Abstract
Cancer is a collection of very complex diseases that share many traits while differing in many ways as well. This makes a universal cure difficult to attain, and it highlights the importance of understanding each type of cancer at a molecular level. Although many strides have been made in identifying the genetic causes for some cancers, we now understand that simple changes in the primary DNA sequence cannot explain the many steps that are necessary to turn a normal cell into a rouge cancer cell. In recent years, some research has shifted to focusing on detailing epigenetic contributions to the development and progression of cancer. These changes occur apart from primary genomic sequences and include DNA methylation, histone modifications, and miRNA expression. Since these epigenetic modifications are reversible, drugs targeting epigenetic changes are becoming more common in clinical settings. Daily discoveries elucidating these complex epigenetic processes are leading to advances in the field of cancer research. These advances, however, come at a rapid and often overwhelming pace. This review specifically summarizes the main epigenetic mechanisms currently documented in solid tumors common in the United States and Europe.
Common Themes in Cancer Epigenetics
DNA methylation
The most well-defined epigenetic modification existing in cancer to date is DNA methylation (Rodriguez-Paredes and Esteller, 2011). The addition of a methyl group to cytosine residues directly preceding guanidine (CpG) residues is a common postreplicative mark. Most often, these modifications occur in areas of high CpG content, termed CpG islands. CpG islands are found in about 60% of human gene promoters (Bird, 2002). Hypermethylation tends to mark a silent gene, while hypomethylation is usually associated with gene activity. In comparison to normal tissue, cancers are traditionally globally hypomethylated, while many tumor suppressor genes within the tumor are often hypermethylated. The genes that are known to be hypo- and hypermethylated in common cancers are summarized in Tables 1 and 2.
Table 1.
Genes Known to Be Hypomethylated in Common Cancers
| Cancer type | Representative hypomethylated genes |
|---|---|
| Breast | BCSG1, CAV1, CHD1, CDH3, NAT1, UPAN (Jovanovic et al., 2010) |
| Prostate | PLAU (Pakneshan et al., 2003); TFF1, TFF3 (Vestergaard et al., 2010) |
| Melanoma | 14-3-3σ, GAGE 1–6, HMW-MAA, MAGE-A1, MAGE-A2, MAGE-A3, MAGE-A4, MAGE-A6, NY-ESO-1, PI5, PRAME, SSX 1–5, (Howell et al., 2009) |
| Pancreatic | CLDN4, LCN2, MSLN, PSCA, S100P, S100A, SERPINB5, SFN, TFF2 (Hong et al., 2011) |
| Lung | 14-3-3σ (Radhakrishnan et al., 2011); C8orf72 (Rauch et al., 2008) |
| Colorectal | LINE-1 (Sunami et al., 2011); MAEL, SFT2D3 (Kim et al., 2011) |
Table 2.
Genes Known to Be Hypermethylated in Common Cancers
| Cancer type | Representative hypermethylated genes |
|---|---|
| Breast | 14-3-3σ, AK5, APC, BCL2, BRCA1, CCND2, CDH13, CDKN1C, CDKN2A, CST6, DAPK, DCC, ER-α, FOXA2, GPC3, HIC1, HOXA5, HOXD11, LAMA3, LAMB3, LAMC2, LDLRAP1, MGMT, MLH1, PGR, RAD9, RAR-β, RASSF1A, ROBO1, RUNX3, SFRP1, SYK, TMS1, TWIST, WIF1, WRN, WT1 (Jovanovic et al., 2010) |
| Prostate | APC, AR, ASC, BCL2, CAV1, CCND2, CD44, CDH1, CDH13, CDKN2A, DAPK, DKK3, EDNRB, ER-α, ER-β, GPX3, GSTM1, GSTP1, HIC, LAMA 3, LAM C2, MDR1, MGMT, PTGS2, RAR-β2, RARRES1, RASSF1A, RPRM, RUNX3, SFN, SFRP1, TIMP3 (Jeronimo et al., 2011) |
| Melanoma | 3-OST-2, APC, ASC, BST2, Caspase 8, CCND2, CD10, CDH1, CDH8, CDKN1B, CDKN1C, CDKN2A, CIITA-PIV, COL1A2, CXCR4, CYP1B1, DAL1, DAPK1, DDIT4L, DNAJC15, DPPIV, E cadherin, ER-α, GATA4, GDF15, HMW-MAA, HOXB13, HSP11, KR18, LOX, LRRC2, LXN, Maspin, MDR1, MINT 17, MINT 31, Megalin, MFAP2, MGMT, MIB2, MTAP, NPM2, p101, P73, PCSK1, PRDX2, PTEN, PTGS2, QPCT, RAR-β2, RASSF1A, Ril, SOCS-1. SOCS-2, SOCS-3, SYK, TERC, TFPI2, THBS1, THBS4, TIMP3, TM, TNFRSF10A, TNRSF10D, TNRSF10D, TPM1, WIF1, WFDC1 (Howell et al., 2009; Schinke et al., 2010) |
| Pancreatic | BNIP3, CCND2, CDKN2A, CDKN1C/p57, CNTNAP2, MDF-1, miR-9-1, NPTX2, PENK, RPRM, SPARC/ON, TFPI2, UCHL1, ZNF415 (Hong et al., 2011) |
| Lung | BARHL2, CBLN4, CDKN2A, CDH1, DAPK, DPP6, FLJ30851, GRIK2, HAND2, HOXA9, HOXD9, KIAA1024, LHX2, MGMT, NR2E1, OSR1, PCDHGA12, RAR-β, RASSF1A, RUNX3, SIM2, SIX6, ST8SIA3, TIMP3 (Palmisano et al., 2000; Tsou et al., 2002; Rauch et al., 2011) |
| Colorectal | ALX4, APC, CDKN2A, CDH4, CHD1, CHD13, CHFR, CRABO1, CXCL12, DAPK, DKK1, EphA1, ER-α, GALR2, GATA4, GATA5, HLTF, ID4, IEF8, MGMT, MLH1, NGX6, p14, p15, PPARG, RASSF1A, RUNX3, SEPT9 SFRP1, SFRP2, SLC5A8, Sox17, TFPI2, THBS1/TSP1, TIMP3, VIM (Jia and Guo, 2011; Lao and Grady, 2011) |
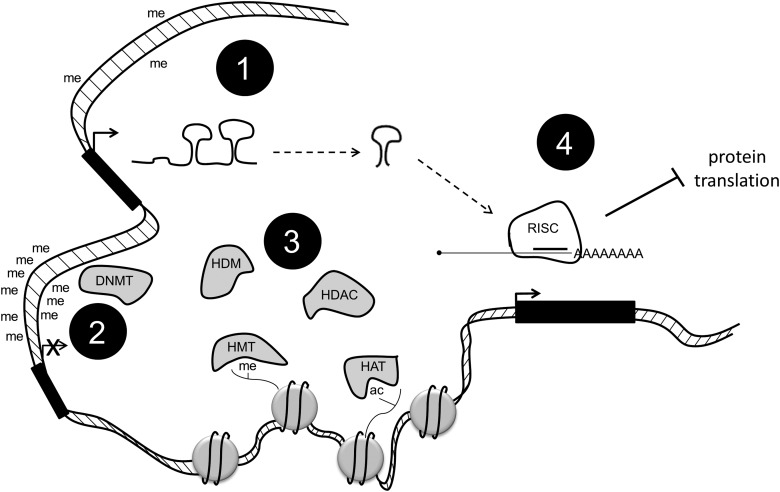
DNA methylation in humans is carried out by three enzymes: DNA methyltransferase 1 (DNMT1), which maintains parental methylation patterns, and DNMT3A and DNMT3B, which regulate de novo methylation (Bestor and Ingram, 1983; Okano et al., 1999; Chen et al., 2007; Chedin, 2011). As detailed next, these enzymes are differentially expressed in certain cancers, but their expression does not always correlate with hyper- or hypomethylation, as one would expect. This may be partially explained by the fact that DNMT expression is sometimes regulated by miRNAs (described next, Fig. 1).
FIG. 1.
DNA methylation, histone modification, and micro RNA (miRNA) expression work together to control the complex environment of epigenetics in cancer. Promoter hypomethylation is linked to the expression of genes, including miRNA, DNA methyltransferases (DNMTs), and histone modifiers (1). DNMTs can, in turn, hypermethylate promoters and turn off these same genes (2). Histone-modifying enzymes can affect gene expression by adding or removing certain marks (3). miRNAs are processed and targeted to mRNA, leading to a decrease in the protein expression of certain enzymes (4).
Active loss of DNA methylation has been observed during development, suggesting the existence of DNA demethylases. Multiple mechanisms for DNA demethylation have been proposed and recently discussed (Wu and Zhang, 2010). In particular, one potential mechanism is through Ten-eleven translocation 1–3 (TET1-3) proteins, which are a group of hydroxylases (Tahiliani et al., 2009; Ito et al., 2011). These enzymes can convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Ito et al., 2010; He et al., 2011; Pfaffeneder et al., 2011). These modified bases may not only serve as intermediates in the DNA demethylation process, but they may also increase the diversity of the epigenetic states of genomic DNA. The TET1 protein was identified as a fusion partner of mixed lineage leukemia (MLL) in patients with acute myeloid leukemia (AML), while mutations of TET2 proteins have been found in patients with various myeloid malignancies (Wu and Zhang, 2011).
Histone modification
Another way that cancers are epigenetically regulated is through alterations in histone modification. In each cell, nucleus DNA is wrapped around an octamer of histone proteins. These proteins have flexible N-terminal tails that are extensively modified (Fig. 1). Certain modifications serve to tighten or loosen DNA from the octamer, thus allowing or disallowing the binding of transcriptional machinery. This enables a tight control of gene expression. Some histone modifications are associated with active genes (i.e., trimethylation on histone H3, lysine 4 [H3K4me3]), and some are associated with repressed genes (i.e., H3K9me3) (reviewed in Fullgrabe et al., 2011). The enzymes responsible for adding and removing these marks are differentially expressed in many cancers (Chi et al., 2010; Blair et al., 2011).
The most common histone modifications are acetylation, methylation, and phosphorylation. Acetyl groups are added to lysine residues by enzymes called histone acetyltransferases (HATs) and are removed by histone deacetylases (HDACs). Methyl groups are added to lysine and arginine residues by histone methyltransferases (HMTs) and are removed by histone demethylases (HDMs). The roles of these histone-modifying enzymes in cancer are still not completely understood, but some have been linked to specific diseases, as discussed next.
Micro RNAs
Recent discoveries have highlighted the importance of microRNAs (miRs) in the regulation of cancer (Braconi et al., 2011; Fanini et al., 2011; Ferracin et al., 2011; Landau and Slack, 2011). miRs are small noncoding RNAs that are highly processed using specific machinery (Fig. 1). Once processed down to mature miRNAs, they are targeted to 3′UTRs of mRNA, where they inhibit the translation of the targeted mRNAs and/or cause those mRNAs to be targeted for degradation. This leads to a decrease in the amount of protein made. Either miRs can be oncogenic or they can have tumor suppressive functions. More recently, a push for diagnostic and prognostic miRs has been established due to the identification of many differentially expressed miRs in cancers as compared with normal tissue (Kasinski and Slack, 2011).
Epigenetic crosstalk
Interestingly, all three types of epigenetic regulation just mentioned are somewhat intertwined (Fig. 1). DNA demethylases are post-transcriptionally regulated by miR-29 and miR-148 (Fabbri et al., 2007; Duursma et al., 2008). DNMTs have also been shown to interact with HDACs (Yang et al., 2001; Bai et al., 2005). In fact, DNMT and HDAC inhibitor combinations are at the forefront of cancer therapies (Zhu and Otterson, 2003; Unoki, 2011). Additionally, miR expression is regulated by DNMT-mediated promoter methylation (So et al., 2011).
Epigenetic markers show promise as biomarkers for many different reasons. Isolating genomic DNA for methylation profiling is easier, and the DNA is more stable than extracting mRNA for gene expression analysis. Strides have been made in profiling clinical samples for histone marks using immunohistochemistry and mass spectrometry. Studying the enzymes that alter DNA methylation and histone modification patterns may be the key to curbing cancer progression. Next, we summarize the advances in the epigenetic mechanisms of DNA methylation, histone modifications, and miRNA expression in six different cancers: breast, prostate, melanoma, pancreatic, lung, and colorectal.
Breast Cancer
Women in the United States have a one in eight chance of being diagnosed with some type of breast cancer in their lifetime. The disease has very personal connotations to many people and, as a result, greater efforts have been dedicated to investigating breast cancer than many other cancers. This has led to a plethora of literature in recent years on the mechanisms of breast cancer. Many of the concepts and mechanisms discovered in breast cancer are being applied to the research of other cancers. Much progress has been made in the education, early detection, and treatment of breast cancer, but a few drugs have been developed to treat specific forms of the disease. One such drug is Herceptin/trastuzumab, which targets human epidermal growth factor receptor 2 (HER2/ERBB2), a receptor tyrosine kinase (Vogel et al., 2002). Many patients have responded to this medication, and it has undoubtedly saved lives. However, there are many different forms of breast cancer, and the development of drugs that are specific to mechanisms within these subtypes will allow for a type of personalized medicine with higher specificity and less severe side effects. Although epigenetics is a relatively young field, much has been published about breast cancer epigenetics.
There are currently six major sub-types of breast cancer and within these sub-types, individual cases differ (Chin et al., 2006; Prat et al., 2010; Stecklein et al., 2012). Breast cancers are often classified by their estrogen receptor (ER/ER-α/ESR1), progesterone receptor (PR), and HER2 status. Patients negative for all three are classified as triple negative, which is a very aggressive and devastating form that is difficult to treat. ER positive tumors often respond to hormone therapy, and HER2+ tumors have shown some success with Herceptin. It is crucial to attain a better understanding of the different types of breast cancer in order to provide patients with the best care possible.
Similar to many tumors, breast cancers have been shown to be globally hypomethylated (Soares et al., 1999). On a more detailed study of CpG islands, however, it becomes clear that many tumor suppressor genes are hypermethylated in cancer compared with their noncancerous counterparts (Hinshelwood and Clark, 2008). The suppression of tumor suppressor genes is a common mechanism that is used in the progression of cancer. DNA hypermethylation is generally associated with gene instability due to the downregulation of tumor suppressors in DNA repair pathways in breast cancer (Widschwendter and Jones, 2002). Some gene-specific methylation may be accounted for by the fact that DNMTs are overexpressed in breast cancer, specifically DNMT3B, but this remains a loose correlation (Girault et al., 2003).
Some gene methylation changes have been linked to specific types of breast cancer. For example, ER promoter methylation is linked to a loss of ER expression (Weigel and deConinck, 1993; Ferguson et al., 1995). Breast cancers overexpressing ER and PR are often treated using hormone therapy involving drugs that mimic estrogen or block these receptors. Methylation patterns have also been used more recently to determine a patient response to a certain treatment. For example, patients using Tamoxifen are profiled for the methylation levels of circulating Ras-association domain family 1 isoform A (RASSF1A). If the levels of RASSF1A methylation are elevated, then it indicates that the patient is not responding to the treatment (Fiegl et al., 2005). This is a valuable tool in determining how to proceed with individual patients.
Since DNA hypermethylation is partially driven by the overexpression of DNMTs, they have also become targets for cancer drugs. Traditionally, DNMT inhibitors are nucleoside analogs that are incorporated into DNA and serve to permanently link the DNMTs to the DNA. These DNMT/DNA hybrid molecules are then targeted to the proteasome for degradation. Some of these inhibitors have been approved for clinical use, although none specifically for breast cancer (Foulks et al., 2011).
The role of histone-modifying enzymes in breast cancer has become more apparent in recent years (reviewed in Huang et al., 2011). The majority of breast cancer treatment advances involving histone-modifying enzymes have come in the form of HDAC inhibitors. In general, histone acetylation is linked to the activation of genes. It is thought that acetylation causes loosening of the DNA around the histones, which allows transcriptional machinery to access the DNA. The removal of acetylation, conversely, is thought to cause the DNA to tighten around the histones and to inhibit the access of transcription machinery and silencing genes. HDACs have been shown to remove these activating marks from tumor suppressor genes, thereby reducing their expression and leading to the development of cancer. HDAC inhibitors have shown some clinical promise in breast cancer patients. For example, Vorinostat has entered phase II clinical trials for metastatic breast cancer in combination with other anticancer drugs (Ramaswamy et al., 2011).
More recently, emphasis has also been placed on determining the role of HMTs and HDMs in cancer. An alteration in the expression and regulation of these genes has been noted in breast cancers. Enhancer of zeste homolog 2 (EZH2) is an H3K27 methyltransferase that has been shown to be up-regulated in breast cancers (Kleer et al., 2003). Lysine-specific demethylase 1 (LSD1) is an H3K4 demethylase that is highly expressed in ER negative tumors and has been suggested as a potential predictive marker for aggressive breast cancers (Lim et al., 2010). LSD1 also interacts with HDACs, which suggests that a combination of demethylase inhibitors and HDAC inhibitors could be a potent cancer treatment (Huang et al., 2011). Interestingly, LSD1 has been shown to regulate the methylation of both p53 and DNMT1, which could suggest a key role in other cancer mechanisms (Huang et al., 2007; Wang et al., 2009).
Since the discovery of LSD1 in 2004, more HDMs have been identified, and many of them appear to play instrumental roles in breast cancer (Blair et al., 2011). For example, lysine demethylase 5B (KDM5B, also known as PLU1 and JARID1B), an H3K4me3 demethylase that is overexpressed in breast cancer, has been shown to repress tumor suppressor genes such as BRCA1 and CAV1 in breast cancer cell lines (Yamane et al., 2007). The Taylor–Papadimitriou lab showed a direct interaction between HDAC4 and KDM5B, which they suggest to be a potential mechanism for KDM5B in cancer (Barrett et al., 2007). KDM5B has also been implicated in mammary development and is becoming a major player in breast cancer research (Catchpole et al., 2011).
There are currently estimated to be many differentially expressed miRNAs in breast cancer, many of which are noted in Table 3 (Veeck and Esteller, 2010). Let-7 is down-regulated in breast cancers, and its overexpression in these cancers leads to a decrease in metastatic potential and proliferation rate, suggesting that Let-7 is a key player in the disease (Yu et al., 2007). miR-125 targets HER2 and is, therefore, thought to be a tumor suppressor in HER2+ breast cancers (Scott et al., 2007). Many miRNAs have been suggested to be potential diagnostic or prognostic markers for breast cancer; for example, circulating levels of miR-195 have been noted to be up-regulated in breast cancer patients (Heneghan et al., 2010; Ferracin et al., 2011). Recent studies by Ma et al. (2010) outline the potential for miR-targeted therapies, so called “antago-miRs,” in treating the disease. They show that antagomiR-10b aids the prevention of lung metastases in a mouse breast cancer model (Ma et al., 2010). Additionally, many miRs have been directly associated with the different stages of metastasis and are, therefore, of interest in all types of cancers, specifically those that are prone to metastasis (reviewed in (Shi et al., 2010).
Table 3.
Micro RNAs Involved in Common Cancers
| Cancer type | Representative miRNAs |
|---|---|
| Breast | Let7, miR-9-1, miR-10b, miR-15/16, miR-17-5p, miR-18a, miR-21, miR-27a, miR-27b, miR-29a-c, miR-125a/b, miR-126, miR-130a, miR-143, miR-145, miR-148, miR-155, miR-200c, miR-205, miR-206, miR-335 (Veeck and Esteller, 2010) |
| Prostate | Let-7a miR-1, miR-7, miR-15a-6, miR-20a, miR-21, miR-24, miR-32, miR-34a, miR-34c, miR-101, miR-106b, miR-107, miR-125b, miR-143, miR-145, miR-146a, miR-148a, miR-205, miR-221, miR-222, miR-331-3P, miR-449a, miR-521, miR-1296, (Catto et al., 2011) |
| Melanoma | Let-7a, let-7b, miR-15b, miR-29c, miR-30b/c, miR-31, miR-34a, miR-34b, miR-124/506, miR-137, miR-148/152, miR-155, miR-182, miR-184, miR-185, miR-193b, miR-196a, miR-204, miR-205, miR-211, miR-214, miR-221, miR-222, miR-340, miR-375, miR-506-514 (Bonazzi et al., 2011) |
| Pancreatic | miR-10a, miR-10b, miR-21, miR-23a, miR-23b, miR-99, miR-100-1/2, miR-103-2, miR-107, miR-125a, miR-125b-1, miR-143, miR-146, miR-155, miR-181a, miR-181b, miR-181c, miR-181d, miR-199a, miR-205, miR-210, miR-213, miR-220, miR-221, miR-222, miR-223, miR-301, miR-376a (Bloomston et al., 2007; Lee et al., 2007) |
| Lung | Let-7, miR-17-92, miR-21, miR-31, miR-34b, miR-34c, miR-42-5p, miR-125b miR-126, miR-127, miR-128b, miR-136, miR-142-3p, miR-143, miR-145, miR-155, miR-150, miR-205, miR-337, miR-376a, miR-379, miR-410, miR-431 (Liu et al., 2011) |
| Colorectal | Let-7 family, miR-17-92, miR-21, miR-31, miR-34a, miR-107, miR-126, miR-141, miR-143, miR-192, miR-194-2, miR-195, miR-196a, miR-200b, miR-200c, miR-215, miR-373, miR-451, miR-491, miR-661 (de Krijger et al., 2011) |
miRNA, micro RNA.
Prostate Cancer
Prostate cancer is one of the most commonly occurring cancers in men in the United States and Europe (Jemal et al., 2010; Siegel et al., 2011). In the absence of metastasis, the disease is often slow progressing and can sometimes be treated by a “watch and wait” procedure. Since this type of prostate cancer is usually androgen regulated, many treatments similar to breast cancer, such as hormone deprivation, are used to control the disease (Shepard and Raghavan, 2010; Corona et al., 2011). However, there is still no effective treatment method against metastatic prostate cancer, and better ways to diagnose and treat the disease are crucial to curing it. Although most cancers have been shown to be globally hypomethylated, prostate cancer is generally an exception to this rule (Jeronimo et al., 2011). More than 50 genes are hypermethylated in the disease, some of which are highlighted in Table 2. Many of these genes are involved in important cellular pathways and present relatively early in the disease stage. Additionally, urokinase plasminogen activator (PLAU) is highly hypomethylated in castration-resistant prostate cancers (CRPCs) (Pakneshan et al., 2003). A thorough understanding of which genes are differentially methylated in which types of prostate cancer could lead to early diagnosis and improve treatment outcomes (Van Neste et al., 2011). Studies focusing on detailing the differential methylation patterns of these diseases are ongoing, and information about the mechanisms behind these aberrant methylation patterns is emerging (reviewed in Park, 2010). Some of these studies outline the effect of certain factors, including age, diet, and environment, on promoter methylation (Venkateswaran and Klotz, 2010). While changing a person's ethnicity or age is not an option, information linking diet and prostate cancer epigenetics has great potential for preventative purposes.
Histone-modifying enzymes have also been implicated as being important in prostate cancer. Similar to breast cancer, the most commonly studied histone modifying enzymes in prostate cancer are EZH2, HDACs, and LSD1. EZH2 overexpression leads to promoter hypermethylation in prostate cancer, which results in the repression of certain developmental and tumor suppressor genes (Hoffmann et al., 2007). HDACs are overexpressed in prostate cancer, particularly so in CRPC (Halkidou et al., 2004). In addition, androgen receptor (AR) in CRPC has recently been shown to mediate its own expression by recruiting LSD1 to the AR gene and suppressing expression through H3K4 demethylation (Cai et al., 2011).
The profiling of prostate cancer cells has led to the discovery of 50 differentially regulated miRs (reviewed in Catto et al., 2011). Only a few of these targets have been extensively studied, such as miR-221/222. The overexpression of miR-221 or miR-222 in LNCaP prostate cancer cells leads to a reduction in prostate-specific antigen (PSA), which is currently the most common biomarker for prostate cancer (Sun et al., 2009). Although testing PSA levels in serum has been useful in diagnosis, the test is not specific and leads to many false positives and false negatives (Croswell et al., 2011). This has led to the push for a better biomarker for prostate cancer screening. As in other cancers, miRNAs are exciting potential targets for such a test. Specifically, miR-200a has been shown to be differentially expressed in men who relapse from radical prostatectomy (Barron et al., 2011). Transient transfection of miR-200a significantly reduces the proliferation of prostate cancer cells, suggesting that it might be a potential target for therapies (Barron et al., 2011).
Apart from serving as prognostic and diagnostic markers, epigenetic marks in prostate cancer also show promise as therapeutic targets. Clinical trials using DNMT inhibitors in prostate cancer have shown minimal benefit, but these trials were performed in patients with advanced stage, hormone independent disease (Thibault et al., 1998). More trials are needed using these inhibitors alone and in combination with other epigenetic inhibitors. HDAC inhibitors have been used in clinical trials for patients with advanced-stage prostate cancer (Munster et al., 2009). Their success has been highest in trials that combine them with conventional chemotherapeutic drugs. HMT and demethylase inhibitors have shown promise in in vitro prostate cancer systems and are currently under development for use in a clinical setting (Tan et al., 2007; Miranda et al., 2009).
Melanoma
Many skin cancers are benign and, if caught early enough, can be removed in a simple procedure, often performed in the doctor's office. In recent years, skin cancer prevention and screening have become more popular, and the public is becoming more aware of the dangers of UV exposure and other causes of the disease. The incidence of melanoma, however, has risen in recent years (Linos et al., 2009). Melanoma is considered the most devastating form of skin cancer due to the high mortality associated with the disease. Early detection is key, because once this type of cancer metastasizes, then a patient's life expectancy is around 2 years (Koon and Atkins, 2007). Cases of melanoma are on the rise around the world, and it has become more important to understand the molecular mechanisms of this disease in order to improve early detection methods and treatments.
Interestingly, hypomethylation has been discovered not only globally but also in certain genes in melanoma (Table 1). These genes tend to be oncogenic in nature; therefore, hypomethylation of their promoters causes overexpression and leads to cancer progression. A recent study outlines the importance of mutant BRAFV600E in hypomethylation and, thus, the activation of the genes associated with cell proliferation and invasion in melanoma cell lines (Hou et al., 2012).
More than 50 genes have been reported as being hypermethylated in melanoma (Table 2). The function of some of these genes remains unknown, but many are associated with processes involved in cancer progression such as apoptosis and cell-cycle regulation (Rothhammer and Bosserhoff, 2007). Promising results point to the use of DNA methylation levels as melanoma biomarkers. For example, circulating levels of ER methylation can be associated with poor prognosis (Mori et al., 2006). Additionally, a methylation signature could be established for diagnosing melanoma. Reports show that suppressor of cytokine signaling 1 and 2 (SOCS1 and SOCS2), retinoic acid receptor β-2 (RARβ2), and tumor necrosis factor receptor superfamily, members 10c and 10d (TNFRSF10C and TNFRSF10D) are frequently methylated in clinical melanoma samples, although further validation of these methylation patterns is needed (Howell et al., 2009). DNMTs have also been targeted in melanoma patients. Studies have shown that DNMT inhibitors can slow the progression of melanoma in nude mouse models (Morita et al., 2006).
Histone-modifying enzymes have also been studied as being possible targets for melanoma treatment. Melanoma tumor suppressor genes inhibitor of growth 3 and 4 (ING3 and ING4) have been linked to lysine acetyltransferase 7 and 8 (KAT7 and KAT8), which are components of a larger histone acetyltransferase complex (Li et al., 2008). In addition, the HAT components p300 and CPB have been linked to senescence in melanoma cells (Bandyopadhyay et al., 2002). These factors act by altering the expression of microphthalmia-associated transcription factor (MITF), a master regulator of melanocyte development and melanomagenesis (Sato et al., 1997; Price et al., 1998; Bandyopadhyay et al., 2002). Although the mechanisms of HDAC inhibitors are not well understood, it is thought that they can allow re-expression of tumor suppressor genes in melanoma and increase the acetylation levels of some nonhistone targets (Howell et al., 2009). One particular HDAC inhibitor, MS-275, has shown promise in clinical trials with melanoma patients (Gore et al., 2008).
Some more recent reports have outlined the importance of KDM5B in melanoma. A population of slow-cycling and stem-like melanoma cell lines expressed high levels of KDM5B, indicating that it is involved in making melanoma cells more “stem-like” (Roesch et al., 2010). The exact mechanism by which KDM5B contributes to the disease is still under investigation, and drugs targeting the enzyme are currently under development.
Several miRNAs have been implicated in melanoma progression. miR-137, for example, is expressed in melanoma cell lines and targets MITF (Levy et al., 2006). MITF acts by regulating the differentiation and cell-cycle genes, and the binding of miR-137 prevents these mRNAs from being translated (Bonazzi et al., 2011). Let-7 is down-regulated in melanomas, and studies show that loss of let-7a is linked to the development and progression of melanoma (Muller and Bosserhoff, 2008; Schultz et al., 2008). miR-221 and miR-222 are regulated by the transcription factor promyelocytic leukemia zinc finger (PLZF) in melanoma. By binding the regulatory regions of miR-221/222, PLZF inhibits their ability to target mRNA, such as c-Kit Receptor and p27/CDK5N1B. By inhibiting the degradation of these tumor suppressors, PLZF suppresses melanoma progression (Felicetti et al., 2008).
Pancreatic Cancer
Due to the lack of symptoms and difficulty in attaining early diagnoses, pancreatic cancer is one of the deadliest forms of cancer. At the time of diagnosis, most patients have an unresectable form of the disease and have a less than 2% 5-year rate of survival (Jemal et al., 2010; Siegel et al., 2011). Although progress has been made in recent years, better ways to detect and treat pancreatic cancer are needed. The only current prevention measures suggested are those recommended for all cancers: smoking cessation, exercise, and a diet that is high in fruits and vegetables. Some cases have been linked to hereditary causes, but the specific genes that are involved have yet to be identified (Matsubayashi et al., 2011).
Due to the correlation between late diagnosis and increased mortality, finding a biomarker associated with the disease is crucial. Therefore, determining the epigenetic mechanisms of the disease is useful. Investigations into DNA methylation have shown that 80% of pancreatic cancers overexpress DNMT1 (Peng et al., 2005). Some tumor suppressor genes such as CDKN2A/p16, MutL homolog 1 (MLH1), and cadherin 1 (CDH1) are hypermethylated in pancreatic cancer (Schutte et al., 1997; Ueki et al., 2000). Proenkephalin (PENK) is aberrantly methylated in 90% of pancreatic cancers and reprimo (RPRM), in 80% (Fukushima et al., 2002; Sato et al., 2006). Tumor suppressor CDKN2A is often used as an example of epigenetics in pancreatic cancer (Lomberk, 2011). It is hypermethylated and, therefore, silenced in pancreatic cancer (Singh and Maitra, 2007). Many genes silenced in pancreatic cancer are also hypermethylated in precursor lesions, indicating that the screening for DNA methylation patterns could be a reliable basis for early diagnosis (Sato et al., 2005). As seen in Table 1, some genes have been shown to be hypomethylated in pancreatic cancer, but the mechanism behind this phenomenon is not well understood (Sato et al., 2003).
Recent studies have outlined the importance of chromatin-associated proteins in pancreatic cancer. The polycomb protein BMI1 is recruited to CDKN2A, where it stalls EZH2, allowing for the hypermethylation of H3K27 at the CDKN2A promoter, leading to the repression of CDKN2A (Bracken et al., 2007). Additionally, Jiao et al. (2011) used exome sequencing to discover common mutations in alpha thalassemia/mental retardation syndrome X-linked (ATRX) and death-domain-associated protein (DAXX) in pancreatic neuroendocrine cancers (panNETs). Both ATRX and DAXX are involved in chromatin remodeling (Jiao et al., 2011). They work together to deposit histone variant H3.3 at regions of silent chromatin (Goldberg et al., 2010). They also discovered mutations in menin (MEN1), which regulates histone methylation by recruiting the HMT MLL (Jiao et al., 2011).
More than 50 miRNAs have been associated with pancreatic cancer (reviewed in Wang and Sen, 2011). Some of these miRs, such as miR-155, miR-221, and miR-222, are also overexpressed in other cancers. However, some unique miRNAs, such as mIR-376a and miR-301, have been discovered as being expressed in pancreatic cancer (Lee et al., 2007). Although follow-up literature on pancreatic cancer is not currently available, other reports show that miR-376a is associated with erythroid differentiation, and miR-301 can promote invasion in breast cancer (Shi et al., 2011; Wang et al., 2011). A recent report by Wang et al. (2011) uses 11 miRNAs deregulated in pancreatic ductal adenocarcinoma (PDAC) to determine the most common cellular pathways that are affected by these changes (Wang and Sen, 2011). They showed that major components of the phosphatase and tensin homolog (PTEN) signaling pathway are affected by the down-regulation of these miRs. PTEN is a tumor suppressor whose function is lost in many cancers, including pancreatic cancer (Krausch et al., 2011). Some miRNA expression profiles, specifically the down-regulation of miR-217 and the overexpression of miR-196a, have even been suggested for their use to diagnose pancreatitis versus PDAC (Bloomston et al., 2007; Lee et al., 2007; Szafranska et al., 2007).
Lung Cancer
The leading cause of cancer deaths worldwide is lung cancer. The outcome for patients with lung cancer is not a good one. The 5-year survival rate is around 13%. Unfortunately, a stigma is attached to having the disease. Many lung cancer patients have smoked for much of their lives, and many continue to even after treatment; however, many lung cancer patents are never-smokers. This stigma has led to a lack of sympathy for these patients as compared with patients with other types of cancer and has translated to a difference in the level of funding for lung cancer research compared with other common cancers.
There are two types of lung cancer: small cell (SCLC) and nonsmall cell (NSCLC). The more common of the two is NSCLC, yet the more aggressive is SCLC. Since lung cancer is cell-type specific and it is often difficult to isolate enough cells for analysis, much epigenetic research is done using cell lines or the entire lung. For this reason, progress in lung cancer epigenetics is not as extensive as in some other cancers.
Environmental factors have a more pronounced contribution to lung cancers than to most other cancers due to the direct interaction of the lungs with the outside environment through respiration. The effect of environmental factors on epigenetics is well established (Bell and Beck, 2010; Kabesch et al., 2010; Galea et al., 2011; Herceg and Paliwal, 2011; Herceg and Vaissiere, 2011; Yang and Schwartz, 2011). Some high-throughput studies of DNA methylation patterns in lung cancer have been recently reported (Ocak et al., 2009; Heller et al., 2010; Toyooka and Gazdar, 2011). A number of promoters were found to be aberrantly methylated, and validation studies are underway to verify these findings in different types of lung cancers. These validation studies aim at detailing the involvement of these aberrant methylation patterns in different types and stages of lung cancer in hopes of finding a diagnostic or prognostic marker or a therapeutic target. As in pancreatic cancer, methylation tends to occur early in lung cancer development. One interesting finding was that CDKN2A and O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation could be used to predict cancer development 3 years before clinical diagnosis (Palmisano et al., 2000). The methylation data collected to date is also being used to develop methylation profiles for diagnosis (Rauch et al., 2011).
Histone-modifying enzymes have also been shown to contribute to different types of lung cancer. Histone hypoacetylation has been recorded in NSCLC and has been linked to a differential expression of HDACs (Osada et al., 2004; Sasaki et al., 2004; Van Den Broeck et al., 2008). As a result, HDAC inhibitors have been suggested for the treatment of NSCLC (Loprevite et al., 2005; Komatsu et al., 2006; Cuneo et al., 2007; Gridelli et al., 2008). A decreased expression in HATs has also been suggested in order to explain hypoacetylation in some lung cancers (LLeonart et al., 2006; Vaquero et al., 2007). Some histone demethylase enzymes have been linked to lung cancer as well. The overexpression of KDM5B, lysine demethylase 4C (KDM4C), and JmjC-domain containing MAPJD have been noted in lung cancer and have been suggested as contributing to aberrant histone methylation levels seen in lung cancer (Italiano et al., 2006; Suzuki et al., 2007; Hayami et al., 2010). More recently, drug resistance by lung cancer cells was reported to be mediated by an increased expression of lysine demethylase 5A (KDM5A, also known as RBP2 and JARID1A) (Sharma et al., 2010).
The role of miRNAs is relatively well established in lung cancer. Some of the more commonly studied lung cancer miRs are noted in Table 3. Environmental factors such as cigarette smoke have been shown to affect miRNA expression (Hou et al., 2011; Russ and Slack, 2012). Currently, no miRNAs are used as biomarkers in the clinic. The main problem with measuring levels of miRNAs is that an accurate and sensitive method for quantitating them has not been developed (Yendamuri and Kratzke, 2011). A further study of the role of these molecules in lung cancer may lead to strides in the cancer field and is ongoing.
Colorectal Cancer
Another leading cause of cancer deaths in the United States and Europe is colorectal cancer (Jemal et al., 2010; Siegel et al., 2011). This cancer affects the large intestine of the digestive system and usually presents in polyp form. Recently, awareness and resulting heightened screening efforts have lowered the mortality rate associated with this cancer. However, it remains a potent killer once it reaches the metastatic stage. Colorectal cancers can be classified as arising from either chromosomal instability or microsatellite instability (Jass 2007; Ogino and Goel, 2008). Some treatments for the disease are available, but much more research into the molecular mechanisms of the different types of colorectal cancer is needed.
DNA methylation patterns in colorectal cancers have been studied to the extent that certain panels of methylated genes have been suggested for colorectal cancer classification (Ahlquist et al., 2008). These classifications are still under debate, but they present promising candidates for diagnostic and prognostic biomarkers. Interestingly, promoter methylation in colorectal cancers can often be associated with Kirsten rat sarcoma viral oncogene homolog (KRAS) and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation status. Cancers containing BRAF and KRAS mutations tend to have higher methylation levels in certain genes (Samowitz et al., 2005; Nosho et al., 2008).
While most colorectal epigenetic studies focus on DNA methylation, certain distinct histone methylation patterns have also been noted in the disease. For example, H3K9me2 levels are elevated in colorectal adenocarcinomas when compared with normal tissue and are a determinant of more aggressive forms of the disease (Nakazawa et al., 2011). Polycomb protein and HMT EZH2 is overexpressed in colorectal tumors associated with shorter survival times (Crea et al., 2011).
Some miRNAs identified as being involved in colorectal cancer include those listed in Table 3. These miRNAs function in various cellular processes that are associated with cancer, such as evasion of apoptosis, angiogenesis, epithelial-to-mesenchymal transition (EMT), differentiation, invasion, and signaling (de Krijger et al., 2011). Data on the molecular mechanisms of miRNAs in colorectal cancer are still limited, but a more extensive study of the roles of these miRNAs could lead to diagnostic and prognostic markers that could aid in the early detection or treatment.
Can We Secure a Cure?
In his recent article “Curing ‘Incurable’ Cancer,” James Watson outlines the importance of epigenetics in deciphering the mechanisms of cancer (Watson, 2011). There is a definite theme that runs through the epigenetic mechanisms of all cancers. We have outlined the link between the six cancers addressed here in Tables 4 and 5. Table 4 highlights the genes that are hypermethylated in three or more of the common cancers covered in this review. Table 5 contains the same information for miRNA expression. Studying the epigenetic links shared by common cancers provides exciting potential for a powerful anticancer drug targeting many forms of the disease. Conversely, profiling the epigenetic differences between certain cancers will allow us to design more specific drugs. Elucidating and detailing these differences and the differences in the epigenetics of diseased versus normal tissue will allow the continued development of drugs that are unique to those diseases.
Table 4.
Genes Hypermethylated in At Least Three Out of Six Common Solid Tumors
| Gene | Breast | Prostate | Melanoma | Pancreatic | Lung | Colorectal |
|---|---|---|---|---|---|---|
| APC | ✓ | ✓ | ✓ | ✓ | ||
| CCND2 | ✓ | ✓ | ✓ | ✓ | ||
| CDH1 | ✓ | ✓ | ✓ | |||
| CDKN2A | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| DAPK | ✓ | ✓ | ✓ | ✓ | ✓ | |
| ER-α | ✓ | ✓ | ✓ | ✓ | ||
| MGMT | ✓ | ✓ | ✓ | ✓ | ✓ | |
| RAR-β/β2 | ✓ | ✓ | ✓ | ✓ | ||
| RASSF1A | ✓ | ✓ | ✓ | ✓ | ✓ | |
| RUNX3 | ✓ | ✓ | ✓ | ✓ | ||
| TFPI2 | ✓ | ✓ | ✓ | |||
| TIMP3 | ✓ | ✓ | ✓ | ✓ |
Table 5.
Micro RNAs Aberrantly Expressed in At Least Three Out of Six Common Solid Tumors
| miRNA | Breast | Prostate | Melanoma | Pancreatic | Lung | Colorectal |
|---|---|---|---|---|---|---|
| let-7 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| miR-21 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| miR-31 | ✓ | ✓ | ✓ | |||
| miR-143 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| miR-205 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| miR-221 | ✓ | ✓ | ✓ | |||
| miR-222 | ✓ | ✓ | ✓ |
Since epigenetics is a relatively young field, there is a flourish of publications daily outlining the mechanisms by which different epigenetic processes work. Some epigenetic mechanisms not mentioned here, such as chromatin remodeling and gene looping, are also being studied more extensively in the current literature and are contributing to the overall knowledge of the mechanisms of cancer not involving direct alterations in the DNA sequence. This information is currently serving and will continue to serve in the development of many different weapons in the fight against cancer. The fact that epigenetic mechanisms are generally reversible makes them excellent targets for clinical therapies. The ease of DNA isolation points to the use of DNA methylation patterns in noninvasive diagnostic and prognostic testing. The field of cancer epigenetics is one of the most logical and promising places to seek to cure the “incurable.”
Acknowledgments
This work was supported in part by the V Scholar Award, Breast Cancer Alliance Young Investigator Grant, Melanoma Research Foundation Career Development Award, the Alexander and Margaret Stewart Trust Fellowship, CTSA Scholar Award from Yale Center for Clinical Investigation (all to Q.Y.), and NIH grant CA16359 (to the Yale Comprehensive Cancer Center). This publication was made possible by the CTSA Grant Number UL1 RR024139 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCATS or NIH. The authors apologize to their colleagues whose work could not be cited due to space limitation.
Disclosure Statement
No competing financial interests exist.
References
- Ahlquist D.A. Sargent D.J. Loprinzi C.L. Levin T.R. Rex D.K. Ahnen D.J. Knigge K. Lance M.P. Burgart L.J. Hamilton S.R. Allison J.E. Lawson M.J. Devens M.E. Harrington J.J. Hillman S.L. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–450. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. Ghoshal K. Datta J. Majumder S. Yoon S.O. Jacob S.T. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of Pc12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bandyopadhyay D. Okan N.A. Bales E. Nascimento L. Cole P.A. Medrano E.E. Down-regulation of P300/Cbp histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62:6231–6239. [PubMed] [Google Scholar]
- Barrett A. Santangelo S. Tan K. Catchpole S. Roberts K. Spencer-Dene B. Hall D. Scibetta A. Burchell J. Verdin E. Freemont P. Taylor-Papadimitriou J. Breast cancer associated transcriptional repressor Plu-1/Jarid1b interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- Barron N. Keenan J. Gammell P. Martinez V.G. Freeman A. Masters J.R. Clynes M. Biochemical relapse following radical prostatectomy and Mir-200a levels in prostate cancer. Prostate. 2011 doi: 10.1002/pros.22469. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bell C.G. Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9:477–485. doi: 10.1093/bfgp/elq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H. Ingram V.M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blair L.P. Cao J. Zou M.R. Sayegh J. Yan Q. Epigenetic regulation by lysine demethylase 5 (Kdm5) enzymes in cancer. Cancers. 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomston M. Frankel W.L. Petrocca F. Volinia S. Alder H. Hagan J.P. Liu C.G. Bhatt D. Taccioli C. Croce C.M. Microrna expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Bonazzi V.F. Stark M.S. Hayward N.K. Microrna regulation of melanoma progression. Melanoma Res. 2011;22:101–113. doi: 10.1097/CMR.0b013e32834f6fbb. [DOI] [PubMed] [Google Scholar]
- Bracken A.P. Kleine-Kohlbrecher D. Dietrich N. Pasini D. Gargiulo G. Beekman C. Theilgaard-Monch K. Minucci S. Porse B.T. Marine J.C. Hansen K.H. Helin K. The polycomb group proteins bind throughout the Ink4a-Arf locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C. Henry J.C. Kogure T. Schmittgen T. Patel T. The role of micrornas in human liver cancers. Semin Oncol. 2011;38:752–763. doi: 10.1053/j.seminoncol.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C. He H.H. Chen S. Coleman I. Wang H. Fang Z. Nelson P.S. Liu X.S. Brown M. Balk S.P. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S. Spencer-Dene B. Hall D. Santangelo S. Rosewell I. Guenatri M. Beatson R. Scibetta A.G. Burchell J.M. Taylor-Papadimitriou J. Plu-1/Jarid1b/Kdm5b is required for embryonic survival and contributes to cell proliferation in the mammary gland and in Er+ breast cancer cells. Int J Oncol. 2011;38:1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- Catto J.W. Alcaraz A. Bjartell A.S. De Vere White R. Evans C.P. Fussel S. Hamdy F.C. Kallioniemi O. Mengual L. Schlomm T. Visakorpi T. Microrna in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- Chedin F. The Dnmt3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
- Chen T. Hevi S. Gay F. Tsujimoto N. He T. Zhang B. Ueda Y. Li E. Complete inactivation of Dnmt1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Chi P. Allis C.D. Wang G.G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K. DeVries S. Fridlyand J. Spellman P.T. Roydasgupta R. Kuo W.L. Lapuk A. Neve R.M. Qian Z. Ryder T. Chen F. Feiler H. Tokuyasu T. Kingsley C. Dairkee S. Meng Z. Chew K. Pinkel D. Jain A. Ljung B.M. Esserman L. Albertson D.G. Waldman F.M. Gray J.W. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Corona G. Rastrelli G. Forti G. Maggi M. Update in testosterone therapy for men. J Sex Med. 2011;8:639–654. doi: 10.1111/j.1743-6109.2010.02200.x. [DOI] [PubMed] [Google Scholar]
- Crea F. Fornaro L. Paolicchi E. Masi G. Frumento P. Loupakis F. Salvatore L. Cremolini C. Schirripa M. Graziano F. Ronzoni M. Ricci V. Farrar W.L. Falcone A. Danesi R. An Ezh2 polymorphism is associated with clinical outcome in metastatic colorectal cancer patients. Ann Oncol. 2011 doi: 10.1093/annonc/mdr1387. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Croswell J.M. Kramer B.S. Crawford E.D. Screening for prostate cancer with Psa testing: current status and future directions. Oncology (Williston Park) 2011;25:452–460. , 463. [PubMed] [Google Scholar]
- Cuneo K.C. Fu A. Osusky K. Huamani J. Hallahan D.E. Geng L. Histone deacetylase inhibitor Nvp-Laq824 sensitizes human nonsmall cell lung cancer to the cytotoxic effects of ionizing radiation. Anticancer Drugs. 2007;18:793–800. doi: 10.1097/CAD.0b013e3280b10d57. [DOI] [PubMed] [Google Scholar]
- de Krijger I. Mekenkamp L.J. Punt C.J. Nagtegaal I.D. Micrornas in colorectal cancer metastasis. J Pathol. 2011;224:438–447. doi: 10.1002/path.2922. [DOI] [PubMed] [Google Scholar]
- Duursma A.M. Kedde M. Schrier M. le Sage C. Agami R. Mir-148 targets human Dnmt3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M. Garzon R. Cimmino A. Liu Z. Zanesi N. Callegari E. Liu S. Alder H. Costinean S. Fernandez-Cymering C. Volinia S. Guler G. Morrison C.D. Chan K.K. Marcucci G. Calin G.A. Huebner K. Croce C.M. Microrna-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanini F. Vannini I. Amadori D. Fabbri M. Clinical implications of micrornas in lung cancer. Semin Oncol. 2011;38:776–780. doi: 10.1053/j.seminoncol.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Felicetti F. Errico M.C. Segnalini P. Mattia G. Care A. Microrna-221 and −222 pathway controls melanoma progression. Expert Rev Anticancer Ther. 2008;8:1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- Ferguson A.T. Lapidus R.G. Baylin S.B. Davidson N.E. Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res. 1995;55:2279–2283. [PubMed] [Google Scholar]
- Ferracin M. Querzoli P. Calin G.A. Negrini M. Micrornas: toward the clinic for breast cancer patients. Semin Oncol. 2011;38:764–775. doi: 10.1053/j.seminoncol.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Fiegl H. Millinger S. Mueller-Holzner E. Marth C. Ensinger C. Berger A. Klocker H. Goebel G. Widschwendter M. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–1145. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- Foulks J.M. Parnell K.M. Nix R.N. Chau S. Swierczek K. Saunders M. Wright K. Hendrickson T.F. Ho K.K. McCullar M.V. Kanner S.B. Epigenetic drug discovery: targeting DNA methyltransferases. J Biomol Screen. 2011;17:2–17. doi: 10.1177/1087057111421212. [DOI] [PubMed] [Google Scholar]
- Fukushima N. Sato N. Ueki T. Rosty C. Walter K.M. Wilentz R.E. Yeo C.J. Hruban R.H. Goggins M. Aberrant methylation of preproenkephalin and P16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J. Kavanagh E. Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- Galea S. Uddin M. Koenen K. The urban environment and mental disorders: epigenetic links. Epigenetics. 2011;6:400–404. doi: 10.4161/epi.6.4.14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault I. Tozlu S. Lidereau R. Bieche I. Expression analysis of DNA methyltransferases 1, 3a, and 3b in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- Goldberg A.D. Banaszynski L.A. Noh K.M. Lewis P.W. Elsaesser S.J. Stadler S. Dewell S. Law M. Guo X. Li X. Wen D. Chapgier A. DeKelver R.C. Miller J.C. Lee Y.L. Boydston E.A. Holmes M.C. Gregory P.D. Greally J.M. Rafii S. Yang C. Scambler P.J. Garrick D. Gibbons R.J. Higgs D.R. Cristea I.M. Urnov F.D. Zheng D. Allis C.D. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore L. Rothenberg M.L. O'Bryant C.L. Schultz M.K. Sandler A.B. Coffin D. McCoy C. Schott A. Scholz C. Eckhardt S.G. A phase I and pharmacokinetic study of the oral histone deacetylase inhibitor, Ms-275, in patients with refractory solid tumors and lymphomas. Clin Cancer Res. 2008;14:4517–4525. doi: 10.1158/1078-0432.CCR-07-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridelli C. Rossi A. Maione P. The potential role of histone deacetylase inhibitors in the treatment of non-small-cell lung cancer. Crit Rev Oncol/Hematol. 2008;68:29–36. doi: 10.1016/j.critrevonc.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Halkidou K. Gaughan L. Cook S. Leung H.Y. Neal D.E. Robson C.N. Upregulation and nuclear recruitment of Hdac1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- Hayami S. Yoshimatsu M. Veerakumarasivam A. Unoki M. Iwai Y. Tsunoda T. Field H.I. Kelly J.D. Neal D.E. Yamaue H. Ponder B.A. Nakamura Y. Hamamoto R. Overexpression of the Jmjc histone demethylase Kdm5b in human carcinogenesis: involvement in the proliferation of cancer cells through the E2f/Rb pathway. Mol Cancer. 2010;9:59. doi: 10.1186/1476-4598-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.F. Li B.Z. Li Z. Liu P. Wang Y. Tang Q. Ding J. Jia Y. Chen Z. Li L. Sun Y. Li X. Dai Q. Song C.X. Zhang K. He C. Xu G.L. Tet-mediated formation of 5-carboxylcytosine and its excision by Tdg in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller G. Zielinski C.C. Zochbauer-Muller S. Lung cancer: from single-gene methylation to methylome profiling. Cancer Metastasis Rev. 2010;29:95–107. doi: 10.1007/s10555-010-9203-x. [DOI] [PubMed] [Google Scholar]
- Heneghan H.M. Miller N. Lowery A.J. Sweeney K.J. Newell J. Kerin M.J. Circulating micrornas as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- Herceg Z. Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res. 2011;727:55–61. doi: 10.1016/j.mrrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Herceg Z. Vaissiere T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011;6:804–819. doi: 10.4161/epi.6.7.16262. [DOI] [PubMed] [Google Scholar]
- Hinshelwood R.A. Clark S.J. Breast cancer epigenetics: normal human mammary epithelial cells as a model system. J Mol Med (Berl) 2008;86:1315–1328. doi: 10.1007/s00109-008-0386-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann M.J. Engers R. Florl A.R. Otte A.P. Muller M. Schulz W.A. Expression changes in Ezh2, but not in Bmi-1, Sirt1, Dnmt1 or Dnmt3b are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther. 2007;6:1403–1412. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- Hong S.M. Park J.Y. Hruban R.H. Goggins M. Molecular signatures of pancreatic cancer. Arch Pathol Lab Med. 2011;135:716–727. doi: 10.5858/2010-0566-ra.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L. Wang D. Baccarelli A. Environmental chemicals and micrornas. Mutat Res. 2011;714:105–112. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P. Liu D. Dong J. Xing M. The Brafv600e causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11:286–295. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell P.M., Jr. Liu S. Ren S. Behlen C. Fodstad O. Riker A.I. Epigenetics in human melanoma. Cancer Control. 2009;16:200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- Huang J. Sengupta R. Espejo A.B. Lee M.G. Dorsey J.A. Richter M. Opravil S. Shiekhattar R. Bedford M.T. Jenuwein T. Berger S.L. P53 is regulated by the lysine demethylase Lsd1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Huang Y. Nayak S. Jankowitz R. Davidson N.E. Oesterreich S. Epigenetics in breast cancer: what's new? Breast Cancer Res. 2011;13:225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Vasilatos S.N. Boric L. Shaw P.G. Davidson N.E. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2011;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A. Attias R. Aurias A. Perot G. Burel-Vandenbos F. Otto J. Venissac N. Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-P24 amplification including Jak2 and Jmjd2c. Cancer Genet Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Ito S. D'Alessio A.C. Taranova O.V. Hong K. Sowers L.C. Zhang Y. Role of Tet proteins in 5mc to 5hmc conversion, Es-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. Shen L. Dai Q. Wu S.C. Collins L.B. Swenberg J.A. He C. Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass J.R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- Jemal A. Siegel R. Xu J. Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jeronimo C. Bastian P.J. Bjartell A. Carbone G.M. Catto J.W. Clark S.J. Henrique R. Nelson W.G. Shariat S.F. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 2011;60:753–766. doi: 10.1016/j.eururo.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Jia Y. Guo M. Epigenetic changes in colorectal cancer. Chin J Cancer. 2011 doi: 10.5732/cjc.5011.10245. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. Shi C. Edil B.H. de Wilde R.F. Klimstra D.S. Maitra A. Schulick R.D. Tang L.H. Wolfgang C.L. Choti M.A. Velculescu V.E. Diaz L.A., Jr. Vogelstein B. Kinzler K.W. Hruban R.H. Papadopoulos N. Daxx/Atrx, Men1, and Mtor pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic J. Ronneberg J.A. Tost J. Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;4:242–254. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabesch M. Michel S. Tost J. Epigenetic mechanisms and the relationship to childhood asthma. Eur Respir J. 2010;36:950–961. doi: 10.1183/09031936.00019310. [DOI] [PubMed] [Google Scholar]
- Kasinski A.L. Slack F.J. Epigenetics and genetics. Micrornas en route to the clinic: progress in validating and targeting micrornas for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H. Lee H.C. Kim S.Y. Yeom Y.I. Ryu K.J. Min B.H. Kim D.H. Son H.J. Rhee P.L. Kim J.J. Rhee J.C. Kim H.C. Chun H.K. Grady W.M. Kim Y.S. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18:2338–2347. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer C.G. Cao Q. Varambally S. Shen R. Ota I. Tomlins S.A. Ghosh D. Sewalt R.G. Otte A.P. Hayes D.F. Sabel M.S. Livant D. Weiss S.J. Rubin M.A. Chinnaiyan A.M. Ezh2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N. Kawamata N. Takeuchi S. Yin D. Chien W. Miller C.W. Koeffler H.P. Saha, a Hdac inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol Rep. 2006;15:187–191. [PubMed] [Google Scholar]
- Koon H.B. Atkins M.B. Update on therapy for melanoma: opportunities for patient selection and overcoming tumor resistance. Expert Rev Anticancer Ther. 2007;7:79–88. doi: 10.1586/14737140.7.1.79. [DOI] [PubMed] [Google Scholar]
- Krausch M. Raffel A. Anlauf M. Schott M. Willenberg H. Lehwald N. Hafner D. Cupisti K. Eisenberger C.F. Knoefel W.T. Loss of Pten expression in neuroendocrine pancreatic tumors. Horm Metab Res. 2011;43:865–871. doi: 10.1055/s-0031-1291333. [DOI] [PubMed] [Google Scholar]
- Landau D.A. Slack F.J. Micrornas in mutagenesis, genomic instability, and DNA repair. Semin Oncol. 2011;38:743–751. doi: 10.1053/j.seminoncol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao V.V. Grady W.M. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.J. Gusev Y. Jiang J. Nuovo G.J. Lerner M.R. Frankel W.L. Morgan D.L. Postier R.G. Brackett D.J. Schmittgen T.D. Expression profiling identifies microrna signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C. Khaled M. Fisher D.E. Mitf: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Li J. Martinka M. Li G. Role of Ing4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–1379. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- Lim S. Janzer A. Becker A. Zimmer A. Schule R. Buettner R. Kirfel J. Lysine-specific demethylase 1 (Lsd1) is highly expressed in Er-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- Linos E. Swetter S.M. Cockburn M.G. Colditz G.A. Clarke C.A. Increasing burden of melanoma in the United States. J Invest Dermatol. 2009;129:1666–1674. doi: 10.1038/jid.2008.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Sempere L.F. Guo Y. Korc M. Kauppinen S. Freemantle S.J. Dmitrovsky E. Involvement of micrornas in lung cancer biology and therapy. Transl Res. 2011;157:200–208. doi: 10.1016/j.trsl.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLeonart M. Vidal F. Gallardo D. Diaz-Fuertes M. Rojo F. Cuatrecasas M. Lopez-Vicente L. Kondoh H. Blanco C. Carnero A. Ramon y Cajal S. New P53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–608. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- Lomberk G.A. Epigenetic silencing of tumor suppressor genes in pancreatic cancer. J Gastrointest Cancer. 2011;42:93–99. doi: 10.1007/s12029-011-9256-2. [DOI] [PubMed] [Google Scholar]
- Loprevite M. Tiseo M. Grossi F. Scolaro T. Semino C. Pandolfi A. Favoni R. Ardizzoni A. In vitro study of Ci-994, a histone deacetylase inhibitor, in non-small cell lung cancer cell lines. Oncol Res. 2005;15:39–48. doi: 10.3727/096504005775082066. [DOI] [PubMed] [Google Scholar]
- Ma L. Reinhardt F. Pan E. Soutschek J. Bhat B. Marcusson E.G. Teruya-Feldstein J. Bell G.W. Weinberg R.A. Therapeutic silencing of Mir-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi H. Maeda A. Kanemoto H. Uesaka K. Yamazaki K. Hironaka S. Miyagi Y. Ikehara H. Ono H. Klein A. Goggins M. Risk factors of familial pancreatic cancer in japan: current smoking and recent onset of diabetes. Pancreas. 2011;40:974–978. doi: 10.1097/MPA.0b013e3182156e1b. [DOI] [PubMed] [Google Scholar]
- Miranda T.B. Cortez C.C. Yoo C.B. Liang G. Abe M. Kelly T.K. Marquez V.E. Jones P.A. Dznep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T. Martinez S.R. O'Day S.J. Morton D.L. Umetani N. Kitago M. Tanemura A. Nguyen S.L. Tran A.N. Wang H.J. Hoon D.S. Estrogen receptor-alpha methylation predicts melanoma progression. Cancer Res. 2006;66:6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S. Iida S. Kato K. Takagi Y. Uetake H. Sugihara K. The synergistic effect of 5-Aza-2′-deoxycytidine and 5-fluorouracil on drug-resistant tumors. Oncology. 2006;71:437–445. doi: 10.1159/000107110. [DOI] [PubMed] [Google Scholar]
- Muller D.W. Bosserhoff A.K. Integrin beta 3 expression is regulated by Let-7a mirna in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- Munster P.N. Marchion D. Thomas S. Egorin M. Minton S. Springett G. Lee J.H. Simon G. Chiappori A. Sullivan D. Daud A. Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br J Cancer. 2009;101:1044–1050. doi: 10.1038/sj.bjc.6605293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T. Kondo T. Ma D. Niu D. Mochizuki K. Kawasaki T. Yamane T. Iino H. Fujii H. Katoh R. Global histone modification of histone H3 in colorectal cancer and its precursor lesions. Human Pathol. 2011 doi: 10.1016/j.humpath.2011.1007.1009. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Nosho K. Irahara N. Shima K. Kure S. Kirkner G.J. Schernhammer E.S. Hazra A. Hunter D.J. Quackenbush J. Spiegelman D. Giovannucci E.L. Fuchs C.S. Ogino S. Comprehensive biostatistical analysis of Cpg island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003698. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocak S. Sos M.L. Thomas R.K. Massion P.P. High-throughput molecular analysis in lung cancer: insights into biology and potential clinical applications. Eur Respir J. 2009;34:489–506. doi: 10.1183/09031936.00042409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S. Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M. Bell D.W. Haber D.A. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Osada H. Tatematsu Y. Saito H. Yatabe Y. Mitsudomi T. Takahashi T. Reduced expression of class Ii histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112:26–32. doi: 10.1002/ijc.20395. [DOI] [PubMed] [Google Scholar]
- Pakneshan P. Xing R.H. Rabbani S.A. Methylation status of Upa promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- Palmisano W.A. Divine K.K. Saccomanno G. Gilliland F.D. Baylin S.B. Herman J.G. Belinsky S.A. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- Park J.Y. Promoter hypermethylation in prostate cancer. Cancer Control. 2010;17:245–255. doi: 10.1177/107327481001700405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D.F. Kanai Y. Sawada M. Ushijima S. Hiraoka N. Kosuge T. Hirohashi S. Increased DNA methyltransferase 1 (Dnmt1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96:403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T. Hackner B. Truss M. Munzel M. Muller M. Deiml C.A. Hagemeier C. Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angewandte Chemie. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Prat A. Parker J.S. Karginova O. Fan C. Livasy C. Herschkowitz J.I. He X. Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. BCR. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price E.R. Ding H.F. Badalian T. Bhattacharya S. Takemoto C. Yao T.P. Hemesath T.J. Fisher D.E. Lineage-specific signaling in melanocytes. C-Kit stimulation recruits P300/Cbp to microphthalmia. J Biol Chem. 1998;273:17983–17986. doi: 10.1074/jbc.273.29.17983. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan V.M. Jensen T.J. Cui H. Futscher B.W. Martinez J.D. Hypomethylation of the 14–3-3sigma promoter leads to increased expression in non-small cell lung cancer. Genes Chromosomes Cancer. 2011;50:830–836. doi: 10.1002/gcc.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy B. Fiskus W. Cohen B. Pellegrino C. Hershman D.L. Chuang E. Luu T. Somlo G. Goetz M. Swaby R. Shapiro C.L. Stearns V. Christos P. Espinoza-Delgado I. Bhalla K. Sparano J.A. Phase I-II study of vorinostat plus paclitaxel and bevacizumab in metastatic breast cancer: evidence for vorinostat-induced tubulin acetylation and Hsp90 inhibition in vivo. Breast Cancer Res Treat. 2011 doi: 10.1007/510540-011-1928-X. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T.A. Wang Z. Wu X. Kernstine K.H. Riggs A.D. Pfeifer G.P. DNA methylation biomarkers for lung cancer. Tumour Biol. 2011;33:287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- Rauch T.A. Zhong X. Wu X. Wang M. Kernstine K.H. Wang Z. Riggs A.D. Pfeifer G.P. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M. Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Roesch A. Fukunaga-Kalabis M. Schmidt E.C. Zabierowski S.E. Brafford P.A. Vultur A. Basu D. Gimotty P. Vogt T. Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer T. Bosserhoff A.K. Epigenetic events in malignant melanoma. Pigment Cell Res. 2007;20:92–111. doi: 10.1111/j.1600-0749.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- Russ R. Slack F.J. Cigarette-smoke-induced dysregulation of microrna expression and its role in lung carcinogenesis. Pulm Med. 20122012:791234. doi: 10.1155/2012/791234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz W.S. Albertsen H. Herrick J. Levin T.R. Sweeney C. Murtaugh M.A. Wolff R.K. Slattery M.L. Evaluation of a large, population-based sample supports a Cpg island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Sasaki H. Moriyama S. Nakashima Y. Kobayashi Y. Kiriyama M. Fukai I. Yamakawa Y. Fujii Y. Histone deacetylase 1 Mrna expression in lung cancer. Lung Cancer. 2004;46:171–178. doi: 10.1016/j.lungcan.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Sato N. Fukushima N. Matsubayashi H. Iacobuzio-Donahue C.A. Yeo C.J. Goggins M. Aberrant methylation of reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma. Cancer. 2006;107:251–257. doi: 10.1002/cncr.21977. [DOI] [PubMed] [Google Scholar]
- Sato N. Maitra A. Fukushima N. van Heek N.T. Matsubayashi H. Iacobuzio-Donahue C.A. Rosty C. Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- Sato N. Matsubayashi H. Abe T. Fukushima N. Goggins M. Epigenetic down-regulation of Cdkn1c/P57kip2 in pancreatic ductal neoplasms identified by gene expression profiling. Clin Cancer Res. 2005;11:4681–4688. doi: 10.1158/1078-0432.CCR-04-2471. [DOI] [PubMed] [Google Scholar]
- Sato S. Roberts K. Gambino G. Cook A. Kouzarides T. Goding C.R. Cbp/P300 as a co-factor for the microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- Schinke C. Mo Y. Yu Y. Amiri K. Sosman J. Greally J. Verma A. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–265. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. Lorenz P. Gross G. Ibrahim S. Kunz M. Microrna Let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Schutte M. Hruban R.H. Geradts J. Maynard R. Hilgers W. Rabindran S.K. Moskaluk C.A. Hahn S.A. Schwarte-Waldhoff I. Schmiegel W. Baylin S.B. Kern S.E. Herman J.G. Abrogation of the Rb/P16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- Scott G.K. Goga A. Bhaumik D. Berger C.E. Sullivan C.S. Benz C.C. Coordinate suppression of Erbb2 and Erbb3 by enforced expression of micro-Rna Mir-125a or Mir-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- Sharma S.V. Lee D.Y. Li B. Quinlan M.P. Takahashi F. Maheswaran S. McDermott U. Azizian N. Zou L. Fischbach M.A. Wong K.K. Brandstetter K. Wittner B. Ramaswamy S. Classon M. Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard D.R. Raghavan D. Innovations in the systemic therapy of prostate cancer. Nat Rev Clin Oncol. 2010;7:13–21. doi: 10.1038/nrclinonc.2009.187. [DOI] [PubMed] [Google Scholar]
- Shi M. Liu D. Duan H. Shen B. Guo N. Metastasis-related mirnas, active players in breast cancer invasion, and metastasis. Cancer Metastasis Rev. 2010;29:785–799. doi: 10.1007/s10555-010-9265-9. [DOI] [PubMed] [Google Scholar]
- Shi W. Gerster K. Alajez N.M. Tsang J. Waldron L. Pintilie M. Hui A.B. Sykes J. P'ng C. Miller N. McCready D. Fyles A. Liu F.F. Microrna-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- Siegel R. Ward E. Brawley O. Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Singh M. Maitra A. Precursor lesions of pancreatic cancer: molecular pathology and clinical implications. Pancreatology. 2007;7:9–19. doi: 10.1159/000101873. [DOI] [PubMed] [Google Scholar]
- So A.Y. Jung J.W. Lee S. Kim H.S. Kang K.S. DNA methyltransferase controls stem cell aging by regulating Bmi1 and Ezh2 through micrornas. PLoS One. 2011;6:e19503. doi: 10.1371/journal.pone.0019503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J. Pinto A.E. Cunha C.V. Andre S. Barao I. Sousa J.M. Cravo M. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- Stecklein S.R. Jensen R.A. Pal A. Genetic and epigenetic signatures of breast cancer subtypes. Front Biosci (Elite Ed) 2012;4:934–949. doi: 10.2741/E431. [DOI] [PubMed] [Google Scholar]
- Sun T. Wang Q. Balk S. Brown M. Lee G.S. Kantoff P. The role of Microrna-221 and Microrna-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami E. de Maat M. Vu A. Turner R.R. Hoon D.S. Line-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C. Takahashi K. Hayama S. Ishikawa N. Kato T. Ito T. Tsuchiya E. Nakamura Y. Daigo Y. Identification of Myc-associated protein with Jmjc domain as a novel therapeutic target oncogene for lung cancer. Mol Cancer Ther. 2007;6:542–551. doi: 10.1158/1535-7163.MCT-06-0659. [DOI] [PubMed] [Google Scholar]
- Szafranska A.E. Davison T.S. John J. Cannon T. Sipos B. Maghnouj A. Labourier E. Hahn S.A. Microrna expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- Tahiliani M. Koh K.P. Shen Y. Pastor W.A. Bandukwala H. Brudno Y. Agarwal S. Iyer L.M. Liu D.R. Aravind L. Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by Mll partner Tet1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. Yang X. Zhuang L. Jiang X. Chen W. Lee P.L. Karuturi R.K. Tan P.B. Liu E.T. Yu Q. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault A. Figg W.D. Bergan R.C. Lush R.M. Myers C.E. Tompkins A. Reed E. Samid D. A phase Ii study of 5-Aza-2’deoxycytidine (decitabine) in hormone independent metastatic (D2) prostate cancer. Tumori. 1998;84:87–89. doi: 10.1177/030089169808400120. [DOI] [PubMed] [Google Scholar]
- Toyooka S. Gazdar A.F. Methylation profiling of lung cancer: a decade of progress. Mol Cancer Ther. 2011;10:2020. doi: 10.1158/1535-7163.MCT-11-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou J.A. Hagen J.A. Carpenter C.L. Laird-Offringa I.A. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21:5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- Ueki T. Toyota M. Sohn T. Yeo C.J. Issa J.P. Hruban R.H. Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- Unoki M. Current and potential anticancer drugs targeting members of the Uhrf1 complex including epigenetic modifiers. Recent Pat Anticancer Drug Discov. 2011;6:116–130. doi: 10.2174/157489211793980024. [DOI] [PubMed] [Google Scholar]
- Van Den Broeck A. Brambilla E. Moro-Sibilot D. Lantuejoul S. Brambilla C. Eymin B. Khochbin S. Gazzeri S. Loss of histone H4k20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res. 2008;14:7237–7245. doi: 10.1158/1078-0432.CCR-08-0869. [DOI] [PubMed] [Google Scholar]
- Van Neste L. Herman J.G. Otto G. Bigley J.W. Epstein J.I. Van Criekinge W. The epigenetic promise for prostate cancer diagnosis. Prostate. 2011 doi: 10.1002/pros.22459. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Vaquero A. Sternglanz R. Reinberg D. Nad+-dependent deacetylation of H4 lysine 16 by Class Iii Hdacs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Veeck J. Esteller M. Breast cancer epigenetics: from DNA methylation to micrornas. J Mammary Gland Biol Neoplasia. 2010;15:5–17. doi: 10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran V. Klotz L.H. Diet and prostate cancer: mechanisms of action and implications for chemoprevention. Nat Rev Urol. 2010;7:442–453. doi: 10.1038/nrurol.2010.102. [DOI] [PubMed] [Google Scholar]
- Vestergaard E.M. Nexo E. Torring N. Borre M. Orntoft T.F. Sorensen K.D. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int J Cancer. 2010;127:1857–1865. doi: 10.1002/ijc.25209. [DOI] [PubMed] [Google Scholar]
- Vogel C.L. Cobleigh M.A. Tripathy D. Gutheil J.C. Harris L.N. Fehrenbacher L. Slamon D.J. Murphy M. Novotny W.F. Burchmore M. Shak S. Stewart S.J. Press M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of Her2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- Wang F. Yu J. Yang G.H. Wang X.S. Zhang J.W. Regulation of erythroid differentiation by Mir-376a and its targets. Cell Res. 2011;21:1196–1209. doi: 10.1038/cr.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Hevi S. Kurash J.K. Lei H. Gay F. Bajko J. Su H. Sun W. Chang H. Xu G. Gaudet F. Li E. Chen T. The lysine demethylase Lsd1 (Kdm1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J. Sen S. Microrna functional network in pancreatic cancer: from biology to biomarkers of disease. J Biosci. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- Watson J.D. Curing “Incurable” cancer. Cancer Discov. 2011;1:477–480. doi: 10.1158/2159-8290.CD-11-0220. [DOI] [PubMed] [Google Scholar]
- Weigel R.J. deConinck E.C. Transcriptional control of estrogen receptor in estrogen receptor-negative breast carcinoma. Cancer Res. 1993;53:3472–3474. [PubMed] [Google Scholar]
- Widschwendter M. Jones P.A. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- Wu H. Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.C. Zhang Y. Active DNA demethylation: many roads lead to rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K. Tateishi K. Klose R.J. Fang J. Fabrizio L.A. Erdjument-Bromage H. Taylor-Papadimitriou J. Tempst P. Zhang Y. Plu-1 is an H3k4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yang I.V. Schwartz D.A. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med. 2011;183:1295–1301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Phillips D.L. Ferguson A.T. Nelson W.G. Herman J.G. Davidson N.E. Synergistic activation of functional estrogen receptor (Er)-alpha by DNA methyltransferase and histone deacetylase inhibition in human Er-alpha-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [PubMed] [Google Scholar]
- Yendamuri S. Kratzke R. Microrna biomarkers in lung cancer: miracle or quagmire? Transl Res. 2011;157:209–215. doi: 10.1016/j.trsl.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Yu F. Yao H. Zhu P. Zhang X. Pan Q. Gong C. Huang Y. Hu X. Su F. Lieberman J. Song E. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhu W.G. Otterson G.A. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anticancer Agents. 2003;3:187–199. doi: 10.2174/1568011033482440. [DOI] [PubMed] [Google Scholar]