Abstract
Though the linkages between germline mutations of BRCA1 and hereditary breast cancer are well known, recent evidence suggests that altered BRCA1 transcription may also contribute to sporadic forms of breast cancer. Here we show that BRCA1 expression is controlled by a dynamic equilibrium between transcriptional co-activators and co-repressors that govern histone acetylation and DNA accessibility at the BRCA1 promoter. Eviction of the transcriptional co-repressor and metabolic sensor, C-terminal-binding protein (CtBP) plays a central role in this regulation. Loss of CtBP from the BRCA1 promoter through either estrogen induction, RNAi depletion or increased NAD+/NADH ratio results in HDAC1 dismissal, elevated histone acetylation, and increased BRCA1 transcription. The active control of chromatin marks, DNA accessibility and gene expression at the BRCA1 promoter by this “metabolic switch” provides an important molecular link between caloric intake and tumor suppressor expression in mammary cells.
Keywords: BRCA1, histone acetylation, epigenetic modifications, tumor suppressor, NADH, CtBP, RB pocket proteins, estrogen
The worldwide mortality from breast cancer is the second leading cause of death in women and the number one cause of death from cancer in females aged 20–59 1. Individuals harboring germline mutations in the breast cancer susceptibility gene BRCA1, carry an 80% lifetime risk of developing breast cancer 2. Though very few cases of non-inherited sporadic forms of breast cancer have been found to be associated with mutation in BRCA1, nearly 40% of these tumors demonstrate a deficiency in BRCA1 expression 3. Since the majority of these cases do not show hypermethylation of the BRCA1 promoter 4, a growing consensus has emerged suggesting that a large percentage of sporadic, non-inherited breast cancers are associated with altered transcriptional regulation of the BRCA1 gene 3, 5. The human BRCA1 promoter is bidirectional, controlling divergent transcription of the BRCA1 and NBR2 genes 6 and many aspects of its regulation have been extensively studied. In addition to methylation of specific CpG residues and islands within the promoter 7, several groups have demonstrated that the BRCA1 promoter is regulated by a complex and dynamic array of DNA binding proteins, transcriptional co-activators and transcriptional co-repressors 8–10.
The protein product of the BRCA1 gene has many important cellular functions including DNA repair, cell cycle regulation, and transcriptional regulation. Accordingly, deficiency in BRCA1 results in accelerated proliferation, aberrant mitosis, increased chromosome instability and tumorigenesis 11, 12. BRCA1 transcription is regulated by diverse types of environmental stimuli including genotoxic agents, hypoxia, and mitogenic hormone stimulation. The best characterized stimulant of BRCA1 expression is estrogen, which induces the highest elevations in BRCA1 mRNA levels, routinely peaking just prior to the onset of DNA synthesis 13, 14. In this way, BRCA1 is thought to provide a feedback control that monitors and restrains the growth and pro-proliferative effects of estrogen in hormone responsive tissues 14–16. Consequently, disruption of this close opposing relationship with estrogen receptor, in combination with decreased genome stability, is believed to account for the remarkably restricted occurrence of inherited BRCA1-related malignancies in hormone regulated tissues like breast, ovary and prostate 16.
The transcriptional co-repressor C-terminal binding proteins (CtBP1 and CtBP2) are members of an evolutionally conserved family of proteins that regulates several different cellular functions in vertebrates 17. Over-expression of these proteins has been linked to epithelial-mesenchymal transition in breast cancer, a process whose gene expression profile shares many similarities with the molecular signature of BRCA1-deficient tumors 17–19. CtBP is a homodimer or heterodimer of CtBP1 and CtBP2 that assembles with a diverse array of factors that regulate chromatin structure. These include, the histone deacetylases (HDACs) HDAC1/2, the histone acetyl-transferases p300/CBP, and the histone methyl-transferase G9a 17. Several studies have shown that CtBP can antagonize the expression of multiple tumor suppressors including CDH1 (E-cadherin), CDKN2A (p16) and PTEN 17. Most notably, CtBP contains a binding site for NADH that regulates its ability to dimerize, thus establishing CtBP as an important nuclear sensor of cellular metabolic status 20, 21. In this report we demonstrate that CtBP assembles at the BRCA1 promoter as part of a dynamic multi-component co-repressor complex containing p130, BRCA1 and HDAC1 that represses local histone acetylation at the BRCA1 promoter and BRCA1 transcription. Disruption of this complex by estrogen stimulation and/or changes in NAD+/NADH ratio, results in CtBP dismissal, HDAC1 eviction, increased histone acetylation and subsequent increased BRCA1 transcription from the BRCA1 promoter. These observations define a direct link between cellular metabolic status and the expression of BRCA1 and suggest that caloric intake may selectively influence the levels of tumor suppressor function in mammary tissues.
Results
A dynamic co-regulatory complex controls the BRCA1 promoter
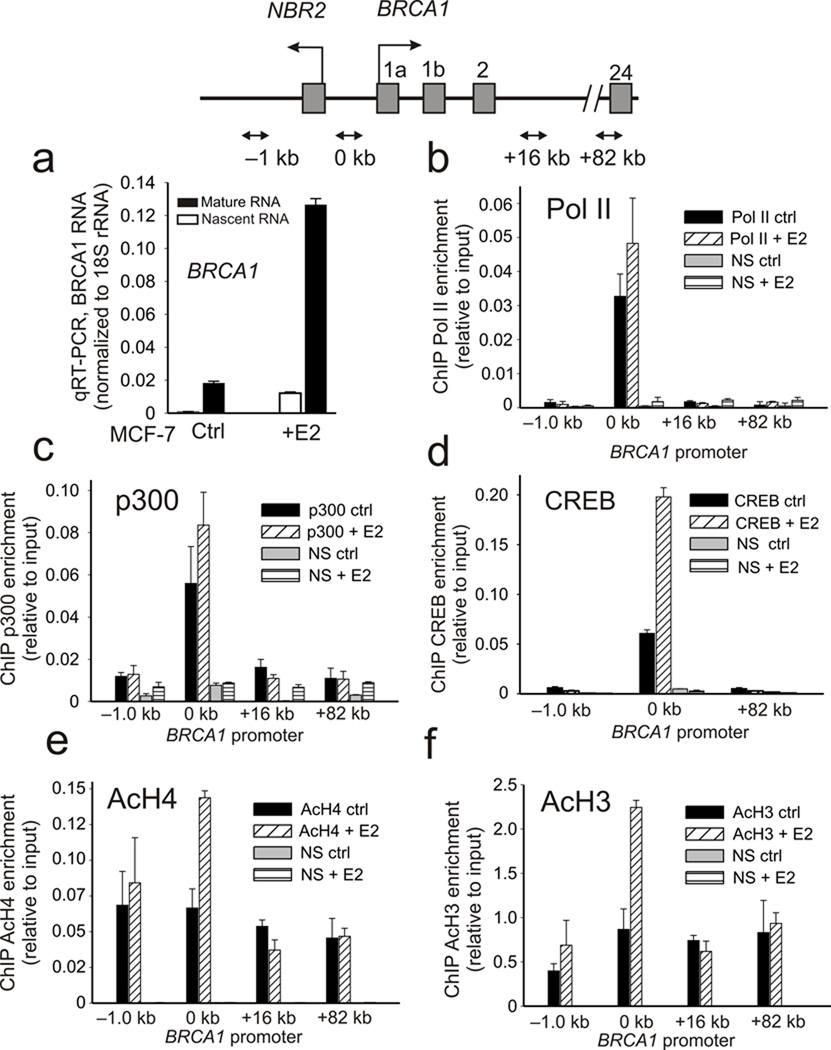
Prior studies have shown that BRCA1 transcription can be readily induced by exposure to estrogen 13, 14. Treatment of MCF-7 cells with 10 nM estradiol (E2) for 24 h produces a 6–7 fold increase in both mature and unspliced (nascent) BRCA1 RNA (Fig. 1a). By chromatin immunoprecipitation (ChIP) the BRCA1 promoter shows preloading by a poised RNA polymerase II (Pol II) and p300 histone acetyl-transferase (HAT) complex in the absence of estrogen stimulation, which is consistent with what has been observed for many promoters in recent genome-wide studies 22. Neither p300/Pol II assembly nor activation-associated histone methylation (H3K4Me3) changes significantly from their elevated levels following estrogen stimulation, though binding by the CREB transcription factor increases more notably (Fig. 1b–d and Supplemental Fig. S1). Interestingly, in contrast to Pol II, p300, and histone H3K4Me3, there is a significant increase in the levels of both histone H4 and H3 acetylation (Fig. 1e,f). These observations suggest that a major regulatory step following estrogen induction at the BRCA1 promoter involves events linked to increased promoter proximal histone acetylation that occur following the initial recruitment of p300 and the basal transcriptional machinery.
Figure 1.
Estrogen induction increases histone acetylation at the BRCA1 promoter. Top panel: a schematic illustration of the bidirectional promoter of the BRCA1/NBR2 gene locus showing positions of the ChIP amplicons. (a) BRCA1 nascent and mature RNA expression in control or MCF-7 cell treated 24 h with 10 nM estradiol (E2). Error bars indicate the s.e.m. of N=3 biological replicates. (b–f) ChIP profiles of resting and E2 stimulated MCF-7 cells using antibodies against Pol II (b), p300 (c), CREB (d), acetylated histone H4 (e), and acetylated histone H3 (f) at the BRCA1 promoter. Error bars represent the s.e.m. for N=3 (Pol II), N=2 (p300), N=2 (CREB), N=3 (AcH4) and N=3 (AcH3) biological replicates.
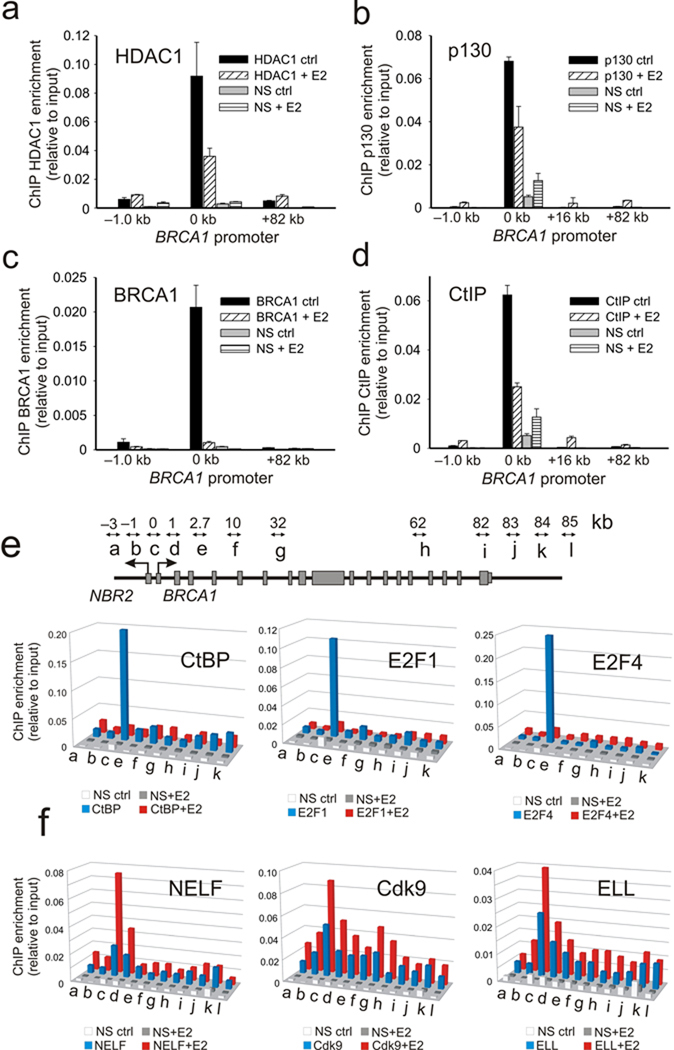
The increase in histone H3 /H4 acetylation at the proximal promoter despite small changes in p300 HAT occupancy suggests that changes in HDAC recruitment may play a role in the estrogen-induced control of BRCA1 expression. Notably, in addition to their direct interactions with the Rb pocket protein family, HDACs can be recruited in the context of several different types of co-repressor complexes including, Sin3A, NuRD and CtBP 23, 24. Previous studies have shown that the BRCA1 promoter is negatively regulated by the dynamic assembly of co-repressor complexes containing, E2F-1, E2F-4, Rb and Rb-related pocket proteins (e.g. p130), and BRCA1 9, 10, 25. Each one of these factors, including BRCA1 itself, have the capacity to form complexes with HDACs either directly or through interactions involving the C-terminal binding protein interacting protein (CtIP) 23, 26 17, 27–29. Accordingly, and consistent with increased histone acetylation at the BRCA1 promoter following estrogen induction, there is a dynamic loss of HDAC1, p130, BRCA1, CtIP, CtBP, E2F-1 and E2F-4 from the BRCA1 proximal promoter following estrogen treatment (Fig. 2a–e). Also, in agreement with the interdependent interactions between these components and BRCA1 expression, gene depletion of BRCA1 impairs recruitment of CtBP (Supplementary Fig. S2a) while gene depletion of E2F-1 impairs both BRCA1 and CtBP recruitment to the BRCA1 promoter (Supplementary Fig. S2b,c). Furthermore, multiple estrogen dependent complexes containing BRCA1, CtBP, E2F-4, p130 and p300 can be detected by co-immunoprecipitation from nuclear extracts derived from E2 treated and untreated MCF-7 cells (Supplementary Fig. S2d). Together, these observations indicate that BRCA1 transcription is regulated by a multi-component co-repressor complex containing CtBP that is linked to HDAC1 through multivalent interactions. Disassembly and dismissal of this complex from the BRCA1 proximal promoter, following estrogen stimulation, is a major regulatory step that governs BRCA1 expression.
Figure 2.
A multi-component co-repressor complex containing CtBP is dismissed and elongation factors are recruited to the BRCA1 promoter following estrogen induction. (a–d). ChIP profiles of MCF-7 cells stimulated 24 h with E2 using antibodies against HDAC1, p130, BRCA1, and CtIP as indicated. Error bars represent the s.e.m. for N=2 biological replicates. (e) Upper panel shows schematic of location of ChIP primer pairs (a–j) across the BRCA1 locus. ChIP profiles of CtBP, E2F1, and E2F4 enrichment across the 85 kb BRCA1 locus before (blue) and after (red) estrogen induction. The mean of N=2 biological replicates is represented and is associated with an average s.e.m. that is 24.6% of the mean. (f) ChIP profiles of NELF, Cdk9, and ELL enrichment across the BRCA1 locus before and after estrogen induction. The mean of N=2 biological replicates is represented and is associated with an average standard error that is 19.5% of the mean.
Finally, consistent with transcriptional regulation by a post recruitment step, this estrogen induced dismissal of repressive factors is associated with increased assembly of known elongation factors 30 at the BRCA1 locus including the negative elongation factor (NELF), the eleven nineteen lysine rich leukemia protein (ELL) and the Cdk9 subunit of the positive transcriptional elongation factor b complex (P-TEFb) (Fig. 2f). With the exception of NELF, whose assembly at mammalian promoters occurs without traveling with the elongating polymerase 31, 32, both ELL and P-TEFb are recruited to the BRCA1 locus and show increased distribution into the transcribed region of BRCA1 in coordination with increased histone lysine 36 trimethylation marks (H3K36Me3) commonly associated with Pol II elongation 30(Fig. 2f and Supplementary Fig. S2e).
CtBP regulates HDAC1 recruitment and BRCA1 promoter acetylation
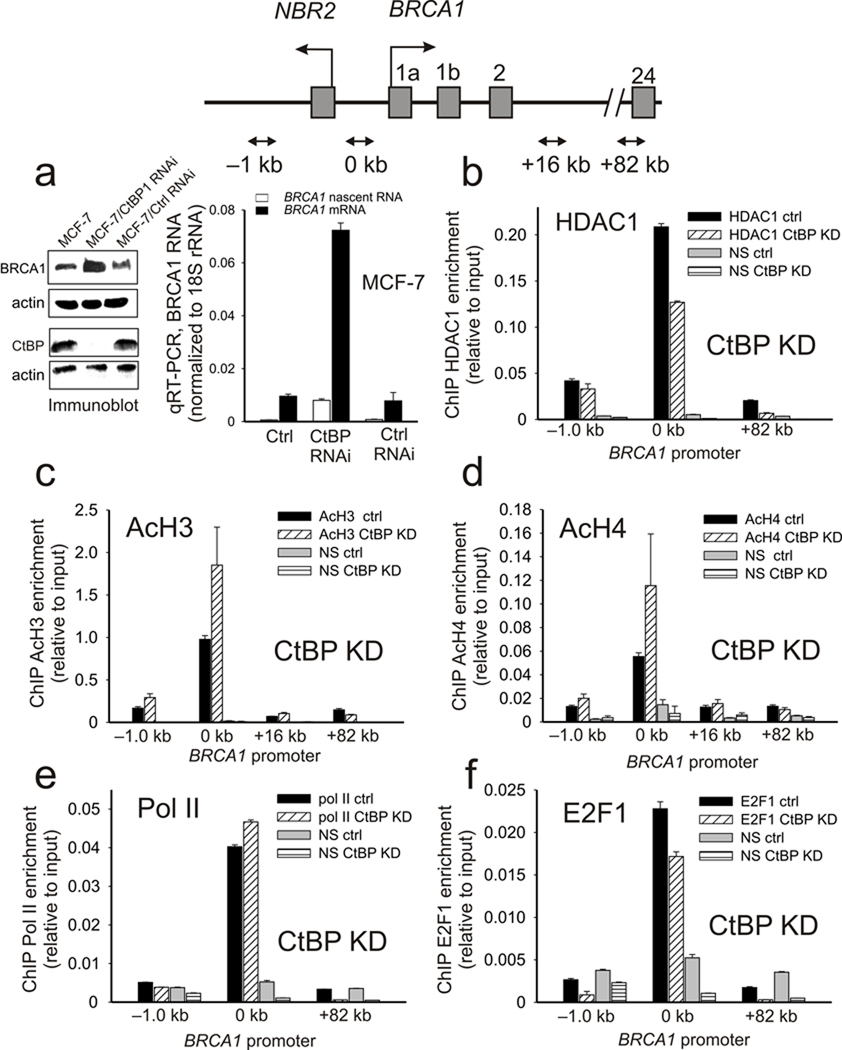
Thus far, the data demonstrate that complexes containing CtBP play a central role in BRCA1 transcriptional regulation through control of the deposition of chromatin marks at the BRCA1 promoter in response to environmental stimuli via regulation of HDAC1 recruitment. To test for the role of CtBP in this process, CtBP expression was suppressed by RNAi inhibition (Fig. 3a). Silencing of CtBP expression in MCF-7 cells results in a significant increase in both BRCA1 protein and BRCA1 RNA message (nascent and mature). The increased BRCA1 transcription following CtBP depletion is associated with loss of HDAC1 from the BRCA1 promoter and a corresponding increase in histone H3 and H4 acetylation (Fig. 3b–3d). Notably these changes occur with minimal alteration in either Pol II or E2F-1 occupancy at the BRCA1 promoter (Fig. 3e,f).
Figure 3.
CtBP regulates BRCA1 expression by influencing histone acetylation at the BRCA1 promoter. (a) (left panel) Immunoblot of CtBP and BRCA1 expression in control and MCF-7 cells depleted of CtBP by RNAi. Actin is shown as an endogenous control. (right panel) Nascent and mature BRCA1 RNA levels in control and CtBP depleted MCF-7 cells. Error bars represent the s.e.m. for N=3 biological replicates. (b–f) Estrogen stimulated enrichment of HDAC1 (b), acetylated histone H3 (c), acetylated histone H4 (d), Pol II (e), and E2F1 (f) at the BRCA1 promoter in control and CtBP depleted MCF-7 cells. Error bars represent s.e.m. for N=2 independent biological replicates.
CtBP specificity requires chromatin structure at the BRCA1 promoter
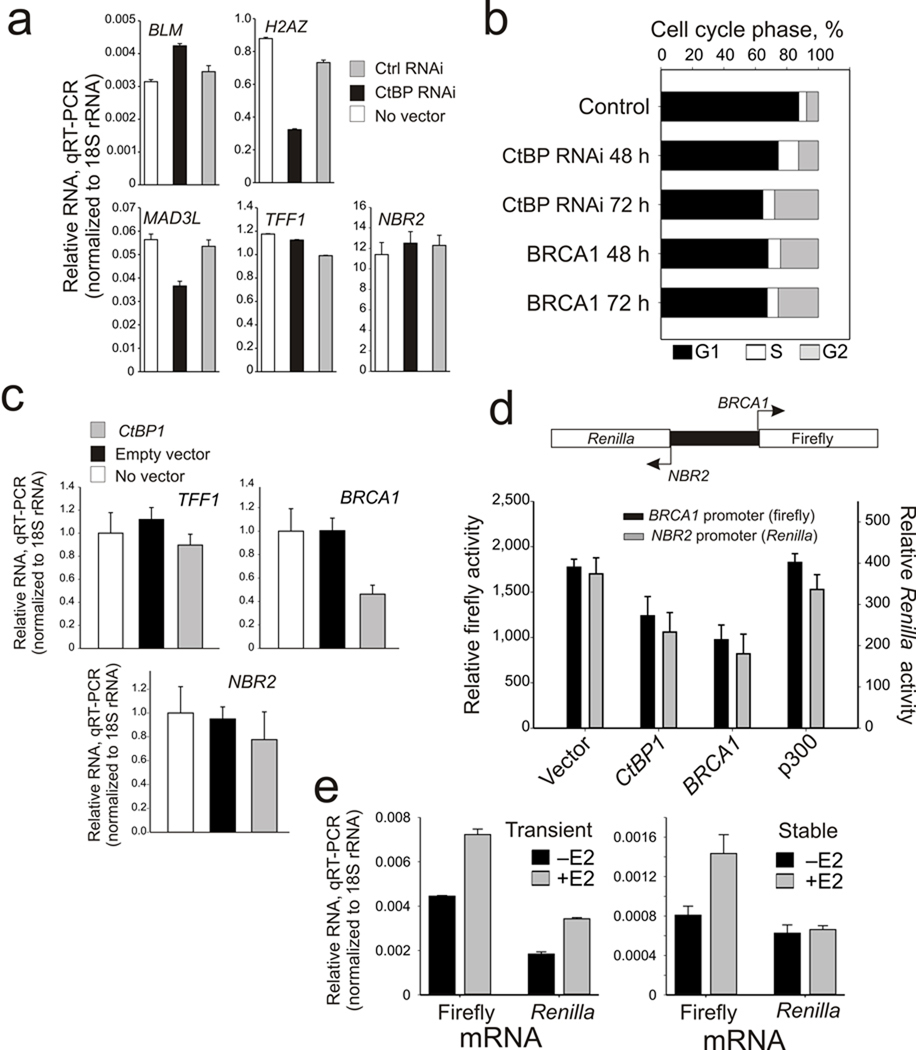
The increased BRCA1 induction by CtBP depletion is both gene and promoter specific since it has minimal effects on BLM expression, inhibits H2AZ and MAD3L expression, fails to induce the estrogen-responsive TFF1 (pS2) gene (as has been shown previously 33), and has insignificant effect on the divergent transcription of the NBR2 gene (Fig. 4a). CtBP depletion also mimics the functional influences of BRCA1 over-expression 12 by inducing a cell cycle block in G2 phase (Fig. 4b). Finally, CtBP depletion also renders the BRCA1 promoter less responsive to estrogen induction with minimal influence on TFF1 (Supplementary Fig. S3) and over-expression of CtBP represses BRCA1 expression without influencing either TFF1 or divergent NBR2 transcription (Fig. 4c).
Figure 4.
CtBP control of BRCA1 is gene specific, functionally influences cell cycle progression and is chromatin dependent. (a) mRNA levels of BLM, H2AZ, MAD3L, TFF1, and NBR2 in control and CtBP depleted MCF-7 cells. Error bars represent the s.e.m. for N=3 biological replicates. (b) Cell cycle profiles (percent distribution in G1, S, and G2/M phases) of MCF-7 cells depleted of CtBP for 48 h and 72 h, or over-expressing BRCA1 for 48 h and 72 h. (c) mRNA profiles of TFF1, BRCA1 and NBR2 in control MCF-7 or cells 48 h after transfection with empty vector or CtBP1 expressing plasmids. Error bars represent the s.e.m. for N=3 independent biological replicates. (d) Upper panel: schematic diagram of the dual NBR2/BRCA1 promoter reporter. Lower panel: Firefly and Renilla luciferase activity profiles of MCF-7 cells co-transfected with the dual NBR2/BRCA1 luciferase reporter and either control or vectors expressing CtBP1, BRCA1 or p300. (e) Firefly and Renilla luciferase mRNA levels in MCF-7 cells expressing a transiently (left) or stably integrated (right) BRCA1 bi-directional firefly/Renilla luciferase reporter after 24 h stimulation with estrogen. Error bars represent the s.e.m. for N=2 biological replicates.
As mentioned previously, the BRCA1 gene is transcribed from a bidirectional promoter 6, 34. Although most bidirectional promoters show highly correlated bidirectional expression 35, the expression at the BRCA1 promoter is primarily unidirectional in response to CtBP depletion (Fig. 3a) and estrogen induction (see below). Therefore, unique aspects of the BRCA1 promoter sequence and chromatin structure may account for the unidirectionality. These possibilities were tested by transient transfection of a bidirectional BRCA1 promoter driving firefly luciferase transcription in the direction of the BRCA1 1st exon and Renilla luciferase transcription in the divergent direction of the NBR2 1st exon (Fig. 4d). Over-expression of either CtBP or BRCA1 caused bidirectional repression of transcription, suggesting that promoter and direction specific repression of BRCA1 transcription by CtBP and BRCA1 requires a structural chromatin context at the endogenous BRCA1 promoter that is not recapitulated by transiently transfected DNA constructs (Fig. 4d). To test this, cells transiently transfected with the bidirectional reporter were compared to cells in which the reporter was stably integrated (Fig. 4e). As shown in Figure 4e, return of the BRCA1 bidirectional promoter to a chromatin context recovers the unidirectional transcriptional response to estrogen induction, thus highlighting the role of chromatin structure in maintaining the fidelity of BRCA1 transcriptional regulation.
HDAC inhibition mimics BRCA1 induction by estrogen or CtBP depletion
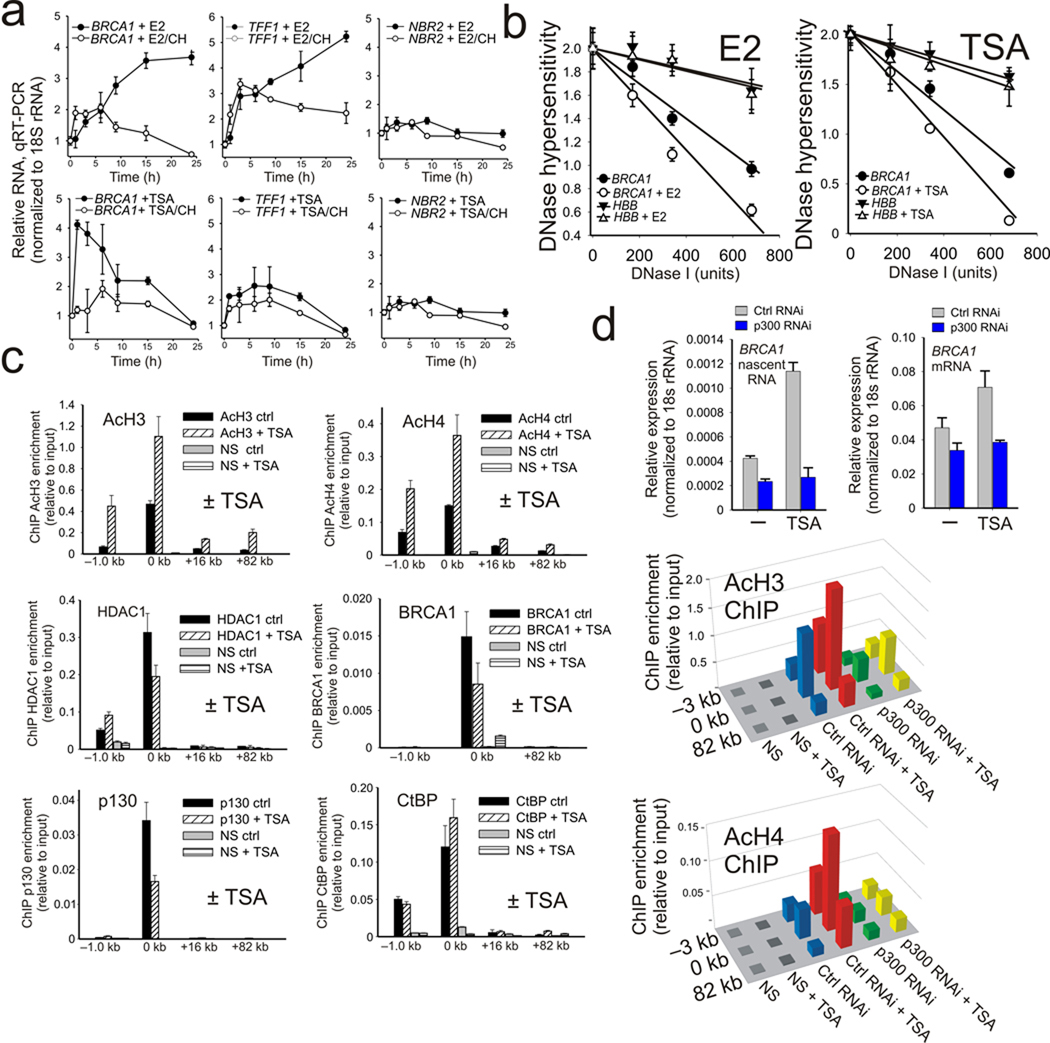
The mechanism through which CtBP is able to control BRCA1 transcription involves direct regulation of the local chromatin marks within the BRCA1 bidirectional promoter by preventing histone H3 and H4 acetylation (Figs. 1e,f; and Fig. 3c, 3d) through HDAC1 recruitment (Fig. 2a; and Fig. 3b). If the key regulatory step in this process is histone acetylation, then a reasonable prediction would be that HDAC inhibition would lead to BRCA1 transcriptional induction. As shown in Fig. 5a this is indeed the case since incubation of MCF-7 with TSA produces a rapid induction of BRCA1 transcription that occurs much earlier than estrogen stimulation for both mature and nascent RNA transcripts (Supplementary Fig. S4). Also, like estrogen, TSA induction is inhibited by treatment with protein synthesis inhibitors. Notably, the influence of HDAC inhibition is both promoter-specific and directional since neither TFF1 nor NBR2 are significantly induced by HDAC inhibition (Fig. 5a). Moreover, both estrogen treatment and HDAC inhibition produce nearly identical increases in chromatin accessibility at the BRCA1 promoter compared to the untreated control (Fig. 5b). As expected HDAC inhibition with TSA results in significantly increased histone H3 and H4 acetylation at the BRCA1 promoter in addition to some alteration in the assembly of HDAC1, BRCA1 and p130 while having insignificant influence on the assembly of CtBP, E2F1 and p300 (Fig. 5c). These differences suggest that, although histone acetylation is a major target, additional protein or factor acetylation may also play a role in the stability of the co-repressor complexes assembled at the BRCA1 promoter (Fig. 5c and Supplementary Fig. S5a,b). Finally, loss of p300 by RNAi depletion renders the BRCA1 promoter unresponsive to TSA treatment and blocks the increase in promoter proximal histone acetylation, thus demonstrating that p300 is primarily responsible for the positive influence of HDAC inhibition on histone acetylation and transcription at the BRCA1 promoter (Fig 5d and Supplementary Fig. S5c).
Figure 5.
TSA mimics estrogen induced activation of BRCA1 by increasing p300 dependent histone acetylation at the BRCA1 promoter. (a) Time course of TFF1, NBR2, and BRCA1 expression in MCF-7 cells treated 0–24 h with either E2, E2 + cycloheximide (10 µg ml−1), TSA (500 ng ml−1), or TSA + cycloheximide as indicated. Error bars represent the s.e.m. for N=2 independent biological replicates. (b) DNase I hypersensitivity profile of the BRCA1 promoter and an (HBB) locus control from MCF-7 cells treated with either estrogen or TSA. The error bars represent the s.e.m. for N=3 biological replicates. (c) Acetylated histone H3, acetylated histone H4, HDAC1, BRCA1, p130, and CtBP ChIP profiles at the BRCA1 promoter in control or MCF-7 cells treated 1 h with 500 ng ml−1 TSA. Error bars represent the s.e.m. for N=2 biological replicates. (d) Upper panel: TSA stimulated expression of BRCA1 nascent and mature RNA levels in either control or p300 depleted MCF-7. Error bars represent the s.e.m. for N=2 biological replicates. Lower panel: ChIP enrichment for H3 and H4 histone acetylation at the BRCA1 locus in control versus p300 depleted MCF-7 cells with or without TSA stimulation. Means from N=2 independent biological replicates are shown.
CtBP acts as a “metabolic switch” to control BRCA1 transcription
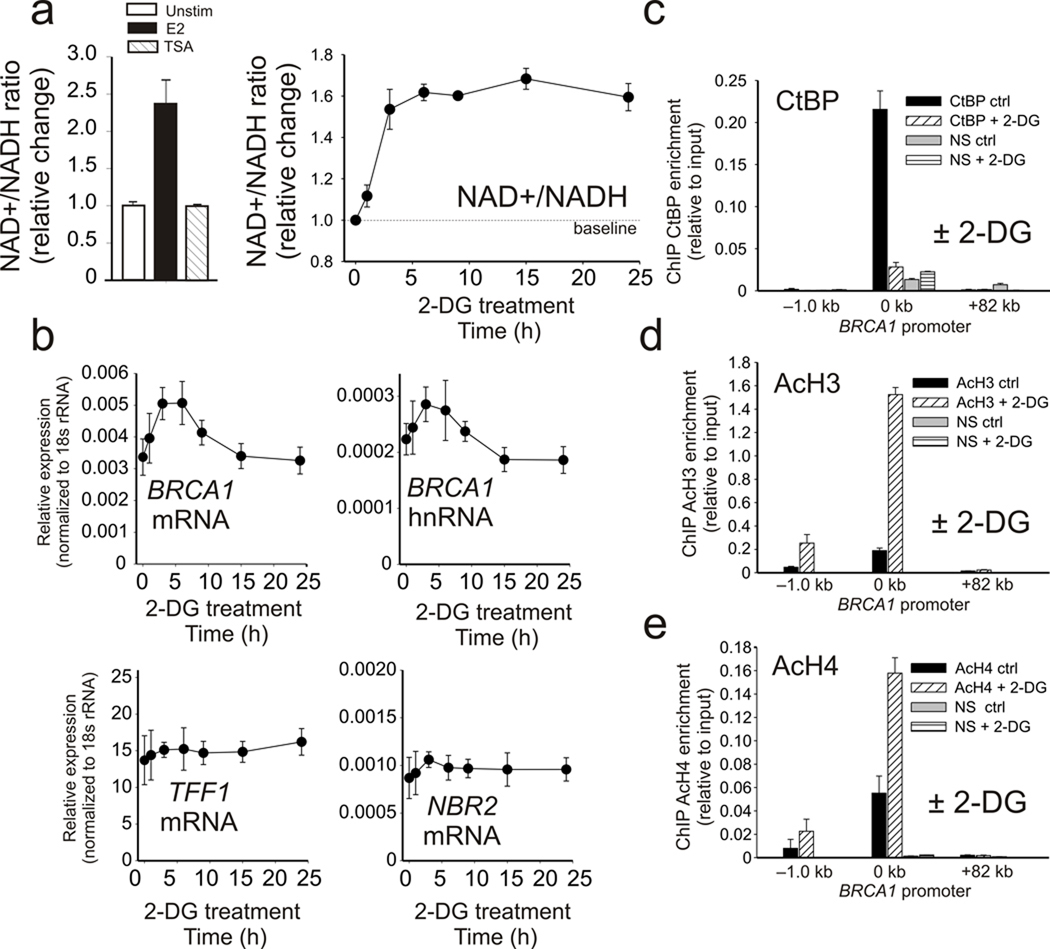
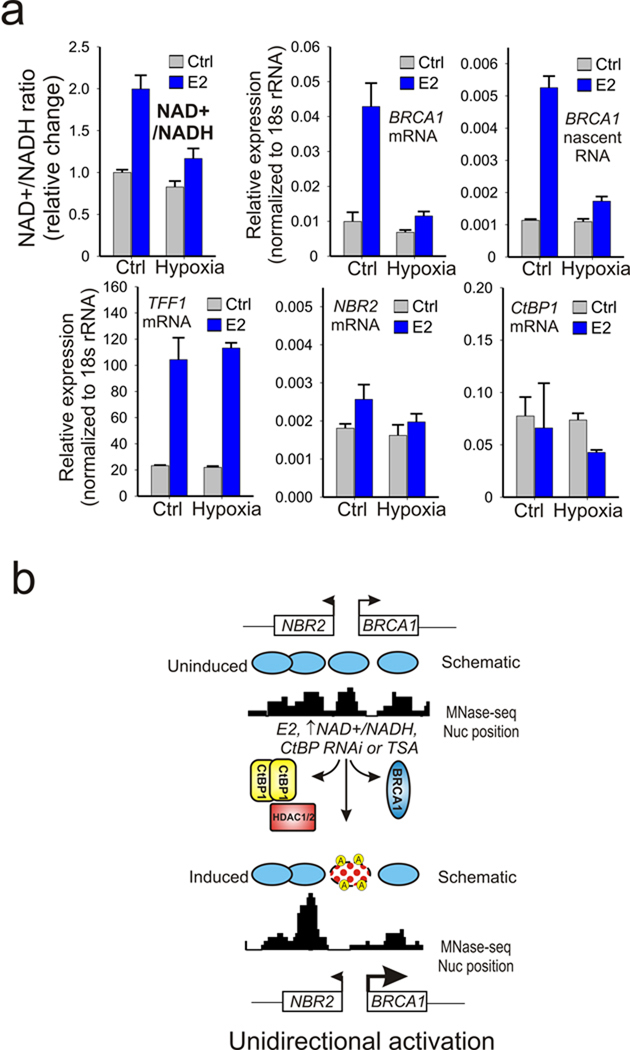
A very unique and biologically important aspect of CtBP is that it is most active as a dimer and its dimerization is promoted by binding to NAD+ and NADH 17. CtBP has a much higher affinity (>100 fold) for NADH compared to NAD+, and the free cellular concentrations of both NAD species approach their CtBP binding affinities. Because of this, CtBP is thought to act as both a sensor and effector of cellular metabolic status 20. Estrogen treatment is a major form of mitogenic stimulation that increases cellular proliferation in normal mammary tissues and enhances cell cycle entry in breast-derived cell lines like MCF-7 (Supplementary Fig. S6). The elevated respiration associated with increased proliferation causes the NAD+/NADH ratio to rise to meet the increased energy demand due to surges in protein synthesis and DNA replication 36, 37. This is clearly demonstrated by comparison of MCF-7 cells stimulated in the presence of estrogen versus TSA, which shows that estrogen stimulation significantly increases the NAD+/NADH ratio (Fig. 6a, left). The NAD+/NADH ratio can also be increased by treatment with the glycolysis inhibitor 2-deoxyglucose (2-DG) (Fig. 6a, right). Most important is the observation that increases in the NAD+/NADH levels by 2-DG treatment selectively results in increased expression of BRCA1 mature and nascent RNA while having no effect on TFF1 expression or divergent transcription from the NBR2 gene (Fig. 6b). Furthermore the 2-DG induction of BRCA1 transcription is associated with loss of CtBP from the BRCA1 promoter and a concomitant increase in histone H3 and H4 acetylation (Fig. 6c–e) without significant influence on CtBP expression or E2F recruitment (Supplementary Fig. S7). Finally, though hypoxia has been well established to block BRCA1 transcription in addition to other factors important in the response to DNA damage 9, 38, its influence on estrogen regulated induction of BRCA1 has not been explored. Since, in contrast to acute 2-DG treatment, hypoxia causes a decrease in the NAD+/NADH ratio, it should block estrogen induction of BRCA1 transcription. As shown in Figure 7a, hypoxia produces a selective block to the estrogen induction of BRCA1 transcription while influencing neither TFF1 induction nor divergent transcription from the NBR2 promoter. These compelling findings demonstrate that CtBP functions as a “metabolic switch” at the BRCA1 promoter that selectively controls the levels of histone acetylation, chromatin structure, and transcription at the BRCA1 promoter in response to the cell’s metabolic status.
Figure 6.
CtBP functions as a metabolic switch to control BRCA1 expression. (a) (Left) Relative change in the NAD+/NADH ratio in lysates from MCF-7 cells treated with vehicle or E2 for 24 h, or TSA for 1 h as indicated. (Right), Time course of the relative change in the NAD+/NADH ratio in MCF-7 cells treated 0–24 h with 10 mM 2-DG. (b) Relative enrichment of TFF1, NBR2, and nascent and mature BRCA1 RNA in MCF-7 cells treated 0–24 h with 2-DG. Error bars represent the s.e.m. for N=2 independent biological replicates. (c–e). ChIP enrichment for CtBP (c), acetylated histone H3 (d) and acetylated histone H4 (e) at the BRCA1 promoter in MCF-7 cells treated 3 h with 2-DG. Error bars represent the s.e.m. for N=2 independent biological replicates.
Figure 7.
Hypoxia inhibits estrogen induced changes in the NAD+/NADH ratio and selectively represses estrogen induction of BRCA1 transcription. (a) Assay of relative change in the NAD+/NADH ratio, and BRCA1, TFF1, NBR2, and CtBP1 expression in control versus hypoxic cells in the presence or absence of estrogen stimulation.(b) Schematic hypothetical model for the mechanism of CtBP control of BRCA1 transcription. The nucleosome positioning is according to Schones et al 2008 39 by the genome-wide sequencing of micrococcal nuclease generated fragments. E2, changing NAD+/NADH ratio, CtBP knockdown or TSA treatment induce removal or inactivation of a repressive complex composed of CtBP, BRCA1 and HDAC1/2 at the dual BRCA1 promoter. Acetylation associated destabilization of the centrally positioned nucleosome, in combination with the asymmetric nucleosome distribution at the BRCA1 locus biases more toward expression of BRCA1 compared to NBR2 in response to the activating signals.
Prior studies that have mapped nucleosome positioning at the BRCA1 promoter in quiescent and proliferating cells demonstrate that there is a pronounced shift in locally distributed nucleosomes that results in dramatic increases in chromatin accessibility 39. Mapping of the position of the 5′ end or TSS of NBR2 and BRCA1 based on their refseq annotation indicates that the intergenic distance between the two NRB2 and BRCA1 TSSs is approximately 133 bp, which is less than the 147 bp occupied by a single nucleosome. These observations indicate that the bidirectional promoter shared by NRB2 and BRCA1 is effectively occluded by a single, dynamically regulated nucleosome (Fig. 7b). Thus a central regulatory event that controls BRCA1 expression is an active and persistent competition between DNA bound transcriptional complexes and the centrally occluding nucleosome which undergoes cycles of targeted disruption and stabilization by the competing activities of co-activators and co-repressors assembled at the BRCA1 promoter. This balance ultimately influences the accessibility of the promoter to additional positive regulators of the transcription cycle that drive BRCA1 expression.
DISCUSSION
The BRCA1 promoter is known to be regulated by a variety of different stimuli including estrogen stimulation, DNA damage, and hypoxia 9, 10, 40. Each of these processes influence the NAD+/NADH ratio. Estrogen increases the NAD+/NADH ratio secondary to increased respiration due to the proliferative response 37, 41. Conversely hypoxia increases NADH levels. DNA damage consumes NAD+ through PARP-1 and some forms of DNA damage activate the HIPK2 kinase which phosphorylates CtBP resulting in its elimination via the proteasome pathway 42, 43. All of these pathways contribute to upregulation of BRCA1 and, consistent with the proposed role of hypoxia and anaerobic glycolysis in promoting tumor formation, suggest a contribution from the downregulation of tumor suppressors in this process 44. The selective inhibition of estrogen induced expression of BRCA1 by hypoxia suggests a direct role for this form of regulation during tumor progression in patients with estrogen receptor positive metastatic breast cancer. Recent reports that PARP-1 assembles at the BRCA1 promoter and that its inhibition represses BRCA1 transcription indicates a potential role for PARP-1 in BRCA1 regulation. Although we detect occupancy of PARP-1 at the BRCA1 promoter in MCF-7 cells this assembly is constitutive and does not appear to be influenced by estrogen or HDAC inhibition (Supplementary Fig. S8).
Post-recruitment regulation of BRCA1 transcription
The BRCA1 promoter is a member of a unique class of bidirectional promoters. As mentioned previously, nearly all genes in this class contain CpG islands, exclude TATA boxes and are enriched for binding site for Myc, GABPA, E2F-1, E2F-4, and the CCAAT box 35, 45, all of which have been characterized and studied in the bidirectional BRCA1 promoter 6, 8–10, 25, 34. A second very common feature of bidirectional promoters is their high enrichment in activating histone marks and poised RNA polymerases suggesting that their chromatin structure is generally more open than other gene classes35. This is certainly consistent with the findings in this current study demonstrating that the resting BRCA1 promoter is already occupied by a poised RNA polymerase II and p300 complex, maintains constitutive histone marks associated with transcriptional activation, and is highly accessible to nuclease digestion in comparison to β-globin in MCF-7 cells. This is also consistent with genome-wide studies that indicate that certain classes of genes containing CpG islands already have destabilized nucleosomes in their proximal promoter and therefore have reduced requirements for chromatin remodeling factors during activation 46. These properties are highly consistent with what we have observed at the BRCA1 promoter, where a central destabilized nucleosome provides a major means of control of BRCA1 transcription through regulation of chromatin marks via histone acetylation. The preloading of Pol II and p300 at the BRCA1 promoter indicates that subsequent post-recruitment steps play an important role in BRCA1 transcriptional regulation. One such step in the transcription cycle is elongation. Recent studies are beginning to link histone acetylation and the recruitment of HAT activity to transcriptional elongation possibly through recruitment of P-TEFb through factors like bromodomain protein 4 (Brd4) 47, 48, or the 14-3-3 adapter proteins that bind to phospho-acetylated histone tails to enhance recruitment of other HAT activity to targeted promoters 49. The fact that we observe recruitment of both P-TEFb and ELL to the BRCA1 promoter and transcribed regions following estrogen induction suggests an intimate association between these factors and chromatin modification during estrogen induction. How estrogen induced chromatin modification facilitates elongation events will be an important area to explore in future studies. Another important area in post-recruitment regulation of BRCA1 will be the role of CtBP in long range changes in chromatin structure at the BRCA1 promoter. Prior studies indicate that lost spatial interactions between the promoter and terminator region of BRCA1 following estrogen stimulation may induce BRCA1 expression 50. Though we do not detect any interaction between CtBP and the terminator region of the BRCA1 locus, a possible role of the CtBP repressor complex in chromatin looping will be an important area for future investigation.
Multiple modes of estrogen stimulation of the BRCA1 promoter
The precise manner in which estrogen stimulates the BRCA1 promoter remains a matter of debate. A general consensus is that estrogen stimulates BRCA1 through an indirect response based on S-phase entry secondary to mitogenic genomic and non-genomic responses caused by estrogen stimulation (e.g. RAS/MAP kinase signaling) 13, 14, 51. Moreover, multiple genome-wide studies of estrogen receptor binding sites by chromatin immunoprecipitation have failed to detect direct binding of ER to the BRCA1 promoter 52. However several groups have proposed that regulation could involve direct association of estrogen receptor via a binding site with weak homology to an estrogen response element (ERE) in the downstream alternate BRCA1 promoter (1b) or through tethering to AP1 or aromatic hydrocarbon receptor binding sites 6, 34, 53, 54. Regardless of these disputed points, none of the mechanisms described above for estrogen stimulation are mutually exclusive and all would be subject to titrated regulation by the assembly and release of the co-activator and co-repressor complexes described in this work.
Does CtBP participate in feedback control of estrogen stimulation?
The estrogen receptor and BRCA1 have a very complex relationship in estrogen responsive tissues 16. Estrogen induces proliferation and the activation of BRCA1 functions in a negative feedback loop to control or restrain the effects of estrogen through targeting estrogen controlled genes, many of which are also controlled by p300 coactivation 55. It is therefore reasonable to imagine that NADH consumption, secondary to estrogen induced proliferation, would serve to activate BRCA1 expression through dismissal of CtBP/HDAC1 complexes from the BRCA1 promoter. This is consistent with the observation that BRCA1 expression is highest in proliferating tissues 56. Interestingly, a high percentage of sporadic breast cancers that show decreased levels of BRCA1 expression also share gene expression profiles that are very similar to those displayed by basal-like subtypes of breast cancer, which express markers normally associated with myoepithelial cells and is the tumor phenotype that most frequently arises in patients with germline mutations of BRCA1 3. A feature that is common to the basal-like phenotype is the loss of markers associated with epithelial differentiation and the acquisition of features that promote motility and invasiveness. Notably this BRCA-like phenotype is very similar to that seen in breast-derived epithelia cells undergoing epithelial-to-mesenchymal transition (EMT) 57, a process that is frequently associated with overexpression of CtBP 17. It is therefore likely that CtBP overexpression may play a role in a variety of malignancies by antagonizing the expression of BRCA1 and other tumor suppressor genes during tumor progression 17.
High caloric intake, estrogen, CtBP and BRCA1: a perfect storm?
There is a strong correlation between pre and post-menopausal high caloric intake, weight gain and obesity and increased risk for breast cancer 58. The physiological factors associated with increased risk involve elevated levels of extra-gonadal production of circulating and mammary estrogen due to aromatase present in fatty tissues of the breast and throughout the body. The elevated expression of estrogen in the context of higher levels of NADH or lower NAD+/NADH levels due to high caloric intake and/or obesity could establish a state where the pro-proliferative effects of estrogen are not completely balanced by the protective functions of BRCA1 that would normally restrain estrogen induced proliferation and heighten genome surveillance. It is reasonable to speculate that the enhanced CtBP activity in mammary tissues with lower NAD+/NADH ratios, secondary to high caloric diet or obesity, may contribute to the increased risk for malignancies of the breast. In this regard, it would also be of interest to ascertain what percentage of postmenopausal breast cancer cases, associated with pre or post-menopausal weight gain or obesity, display the basal-like phenotype associated with BRCA1 deficiency and/or germline mutation.
MATERIALS AND METHODS
Chemicals and reagents
E2 (estradiol), TSA (trichostatin A), CHX (cycloheximide) were from Sigma Aldrich. 2-DG (2-Deoxy-D-glucose), anti-E2F1, anti-E2F4, anti-p107, anti-p130, anti-CtBP, and anti-CtIP antibodies were from Santa Cruz Biotechnology. Anti-CtBP antibodies are cross-reactive with both CtBP1 and CtBP2. Anti-acetylated histone H3 and anti-acetylated histone H4 antibodies were from Millipore. Anti-HDAC1 antibody was from ABR (Affinity BioReagents). DNase I was from Roche.
Cell culture
MCF-7 cells were maintained in regular DMEM (Dulbecco's Modified Eagle Medium) supplemented with 10% (v/v) FBS (fetal bovine serum), penicillin/streptomycin (Invitrogen) and insulin. Prior to treatment, MCF-7 cells were grown in phenol-red free DMEM medium supplemented with 5% (v/v) charcoal filtered FBS, penicillin/streptomycin, 1 mM pyruvate and insulin for at least 3 days. Generally, 10 nM E2, 500 ng ml−1 TSA and 10 µg ml−1 CHX, were used to treat the cells and 95% (v/v) ethanol was used as a vehicle control. 2-DG was dissolved in ddH2O and used at 10 mM final concentration.
Chromatin immunoprecipitation
All ChIP experiments were performed as previously described 47, with minor revisions. In brief, cells were crosslinked with 1% formaldehyde (w/v) for 5 min at room temperature. The cross-linking was quenched by 0.125 M glycine for 15 min. Then the cells were washed twice with PBS and collected. Approximately 1 × 107 crosslinked cells, resuspended in 1ml immunoprecipitation (IP) buffer (150 mM NaCl, 50 mM Tris-HCl pH7.5, 5 mM EDTA, 0.5% (v/v) NP-40, 1.0% (v/v) Triton X-100, and freshly added proteinase inhibitor cocktail), were sonicated for 13×20 s with 30 s break. Then the sonicated cells were centrifuged and the supernatant was used for IP. In most cases, the lysate from at least 2 million cells (up to 10 million) was incubated with each antibody overnight with rotation at 4 °C. The pre-blocked protein G beads were added to the lysate with rotation for 2 h at 4 °C. The beads were washed with IP buffer supplemented with 500 mM NaCl, IP buffer and then TE pH8.0 buffer. Finally, the precipitated DNA-protein complex was eluted 10 min at 100 °C with chelex-100 or overnight incubation with SDS and proteinase K at 65 °C and used directly for qPCR. Alternatively protein and SDS was removed through standard phenol-chloroform extractions and ethanol precipitation. The qPCR was performed using the SybrGreen® qPCR kit by Invitrogen. The sequences of all primers are provided in the supplemental materials and methods.
Luciferase reporter assays
After 3 days growth in phenol-red free DMEM medium MCF-7 cells were trypsinized, washed twice with PBS and resuspended in DMEM medium with 2.5% FBS (v/v). The plasmid harboring the bidirectional promoter of the BRCA1 locus driving Renilla luciferase transcription from the NBR2 TSS and firefly luciferase transcription from the BRCA1 TSS 9 was kindly provided by P.M. Glazer. The CtBP expression vector was purchased from Origene. The BRCA1 and p300 expression vectors were previously described 10. pcDNA 3.1 is from Invitrogen. In brief, approximately 5 × 106 MCF-7 cells were transfected with 6 µg reporter plasmid and 10 µg expression vector or control pcDNA 3.1 empty vector. Electroporation was performed using the ElectrosquarePorator ECM T820 according to the manufacturer’s instructions. After electroporation, the cells were again seeded to plates with the regular phenol red-free DMEM medium. By 48 h, the cells were collected for luciferase assay by using a Dual Luciferase Reporter assay system kit (Promega) according to the manufacturer’s instructions. Both firefly and Renilla luciferase activity were normalized to total protein levels.
Transfection, qRT-PCR and western blotting
MCF-7 cells were grown in phenol-red free DMEM medium for 3 days. The cells were split and seeded to 80% confluency. The transient transfection of CtBP to MCF-7 cells was performed on the next day using Lipofectamine LTX Reagent (Invitrogen). After 48 h, the cells were collected for further assays. The total RNA was prepared using the RNAeasy kit (Qiagen) following the manufacturer’s protocol. Reverse transcription of 1 µg RNA was carried out by following the QuantiTect® Reverse Transcription procedure (Qiagen). For western blotting, the cells were resuspended in the lysis buffer (50 mM Tris pH7.5, 1 mM EDTA, 150 mM NaCl, 0.1% (w/v) SDS, 1% (v/v) TritonX-100, 1% (w/v) sodium deoxycholate and freshly added proteinase inhibitor cocktail) for 30 min on ice. The lysates were centrifuged for 20 min at 12,000 × g at 4 °C and the supernatants were analyzed by western blotting.
CtBP knockdown
siRNA oligonucleotides specific for CtBP 43 were synthesized by Dharmacon Research, Inc. The Scramble II Duplex was used as a negative control. MCF-7 cells were transfected with 100 nM oligonucleotides and, 48 h later, the expression of CtBP was analyzed by either qRT-PCR or western blotting using anti-CtBP antibody.
NAD+/NADH ratio determination
Determination of the NAD+/NADH ratio in cellular lysates was performed using a Biovision NAD+ and NADH quantitation kit according to the manufacturer’s specifications.
Flow cytometry
For determination of DNA content, all floating and attached cells were collected and combined for analysis. The cells were fixed with cold 70% (v/v) ethanol and stored at −20 °C for at least 24 h. The cells were then washed twice with 1X PBS and once with 1X PBS supplemented with 0.1% (v/v) TritonX-100 and resuspended in 50 µg ml−1 PI (propidium iodide) staining buffer in the presence of 300 µg ml−1 RNase A for 30 min at room temperature. Flow cytometry was performed using FACScalibur (Becton Dickinson) equipped with CellQuest software (Becton Dickinson).
DNase I hypersensitivity assay
MCF-7 cells were collected from plates by trypsinization and washed twice with ice-cold PBS. To isolate nuclei, cells harvested at 250 × g for 5 min at 4 °C were resuspended in ice-cold Buffer A (15 mM Tris-HCl (pH8.0), 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, and freshly added proteinase inhibitor cocktail) followed by addition of equal volume of Buffer A containing 0.04% (v/v) NP-40. Nuclei were washed three times with ice-cold Buffer A and resuspended again in Buffer A. For each DNase I digestion, approximately 1×106 nuclei were harvested and resuspended in 200 ul of pre-warmed (37 ° C) Buffer A, supplemented with 6 mM CaCl2, 75 mM NaCl, and the DNase I (0, 170, 340, and 680 units). Digestions were performed for 6 min at 37 °C, quenched by addition of stop buffer (50 mM Tris-HCl (pH8.0), 100 mM NaCl, 0.1% (w/v) SDS, 100 mM EDTA, and 50 µg ml−1 RNase A) and incubated 1.5 h at 55 °C. Samples were deproteinized at 55 °C overnight in the presence of 50 µg ml−1 proteinase K prior to qPCR analysis.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, the National Cancer Institute and the National Institute on Aging, and the Argentinean Agency of Science and Technology (ANPCyT, PICT 2006-00228). We would like to thank Peter M. Glazer (Yale University) for the gift of the BRCA1 promoter dual reporter.
Footnotes
AUTHOR CONTRIBUTIONS
L-J.D. and A.G.F. performed the experiments. A.D. and D.L.L. helped write the paper and contributed valuable reagents. L-J.D, and K.G. designed experiments and wrote the paper.
REFERENCES
- 1.Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1999. CA Cancer J. Clin. 1999;49:8–31. doi: 10.3322/canjclin.49.1.8. 1. [DOI] [PubMed] [Google Scholar]
- 2.Ford D, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner NC, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 4.Catteau A, Morris JR. BRCA1 methylation: a significant role in tumour development? Semin. Cancer Biol. 2002;12:359–371. doi: 10.1016/s1044-579x(02)00056-1. [DOI] [PubMed] [Google Scholar]
- 5.Wilson CA, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat. Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 6.Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J. Biol. Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- 7.Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 8.Mueller CR, Roskelley CD. Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res. 2003;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bindra RS, et al. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 10.De Siervi A, et al. Transcriptional autoregulation by BRCA1. Cancer Res. 2010;70:532–542. doi: 10.1158/0008-5472.CAN-09-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 13.Spillman MA, Bowcock AM. BRCA1 and BRCA2 mRNA levels are coordinately elevated in human breast cancer cells in response to estrogen. Oncogene. 1996;13:1639–1645. [PubMed] [Google Scholar]
- 14.Marks JR, et al. BRCA1 expression is not directly responsive to estrogen. Oncogene. 1997;14:115–121. doi: 10.1038/sj.onc.1200808. [DOI] [PubMed] [Google Scholar]
- 15.Lane TF, et al. Expression of Brca1 is associated with terminal differentiation of ectodermally and mesodermally derived tissues in mice. Genes Dev. 1995;9:2712–2722. doi: 10.1101/gad.9.21.2712. [DOI] [PubMed] [Google Scholar]
- 16.Gorski JJ, Kennedy RD, Hosey AM, Harkin DP. The complex relationship between BRCA1 and ERalpha in hereditary breast cancer. Clin. Cancer Res. 2009;15:1514–1518. doi: 10.1158/1078-0432.CCR-08-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 19.Gorski JJ, et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2009 doi: 10.1007/s10549-009-0565-0. [DOI] [PubMed] [Google Scholar]
- 20.Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 22.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr. Opin. Genet. Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG. Regulation of BRCA1 expression by the Rb-E2F pathway. J. Biol. Chem. 2000;275:4532–4536. doi: 10.1074/jbc.275.6.4532. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 28.Aprelikova ON, et al. BRCA1-associated growth arrest is RB-dependent. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11866–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan S, et al. Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding. Oncogene. 2001;20:4827–4841. doi: 10.1038/sj.onc.1204666. [DOI] [PubMed] [Google Scholar]
- 30.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 31.Aiyar SE, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol. Cell Biol. 2009;29:1749–1759. doi: 10.1128/MCB.01476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu CF, et al. Distinct transcription start sites generate two forms of BRCA1 mRNA. Hum. Mol. Genet. 1995;4:2259–2264. doi: 10.1093/hmg/4.12.2259. [DOI] [PubMed] [Google Scholar]
- 35.Lin JM, et al. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007;17:818–827. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks RF. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell. 1977;12:311–317. doi: 10.1016/0092-8674(77)90209-4. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Enriquez S, et al. Control of cellular proliferation by modulation of oxidative phosphorylation in human and rodent fast-growing tumor cells. Toxicol. Appl. Pharmacol. 2006;215:208–217. doi: 10.1016/j.taap.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Meng AX, et al. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother. Oncol. 2005;76:168–176. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andres JL, et al. Regulation of BRCA1 and BRCA2 expression in human breast cancer cells by DNA-damaging agents. Oncogene. 1998;16:2229–2241. doi: 10.1038/sj.onc.1201752. [DOI] [PubMed] [Google Scholar]
- 41.Stockl P, Hutter E, Zwerschke W, Jansen-Durr P. Sustained inhibition of oxidative phosphorylation impairs cell proliferation and induces premature senescence in human fibroblasts. Exp. Gerontol. 2006;41:674–682. doi: 10.1016/j.exger.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 44.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2009 doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byun JS, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc. Natl. Acad. Sci. U. S. A. 2009 doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 49.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Driggers PH, Segars JH. Estrogen action and cytoplasmic signaling pathways. Part II: the role of growth factors and phosphorylation in estrogen signaling. Trends Endocrinol. Metab. 2002;13:422–427. doi: 10.1016/s1043-2760(02)00634-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeffy BD, et al. An estrogen receptor-alpha/p300 complex activates the BRCA-1 promoter at an AP-1 site that binds Jun/Fos transcription factors: repressive effects of p53 on BRCA-1 transcription. Neoplasia. 2005;7:873–882. doi: 10.1593/neo.05256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hockings JK, Degner SC, Morgan SS, Kemp MQ, Romagnolo DF. Involvement of a specificity proteins-binding element in regulation of basal and estrogen-induced transcription activity of the BRCA1 gene. Breast Cancer Res. 2008;10:R29. doi: 10.1186/bcr1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan S, et al. p300 Modulates the BRCA1 inhibition of estrogen receptor activity. Cancer Res. 2002;62:141–151. [PubMed] [Google Scholar]
- 56.Blackshear PE, et al. Brca1 and Brca2 expression patterns in mitotic and meiotic cells of mice. Oncogene. 1998;16:61–68. doi: 10.1038/sj.onc.1201506. [DOI] [PubMed] [Google Scholar]
- 57.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 58.Cleary MP, Grossmann ME. Minireview: Obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150:2537–2542. doi: 10.1210/en.2009-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.