SUMMARY
The relationships between mitochondrial respiration, reactive oxygen species (ROS), and life span are complex and remain controversial. Inhibition of the target of rapamycin (TOR) signaling pathway extends life span in several model organisms. We show here that deletion of the TOR1 gene extends chronological life span in Saccharomyces cerevisiae, primarily by increasing mitochondrial respiration via enhanced translation of mtDNA-encoded oxidative phosphorylation complex subunits. Unlike previously reported pathways regulating chronological life span, we demonstrate that deletion of TOR1 delays aging independently of the antioxidant gene SOD2. Furthermore, wild-type and tor1 null strains differ in life span only when respiration competent and grown in normoxia in the presence of glucose. We propose that inhibition of TOR signaling causes derepression of respiration during growth in glucose and that the subsequent increase in mitochondrial oxygen consumption limits intracellular oxygen and ROS-mediated damage during glycolytic growth, leading to lower cellular ROS and extension of chronological life span.
INTRODUCTION
Mitochondrial respiration is the main source of reactive oxygen species (ROS) production in most cells (Boveris et al., 1972; Turrens, 1997). ROS, in turn, can damage cellular macromolecules such as DNA and proteins and are widely believed to contribute to functional decline and increased mortality of cells and organisms over time (Balaban et al., 2005; Finkel and Holbrook, 2000). However, while it is often incorrectly assumed that respiration and ROS production are directly proportional, the biological consequences of endogenous or exogenous changes in respiration with respect to ROS production and life span are complex and far from completely understood (Balaban et al., 2005; Speakman et al., 2004). For example, lifespan extension has been attributed to both increases and decreases in respiration (Barros et al., 2004; Bonawitz et al., 2006b; Dillin et al., 2002; Feng et al., 2001; Lin et al., 2002; Speakman et al., 2004), and ROS production by the mitochondrial electron transport chain can be either increased or decreased with pharmacological inhibitors of respiration depending on the site and mode of action of the drug (Kushnareva et al., 2002).
Mitochondria harbor their own DNA (mtDNA), containing a small but essential subset of genes encoding the oxidative phosphorylation (OXPHOS) machinery. Transcription of these mitochondrial genes is carried out by a dedicated RNA polymerase and its associated transcription factors (Bonawitz et al., 2006a). Though these proteins are distinct from those responsible for transcription of nuclear genes, they are themselves encoded in the nucleus and imported into the mitochondrion. More complex still is the mitochondrial translation system. Again, this system is distinct from that used in the expression of nuclear genes, but in this case, the protein components (ribosomal proteins, elongation factors, etc.) are encoded in the nucleus, whereas the RNA components (rRNAs and tRNAs) are encoded by mtDNA. Thus, mitochondrial ribosome biogenesis (and OXPHOS complex assembly as well) requires precise coordination of mitochondrial and nuclear gene expression, the mechanisms of which are largely unknown. Interestingly, a link exists between life span and mitochondrial translation, as we have previously shown that yeast mutants with globally reduced or imbalanced translation of OXPHOS components exhibit increased ROS and abbreviated chronological life span (Bonawitz et al., 2006b).
The TOR (target of rapamycin) kinases are highly conserved and regulate cell growth by phosphorylating a number of downstream targets in response to favorable nutrient conditions and/or growth-factor signals (Harris and Lawrence, 2003; Martin and Hall, 2005; Schmelzle and Hall, 2000). Arguably the most substantial mechanism by which TOR controls cell growth is via its influence on cytoplasmic translation. In the presence of favorable nutrients, TOR not only greatly promotes the production of new ribosomes but also increases translation from existing ribosomes (Barbet et al., 1996; Schmelzle and Hall, 2000). However, whether and how TOR regulates mitochondrial translation has not been addressed.
The TOR pathway has warranted increased attention from the aging-research community due to its apparently conserved influence on life span in a number of organisms. Decreased TOR signaling (using either RNAi or a hypomorphic TOR mutant) has been shown to extend life span in the nematode Caenorhabditis elegans (Vellai et al., 2003). Likewise, overexpression of a dominant-negative allele of TOR or inhibitors of TOR (Tsc1 and Tsc2) extends life span in Drosophila (Kapahi et al., 2004). Deletion of the Saccharomyces cerevisiae TOR1 gene (encoding one of two partially redundant TOR kinases in yeast) was shown to increase replicative life span, the number of cell divisions that a mother cell can support before senescence (Kaeberlein et al., 2005). It should be pointed out that this measure of cellular life span is distinguishable from chronological life span, defined as the amount of time that a nondividing cell can remain viable (Fabrizio and Longo, 2003). A recent high-throughput screen for gene deletions that extend chronological life span yielded a number of genes involved in nutrient sensing and influenced in part by the TOR pathway (Powers et al., 2006). In addition, it was shown that treatment of stationary-phase yeast cultures with rapamycin, a specific inhibitor of TOR, also extends chronological life span (Powers et al., 2006). However, the TOR1 gene was not identified in this screen, even though its deletion decreases TOR signaling.
The determination of the mechanism by which TOR signaling limits life span is of critical importance. It has been claimed that the chronological life-span extension resulting from TOR inhibition in yeast is dependent on upregulation of stress-response genes by the Msn2/4 transcription factors. However, it was shown that deletion of GLN3 (taken as a proxy for inhibition of TOR) dramatically extends life span even in an msn2/msn4 double mutant (Powers et al., 2006). Thus, although the Msn2/4 factors and their downstream targets are undoubtedly involved in life-span regulation (Fabrizio and Longo, 2003; Fabrizio et al., 2001), it appears that the majority of the life-span extension afforded by inhibition of TOR is Msn2/4 independent. Furthermore, we have previously shown that respiration and chronological life span are positively correlated in yeast, with those strains that have the highest level of respiration showing the longest life span (Bonawitz et al., 2006b). Therefore, we set out with the goal of determining the mechanism of life-span extension afforded by decreased TOR signaling and whether alterations in mitochondrial respiration are involved.
RESULTS
Reduced TOR Signaling via Deletion of TOR1 Extends Chronological Life Span and Increases Respiration
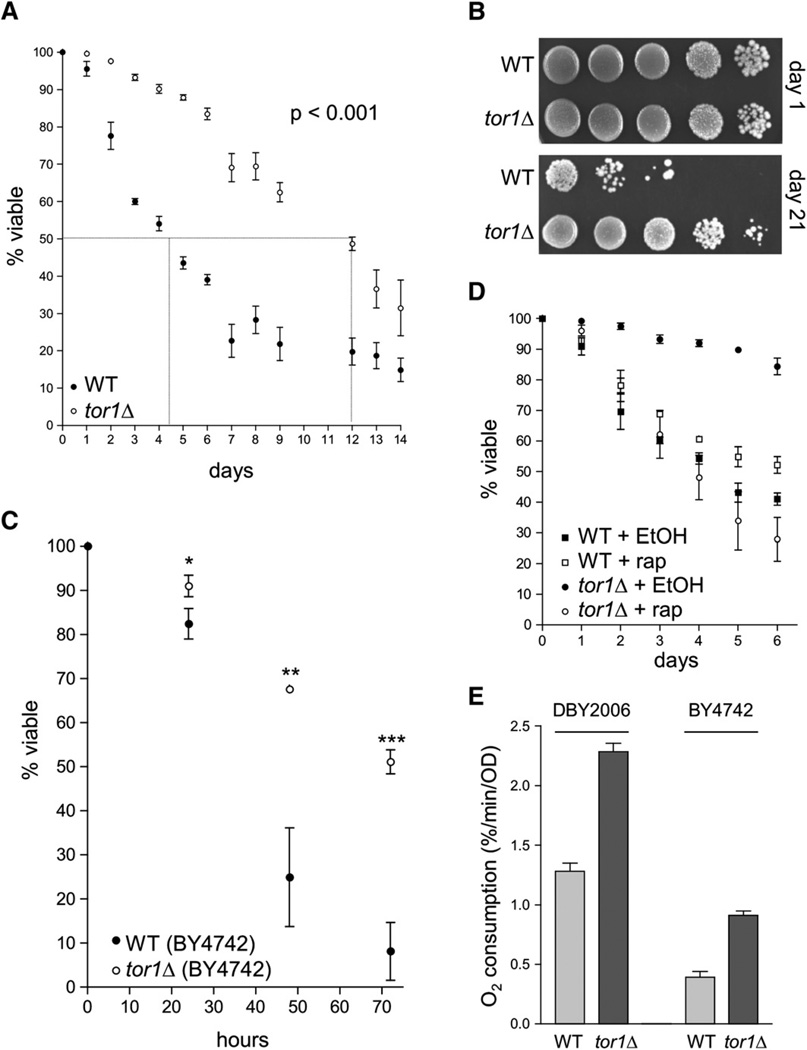
The S. cerevisiae genome encodes two partially redundant TOR proteins called Tor1p and Tor2p that are interchangeable as components of the nutrient-sensing TOR complex I. However, Tor2p is also found in another complex (TOR complex II), which is involved in cytoskeletal remodeling. The functions of both TOR complexes are indispensable; therefore, the TOR2 gene is essential, while the TOR1 gene is not (Loewith et al., 2002). Although it has been shown that deletion of TOR1 postpones replicative aging (Kaeberlein et al., 2005), its effect on chronological life span has not been reported. To address this, we knocked out the TOR1 gene in S. cerevisiae strain DBY2006, which we have used for aging studies previously (Bonawitz et al., 2006b). Deletion of TOR1 substantially extended chronological life span, increasing median survival almost 3-fold (wild-type ~4.5 days, tor1 null ~12 days) (Figure 1A). By 21 days in culture, the vast majority of wild-type cells had died (>99.9%), whereas many tor1 null cells remained viable (Figure 1B). Deletion of TOR1 also extended the chronological life span of the relatively short-lived strain BY4742 (Figure 1C), one of the two haploid genetic backgrounds of the widely used Yeast Knockout Collection available from Open Biosystems. Analysis of growth curves of all of the strains in Figure 1 demonstrates that, while deletion of TOR1 resulted in a slightly slower growth rate in log phase as predicted, the TOR1 and tor1 null strains both reached stationary phase at day 1 postinoculation (see Experimental Procedures and Figure S3 in the Supplemental Data available with this article online). We also compared the life span of tor1 null cells to the life span of wild-type cells treated with rapamycin, a specific inhibitor of the TOR pathway that has previously been reported to extend chronological life span (Powers et al., 2006). Treatment of stationary phase cultures of wild-type cells with rapamycin extended chronological life span compared to treatment with drug vehicle (ethanol), albeit to a much lesser extent than deletion of TOR1 (Figure 1D). Treatment of tor1 null cultures with rapamycin actually decreased life span (Figure 1D). This effect was partially suppressed by expressing a rapamycin-resistant allele of Tor2p (data not shown), suggesting that optimal life-span extension is achieved in tor1 null strains by decreasing overall TOR signaling while leaving Tor2p-specific signaling intact.
Figure 1. Deletion of TOR1 Extends Chronological Life Span and Increases Respiration in Two Different Yeast Strains.
(A) Chronological life-span curves (i.e., % viability versus days in stationary phase) of wild-type (closed circles) and tor1 null cells (open circles) over two weeks, as assessed by staining with trypan blue. The curves shown were generated by compiling survival measurements from 11 different samples performed in 5 independent experiments. For clarity, we have marked the approximate median survival point of each curve with a dotted line. In this and all other figures, error bars represent standard deviation unless otherwise noted.
(B) Serial 10-fold dilutions of stationary-phase cultures plated onto rich medium (YPD) at day 1 (upper panel) and day 21 (lower panel).
(C) Chronological life-span measurements of wild-type and tor1 null derivatives of the short-lived strain BY4742 plotted as described in (A). *p < 0.05; **p < 0.005; ***p < 0.0005.
(D) Chronological life-span curves of wild-type and tor1 null cells treated with 50 nM rapamycin or drug vehicle (ethanol). Survival was monitored by trypan blue staining.
(E) Mitochondrial oxygen consumption of wild-type and tor1 null derivatives of strains DBY2006 (left) and BY4742 (right) at day 1 postinoculation in SD medium.
In a previous study, we observed a striking correlation between respiration at day 1 postinoculation and chronological life span in a number of strains, with those that have the highest levels of respiration exhibiting the longest life span (Bonawitz et al., 2006b). Therefore, we tested whether this correlation held true in the tor1 null strain. At day 1 postinoculation in glucose medium, tor1 null strains in both the DBY2006 and BY4742 backgrounds exhibited a reproducible ~2-fold increase in oxygen consumption in comparison to their respective wild-type parent strains (Figure 1E). Interestingly, the short-lived BY4742 strain showed substantially lower rates of oxygen consumption than DBY2006, with both wild-type and tor1 null strains of BY4742 exhibiting only ~30% of the oxygen consumption of the corresponding DBY2006 strains (Figure 1E). This observation extends the correlation between respiration and life span noted above (Bonawitz et al., 2006b). Treatment of wild-type cells with rapamycin likewise led to increased respiration, though to a lesser degree than deletion of TOR1 (Figure S1). In contrast, treatment of tor1 null cells with rapamycin effected a substantial decrease in respiration (Figure S1). The reasons for this decrease are unclear; however, coupled with the observation that rapamycin decreases life span in tor1 null cells, it appears likely that excessive inhibition of TOR signaling leads to deleterious consequences for both the maintenance of respiration and life span.
Respiration Is Required for Life-Span Extension in tor1 Null Strains
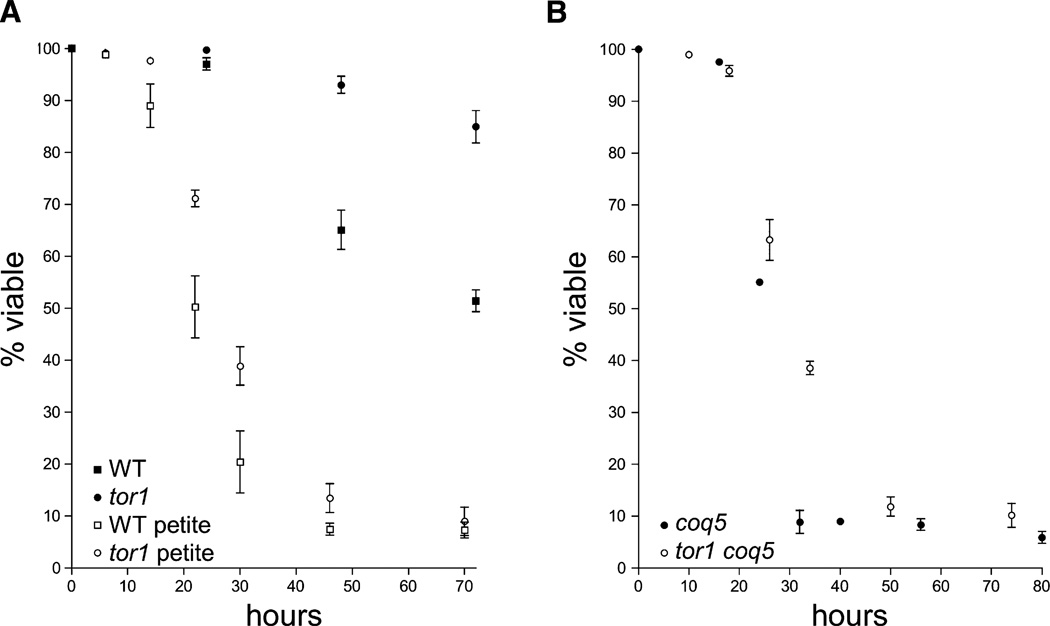
Based on our extensive observations of a correlation between oxygen consumption and life span as well as previous suggestions that high levels of respiration correlate with long life (Barros et al., 2004; Lin et al., 2002; Speakman et al., 2004), we hypothesized that the increased respiration exhibited by tor1 null cells was directly responsible for their enhanced longevity. To test this, we generated respiration-defective (petite) isolates of our wild-type and tor1 null strains using a standard ethidium bromide treatment that induces deletion and/or loss of mtDNA. Since mtDNA encodes essential OXPHOS components, these strains are unable to respire. We found that the life span of both wild-type and tor1 null petite derivatives of strain DBY2006 was similar to the short-lived strain BY4742 (Figure 1C) and much briefer than their respiration-competent parent strains, consistent with an important role for mitochondrial respiration in the maintenance of life span (Figure 2A). However, in contrast to the life-span extension seen in respiration-competent BY4742 cells, deletion of TOR1 in petite strains extended life span only very minimally, and only at early time points. To confirm that this was due to the elimination of respiration and not other unexpected consequences resulting from the inactivity or elimination of mtDNA, we also measured the life span of wild-type and tor1 null strains that are petite due to deletion of the nuclear COQ5 gene. Coq5p is essential for the synthesis of the mitochondrial electron carrier coenzyme Q, and although coq5 mutants are unable to respire, mtDNA is maintained. The life span of the coq5 petite mutant mirrored that of the mitochondrial petites described above (Figure 2B; see also growth curves in Figure S3). Therefore, we conclude that mitochondrial respiration is required for the majority of the ability of TOR1 deletion to extend life span in yeast.
Figure 2. Deletion of TOR1 Fails to Extend Life Span in Petite Strains that Are Unable to Respire.
Life-span curves of mitochondrial petite derivatives of wild-type and tor1 null strains due to inactivated mtDNA (A) or deletion of the COQ5 gene (B) plotted as described in Figure 1, except that time is reported in hours.
Differences in Both Respiration and Life Span between Wild-Type and tor1 Null Strains Are Glucose Dependent
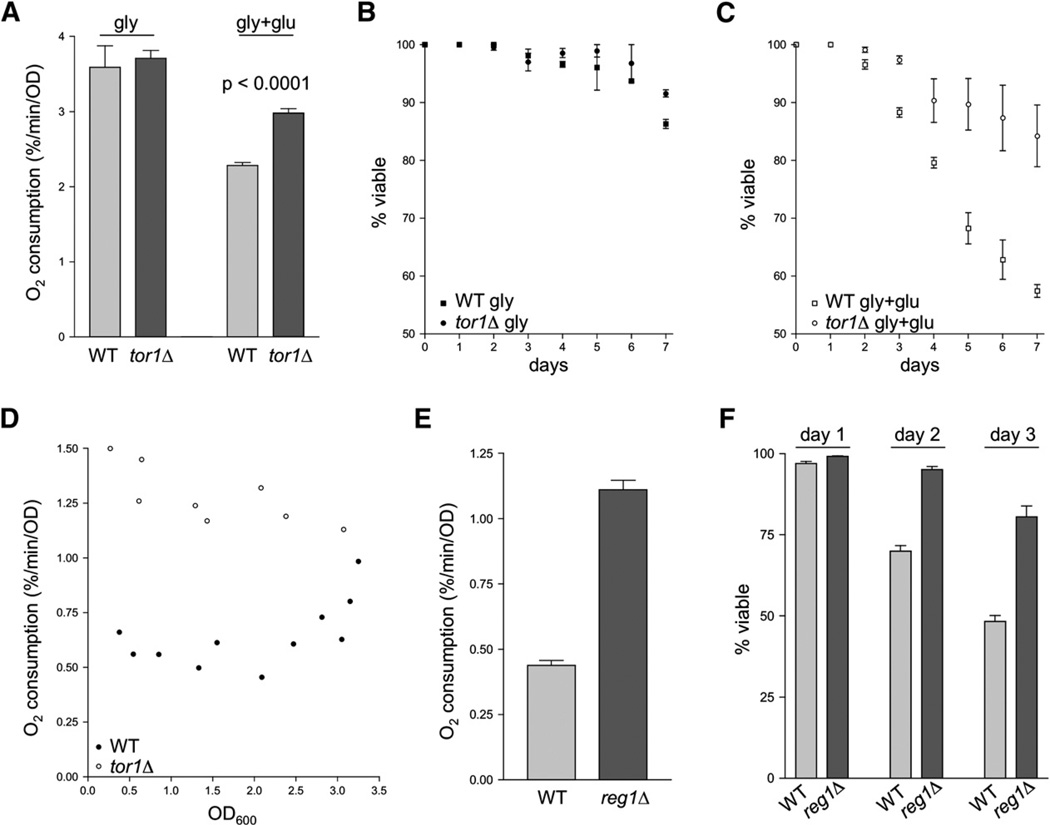
The main function of the TOR pathway is nutrient sensing, the coordination of cellular metabolism with nutrient availability. Furthermore, TOR has been shown to be involved in modulating the expression of at least some genes in response to glucose (Tomas-Cobos et al., 2005). Since respiratory activity in facultative anaerobes like S. cerevisiae is contingent on the availability of glucose, we tested whether the tor1 null phenotypes we observed were glucose dependent. We found that oxygen consumption levels of wild-type and tor1 null cells at day 1 postinoculation in growth medium containing glycerol as a sole carbon source (SG medium) were indistinguishable from one another (Figure 3A, left) and, as expected, were significantly higher than those exhibited by day 1 glucose cultures (compare Figure 3A with Figure 1E). Upon addition of glucose to glycerol cultures, a difference in the oxygen consumption of wild-type and tor1 null cells was observed within 4 hr (Figure 3A, right). Consistent with the correlation between respiration and life span described above, the life spans of wild-type and tor1 null cultures grown in glycerol were also indistinguishable, and both strains showed longer life span in glycerol medium than in glucose (compare Figure 3B with Figure 1A). Addition of glucose to glycerol cultures at day 1 postinoculation was sufficient to restore a difference in the life span of wild-type versus tor1 null cultures by causing a greater decrease in life span in wild-type than in tor1 null cells (Figure 3C). We observed similar results for cells grown in media containing raffinose as the sole carbon source (data not shown). When oxygen consumption measurements were taken throughout a typical growth curve in glucose-containing medium, it became clear that as wild-type cultures approach saturation (where glucose is exhausted), their oxygen consumption increases, whereas respiration in tor1 cultures remains high at all stages of growth (Figure 3D). After extended amounts of time in saturation (2 days and beyond), oxygen consumption of wild-type and tor1 null cultures was identical when corrected for viability (data not shown). We conclude from these data that the inhibitory function of the TOR pathway on respiration and life span is only active in the presence of glucose.
Figure 3. The Increased Respiration and Life-Span Phenotypes of tor1 Null Cells Are Glucose Dependent, and a Known Glucose-Repression Mutant, reg1, Phenocopies tor1 Null.
(A) Mitochondrial oxygen consumption of wild-type and tor1 null cultures growing in glycerol medium before (left) and 4 hr after (right) addition of glucose (to 2%). Error bars in all panels of this figure represent the standard deviation of three replicates.
(B) Chronological life-span curves of wild-type and tor1 null cells grown in glycerol medium.
(C) As in (B), except that glucose was added (to 2%) at day 1.
(D) Mitochondrial oxygen consumption of wild-type and tor1 null cells, graphed as a function of culture density (OD600). The points shown are the combination of three independent experiments, and each represents an individual oxygen consumption reading.
(E) Oxygen consumption of reg1 null cells and their isogenic wild-type control (BY4741) at day 1.
(F) Chronological life span measurements of the cultures described in (E) over three days in stationary phase.
Increased Respiration Due to Faulty Glucose Repression in reg1 Null Strains Mimics the Extension of Life Span by TOR1 Deletion
Having shown that eliminating respiration severely attenuates the effect of TOR signaling on life span and that increasing respiration via growth in glycerol extends life span, we sought to test whether another genetic manipulation reported to increase respiration would influence life span in a manner similar to TOR inhibition. As described above, wild-type yeast cells prefer glycolysis over respiration and repress respiration in the presence of glucose. We therefore tested a known glucose-repression-defective mutant, reg1Δ (Matsumoto et al., 1983), for respiration and stationary-phase survival phenotypes. Like deletion of TOR1, deletion of REG1 led to substantially higher respiration levels at day 1, increasing oxygen consumption more than 2-fold compared to the isogenic parent strain (Figure 3E). As predicted, the higher level of oxygen consumption seen in reg1 null cells correlated with increased stationary-phase survival compared to control cells (Figure 3F), thus adding additional weight to our conclusion that increased respiration is responsible for life-span extension in tor1 null cells.
Life-Span Extension by TOR1 Deletion Is Independent of SOD2 despite the Upregulation of Sod2p Abundance
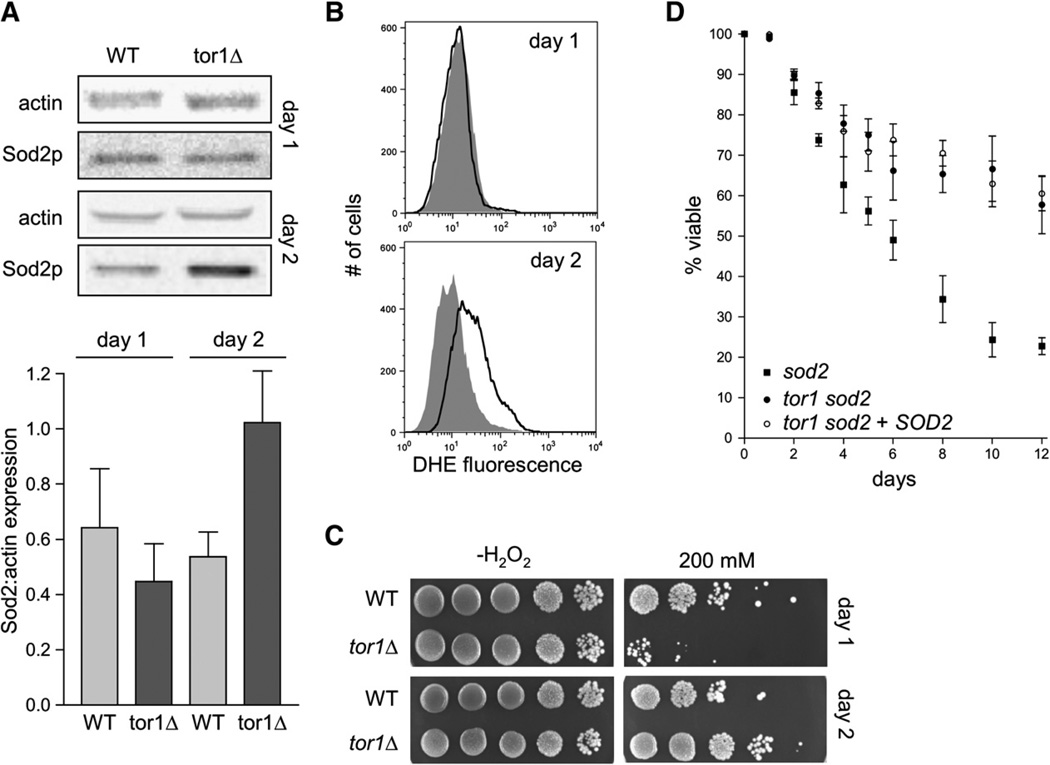
Some of the life-span extension resulting from TOR inhibition has been previously attributed to increased stress resistance (Powers et al., 2006). Also, it has been shown that the life-span extension resulting from inhibition of the well-characterized Sch9p pathway is completely eliminated by deletion of SOD2, the gene encoding the mitochondrial-matrix-localized manganese-dependent superoxide dismutase (Fabrizio et al., 2003). We therefore investigated the potential role of SOD2 in the phenotype of tor1 null cells. We first measured steady-state abundance of Sod2p by western blot analysis at both day 1 and day 2 postinoculation in wild-type and tor1 null cells. Surprisingly, we found that Sod2p levels were identical in wild-type and tor1 null cells at day 1 and were increased in tor1 null cells only at day 2 of stationary phase (Figure 4A). (One might predict that if TOR signaling influences SOD2 expression, then deletion of TOR1 would have the greatest effect during growth, when TOR signaling is most active.) Consistent with this observation, we found that ROS abundance in wild-type and tor1 null cells was identical at day 1 postinoculation but diverged at day 2, with wild-type cells, but not tor1 null cells, exhibiting significantly more ROS at day 2 than at day 1 (Figure 4B). We also measured resistance of wild-type and tor1 null cells at day 1 and day 2 to hydrogen peroxide, an inducer of oxidative stress. We found that at day 1, tor1 null cells were more sensitive to hydrogen peroxide than wild-type cells were, but, interestingly, the situation was reversed at day 2 (Figure 4C). We next deleted the SOD2 gene in wild-type and tor1 null cells to test whether life-span extension via inhibition of TOR signaling was dependent on the presence of Sod2p. Neither deletion of SOD2 nor its replacement with a plasmid-borne copy had any measurable impact on the ability of TOR1 deletion to extend chronological life span (Figure 4D). Taken together, these data indicate that, in contrast to previously characterized pathways involved in the regulation of chronological life span, the expression of superoxide dismutase is unlikely to contribute substantially to the increased chronological life span resulting from TOR1 deletion. Nonetheless, our results do implicate TOR signaling in the regulation of expression and/or stability of Sod2p.
Figure 4. Sod2p Is Upregulated Specifically in Stationary Phase in tor1 Null Cells and Contributes to Stress Resistance, but Not to Life-Span Extension.
(A) Western blot analysis of Sod2p levels in wild-type and tor1 null cells at day 1 and day 2 of stationary phase. Shown at top is a pair of representative samples, and shown below is quantification of Sod2p relative to actin from three biological replicates.
(B) ROS levels of wild-type and tor1 null cells at day 1 postinoculation, measured with the ROS-sensitive fluorescent dye dihydroethidium (DHE) and flow cytometry. The y axis shows the number of cells measured at a given point on the x axis, which is a logarithmic scale of arbitrary fluorescence units. The black line represents the fluorescence profile of the wild-type strain, and the filled gray curve represents that of the tor1 null strain.
(C) Serial 10-fold dilutions of wild-type and tor1 null cells treated with the indicated levels of hydrogen peroxide (H2O2) at either day 1 or day 2 of stationary phase.
(D) Life-span curves of sod2 mutants (closed squares), tor1 sod2 double mutants (closed circles), and tor1 sod2 double mutants transformed with a plasmid bearing wild-type SOD2 (open circles) compared to appropriate empty-vector control strains.
Limiting Culture Aeration Early in Growth Mimics TOR1 Deletion
One potential consequence of the increased respiration observed in the tor1 null strain is that intramitochondrial oxygen levels could be lower than in the wild-type strain, a situation that is unfavorable for ROS production (Vinogradov and Grivennikova, 2005). We reasoned that if this is the case, then limiting oxygen availability to wild-type cells should be beneficial for life span. We grew wild-type and tor1 null cultures under hypoxic conditions by limiting culture aeration and measured viability compared to normal growth conditions. Wild-type cultures grown for 4 or 5 days in hypoxic conditions contained significantly more viable cells than cultures grown normally. In contrast, hypoxia had no effect on the viability of tor1 null cultures (Figure S2A).
As described above, we observed a difference between the respiratory rate of wild-type and tor1 null cells only during the initial stages of growth (i.e., by day 2, both strains showed similar levels of respiration). If this difference in respiration is responsible for the extended life span in tor1 null cells, and if limiting oxygen mimics this effect, then we reasoned that it should be possible to effect a substantial increase in viability in wild-type cultures by limiting aeration only during the first 2 days in culture. Remarkably, we found that growing wild-type cells for just 2 or 4 days in hypoxia and then switching to normoxia rendered their viability indistinguishable from tor1 null cells at day 21 (compare Figure S2B with Figure 1B). We conclude that an oxygen-dependent process that occurs early in the course of normally growing wild-type cultures is responsible for their rapid loss of viability later in stationary phase and that tor1 null cells are resistant to this process.
TOR Regulates Mitochondrial Translation
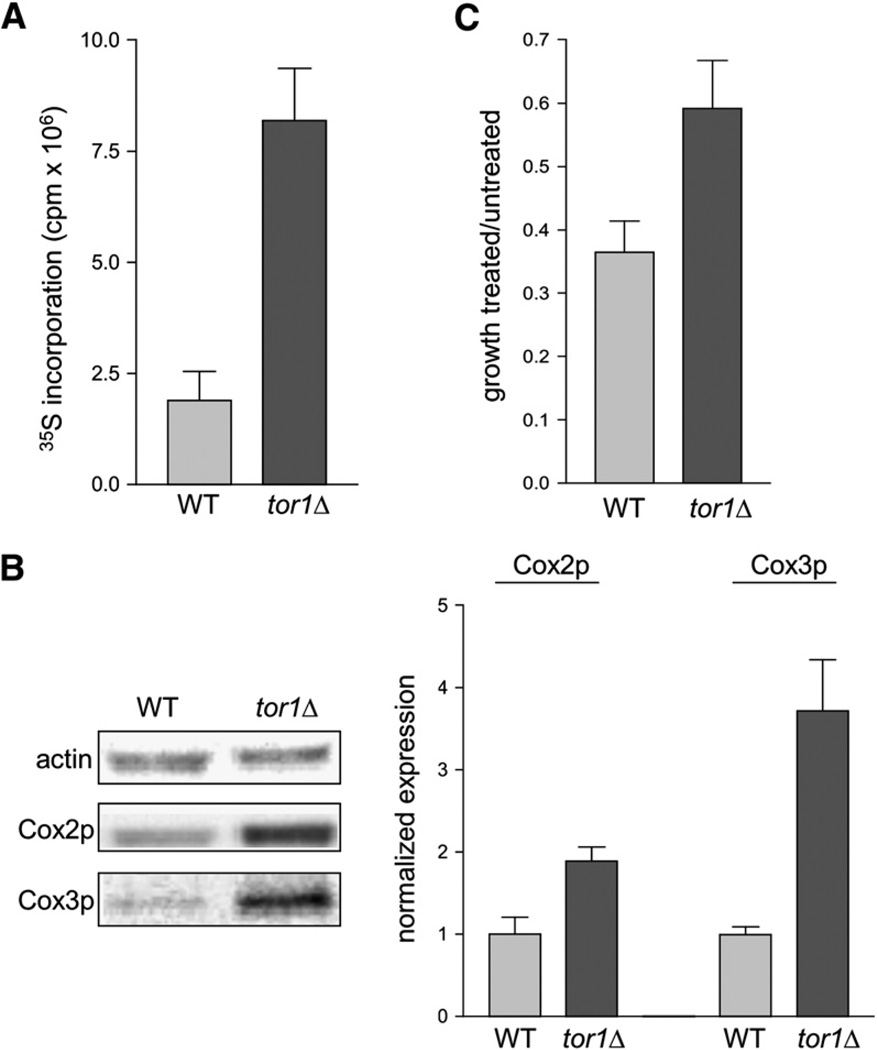
Given the known role of the TOR pathway in regulating cytoplasmic translation of nucleus-encoded genes, we tested the hypothesis that the increase in mitochondrial respiration that we observed in tor1 null strains was mediated through an effect on mitochondrial translation. To address this, we first quantified mitochondrial translation rates using a standard in vivo labeling approach that we have used previously to measure mitochondrial protein synthesis (Rodeheffer and Shadel, 2003). Cultures of wild-type and tor1 null cells at day 1 postinoculation were treated with emetine (to inhibit cytoplasmic translation) and then incubated with 35S-labeled methionine to measure incorporation into mtDNA-encoded proteins. In this assay, tor1 null cells incorporated ~3-fold more radiolabel than wild-type cells, indicating a greater rate of mitochondrial translation (Figure 5A).
Figure 5. tor1 Null Cells Exhibit Increased Translation and Steady-State Levels of Mitochondrially Encoded Proteins.
(A) Specific incorporation of 35S-labeled methionine into mtDNA-encoded OXPHOS subunits in wild-type and tor1 null strains (DBY2006 genetic background), plotted in millions of counts per minute (cpm × 106).
(B) Western blot analysis of two mitochondrially encoded subunits of cytochrome c oxidase, Cox2p and Cox3p, at day 1 postinoculation. The left panel shows levels of actin (a loading control), Cox2p, and Cox3p in two representative samples, and the right panel shows quantification of Cox2p and Cox3p in three independent samples. For quantification, the ratio of actin:Cox2p or actin:Cox3p was determined for all samples and then normalized to the average wild-type ratio.
(C) Chloramphenicol resistance of wild-type and tor1 null strains. Shown is the ratio for both wild-type and tor1 null cells of growth in glycerol medium containing 0.25 mg/ml chloramphenicol to growth in glycerol without chloramphenicol.
Consistent with the results described above, we found by western blotting that the steady-state levels of the mitochondrially encoded cytochrome c oxidase (complex IV) subunits Cox2p and Cox3p were ~2-fold and ~3.5-fold greater, respectively, in tor1 null strains (Figure 5B). Finally, we measured the resistance of wild-type and tor1 null cells to chloramphenicol, an inhibitor of mitochondrial ribosomes, hypothesizing that if tor1 null cells do indeed have greater rates of mitochondrial translation due to increased mitochondrial ribosome biogenesis, they would then exhibit resistance to this drug. Wild-type and tor1 null cells were grown in glycerol medium (which requires respiration and therefore mitochondrial gene expression) containing either 0.25 mg/ml chloramphenicol or drug vehicle. Chloramphenicol treatment of wild-type cells led to an ~65% decrease in growth rate compared to untreated cells, whereas treatment of tor1 null cells inhibited growth by only ~40% (Figure 5C). Taken together, these results indicate that the TOR pathway normally serves to inhibit respiration by repression of synthesis of mtDNA-encoded proteins. Conversely, deletion of the TOR1 gene increases respiration via the derepression of these genes and perhaps also by increasing mitochondrial ribosome biogenesis.
DISCUSSION
Due to its involvement in many critical biological processes, interest in the TOR signaling pathway has grown at an astounding rate since its discovery in the early 1990s. Recently it has become clear that TOR plays an important role in the regulation of life span in both metazoans and yeast, though the precise mechanisms by which this regulation occurs has been unclear. The main conclusion we draw from our experimental results is that inhibition of TOR signaling extends life span in yeast by increasing respiration via enhanced mitochondrial gene expression. First, deletion of the TOR1 gene increases respiration (Figure 1E) and extends life span (Figure 1A) in two different yeast strains, DBY2006 and BY4742. Both wild-type and tor1 null strains of BY4742 show lower respiration than their DBY2006 counterparts, as well as shorter life span. Treatment of wild-type cells with rapamycin, which has previously been reported to extend life span, also increases respiration (Figure S1), though to a lesser degree than deletion of TOR1. Growth in glycerol medium renders both respiration and life span indistinguishable between wild-type and tor1 null cells, increasing both substantially in comparison to growth in glucose (Figures 3A–3C). Furthermore, another genetic manipulation that results in higher rates of respiration (REG1 deletion) similarly extends life span (Figures 3E and 3F). And finally, elimination of respiration in strains lacking functional mtDNA (Figure 2A) or via deletion of COQ5 (Figure 2B) (to eliminate production of the essential electron carrier coenzyme Q) almost completely abolishes life-span extension in a tor1 null strain, showing that respiration is responsible for the vast majority of the influence of the TOR pathway on life span. It is possible that there is a slight (i.e., several hour) extension of life span in petite tor1 null strains at early time points (Figures 2A and 2B). Though it is arguable whether this effect is of biological significance, it is possible that eliminating respiration unmasks a slight difference between wild-type and tor1 null cells due to influences on stress resistance.
As mentioned above, the increase in both respiration and life span in tor1 null cells is dependent on the presence of glucose. Thus, in wild-type cells, the TOR pathway normally plays a role in glucose-dependent inhibition of respiration, extending further the extensive list of cellular functions influenced by TOR. We do not yet know whether TOR is involved in the well-characterized process of canonical glucose repression or represents a previously unknown input to the regulation of respiration. However, we have presented evidence that the mechanism by which TOR inhibition serves to increase respiration in the presence of glucose is via upregulation of mitochondrial gene expression (Figure 5). Although inhibition of the TOR pathway leads to decreased global (cytoplasmic) translation (Barbet et al., 1996; Gingras et al., 2001; Schmelzle and Hall, 2000), we have shown that tor1 null cells exhibit a higher rate of mitochondrial translation (Figure 5A) and steady-state abundance of several mitochondrially encoded OXPHOS components (Figure 5B) and are resistant to an inhibitor of mitochondrial ribosomes (Figure 5C). We therefore conclude that TOR influences the reciprocal allocation of resources between cytoplasmic and mitochondrial translation. Consistent with our results showing a direct effect of TOR on mitochondrial translation is the report by Shamji et al. (2000) showing that treatment of yeast with rapamycin increases the expression of a number of genes involved in the TCA cycle, mitochondrial ribosome biogenesis, and assembly of the OXPHOS complexes. Thus, the TOR signaling pathway apparently has a global and pivotal role in regulating mitochondrial gene expression and oxidative metabolism.
Stress-resistance pathways, and specifically the Sod2p protein, have been shown to be very important players in the maintenance of chronological life span (Fabrizio and Longo, 2003; Fabrizio et al., 2001). Deletion of the SOD2 gene, for example, completely eliminates the life-span extension observed in a sch9 null strain (Fabrizio et al., 2003). However, it is clear from our results that inhibition of TOR is acting in another fashion. We found that deletion of SOD2 had no effect on the ability of TOR1 deletion to extend life span (Figure 4D). Interestingly, although conventional wisdom states that inhibition of TOR signaling should lead to generally increased stress resistance, several studies have shown that deletion of TOR1 in S. cerevisiae actually sensitizes cells to a number of different stresses, including heat, salt, and cell-wall stresses (Crespo et al., 2001; Reinke et al., 2004). Similarly, tor1 null strains of Schizosaccharomyces pombe exhibit increased sensitivity to heat, oxidative, pH, and osmotic stresses (Kawai et al., 2001; Weisman and Choder, 2001). Consistent with these results, we found that tor1 null strains are sensitive to hydrogen peroxide at day 1 (Figure 4C). Surprisingly, we found that deletion of TOR1 influenced SOD2 levels only in later stationary phase, causing its upregulation (and increased resistance to oxidative stress) only at day 2 postinoculation (Figures 4A and 4C). Based on our oxygen consumption data (where we see effects only at day 1), as well as the generally accepted fact that TOR signaling is largely inactivated during stationary phase/starvation, we had predicted that any effect of TOR inhibition on SOD2 would be manifest sooner rather than later. These data lead us to speculate the existence of alternative TOR signaling that occurs only during stationary phase. Indeed, this hypothesis is consistent with our observation that the treatment of tor1 null cells with rapamycin actually causes them to die (Figure 1D), indicating that some level of TOR signaling in stationary phase is essential to maintain viability.
Although Sod2p is not required for life-span extension in tor1 null cells (Figure 4D), it is still quite probable that ROS are involved in the life-span extension by TOR given that the presence of oxygen (the substrate for ROS) is required to observe a difference in life span between wild-type and tor1 null strains (Figure S2). We previously showed that inhibition of respiration via imbalanced production of OXPHOS components leads first to ROS production and subsequently to the complete inactivation of respiration (Bonawitz et al., 2006b). We interpreted these data as evidence for the often cited “vicious cycle” of oxidative damage, wherein oxidatively damaged components of the OXPHOS machinery result in higher levels of ROS production, which then cause more damage, etc. Conversely, the increased respiration in tor1 null cells could result in decreased production of ROS by the mitochondrial electron transport chain. Although we do not see a difference in the amount of ROS at day 1 postinoculation, it should be pointed out that our ROS data are measurements of steady-state abundance. Thus, if hydrogen peroxide sensitivity in a tor1 null strain is taken as an indication that its antioxidant capacity is lower than that of wild-type, then the observation of equivalent ROS steady-state levels in the two strains implies that tor1 null cells produce less ROS. Consistent with this hypothesis, we have observed fewer numbers of mitochondrial petites in tor1 null cultures (data not shown), suggesting lower amounts of damage to mitochondrial DNA (Shadel, 1999). Alternatively, it is possible that there is a relevant difference in ROS levels between wild-type and tor1 null cells but that it was undetectable by the methods used.
Although it is often casually stated that respiratory rate should correlate with ROS production, there is mounting evidence from a number of groups suggesting that it is not the absolute number of electrons flowing through the electron transport chain but rather the state of the chain that determines the rate of ROS production. It has been shown, for example, that high membrane potential favors the formation of ROS (Korshunov et al., 1997; Papa and Skulachev, 1997), as does high oxygen concentration (Vinogradov and Grivennikova, 2005), in both cases at levels found in vivo. Also, when isolated mitochondria are shifted from state IV (ADP-limited) to state III (substrate-limited) respiration, they show a substantial increase in the amount of oxygen they consume without a concomitant increase in ROS (Barja, 1999). The emerging model posits, in fact, that conditions of rest (low metabolic rate) can actually favor ROS production (Papa and Skulachev, 1997; Wallace, 2005). In the presence of excess oxidizable substrates (i.e., calories), respiration is limited only by the availability of ADP and approaches state IV. Since ADP is required for complex V (ATP synthase) to allow protons to flow back into the matrix, membrane potential climbs, electron transport stalls as all sites become reduced, and the dwell time of radical-generating intermediates increases. These radical intermediates then react with the growing concentration of oxygen not being utilized by the stalled respiratory chain. Consistent with this model, caloric restriction, which extends life span in mice (Masoro, 2003), has been shown to increase respiratory rate (Lambert and Merry, 2004). Thus, nutrient sensing, caloric restriction, mitochondrial respiration, and life span all appear to be linked. Furthermore, we previously showed that a partial block in the respiratory chain due to imbalanced production of mitochondrially encoded components generates ROS and shortens life span in yeast (Bonawitz et al., 2006b), and Barros et al. (2004) showed the converse, that ROS production could be inhibited by activating respiration (Barros et al., 2004). Taken together with the results of this study, it is tempting to speculate that the measurable mitochondrial respiration occurring in yeast growing glycolytically is similarly acting as a sort of “release valve,” oxidizing the electron transport chain and reducing unwanted oxygen as a means to limit ROS production, and that this activity is increased in strains with an inhibited TOR pathway.
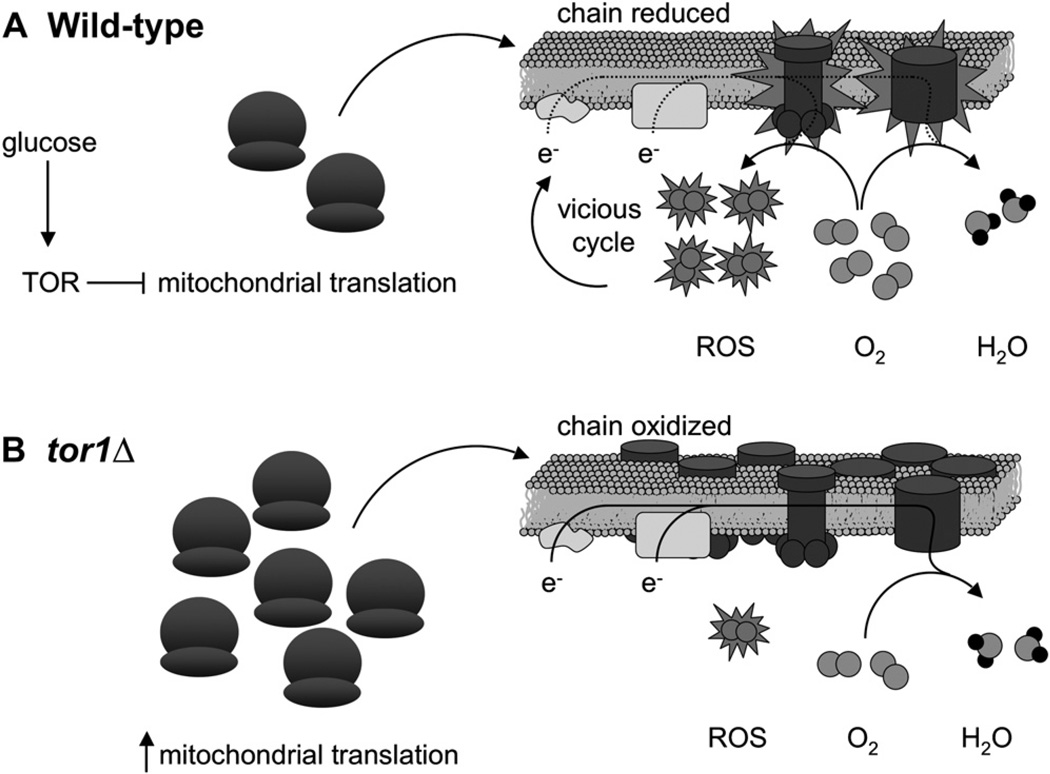
We propose the following model to describe all of our results taken together (Figure 6). In wild-type cells in the presence of glucose, TOR inhibits mitochondrial translation and/or ribosome biogenesis. This inhibition allows cells to grow more quickly, but as a result of the low number or activity of respiratory complexes, oxygen concentration is high and the transit time of electrons through the chain is protracted, reducing the chain and favoring the formation of ROS. These ROS can damage cellular components including mtDNA and OXPHOS components, setting into motion the vicious cycle of oxidative stress that eventually results in death. However, the evolutionary benefits for a facultative anaerobe of quick growth and utilization of resources while they are available outweigh the cost of short life span; thus, inhibition of respiration by TOR is favored by evolution and is retained. Disruption of TOR signaling, via deletion of TOR1, derepresses respiration by increasing mitochondrial translation and generates less ROS as a result of the presence of more OXPHOS complexes. This postpones the initiation of the vicious cycle and extends life span but comes at the cost of slowing growth. Thus, we propose that oxidative stress experienced during the initial stages of culture can manifest itself much later by damaging cellular components, particularly in stationary phase, where the ability to make new OXPHOS complexes and other mitochondrial components is likely compromised due to the downregulation of cytoplasmic translation. Supporting this model are our results showing that limiting culture aeration (i.e., oxygen concentration) mimics the effects of reduced TOR signaling, even if administered only during the initial stages of culture growth.
Figure 6. A Speculative Model Describing the Influence of TOR Signaling on Chronological Life Span through Effects on Mitochondrial Translation, Respiration, and ROS Production.
Based on our results, TOR normally negatively influences mitochondrial gene expression by repressing mitochondrial translation and/or ribosome biogenesis (depicted as the number of mitochondrial ribosomes) during growth in glucose (A). While likely optimal for growth on glucose via glycolysis and subsequent fermentation, this has a deleterious effect on life span by promoting ROS production. We propose that this occurs because a less active electron transport chain (consisting of the OXPHOS complexes shown embedded in the inner mitochondrial membrane) first accumulates reduced and reactive redox components (shown as OXPHOS components with explosions) that can more readily transfer electrons (e−) to oxygen (O2; gray double circles) prior to complex IV to generate ROS (O2 molecules with explosions) instead of water (H2O; gray circles with black circles attached) and additionally allows more oxygen, the substrate for ROS, to accumulate in mitochondria. In other words, the electron transport chain becomes reduced and mitochondrial oxygen concentration rises, favoring ROS production and initiating the vicious cycle of mitochondrial and cellular damage that further reduces mitochondrial respiration and limits life span. In tor1 null cells (B), mitochondrial translation and/or ribosome biogenesis is derepressed, leading to a greater steady-state level of OXPHOS complexes in the inner mitochondrial membrane. This scenario is proposed to facilitate oxidization of the mitochondrial electron transport chain and keep intramitochondrial oxygen levels low, limiting ROS production and accumulation and delaying the onset of the life-span-limiting vicious cycle.
Although our results apparently contradict a recent publication showing that TOR activates respiration in human cells (Schieke et al., 2006), we feel that this can be simply explained by differences in the nature of glucose metabolism between humans and yeast, with the former favoring aerobic and the latter anaerobic metabolism. That is, both sets of results are consistent with the idea that the pro-growth TOR pathway has a hand in increasing glucose metabolism, but in humans, this glucose is ultimately fed into mitochondrial respiration, whereas in yeast, respiration is inhibited and anaerobic fermentation predominates. A discrepancy also exists between our model and that of Powers et al. (2006), who claim that life-span extension by TOR inhibition is due in part to Msn2/4-mediated stress resistance. However, we feel that this conclusion does not follow from their results and furthermore suggest that deletion of one downstream target of a signaling pathway as intricate as TOR cannot be expected to recapitulate the phenotype of inhibiting the entire pathway. Finally, although it has been shown that RNAi or mutation of mitochondrial OXPHOS components extends life span in worms (Dillin et al., 2002; Feng et al., 2001; Lee et al., 2003), providing important insight into aging in metazoans, there are indications that this is not simply due to the “less respiration, less ROS” explanation. Dillin et al. (2002), for example, showed that inhibition of respiration only extends life span if imposed during development, suggesting some sort of “metabolic reprogramming”; alternatively, it has been proposed that the effects of respiration inhibition in C. elegans could be due to the use of an alternate anaerobic mode of respiration utilizing fumarate as an alternative electron sink and producing fewer ROS (Rea and Johnson, 2003).
In summary, we have revealed important connections among the TOR pathway, mitochondrial respiration, and life span that provide insight into how these likely synergize to influence aging and longevity. Of particular note is our discovery that mitochondrial translation and respiration are downstream targets of TOR, indicating that the regulation of mitochondrial metabolism is critical for balancing cell growth and life span in response to nutrients. It is already documented that there is crosstalk between the TOR pathway and the retrograde pathway (Butow and Avadhani, 2004; Crespo et al., 2002), which relays signals from the mitochondria to the nucleus. This, coupled with our results showing that TOR is signaling to mitochondria, implies the existence of a homeostatic regulatory circuit operating in cells whereby mitochondrial respiratory capacity and ROS production are both sensed and controlled by the TOR pathway. Determining precisely how this circuit operates and influences aging and life span is fertile ground for future investigation.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
Media and techniques for culturing and genetic manipulation of yeast were used as described in Sherman (1991). Synthetic dextrose (SD), synthetic glycerol (SG), rich dextrose (YPD), and rich glycerol (YPG) media were used as indicated. Unless otherwise indicated in the text, experiments were carried out with strain DBY2006 (MATα his3-Δ200 leu2-3,-112 ura3-52 trp1-Δ1 ade2-1) or a derivative. The tor1 null derivative of DBY2006 was generated by transformation with a PCR-amplified tor1∷kanMx4 deletion cassette from the tor1 null strain of the Yeast Knockout Collection available from Open Biosystems. The reg1 null strain and its isogenic wild-type control were derivatives of BY4742 (MATα his3-Δ1 leu2Δ0 lys2-Δ0 ura3-Δ0), obtained directly from the Yeast Knockout Collection. The sod2 and coq5 null strains of DBY2006 and DBY2006 tor1 null were generated by PCR amplification and transformation with TRP1 and HIS3 knockout cassettes, respectively. Trp+ and His+ transformants were screened by PCR to confirm correct insertion of the knockout cassettes, and the coq5 null strain was further verified by its inability to grow on YPG medium. The SOD2 overexpression plasmid used was described previously (Bonawitz et al., 2006b). Mitochondrial petite strains were generated using a standard ethidium bromide method based on that described in Slonimski et al. (1968). Inability to respire was confirmed by lack of growth on YPG medium.
Chronological Life Span
Our method for the determination of chronological life span has been described previously (Bonawitz et al., 2006b). Briefly, cultures were inoculated to an OD600 of ~0.05 in 50 ml total volume in 125 ml flasks and incubated at 30°C in an orbital shaker moving at 200 rpm. Viability was assayed either by staining a 100 µl sample of the culture with 100 µl of trypan blue (0.4 mg/ml), incubating at 30°C for 5 min, and counting clear versus blue cells or by plating serial dilutions onto YPD plates, as indicated. Growth of TOR1 and tor1 null cells was measured to ensure that the absence of the TOR1 gene did not significantly change the amount of time taken to grow from inoculation to stationary phase (Figure S3). This held true for all strains except coq5 null and coq5 tor1 null. In this case, coq5 tor1 null cells took at least 6 hr longer than coq5 null cells to reach stationary phase. The viability curve shown in Figure 2B takes this 6 hr lag into account. For the life-span experiments shown in Figure 3, we supplemented both BY4742 and reg1 null strains with 0.34% yeast nitrogen base without amino acids each day, as reg1 null strains are known to be intolerant to nitrogen starvation (Frederick and Tatchell, 1996). Culture aeration was limited to establish hypoxic conditions by replacing the typical loose-fitting metal cap of 125 ml flasks with rubber stoppers and limiting gas exchange to a small-gauge needle pushed through the stopper.
Oxygen Consumption
Oxygen consumption assays were performed as described in Bonawitz et al. (2006b).
Mitochondrial Translation Assay
Mitochondrial translation assays were performed essentially as described (Rodeheffer and Shadel, 2003). Five milliliter SD cultures were inoculated to an OD600 of 0.1 and grown ~24 hr at 30°C, after which 4 × 108 cells were harvested from each culture. Cells were incubated with 250 µg/ml of emetine (Sigma) for 5 min at 30°C to inhibit cytoplasmic translation. Mitochondrial translation products were labeled by incubation with 50 µCi of [35S]methionine for 15 min at 30°C, followed by a 10 min chase with 4 ml of chase solution (15 mM (NH4)2SO4, 1% casamino acids). Cells were washed twice with 4 ml of chase solution, pelleted and resuspended in 1 ml H2O, and added to 10 ml Ultima Gold scintillation fluid (PerkinElmer), and [35S]methionine incorporation was quantified as counts per minute by liquid scintillation.
Western Blotting
Cells were grown as described for chronological life-span assays described above and harvested at either day 1 (24 hr) or day 2 (48 hr) postinoculation. Protein (20 µg) was extracted using a standard TCA precipitation protocol, run on a 12% SDS-PAGE gel, and transferred to a PVDF membrane (Millipore Immobilon). Both blocking and antibody incubations were carried out in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 5% nonfat dry milk. Antibodies used were actin (Chemicon, 1:1000), Cox2p (Molecular Probes, 1:1000), Cox3p (Molecular Probes, 1:1000), Sod2p (Stressgen, 1:1000), HRP-conjugated goat α-mouse IgG (Molecular Probes, 1:5000), and HRP-conjugated donkey α-rabbit IgG (Santa Cruz, 1:5000). Secondary antibodies were detected with Western Lighting Chemiluminescent Reagent Plus (PerkinElmer).
Chloramphenicol Resistance Assay
Five milliliter cultures were inoculated to an OD600 of 0.01 in YPG medium containing either 0.25 mg/ml chloramphenicol (Cellgro, Mediatech) or an equivalent volume of drug vehicle (ethanol). The OD600 of all cultures was then measured after ~24 hr of growth at 30°C, and the number reported is the OD600 ratio for the growth of each strain in the presence versus absence of chloramphenicol.
Hydrogen Peroxide Sensitivity
Samples (0.5 ml) of a saturated culture at the time point indicated were added to 0.5 ml of hydrogen peroxide in water at twice the final concentration desired and incubated at 30°C for 90 min. Cells were then collected by centrifugation and resuspended in sterile water. Serial dilutions were plated onto YPD plates to determine survival. SD cultures were used in all cases.
Flow Cytometry
Use of dihydroethidium (DHE) to stain for ROS was performed as described in Bonawitz et al. (2006b).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant DAAD19-00-1-0560 from the Army Research Office to G.S.S. M.C.-L. is supported by NIH Genetics Training Grant 5 T32 GM07499-30. We would like to thank P. Doetsch for supplying the sod2∷trp1 strain and J. Cotney for help with the figures.
Footnotes
Supplemental Data
Supplemental Data include three figures and can be found with this article online at http://www.cellmetabolism.org/cgi/content/full/5/4/265/DC1/.
REFERENCES
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J. Bioenerg. Biomembr. 1999;31:347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006a;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell. Biol. 2006b;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem. J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Daicho K, Ushimaru T, Hall MN. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, SelverstoneValentine J, Gralla EB, Longo VD. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Frederick DL, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol. Cell. Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Harris TE, Lawrence JC., Jr TOR signaling. Sci. STKE. 2003;2003:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Nakashima A, Ueno M, Ushimaru T, Aiba K, Doi H, Uritani M. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 2001;39:166–174. doi: 10.1007/s002940100198. [DOI] [PubMed] [Google Scholar]
- Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Merry BJ. Effect of caloric restriction on mitochondrial reactive oxygen species production and bioenergetics: reversal by insulin. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R71–R79. doi: 10.1152/ajpregu.00341.2003. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Martin DE, Hall MN. The expanding TOR signaling network. Curr. Opin. Cell Biol. 2005;17:158–166. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci. Aging Knowledge Environ. 2003;2003:RE2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Yoshimatsu T, Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J. Bacteriol. 1983;153:1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa S, Skulachev VP. Reactive oxygen species, mitochondria, apoptosis and aging. Mol. Cell. Biochem. 1997;174:305–319. [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Johnson TE. A metabolic model for life span determination in Caenorhabditis elegans. Dev. Cell. 2003;5:197–203. doi: 10.1016/s1534-5807(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mTOR pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Yeast as a model for human mtDNA replication. Am. J. Hum. Genet. 1999;65:1230–1237. doi: 10.1086/302630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Slonimski PP, Perrodin G, Croft JH. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites. ”. Biochem. Biophys. Res. Commun. 1968;30:232–239. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, Redman P, Krol E, Jackson DM, Johnson MS, Brand MD. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Tomas-Cobos L, Viana R, Sanz P. TOR kinase pathway and 14-3-3 proteins regulate glucose-induced expression of HXT1, a yeast low-affinity glucose transporter. Yeast. 2005;22:471–479. doi: 10.1002/yea.1224. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Superoxide production by the mitochondrial respiratory chain. Biosci. Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vinogradov AD, Grivennikova VG. Generation of superoxide-radical by the NADH:ubiquinone oxidoreductase of heart mitochondria. Biochemistry (Mosc.) 2005;70:120–127. doi: 10.1007/s10541-005-0090-7. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 2001;276:7027–7032. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.