Summary
Hemimegalencephaly (HMG) is a developmental brain disorder characterized by an enlarged, malformed cerebral hemisphere, typically causing epilepsy that requires surgical resection. We studied resected HMG tissue to test whether the condition might reflect somatic mutations affecting genes critical to brain development. We found that 2/8 HMG samples showed trisomy of chromosome 1q, encompassing many genes, including AKT3, which is known to regulate brain size. A third case showed a known activating mutation in AKT3 (c.49G→A, creating p.E17K) that was not present in the patient’s blood cells. Remarkably, the E17K mutation in AKT3 is exactly paralogous to E17K mutations in AKT1 and AKT2 recently discovered in somatic overgrowth syndromes. We show that AKT3 is the most abundant AKT paralogue in brain during neurogenesis and that phosphorylated AKT is abundant in cortical progenitor cells. Our data suggest that somatic mutations limited to brain could represent an important cause of complex neurogenetic disease.
Introduction
Hemimegalencephaly (HMG, literally, enlargement of one brain hemisphere) is a developmental brain disorder characterized by an enlarged, malformed cerebral hemisphere (Flores-Sarnat et al., 2003). The clinical presentation typically includes intellectual disability and severe, intractable epilepsy often necessitating surgical removal or disconnection of the abnormal hemisphere for seizure control (Gowda et al., 2010). Although no specific genetic causes have been identified for isolated HMG, HMG has been reported in association with Proteus syndrome (Griffiths et al., 1994)—another multi-system overgrowth disorder that has recently been associated with somatic activating mutations in the gene AKT1 (Lindhurst et al., 2011)—as well as other rare neurocutaneous syndromes (Mochida et al., 2012 (in press)). There are rare reports of HMG associated with tuberous sclerosis complex (TSC) (Cartwright et al., 2005), a syndrome in which multiple organ systems display disordered and sometimes cancerous growths. While histopathological similarities between HMG and TSC led to hypotheses that there is a shared genetic etiology for these disorders, there are subtle immunohistochemical and ultrastructural features that suggest that the two are separate entities (Arai et al., 1999).
The striking asymmetry of the brain in individuals with HMG has long suggested that HMG reflects spontaneous, somatic, clonal mutation limited to the brain, analogous to cancer but without cellular transformation and ongoing proliferation. Other neurodevelopmental disorders occasionally result from somatic mutations, including DCX (Gleeson et al., 2000) and NF1 (Messiaen et al., 2011; Vogt et al., 2011), but these have generally not been mutations limited to brain. In addition, some tubers from individuals with TSC, who carry germline mutations in TSC1 or TSC2, have been demonstrated to have somatic “second hits’ in the TSC2 gene (Qin et al., 2010). We hypothesized that the somatic mutations causing HMG might be essentially restricted to the brain and detectable by direct study of affected brain tissue. Here we show that 3/8 HMG samples studied showed somatic mutations involving AKT3: two with large duplications of chromosome 1q encompassing AKT3, as well as many other genes, and a third carrying a known activating mutation in AKT3. Moreover, we demonstrate that at least two of three of these mutations are not detectable in blood of the same individuals, reflecting somatic mutations affecting brain preferentially or exclusively.
Results and Discussion
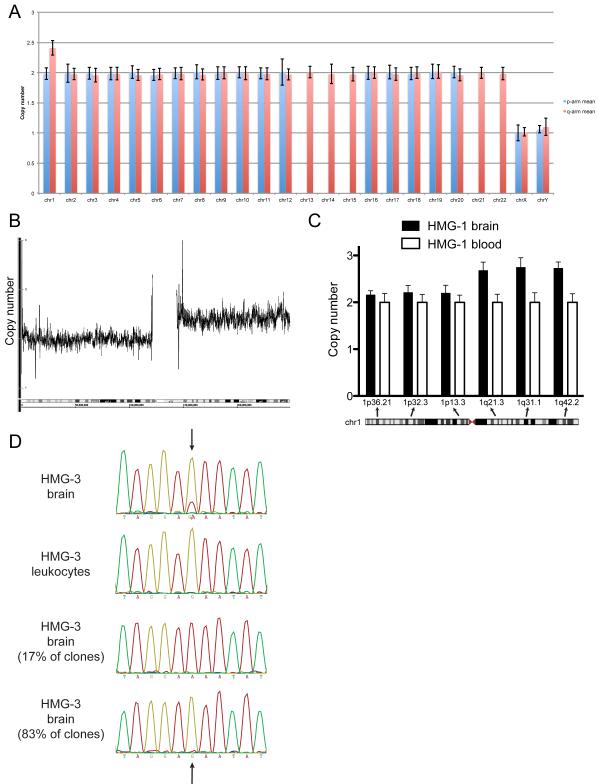
We studied 8 samples of brain tissue resected at the time of epilepsy surgery and identified two that showed trisomy of chromosome 1q. The first partial trisomy case (HMG-1) was a non-dysmorphic boy requiring hemispherectomy at 15 months of age for treatment of epilepsy due to HMG. He had no clinical evidence of non-nervous system involvement. Magnetic resonance imaging (MRI) showed left-sided HMG, with the extent of the lesion reflected in the large amount of brain removed in order to control his seizures (Figure 1C and D show the left HMG before surgery, and E and F show only the normal right hemisphere remaining after surgery). After surgery, seizures were dramatically reduced from ~10/day to 1-4/month. At age 6, he had right-sided weakness but could walk independently; he had good language comprehension, though his speech production was limited to a few words, and he attended school with special services. Neuropathological analysis from the affected hemisphere revealed diffuse abnormalities of cortical development (cortical dysplasia) with irregular cortical architecture, ectopic bands of gray matter in the subcortical white matter, scattered proliferating cells and abnormal neurons consistent with previous reports of HMG (Figure 2) (Flores-Sarnat et al., 2003). Copy number evaluation of single nucleotide polymorphism (SNP) data showed increased signal for the entire q arm of chromosome 1 in the brain sample (Figure 3A and B and Figure S1). The estimated copy number was 2.41 (S.D. 0.12), suggesting a copy number of 3 in the setting of mosaicism, with ≈40% of the cells containing the trisomy chromosome 1q. No other chromosomes displayed abnormal copy number (Figure 3A). Quantitative PCR (qPCR) confirmed the 1q trisomy, generating a calculated copy number of 2.39 (S.D. 0.30) from one brain sample; from a second sample, the calculated copy number was 2.68 (S.D. 0.16), 2.76 (S.D. 0.20), and 2.73 (S.D. 0.13) at 1q21.3, 1q31.1, and 1q42.2, respectively (Figure 3C). This intermediate copy number, between 2 and 3, suggests a mixture of normal and trisomic cells in the brain regions sampled, and together these results suggest that the ratio of normal and abnormal cells varied somewhat in different parts of the resected tissue. High-resolution karyotype and qPCR of peripheral blood cells in the patient did not reveal any evidence of trisomy 1q in these non-brain cells (Figure 3C and data not shown).
Figure 1. MRIs of patients with hemimegalencephaly due to somatic mutations.
(A, B) The first column shows an example of coronal T2-weighted and axial T2-weighted MRI images showing the brain of a normal 1-year-old. Note the symmetric size of the right and left hemispheres, labeled R and L to denote standard MRI convention. (C-J) Representative images from the brain MRIs of two patients with HMG, before and after surgical removal of the abnormal hemisphere, are shown. (C, D) HMG-1 has somatic trisomy of chromosome 1q. MRI before surgery showed left-sided hemispheric enlargement, abnormal cortical thickness and configuration, and enlarged left lateral ventricle in the coronal T2-weighted and axial T2-weighted images. The right hemisphere is smaller and appears normal. (E, F) Following left hemispherectomy surgery, there is cerebrospinal fluid (CSF) where the abnormal hemisphere had been, seen as bright signal in coronal and axial images taken at approximately the same plane as the pre-operative images. (G, H) HMG-3 has a somatic mosaic mutation in AKT3. Coronal T2-weighted and axial T2-weighted MRI images show right-sided hemispheric enlargement, abnormal cortical thickness and signal, abnormal white matter signal, and an enlarged lateral ventricle. (I, J) Following right hemispherectomy surgery, as in the previous case, CSF is visible as bright signal in place of the resected abnormal hemisphere.
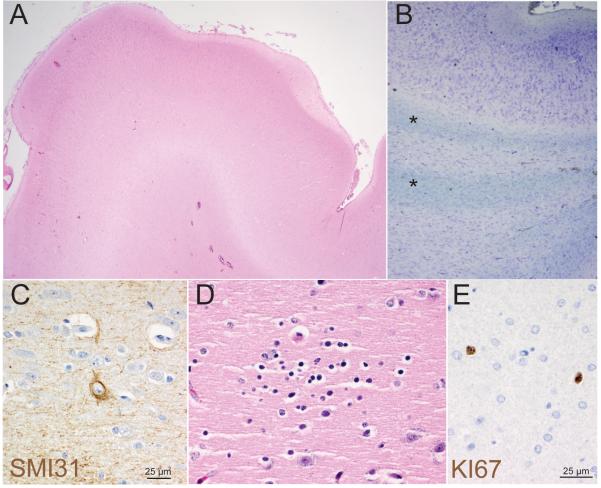
Figure 2. Abnormal cortical development in hemimegalencephaly case HMG-1 with trisomy of chromosome 1q.
(A) Low-power view (20x mag) of a gyrus from the cerebral cortex stained with haematoxylin and eosin (H&E) shows an abnormally contoured surface and variably thick cortical ribbon and molecular layer. (B) Analysis of subcortical white matter using Cresyl violet and Luxol Fast Blue (LFB) highlights numerous subcortical bands/islands of ectopic gray matter containing neurons and glia (*). (C) Immunohistochemical staining for phosphorylated neurofilament, SMI31, highlights scattered abnormal large neurons. (D) Rare small collections of neuroblast-like cells (microdysplasia) were present on H&E. (E) Immunohistochemical staining also demonstrated an abnormal number of proliferating Ki67 positive cells scattered throughout gray and white matter which had an atypical nuclear morphology (e). (A) 20x, (B) 200x (C, D, E) 600x.
Figure 3. Mosaic mutations in hemimegalencephaly: trisomy of chromosome 1q and an activating point mutation in AKT3.
(A) Copy number for all of the chromosomes is shown for HMG-1; the estimated copy number for 1q is 2.41 (S.D. 0.12), consistent with mosaic trisomy 1q. Chromosome 1p as well as the other autosomes have normal copy number of 2, and chromosomes X and Y each show copy number of 1. (B) Copy number evaluation of Affymetrix 6.0 data shows the gain in copy number at chromosome 1q for HMG-1, with the x-axis representing nucleotide position along chromosome 1 and the y-axis denoting copy number. (C) Assuming a copy number of 2 for all regions in the DNA derived from leukocytes (white columns), the calculated copy number from the brain tissue (black columns) was 2.68 (S.D. 0.16) at 1q21.3, 2.76 (S.D. 0.20) at 1q31.1, and 2.73 (S.D. 0.13) at 1q42.2. (D) The AKT3 c.49G→A, p.E17K heterozygous mutation is present in the sequencing traces from brain-derived DNA (first row) and absent in the traces from leukocyte-derived DNA from HMG-3 (second row). The arrows point to AKT3 nucleotide position 49. Cloning results indicate that the mutation is present in 8/46 (17.4%) of the DNA reads from a brain tissue sample, suggesting that the mutation exists in the heterozygous state in 35% of the cells; traces from two clones are shown in the third and fourth rows, the trace in the third row showing the results of sequencing from a clone with the AKT3 c.49G→A mutation present (A) and the bottom row showing the results from a clone without the mutation but rather with the reference allele present (G).
We identified a second case of partial gain of chromosome 1, again involving the entire 1q arm, based on SNP data from the brain sample of an individual (HMG-2) reported to have isolated HMG on MRI, similar but somewhat milder neuropathological findings of mild dysplasia (manifest primarily as a thickened cortical ribbon), and no other medical problems. The gain in copy number is shown in Supplemental Figure 1. Copy number at 1q assessed by qPCR was 2.75 (S.D. 0.28), again consistent with mosaic partial trisomy. Leukocytes or other tissues were not available from this individual, so the somatic nature of the mutation could not be directly tested. Inspection of the published literature and the Database of Genomic Variants (http://projects.tcag.ca/variation/), a large database of copy number variation, suggest that there are no known control individuals with large constitutional duplications of 1q (Iafrate et al., 2004). Wintle and colleagues recently conducted a sensitive copy number analysis on brain tissue from 52 individuals without HMG (Wintle et al., 2011) and reported no duplications of chromosome 1q larger than 1Mb (whereas the 1q region spans nearly 250 Mb), demonstrating that our finding of 2/8 cases with trisomy of 1q is clearly not a common variant (Wintle et al., 2011).
Chromosome 1q contains many genes, but among them AKT3 is a particularly strong candidate for HMG because deletions that encompass AKT3 have been associated with microcephaly, suggesting a role for AKT3 in control of brain size (Ballif et al., 2012; Boland et al., 2007; Hill et al., 2007). Furthermore, somatic activating mutations in AKT1 have recently been reported to cause Proteus syndrome, and somatic activating mutations in AKT2 have been reported to cause hypoglycemia and asymmetrical somatic growth (Hussain et al., 2011; Lindhurst et al., 2011). Earlier screening for candidate mutations in cancer-associated genes did not reveal any mutations in our cases (data not shown); AKT3 was not included among the genes screened. We sequenced AKT3 as a candidate gene in the 6 remaining non-trisomy cases of HMG and identified 1/6 with a somatic point mutation in AKT3. This case (HMG-3) was a non-dysmorphic boy requiring hemispherectomy at 5 months of age for seizures beginning in the first week of life due to right-sided HMG (MRI before surgery is shown in Figure 1G and H, and after surgery in Figure 1I and J). Following surgery, he had two periods of breakthrough seizures but has been seizure-free for 6 years at 10 years of age. He has left-sided weakness but walks independently, speaks fluently, is able to read, and attends school with special education services. DNA sequencing revealed the mutation AKT3 c.49G→A, p.E17K in the DNA derived from the brain; this mutation was not detectable in DNA derived from the patient’s leukocytes (Figure 3D). To confirm the presence of the mutation in brain cells, we cloned the PCR product from brain and re-sequenced multiple clones (Figure 3D). Forty-six individual clones showed either the mutant sequence only (8/46, or 17.4%) or the normal sequence only (38/46, or 82.6%) (examples shown in Figure 3D), suggesting that the mutation exists in the heterozygous state in ≈35% of the cells.
The activating nature of the AKT3 E17K mutation has been shown previously biochemically (Davies et al., 2008). Evaluation of data from the Exome Variant Server revealed that the AKT3 c.49G→A point mutation is not present in >5000 control individuals (http://evs.gs.washington.edu/). Published estimates suggest a somatic mutation frequency on the order of 10−9 per cell division (Lynch, 2010b), making it highly unlikely that we have observed our AKT3 c.49G→A mutation by chance. Published mutation rates from exome sequencing in humans, coupled with extrapolation of somatic mutation rates in mouse, suggest a <1 × 10−7 chance that the specific AKT3 c.49G→A mutation would occur by chance (Awadalla et al., 2010; Lynch, 2010a).
Somatic mutations in AKT3, which encodes the serine-threonine kinase protein kinase B-gamma, have been reported in cancers, including a p.G171R substitution mutation in a glioma (Bamford et al., 2004). The AKT3 c.49G→A E17K mutation itself has been observed in melanoma and lung cancer, and melanoma cell lines overexpressing this exact missense mutation have been demonstrated to show increased AKT phosphorylation (Davies et al., 2008; Do et al., 2010). Most remarkably though, the somatic AKT3 mutation we report is precisely paralogous to the recurrent E17K mutations in AKT1 associated with Proteus syndrome, and E17K mutations in AKT2 associated with hypoglycemia and left-sided overgrowth, each also with varying degrees of mosaicism (Hussain et al., 2011; Lindhurst et al., 2011). Interestingly, despite prior reports of Proteus-associated HMG (Griffiths et al., 1994), no brain malformations are reported in the patients with AKT1 and AKT2 mutations, consistent with the observation in mouse that AKT3 may be the predominant functional member of the AKT family in the human brain (Easton et al., 2005).
AKT3 expression in human fetal brain is higher than AKT3 expression in any other tissue sampled (Wu et al., 2009), suggesting that its primary role is in brain development. In contrast, AKT1 and AKT2 show levels of fetal brain expression comparable to or lower than those seen in other tissues (Wu et al., 2009). We compared the expression levels of AKT1, AKT2, and AKT3 by RNAseq analysis of perisylvian cortex of human brain at 9 weeks gestation, during active neurogenesis, and found that AKT3 is expressed at higher levels than AKT1 and AKT2 (normalized read depth, reads per kilobase-exon per million mapped reads: AKT1=51.90, AKT2=18.50, AKT3=90.52). Examination of published datasets reveals that AKT3 is expressed at a higher level than AKT1, and both are expressed at higher levels than AKT2, starting at 8 weeks and for the duration of human embryonic cortical development (Kang et al., 2011). To determine the cell types in the brain that would likely be affected by activation of AKT3, we performed immunohistochemistry in sections of mouse brain, using an antiserum that recognizes all three phosphorylated forms of AKT (P-Akt). We observed widespread P-Akt localization in the developing cortex, with notable enrichment in apical progenitor cells in the ventricular zone (VZ). For example, a subset of cells marked by P-Akt also showed the presence of P-Vimentin 4A4, illustrating the presence of Akt activity in dividing radial glial cells (Figure 4).
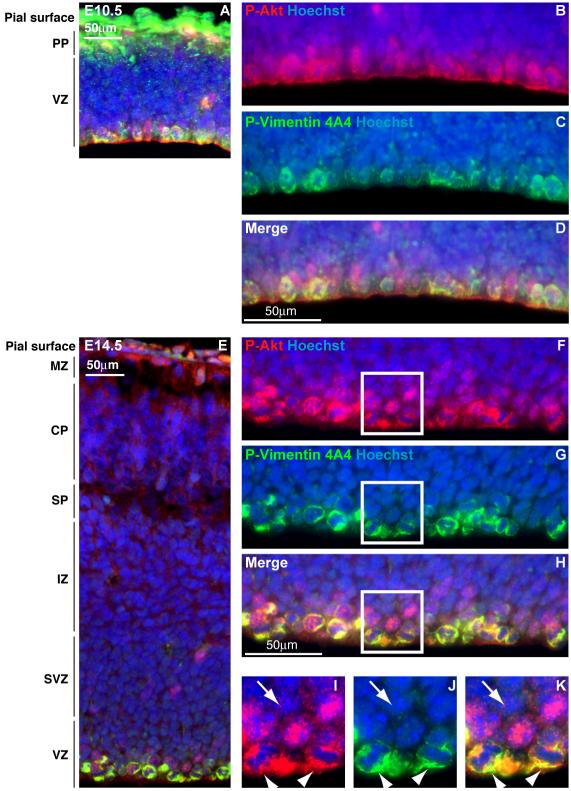
Figure 4. Active Akt signaling in the developing cortex is enriched in apical progenitor cells and the cortical plate.
(A) Immunohistochemistry of cortical sections at embryonic day (E)10.5 reveals Akt activity as assessed by pan-phospho(P)-Akt immunostaining (red) in the cortical plate and ventricular zone. (B-D) Higher magnification images of ventricular zone at E10.5 are shown. Overlay of P-Akt with P-Vimentin 4A4 (green) shows that dividing radial glial cells, which generate cortical pyramidal neurons and glial cells, are P-Akt positive. (E-H) At E14.5, dividing radial glial cells show a similar pattern of immunostaining for P-Akt and P-Vimentin 4A4. (I-K) High-magnification images of the areas delineated by white boxes in F-H demonstrate that P-Akt activity (marked by arrowheads) is not restricted to the P-Vimentin 4A4-positive-staining, M phase cells in the ventricular zone. The arrows indicate an example of a P-Akt-positive, P-Vimentin 4A4-negative cell. Nuclei are labeled with Hoechst. Scale bars, 50μm. MZ: marginal zone, CP: cortical plate, SP: subplate, IZ: intermediate zone, SVZ: subventricular zone, VZ: ventricular zone.
Similarly, P-Akt colocalized with phospho-Histone H3, a marker of M phase, in dividing apical progenitors as well as GLAST, a marker of radial glial cells (data not shown). Since these apical progenitor cells give rise to both neurons and glia, this localization is consistent with activation of AKT3 in both neurons and glia. Abnormal AKT function would be consistent with the MRI patterns and neuropathological studies (Figures 1 and 2), which show abnormal organization of neurons in the cortex and abnormal MRI signal characteristics of white matter.
Our data suggest that activation of AKT3, either by duplication or by point mutation, contributes to hemispheric brain overgrowth. Two of our cases (the point mutation and one partial trisomy) are confirmed to be de novo, somatic mutations, undetectable in blood, and although non-brain tissues were not available from the other partial trisomy case, this is likely to be a somatic mutation as well since individuals reported with constitutional trisomy 1q, even a portion of 1q, show dysmorphic features and in nearly all cases early lethality (Mark et al., 2005; Mefford et al., 2008; Patel et al., 2009). We postulate that increasing AKT3 dosage and activation of AKT3 would have the same effect in the setting of a somatic mutation. Interestingly, HMG has not been reported in the constitutional trisomy cases, even those that have partial trisomy including AKT3. It is possible that HMG might not be present in the cases with early lethality; perhaps more important, since all of the constitutional trisomy 1q cases were de novo, the trisomy may not be present in all tissues. Though we have not sampled other tissues, there was no clinical evidence of extra-cerebral involvement phenotypically in any of the three cases, suggesting either that the mutation was limited to brain, or that activation of AKT3 in other tissues does not have phenotypic consequences. Increased rates of brain cancer are not reported in the setting of isolated HMG. In the cases we report here, who have not shown any form of cancer, it is likely that activation of AKT3 disrupts normal cortical development but does not result in continued dysregulated growth outside the setting of cortical progenitor cells.
Further support for the role of AKT3 in controlling brain size comes from animal studies. A mouse Akt3 knockout model shows selective reduction in brain size due to decreased neuronal number and size (Easton et al., 2005), whereas mice with an activating mutation in the kinase domain of Akt3 show larger hippocampal size and abnormal Ki67-positive ectopic neurons in the hippocampus (Tokuda et al., 2011). Additionally, in zebrafish, overexpression of wild-type AKT3 produces increased embryonic brain thickness (Chen, 2011). All of these results strongly suggest that AKT3 activity dynamically regulates brain size and that increased dosage of AKT3 might increase brain size in humans.
Somatic mosaicism refers to the presence of more than one genetically distinct population of cells in an individual. Somatic mutations are thought to arise not infrequently during development (Youssoufian and Pyeritz, 2002), and some chromosomal rearrangements and mutations that may be lethal if present in the entire embryo could be sustained in clonal populations of cells and produce localized abnormalities. The size and architecture of HMG may be determined in part by the stage at which the mutation occurs relative to the period of neurogenesis, which is when AKT3 normally becomes the predominant AKT form in brain. As better techniques emerge for copy number and whole exome/genome sequencing on smaller and smaller amounts of DNA, somatic mutations in other genes might emerge as causes of other neurogenetic disorders not associated with obvious morphological phenotypes like HMG. For example, de novo CNVs are an important cause of autism spectrum disorders and schizophrenia (Sanders et al., 2011), and hence may also occur somatically. In epilepsy, at least one third of individuals with imaging-negative, refractory, focal seizures show pathological evidence of dysplasia (Porter et al., 2003) that may also be due to somatic mutations. Therefore, more detailed exploration of somatic mosaicism in may allow for better genetic understanding of many neurogenetic disorders, especially those for which de novo mutations are known to play a role.
Experimental Procedures
Brain sample ascertainment
Tissue samples for molecular analysis were available through two sources: (1) patients enrolled in clinical research in accordance with requirements of the Institutional Review Boards of Children’s Hospital Boston (CHB) and Beth Israel Deaconess Medical Center (6 cases, including HMG-1 and HMG-3); and (2) excess tissue obtained from the Brigham and Women’s Hospital Department of Neurosurgery Tissue Bank along with limited clinical information (2 cases, including HMG-2).
Phenotypic assessment
Detailed clinical information and leukocyte-derived DNA were available for 6 cases enrolled in human subjects research, including HMG-1 and HMG-3. We reviewed the history and examination of each case reported (AP, BB, JJR) and the MRI (AP, AJB, CAW). Supplemental Table 1 summarizes the imaging and neuropathological findings of the three cases with mutations.
Neuropathological analysis
Formalin fixed paraffin embedded sections from the clinical resection specimens were obtained from the CHB pathology archives for pathological re-review by a board-certified neuropathologist (KLL). Slides were stained with hematoxylin and eosin, Cresyl violet/Luxol fast blue according to standard methods. Immunohistochemistry was performed using phosphorylated neurofilament (SMI31, Covance Inc.) and Ki67 (DAKO, Clone MIB1) using DAKO Envision Plus and DAB development.
Copy number assessment
We obtained 8 samples of flash-frozen brain tissue resected during focal epilepsy surgery for HMG. DNA was extracted using standard methods and then digested, amplified, and hybridized to Affymetrix 100K SNP arrays for 6 of the samples (Affymetrix, Cleveland, OH). On the original arrays (e.g., 100K), copy number was assessed based on intensity of signal from each SNP. For the Affymetrix 6.0 arrays, copy number probes are included in addition to the full array of SNPs, and both are used for quantitation. The Gaussian smoothed signal log2-ratio of all probe intensities normalized to a reference of 270 normal Hapmap samples was calculated by Affymetrix Genotyping Console (AGC) with standard settings. Additional DNA from HMG-1 and two other samples were assessed using the Affymetrix 6.0 SNP array. The software dChipSNP was used for analysis.
For HMG-1 and HMG-2, we performed qPCR in cases in which copy number change was detected. Primers were designed to 1q44 and 1p21.1. DNA from two control individuals (Promega, Madison, WI) was used for comparison. We repeated qPCR in an additional specimen from HMG-1 for confirmation using primers targeting 1p (1p13.3, 1p32.3, and 1p36.2) and 1q (1q21.3, 1q31.1, and 1q42.2).
Leukocytes were obtained from 6 of the cases; DNA was extracted using standard methods and used for SNP analysis as above. For HMG-1, we performed SNP analysis and clinical karyotype to assess for the presence of the trisomy 1q in peripheral blood leukocytes (evaluating 50 cells to detect even a low level of mosaicism).
Screening for Candidate Mutations in Oncogenes
Based on the hypothesis that our cases harbor somatic mutations in genes that result in dysregulated growth, we screened the DNA from the brain samples for a panel of known point mutations in cancer-associated genes (OncoMap Project, Dana Farber Cancer Institute) (MacConaill et al., 2009). This panel did not include AKT3; genes included in the 1q region were ABL2, DDR2, and NTRK1.
Evaluation for AKT3 c.49G→A activating mutations
We designed primers using Primer 3 software (http://primer3.sourceforge.net/) for the second exon of AKT1, AKT2, and AKT3 in order to evaluate nucleotide position 49. In cases without trisomy 1q, we sequenced DNA from brain tissue (6 HMG cases) and leukocytes from the same cases (5 cases).
Evaluation for mosaicism of the AKT3 mutation in HMG-3
To determine the degree of mosaicism in the brain tissue specimen of HMG-3, we performed TOPO TA cloning using standard methods (Invitrogen, Carlsbad, CA), successfully analyzing 46 clones for the AKT3 c.49G→A mutation.
Estimation of the likelihood that the AKT3 mutation would occur by chance
Published sequencing data indicate that each individual has approximately 1-2 de novo nonsynonymous variants per diploid genome generation (Awadalla et al., 2010). The likelihood that this would affect this one base pair in all of the 6 × 107 base pairs of the diploid genome is therefore 2-3 × 10−8. Correcting by a factor of 10 to reflect the increased somatic vs. germline mutation rate (Lynch, 2010a) and accounting for three potential mutations at a given nucleotide position, the estimated likelihood that our mutation would occur by chance is at most 1 × 10−7.
Localization of P-AKT in the developing cortex
We labeled embryonic mouse cortex at E10.5, E12.5, E14.5, E16.5, and E18.5 with the following antibodies: rabbit anti-phospho-Akt 1:50 (#4060S, Cell Signaling), mouse anti-phospho-Vimentin 4A4 1:100 (Assay Designs), rabbit anti-phospho-Histone H3 1:400 (Upstate), and anti-GLAST 1:5000 (Chemicon).
RNA-Seq analysis of AKT isoforms in the developing human cortex
We obtained snap-frozen brain tissue from a human fetus at roughly 9 weeks’ gestation from the Institute of Human Genetics at Newcastle University. RNA was isolated from several regions of cortex, including the perisylvian region, and purified using standard methods. We purified polyA-tailed mRNA using an Oligotex mRNA minikit (Qiagen, Valencia, CA) and prepared a barcoded sequencing library using the SOLiD Whole Transcriptome Analysis Kit (Applied Biosystems, Foster City, CA). We sequenced the library on the SOLiD v3 Plus system (read depth 105 million reads), mapped the reads with Bioscope v1.2 (Applied Biosystems) to the hg18 human genome reference, and normalized coverage of uniquely mapping reads to the number of million mapped reads.
Supplementary Material
Acknowledgments
The authors thank the patients and families who have participated in this research. We thank Rona Carroll in the Brigham and Women’s Hospital Department of Neurosurgery Tissue Bank, Abha Gupta in the Cytogenetics Laboratory at Brigham and Women’s Hospital, Laura MacConaill and Levi Garraway at the Dana Farber Cancer Institute Oncomap Project, and Elizabeth Bundock, formerly in the CHB Department of Pathology. Annapurna Poduri was supported by the American Academy of Neurology Clinical Research Training Fellowship, the Milken Family Foundation and American Epilepsy Society, and the NINDS (K23NS069784). Maria K. Lehtinen is supported by a Shore Fellowship and a K99/R00 from the NINDS (R00 NS072192). Keith L. Ligon is supported by grants from NCI (P01 CA142536), NINDS (K08 NS047213) and the Sontag Foundation. Christopher A. Walsh is an Investigator of the Howard Hughes Medical Institute and is supported by grants from the NINDS (R01 NS35129).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai Y, Edwards V, Becker LE. A comparison of cell phenotypes in hemimegalencephaly and tuberous sclerosis. Acta Neuropathol. 1999;98:407–413. doi: 10.1007/s004010051101. [DOI] [PubMed] [Google Scholar]
- Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, Cote M, Henrion E, Spiegelman D, Tarabeux J, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Rosenfeld JA, Traylor R, Theisen A, Bader PI, Ladda RL, Sell SL, Steinraths M, Surti U, McGuire M, et al. High-resolution array CGH defines critical regions and candidate genes for microcephaly, abnormalities of the corpus callosum, and seizure phenotypes in patients with microdeletions of 1q43q44. Hum Genet. 2012;131:145–156. doi: 10.1007/s00439-011-1073-y. [DOI] [PubMed] [Google Scholar]
- Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland E, Clayton-Smith J, Woo VG, McKee S, Manson FD, Medne L, Zackai E, Swanson EA, Fitzpatrick D, Millen KJ, et al. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am J Hum Genet. 2007;81:292–303. doi: 10.1086/519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright MS, McCarthy SC, Roach ES. Hemimegalencephaly and tuberous sclerosis complex. Neurology. 2005;64:1634. doi: 10.1212/01.WNL.0000154465.87255.78. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou J, Ling Y, Li Y. Molcular Cloning, Expression and Overexpression Analysis of AKT3 (PKBγ) in Zebrafish. Acta Hydrobiologica Sinica. 2011;35:717–726. [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Salemi R, Murone C, Mitchell PL, Dobrovic A. Rarity of AKT1 and AKT3 E17K mutations in squamous cell carcinoma of lung. Cell Cycle. 2010;9:4411–4412. doi: 10.4161/cc.9.21.13654. [DOI] [PubMed] [Google Scholar]
- Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VM, Szabolcs M, de Jong R, Oltersdorf T, et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol Cell Biol. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sarnat L, Sarnat HB, Davila-Gutierrez G, Alvarez A. Hemimegalencephaly: part 2. Neuropathology suggests a disorder of cellular lineage. Journal of child neurology. 2003;18:776–785. doi: 10.1177/08830738030180111101. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Minnerath S, Kuzniecky RI, Dobyns WB, Young ID, Ross ME, Walsh CA. Somatic and germline mosaic mutations in the doublecortin gene are associated with variable phenotypes. Am J Hum Genet. 2000;67:574–581. doi: 10.1086/303043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S, Salazar F, Bingaman WE, Kotagal P, Lachhwani DL, Gupta A, Davis S, Niezgoda J, Wyllie E. Surgery for catastrophic epilepsy in infants 6 months of age and younger. J Neurosurg Pediatr. 2010;5:603–607. doi: 10.3171/2010.1.PEDS08301. [DOI] [PubMed] [Google Scholar]
- Griffiths PD, Welch RJ, Gardner-Medwin D, Gholkar A, McAllister V. The radiological features of hemimegalencephaly including three cases associated with proteus syndrome. Neuropediatrics. 1994;25:140–144. doi: 10.1055/s-2008-1073012. [DOI] [PubMed] [Google Scholar]
- Hill AD, Chang BS, Hill RS, Garraway LA, Bodell A, Sellers WR, Walsh CA. A 2-Mb critical region implicated in the microcephaly associated with terminal 1q deletion syndrome. American journal of medical genetics Part A. 2007;143A:1692–1698. doi: 10.1002/ajmg.a.31776. [DOI] [PubMed] [Google Scholar]
- Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout D. Somatic mosaicism as a basic epileptogenic mechanism? Brain. 2008;131:900–901. doi: 10.1093/brain/awn056. [DOI] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, Turner J, Cannons JL, Bick D, Blakemore L, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010a;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010b;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, Davis M, Yao K, Hanna M, Mondal C, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark HF, Wyandt H, Pan A, Milunsky JM. Constitutional partial 1q trisomy mosaicism and Wilms tumor. Cancer Genet Cytogenet. 2005;162:166–171. doi: 10.1016/j.cancergencyto.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L, Vogt J, Bengesser K, Fu C, Mikhail F, Serra E, Garcia-Linares C, Cooper DN, Lazaro C, Kehrer-Sawatzki H. Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1) Human mutation. 2011;32:213–219. doi: 10.1002/humu.21418. [DOI] [PubMed] [Google Scholar]
- Mochida GH, Poduri A, Walsh CA. Genetic disorders of cerebral cortical development. In: Rimoin DL, Pyeritz RE, Korf B, editors. Emery and Rimoin’s principles and practices of medical genetics. 6th edition. Elsevier; UK: 2012. in press. [Google Scholar]
- Patel C, Hardy G, Cox P, Bowdin S, McKeown C, Russell AB. Mosaic trisomy 1q: The longest surviving case. American journal of medical genetics Part A. 2009;149A:1795–1800. doi: 10.1002/ajmg.a.32959. [DOI] [PubMed] [Google Scholar]
- Porter BE, Judkins AR, Clancy RR, Duhaime A, Dlugos DJ, Golden JA. Dysplasia: a common finding in intractable pediatric temporal lobe epilepsy. Neurology. 2003;61:365–368. doi: 10.1212/01.wnl.0000076487.28227.6e. [DOI] [PubMed] [Google Scholar]
- Qin W, Chan JA, Vinters HV, Mathern GW, Franz DN, Taillon BE, Bouffard P, Kwiatkowski DJ. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol. 2010;20:1096–1105. doi: 10.1111/j.1750-3639.2010.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S, Mahaffey CL, Monks B, Faulkner CR, Birnbaum MJ, Danzer SC, Frankel WN. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum Mol Genet. 2011;20:988–999. doi: 10.1093/hmg/ddq544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt J, Kohlhase J, Morlot S, Kluwe L, Mautner VF, Cooper DN, Kehrer-Sawatzki H. Monozygotic twins discordant for neurofibromatosis type 1 due to a postzygotic NF1 gene mutation. Human mutation. 2011;32:E2134–2147. doi: 10.1002/humu.21476. [DOI] [PubMed] [Google Scholar]
- Wintle RF, Lionel AC, Hu P, Ginsberg SD, Pinto D, Thiruvahindrapduram B, Wei J, Marshall CR, Pickett J, Cook EH, et al. A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism Res. 2011;4:89–97. doi: 10.1002/aur.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.