Abstract
This study investigated and compared the antimicrobial resistance patterns and ribotypes of Staphylococcus aureus isolated from pig tonsils and cow’s milk in China. A total of 90 isolates of S. aureus was included: 42 strains were isolated from tonsils of pigs and 48 from half-udder milk. The broth microdilution method and the double-disc diffusion test (D test) were used for antimicrobial susceptibility testing. The mecA gene for methicillin-resistant S. aureus (MRSA) and the ermA, ermB, ermC, and msrA genes for erythromycin-resistant strains were detected by polymerase chain reaction (PCR). The isolates were ribotyped with the Riboprinter system. The highest frequency of resistance was observed with clindamycin (91.1%), followed by penicillin (90.0%), and erythromycin (85.6%). All strains were susceptible to vancomycin and trimethoprim-sulfamethoxazole. The D test showed that 54.5% (42/77) of erythromycin-resistant isolates had the constitutive resistance phenotype and 45.5% (35/77) had the inducible resistance phenotype to clindamycin. A higher proportion of resistance to cephalosporins, macrolides, fluoroquinolones, and pleuromutilins was observed in pig isolates than in milk isolates (P < 0.05). The mecA gene was detected in all MRSA isolates; 89.6% of erythromycin-resistant strains harbored the ermC gene and 16.9% harbored the ermB gene. A total of 35 different ribogroups was found among the isolates investigated; 83.3% of pig strains belonged to 1 cluster with a similarity coefficient of 0.84. In contrast, 3 main clusters were observed among 68.8% of milk strains, which indicates a high degree of host specificity.
Résumé
Ce travail visait à étudier et comparer les patrons de résistance antimicrobienne et les ribotypes d’isolats de Staphylococcus aureus provenant des amygdales de porc et du lait de vache en Chine. Un total de 90 isolats de S. aureus était inclus : 42 souches provenaient des amygdales de porcs et 48 provenant de lait de 2 des 4 quartiers. La méthode de microdilution en bouillon et l’épreuve de double diffusion en disque (test D) ont été utilisées pour déterminer la sensibilité aux antibiotiques. Le gène mecA des S. aureus résistants à la méthicilline (MRSA) et les gènes ermA, ermB, ermC, et msrA des souches résistantes à l’érythromycine ont été détectés par réaction d’amplification en chaîne par la polymérase (PCR). Les isolats ont été ribotypés avec le système Riboprinter. La fréquence de résistance la plus élevée a été observée avec la clindamycine (91,1 %), suivie de la pénicilline (90,0 %) et l’érytrhomycine (85,6 %). Toutes les souches étaient sensibles à la vancomycine et au trimethoprime-sulfaméthoxazole. Le test D a montré que 54,5 % (42/77) des isolats résistants à l’érythromycine avaient le phénotype constitutif de résistance et 45,5 % (35/77) avaient le phénotype de résistance inductible à la clindamycine. Une fréquence plus élevée de résistance aux céphalosporines, aux macrolides, aux fluoroquinolones, et aux pleuromutilines était observée chez les isolats porcins comparativement aux isolats laitiers (P < 0,05). Le gène mecA a été détecté à partir de tous les isolats MRSA; 89,6 % des souches résistantes à l’érythromycine portaient le gène ermC et 16,9 portaient le gène ermB. Un total de 35 ribogroupes différents a été trouvé parmi les isolats étudiés; 83,3 % des souches porcines appartenaient à 1 regroupement avec un coefficient de similarité de 0,84. En contrepartie, 3 regroupements principaux ont été observés parmi les 68,8 % d’isolats provenant de lait, ce qui indique un degré élevé de spécificité d’hôte.
(Traduit par Docteur Serge Messier)
Introduction
Staphylococcus aureus (S. aureus) is a common pathogen in domestic livestock. It can be harbored in pigs’ tonsils and cows’ udders and causes a wide variety of diseases as well as potential economic losses. Strains of S. aureus can also cause human food poisoning through the production of an enterotoxin (1). Researchers have recently discovered that animals can be sources of S. aureus infections in humans (2).
Staphylococcus aureus has become resistant to many commonly used antibiotics, which have been widely used to treat S. aureus infections. Multiple antimicrobial susceptibility patterns of S. aureus have been observed in various animals (3,4). At one time, β-lactam antibiotics were one of the most effective drugs against S. aureus infection, but many studies have now reported that S. aureus is resistant to penicillin and methicillin (5,6). The former is mediated by penicillinase, whereas the latter, methicillin-resistant S. aureus (MRSA), is conferred by the mecA gene. This gene codes for an altered penicillin-binding protein (PBP2a or PBP2′), which has a lower affinity for binding β-lactams and leads to increased resistance to other β-lactams.
In recent years, the fight against MRSA has become an important public health issue. Knowledge of molecular typing of S. aureus is important in order to monitor the source and transmission routes of contamination and to establish effective control measures. To date, the following molecular subtyping methods have been used to study the clonal relationship among S. aureus isolates: multilocus sequence typing (MLST) (7), staphylococcal chromosome cassette (SCCmec) typing (8), pulsed-field gel electrophoresis (PFGE) (9), and ribotyping (10). Of these methods, PFGE is considered the gold standard for genotyping of S. aureus, but its use has been limited by the need for fastidious operations and highly skilled technicians. In contrast, ribotyping methods using the Riboprinter system can rapidly and easily identify and distinguish S. aureus from different sources of infection and identical hierarchical typing results of the strains for PFGE and ribotyping methods have also been observed (11).
The objectives of this study were to determine the antimicrobial susceptibility and genetic patterns of S. aureus isolated from pig tonsils and cow’s milk and to compare the differences among isolates, since information on this important zoonotic pathogen is limited in veterinary medicine in China.
Materials and methods
Bacterial isolates
A total of 90 isolates identified as S. aureus in coagulase tests and biochemical tests by our lab were used in this study. Of these, 42 strains were isolated from tonsil of the soft palate of swine at slaughter and 48 strains were isolated from half-udder milk samples of cows with clinical or subclinical bovine mastitis. Samples were collected from 8 regions in China (Shandong, Guangdong, Beijing, Dalian, Hebei, Henan, Guangxi, and Tianjin).
Antimicrobial susceptibility testing
Susceptibility to antimicrobial agents was determined by the broth microdilution method applied according to the Clinical Laboratory Standard Institute (CLSI) standard (12,13). The following 11 antimicrobials were used as test drugs: penicillin (0.06 to 2 μg/mL), cefoxitin (0.5 to 16 μg/mL), ceftiofur (0.12 to 32 μg/mL), erythromycin (0.12 to 32 μg/mL), vancomycin (0.25 to 16 μg/mL), sulfisoxazole (16 to 512 μg/mL), trimethoprim-sulfamethoxazole (0.5/9.5 to 4/76 μg/mL), enrofloxacin (0.015 to 16 μg/mL), ofloxacin (0.06 to 16 μg/mL), tiamulin (0.25 to 256 μg/mL), and clindamycin (0.03 to 256 μg/mL). The reference strain used for antibiotic susceptibility testing was S. aureus ATCC 29213. The results were recorded as resistant or susceptible according to the interpretive standards of the CLSI (12,13). The isolate was defined as an MRSA-phenotype that was found to be resistant to both penicillin and cefoxitin, while the isolates resistance to only penicillin, but susceptibility to ceftoxitin, represented the penicillinase-producing S. aureus. Moreover, strains that were resistant to both erythromycin and clindamycin were defined as showing constitutive MLSB resistance.
D-test
The double-disk diffusion test (D-test) was performed according to the CLSI guideline (13) to determine if macrolide-resistant, clindamycin-susceptible isolates have inducible resistance to lincosamides. Briefly, a 2-μg clindamycin disk was placed 15 mm from the edge of a 15-μg erythromycin disk as part of the normal disk diffusion procedure. After incubation, organisms that showed flattening of the clindamycin zone adjacent to the erythromycin disk (“D” zone) indicated inducible clindamycin resistance, defined as having inducible macrolides-lincosamides-streptogramin B (MLSB) resistance. Those strains that did not show flattening of the clindamycin zone (no induction) were considered to have the MS phenotype.
Detection of mecA gene and erythromycin-resistant gene by polymerase chain reaction (PCR)
Possible MRSA colonies were subcultivated and identified for the mecA gene and erythromycin-resistant isolates were tested for the presence of ermA, ermB, ermC, and msrA genes by polymerase chain reaction (PCR). The temple DNA was extracted by the boiling method and either used immediately for PCR or stored at −20°C. The oligonucleotide primer sequences (14–15) are listed in Table I. Negative controls for all PCRs consisted of the PCR mixes without template deoxyribonucleic acid (DNA). Polymerase chain reaction (PCR) products were analyzed by agarose gel electrophoresis. Confirmation of the amplicons was determined by DNA sequencing and the results were analyzed using BLAST on-line software (http://www.ncbi.nlm.nih.gov).
Table I.
Oligonucleotide primer sequences used in this study
| Gene | Primer sequence (5′–3′) | PCR product size (bp) | Reference number |
|---|---|---|---|
| mecA | GTT GTA GTT GTC GGG TTT GG CTT CCA CAT ACC ATC TTC TTT AAC |
163 | (14) |
| ermA | TAT CTT ATC GTT GAG AAG GGA TT CTA CAC TTG GCT TAG GAT GAA A |
139 | (15) |
| ermB | CTA TCT GAT TGT TGA AGA AGG ATT GTT TAC TCT TGG TTT AGG ATG AAA |
142 | (15) |
| ermC | CTT GTT GAT CAC GAT AAT TTC C ATC TTT TAG CAA ACC CGT ATT C |
190 | (15) |
| msrA | TCC AAT CAT TGC ACA AAA TC AAT TCC CTC TAT TTG GTG GT |
163 | (15) |
Ribotyping
The study was conducted with the RiboPrinter (DuPont Qualicon, Wilmington, Delaware, USA), an automated ribotyping system, following the manufacturer’s operating instructions. Briefly, the strains were grown on nutrient agar plates overnight at 37°C and pure colonies picked were inactivated by heat treatment. DNA was extracted from cell lysate and cut into fragments by the restriction enzyme EcoRI. These fragments were separated according to size using gel electrophoresis and transferred to a membrane, which was followed by hybridization. The genetic fingerprint was visualized and then captured by a digitizing camera. Similarities between restriction endonuclease digestion profiles were analyzed using BioNumerics software (Applied Maths, Kortrijk, Belgium). Clustering was performed by the unweighted pair group method with arithmetic mean (UPGMA). The dendrogram of ribotyping patterns of isolates tested was drawn with a 1.0% position tolerance and 0.5% optimization. Strains were assigned to a special ribogroup if the similarity coefficient between their patterns was ≥ 0.85 and judged as epidemiology correlation.
Statistical analysis
Version 16.0 of SPSS statistics software for Windows was used for statistical analysis. The categorical variables were compared using a Pearson Chi-squared or Fisher’s exact test, as appropriate. Differences were considered significant when the 2-sided P-value was < 0.05.
Results
Antibiograms
The antimicrobial resistance profile of S. aureus strains isolated from pig tonsils and cow’s milk is summarized in Table II. Intermediate resistance was observed only for enrofloxacin among isolates from pigs (32/42, 76.2%) and milk (2/48, 4.2%) and is therefore not shown in the table. The highest frequency of resistance was observed with clindamycin (91.1%, 82/90), followed by penicillin (90.0%, 81/90), erythromycin (85.6%, 77/90), tiamulin (48.9%, 44/90), ofloxacin (45.6%, 41/90), ceftiofur (38.9%, 35/90) and cefoxitin (38.9%, 35/90). All strains were susceptible to vancomycin and trimethoprim-sulfamethoxazole. Among the 77 erythromycin-resistant isolates, 54.5% (42/77) showed the constitutive and 45.5% (35/77) showed the inducible resistance phenotype to clindamycin, while there was no MS phenotype based on the double-disk test.
Table II.
Antimicrobial resistance profile of S. aureus isolated from pig tonsils and cow’s milk
| Antimicrobial agent (μg/mL) | MIC resistant breakpointa | Pig origin (n = 42)
|
Milk origin (n = 48)
|
Total (n = 90)
|
P-valueb | |||
|---|---|---|---|---|---|---|---|---|
| MIC range | NR (%) | MIC range | NR (%) | MIC range | NR (%) | |||
| Penicillin (0.06 to 2) | 0.25 | ≤ 0.06 to > 2 | 39 (92.9) | 0.5 to 2 | 42 (87.5) | ≤ 0.06 to > 2 | 81 (90.0) | 0.314 |
| Cefoxitin (0.5 to 16) | 8 | 2 to > 16 | 35 (83.3) | 2 to 4 | 0 | 2 to > 16 | 35 (38.9) | < 0.01 |
| Ceftiofur (0.12 to 32) | 8 | 0.5 to > 32 | 35 (83.3) | 0.5 to 4 | 0 | 0.5 to > 32 | 35 (38.9) | < 0.01 |
| Erythromycin (0.12 to 32) | 8 | 1 to > 32 | 40 (95.2) | 0.5 to > 32 | 37 (77.1) | 0.5 to > 32 | 77 (85.6) | < 0.05 |
| Vancomycin (0.25 to 16) | 16 | 0.5 to 1 | 0 | 0.25 to 1 | 0 | 0.25 to 1 | 0 | — |
| Sulfisoxazole (16 to 512) | 512 | 64 to > 512 | 1 (2.4) | 16 to 128 | 4 (8.3) | 16 to 512 | 5 (5.6) | 0.219 |
| Trimethoprim-sulfamethoxazole (0.5/9.5 to 4/76) | 4/76 | ≤ 0.5/9.5 | 0 | ≤ 0.5/9.5 | 0 | ≤ 0.5/9.5 | 0 | — |
| Enrofloxacin (0.015 to 16) | 4 | 0.25 to > 4 | 6 (14.3) | 0.03 to 0.12 | 1 (2.1) | 0.03 to > 4 | 7 (7.8) | < 0.01 |
| Ofloxacin (0.06 to 16) | 4 | 0.5 to > 16 | 38 (90.5) | 0.5 to 1 | 3 (6.2) | 0.5 to > 16 | 41 (45.6) | < 0.01 |
| Tiamulin (0.25 to 256) | 32 | 1 to > 256 | 42 (100) | 2 to > 256 | 2 (4.2) | 1 to > 256 | 44 (48.9) | < 0.01 |
| Clindamycin (0.03 to 256) | 4 | 1 to > 256 | 39/40 (92.9/95.2)c | 1 to > 256 | 8/42 (16.7/87.5)c | 1 to > 256 | 47/82 (52.2/91.1)c | < 0.01/0.36d |
NR (%) — number (%) of resistant isolates.
MIC (minimum inhibitory concentration) resistant breakpoint taken from the Clinical and Laboratory Standards Institute (CLSI) (13).
P-value accounts for the differences between the number of resistant strains isolated from pig tonsils and cow’s milk.
For 39/40 (92.9/95.2) and 8/42 (16.7/87.5), 39 (92.9) or 8 (16.7) refers to the number of resistant strains (resistant rate in percent) to clindamycin by broth microdilution method, whereas 40 (95.2) or 42 (87.5) means the number of all resistant strains (resistant rate in percent) after D-test according to CLSI.
< 0.01 and 0.36 are the results of P-value after broth microdilution testing and D-test, respectively.
In contrast, resistances to cephalosporins, macrolides, fluoroquinolones, and pleuromutilins were significantly higher in pig isolates than in milk isolates (P < 0.05). A higher proportion of MRSA phenotype was observed in pig isolates (83.3%, 35/42) than in milk strains (0, 0/48), although the levels of resistance to penicillin were similar (92.9% versus 87.5%). Furthermore, a higher proportion of isolates from pigs (92.9%, 39/42) was constitutive MLSB resistance phenotype than that of milk strains (10.4%, 5/48), although resistant rates similar to clindamycin were observed in isolates from pigs and those from milk (95.2% versus 91.1%).
There was no significant difference in resistance to penicillin, sulfisoxazole, and clindamycin between the isolates from pigs and those from milk.
Harboring of mecA, ermA, ermB, ermC, and msrA genes
All 35 MRSA isolates from pigs were positive for the mecA gene. Among the erythromycin-resistant isolates, 69 strains (89.6%) were found to harbor the ermC and 13 strains (16.9%) harbored the ermB gene. Strains with inducible MLSB resistance harbored only the ermC gene, whereas isolates with constitutive MLSB phenotype may have contained the ermB and/or ermC gene. The ermA and msrA genes were not detected.
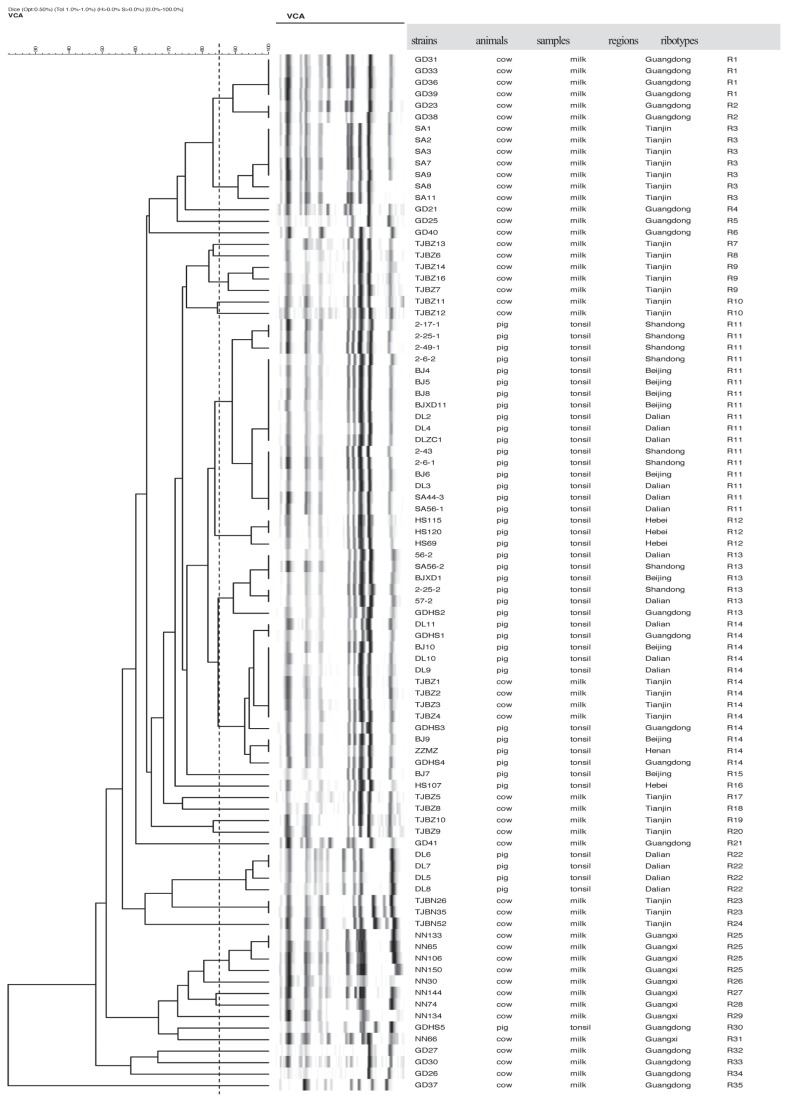
Ribotyping
All 90 S. aureus strains used in this study were typed into 35 different ribogroups by the RiboPrinter and were tentatively designated as R1 to R35. Each of the patterns contained 8 to 15 fragments (Figure 1). The dendrogram presented in Figure 1 shows that the genetic diversity of strains isolated from milk was greater than that of strains isolated from tonsils. Most pig strains (83.3%, 35/42) belonged to 1 cluster (including 4 ribogroups, R11 to R14), which showed high genetic relatedness with a similarity coefficient of 0.84. Four pig isolates (9.5%) from Dalian belonged to R22 with a similarity coefficient of higher than 0.94. Another 3 pig strains (BJ7, HS107, and GDHS5) were classified as R15, R16, and R30, respectively. In contrast, 48 milk strains belonged to the rest of the 27 ribotypes, with 3 main clusters (33/48 milk strains, 68.8%). One cluster (R1 to R6) included 16 strains from Guangdong and Tianjin with the coefficient higher than 0.72. The other 2 clusters contained 7 (R7 to R10) and 10 (R25 to R31) milk strains from Guangdong and Guangxi with the coefficient higher than 0.66. Moreover, 4 milk isolates and 9 pig strains were classified as ribogroup R14 with a relatedness higher than 0.94, which was thought to be an epidemiology correlation.
Figure 1.
Dendrogram of ribotyping patterns of 90 S. aureus strains from pig tonsils and cow’s milk was drawn by BioNumerics software with a 1.0% position tolerance and 0.5% optimization. Percentage similarity was determined with the use of Dice coefficient and clustering was performed by unweighted pair group method with arithmetic mean (UPGMA).
Discussion
Antimicrobial resistance and resistant genes
Most of the S. aureus strains in this study were resistant to clindamycin (91.1%), penicillin (90%), and erythromycin (85.6%). This is similar to findings obtained in other countries (16,17), which reflects the predominant use of these drugs in pig and dairy cattle husbandry worldwide.
Among the penicillin-resistant isolates, 35/39 strains from pigs were MRSA in this study. This was further confirmed by the carriage of the mecA gene; and only 4/39 penicillinase-producing strains were detected. In contrast, all 42 penicillin-resistant isolates from milk were penicillinase-producing strains. Methicillin-resistant Staphylococcus aureus (MRSA) among pig isolates was more prevalent than among strains isolated from milk, similar results were observed by other researchers (16,18,19). This may be associated with the variable use of antibiotics in different animals. For example, ceftiofur, which is for veterinary use only, is widely used in pigs and seldom used in dairy cattle, while penicillin is still the most clinically used antimicrobial agent in China to control bovine mastitis.
Moreover, the results obtained in this study showed that all 77 erythromycin-resistant strains (85.6%) had the MLSB phenotype. Of these, 54.5% had constitutive resistance, 45.5% had inducible resistance, and no strains had the MS phenotype. Similar results were observed in human clinical isolates. Delialioglu et al (20) reported that 56 strains (75.7%) had constitutive resistance, 18 strains (24.3%) had inducible MLSB resistance, and no MS phenotype was found. Aktas et al (21) found that 58.3% of S. aureus isolates were constitutive MLSB and 20.8% were inducible phenotypes.
Results of PCRs showed that the ermC gene was the most prevalent found (89.6%), followed by the ermB gene (16.9%) in MLSB isolates. This finding reveals that target-site modification, encoded by the erm gene, is responsible for the resistance to macrolides and further leads to cross-resistance among macrolides, lincosamides, and streptogramin B, which is the most widespread mechanism of resistance to MLSB. Findings from Turkey showed an incidence of 62.5% for the ermC and 8.3% for the ermB gene among the erythromycin-resistant S. aureus (21). Schmitz et al (22) reported that ermC was responsible for erythromycin resistance in 50.7% of the strains, followed by ermA (38.8%).
No strains were resistant to vancomycin or trimethoprim-sulfamethoxazole. The same results for S. aureus from milk and dairy products were obtained in other studies (19,23,24). Although vancomycin-resistant MRSA (VRSA) from clinical patients was increasingly reported in different countries (25,26), no VRSA has been separated from food production animals to date.
Molecular subtype of S. aureus
A total of 35 different ribotypes was detected in this study, which indicates considerable genetic diversity within the species S. aureus, as previously observed (18,27). The differences in the genotypes of S. aureus found among various animals, based on EcoRI ribotyping, however, show a high degree of host specificity. The high prevalence (83.3%) of ribotypes R11 to R14 in pig strains from different regions indicates that the predominant S. aureus genotypes can exhibit high colonization and survival in tonsil of pigs, which favors a benign relationship between the microbe and the host animal (28,29). Moreover, the results given in Figure 1 showed that most S. aureus strains from milk consisted of the major genotypes, which were relative to the regions and herds isolated. The number of genotypes of strains from Tianjin, which included several herds, was higher than that from Guangxi (only 1 herd). Similar results were observed by Haveri et al (30), who found more genotypes among S. aureus isolates from an open herd than from a closed herd.
Complex genetic diversity of strains from pigs and milk was observed as 13 S. aureus isolates (4 from pigs and 9 from milk) belonged to the same ribotype (R14) with a similarity coefficient of 0.94. This indicated a possible host-unspecificity or cross-dissemination of ribotype R14 between these 2 types of animals considering the presence of mixed pig and cow farms and S. aureus carriers (31,32). Further research is required in order to elucidate transmission routes or risk factors.
Although we tried to determine whether or not the antimicrobial susceptibility patterns are associated with the molecular typing of S. aureus, unfortunately, no association was established in this study.
In conclusion, the data acquired in this study confirm the wide antimicrobial resistance phenotype and genotype diversity of S. aureus from pigs and milk. A high prevalence of MRSA was observed in pig isolates. All erythromycin-resistant strains were MLSB in spite of the different distribution of constitutive and inducible resistance among pig isolates and milk strains. The ribotypes of S. aureus isolates were associated with host specificity. It is interesting to note, however, that there was no evident correlation between the observed strain variability and antimicrobial susceptibility. This information may be useful in planning strategies to prevent and control the emergence and spread of S. aureus within and between herds.
Acknowledgments
This study was supported by the Beijing NOVA Program (2009 B 52) and a grant from the Ministry of Agriculture of the People’s Republic of China.
References
- 1.Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003;2:63–76. [PubMed] [Google Scholar]
- 2.Lewis HC, Mølbak K, Reese C, et al. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg Infect Dis. 2008;14:1383–1389. doi: 10.3201/eid1409.071576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoekstra KA, Paulton RJ. Clinical prevalence and antimicrobial susceptibility of Staphylococcus aureus and Staph. intermedius in dogs. J Appl Microbiol. 2002;93:406–413. doi: 10.1046/j.1365-2672.2002.01708.x. [DOI] [PubMed] [Google Scholar]
- 4.Huber H, Koller S, Giezendanner N, Stephan R, Zweifel C. Prevalence and characteristics of methicillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland. Euro Surveill. 2009;20;15:1–4. [PubMed] [Google Scholar]
- 5.Virdis S, Scarano C, Cossu F, Spanu V, Spanu C, De Santis EP. Antibiotic resistance in Staphylococcus aureus and coagulase negative staphylococci isolated from goats with subclinical mastitis. Vet Med Int. 2010;517060 doi: 10.4061/2010/517060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- 7.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boye K, Bartels MD, Andersen IS, Møller JA, Westh J. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect. 2007;13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 9.Thong KL, Junnie J, Liew FY, Yusof MY, Hanifah YA. Antibiograms and molecular subtypes of methicillin-resistant Staphylococcus aureus in local teaching hospital, Malaysia. J Microbiol Biotechnol. 2009;19:1265–1270. [PubMed] [Google Scholar]
- 10.Ross TL, Fuss EP, Harrington SM, Cai M, Perl TM, Merz WG. Methicillin-resistant Staphylococcus caprae in a neonatal intensive care unit. J Clin Microbiol. 2005;43:363–367. doi: 10.1128/JCM.43.1.363-367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito Y, Iinuma Y, Baba H, et al. Evaluation of automated ribo-typing system for characterization and identification of verocytotoxin-producing Escherichia coli isolated in Japan. Jpn J Infect Dis. 2003;56:200–204. [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, Approved Standard. 3rd ed. Wayne, Pennsylvania: CLSI; 2008. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Testing 18th informational supplement. Wayne, Pennsylvania: CLSI; 2008. Performance Standards for Antimicrobial Susceptibility. [Google Scholar]
- 14.Wielders CL, Fluit AC, Brisse S, Verhoef J, Schmitz FJ. mecA gene is widely disseminated in Staphylococcus aureus population. J Clin Microbiol. 2002;40:3970–3975. doi: 10.1128/JCM.40.11.3970-3975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–238. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Neeling AJ, van den Broek MJ, Spalburg EC, et al. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2007;122:366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Guven K, Mutlu MB, Gulbandilar A, Cakir P. Occurrence and characterization of Staphylococcus aureus isolated from meat and dairy products consumed in Turkey. J Food Safety. 2010;30:196–212. [Google Scholar]
- 18.Cui S, Li J, Hu C, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother. 2009;64:680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
- 19.Morandi S, Brasca M, Andrighetto C, Lombardi A, Lodi R. Phenotypic and genotypic characterization of Staphylococcus aureus strains from Italian dairy products. Int J Microbiol. 2009;2009:501362. doi: 10.1155/2009/501362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delialioglu N, Aslan G, Ozturk C, Baki V, Sen S, Emekdas G. Inducible clindamycin resistance in staphylococci isolated from clinical samples. Jpn J Infect Dis. 2005;58:104–106. [PubMed] [Google Scholar]
- 21.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococci isolated in Istanbul, Turkey. J Microbiol. 2007;45:286–290. [PubMed] [Google Scholar]
- 22.Schmitz FJ, Petridou J, Fluit AC, Hadding U, Peters G, von Eiff C. Distribution of macrolide-resistance genes in Staphylococcus aureus blood-culture isolates from fifteen German university hospitals. M.A.R.S. Study Group. Multicentre Study on Antibiotic Resistance in Staphylococci. Eur J Clin Microbiol Infect Dis. 2000;19:385–387. doi: 10.1007/s100960050500. [DOI] [PubMed] [Google Scholar]
- 23.Gianneechini RE, Concha C, Franklin A. Antimicrobial susceptibility of udder pathogens isolated from dairy herds in the west littoral region of Uruguay. Acta Vet Scand. 2002;43:31–41. doi: 10.1186/1751-0147-43-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunz F, Corti S, Giezendanner N, Stephan R, Wittenbrink M, Zweifel C. Antimicrobial resistance of Staphylococcus aureus and coagulase negative staphylococci isolated from mastitis milk samples from sheep and goats. Schweiz Arch Tierheilkd. 2011;153:63–69. doi: 10.1024/0036-7281/a000152. [DOI] [PubMed] [Google Scholar]
- 25.Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. Vancomycin-resistant Staphylococcus aureus in the United States, 2002–2006. Clin Infect Dis. 2008;46:668–674. doi: 10.1086/527392. [DOI] [PubMed] [Google Scholar]
- 26.Loomba PS, Taneja J, Mishra B. Methicillin and vancomycin resistant S. aureus in hospitalized patients. J Glob Infect Dis. 2010;2:275–283. doi: 10.4103/0974-777X.68535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aarestrup FM, Wegener HC, Jensen NE, et al. A study of phage- and ribotype patterns of Staphylococcus aureus isolated from bovine mastitis in the Nordic countries. Acta Vet Scand. 1997;38:243–252. doi: 10.1186/BF03548487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben Zakour NL, Sturdevant DE, Even S, et al. Genome-wide analysis of ruminant Staphylococcus aureus reveals diversification of the core genome. J Bacteriol. 2008;190:6302–6317. doi: 10.1128/JB.01984-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haveri M, Hovinen M, Roslöf A, Pyörälä S. Molecular types and genetic profiles of Staphylococcus aureus strains isolated from bovine intramammary infections and extramammary sites. J Clin Microbiol. 2008;46:3728–3735. doi: 10.1128/JCM.00769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Giessen AW, van Santen-Verheuvel MG, Hengeveld PD, Bosch T, Broens EM, Reusken CB. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev Vet Med. 2009;91:270–273. doi: 10.1016/j.prevetmed.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Moodley A, Nightingale EC, Stegger M, Nielsen SS, Skov RL, Guardabassi L. High risk for nasal carriage of methicillin-resistant Stapylococcus aureus among Danish veterinary practitioners. Scand J Work Environ Health. 2008;34:151–157. doi: 10.5271/sjweh.1219. [DOI] [PubMed] [Google Scholar]