Abstract
Trimeric sodium channels of the DEG/ENaC family have important roles in neurons, but the specific functions of different subunits present in heteromeric channels are poorly understood. We previously reported that the Drosophila DEG/ENaC subunit Ppk25 is essential in a small subset of gustatory neurons for activation of male courtship behavior, likely through detection of female pheromones. Here we show that, like mutations in ppk25, mutations in another Drosophila DEG/ENaC subunit gene, nope, specifically impair male courtship of females. nope regulatory sequences drive reporter gene expression in gustatory neurons of the labellum wings, and legs, including all gustatory neurons in which ppk25 function is required for male courtship of females. In addition, gustatory-specific knockdown of nope impairs male courtship. Further, the impaired courtship response of nope mutant males to females is rescued by targeted expression of nope in the subset of gustatory neurons in which ppk25 functions. However, nope and ppk25 have nonredundant functions, as targeted expression of ppk25 does not compensate for the lack of nope and vice versa. Moreover, Nope and Ppk25 form specific complexes when coexpressed in cultured cells. Together, these data indicate that the Nope and Ppk25 polypeptides have specific, nonredundant functions in a subset of gustatory neurons required for activation of male courtship in response to females, and suggest the hypothesis that Nope and Ppk25 function as subunits of a heteromeric DEG/ENaC channel required for gustatory detection of female pheromones.
Introduction
The study of the elaborate courtship displayed by Drosophila males toward females presents a unique opportunity for understanding pheromonal control of a complex behavior at all levels, from the molecular events underlying specific chemosensory detection of pheromones, to the neuronal circuitry that integrates multiple signals, and ultimately controls behavior. The advantages of Drosophila as a model organism have resulted in a sophisticated understanding of the general mechanisms involved in both taste and smell (Dahanukar and Ray, 2011; Tunstall and Warr, 2012), as well as the genetic control and brain circuitry underlying male courtship (Billeter et al., 2006; Villella and Hall, 2008; Siwicki and Kravitz, 2009; Dauwalder, 2011). Furthermore, the chemical structure of a number of pheromones that either stimulate or inhibit male courtship (Ferveur, 2005; Wicker-Thomas, 2007; Levine and Millar, 2009), and both receptors and neurons involved in both olfactory and gustatory modulation of courtship have been described (Benton, 2007; Dickson, 2008; Vosshall, 2008; Ronderos and Smith, 2009; Stowers and Logan, 2010). Activation of courtship behavior involves both olfaction and taste, but while loss of olfaction decreases male courtship by approximately twofold (Stockinger et al., 2005; Krstic et al., 2009), males with impaired gustatory function display almost no courtship of females (Krstic et al., 2009). However, the neuronal circuitry and molecular mechanisms involved in gustatory activation of courtship have remained elusive. Recently, we reported that ppk25, a DEG/ENaC subunit gene with an essential role in activating male courtship (Lin et al., 2005), functions specifically in a small subset of peripheral taste neurons on the legs and wings of males (Starostina et al., 2012). In addition to identifying for the first time a subset of gustatory neurons required for activation of male courtship behavior, most likely through detection of female pheromones, this work demonstrated that a DEG/ENaC subunit is specifically required for the function of those neurons, but not for gustatory detection of sugars, or of pheromones that inhibit rather than activate courtship behavior. Here, we describe the discovery of nope, another DEG/ENaC subunit gene specifically required for male courtship of females. Normal male response to females requires expression of both nope and ppk25 in a common subset of gustatory neurons in legs and wings, indicating that each of these subunits has a unique, nonredundant role. Furthermore, Ppk25 and Nope form specific complexes when coexpressed in cultured cells, suggesting that they also interact in the subset of gustatory neurons in which they are coexpressed. These data provide further evidence that a specific subset of gustatory neurons is required to activate male courtship, support a critical role for DEG/ENaC channels in these neurons, and suggest that Ppk25 and Nope are subunits of a common heteromeric channel required for gustatory activation of male courtship in response to female pheromones.
Materials and Methods
Generation of the nope-Gal4 and UAS-nope transgenes and immunostaining.
The nope-Gal4 construct was generated by cloning a 3020 bp fragment upstream of the nope start codon flanked by PmeI and BglII sites into a derivative of the pP{CaSpeR-4} vector (Qian et al., 1991; Starostina et al., 2012) digested with PmeI and BamHI. In the resulting construct, the nope initiator ATG is fused in frame with the coding sequence of the yeast transcriptional activator Gal4, followed by SV40 polyadenylation sequences obtained from the pGATB vector (Brand and Dormand, 1995). Transgenic lines for nope-Gal4 were generated using standard procedures for random integration of P-element vectors (Rubin and Spradling, 1982). For generation of UAS-nope and UAS-nopealt, cDNAs were generated from RNA extracted from male front legs. Either the full-length 1496 bp nope cDNA or a 5 nt shorter form in which intron 10 is spliced out using an alternative 5′ splice site was cloned into pUASattB (Bischof et al., 2007) and integrated by Bestgene using site-specific integration at the attP2 locus on chromosome 3 (Bischof et al., 2007; Markstein et al., 2008). Immunostaining was performed as described by Starostina et al. (2012) based on a previous report (Laissue et al., 1999). Rabbit anti-GFP (Invitrogen) was used at a dilution of 1:200, and anti-nc82 (Developmental Studies Hybridoma Bank, supported by the NICHD at the University of Iowa, Iowa City, IA) at a dilution of 1:40.
Sequence analysis.
Unannotated Nope orthologs were initially identified in most Drosophila species by searching genomic sequences using TBLASTN (Johnson et al., 2008). Analysis of the corresponding genomic sequences yielded predicted spliced transcripts encoding proteins with extensive similarity to Drosophila melanogaster Nope throughout their sequence, confirming that they are likely Nope orthologs (our unpublished data). Sequences encoding Nope orthologs in Drosophila ananassae and Drosophila grimshawii overlap putative annotated genes GF13482 and GH20163, respectively. Sequence alignments were performed using ClustalW (Larkin et al., 2007) and displayed using the Boxshade 3.21 server at http://www.ch.embnet.org/.
Cell transfection and coimmunoprecipitation.
cDNAs encoding full-length Ppk25 and Nope were cloned into pCMV5 with C-terminal 3xFlag and 3xHA epitopes. A similar β-ENaC-HA construct with a 3xHA epitope was generated using a 1924bp β-ENaC human cDNA (Thermo Scientific Dharmacon). The Arm-2XHA pCMV5 clone was a kind gift from Hassina Benchabane (Geisel School of Medicine at Dartmouth, Hanover, NH) (Benchabane et al., 2011). Transfections and immunoprecipitations were according to published protocols (Meltzer et al., 2007) with the following modifications. HEK293T human embryonic kidney cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Transfected cells were lysed at 4°C in Lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1 mm EDTA) in the presence of protease inhibitors (Roche Diagnostics) and 1% Triton X-100 for 1 h. Cell lysates were collected, clarified by microcentrifugation for 15 min at 4°C and incubated for 1 h at 4°C with a 1:350 dilution of a 5 μg/μl solution of mouse monoclonal anti-Flag M2 antibody (Sigma-Aldrich). Twenty microliters of Protein A/G PLUS Agarose beads (Santa Cruz Biotechnology) was added to the extract and incubated for 1 h at 4°C. The beads were then pelleted and washed three times in Lysis buffer with 0.2% Triton X-100. Immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotted with rat anti-HA antibody (Roche Diagnostics).
Behavioral assays.
Conditions for fly rearing and courtship assays were as described by Starostina et al. (2012) with the following modifications. Courtship directed at females was assayed under infrared lights using 5- to 7-d-old w1118 virgin females that stood upright and preened after having been anesthetized and decapitated. Courtship directed at males was assayed using intact w1118 males under visible laboratory lights. Behavior was recorded with a digital Mini-DV camera for 10 min, scored blind, and analyzed using the LIFESONG X software (version 0.8) (Villella et al., 2005). The Courtship Index (CI) is the fraction of time the male spends performing any courtship behavior ×100 (Hall, 1978), and the Total Behavioral Index (TBI) is the fraction of the same 10 min period during which the male courts, walks, or preens ×100 (Starostina et al., 2012). For CI and TBI, error bars represent the SEM, and the Wilcoxon rank-sum test was used to determine statistical significance. For the fraction of males initiating courtship (number of males with nonzero CI divided by the total number of males tested), error bars indicate 95% confidence intervals for binomial distributions, and statistical significance was calculated using a χ2 test for equality of distributions.
Drosophila stocks.
Most fly lines used were obtained from the Bloomington Stock Center. The UAS-nopeRNAi line (stock number 27241) was obtained from the Transgenic RNAi Project at Harvard Medical School (Boston, MA) (Ni et al., 2009).
Results
A second DEG/ENaC subunit, Nope, is required for male response to females
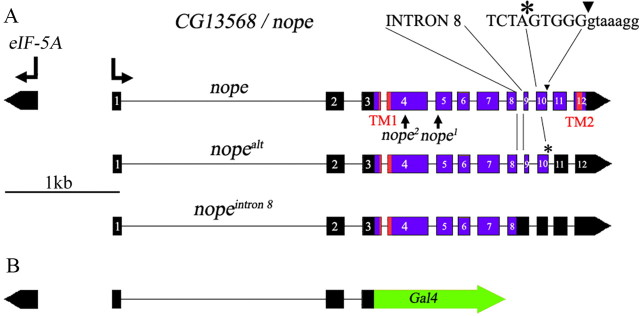
Our previous work demonstrated that the Ppk25 DEG/ENaC subunit is required specifically in gustatory neurons that activate male courtship behavior in response to females (Lin et al., 2005; Starostina et al., 2012). As DEG/ENaC channels are often composed of several distinct subunits, our findings suggested that other Drosophila DEG/ENaCs may also be required for activation of male courtship by females. We therefore tested whether males homozygous for mutations in other Drosophila DEG/ENaC subunit genes respond normally to females (Fig. 1A). Male courtship was analyzed in the presence of decapitated females to prevent behavioral feedback, and under infrared light, thereby preventing visual stimulation (Frank and Zimmerman, 1969; Lin et al., 2005). Courtship was quantitated using the CI, the fraction of a 10 min observation period in which a male displays any courtship behavior toward a female, multiplied by 100, as well as the fraction of males that initiate courtship during the observation period (Villella and Hall, 2008; Starostina et al., 2012). While most mutants tested court females at normal levels, males homozygous for the f06838 mutation displayed significant reductions in both CI and the fraction of males initiating courtship (Fig. 1A). The f06838 mutation corresponds to the insertion of a transposable element in exon 5 of a previously uncharacterized gene, CG13568, which we have named nope (for not passionate; Fig. 2A), which encodes a 425 aa protein. Interestingly, RT-PCR analysis indicates that, in addition to nope transcripts encoding a full-length protein, alternative splicing patterns result in nope transcripts that either retain intron 8 or use a different 5′ splice site for intron 10, in both cases resulting in shorter open reading frames encoding truncated Nope proteins (Fig. 2A) (data not shown). A multiple sequence alignment shows extensive sequence similarity between Nope, two likely Nope orthologs from D. ananassae and D. grimshawii, and other members of the family of DEG/ENaC sodium channel subunits (Fig. 3). In particular, clusters of conserved residues are found in and around both transmembrane domains and in a neurotoxin-related cysteine-rich stretch within the long extracellular region (Tavernarakis and Driscoll, 2000; Kellenberger and Schild, 2002). Surprisingly, however, Nope and its orthologs lack a highly conserved N-terminal motif that is required for the function of acid-sensing ion channel 3 (ASIC3) (Salinas et al., 2009), of the epithelial sodium channel ENaC (Gründer et al., 1997), and of the Caenorhabditis elegans degenerin, Mec-4 (Hong et al., 2000). These data indicate that, like ppk25, the nope gene encodes a DEG/ENaC subunit.
Figure 1.
Mutations in the nope DEG/ENaC subunit gene specifically block male courtship response to females. A, CI and fraction of males initiating courtship in the presence of decapitated females are shown for males homozygous for mutations in seven different Drosophila genes that code for DEG/ENaC subunits. The lines used were as follows: ppk11, f02053; ppk12, MB11059; ppk19, MB05382; ppk20, MB01352; CG13120, MB07822; CG33289, EY00388; CG13568/nope, f06838 (nope1), and f04205 (nope2). B, Male–female (M-F) and male–male (M-M) indicate courtship of test males directed toward females and males, respectively. Male–female courtship was tested under infrared light and in the presence of decapitated females. Courtship directed at other males was measured under visible light using live “object” males (Liu et al., 2008). The CI and the fraction of males initiating courtship are shown for the genotypes indicated. Δnope is BSC136, a deletion that includes nope and several neighboring genes. As a control, we used f02213 males homozygous for a PiggyBac insertion in CG13563, a gene neighboring nope that has no known role in chemical senses. Error bars show the SEM, N > 24; ***p < 0.001 between (Wilcoxon rank-sum test for CI, and χ2 test for equality of distribution for the fraction of males initiating courtship). In the case of male–male courtship by nope1 and nope2 mutants, neither the distribution of CI values nor the fraction of males initiating courtship are significantly different from 0.
Figure 2.
Schematic representation of the nope gene. A, The genomic region that includes nope and the divergently transcribed eIF5A is shown. A fully spliced nope transcript encodes a DEG/ENaC subunit of 425 aa with two conserved transmembrane domains (TM1 and TM2) (Tavernarakis and Driscoll, 2000; Kellenberger and Schild, 2002). Two other nope transcripts have alternative splicing patterns resulting in shorter open reading frames that encode truncated proteins lacking part of the extracellular domain, the second transmembrane domain and the C-terminal intracellular domain (Berkeley Drosophila Genome Project and data not shown). In nopealt, intron 10 is spliced to an alternative 5′ splice site, 5 nt upstream of the normal 5′ splice junction, and intron 8 is retained in the nopeintron 8 transcript. The nope1 and nope2 alleles contain insertions of PiggyBac elements in exons 5 and 4, respectively, and are therefore also predicted to encode truncated Nope proteins. B, The nope-Gal4 transgene was generated by fusing the open reading frame of the yeast Gal4 transcriptional activator gene at the nope ATG. It contains ∼3 kb of genomic sequences: 0.7 kb of intergenic region between eIF5A and nope, and 2.3 kb of 5′ untranslated sequences of the nope precursor mRNA, up to the nope ATG, followed by the Gal4 open reading frame and the polyadenylation site of SV40 (Elliott and Brand, 2008).
Figure 3.

Nope is a member of the DEG/ENaC family of sodium channel subunits. This multiple sequence alignment displays sequence conservation between Nope from D. melanogaster, predicted Nope orthologs from other Drosophila species and other members of the family of DEG/ENaC sodium channel subunits from Drosophila and vertebrates. Identical and similar residues present in three or more of the aligned sequences are shaded in black and gray, respectively. Sequence conservation between Nope and other DEG/ENaCs is present throughout the protein, particularly in the first and second transmembrane domains (TM1 and TM2), and in a cysteine-rich extracellular domain related to neurotoxins (Tavernarakis and Driscoll, 2000; Kellenberger and Schild, 2002). However, Nope and its orthologs in other Drosophila species lack a highly conserved and functionally important N-terminal domain found in other DEG/ENaCs (Gründer et al., 1997, 1999; Coscoy et al., 1999; Salinas et al., 2009; Carattino, 2011). Interestingly, as was observed for some other Drosophila DEG/ENaC subunits (Liu et al., 2003), Nope and its orthologs, as well as Ppk25 and its orthologs have bulky amino acids (Valine or Methionine) at the functionally important degenerin site, just N-terminal to TM2. In vertebrate and C. elegans DEG/ENaC subunits, as well as in other Drosophila DEG/ENaCs such as Ppk28, this position always encodes residues with a small side chain such as Gly, Ala, or Ser, and mutations to amino acids with a larger side chain, such as Leu, Thr, or Lys result in constitutively open channels (Kellenberger and Schild, 2002). Nopealt indicates the last amino acid of Nope that is retained in Nopealt, a translation product resulting from alternative splicing of intron 10 that lacks TM2 and some preceding extracellular sequences, including the degenerin site. Sequences and accession numbers: ASIC1a, gi|94957760; β-ENaC, gi|8928561; γ-ENaC, gi|6755414|. Ppk28-Dere, Ppk28-Dgri and Ppk28-Daaed correspond to annotated Ppk28 orthologs in Drosophila erecta (GG19083), D. grimshawii (GH11924), and Aedes aegypti (AAEL000582). Ppk25-Dvir and Ppk25-Agam are annotated Ppk25 orthologs in Drosophila virilis (GJ17577), and Anopheles gambiae (AGAP005516). Nope-Dana and Nope-Dgri are previously unannotated Nope orthologs encoded by the genomes of D. ananassae and D. grimshawii, respectively.
To confirm that the defective courtship of males homozygous for the f06838 insertion results from disruption of nope, we tested the behavior of males homozygous for an independent nope mutation, f04205, in which a transposon is inserted in the fourth exon of nope (Fig. 2A). Males homozygous for either f06838 (nope1) or f04205 (nope2), and males transheterozygous for either nope1 or nope2 and a deletion of the entire locus court females significantly less than control males, confirming that the defect in courtship behavior results from disruption of nope (Fig. 1B). In contrast, nope1 or nope2 heterozygous males court females normally, indicating that both alleles of nope are recessive. Furthermore, the TBI, which is the fraction of the 10 min observation period in which the male performs any of the most frequent behaviors (walking, courting, preening), multiplied by 100, is not significantly different between nope mutant males and controls (data not shown), indicating that mutations in nope specifically affect courtship behavior. Finally, as is the case for wild-type males (data not shown), nope1 or nope2 homozygous males do not display significant levels of courtship toward other males; even in the presence of visible light, which typically results in higher levels of courtship and has been used in other studies of male–male courtship (Liu et al., 2008), the CI of nope mutants is not significantly different from 0 (Fig. 1B). Therefore, similar to ppk25 (Starostina et al., 2012), but in contrast to fruitless (fru), a master regulator of sex-specific behavior (Billeter et al., 2006; Villella and Hall, 2008; Siwicki and Kravitz, 2009), nope is not required for suppression of male–male courtship. Instead, nope function is specifically required for activation of male courtship in response to females.
nope is expressed specifically in a small subset of taste neurons that includes taste neurons required for male response to females
To determine the expression pattern of the nope gene, and given the impossibility of detecting low-abundance mRNAs in Drosophila legs using in situ hybridization, we generated a transgene in which the open reading frame of the yeast Gal4 transcriptional activator (Elliott and Brand, 2008) is placed at the predicted initiation of translation of the Nope protein (Fig. 2B). This transgene contains 3 kb of putative nope regulatory sequences upstream of the Gal4 ATG: 700 bp of intergenic DNA between the divergent transcription initiation sites of nope and eIF5a, the nearest upstream gene, and 2.3 kb of the nope 5′ UTR, including two large introns, noncoding exons 1 and 2, and a noncoding segment of exon 3 (Fig. 2B). Flies containing nope-Gal4 and a UAS-mCD8-GFP transgene express mCD8-GFP specifically in gustatory neurons of the legs, wings, and proboscis in both males and females (Fig. 4), with a greater number of cells in the front legs of males than females (data not shown), consistent with the sexually dimorphic number of taste hairs on that appendage (Possidente and Murphey, 1989). On the legs of males, expression is observed in taste neurons distributed on the tibia and all five tarsal segments, in a pattern very similar to that obtained with the ppk25-Gal4 driver (Starostina et al., 2012) (Fig. 4A,B). Furthermore, as documented previously for ppk25-Gal4 (Starostina et al., 2012), nope-Gal4 does not drive expression of mCD8-GFP in the two taste hairs located at the distal tip of the leg, which are the only taste hairs on the legs of males that respond to sugars and salts (Meunier et al., 2000, 2003). Finally, expression of nope-Gal4 and ppk25-Gal4 is not detectably altered in mutants homozygous for nope or ppk25, respectively, indicating that neither gene is essential for development or survival of the neurons in which it is expressed (data not shown). An important difference between the expression of ppk25-Gal4 and nope-Gal4 in gustatory neurons of the legs is that while ppk25-Gal4 expression is invariably associated with a single neuron per taste hair (Starostina et al., 2012), in some cases the dendrites of two nope-Gal4-expressing gustatory neurons project into the lumen of a single taste hair (Fig. 4C, “p” indicates a pair of neurons projecting their dendrites into a single taste hair). nope-Gal4 is also associated with taste neurons on the labellum (Fig. 4D) and wings (Fig. 4E), while ppk25-Gal4 is expressed in wings, but not in the labellum (Starostina et al., 2012). Finally, whole-mount staining with anti-GFP allows visualization of the axonal projections of taste neurons in the brain and ventral nerve chord (Fig. 4F–I). nope-Gal4-expressing gustatory neurons on the labellum project to discrete medial areas in the subesophageal ganglion, the site of the first synaptic relay for taste neurons in the head (Vosshall and Stocker, 2007) (Fig. 4F,G). In contrast, nope-Gal4-expressing taste neurons on the legs and wings project to each of the thoracic ganglia in the central nerve cord (Fig. 4H,I). Of particular interest, projections from the front legs onto the first thoracic ganglia are sexually dimorphic, crossing the midline in males more frequently than in females (Fig. 4H,I). This pattern is identical to that seen using fru-LexA (Mellert et al., 2010), a driver expressing the transcriptional activator LexA in the pattern of fru (Billeter et al., 2006; Siwicki and Kravitz, 2009) or ppk25-Gal4 (Starostina et al., 2012). These findings suggest that, like ppk25-Gal4 (Starostina et al., 2012), nope-Gal4 expression may occur in a subset of fru-expressing neurons. Indeed, double staining of gustatory neurons expressing nope-Gal4 and fru-LexA in the legs with different fluorescent proteins indicates that nope-Gal4-expressing nuclei also express fru-LexA (Fig. 4J,K). In contrast, gustatory neurons that express nope-Gal4 on the labellum do not express fru (Fig. 4L). fru's role in courtship is mediated by the male-specific FruM transcription factor (Siwicki and Kravitz, 2009; Dauwalder, 2011). However, FruM is unlikely to directly regulate expression of either ppk25 or nope, since, with the exception of the greater number of fru-expressing taste neurons on the front legs of males, which are under dsx control (Mellert et al., 2010), both DEG/ENaC subunit genes are expressed in similar patterns and at comparable levels in males and females (data not shown). Indeed, quantitative real-time PCR indicates that expression of the ppk25 mRNA is not affected by a mutation in fru (unpublished data). Together, these findings indicate that nope-Gal4 and ppk25-Gal4 are expressed in similar subsets of gustatory neurons in the legs and wings.
Figure 4.

The nope-Gal4 transgene is expressed specifically in gustatory neurons that also express fru, including all gustatory neurons that express ppk25-Gal4. GFP fluorescence was analyzed in males expressing mCD8-GFP under control of ppk25-Gal4, nope-Gal4 or both drivers as indicated. A, B, nope-Gal4 (A) and ppk25-Gal4 (B) are expressed in very similar patterns in gustatory neurons in the front legs of males (only the tarsi are shown, TA1-TA5 indicate tarsal segments 1–5). C, Higher-magnification image of three gustatory neurons expressing mCD8-GFP under control of nope-Gal4 (arrows). Each neuron has a single sensory dendrite (arrowhead) that can be traced to the base of a taste hair (h). In the case of two neighboring neurons (labeled “p” for paired), the two dendrites run in parallel toward the base of the same taste hair. D, E, expression of UAS-mCD8-GFP under control of nope-Gal4 shows gustatory neurons in the labellum (D) and wings (E, superimposed onto bright-field image). F, G, Whole-brain staining of male expressing mCD8-GFP under control of nope-Gal4 using anti-GFP shows axonal projections of taste neurons onto the subesophageal ganglion. In G, anti-GFP staining is superimposed to staining with nc82, a monoclonal antibody that allows visualization of brain structures (Laissue et al., 1999). H, I, Whole-mount staining of the ventral nerve chord with anti-GFP shows the axonal projections labeled by UAS-mCD8-GFP under control of nope-Gal4 in males and females, respectively. Gustatory neurons on all three pairs of legs (l1, l2, l3) and on the wings (w) project to specific thoracic ganglia. Projections from the front legs onto the first thoracic ganglia cross the midline much more frequently in males than females (arrowheads). J, K, Imaging of taste neurons on the front legs of males expressing two nuclear fluorescent proteins shows that all nope-Gal4 expressing neurons express fru; expression of nuclear GFP is controlled by nope-Gal4 (green arrowheads), and expression of nuclear tdTomato is directed by fru-LexA (Mellert et al., 2010) (blue arrows). While fru is expressed in two gustatory neurons per taste hair (Mellert et al., 2010), nope-Gal4 is expressed in both fru-expressing neurons in some hairs (J), or only one in others (K). L, Double labeling of the neurons on the labellum with membrane-bound mCD8GFP (green arrows) under control of nope-Gal4, and nuclear tdTomato (red arrowheads) under control of fru-LexA (Mellert et al., 2010). In contrast to the male front legs, expression of the two drivers on the labellum does not overlap. M–O, Comparison of the number of taste neurons expressing mCD8-GFP under control of ppk25-Gal4 (M), nope-Gal4 (N), or both transgenes (O) on the third tarsal segment of male front legs (“p” indicates a pair of mCD8-GFP-expressing neurons associated with a single taste hair).
Given the similar expression patterns of ppk25-Gal4 and nope-Gal4 in the legs, we tested for overlapping expression of the two drivers in the front legs of males by counting the number of cells expressing mCD8-GFP in each leg segment for males containing ppk25-Gal4, nope-Gal4, or both drivers (Fig. 4M–O; Table 1). Under control of ppk25-Gal4, we consistently observe mCD8-GFP expression in 43 gustatory neurons on the tibia and the five tarsal segments. However, in the presence of either nope-Gal4 alone or when both drivers are present, mCD8-GFP expression is observed in 54 taste neurons. These results indicate that while all neurons that express ppk25-Gal4 also express nope-Gal4, 11 neurons that express nope-Gal4 do not express ppk25-Gal4. Furthermore, two lines of evidence suggest that in most cases nope-Gal4-expressing neurons that do not express ppk25-Gal4 are paired within a single taste hair with a neuron that expresses both drivers. First, on each leg, 10 taste hairs containing two nope-Gal4-expressing neurons account for all but one of the 11 nope-Gal4-expressing neurons that do not express ppk25-Gal4 (Table 1). Second, the position of taste hairs containing either one or two nope-Gal4-expressing neurons on the surface of the tarsal segment always closely matches that of taste hairs containing a single ppk25-Gal-expressing neuron (Fig. 4, compare M–O). In one exception to this rule, a single nope-Gal4-expressing taste neuron is typically present on the second tarsal segment at a position where ppk25-Gal4 is not expressed (data not shown). These data reveal that in ∼33 taste hairs located at stereotypic positions on the tibia and tarsi of male front legs, only one of the two fru-expressing neurons (Mellert et al., 2010) expresses nope-Gal4, and the same neuron also expresses ppk25-Gal4. In another 10 taste hairs, one of the fru-expressing taste neurons expresses both nope-Gal4 and ppk25-Gal4, while the other expresses only nope-Gal4.
Table 1.
Number of gustatory neurons in the legs of males expressing the UAS-mCD8-GFP transgene under control of nope-Gal4 or ppk25-Gal4, or in the presence of both drivers
| Driver |
ppk25-Gal4 |
nope-Gal4 |
ppk25-Gal4 + nope-Gal4 |
|||
|---|---|---|---|---|---|---|
| Neurons | Neurons pairs | Neurons | Neurons pairs | Neurons | Neurons pairs | |
| Tibia | 10 | 0 | 11 | 1 | 11 | 1 |
| TA1 | 11 | 0 | 13 | 2 | 13 | 2 |
| TA2 | 7 | 0 | 11 | 3 | 11 | 3 |
| TA3 | 6 | 0 | 8 | 2 | 8 | 2 |
| TA4 | 7 | 0 | 9 | 2 | 9 | 2 |
| TA5 | 2 | 0 | 2 | 0 | 2 | 0 |
| Total | 43 | 0 | 54 | 10 | 54 | 10 |
For males of each genotype, the total number of taste neurons as well as the number of neuron pairs associated with a single taste hair is shown for the tibia and tarsal segments 1 through 5 (TA1 to TA5). The numbers shown are typical for the majority of legs examined for each genotype (N > 10), with occasional exceptions displaying an extra cell at a variable location.
Nope functions in a small subset of taste neurons
The expression pattern of nope-Gal4 suggests that, like ppk25 (Starostina et al., 2012), nope may function specifically in gustatory neurons. To test this possibility, we targeted RNAi-mediated knockdown of nope using Poxn-Gal4, a driver expressed in all taste neurons (Boll and Noll, 2002) (Fig. 5). Indeed, gustatory-specific knockdown of nope results in significant decreases, both in CI and in the fraction of males initiating courtship relative to control males in which Poxn-Gal4 drives expression of GFP, or of an RNAi targeting expression of CG13895, a gene with no known role in courtship behavior (Benchabane et al., 2011; Starostina et al., 2012).
Figure 5.
Gustatory-specific RNAi knockdown of nope impairs male courtship response to females. The gustatory-specific driver Poxn-Gal4 (Boll and Noll, 2002) was used to drive expression of GFP, of an RNAi targeting nope (nopeRNAi), or of a control RNAi (ctrlRNAi) targeting CG13895, a gene with no known involvement in courtship behavior or chemical senses (Benchabane et al., 2011). Error bars are SEM; N > 30; *p < 0.05 (Wilcoxon rank-sum test for CI, and χ2 test for equality of distribution for the fraction of males initiating courtship).
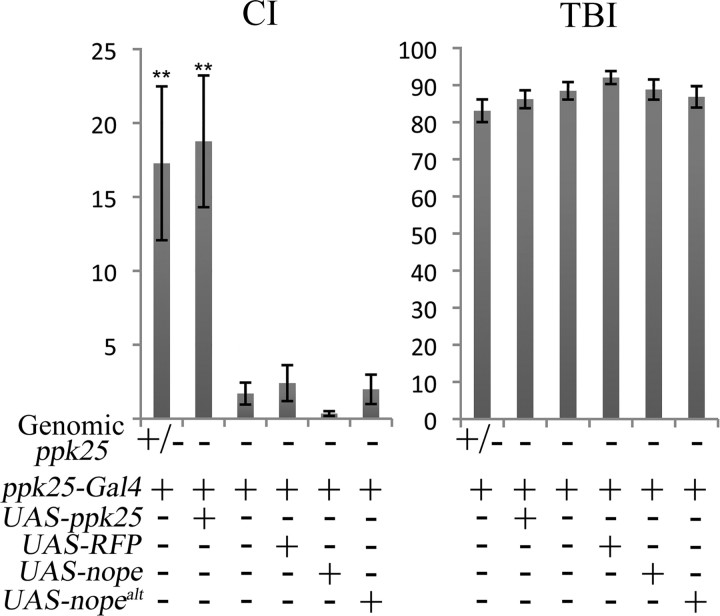
To further define the taste neurons in which nope functions, we used nope-Gal4 to drive expression of nope in nope1/nope2 mutant males (Fig. 6). For this purpose, we generated constructs in which UAS, the target sequence of Gal4, controls expression of two different nope transcripts (Fig. 2A). UAS-nope directs expression of a fully spliced mRNA with a single open reading frame encoding the full-length Nope subunit. UAS-nopealt expresses a transcript in which intron 10 is spliced out using an alternative 5′ splice site and encodes a truncated Nope lacking most of the extracellular domain, the second transmembrane domain, and the C-terminal intracellular domain. To focus on activation of courtship by gustatory circuits, the courtship response of nope1/nope2 males with various combinations of transgenes was tested after removal of the third antennal segment, the main Drosophila olfactory organ (Dahanukar and Ray, 2011; Tunstall and Warr, 2012). The courtship levels of nope1/nope2 males containing both nope-Gal4 and UAS-nope are significantly higher than in the presence of only one of these transgenes, and are indistinguishable from those of males with a wild-type nope gene. In contrast, pairing nope-Gal4 with either UAS-nopealt or UAS-RFP does not rescue the courtship of nope1/nope2 mutant males. We also find that while nope1/nope2 mutant males carrying only the UAS-nope transgene display higher levels of courtship relative to nope1/nope2 males or nope1/nope2 males with control transgenes, their courtship remains significantly lower than that of control males. In contrast, the presence of UAS-nopealt at the same genomic site has no effect. These results suggest that UAS constructs inserted at this genomic site result in some Gal4-independent expression in cells in which nope function is required for courtship. Furthermore, the impaired courtship of nope mutants can be rescued by expression of the nope transcript that encodes a full-length Nope protein, but not of the alternatively spliced nopealt transcript encoding a truncated Nope. Finally, nope expression has no effect on the TBI, further confirming nope's specific role in activation of courtship behavior. Together, these data indicate that expression of full-length Nope protein in the small subset of gustatory neurons identified by expression of nope-Gal4 is sufficient for normal male courtship behavior.
Figure 6.
Targeted expression of nope but not of ppk25 in a small subset of taste neurons in the legs and wings rescues the courtship defect of nope mutant males. The CI and TBI are shown for antennaless males containing the transgenes indicated and with the following genotypes at the endogenous nope gene: heterozygous males carrying one mutant and one wild-type copy of the nope gene, +; and nope1/nope2 mutant males, −. nope mutant males carrying the UAS-nope transgene display significantly more courtship than nope mutant males with other transgenes, but significantly less than male wild-type for nope. nope mutant males expressing nope under control of either nope-Gal4 or ppk25-Gal4 have significantly higher CIs relative to nope mutants with any other combination of transgenes. Vertical arrow 1 represents a statistically significant difference in CI between nope1/nope2 males carrying the UAS-nope transgene and other nope1/nope2 mutant males, while vertical arrow 2 represents a statistically significant difference between the latter and nope1/+ controls. Vertical arrows 3 and 4 represent statistically significant differences in CI between nope1/nope2 males carrying the UAS-nope transgene and males of identical genotype except for the additional presence of either nope-Gal4 or ppk25-Gal4, respectively. Error bars are SEM; N > 30; **p < 0.01, *p < 0.05 (Wilcoxon rank-sum test).
The Nope and Ppk25 DEG/ENaC subunits have distinct, nonredundant functions
nope mutant males display a severe courtship phenotype despite the presence of the wild-type ppk25 gene. To test the possibility that this apparent absence of functional redundancy reflects differences in expression of the two genes, we targeted expression of ppk25 in nope1/nope2 males using nope-Gal4 and a UAS-ppk25 transgene that, in the presence of ppk25-Gal4, rescues the phenotype of ppk25 mutant males (Starostina et al., 2012). Contrary to expression of nope in the same cells, expression of ppk25 does not rescue the phenotype of nope1/nope2 males, indicating that the absence of Nope is not compensated by expression of Ppk25 in the cells where Nope normally functions (Fig. 6). In a reciprocal experiment, we tested whether expression of nope can rescue the function of ppk25 mutants (Fig. 7). Confirming our previous report, expression of ppk25 under control of ppk25-Gal4 rescues the deficient courtship of ppk25 mutant males (Starostina et al., 2012); in contrast, expression of nope in the same cells has no effect. Together, these experiments indicate that the lack of redundancy between the ppk25 and nope genes does not result from differences in their expression patterns but from distinct properties of the DEG/ENaC subunits they encode.
Figure 7.
Targeted expression of ppk25 but not nope rescues the courtship defects of ppk25 mutant males. CI and TBI are shown for antennaless males containing the transgenes indicated and with the following genotypes at the endogenous ppk25 gene: heterozygous males carrying one mutant and one wild-type copy of the ppk25 gene, +; and males heterozygous for two different deletions of the ppk25 gene, −. As shown previously (Lin et al., 2005; Starostina et al., 2012), ppk25 mutant males display significantly lower CIs than controls, and the presence of both ppk25-Gal4 and UAS-ppk25 restores normal courtship levels. In contrast, the courtship of ppk25 mutant males is not improved in the presence of both ppk25-Gal4 and UAS-nope transgenes. N > 30. Statistical significance relative to the CI of ppk25 mutant males calculated with the Wilcoxon rank-sum test: **p < 0.01.
Ppk25 and Nope function in a common subset of gustatory neurons required for male response to females
To test whether nope expression in the subset of nope-expressing taste neurons that also express ppk25 is sufficient for normal courtship, we used ppk25-Gal4 to drive nope expression in nope1/nope2 mutants (Fig. 6). As with nope-Gal4, ppk25-Gal4-directed expression of fully spliced nope, but not of nopealt, ppk25, or RFP rescues courtship response of nope1/nope2 mutants. Therefore, expression of nope in the small subset of taste neurons on the wings and legs that express both nope-Gal4 and ppk25-Gal4 is sufficient for normal male response to females. Since expression of ppk25 in the same neurons restores the courtship of ppk25 mutant males (Fig. 7) (Starostina et al., 2012), these data indicate that Ppk25 and Nope function in a common subset of gustatory neurons and confirm that each of these DEG/ENaC subunits has a specific, nonredundant role.
Nope interacts specifically with Ppk25
Their nonredundant functions in a common subset of gustatory neurons suggest that, like other DEG/ENaC subunits (Carattino, 2011; Kashlan and Kleyman, 2011), Nope and Ppk25 may function within a heteromeric channel. To test whether Nope and Ppk25 interact when coexpressed in cultured cells, and given the role of the N terminus of other DEG/ENaC subunits in channel function and trafficking (Tavernarakis et al., 2001; Wesch et al., 2012), we performed immunoprecipitations with C-terminal epitope-tagged versions of the two proteins (Fig. 8). Cells were simultaneously transfected with multiple plasmids encoding epitope-tagged proteins. In a first set of experiments, we tested whether Ppk25-Flag interacts with Nope-HA using as an internal control HA-tagged Armadillo (Arm-HA), a protein that is present in both cytoplasmic and membrane-associated forms (Riggleman et al., 1989) (Fig. 8A). Cellular extracts were immunoprecipitated with an anti-Flag antibody, and extracts, supernatants, and precipitates were subsequently analyzed by immunoblotting with an anti-HA antibody (Fig. 8A). In the presence of Ppk25-Flag but not in its absence, immunoprecipitation with anti-Flag results in coprecipitation of Nope-HA but not of Arm-HA, revealing a specific interaction between Ppk25-Flag and Nope-HA. Furthermore, Nope-HA levels are depleted in the supernatant relative to the extract, indicating that most Nope-HA in the extract is bound to Ppk25-Flag. Similarly, in the presence of Nope-Flag, Ppk25-HA but not Arm-HA is coprecipitated by anti-Flag antibody, indicating that the interaction between Ppk25 and Nope does not depend on a particular tagged version of either protein (Fig. 8B). To further test the specificity of the interaction between Nope and Ppk25, we conducted competition experiments in the presence of another DEG/ENaC subunit, human β-ENaC (Fig. 8C). While in the presence of Nope-Flag, some β-ENaC-HA is coprecipitated with anti-Flag, coprecipitation of Ppk25-HA is significantly more efficient (Fig. 8C, compare the ratios of β-ENaC-HA and Ppk25-HA in the pellet and supernatant). Similarly, Flag immunoprecipitation of extracts from cells expressing Ppk25-Flag, Nope-HA, and β-ENaC-HA results in preferential coprecipitation of Nope-HA relative to β-ENaC-HA (Fig. 8D, compare the ratios of β-ENaC-HA and Nope-HA in the pellet and supernatant). Finally, as assembly of heteromeric DEG/ENaC channels occurs during transit of the newly synthesized subunits to the membrane surface (Butterworth et al., 2008), we tested whether expression of Nope and Ppk25 in the same cells is required for their interaction. Indeed, when Ppk25-Flag and Nope-HA are expressed in different cells, mixing cellular extracts does not result in coprecipitation (data not shown), further confirming the specificity of the interaction between these two DEG/ENaC subunits. Together, these data indicate that Nope and Ppk25 form specific complexes when coexpressed in cultured cells, suggesting that, in the courtship-activating gustatory neurons where they are both expressed, these two subunits assemble into a heteromeric DEG/ENaC channel. In an indirect test of this hypothesis, we asked whether the nope and ppk25 genes interact genetically, resulting in lower courtship levels for males heterozygous for both nope and ppk25, but find that such males court at normal levels (CI = 53 ± 5 and 89% of males initiate courtship; those numbers are comparable to those of control males, not shown). Since such a lack of genetic interaction does not necessarily reflect an absence of direct interaction between the two encoded proteins, more direct approaches will be required to test whether the function of Nope and Ppk25 in gustatory activation of courtship requires formation of a specific complex between these two subunits in pheromone-sensing neurons.
Figure 8.
Nope and Ppk25 interact specifically in cultured cells. A, HEK293 cells were transfected with plasmids encoding Nope-HA, Arm-HA, and, where indicated, Ppk25-Flag, and were immunoprecipitated using anti-Flag antibody. Extracts, supernatants (S), and precipitates (P) were analyzed on Western blots with anti-HA antibody. B, Same as in A, except that Nope was tagged with Flag and Ppk25 with HA. C, Extracts of cells expressing Nope-Flag, Ppk25-HA, and human β-ENaC-HA were immunoprecipitated with anti-Flag, and both supernatant and pellet were immunoblotted with anti-HA. The increased ratio of Ppk25-HA to β-ENaC-HA in the pellet compared with the supernatant indicates that Nope-Flag interacts preferentially with Ppk25-HA. D, Same as in C, except that Ppk25 was tagged with Flag and Nope with HA. Here also, the ratio of Nope-HA to β-ENaC-HA is higher in the pellet than in the supernatant.
In summary, our data indicate that two DEG/ENaC subunits, Nope and Ppk25, are coexpressed and have specific, nonredundant functions in a common subset of courtship-activating gustatory neurons in the wings and legs of males. In addition, Nope and Ppk25 interact specifically when coexpressed in cultured cells. Together, these data suggest the hypothesis that Ppk25 and Nope are subunits of a heteromeric DEG/ENaC channel required for gustatory activation of courtship in the presence of females, most likely in response to female-specific pheromones.
Discussion
Nope and Ppk25 function in a common subset of gustatory neurons in the legs and wings of males that are required for activation of male courtship
We previously discovered that mutations in ppk25, a DEG/ENaC subunit gene, specifically impair male courtship of females, but not other behaviors (Lin et al., 2005). Recently, we reported that expression of ppk25 in a small subset of taste neurons on the legs and wings of males is sufficient for a normal male courtship response to females (Starostina et al., 2012). Here, we describe a second DEG/ENaC subunit gene, nope, that is also specifically required for male response to females. Several lines of evidence indicate that both genes are expressed in, and are required for the function of a common subset of gustatory neurons that activate courtship behavior. First, mutations in ppk25 and nope have indistinguishable effects on male courtship. Like mutations in ppk25 (Lin et al., 2005), mutations in nope specifically impair the courtship response of males to females but have no general effect on other behaviors, such as walking or preening. Furthermore, like mutations in ppk25 (Starostina et al., 2012), but in contrast to mutations in fru (Billeter et al., 2006; Villella and Hall, 2008; Siwicki and Kravitz, 2009) or in the Gr32a gustatory receptor gene (Miyamoto and Amrein, 2008), mutations in nope do not increase homosexual behavior, suggesting that neither ppk25 nor nope is required for gustatory detection of inhibitory pheromones, such as the male-enriched 7-tricosene. Finally, consistent with gustatory function being required not only to initiate courtship but also to progress to late steps (Krstic et al., 2009), mutations in either ppk25 (Starostina et al., 2012) or nope result in significant decreases in both the fraction of males that initiate courtship and the total time spent courting. Second, expression of ppk25-Gal4 and nope-Gal4 transgenes overlaps in a small subset of taste neurons on the legs and wings of males. In the front legs in particular, both transgenes are expressed in fru-expressing taste neurons that make sexually dimorphic projections onto the thoracic ganglia (Mellert et al., 2010), and all neurons that express ppk25-Gal4 also express nope-Gal4. Third, as in the case of ppk25 (Starostina et al., 2012), targeted RNAi-mediated knockdown of nope in gustatory neurons specifically impairs the male response to females, indicating that both genes are required for normal gustatory response to females. Finally, and most importantly, using the same ppk25-Gal4 driver, targeted expression of nope and ppk25 rescues the courtship of nope and ppk25 mutant males, respectively, indicating that for both DEG/ENaC subunits, expression in a common subset of gustatory neurons on the legs and wings of males is sufficient for normal response to females. Coupled with our previous work (Starostina et al., 2012), these findings further demonstrate that this specific subset of gustatory neurons on the legs and wings has an essential role in activating male courtship, starting from the earliest steps.
In addition to the subset of neurons defined by coexpression of ppk25-Gal4 and nope-Gal4, some chemosensory neurons express only one of the two drivers; ppk25-Gal4 is expressed in two subsets of olfactory neurons implicated in activating courtship (Starostina et al., 2012), and nope-Gal4 is expressed in gustatory neurons on the legs and labellum. Furthermore, targeted expression of nope using the ppk25-Gal4 driver is sufficient to rescue the courtship of nope mutant males. This result indicates that, under our experimental conditions at least, gustatory neurons that express nope-Gal4 but not ppk25-Gal4 are not essential for activation of courtship in response to females, and suggests that expression of nope-Gal4 may occur in functionally distinct subsets of gustatory neurons. In summary, our data indicate that nope and ppk25 function in a common subset of gustatory neurons on the front legs and wings of males that is required for activation of male courtship in the presence of females.
Their nonredundant functions and specific interaction suggest the hypothesis that Nope and Ppk25 function within a heteromeric DEG/ENaC channel required specifically for gustatory activation of courtship
While ppk25 and nope function in the same subset of gustatory neurons, mutants lacking either subunit display severely reduced male response to females, indicating that the ppk25 and nope genes have nonredundant functions. Furthermore, the phenotype of nope mutant males is rescued by targeted expression of nope, but not of ppk25, and vice versa, indicating that the lack of redundancy is due to intrinsic properties of the proteins encoded by each gene rather than differences in their expression patterns. In addition, when coexpressed in cultured cells, Nope forms specific complexes with Ppk25 more efficiently than with human β-ENaC, another DEG/ENaC subunit. Together, these data support the hypothesis that Ppk25 and Nope assemble into a heteromeric DEG/ENaC channel with a specific and essential function in gustatory activation of courtship behavior. The crystal structure of the vertebrate DEG/ENaC acid-sensing channel, ASIC1a, reveals that it is a homotrimer, suggesting that most if not all DEG/ENaC channels are also trimers (Jasti et al., 2007). Furthermore, ENaC, the vertebrate epithelial sodium channel is a heterotrimer containing three types of subunits (α, β, and γ), all of which must be present for assembly of a functional channel. Finally, heteromeric assemblies of C. elegans Mec-4 and Mec-10 are required for response to gentle touch (Bianchi, 2007). By analogy, Ppk25 and Nope are likely to assemble into a heterotrimeric DEG/ENaC channel, perhaps together with a third subunit yet to be discovered. In addition, while ppk25-Gal4 and nope-Gal4 have overlapping expression patterns, each driver is also expressed in cells where the other is not; ppk25-Gal4 is expressed in two subsets of olfactory neurons involved in activation of male courtship (Root et al., 2008; Grosjean et al., 2011; Starostina et al., 2012), and nope-Gal4 is expressed in gustatory neurons of unknown function on the front legs and labellum of males (this study). Ppk25 may therefore also participate in trimeric channels lacking Nope, and vice versa. By analogy with human ASICs, for which channels of different subunit composition have different functional properties (Sherwood et al., 2011), combinatorial subunit composition may confer functional variety to DEG/ENaC channels in gustatory and olfactory neurons.
Finally, the specific phenotypes of ppk25 and nope mutants coupled with their expression in a small subset of gustatory neurons required for response to females suggest that both DEG/ENaC subunits function specifically in detection of courtship-stimulating pheromones. What is the molecular role of DEG/ENaC channels in pheromone response? In the same way that other DEG/ENaCs channels function as gustatory receptors for water (Cameron et al., 2010; Chen et al., 2010) or sodium ions (Chandrashekar et al., 2010), heteromeric channels containing Ppk25 and Nope may be directly gated by pheromones or by other courtship-activating compounds. Alternatively, heteromeric DEG/ENaC channels containing Nope and Ppk25 may have a less direct role in modulating courtship, similar to the function of C. elegans ASIC1 in learning modulation through presynaptic facilitation of dopamine release (Voglis and Tavernarakis, 2008). Our work lays the foundation for the dissection of the molecular roles of DEG/ENaC channels and the contribution of subunit composition in gustatory activation of male courtship, with relevance to the function of DEG/ENaC channels involved in a number of physiological and pathological processes including hypertension, cystic fibrosis, touch and pain, memory formation, evoked fear, and neuronal cell death in stroke (Kellenberger and Schild, 2002; Bianchi, 2007; Cameron et al., 2010; Chandrashekar et al., 2010; Chen et al., 2010; Qadri et al., 2012).
Note added in proof.
As this manuscript was in revision, independent findings of nope's expression and role in the detection of a courtship-stimulating pheromone were published by others (Thistle et al., 2012), and several reports also implicated a third DEG/ENaC subunit gene, ppk23, in pheromone response (Lu et al., 2012; Thistle et al., 2012; Toda et al., 2012), further supporting the central role of DEG/ENaC channels in the gustatory detection of pheromones.
Footnotes
This work was supported by Grant R01DC007911 from the NIDCD to C.W.P., and through an award from the Biomedical Research Support Program for Medical Schools from the Howard Hughes Medical Institute to Geisel School of Medicine at Dartmouth (76200-560801). The authors are grateful to Yashi Ahmed for suggestions on the manuscript. We thank the Bloomington Stock Center (Indiana University), the Transgenic RNAi project at Harvard Medical School, and the Vienna Drosophila RNAi Center for fly lines, and the Developmental Studies Hybridoma bank for a monoclonal antibody used in this study.
References
- Benchabane H, Xin N, Tian A, Hafler BP, Nguyen K, Ahmed A, Ahmed Y. Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating beta-catenin-TCF activity. EMBO J. 2011;30:1444–1458. doi: 10.1038/emboj.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R. Sensitivity and specificity in Drosophila pheromone perception. Trends Neurosci. 2007;30:512–519. doi: 10.1016/j.tins.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Bianchi L. Mechanotransduction: touch and feel at the molecular level as modeled in Caenorhabditis elegans. Mol Neurobiol. 2007;36:254–271. doi: 10.1007/s12035-007-8009-5. [DOI] [PubMed] [Google Scholar]
- Billeter JC, Rideout EJ, Dornan AJ, Goodwin SF. Control of male sexual behavior in Drosophila by the sex determination pathway. Curr Biol. 2006;16:R766–R776. doi: 10.1016/j.cub.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- Brand AH, Dormand EL. The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- Butterworth MB, Weisz OA, Johnson JP. Some assembly required: putting the epithelial sodium channel together. J Biol Chem. 2008;283:35305–35309. doi: 10.1074/jbc.R800044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P, Hiroi M, Ngai J, Scott K. The molecular basis for water taste in Drosophila. Nature. 2010;465:91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino MD. Structural mechanisms underlying the function of epithelial sodium channel/acid-sensing ion channel. Curr Opin Nephrol Hypertens. 2011;20:555–560. doi: 10.1097/MNH.0b013e328348bcac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, Zuker CS. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang Q, Wang Z. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for Drosophila gustatory water reception. J Neurosci. 2010;30:6247–6252. doi: 10.1523/JNEUROSCI.0627-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy S, de Weille JR, Lingueglia E, Lazdunski M. The pre-transmembrane 1 domain of acid-sensing ion channels participates in the ion pore. J Biol Chem. 1999;274:10129–10132. doi: 10.1074/jbc.274.15.10129. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Ray A. Courtship, aggression and avoidance: pheromones, receptors and neurons for social behaviors in Drosophila. Fly (Austin) 2011;5:58–63. doi: 10.4161/fly.5.1.13794. [DOI] [PubMed] [Google Scholar]
- Dauwalder B. The roles of fruitless and doublesex in the control of male courtship. Int Rev Neurobiol. 2011;99:87–105. doi: 10.1016/B978-0-12-387003-2.00004-5. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Wired for sex: the neurobiology of Drosophila mating decisions. Science. 2008;322:904–909. doi: 10.1126/science.1159276. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Brand AH. The GAL4 system: a versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- Ferveur JF. Cuticular hydrocarbons: their evolution and roles in Drosophila pheromonal communication. Behav Genet. 2005;35:279–295. doi: 10.1007/s10519-005-3220-5. [DOI] [PubMed] [Google Scholar]
- Frank KD, Zimmerman WF. Action spectra for phase-shifts of a circadian rhythm in Drosophila. Science. 1969;163:688–689. doi: 10.1126/science.163.3868.688. [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- Gründer S, Firsov D, Chang SS, Jaeger NF, Gautschi I, Schild L, Lifton RP, Rossier BC. A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 1997;16:899–907. doi: 10.1093/emboj/16.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer S, Jaeger NF, Gautschi I, Schild L, Rossier BC. Identification of a highly conserved sequence at the N-terminus of the epithelial Na+ channel alpha subunit involved in gating. Pflugers Arch. 1999;438:709–715. doi: 10.1007/s004249900119. [DOI] [PubMed] [Google Scholar]
- Hall JC. Courtship among males due to a male-sterile mutation in Drosophila melanogaster. Behav Genet. 1978;8:125–141. doi: 10.1007/BF01066870. [DOI] [PubMed] [Google Scholar]
- Hong K, Mano I, Driscoll M. In vivo structure-function analyses of Caenorhabditis elegans MEC-4, a candidate mechanosensory ion channel subunit. J Neurosci. 2000;20:2575–2588. doi: 10.1523/JNEUROSCI.20-07-02575.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Kleyman TR. ENaC structure and function in the wake of a resolved structure of a family member. Am J Physiol Renal Physiol. 2011;301:F684–F696. doi: 10.1152/ajprenal.00259.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- Krstic D, Boll W, Noll M. Sensory integration regulating male courtship behavior in Drosophila. PLoS One. 2009;4:e4457. doi: 10.1371/journal.pone.0004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Levine JD, Millar JG. Chemical signalling: laser on the fly reveals a new male-specific pheromone. Curr Biol. 2009;19:R653–R655. doi: 10.1016/j.cub.2009.06.051. [DOI] [PubMed] [Google Scholar]
- Lin H, Mann KJ, Starostina E, Kinser RD, Pikielny CW. A Drosophila deg/ENaC channel subunit is required for male response to female pheromones. Proc Natl Acad Sci U S A. 2005;102:12831–12836. doi: 10.1073/pnas.0506420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Leonard AS, Motto DG, Feller MA, Price MP, Johnson WA, Welsh MJ. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 2003;39:133–146. doi: 10.1016/s0896-6273(03)00394-5. [DOI] [PubMed] [Google Scholar]
- Liu T, Dartevelle L, Yuan C, Wei H, Wang Y, Ferveur JF, Guo A. Increased dopamine level enhances male–male courtship in Drosophila. J Neurosci. 2008;28:5539–5546. doi: 10.1523/JNEUROSCI.5290-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, LaMora A, Sun Y, Welsh MJ, Ben-Shahar Y. ppk23-dependent chemosensory functions contribute to courtship behavior in Drosophila melanogaster. PLoS Genet. 2012;8:e1002587. doi: 10.1371/journal.pgen.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert DJ, Knapp JM, Manoli DS, Meissner GW, Baker BS. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development. 2010;137:323–332. doi: 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer RH, Kapoor N, Qadri YJ, Anderson SJ, Fuller CM, Benos DJ. Heteromeric assembly of acid-sensitive ion channel and epithelial sodium channel subunits. J Biol Chem. 2007;282:25548–25559. doi: 10.1074/jbc.M703825200. [DOI] [PubMed] [Google Scholar]
- Meunier N, Ferveur JF, Marion-Poll F. Sex-specific non-pheromonal taste receptors in Drosophila. Curr Biol. 2000;10:1583–1586. doi: 10.1016/s0960-9822(00)00860-5. [DOI] [PubMed] [Google Scholar]
- Meunier N, Marion-Poll F, Rospars JP, Tanimura T. Peripheral coding of bitter taste in Drosophila. J Neurobiol. 2003;56:139–152. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Amrein H. Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci. 2008;11:874–876. doi: 10.1038/nn.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possidente DR, Murphey RK. Genetic control of sexually dimorphic axon morphology in Drosophila sensory neurons. Dev Biol. 1989;132:448–457. doi: 10.1016/0012-1606(89)90241-8. [DOI] [PubMed] [Google Scholar]
- Qadri YJ, Rooj AK, Fuller CM. ENaCs and ASICs as therapeutic targets. Am J Physiol Cell Physiol. 2012;302:C943–C965. doi: 10.1152/ajpcell.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggleman B, Wieschaus E, Schedl P. Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- Ronderos DS, Smith DP. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly (Austin) 2009:3. doi: 10.4161/fly.9801. [DOI] [PubMed] [Google Scholar]
- Root CM, Masuyama K, Green DS, Enell LE, Nässel DR, Lee CH, Wang JW. A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron. 2008;59:311–321. doi: 10.1016/j.neuron.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable elements. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Salinas M, Lazdunski M, Lingueglia E. Structural elements for the generation of sustained currents by the acid pain sensor ASIC3. J Biol Chem. 2009;284:31851–31859. doi: 10.1074/jbc.M109.043984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood TW, Lee KG, Gormley MG, Askwith CC. Heteromeric acid-sensing ion channels (ASICs) composed of ASIC2b and ASIC1a display novel channel properties and contribute to acidosis-induced neuronal death. J Neurosci. 2011;31:9723–9734. doi: 10.1523/JNEUROSCI.1665-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki KK, Kravitz EA. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr Opin Neurobiol. 2009;19:200–206. doi: 10.1016/j.conb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina E, Liu T, Vijayan V, Zheng Z, Siwicki KK, Pikielny CW. A Drosophila DEG/ENaC subunit functions specifically in gustatory neurons required for male courtship behavior. J Neurosci. 2012;32:4665–4674. doi: 10.1523/JNEUROSCI.6178-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Stowers L, Logan DW. Sexual dimorphism in olfactory signaling. Curr Opin Neurobiol. 2010;20:770–775. doi: 10.1016/j.conb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Driscoll M. Caenorhabditis elegans degenerins and vertebrate ENaC ion channels contain an extracellular domain related to venom neurotoxins. J Neurogenet. 2000;13:257–264. doi: 10.3109/01677060009084497. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Everett JK, Kyrpides NC, Driscoll M. Structural and functional features of the intracellular amino terminus of DEG/ENaC ion channels. Curr Biol. 2001;11:R205–R208. doi: 10.1016/s0960-9822(01)00106-3. [DOI] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149:1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda H, Zhao X, Dickson BJ. The Drosophila female aphrodisiac pheromone activates ppk23+ sensory neurons to elicit male courtship behavior. Cell Rep. 2012;1:599–607. doi: 10.1016/j.celrep.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Tunstall NE, Warr CG. Chemical communication in insects: the peripheral odour coding system of Drosophila melanogaster. Adv Exp Med Biol. 2012;739:59–77. doi: 10.1007/978-1-4614-1704-0_4. [DOI] [PubMed] [Google Scholar]
- Villella A, Hall JC. Neurogenetics of courtship and mating in Drosophila. Adv Genet. 2008;62:67–184. doi: 10.1016/S0065-2660(08)00603-2. [DOI] [PubMed] [Google Scholar]
- Villella A, Ferri SL, Krystal JD, Hall JC. Functional analysis of fruitless gene expression by transgenic manipulations of Drosophila courtship. Proc Natl Acad Sci U S A. 2005;102:16550–16557. doi: 10.1073/pnas.0507056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglis G, Tavernarakis N. A synaptic DEG/ENaC ion channel mediates learning in C. elegans by facilitating dopamine signalling. EMBO J. 2008;27:3288–3299. doi: 10.1038/emboj.2008.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB. Scent of a fly. Neuron. 2008;59:685–689. doi: 10.1016/j.neuron.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wesch D, Althaus M, Miranda P, Cruz-Muros I, Fronius M, González-Hernández T, Clauss WG, Alvarez de la Rosa D, Giraldez T. Differential N termini in epithelial Na+ channel delta-subunit isoforms modulate channel trafficking to the membrane. Am J Physiol Cell Physiol. 2012;302:C868–C879. doi: 10.1152/ajpcell.00255.2011. [DOI] [PubMed] [Google Scholar]
- Wicker-Thomas C. Pheromonal communication involved in courtship behavior in Diptera. J Insect Physiol. 2007;53:1089–1100. doi: 10.1016/j.jinsphys.2007.07.003. [DOI] [PubMed] [Google Scholar]