. Variable expression of P-glycoprotein in the human placenta and its association with mutations of the multidrug resistance 1 gene (MDR1, ABCB1) Pharmacogenetics. 2004;14:309–18. doi: 10.1097/00008571-200405000-00006. [DOI] [PubMed] [Google Scholar]
- 40.Hampras SS, Sucheston L, Weiss J, et al. Genetic polymorphisms of ATP-binding cassette (ABC) proteins, overall survival and drug toxicity in patients with Acute Myeloid Leukemia. Int J Mol Epidemiol Genet. 2010;1:201–7. [PMC free article] [PubMed] [Google Scholar]
- 41.Ansari M, Sauty G, Labuda M, et al. Polymorphisms in multidrug resistance-associated protein gene 4 is associated with outcome in childhood acute lymphoblastic leukemia. Blood. 2009;114:1383–6. doi: 10.1182/blood-2008-11-191098. [DOI] [PubMed] [Google Scholar]
- 42.Jamroziak K, Robak T. Do polymorphisms in ABC transporter genes influence risk of childhood acute lymphoblastic leukemia? Leuk Res. 2008;32:1173–5. doi: 10.1016/j.leukres.2008.01.009. [DOI] [PubMed] [Google Scholar]