Abstract
Candida albicans remains the main etiological agent of candidiasis, which currently represents the fourth most common nosocomial bloodstream infection in US hospitals1. These opportunistic infections pose a growing threat for an increasing number of compromised individuals, and carry unacceptably high mortality rates. This is in part due to the limited arsenal of antifungal drugs, but also to the emergence of resistance against the most commonly used antifungal agents. Further complicating treatment is the fact that a majority of manifestations of candidiasis are associated with the formation of biofilms, and cells within these biofilms show increased levels of resistance to most clinically-used antifungal agents2. Here we describe the development of a high-density microarray that consists of C. albicans nano-biofilms, which we have named CaBChip3. Briefly, a robotic microarrayer is used to print yeast cells of C. albicans onto a solid substrate. During printing, the yeast cells are enclosed in a three dimensional matrix using a volume as low as 50 nL and immobilized on a glass substrate with a suitable coating. After initial printing, the slides are incubated at 37 °C for 24 hours to allow for biofilm development. During this period the spots grow into fully developed "nano-biofilms" that display typical structural and phenotypic characteristics associated with mature C. albicans biofilms (i.e. morphological complexity, three dimensional architecture and drug resistance)4. Overall, the CaBChip is composed of ~750 equivalent and spatially distinct biofilms; with the additional advantage that multiple chips can be printed and processed simultaneously. Cell viability is estimated by measuring the fluorescent intensity of FUN1 metabolic stain using a microarray scanner. This fungal chip is ideally suited for use in true high-throughput screening for antifungal drug discovery. Compared to current standards (i.e. the 96-well microtiter plate model of biofilm formation5), the main advantages of the fungal biofilm chip are automation, miniaturization, savings in amount and cost of reagents and analyses time, as well as the elimination of labor intensive steps. We believe that such chip will significantly speed up the antifungal drug discovery process.
Keywords: Biomedical Engineering, Issue 65, Bioengineering, Immunology, Infection, Molecular Biology, Candida albicans, Biofilm, High-throughput screening
Protocol
1. Preparation of Functionalized Slides
Place the microscope slides in a removable slide rack, and wash twice by immersing in a staining jar containing 99% ethanol (histological grade). Wipe the slides clean using paper towels (ensuring not to generate paper dust), and dry using a jet of compressed nitrogen gas.
NOTE: Do not use Kim-Wipes to wipe the slides as it would generate fine paper dust.
Immerse the slide rack containing the slides in a staining jar filled with concentrated sulphuric acid and incubate at room temperature overnight.
NOTE: To avoid contact with skin, use chemical-resistant gloves and safety goggles while working with concentrated acids and toxic and corrosive chemicals.
Sonicate the slides for 30 min and wash with Milli-Q (18 MΩ) water for 30 min, follow with another wash in acetone for 5 min. This treatment exposes the silanol groups on the glass surface.
Coat the clean slides with 2.5% (vol/vol in water) of 3-aminopropyltriethoxysilane (APTES) solution, by immersing the slide rack in APTES for 30 min, washing 3 times in Milli-Q water for 15 min each wash, and baking the slides in a furnace at 110 °C for 15 min. Baking allows cross-linking of the APTES, resulting in -NH2-functionalization of the surface.
NOTE: Make the APTES solution in a plastic container since APTES deposits preferentially on the walls of glass container.
Using a spin coater, coat the slides with 1% (wt/vol in toluene) Polystyrene-Co-Maleic Anhydride (PS-MA) (Sigma) to achieve a mono-layer of hydrophobic coating 6. Add 2.0 mL of PS-MA onto a clean glass slide mounted on a spin coater and coat at 3,000 rpm for 30 s. These conditions may vary based on spin coating parameters such as coating solution, substrate, thickness of coating and governed by the following equation 7
![]() where h is thickness of film coating, e is rate of evaporation, η , C and ρ are viscosity, concentration and density of the coating solution, respectively, and ω is angular velocity.
where h is thickness of film coating, e is rate of evaporation, η , C and ρ are viscosity, concentration and density of the coating solution, respectively, and ω is angular velocity.
NOTE: Toluene is harmful when inhaled for a long time and use of a fume hood is recommended.
The slides can be stored in a slide rack for up to one month in dry, dust-free conditions at 2- 8 °C.
2. Preparation of Yeast Inocula and Collagen Encapsulation
Prepare an overnight culture of C. albicans strain SC5314, in Yeast Peptone Dextrose [YPD; 10 g/L yeast extract, 20 g/L peptone and 20 g/L dextrose] liquid medium by inoculating a single colony of C. albicans into 10 - 20 mL of YPD8. Incubate culture in an orbital shaker (about 150 - 200 rpm) at 30 °C overnight. Harvest 1 mL Candida albicans yeast cells from overnight YPD cultures (by centrifugation at 5,000 rpm for 5-10 minutes) and wash twice for 10 min in 1 mL of sterile phosphate buffered saline (PBS; 10 mM phosphate buffer, 2.7 mM potassium chloride, 137 mM sodium chloride, pH 7.4) (Sigma) per wash. Harvest washed cells also by centrifugation at 1900 rpm for 10 minutes. Resuspend the washed cells in 1 mL reconstruction buffer (0.2 N NaOH solution with 2.2% (wt/vol) sodium bicarbonate and 4.8% (wt/vol) HEPES, pH 7.2).
NOTE: C. albicans is a Risk Group 1/BSL1 microorganism. Always remember to use good aseptic/sterile techniques for work with this microorganism and follow institutional procedures for proper disposal of biohazardous materials.
Count cells using a hemocytometer on a bright field microscope and adjust to a cell density of 5×107 cells/mL.
Further dilute the suspension ten times by addition of 10× RPMI-1640 supplemented with L-glutamine and buffered with morpholinepropanesulfonic (MOPS) acid (pH 7.2).
Encapsulate the yeast cells in collagen by mixing the cell suspension in RPMI-1640 with collagen (1.8 mg/mL) (Type 1 from rat tail, BD Biosciences, Bedford, MA), to obtain a final concentration of 4× 106 cells/mL. Keep the collagen-cell suspension on ice to prevent the gelation of collagen before printing.
3. Preparation of CaBChip
Clean and disinfect all surfaces of the microarrayer, including the source plate station, wash and vacuum station, vacuum slide platter and printing chamber, by wiping with 70% isopropanol.
Place and hold by vacuum the desired number of PS-MA-coated glass slides on the slide deck of the microarrayer.
Vortex the cell suspension in collagen vigorously and aspirate 100 μL of the well-mixed suspension into a well of a 96-well plate, just before printing. Place this source plate in the loading station.
Switch on the humidifier to maintain a relative humidity of 100% in the microarray chamber throughout the printing process.
Print 50 nL of cell suspension in an array of 48 rows and 16 columns with spots spaced 1.2 mm apart using a microarray spotter (Omnigrid Micro, Digilab Inc., Holliston, MA) by non-contact deposition using conically tapered 190 μm orifice ceramic tips (Digilab).
Prime, rinse and vacuum-dry the tips twice after each round of printing.
Immediately after printing, place the slide in an air-tight, humidified chamber (Hybridization cassette, ArrayIt Corporation, Sunnyvale, CA) and incubate at 37 °C for 24 hr to allow for biofilm formation.
4. Susceptibility Testing of Preformed Biofilms in CaBChip Against Antifungal Agents
From stock solutions or powder of antifungal drugs, prepare a working solution in RPMI- 1640 medium. Typical maximum concentrations of the working solution are 1,024 μg/mL for fluconazole and 16 μg/mL for amphotericin B5. Other concentrations may be used for different agents.
Prepare eight different concentrations of the compound by diluting the stock solution in two-fold dilutions, spanning a range with the estimated IC50 of the compound in the middle.
After 24 hr of biofilm growth, use the microarrayer to print 50 nL of eight different concentrations of the drugs, along with positive (no drug i.e. cells in only media) and negative (treated with sodium hypochlorite for 20 min) controls, in at least six replicates on top of the biofilms.
NOTE: Maintain a relative humidity of 100% to prevent the drying of the spots while adding the drugs.
Soon after adding the drugs, incubate the chip with the drugs in a humidified chamber at 37 °C for 24 hr.
Wash the drugs off by dunking the CaBChip 3-5 times for 2 min every time in PBS and stain by incubating the CaBChip with 0.5 μM FUN1 at 37 °C for 30 min9.
Wash the stain off by dunking the CaBChip 3-5 times in PBS for 1-3 min every dunk, and dry the CaBChip using a nitrogen stream. Read fluorescence intensity using a microarray scanner (GenePix 4100A, Axon Instruments, CA) at a wavelength of 532 nm with a PMT gain of 380.
Set the fluorescence intensity of the positive control (no drug) and dead (bleach-treated) biofilm spots at 100% and 0%, respectively.
Determine the percentage inhibition by the reduction in the fluorescence intensity (F) relative to the average of the control spots using the following equation
![]() where F, Fmax and Fo are raw fluorescence intensities of drug-treated, no-drug control and bleach-treated spots, respectively. The scanner settings were adjusted to obtain Fmax and Fo of 30000 and 4000 RFU, respectively.
where F, Fmax and Fo are raw fluorescence intensities of drug-treated, no-drug control and bleach-treated spots, respectively. The scanner settings were adjusted to obtain Fmax and Fo of 30000 and 4000 RFU, respectively.
Calculate the 50% inhibitory concentration or IC50 by fitting the variable slope Hill equation using GraphPad Prism Software (La Jolla, CA).
5. Representative Results
A representative CaBChip, consisting of a 48×16 array of nano-biofilms of C. albicans, is shown in Figure 2. The bright field microscopy shows an overall architectural feature of the nano-biofilm. The scanning electron microscopic images of the biofilm show that the fungal hyphae are embedded within the matrix of collagen fibers, which are approximately 2 μm and 100 nm in diameter, respectively. The FUN1-stained microarray scanner images show the yeast and hyphal forms, which are characteristic of fungal biofilms. The 2D- and 3D-confocal fluorescence images of FUN1-stained biofilms can be seen to have spatial heterogeneity, with regions of metabolically active cells interspersed within the extracellular matrix (composed of collagen as the encapsulating material and most likely also of exopolymeric material produced by biofilm cells), which is not stained by the metabolic dye. The thickness of the biofilm was estimated to be approximately 50 μm. The CaBChip was used to estimate the antifungal susceptibility of two drugs, fluconazole and amphotericin B, and the results are shown in Figure 3. Consistent with published reports on industry standard 96-well plate assays, the biofilms are resistant against fluconazole10 and the calculated IC50 for amphotericin B is 0.3 μg/mL11.
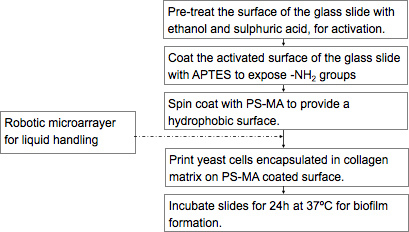 Figure 1. Flow chart for the fabrication of CaBChip.
Figure 1. Flow chart for the fabrication of CaBChip.
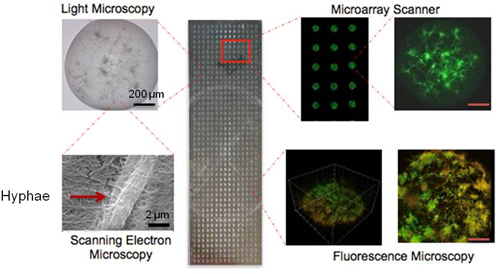 Figure 2. A picture of the high-thoughput CaBChip, printed using a robotic microarrayer and containing 768 spots on PS-MA-coated slides. Each hemispherical spot is 50 nL in volume, approximately 700 μm in diameter, and with a 1.2 mm separation between spots. Also shown (clockwise) are light microscopy, microarray scanner, 2D- and 3D-fluorescence microscopy, and scanning electron microscopy images of the individual biofilms on the CaBChip. The SEM figure at high magnification of 25,000x shows a hyphal filament of width 2 μm.
Figure 2. A picture of the high-thoughput CaBChip, printed using a robotic microarrayer and containing 768 spots on PS-MA-coated slides. Each hemispherical spot is 50 nL in volume, approximately 700 μm in diameter, and with a 1.2 mm separation between spots. Also shown (clockwise) are light microscopy, microarray scanner, 2D- and 3D-fluorescence microscopy, and scanning electron microscopy images of the individual biofilms on the CaBChip. The SEM figure at high magnification of 25,000x shows a hyphal filament of width 2 μm.
 Figure 3. Results of antifungal susceptibility testing and determination of IC50 values for (A) fluconazole and (B) amphotericin B using CaBChip. The fluorescence microscope images of amphotericin B-treated biofilms are shown in (C). The results are mean ± standard error mean for two separate chips containing 10 replicates for each condition.
Figure 3. Results of antifungal susceptibility testing and determination of IC50 values for (A) fluconazole and (B) amphotericin B using CaBChip. The fluorescence microscope images of amphotericin B-treated biofilms are shown in (C). The results are mean ± standard error mean for two separate chips containing 10 replicates for each condition.
Discussion
We have developed a cell-based high-density microarray, CaBChip, consisting of nanoliter volumes of Candida albicans biofilms. The microarray was printed on modified glass substrates, which allowed for robust attachment of collagen gel spots while providing hydrophobicity necessary for a non-spreading, hemispherical 3D gel. A single CaBChip can replace approximately eight 96-well plates, and several chips can be printed and processed at the same time. The chip utilizes nano-scale cultures, allows for easy and fast handling, is amenable to automation, minimizes manual labor and costs and is fully compatible with standard microarray technology and equipment. Moreover, the technology is flexible and can be easily adapted to other microorganisms. Despite miniaturization, the nano-biofilms formed on CaBChip display properties that are characteristic of the C. albicans biofilm lifestyle, including 3D architecture and drug resistance. Thus, the CaBChip allows for true high-throughput screening, and has the potential for a paradigm shift in antimicrobial drug discovery.
Disclosures
Jose L. Lopez-Ribot and Anand K. Ramasubramanian own equity in MicrobeHTS Technologies, Inc., which is developing antifungal agents. MicrobeHTS Technologies, Inc. provided no financial support for these studies.
Acknowledgments
This work was funded in part by grants from the South Texas Technology Management (POCrr 2009.041), the Institute for Integration of Medicine and Science from the National Center for Research Resources (UL 1RR025767), and from the National Institute of Dental & Craniofacial Research (5R21DE017294).
References
- Edmond MB. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Uppuluri P, Lopez-Ribot J, Ramasubramanian AK. Development of a High-Throughput Candida albicans Biofilm Chip. PLoS ONE. 2011;6:19036–19036. doi: 10.1371/journal.pone.0019036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 2001;18:163–170. [PubMed] [Google Scholar]
- Pierce CG. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008;3:1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl. Acad. Sci. U.S.A. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhofer D. Characteristics of resist films produced by spinning. Journal of Applied Physics. 1978;49 [Google Scholar]
- Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 2001;45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. Journal of Bacteriology. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk MA, Falkler WA, Meiller TF. Fungal biofilms and drug resistance. Emerg. Infect. Dis. 2004;10:14–19. doi: 10.3201/eid1001.030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobudic S, Lassnigg A, Kratzer C, Graninger W, Presterl E. Antifungal activity of amphotericin B, caspofungin and posaconazole on Candida albicans biofilms in intermediate and mature development phases. Mycoses. 2010;53:208–214. doi: 10.1111/j.1439-0507.2009.01690.x. [DOI] [PubMed] [Google Scholar]


