Abstract
Induced pluripotent stem cell (iPSC) therapeutics are a promising treatment for genetic and infectious diseases. To assess engraftment, risk of neoplastic formation, and therapeutic benefit in an autologous setting, testing iPSC therapeutics in an appropriate model, such as the pigtail macaque (Macaca nemestrina; Mn), is crucial. Here, we developed a chemically defined, scalable, and reproducible specification protocol with bone morphogenetic protein 4, prostaglandin-E2 (PGE2), and StemRegenin 1 (SR1) for hematopoietic differentiation of Mn iPSCs. Sequential coculture with bone morphogenetic protein 4, PGE2, and SR1 led to robust Mn iPSC hematopoietic progenitor cell formation. The combination of PGE2 and SR1 increased CD34+CD38−Thy1+CD45RA−CD49f+ cell yield by 6-fold. CD34+CD38−Thy1+CD45RA−CD49f+ cells isolated on the basis of CD34 expression and cultured in SR1 expanded 3-fold and maintained this long-term repopulating HSC phenotype. Purified CD34high cells exhibited 4-fold greater hematopoietic colony-forming potential compared with unsorted hematopoietic progenitors and had bilineage differentiation potential. On the basis of these studies, we calculated the cell yields that must be achieved at each stage to meet a threshold CD34+ cell dose that is required for engraftment in the pigtail macaque. Our protocol will support scale-up and testing of iPSC-derived CD34high cell therapies in a clinically relevant nonhuman primate model.
Introduction
New therapies are needed to treat disease attributed to genetic (ie, Fanconi anemia, hemoglobinopathies) and infectious (ie, HIV/AIDS) hematologic diseases. Autologous or allogeneic HSC transplantation in which the graft has been gene-modified offers a potential cure for hematopoietic or immune system failure. However, appropriate autologous HSCs for genetic modification often are unavailable, and the limited availability of MHC-matched donors significantly restricts allogeneic treatment options.1 In addition, patients receiving allogeneic grafts also may experience acute and chronic life-threatening complications.2
Induced pluripotent stem cell (iPSC) therapies are a tractable alternative to allogeneic cell transplantation because they represent an unlimited source of autologous, patient-specific cells.3–7 However, in vivo preclinical models for long-term functional and safety analysis of iPSC-related therapeutics are limited by model organism life span and further compromised in that available infectious disease models fail to recapitulate human disease. Studies in nonhuman primates (NHP) permit the longitudinal evaluation of iPSC-derived hematopoietic progenitor cells (HPCs) in an autologous setting and provide assays for safety validation, therapeutic benefit, and terminal cell fate and function. Furthermore, findings in NHP models generally translate well to human applications.8 Thus, to critically evaluate the longitudinal therapeutic benefits, risks, and fate of iPSC-derived HPCs in vivo, an appropriate NHP model is required.
Toward modeling novel therapies for blood-related diseases and HIV/AIDS in the pigtail macaque, we have shown previously stable engraftment of bone marrow and peripheral blood–derived autologous CD34+ cells expressing therapeutic and anti-HIV transgenes.9–11 The CD34-enriched fraction from mobilized blood or bone marrow reconstitutes the hematopoietic system, indicating that enriched CD34high fraction contains a subset of cells that are long-term repopulating HSCs (LT-HSCs). We also have shown recently that transplantation with HSCs modified to express the anti-HIV protein C46 reduces viral load in simian/human immunodeficiency virus (SHIV)–infected monkeys and supports partial recovery of immune function (H.-P.K., unpublished results, May 2012). Because iPSCs represent an inexhaustible source for the generation of virus-resistant, gene-modified CD34+ cells and hematopoietic progeny, the pigtail macaque allows us to test SHIV-specific iPSC-derived hematopoietic cell therapeutics in vivo as well as assess the long-term safety of iPSC hematopoietic progeny.
To address safety concerns, we previously generated and characterized several MniPSC lines12,13 and engineered MniPSCs to express an inducible suicide gene.13 Scale-up and quality control of therapeutic cell production to attain an appropriate cell dose for transplantation in the monkey or a patient must meet the following criteria: (1) a relatively efficient hematopoietic induction protocol, (2) a well-established purification method to enrich for the target therapeutic population (CD34+ cells) and simultaneously deplete residual undifferentiated cells, and (3) a strategy to expand primitive CD34+ cells without loss of multipotency.
The production of sufficient iPSC-derived HPCs for engraftment will likely require ex vivo expansion. To this end, novel factors that maintain and expand primitive HSCs ex vivo have been identified, including StemRegenin 1 (SR1), an aryl hydrocarbon receptor (AhR) antagonist,14 and prostaglandin-E2 (PGE2),15 both of which are involved in HSC homeostasis. Several groups have shown that bone morphogenetic protein (BMP) and Wnt pathways specify hematopoietic fate and that activation of these pathways impacts lineage development from mouse and human PSCs.16–19 Here we describe a novel, chemically defined differentiation, expansion, and purification strategy that meets scale-up and quality control criteria and relies on sequential modulation of the BMP, Wnt, and AhR pathways to generate HSC-like cells from MniPSCs. We also describe a method for expansion of enriched CD34high cells with a putative LT-HSC phenotype and show that these cells exhibit both myeloid and lymphoid differentiation potential ex vivo.
Methods
Macaque iPSC culture
Derivation of MniPSC lines 3 and 7 has been described.12 Before differentiation, normal karyotype was validated and phenotype confirmed by flow cytometry and visual assessment of colony appearance. MniPSCs were passaged weekly at ratios of 1:2 to 1:4 on the basis of colony density onto irradiated mouse embryonic fibroblast feeder layers (iMEFs) plated at a density of 6 × 105 cells per well of a 6-well plate. MniPSCs on iMEFs were fed daily with PSC media consisting of 1:1 DMEM/F12 supplemented with 20% knockout serum replacement, 1% MEM nonessential amino acids, 2 mM l-glutamine (all from Life Technologies), 0.1mM 2-mercaptoethanol (Sigma-Aldrich), and 20 ng/mL human basic fibroblast growth factor (bFGF; R&D Systems), and passaged weekly with 200 U/mL collagenase type IV (Life Technologies) in DMEM/F12 containing 100 U/mL Dnase I (Sigma-Aldrich). All undifferentiated iPSC cultures were maintained at 37°C in a humidified incubator (5% CO2, 20% O2).
Hematopoietic mesoderm induction and differentiation
Before passage of MniPSCs to feeder-free conditions, MniPSCs on iMEFs were treated with 10μM Y-27632 rock inhibitor (Calbiochem) for 1 hour and dissociated with 200 U/mL collagenase type IV plus Dnase I. Cells were passaged at ratios of 1:2 to 1:3 onto 6-well plates or 10-cm dishes (Costar) coated with 1:2 GFR matrigel (BD Bioscience) and cultured overnight in PSC media plus 20 ng/mL bFGF and 10μM Y-27632 (Figure 1A). On day −1 relative to differentiation, cells were fed with PSC media plus 20 ng/mL bFGF. On day 0, 75%-90% confluent MniPSC colonies were dissociated with 200 U/mL collagenase type IV plus 200 U/mL Dnase I, washed with DMEM:F12 media, and scraped into clusters. Basal media formulation and BMP4-mediated mesoderm induction has been previously described for hESC differentiation.20,21 MniPSC aggregates were suspended in complete StemPro (cStemPro) media (StemPro-34 plus supplement [Life Technologies], 2mM l-glutamine, 50 μg/mL ascorbic acid [Sigma-Aldrich], 4 × 10−4 M 1-thioglycerol [Sigma-Aldrich], and 150 μg/mL transferrin [Roche]), supplemented with 10, 20, or 50 ng/mL BMP4 (Figure 1A), and cultured in low-cluster plates (Costar). cStemPro was supplemented with human cytokines, indicated below and in panel A of each figure. All cytokines were recombinant human cytokines purchased from R&D Systems unless indicated. On day 1, embryoid bodies (EBs) were settled by gravity and suspended in cStemPro with BMP4 (20 ng/mL) and bFGF (10 ng/mL). On day 4, EBs were settled by gravity and suspended in cStemPro with 10 ng/mL each of VEGF and bFGF.
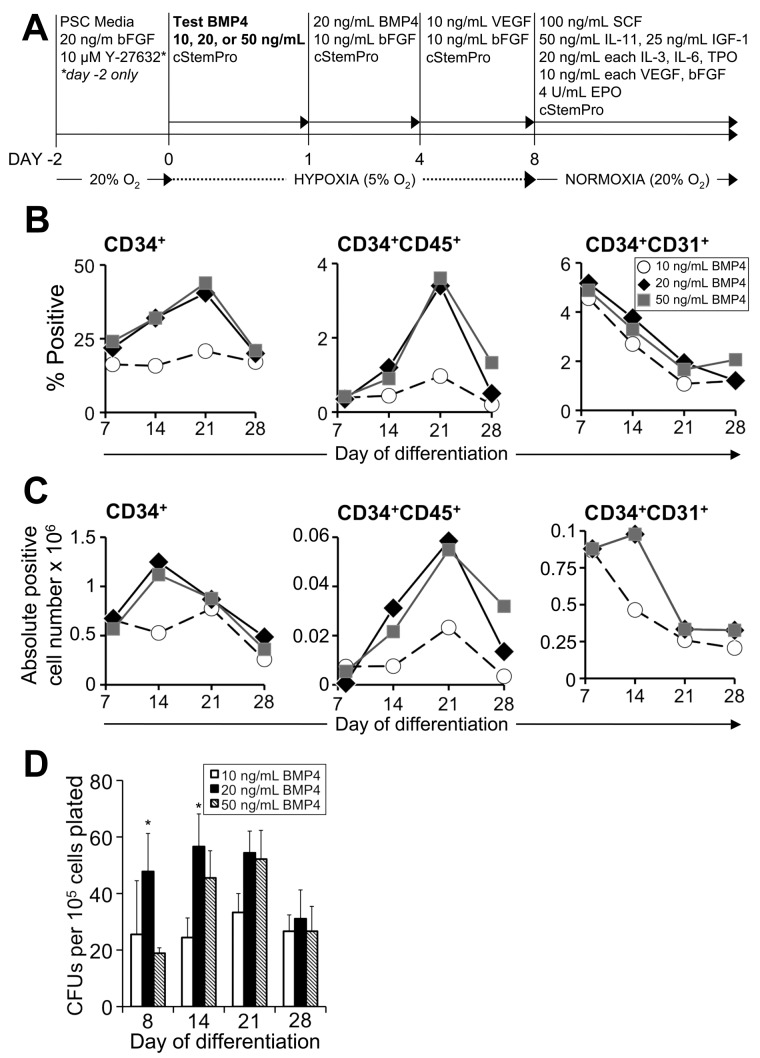
Figure 1.
Optimization of BMP4 concentration for hematopoietic specification of nonhuman primate iPSCs. (A) Schematic of EB method of hematopoietic differentiation specifying titration of BMP4 on day 0 of induction. On day −2, MniPSC line 3 colonies (passage 47) on iMEF feeders were treated with rock inhibitor (10μM Y-27632) and then passaged as clusters 1:2 onto GFR-matrigel in standard PSC media supplemented with bFGF and Y-27632. On day 0, MniPSCs clusters aggregated in media consisting of complete StemPro (cStemPro; StemPro-34 plus StemPro supplement, 2mM l-glutamine, 150 μg/mL transferrin, 50 μg/mL ascorbic acid, and 4 × 10−4M 1-thioglycerol) with different concentrations of BMP4 (10, 20, or 50 ng/mL, test conditions in bold type) and transferred to hypoxia (5% O2). On day 1, EBs were induced with cStemPro supplemented with BMP4 and bFGF. On day 4, media was changed to cStemPro containing VEGF and bFGF for induction. To induce hematopoietic cell expansion, EBs were suspended in cStemPro containing a 10-cytokine cocktail as indicated and placed in normoxia. (B) Comparative kinetics of hematopoietic specification as determined by flow cytometry analysis. On the indicated days of differentiation, EBs were dissociated, counted, stained with fluorophore-conjugated antibodies and analyzed by flow cytometry. (C) Absolute cell number of fluorophore cells. Absolute cell yields corresponding to the indicated hematoendothelial subsets were calculated (absolute positive cell yield = 100 × [% flurophore+ × viable cell yield]). Each number shown is ×106 and is representative of the total fluorpohore+ cell yield obtained from an input undifferentiated MniPS cell number of 1 million. (D) Hematopoietic colony-forming cell assays plated on the indicated days of differentiation. MniPSC-derived cells were dissociated and single cells plated in Methocult H4435. CFUs were enumerated and scored as a function of input cell number (CFUs per 105 cells plated). The results from 1 representative experiment of 3 conducted are shown. Error bars indicate SD of mean of triplicates. ANOVA significance levels: *P = .039 (day 8), *P = .016 (day 14).
For hematopoietic specification, from day 8 onward cStemPro media were supplemented with SCF (100 ng/mL; Miltenyi Biotech), IL-3 (20 ng/mL; Peprotech), IL-6 (20 ng/mL; Peprotech), IL-11 (50 ng/mL), erythropoietin (EPO, 4 U/mL; Peprotech), thrombopoietin (TPO, 20 ng/mL; Peprotech), and insulin-like growth factor-1 (IGF-1; 25 ng/mL). Cultures were maintained in hypoxia (5% CO2/5% O2/90% N2). In some experiments, 0.75μM SR1 (Cellagen Technology, days 1-21 or 8-21) or 2μM PGE2 (Cayman Chemical, days 1-21 or 1-8) were added at indicated time points (Figures 2A, 3A, 4A, and 5A). The PGE2 concentration used (2μM) has been tested in the context of hESC hematopoietic differentiation.19 The SR1 concentration (0.75μM) was selected on the basis of our recent optimization studies expanding macaque cord blood, as well as bone marrow and peripheral blood–derived CD34+CD38− HSCs ex vivo (H.-P.K., unpublished results, September 2011).
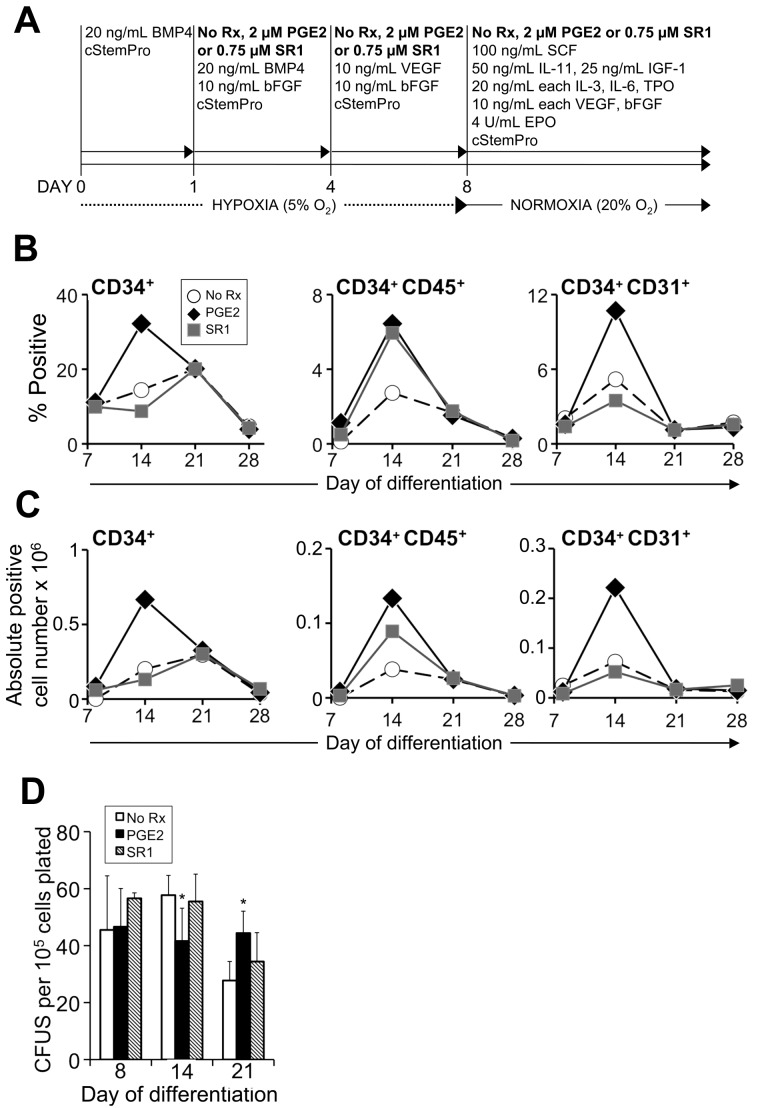
Figure 2.
PGE2 and SR1 alter the kinetics and efficiency of hematopoietic differentiation of MniPSCs. (A) Schematic of hematopoietic differentiation with or without the addition of PGE2 or SR1. On day 0, MniPSC line 3 colonies (passage 50) were aggregated into EBs in cStemPro medium with BMP4. On day 1, media were supplemented with 2μM PGE2, 0.75μM SR1, or untreated (No Rx). For each subsequent induction and media change until the end of the experiment, cStemPro was supplemented with PGE2 or SR1. (B) Comparative kinetics of hematopoietic specification for PGE2, SR1, or untreated cultures as determined by flow cytometry analysis. On the indicated days of differentiation, EBs were dissociated, counted, stained with fluorophore-conjugated antibodies and analyzed by flow cytometry. (C) Absolute cell number of fluorphore+ cells. Absolute cell yields corresponding to the indicated hematoendothelial subsets were calculated (absolute positive cell yield = 100 × [% fluorphore+ × viable cell yield]). Each number shown is ×106 and is representative of the total fluorphore+ cell yield obtained from an input undifferentiated MniPS cell number of 1 million. (D) Hematopoietic colony-forming cell assays plated on the indicated days of differentiation. MniPSC-derived cells were dissociated and single cells plated in Methocult H4435. CFUs were enumerated and scored as a function of input cell number (CFUs per 105 cells plated). The results from 1 representative experiment of 3 conducted are shown. Error bars indicate SD of mean of triplicates. ANOVA significance levels: *P = .042 (day 14), *P = .039 (day 21).
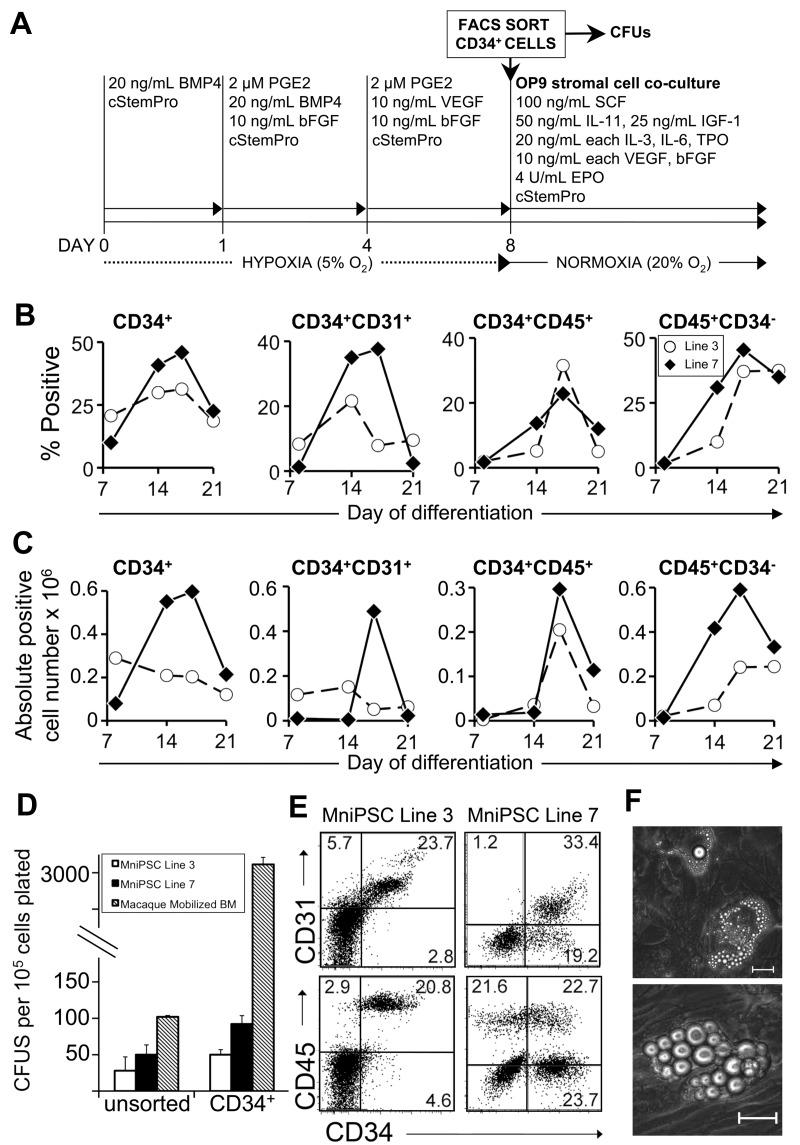
Figure 3.
FACS-sorted MniPSC-CD34+ cells give rise to CD45+CD34− hematopoietic progeny. (A) Schematic of differentiation of MniPSC lines 3 and 7. Both MniPSC lines were induced days 0 to 8 with the optimized protocol outlined in Figure 2 with 2μM PGE2 added to EBs on days 1-8 of differentiation. On day 8, EBs were dissociated into single cells, immunostained with PE-conjugated anti-CD34 antibody (clone 563), and sorted by FACS for the CD34high fraction. CD34high cells were plated in cStemPro plus cytokines (50 000 CD34high cells per well of a 6-well plate) on irradiated OP9 stromal cells. (B) Percentages of CD34high cells that coexpress hematoendothelial markers on the indicated days of differentiation, which include presorted (day 8) and postsorted populations (days 14, 21). (C) Absolute number of fluorphore+ cells for MniPSC lines 3 and 7 that were induced toward hematopoietic differentiation, sorted for the CD34high fraction on day 8, and plated in OP9 coculture. Absolute cell yields corresponding to the indicated hematoendothelial subsets were calculated (absolute positive cell yield = 100 × [% fluorphore+ × viable cell yield]). Each number shown is ×106 and is representative of the total fluorphore+ cell yield obtained from an input undifferentiated MniPS cell number of 1 million. (D) Comparison of hematopoietic colony-forming potential (CFUs) of unsorted and CD34-enriched cells from day 8 MniPSC line 3 and line 7 HPCs and pigtail macaque mobilized bone marrow. To evaluate MniPSC-derived HPCs, EBs were dissociated on day 8, kept in bulk (unsorted), or sorted by FACS and then plated in Methocult H4435. Error bars indicate SD of mean of 3 separate experiments. (E) Representative flow cytometry dot plots of MniPSC-derived HPCs on day 18 of differentiation. (F) Representative images of MniPSC Line 7-derived CD34high cell-derived hematopoietic zones on OP9 stromal cells on day 21 of differentiation. Similar results were obtained from MniPSC line 3 CD34high cells. Colonies were imaged on a Nikon Eclipse Ti-M (TiSR) microscope and photographed with a Nikon camera, in cStemPro media (10× [left] and 20× [right] objectives). Scale bar = 100 μm.
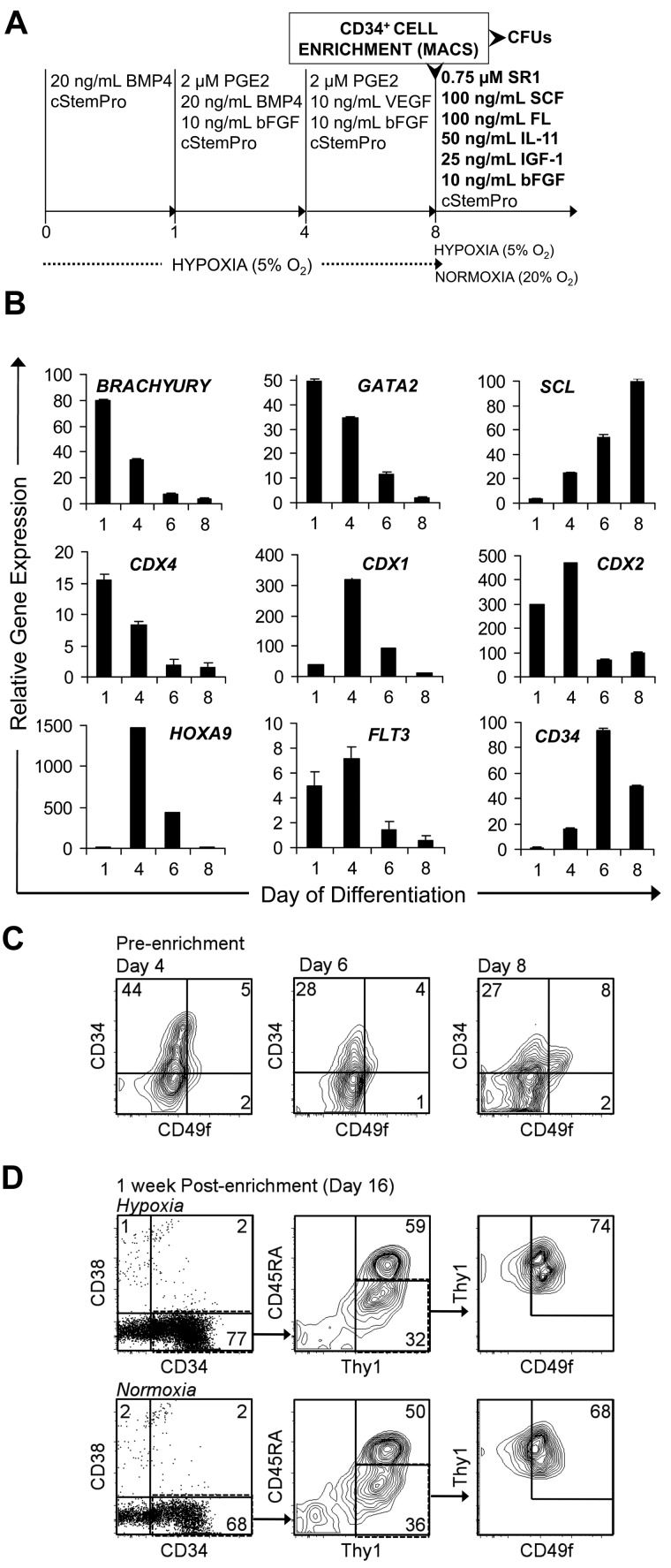
Figure 4.
SR1 maintains and expands MniPSC-derived CD34+ HPCs purified by MACS enrichment. (A) Schematic of MniPSC HPC cell differentiation, enrichment, and expansion. MniPSC Line 7 (Passage 54) cells were aggregated and induced toward differentiation as shown in Figure 3. On day 9 of induction, cells were purified by MACS with anti–human CD34 antibody (clone 12.8). The CD34+ fraction was then cultured under hypoxic (5% O2) or normoxic (20% O2) conditions in cStemPro containing the indicated cytokines and 0.75μM SR1 for HPC expansion. (B) qRT-PCR–based expression of hematopoietic specific genes on days 1, 4, 6, and 8 of hematoendothelial differentiation. Expression levels are relative to β-actin and calibrated to undifferentiated MniPSCs. Error bars indicate SD of mean of triplicate samples from 1 representative experiment of 3. (C) Flow cytometry analysis of CD34 and CD49f coexpression on days 4, 6, and 8 of differentiation before enrichment by MACS. (D) Flow cytometry analysis of putative LT-HSCs (CD34+CD38−Thy1+CD45RA−CD49f+) after a 1-week expansion in SR1 under hypoxic (top) or normoxic (bottom) conditions. Fold expansion of total CD34+CD38−Thy1+CD45RA−CD49f+ cells: 1.1 (hypoxia), 3 (normoxia).
Figure 5.
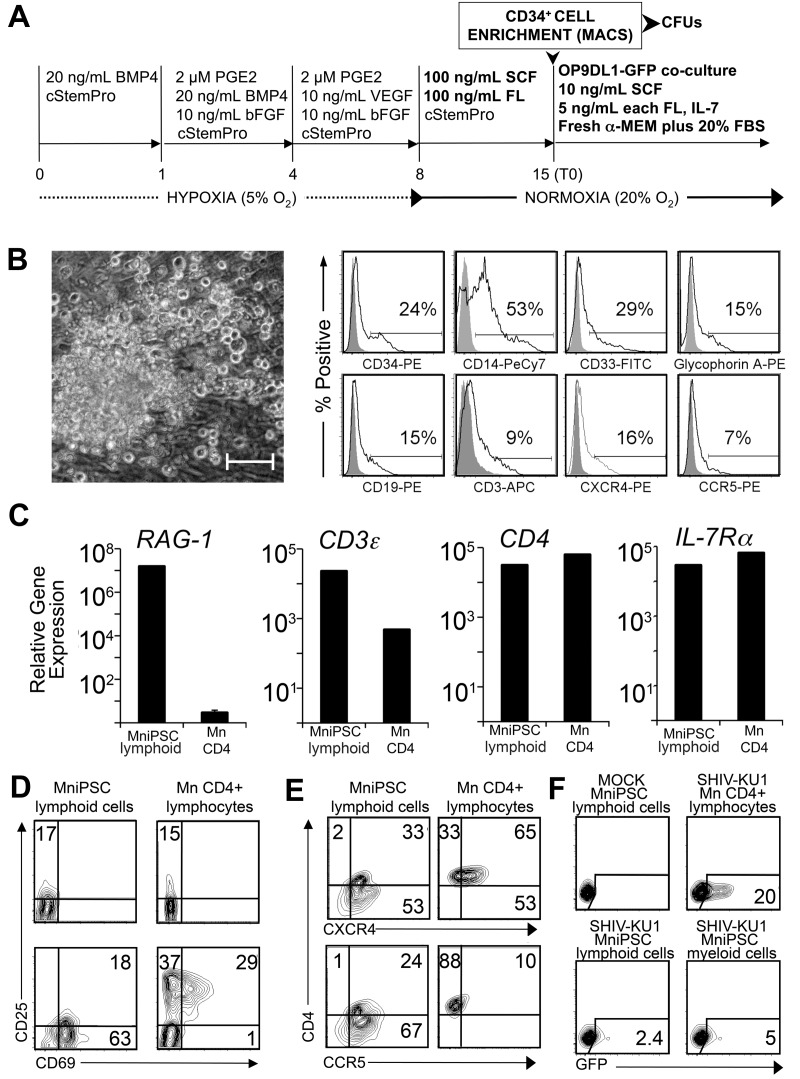
MniPSC-derived CD34+ HPCs exhibit myeloid and lymphoid differentiation potential. (A) Schematic of differentiation. MniPSC Line 7 cells (Passage 18) were induced according to the optimize protocol outlined in the previous figure schema. On day 8, EBs were replated in cStemPro plus SCF and FL for HPC expansion. On day 15, CD34+ cells were MACS purified and plated into Methocult (myeloid CFU assays) or into stromal cell coculture with OP9 cells that express GFP and the Notch ligand Delta-Like 1 (OP9DL1-GFP). To induce lymphoid differentiation, cells were cultured in freshly prepared α-MEM containing FBS, SCF, FL, and IL-7. T0 indicates the starting time point for lymphocyte differentiation. (B) Myeloid phenotype of MniPSC-derived hematopoietic progeny. Left, Representative CFU-M colony differentiated from day 14 CD34+ HPC cells. Colonies were imaged on a Nikon Eclipse Ti-M (TiSR) microscope and photographed with a Nikon camera, in Methocult (20× objective). Scale bar = 100 μm. Right, Flow cytometry analysis of myelomonocytic hematopoietic colonies isolated from Methocult. Myelomonocytic colonies from day 14 MniPSC line 7 CD34high HPCs were isolated from Methocult after CFU scoring on day 24 and analyzed. Gray histograms indicate isotype controls for the indicated fluorophores. (C) qRT-PCR–based expression analysis of T lymphocyte–specific genes in MniPSC-derived lymphoid cocultures (day 30 of lymphoid differentiation). Gene expression also was assessed in CD4+ lymphocytes purified from macaque peripheral blood mononuclear cells and corrected for OP9DL1 stromal cell gene expression. Gene expression levels are relative to β-actin and calibrated to levels detected in undifferentiated MniPSCs. Error bars indicated the SD of the mean of quadruplicate samples. (D) Up-regulation of early activation markers in MniPSC lymphoid cells. Macaque (Mn) CD4+ lymphocytes and day 45 MniPSC lymphoid cultures were untreated or activated for 24 hours with hIL-2 and CD3/CD28 beads and then analyzed by flow cytometry. (E) Expression of HIV entry coreceptors in activated macaque (Mn) CD4+ lymphocytes and day 45 MniPSC-derived lymphoid cultures. (F) MniPSC-derived myeloid and lymphoid cells support SHIVKU-1 entry and viral replication. GFP expression in Ghost cells cultured with supernatant from CD4+ lymphocytes, MniPSC myeloid and lymphoid cultures infected with SHIV-KU1 after virus washout.
Purification of CD34high MniPSC-derived HPCs
Day 8 EBs were dissociated into single cells with Accutase (Life Technologies). Cells were resuspended in DPBS (Life Technologies) containing 100 U/mL Dnase I, 1% FBS, and 2% chick serum; stained with PE conjugated anti–human CD34 antibody (clone 563; BD Bioscience); and sorted by FACS on a FACS Aria II (BD; Figure 3A) or by magnetic-activated cell-sorting system (MACS; Miltenyi Biotech; see Figure 4A), as described.22
Myeloid-type differentiation of CD34high MniPSC-derived hematopoietic progenitors
On day 8, sorted CD34+ cells were plated with or without irradiated mouse OP9 stromal cells23 in 6-well plates and cultured in cStemPro media containing SCF (100 ng/mL), IL-3 and IL-6 (20 ng/mL each), IL-11 (50 ng/mL), EPO (4 U/mL), TPO (20 ng/mL), and IGF-1 (25 ng/mL).
Expansion of CD34high MniPSC-derived hematopoietic progenitors
CD34+ cells were replated at a density of 40 000 cells per well in 12-well plates (nontissue culture treated; Costar) in cStemPro media supplemented with 0.75μM SR1, IL-11 (50 ng/mL), IGF-1 (25 ng/mL), SCF, and fms-like tyrosine kinase receptor-3 ligand (FL; 100 ng/mL each; see Figure 4A). Cultures were maintained in hypoxia (5% CO2/20% O2/90% N2) or normoxia (5% CO2/20% O2) for 1-2 weeks. Media was replaced every 3-4 days.
Colony formation in methylcellulose
For short-term colony-forming cell assays, EBs were dissociated with Accutase and suspended in IMDM and single-cell suspensions from unsorted EBs, or sorted CD34high cells were plated in triplicate in Methocult H4435, H4230, or H4100 (all from StemCell Technologies) at densities of 100 000 or 3000 cells per 3.5-cm2 dish, respectively. For H4100 formulation, IMDM was supplemented with 1% H4100 Methocult, 15% plasma-derived serum, 10% protein-free hybridoma media (Gibco), 2mM l-glutamine, 150 μg/mL transferrin, SCF (100 ng/mL), IL-11 (50 ng/mL), IGF-1 (25 ng/mL), TPO, IL-3, IL-6 (20 ng/mL each), bFGF, VEGF (10 ng/mL each), EPO (4 U/mL), and GM-CSF (1 ng/mL). After 14 days, methylcellulose plates were evaluated for colonies by microscopy (Nikon TMS), and myeloid colony-forming units (CFUs) calculated per input 105 cells.
Lymphoid-type differentiation in OP9DL1 stromal cell coculture
Day 14 HPCs were dissociated and enriched for CD34+ cells as described.33 CD34+ cells were plated on mouse OP9 stromal cells that stably express the Notch ligand Delta-Like 1 and green fluorescent protein (OP9DL1-GFP) in freshly prepared α-MEM media (Life Technologies) containing 50 μg/mL ascorbic acid, 100μM 1-monothioglycerol, 20% FBS, 10 ng/mL SCF, 5 ng/mL each of FL and IL-7, maintained, passaged, and analyzed as previously described (see Figure 5A).24–26
HIV-1 and SHIV infection, Ghost(3) cell, and antigen ELISA assays
T lymphocytes were activated overnight with IL-2 plus CD3/CD28 magnetic beads (Life Technologies). The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HIV-1 89.627 from Dr Ronald Collman (University of Pennsylvania, Philadelphia, PA); and SHIV KU-128 from Dr Opendra Narayan and Dr Sanjay Joag (University of Kansas Medical Center, Kansas City, KS); and pNL4-329 from Dr Malcolm Martin. NL4.3 virus was generated in HEK293T cells as described.29
Myeloid cells isolated from Methocult, unsorted lymphoid-like cells (both derived from MniPSC CD34+ cells), Mn CD4+ lymphocytes from blood MNCs, and OP9DL1-GFP cells were inoculated with HIV 89.6 (50 μL, primary stock 9.12 × 103 TCID50/mL), NL4.3 (20 μL, multiplicity of infection of 5), or SHIV KU-1 (50 μL, primary stock 1.04 × 102 TCID50/mL) and incubated for 24 hours. Media was changed 24 hours after inoculation. Then, 2 days later, cell-free supernatant was collected and transferred to Ghost(3) indicator cells. At 3 days after Ghost(3) cell inoculation, cells were collected and analyzed by flow cytometry as described.30 Background GFP levels detected in cells exposed to OP9DL1-GFP cell supernatant were subtracted from GFP percentages observed in MniPSC lymphoid cultures. For HIV p24 and SIV p27 tests, cell-free supernatants were diluted 1:6 or 1:100, respectively, in media and assayed with the p24 Antigen ELISA kit (PerkinElmer) or the p27 Antigen ELISA kit (ZeptoMetrix Corporation), per the manufacturer's instructions, on the Varioskan Flash Multimode Reader with SkanIt RE for Varioskan Flash, Version 2.4.3 software (490 nm; Thermo Scientific).
Flow cytometry
MniPSC-derived EBs, HPCs, and differentiated cells were dissociated and stained with the following antibodies: CD14-PeCy7, CD31-PE, -APC, CD33-PE, CD34-PE, -APC, CD45-APC, CD45RA-APC-Cy7, CD49f-PerCP Cy5.5, CD90-PeCy7, CCR5-PE, CD235a-PE (Glycophorin A; all from BD); KDR-PE (R&D Systems); CD43-APC-Cy7 (Beckman Coulter), CD38-FITC (StemCell Technologies); and CXCR4-PE (Caltag). The ALDEFLUOR kit (StemCell Technologies) was used to detect aldehyde dehydrogenase. Lymphoid cells were stained with the following antibodies: CD3-PE, -APC, CD4-APC, CD8-PE, CD1a-PE, CD7-PE, CD25-APC (all from BD); CD69-PE, CD27–Pacific Blue (eBioscience); and TCRγδ-PE and TCRαβ-PE (Biolegend). Viability was determined by 7-aminoactinomycin D (eBioscience) staining. Flow cytometry was performed on an LSR-II (BD) and data analyzed with FlowJo Version 9.4.1 (TreeStar) software.
RNA isolation and quantitative real-time RT-PCR
Total RNA was prepared with the RNeasy Plus Kit and treated with RNase-free DNase (all from QIAGEN). RNA was reverse transcribed into cDNA with the use of the Superscript III First-strand Synthesis System kit (Life Technologies) according to the manufacturer's protocol. Macaque-specific (human cross-reactive) primers (Table 1) were designed on the basis of the annotated rhesus macaque genome (synthesized by IDT). For target sequences that lacked annotated confirmed or predicted sequences, oligonucleotides specific for the human sequences were used, with human SCL- and Brachyury-specific primers already described.3 Quantitative PCR (qPCR) was performed with 2X Power SYBR green master mix (Applied Biosystems) on a real-time PCR system machine (Applied Biosystems 7500).
Table 1.
Sequences of macaque-specific oligonucleotides for quantitative RT-PCR assays
| Gene | Forward primer | Reverse primer |
|---|---|---|
| ACTB | TGAGGGGTATGCCCTCCCCCAT | AGGACTCCATGCCCAGGAAGGA |
| BRACHYURY | ACA AAGAGATGATGGAGGAACCCG | AGGATGAGGATTTGCAGGTGGACA |
| CD3ϵ | TGGCGTTTGGGGGCAAGATGGTA | AGTAAACCAGCAGCAGCAAGCCC |
| CD34 | GACCTCCAGCTGTGCGGAGT | GCTCCAGCTGCGGCGATTCA |
| CD4 | AGAAGGTGGAACGCACCCAGGA | ACTGCTGGGAGTAGCGCCAGTT |
| CDX1 | AGTGGCAGCGGTAAGACTCGGA | TGGCCAGGAGGCTAGTGTTGCT |
| CDX2 | TCACCATCCGGAGGAAAGCCGA | GTCCACGCTCCTCATGGCTCAG |
| CDX4 | CCCTATGCATGGATGCGCAAG | CAGAGTCACTTTGCACCGAGCC |
| FLT3 | GCCGATCGTGGAATGGGTGCTT | TTTCTAATTCCCAGGTGAGCCCGA |
| GATA2 | AGCCCAAGCGAAGACTGACGAC | ACAGGTGCCATGTGTCCAGCCA |
| HOXA9 | GATCCCAATAACCCAGCGGCCA | TTTAGAGCCGCTTTGTGCGGGG |
| IL7αR | TGAAATATGTGGGGCCCTCGTGGA | CTGGCGGTAAGCTACATCGTGC |
| RAG-1 | TGGCAGGCCACACTGGACAA | ATGCCCCAATGGAGCCATCCCT |
| SCL | ATGCCTTCCCTATGTTCACCACCA | TGAAGATACGCCGCACAACTTTGG |
Statistical methods
Means and SDs were calculated, and Student t tests and ANOVA statistical tests performed where indicated with the use of Microsoft Excel Version 12.3.2 software.
Results
Titration of BMP4 for efficient hematopoietic mesoderm induction of MniPSCs
Toward scaling up CD34+ cell production, we titrated BMP4 added during day 0 aggregation, which impacts efficiency of mesoderm induction (Figure 1A).31 Because the optimal BMP4 concentration may vary depending on the human ESC/iPSC line, we tested 3 BMP4 concentrations (10, 20, 50 ng/mL) on MniPSC lines 3 and 7, which we previously used for inducing hematopoietic mesoderm in hESCs (data not shown). For MniPSC line 3, high BMP4 on day 0 led to robust expression of CD34 (30%-45%) in day 14 and 21 cultures (Figure 1B). For day 14 MniPSC EBs induced with 20 ng/mL BMP4, 1.5 million viable CD34+ cells were generated from 1 million undifferentiated MniPSCs (Figure 1C). Although the total percentage of CD34+ cells was greater on day 21, the best viable cell yield was achieved on day 14. Only ∼ 5% (< 100 000) CD34+ cells coexpressed hematopoietic markers (CD31, CD45). Importantly, CD31, which is expressed on early hematoendothelial precursors, emerged before CD45, which is induced later and is associated with lineage commitment.
We next assessed myeloid and short-term hematopoietic potential in CFU assays. To determine the optimal Methocult formulation for MniPSC-HPCs, we tested 3 different formulations: H4230, historically used with human and pigtail macaque CD34+ cells; H4435, used with human ESC-derived hematopoietic progenitors; and a noncommercial formulation used with hESCs that has been previously described.20,21 MniPSCs induced with 20 ng/mL BMP4 gave rise to significantly more CFUs, displaying a 90% CFU-M and 10% CFU-GM phenotype. Similar CFU numbers and morphologies were noted for the 3 semisolid media tested; therefore, CFU data for the H4435 formulation only is shown (Figure 1D). CFUs peaked on day 14 and were significantly increased for cells induced in 20 ng/mL BMP4, coinciding with the greatest CD34+ viable cell yield. BMP4 titration also was tested for hematopoietic induction of MniPSC line 7, and similar results were obtained (data not shown).
Treatment with PGE2 and SR1 enhances hematopoietic progenitor emergence and expansion
We next tested whether PGE232 or SR1,14 which activate downstream pathways implicated in HSC emergence and homeostasis, respectively, would improve HPC formation from macaque iPSCs. We first evaluated the effect of PGE2 or SR1 on hematopoietic differentiation of MniPSC lines 3 and 7 (Figure 2A). We selected a previously published PGE2 concentration used in the context of hESC hematopoietic specification19 and a SR1 concentration determined to be optimal for expansion of NHP CD34+ cells obtained from cord blood, peripheral blood, and bone marrow (data not shown). PGE2 treatment nearly tripled the viable CD34+ cell yield in day 14 HPCs generated from MniPSC line 3 cultures (Figure 2B-C). Primitive hematopoietic cell (CD34+CD45+) yields increased 4- and 2-fold, respectively, in PGE2- and SR1-treated MniPSC cultures with no negative impact on total CD34+ cell yield. Importantly, PGE2 treatment restricted to the first 8 days increased CD34+ cell yield without compromising CFUs (Figure 2D). Prolonged treatment with PGE2 (14 days) significantly reduced hematopoietic colony-forming potential compared with untreated controls. In contrast, SR1 treatment increased CFUs compared with controls. Similar results were obtained for MniPSC line 7. These findings indicate that short-term treatment with either PGE2 or SR1 improves CD34+ cell generation from MniPSCs, but PGE2 is superior on the basis of viable cell yields.
Purification of CD34high cells enhances hematopoietic specification and lineage commitment
Although PGE2 increased hematopoietic lineage commitment compared with untreated controls, differentiation was relatively inefficient. Flow cytometry analysis revealed persistence of 10%-30% undifferentiated cells after induction. In addition, hematopoietic maturation as indicated by a loss of CD34 in the CD45+ population was very low (< 2% CD45+CD34−). On the basis of these findings, we hypothesized that removal of undifferentiated cells would improve hematopoietic commitment and maturation. To determine whether hematopoietic specification could be improved by isolation of the CD34+ fraction, day 8 CD34+ cells generated from MniPSC lines 3 and 7 were FACS sorted and replated with or without OP9 stromal cells in hematopoietic expansion medium (Figure 3A). Purified CD34high cells increased production of primitive hematopoietic cells (30% or 0.3 million CD34+CD45+ generated from 1 million undifferentiated MniPSCs) and coexpression of hemogenic markers compared with unsorted hematopoietic EBs (Figure 3B-C). MniPSC line 7 CD34high cells gave rise to more CD45+CD34− cells compared with MniPSC line 3 (Figure 3B-C). Hematopoietic colony-forming potential doubled for sorted compared with unsorted HPCs (Figure 3D). CFUs generated from sorted CD34+ cells were 2-fold greater for line 7 compared with line 3, consistent with their CD45+CD34+CD31low phenotype, which is associated with erythro-myeloid potential33 (Figure 3E). In contrast, line 3-derived cells had a CD45−CD34+CD31high phenotype. Sorted CD34+ cells gave rise to ∼ 4-fold more hematopoietic colonies compared with unsorted cells. Purified CD34+ cells derived from MniPSC line 7 approached CFU counts obtained from unsorted macaque mobilized bone marrow (Figure 3D). Close to 90% of the hematopoietic colonies had CFU-M morphology, with only 10% of colonies displaying CFU-GM morphology. MniPSC CD34+ cells formed hematopoietic zones25 in OP9 stromal cell coculture, which resemble cobblestone area–forming cells (Figure 3F).
An optimized 4-step protocol supports the expansion of MniPSC-derived HPCs with a putative LT-HSC phenotype
On the basis of stage-specific biologic roles of the prostaglandin pathway in early hematopoiesis and AhR signaling in HSC homeostasis, we wanted to determine whether sequential modulation of these pathways with PGE2 and SR1 would enhance the emergence and expansion of putative HSCs. To clarify the effect of SR1 on HPC homeostasis, we tested whether a novel multistep approach to HSC generation, purification, and expansion would increase HPC yield. We hypothesized that early treatment with PGE2 would boost viable CD34+ cell yield, purification of CD34high cells would enhance hematopoietic commitment by eliminating residual undifferentiated iPSCs, and subsequent SR1 treatment would expand the CD34+ putative HSCs (Figure 4A). We also developed a progenitor expansion media, which excludes endothelial (VEGF)–, erythroid (TPO, EPO)–, and myeloid (IL-3, IL-6)–specific cytokines. To expand HPCs, we supplemented basal media with SCF, FL, IL-11, bFGF, and SR1 (Figure 4A).
As the results shown in Figure 3 indicate, MniPSC line 7–derived CD34high cells have greater hematopoietic mesoderm potential compared with line 3; thus, subsequent studies were conducted with MniPSC line 7. Toward scaling up HPC production, we also switched from a 6-well plate format to 10-cm dishes. To clarify the phenotype of MniPSC-derived primitive HPCs, we included hematopoietic cell-specific gene expression by qRT-PCR (Figure 4B and Table 1) and a multiparametric LT-HSC like phenotype, which has been shown to identify human cord blood–derived SCID-repopulating cells (SRCs; CD34+CD38−Thy1+CD45RA−CD49f+).34
Gene expression assays for mesoderm and hematopoietic transcription factors showed that Brachyury, CDX4, and GATA2 mRNA peaked on day 1, whereas CDX1, CDX2, and HOXA9 peaked on day 4, followed by SCL on day 8. Cell-surface markers Flt3 and CD34 peaked on days 4 and 6, respectively. On days 4, 6, and 8 of differentiation, all CD34+ cells also were CD38− and a fraction of the CD34+ cells coexpressed CD49f as determined by flow cytometry (Figure 4C). CD34+ cell yield was greatest on day 4 (4.5 million CD34+ cells per 10-cm plate) and decreased on days 6 and 8 (2.4 million CD34+ cells). The CD34high fraction was isolated on day 8 and maintained in our novel expansion media. After a 1-week expansion in SR1, the enriched cell fraction maintained a putative LT-HSC phenotype (Figure 4D). However, CD34+ cell number cultured under normoxic conditions expanded 3-fold, whereas cells grown in hypoxia maintained their phenotype but did not increase in total cell number. These data clarify a 4-step protocol that expands MniPSC-derived HPCs ex vivo. This optimized hematopoietic specification protocol for MniPSCs includes the following steps: (1) BMP4 mesoderm induction (20 ng/mL BMP4, days 0 and 1); (2) hematopoietic specification with PGE2 (2μM PGE2, days 1-7); (3) purification of CD34high cells; and (4) CD34+ cell expansion in normoxia with SR1.
MniPSC-derived CD34high cells give rise to CD45+CD34− hematopoietic progeny with bilineage differentiation potential
We next evaluated MniPSC-derived CD34+ cell differentiation potential. HPCs were expanded for 14 days and MACS enriched for CD34+ cells (Figure 5A). As in previous experiments, the majority of the hematopoietic colonies had a CFU-M phenotype (Figure 5B). Colonies isolated and evaluated by flow cytometry were heterogeneous in phenotype, expressing HPC (CD34, CXCR4)–, myeloid (CD33, CD14, CCR5)–, erythroid (GlyA)–, and lymphoid (CD3, CD19)–specific cell-surface markers (Figure 5B).
To assess lymphoid potential of the MniPSC-derived CD34high cells, we induced lymphoid differentiation by OP9DL1-GFP coculture (Figure 5A). GFP− colonies appeared 1 week later (supplemental Figure 1A, see the Supplemental Materials link at the top of the article). CD1a, expressed on early lymphocytes, peaked on day 28 whereas CD3, CD4, CD8, and CD7 expression, acquired later, increased over time. Intracellular CD3ϵ increased to 60% by day 35, with surface expression of CD3ϵ with TCRαβ and TCRγδ represented (supplemental Figure 1B). To validate lymphoid phenotype, RAG-1, CD3ϵ, CD4, and IL7-Rα mRNA levels were compared in day 30 MniPSC lymphoid/OP9DL1-GFP coculture and macaque peripheral blood CD4+ cells (Mn CD4) with primers specific for Mn orthologs (Figure 5C and Table 1). Importantly, RAG-1 mRNA was up-regulated 107-fold in MniPSC lymphoid-like cells, suggesting an early lymphoid progenitor state had been achieved (Figure 5C). In contrast, relatively low levels of RAG-1 were detected in Mn CD4+-derived mRNA. These findings are consistent with the 2 waves of RAG-1 gene expression previously reported, in which RAG genes are highly expressed in CD25+CD4−CD8−CD3− T lymphocyte progenitors, when TCR loci undergo rearrangement, and then later at the CD4+CD8+ stage.35,36
Given that ∼ 4% of the Mn CD4+ cells were also CD8+, we expected to detect a low level of RAG-1. Relative expression of CD3ϵ, CD4, and IL7-Rα mRNA in MniPSC lymphoid cultures and Mn CD4+ cells was 103-105-fold greater compared with undifferentiated MniPSCs. Consistent with an immature phenotype, 17% of resting MniPSC lymphoid cultures also expressed CD25 (Figure 4D top). Mn CD4+ and day 45 MniPSC lymphoid cultures were then costimulated with CD3/CD28 beads and assayed for up-regulation of the early activation marker CD69 in combination with CD25 (Figure 4D bottom). In costimulated Mn CD4+ cells, CD25 increased almost 5-fold with 29% of the cells coexpressing CD69, and CD69 expression was restricted to the CD25+ fraction. In contrast, 81% of costimulated day 45 MniPSC lymphoid cultures expressed CD69, a fraction of which expressed CD25. Differential expression of CD69 (63% CD69+CD25−, 18% CD69+CD25+) in MniPSC CD4+ lymphoid-like cells may indicate emergence of 2 distinct lymphoid subsets, the former consistent with a recently identified nontraditional FoxP3-null regulatory T-cell population (CD69+CD4+CD25−) implicated in tumor immune escape.37,38
Myeloid and lymphoid cells generated from MniPSC-derived CD34high cells support HIV-1 and SHIV entry and viral replication ex vivo
To develop HIV-directed PSC therapeutics and validate bilineage differentiation potential, we next assessed whether MniPSC lymphoid-like cells were susceptible to SHIV infection. Expression of HIV-1 entry coreceptors CXCR4 and CCR5 with CD4 were evaluated by flow cytometry on day 45 (Figure 5E). Costimulated Mn CD4+ and MniPSC lymphoid-like cells coexpressed CXCR4 and CD4 (65% and 33%, respectively). Consistent with an activated phenotype,39 10% of costimulated Mn CD4+ cells and 24% of MniPSC lymphoid-like cells coexpressed CD4 with CCR5. Given the limited yield of lymphoid cells and differences between ex vivo and in vivo functionality, we focused on testing SHIV infectivity as an indirect measure of hematopoietic maturation and potential in vivo application and did not conduct additional ex vivo functional assays (cytotoxicity, cytokine production). MniPSC-CD34high derived lymphoid-like and myeloid-like cells were challenged with R5- and X4-tropic 89.6 and X4-tropic NL4.3 HIV-1 isolates.27,29 Mn CD4+ cells, unsorted MniPSC-derived myeloid and lymphoid cells were susceptible to low levels (∼ 1%) of HIV-1 infection, as determined in Ghost(3) cell assays.40 HIV p24 was detected in media of infected cells after virus washout and culture, confirming HIV-1 entry and replication (supplemental Figure 1C).
Given the cellular restriction factors (ie, TRIM5α) that block the HIV life cycle in NHP cells,41 we challenged cells with R4-tropic SHIV-KU1, to which NHP cells should be more permissive.28 Day 45 MniPSC lymphoid-like cells supported SHIV infection (2.4%), at levels 2- and 8-fold less than in MniPSC myeloid cells (5%), and Mn CD4+ cells (20%), respectively (Figure 5F). The SIV p27 antigen capture ELISA, which measures viral replication, indicated high p27 levels in the media of both Mn CD4+ (2.2 × 104 pg/mL) and MniPSC lymphoid-like cells (2.4 × 103 pg/mL; ∼ 9-fold difference; supplemental Figure 1D). These results indicate that NHP iPSCs give rise to lymphoid-like cells that respond to costimulation and support SHIV entry and replication ex vivo.
Discussion
We have developed a novel hematopoietic differentiation, enrichment, and expansion protocol to facilitate the emergence and expansion of HPCs from MniPSCs. We report that sequential treatment with PGE2 followed by SR1 increases production of CD34high cells, which display a cell-surface marker phenotype similar to putative LT-HSCs. Purified CD34high cells have bilineage differentiation and substantial hematopoietic colony-forming potential. This protocol is translatable to human PSC differentiation because BMP4,20 PGE2,19 and SR114 interact with BMP, prostaglandin, Wnt, and AhR signaling in human stem cells. To the best of our knowledge, this is the first reported use of SR1 to expand iPSC-derived HPCs.
Transplantation of gene-modified iPSC derivatives has potential for the treatment of hematologic diseases. To test iPSC-derived cell engraftment in an autologous large animal model, we and other groups have generated NHP iPSCs, including pigtail and rhesus macaque,42 cynomolgus monkey,43,44 and common marmoset.45 Toward scaling up hematopoietic differentiation for HPC transplantation, we developed a strategy to expand and purify HPCs from MniPSCs. We present a chemically defined induction protocol that will permit production scale-up of CD34high cells that have a phenotype associated with SRCs (CD34+CD38−Thy1+CD45RA−CD49f+).
We show that sequential treatment with BMP4 (days 0-1), PGE2 (days 1-7), and SR1 (days 8-17) supports the emergence and expansion of HPCs with an SRC phenotype. We also show that MniPSC-derived CD34+ cells can be purified with the MACS system used for CD34+ purification in autologous transplantation studies in our laboratory. This technology allows us to obtain a > 98% pure population of MniPSC-derived CD34high cells. Second, when sorted CD34+ cells are extracted from heterogeneous culture containing undifferentiated MniPSCs, they expand by coculture with SR1 in normoxia and maintain an HSC-like cell surface phenotype. This finding is novel and significant because translation from ex vivo small-scale CD34+ cell production to transplantation in a large animal will require extensive expansion of the input cell population. On the basis of our experience transplanting gene-modified autologous HSCs enriched from mobilized blood or bone marrow in the macaque, the minimum threshold CD34+ cell dose required for long-term, multilineage repopulation after myeloablative preconditioning is 2 million cells per kilogram of body weight. Our test scale-up in 10-cm plates indicates that to generate 10 million CD34+ cells to transplant into a 5-kg monkey, 400 million cells would be required for aggregation.
Although the scale-up for such a transplantation will be a challenge, other issues must be addressed to translate iPSC technology to a larger physiologic system, including selection of the specific iPSC line, the passage number to be used in transplantation, and quality control of the reagents used for differentiation. In our studies, both the passage number and clonal line of iPSCs impacted efficiency of differentiation and cell yield. For each line, it is likely that the expansion protocol will have to be further optimized to reach minimum cell doses for transplantation. In addition, for the small-scale studies presented herein, we have used previously validated batches of reagents. Scale-up for the monkey transplantation will require liters of StemPro media and several grams of cytokines for differentiation.
If human/mouse xenograft data are predictive of engraftment potential of HSCs derived from various sources (ie, umbilical cord blood, bone marrow, iPSCs), then chemical modification, coculture methods, genetic manipulation, and/or greater cell doses will likely be required to approach the therapeutic benefit that is currently achievable with umbilical cord blood and bone marrow–derived HSC transplantation. To this end, in vivo selection or expansion will likely be required for long-term engraftment and contribution to hematopoiesis, which raises additional concerns. Although it is straightforward to gene modify iPSCs, we have observed loss of transgene expression after serial passaging and lineage commitment, which could impede iPSC-related gene therapy applications. Finally, given the somewhat-undefined repopulation potential of PSC-derived putative HSCs, transplantation of a more phenotypically distinct hematopoietic lineage, such as a purified lymphocyte population, may be more easily translatable to a large animal model and eventual patient population, including HIV/AIDS patients.
Although gene modification of somatic HSCs and T lymphocytes from HIV+ patients may be complicated because of potential infection, iPSCs can be generated from patient cells that are not of hematopoietic origin (ie, fibroblasts) and not susceptible to HIV infection. Transplantation of iPSC-derived HIV-resistant HPCs and lymphocytes would likely decrease toxicity associated with conventional HSC transplantation because only a low-dose conditioning regimen would be required for successful HIV-resistant autologous cell engraftment.46 Given the worldwide impact of HIV infection on human health, iPSC-derived, patient-specific, and HIV-resistant cells for hematopoietic reconstitution is an exciting treatment option. This potential application stems from successful treatment of an HIV+ leukemia patient after radiation and transplantation of donor HSC with a mutated HIV coreceptor (CCR5Δ32).47 Because matched CCR5Δ32 donors are rare, autologous genetically engineered knockout CCR5 iPSC-derived cells may provide an alternative option for HIV+ patients. Nonhuman primate iPSCs provide a preclinical model to test transplantation of virus-resistant cells and other novel therapies.
In summary, our findings establish a reproducible protocol for induction, expansion, and isolation of NHP iPSC-derived HPCs with bilineage differentiation potential, which we will now use to test novel iPSC-based therapeutics for the treatment of hematologic genetic and infectious disease in the pigtail macaque. This protocol may be more broadly applicable to mouse and human PSC systems. Because we engineered our MniPSC lines to express inducible suicide genes (ie, iCaspase9) and selectable markers (ie, MGMTP140K), we now can test the engraftment of MniPSC-CD34high cells and control their in vivo expansion and elimination in a clinically relevant large animal model.
Supplementary Material
Acknowledgments
The authors thank Helen Crawford, Laura Farren, and Bonnie Larson for manuscript preparation; Veronica Nelson and University of Washington National Primate Research Center staff; members of H.-P.K.'s laboratory; UW Department of Cytogenetics for karyotype assessment; FHCRC Flow Cytometry Core Facility; and MSSM hESC/iPSC Shared Resource Facility. H.-P.K. is a Markey Molecular Medicine Investigator and the recipient of the Jose Carreras/E.D. Thomas Endowed Chair for Cancer Research.
This work was supported by National Institutes of Health grants HL098489, HL084345, AI080326, and DK56465. S.L.D. and the hESC/iPSC Shared Resource Facility at Mount Sinai School of Medicine are supported by a New York State Department of Health/NYSTEM grant (C024176).
Footnotes
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.L.G. and H.-P.K. designed the studies; J.L.G. performed the experiments, data analysis and interpretation, and statistical analyses and wrote the manuscript; D.C. contributed to experimental design and performed experiments and data analysis; J.P.K. prepared NL4.3 HIV-1 virus stock and performed p24 and p27 assays; B.C.B. performed (S)HIV infections and Ghost(3) cell assays; S.L.D. contributed to the development of the protocol; and J.L.G., D.C., J.E.A., B.C.B., S.L.D., and H-P.K. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans-Peter Kiem, PO Box 19024, M/S D1-100, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: hkiem@fhcrc.org.
References
- 1.Rudge C, Matesanz R, Delmonico FL, Chapman J. International practices of organ donation. Br J Anaesth. 2012;108(Suppl 1):i48–i55. doi: 10.1093/bja/aer399. [DOI] [PubMed] [Google Scholar]
- 2.Tomblyn M, Chiller T, Einsele H, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective [erratum appears in Biol Blood Marrow Transplant. 2010;16(2):294]. Biol Blood Marrow Transplant. 2009;15(10):1143–1238. doi: 10.1016/j.bbmt.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengerke C, Daley GQ. Autologous blood cell therapies from pluripotent stem cells [review]. Blood Rev. 2010;24(1):27–37. doi: 10.1016/j.blre.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKnight KD, Wang P, Kim SK. Deconstructing pancreas development to reconstruct human islets from pluripotent stem cells. Cell Stem Cell. 2010;6(4):300–308. doi: 10.1016/j.stem.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanal MG. Future of liver transplantation: non-human primates for patient-specific organs from induced pluripotent stem cell [review]. World J Gastroenterol. 2011;17(32):3684–3690. doi: 10.3748/wjg.v17.i32.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laflamme MA, Murry CE. Regenerating the heart [review]. Nat Biotechnol. 2005;23(7):845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 7.Ronaghi M, Erceg S, Moreno-Manzano V, Stojkovic M. Challenges of stem cell therapy for spinal cord injury: human embryonic stem cells, endogenous neural stem cells, or induced pluripotent stem cells? [review]. Stem Cells. 2010;28(1):93–99. doi: 10.1002/stem.253. [DOI] [PubMed] [Google Scholar]
- 8.Trobridge GD, Kiem H-P. Large animal models of hematopoietic stem cell gene therapy. Gene Ther. 2010;17(8):939–948. doi: 10.1038/gt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beard BC, Trobridge GD, Ironside C, McCune JS, Adair JE, Kiem H-P. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J Clin Invest. 2010;120(7):2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trobridge GD, Beard BC, Gooch C, et al. Efficient transduction of pigtailed macaque hemtopoietic repopulating cells with HIV-based lentiviral vectors. Blood. 2008;111(12):5537–5543. doi: 10.1182/blood-2007-09-115022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trobridge GD, Wu RA, Beard BC, et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PLoS One. 2009;4(11):e7693. doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong B, Trobridge GD, Zhang X, et al. Efficient generation of nonhuman primate induced pluripotent stem cells. Stem Cells Dev. 2011;20(5):795–807. doi: 10.1089/scd.2010.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong B, Watts KL, Gori JL, et al. Safeguarding nonhuman primate iPS cells with suicide genes. Mol Ther. 2011;19(9):1667–1675. doi: 10.1038/mt.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.North TE, Goessling W, Walkley CR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goessling W, North TE, Loewer S, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136(6):1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengerke C, Schmitt S, Bowman TV, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2(1):72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 18.Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2(1):60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods NB, Parker AS, Moraghebi R, et al. Brief report: efficient generation of hematopoietic precursors and progenitors from human pluripotent stem cell lines. Stem Cells. 2011;29(7):1158–1164. doi: 10.1002/stem.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109(7):2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigoriadis AE, Kennedy M, Bozec A, et al. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115(14):2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn PA, Topp MS, Morris JC, Riddell SR, Kiem H-P. Highly efficient gene transfer into baboon marrow repopulating cells using GALV-pseudotype oncoretroviral vectors produced by human packaging cells. Blood. 2002;100(12):3960–3967. doi: 10.1182/blood-2002-05-1359. [DOI] [PubMed] [Google Scholar]
- 23.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105(2):617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 24.Van Coppernolle S, Verstichel G, Timmermans F, et al. Functionally mature CD4 and CD8 TCRalphabeta cells are generated in OP9-DL1 cultures from human CD34+ hematopoietic cells. J Immunol. 2009;183(8):4859–4870. doi: 10.4049/jimmunol.0900714. [DOI] [PubMed] [Google Scholar]
- 25.Timmermans F, Velghe I, Vanwalleghem L, et al. Generation of T cells from human embryonic stem cell-derived hematopoietic zones. J Immunol. 2009;182(11):6879–6888. doi: 10.4049/jimmunol.0803670. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Pierce LJ, Spangrude GJ. Distinct roles of IL-7 and stem cell factor in the OP9-DL1 T-cell differentiation culture system. Exp Hematol. 2006;34(12):1730–1740. doi: 10.1016/j.exphem.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collman R, Balliet JW, Gregory SA, et al. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66(12):7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens EB, Joag SV, Atkinson B, et al. Infected macaques that controlled replication of SIVmac or nonpathogenic SHIV developed sterilizing resistance against pathogenic SHIV(KU-1). Virology. 1997;234(2):328–339. doi: 10.1006/viro.1997.8662. [DOI] [PubMed] [Google Scholar]
- 29.Adachi A, Gendelman HE, Koenig S, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vödrös D, Tscherning-Casper C, Navea L, Schols D, De Clercq E, Fenyo EM. Quantitative evaluation of HIV-1 coreceptor use in the GHOST3 cell assay. Virology. 2001;291(1):1–11. doi: 10.1006/viro.2001.1163. [DOI] [PubMed] [Google Scholar]
- 31.Irion S, Clarke RL, Luche H, et al. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development. 2010;137(17):2829–2839. doi: 10.1242/dev.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113(22):5444–5455. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama N, Lee J, Chiu L. Vascular endothelial growth factor synergistically enhances bone morphogenetic protein-4–dependent lymphohematopoietic cell generation from embryonic stem cells in vitro [erratum appears in Blood. 2000;96(4):1273]. Blood. 2000;95(7):2275–2283. [PubMed] [Google Scholar]
- 34.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 35.Wilson A, Held W, MacDonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179(4):1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alt FW, Rathbun G, Oltz E, Taccioli G, Shinkai Y. Function and control of recombination-activating gene activity [review]. Ann NY Acad Sci. 1992;651:277–294. doi: 10.1111/j.1749-6632.1992.tb24626.x. [DOI] [PubMed] [Google Scholar]
- 37.Han Y, Guo Q, Zhang M, Chen Z, Cao X. CD69+ CD4+ CD25− T cells, a new subset of regulatory T cells, suppress T cell proliferation through membrane-bound TGF-beta 1. J Immunol. 2009;182(1):111–120. doi: 10.4049/jimmunol.182.1.111. [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Wang FS. Are non-traditional CD4(+) CD69(+) CD25(−) regulatory T cells involved in disease progression of human hepatocellular carcinoma? J Gastroenterol Hepatol. 2011;26(10):1469–1470. doi: 10.1111/j.1440-1746.2011.06845.x. [DOI] [PubMed] [Google Scholar]
- 39.Palmer LA, Sale GE, Balogun JI, et al. Chemokine receptor CCR5 mediates alloimmune responses in graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16(3):311–319. doi: 10.1016/j.bbmt.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vödrös D, Fenyo EM. Quantitative evaluation of HIV and SIV co-receptor use with GHOST(3) cell assay. Methods Mol Biol. 2005;304:333–342. doi: 10.1385/1-59259-907-9:333. [DOI] [PubMed] [Google Scholar]
- 41.Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280(29):26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Zhu F, Yong J, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3(6):587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto S, Takahashi M. Induction of retinal pigment epithelial cells from monkey iPS cells. Invest Ophthalmol Visual Sci. 2011;52(12):8785–8790. doi: 10.1167/iovs.11-8129. [DOI] [PubMed] [Google Scholar]
- 44.Okahara-Narita J, Umeda R, Nakamura S, Mori T, Noce T, Torii R. Induction of pluripotent stem cells from fetal and adult cynomolgus monkey fibroblasts using four human transcription factors. Primates. 2012;53(2):205–213. doi: 10.1007/s10329-011-0283-1. [DOI] [PubMed] [Google Scholar]
- 45.Tomioka I, Maeda T, Shimada H, et al. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells. 2010;15(9):959–969. doi: 10.1111/j.1365-2443.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiem H-P, Jerome KR, Deeks SG, McCune JM. Hematopoietic-stem-cell-based gene therapy for HIV disease [review]. Cell Stem Cell. 2012;10(3):137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.