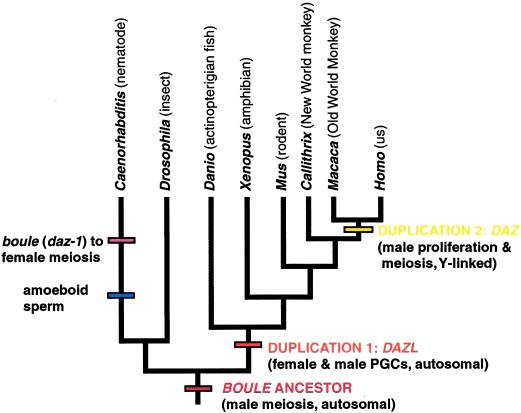
The presence of a dedicated germline, a population of cells that give rise exclusively to gametes, is a unique feature of animal development. Despite variation in the timing of germ cell determination (1, 2) and in the details of lineage and migration by which they populate the gonad, numerous germline genes are conserved across the bilaterian phyla. It is therefore likely that biologists of today are studying variations on a theme first played by a common ancestor of the protostomes and deuterostomes, roughly 600 million years ago (3). In this issue of PNAS , Xu et al. (4) provide further support for deep conservation of germline function with their discovery in mammals of a homologue of another invertebrate gene encoding the RNA-binding protein Boule (5, 6). For some time, however, researchers had believed that they already had the mammalian boule homologues in the Y-linked gene DAZ (deleted in Azoospermia), only in humans and old world monkeys (7–9), and its autosomal ancestor, DAZL (DAZ-like, also known as DAZH; refs. 10–12). The work of Xu et al. suggests that DAZL and DAZ are in fact the divergent products of two nested gene-duplication events, and that the slower evolving progenitor, BOULE, remains a distinct gene in mammals.
A phylogenetic perspective reveals that each duplication event generated one paralog retaining the ancestral function and one that is divergent. The solitary DAZ relative in Drosophila, boule, is expressed only during and is required for male meiosis (5, 13), and the expression data of Xu et al. (4) are consistent with conservation of this role for mouse and human BOULE. Thus, it is likely that the ancestral Boule gene functioned only in male meiosis (Fig. 1). DAZL was generated by a duplication of BOULE in an ancestor of the vertebrates but was expressed in both male and female primordial germ cells, where it was required for their development in Xenopus and mice (14, 15). BOULE itself has not been isolated from fish or amphibians but is predicted to exist (see ref. 4 for discussion). Finally, DAZ was born in primates as a Y-translocated duplicate of the autosomal DAZL but is expressed only in male germ cells (like BOULE). DAZL is expressed before meiosis and is partially redundant with other family members, as judged by the variable penetrance of DAZ deletions in men (7). The phylogenetic view also indicates that in nematodes, BOULE has changed from an ancestral male role to be oocyte-specific, an anomaly that coincides with the invention in nematodes of amoeboid sperm. It is tempting to think that these two may be related.
Figure 1.
Summary of major events in the evolution of the DAZ family. Phylogenetic tree is based on refs. 39–41. PGC, primordial germ cells.
The bursts of functional divergence among DAZ family members also have been accompanied by variable rates of sequence evolution, which can be seen readily in the nucleotide sequence divergence among mammalian DAZ family members (Table 1). For example, the mouse DAZL gene is separated equally in time from each of the human and cynomolgus monkey DAZ/DAZL sequences. Nevertheless, the DAZ genes are 50% more divergent relative to the mouse standard than their autosomal counterparts (Table 1 mDAZL). This discrepancy is even more apparent when comparing the degree of sequence change in DAZ and DAZL since the separation of the human lineage from that of the cynomolgus monkey. Although their DAZL orthologues are a mere 1.5% different at the nucleotide level, the DAZ genes are 10.9% diverged (Table 1 hDAZL), a difference in substitution rate of nearly 7-fold. As relatively conservative as DAZL seems to be, BOULE is even more so (Table 1 mBOULE vs. hBOULE). The higher rates of sequence change seen in DAZ genes also are accompanied by modest increases in the inferred proportion of amino acid-altering nucleotide substitutions (Ka:Ks) in pairwise comparisons involving them (Table 1 decimal number in each row). This phenomenon is consistent with some selection acting to change the DAZ sequence, although a lack of constraint on amino acid sequence or an inability to repair Y-linked mutations also may explain this trend.
Table 1.
Pairwise divergence and nonsynonymous substitution ratios for mammalian DAZ family members
| hDAZL | cynDAZL | hDAZ | cynDAZ | |
|---|---|---|---|---|
| mDAZL | 10.4%* | 11.0%* | 16.1%† | 15.8%† |
| 0.216 | 0.197 | 0.308 | 0.223 | |
| hDAZL | — | 1.5% | 9.1%† | 9.6%† |
| 0.340 | 0.684 | 0.354 | ||
| cynDAZL | — | — | 10.9%† | 9.5%† |
| 0.476 | 0.274 | |||
| hDAZ | — | — | — | 10.9%† |
| 0.530 | ||||
| mBOULE vs. hBOULE | 8.3%‡ | — | — | — |
| 0.128 |
The percentage number in each row is the uncorrected percent divergence in nucleotide sequence; the decimal number is the ratio of nonsynonymous substitutions per nonsynonymous site (Ka) to synonymous substitutions per synonymous site (Ka). Values were obtained using the gap and diverge programs of the GCG Wisconsin Package. m, mouse; h, human; cyn, cynomolgus monkey (Macaca fascicularis). GenBank accession nos. are U21663 (hDAZ), U65918 (hDAZL), U38690 (mDAZL), X99971 (cynDAZL), AJ012216 (cynDAZ), AF272858 (hBOULE), and AF272859 (mBOULE).
Alignment omits last 4 codons of mDAZL and the stop codon of the primate DAZL.
Based only on alignment of exons 1–7.
Alignment omits last 8 codons.
Such rate heterogeneity complicates attempts to reconstruct gene family evolution but it also creates a more general problem—by standard cladistic logic, a gene duplication (like a speciation event) produces two new taxa, neither of which can be, strictly speaking, regarded as the ancestor. However, this view cannot accommodate fully situations in which one gene remains essentially unchanged while its sister becomes modified, and such situations may be especially common in the evolution of male reproduction. For example, consider the Drosophila gene Odysseus (Ods), a homeobox gene expressed in the testis that is evolving so fast it alone produces hybrid sterility (16). Ods is derived by a gene duplication from Dunc-4, which encodes a highly conserved protein involved in neural differentiation (17–20) that lies only 15 kb away from Ods and retains the ancestral function (ref. 16; C. T. Ting and C. I. Wu, personal communication). In such cases, calling the more conservative copy “ancestral” is reasonable, so long as we are aware that this is an approximation.
Progress has been made in clarifying the function of the DAZ family proteins. Drosophila boule has been implicated in the translational activation of twine, a Cdc25-like cell-cycle phosphatase (21). Thus, BOULE may be required for meiosis in male flies because it promotes cell-cycle progression, perhaps by directly interacting with specific mRNAs. This idea has been boosted by the recent observation that murine DAZL protein binds RNA in vitro, and is associated in vivo with actively translating polyribosomes and with poly(A)+ RNA (22). These experiments suggest that the broad role of DAZ family members is to regulate germline RNAs, making them part of a growing group of RNA-binding proteins implicated in this process (23–26). Unresolved matters for the DAZ family include the mechanism of translational activation and the set of RNA targets. In addition, are these targets the same across phylogeny or across the DAZ paralogs within a species?
Although not the focus of their paper, it may turn out to be important that Xu et al. (4) discovered the human BOULE gene by virtue of its interaction with DAZ/DAZL in a yeast two-hybrid screen. Although such homotypic interactions can be artifacts of the two-hybrid system, there was already evidence that mouse DAZL can form homodimers both in yeast and in vitro, and that it can form heterodimers with human DAZ in yeast (27). It is thus possible that DAZ family proteins, including BOULE, may be present in RNA-protein complexes composed of multimerized DAZ proteins. The observation that DAZL is localized to punctate cytoplasmic accumulations in mammals (14, 28) and to the germ plasm in Xenopus (15, 29) is consistent with the participation of DAZ-family proteins in the formation of larger complexes. The isolation of two DAZ-interacting proteins that are not DAZ family members has been reported recently (30).
As revealing as these data about the function of DAZ proteins are, they also raise a perplexing question in light of the evolution of the gene family. If their functions are in the translational regulation of germline mRNAs, why is BOULE sex-specific whereas DAZL is not? Why did DAZ revert to a BOULE-like function in males only? Given their similarities in sequence and ability to form heterodimers, what can DAZ do that BOULE cannot? One intriguing possibility is that DAZ represents a weapon in a sperm competition arms race that has gone from optional to essential during primate evolution. Theoretical considerations of the evolution of male reproductive genes predict that such weapons may accumulate on the Y chromosome if they have collateral deleterious effects in females, because only males inherit them (31–33). One could test this possibility to a first approximation by placing the normally Y-linked DAZ into an autosome or the X chromosome in mice, a circumstance that might allow expression of a DAZ gene during female development. It also may be possible to examine DAZ expression in XY humans that suffer sex reversal caused by deletions of SRY (34–36) or duplication of DAX1 (37, 38).
Acknowledgments
I thank Judith Kimble and Shizuka Hsieh for helpful comments on the manuscript, and Renee Reijo Pera for access to unreleased sequence data. This work was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
See companion article on page 7414.
References
- 1.Dixon K E. Ciba Found Symp. 1994;182:92–110. doi: 10.1002/9780470514573.ch6. [DOI] [PubMed] [Google Scholar]
- 2.Ransick A, Cameron R A, Davidson E H. Proc Natl Acad Sci USA. 1996;93:6759–6763. doi: 10.1073/pnas.93.13.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erwin D H. Am Zool. 1999;39:617–629. [Google Scholar]
- 4.Xu E Y, Moore F L, Reijo Pera R A. Proc Natl Acad Sci USA. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. . (First Published June 5, 2001; 10.1073/pnas.131090498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberhart C G, Maines J Z, Wasserman S A. Nature (London) 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 6.Karashima T, Sugimoto A, Yamamoto M. Development (Cambridge, UK) 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- 7.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 8.Seboun E, Barbaux S, Bourgeron T, Nishi S, Agulnik A, Egashira M, Nikkawa N, Bishop C, Fellous M, McElreavey K, et al. Genomics. 1997;41:227–235. doi: 10.1006/geno.1997.4635. [DOI] [PubMed] [Google Scholar]
- 9.Gromoll J, Weinbauer G F, Skaletsky H, Schlatt S, Rocchietti-March M, Page D C, Nieschlag E. Hum Mol Genet. 1999;8:2017–2024. doi: 10.1093/hmg/8.11.2017. [DOI] [PubMed] [Google Scholar]
- 10.Saxena R, Brown L G, Hawkins T, Alagappan R K, Skaletsky H, Reeve M P, Reijo R, Rozen S, Dinulos M B, Disteche C M, et al. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 11.Reijo R, Seligman J, Dinulos M B, Jaffe T, Brown LG, Disteche C M, Page D C. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 12.Cooke H J, Lee M, Kerr S, Ruggiu M. Hum Mol Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- 13.Castrillon D H, Gonczy P, Alexander S, Rawson R, Eberhart C G, Viswanathan S, DiNardo S, Wasserman S A. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruggiu M, Speed R, Taggart M, McKay S J, Kilanowski F, Saunders P, Dorin J, Cooke H J. Nature (London) 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 15.Houston D W, King M L. Development (Cambridge, UK) 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 16.Ting C T, Tsaur S C, Wu M L, Wu C I. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 17.Miller D M, Shen M M, Shamu C E, Burglin T R, Ruvkun G, Dubois M L, Ghee M, Wilson L. Nature (London) 1992;355:841–845. doi: 10.1038/355841a0. [DOI] [PubMed] [Google Scholar]
- 18.White J G, Southgate E, Thomson J N. Nature (London) 1992;355:838–841. doi: 10.1038/355838a0. [DOI] [PubMed] [Google Scholar]
- 19.Tabuchi K, Yoshikawa S, Yuasa Y, Sawamoto K, Okano H. Neurosci Lett. 1998;257:49–52. doi: 10.1016/s0304-3940(98)00799-x. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, Lo L, Anderson D J, Mikoshiba K. Dev Biol. 1996;180:143–155. doi: 10.1006/dbio.1996.0291. [DOI] [PubMed] [Google Scholar]
- 21.Maines J Z, Wasserman S A. Nat Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 22.Tsui S, Dai T, Warren S T, Salido E C, Yen P H. Biol Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M P. Nature (London) 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 24.Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- 25.Dahanukar A, Walker J A, Wharton R P. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- 26.Jan E, Motzny C K, Graves L E, Goodwin E B. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruggiu M, Cooke H J. Gene. 2000;252:119–126. doi: 10.1016/s0378-1119(00)00219-5. [DOI] [PubMed] [Google Scholar]
- 28.Ruggiu M, Saunders P T, Cooke H J. J Androl. 2000;21:470–477. [PubMed] [Google Scholar]
- 29.Houston D W, Zhang J, Maines J Z, Wasserman S A, King M L. Development (Cambridge, UK) 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 30.Tsui S, Dai T, Roettgen S, Schempp W, Salido E C, Yen P H. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 31.Bull J J. Evolution of Sex Determination Mechanisms. Menlo Park, CA: Benjamin-Cummings; 1983. [Google Scholar]
- 32.Rice W R. Evolution (Lawrence, Kans) 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 33.Fisher R A. Biol Rev. 1931;6:345–368. [Google Scholar]
- 34.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. Nature (London) 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 35.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 36.Berta P, Hawkins J R, Sinclair A H, Taylor A, Griffiths B L, Goodfellow P N, Fellous M. Nature (London) 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- 37.Bardoni B, Zanaria E, Guioli S, Floridia G, Worley K C, Tonini G, Ferrante E, Chiumello G, McCabe E R, Fraccaro M, et al. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- 38.Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R. Nature (London) 1998;391:761–767. doi: 10.1038/35799. [DOI] [PubMed] [Google Scholar]
- 39.Aguinaldo A M, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 40.Metscher B D, Ahlberg P E. Dev Biol. 1999;210:1–14. doi: 10.1006/dbio.1999.9230. [DOI] [PubMed] [Google Scholar]
- 41.Goodman M. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]