Abstract
Background
Cystic Fibrosis (CF) is a monogenic disease with complex expression because of the action of genetic and environmental factors. We investigated whether the ACE gene D/I polymorphism is associated with severity of CF.
Methods
A cross-sectional study was performed, from 2009 to 2011, at University of Campinas – UNICAMP. We analyzed 180 patients for the most frequent mutations in the CFTR gene, presence of the ACE gene D/I polymorphism and clinical characteristics of CF.
Results
There was an association of the D/D genotype with early initiation of clinical manifestations (OR: 1.519, CI: 1.074 to 2.146), bacterium Burkholderia cepacia colonization (OR: 3.309, CI: 1.476 to 6.256) and Bhalla score (BS) (p = 0.015). The association was observed in subgroups of patients which were defined by their CFTR mutation genotype (all patients; subgroup I: no mutation detected; subgroup II: one CFTR allele identified to mutation class I, II or III; subgroup III: both CFTR alleles identified to mutation class I, II and/or III).
Conclusion
An association between the D allele in the ACE gene and the severity of CF was found in our study.
Keywords: Genotype, Phenotype, Variability, Genetic modulation, Angiotensin-converting Enzyme
Background
CFTR gene mutations are crucial in modulating the severity of cystic fibrosis (CF), along with environmental factors and modifier genes [1-7]. CF occurs with heterogeneous clinical presentation. Among the clinical symptoms, that of highest variability is lung disease [5], and modifier genes have been analyzed and associated as possible factors that influence this clinical response [3,7].
The ACE gene codifies the Angiotensin Converting Enzyme (ACE). Based on the pro-inflammatory property of the ACE protein [8,9], the ACE gene was selected as a possible genetic marker for clinical denotation in CF. The ACE enzyme catalyzes the conversion of angiotensin I to angiotensin II peptide, acting in the blood pressure control and the electrolyte balance of blood, being an important vasoconstrictor and stimulant of aldosterone [8,10].
The ACE gene is located on region 17q23.3 [11]. A biallelic polymorphism, named as I (insertion) and D (deletion), with D allele characterized by a deletion of the 287 pb DNA fragment in intron 16, affects the level of the ACE enzyme. The polymorphism determines the amount of ACE enzyme in the plasma and tissues [8,10,12]. Individuals with I/I genotype have low concentrations of ACE; with D/D genotype, higher concentrations; and with D/I genotype, intermediate.
The aim of this study was to investigate the association of the ACE gene D/I polymorphism and CFTR genotype with the severity of CF, determined by twenty four clinical markers of the disease.
Methods
We conducted a cross-sectional study with patients from the CF Specialized Center at the University of Campinas - UNICAMP, in a period from 2009 to 2011. Diagnosis of CF was confirmed in patients through two doses of sodium and chloride from the sweat with values greater than 60 mEq/L. In a cohort of patients we identified two mutations in the CFTR gene. No patient had received the neonatal screening test performed for CF.
Two hundred and fifteen patients were selected for the study. Thirty five patients without clinical data for statistical analysis and those who did not sign the consent form were excluded. Patients' DNA was obtained by phenol-chloroform extraction. The concentration of DNA used for analysis was 50 ng/mL, evaluated using GE NanoVueTM Spectrophotometer (GE Healthcare Biosciences, United States of America, Pittsburgh).
Determination of mutations in the CFTR gene
Determination of mutations in the CFTR gene was performed in the Laboratory of Molecular Genetics for mutations by polymerase chain reaction (F508del) and restriction fragment length polymorphism method (G542X, R1162X, R553X, G551D and N1303K). Some mutations in patients with CF were obtained by sequencing or MLPA (Multiplex Ligation-dependent Probe Amplification) analysis: S4X, 2183A > G, 1717-G > A and I618T. For sequencing and MLPA, we used the same MegaBace1000® property (GE Healthcare Biosciences, United States of America, Pittsburgh) [13]. The CFTR genotype was used as a correction factor for statistical analysis. All mutations identified were included in classes one, two or three of the CFTR gene. Others identified mutations as class IV (P205S e R334W) were included in the statistical analysis in the not identified mutation subgroup, to minimize the associated factor with the mutation classes in the CFTR gene, being that the class IV is associated with a minor severity.
ACE gene D/I polymorphism analysis
ACE*D and ACE*I were identified by amplifying the respective fragments from intron 16 of the ACE gene. The PCR reaction contained 25μL with 100 ng of DNA, 1 μM of each primer, 200 μM of deoxynucleotide triphosphate, 1.3 mM of MgCl2, 50 mM of KCl, 10 mM of Tris–HCl (pH 8.4 at 25°C), 0.1% of Triton X-100 and 0.35U of Taq DNA polymerase. A pair of primers (hace3s, 5'-GCC CTG CAG GTG TCT GCA GCATGT-3'; hace3as, 5'-GGA TGG CTC TCC CCG CCT TG TCTC-3') was used to amplify ACE*D and ACE*I, resulting in 319 bp and 597 bp, respectively [8,10,12,14]. The procedure for thermal cycling consisted of initial denaturation at 94°C for 7 min, subsequent denaturation at 94°C for 30 min, annealing at 56°C for 45 min, and extension at 72°C for 2 min, repeated for 35 cycles followed by a final extension at 72°C for 7 min. After the addition of 5 μL of glycerol-based loading buffer, 7 μL of the reaction was applied in agarose gel containing 1.5% of acetate, 40 mM of TRIS, 2 mM of EDTA and 1 μg of ethidium bromide per milliliter [8,10,12,14].
Due to the preferential amplification of the ACE*D in heterozygous individuals, each initial sample with a D/D genotype was passed through a second PCR reaction. Primers used: Hace 5a, 5'-TGG GAC CAC-AGC GCC CGC CAC TAC-3' and hace 5c, 5'-TCG CCA GCC CTC CCA TGC CCA TAA-3'. The PCR conditions were identical, except for the annealing temperature of 67°C. A 335 bp sequence was amplified in the presence of at least one allele [8,10,12,14].
Clinical markers of severity of disease
Clinical scores of Kanga, Shwachman-Kulczycki and Bhalla (BS) were performed blindly by three qualified professionals from UNICAMP. These scores measure the pulmonary exacerbation, severity of CF and structural impairment of the lung, respectively [15]. Nutritional status was obtained by calculating the Body Mass Index (BMI) for age using the programs WHO Anthro [16], for patients up to five years old, and WHO Anthro Plus [17], for patients aged 5 to 19 years old. For patients older than 19 years old, the BMI was calculated [18]. Age of the diagnosis, the onset of pulmonary and digestive symptoms, and the first isolation of Pseudomonas aeruginosa, were used as markers of initiation of the disease. Results of cultures of sputum, performed during routine diagnosis for the mucoid and non-mucoid bacteria P. aeruginosaStaphylococcus aureusBurkolderia cepacia (BC) and Achromobacter xylosoxidans were included. Spirometry was performed in the Laboratory of Pulmonary Physiology (LAFIP) according the standards of the American Thoracic Society [19]. Parameters analyzed were: forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), FEV1/FVC ratio, forced expiratory flow between 25-75% and transcutaneous oxygen saturation (SaO2). For analysis of the spirometry data, we used the predicted value in%. Comorbidities (nasal polyposis, osteoporosis, diabetes mellitus, pancreatic insufficiency and meconium ileus) were also analyzed.
Statistical analysis
Variables described for the onset of illness (age at diagnosis, onset of pulmonary and digestive symptoms and first isolation of P. aeruginosa) were categorized into two groups according to the median of the data, due to a non-normal distribution of data. Data categorized by the median are divided into two cohorts with similar sample size. For clinical evaluation of the scores from the SaO2 and spirometry tests, analyses were performed without adjusting the data. Bacteria isolated from the culture of airway secretions were used as markers according to the presence or absence of bacteria in three consecutive cultures in the past two years. Comorbidities were compared in terms of presence or absence. Statistical analyses was performed using the Statistical Package for the Social Sciences (SPSS) v.17.0 [20] and the R program version 2.12 (Comprehensive R Archive Network, 2011). In order to avoid spurious data due to the problem of multiple testing [21], the level of significance α, was adjusted using the Bonferroni correction for four tests. Calculation of statistical power for the sample, carried out by software GPOWER 3.0.5 [22], showed a statistical power above 80% for the analysis performed.
Data were compared by the linear and logistic regression analysis. For comparison between genotypes and the variables with numerical distribution, T-student test was applied to normal data distribution or Mann–Whitney test to non-normal data distribution. Genotyped data for the CFTR gene was used to establish an association between the CFTR gene, ACE gene and clinical variables. All mutations analyzed in our study were included in classes I, II or III. In the analyzed sample, four different analyses were performed in order to detail the effect of the genotype of the CFTR gene in clinical severity. The analyses were performed in the cohorts: (1) all patients with CF (180 patients); (2) patients with no identified mutation in the CFTR gene (44 patients); (3) patients with a mutant allele identified in the CFTR gene belonging to class I, II or III mutation(51 patients); and (4) patients with two mutations identified in the CFTR gene belonging to class I, II and/or III (85 patients) - main cohort to analyze the influence of modifier genes associated with clinical variation in CF.
This study was approved by the Institutional Ethics Committee from University of Campinas (Faculty of Medical (No. 528/2008), and all patients signed a consent form before beginning the study.
Results and discussion
From the sample of 180 analyzed patients, 90 (50%) were male, 165 (91.7%) were European-Caucasian derived and 15 (8.3%) were African-derived individuals. The patients’ CFTR genotypes were: 44 patients (24.44%) without identified mutation, 51 (28.33%) with one identified mutation (25% F508del/-, 2.78% G542X/-, 0.56% R1162X/-) and 85 (47.22%) patients with two identified mutations (31.67% F508del/F508del, 6.67% F508del/G542X, 2.78% F508del/R1162X, 2.22% F508del/N1303K, 0.56% F508del/R553X, 0.56% F508del/S4X, 0.56% F508del/1717-1 G > A, 0.56% G542X/R1162X, 0.56% G542X/I618T, 0.56% G542X/2183A > G and 0.56% R1162X/R1162X).
The spectrum of isolated Bacteria in secretion was: 76 (42.2%) with mucoid and 101 (56.1%) with non-mucoid P. aeruginosa; 141 (78.3%), S. aureus; 25 (13.9%), B. cepacia; and 18 (10%), A. xylosoxidans. Comorbidities associated with CF severity were: 143 (79.4%) with pancreatic insufficiency; 33 (18.3%), nasal polyps; 33 (18.3%), diabetes mellitus; 29 (16.1%), osteoporosis; and 27 (15%), meconium ileus. For the variables with their numerical distribution, see data listed in Table 1.
Table 1.
Description of quantitative variables (in months) of CF patients treated at the pediatric clinic at UNICAMP
| Variable | N* | Minimum | Maximum | Median | Mean | Standard error | Standard deviation |
|---|---|---|---|---|---|---|---|
| Age |
179 |
7 |
288 |
154 |
212.64 |
14.13 |
189.04 |
| Onset of the manifestation |
170 |
0 |
156 |
3 |
34.69 |
8.33 |
108.54 |
| Age at diagnosis |
173 |
0 |
170 |
24 |
91.47 |
12.44 |
163.60 |
| Onset of digestive symptons |
150 |
0 |
150 |
3 |
40.69 |
8.93 |
109.32 |
| Onset of lungs symptons |
165 |
0 |
156 |
6 |
42.88 |
9.24 |
118.68 |
| 1st. P. aeruginosa | 131 | 6 | 180 | 31 | 102.60 | 15.16 | 173.47 |
*N- number of patients.
The ACE gene D/I polymorphism showed a higher frequency for ACE*D (228/360 alleles) compared with ACE*I (132/360 alleles). The genotype frequencies were: 72 (40.0%) with D/D; 84 (46.67%) with D/I; and 24 (13.3%) with I/I. The population is in Hardy-Weinberg equilibrium (p > 0.05). Analysis of 70 healthy control subjects in UNICAMP demonstrated the genotype frequency: 20 (29%) with D/D, 37 (53%) of D/I, and 13 (18%) I/I [23]. There was no difference in frequency of genotypes in relation to our study (p = 0.210). The analyses of the ACE gene D/I polymorphism with the clinical variables are denoted in Table 2, where every association possible between the clinical trial, CFTR mutation identified and ACE gene D/I polymorphism can be observed.
Table 2.
Association ofACEgene D/I polymorphism with variables used as markers of severity of CF, patients followed at the pediatric center in UNICAMP distributed byCFTRgene mutation identified divided into cohorts
| Variable |
Without taking CFTR mutation into account |
No identified mutation |
One identified mutation |
Two identified mutation |
|||||
|---|---|---|---|---|---|---|---|---|---|
| E | p | E | p | E | p | E | p | ||
| Patients age |
W:0.791 |
0.374 |
W:3x10-5 |
0.995 |
W:2.969 |
0.085 |
W:0.001 |
0.984 |
|
| Onset of clinical manifestations |
W:0.116 |
0.733 |
W:0.162 |
0.687 |
W:4.29 |
0.038 |
W:0.937 |
0.333 |
|
| Diagnostic |
W:0.11 |
0.74 |
W:0.047 |
0.83 |
W:0.099 |
0.753 |
W:0.087 |
0.768 |
|
| Onset of digestive symptons |
W:1.494 |
0.221 |
W:0.148 |
0.7 |
W:0.297 |
0.586 |
W:0.979 |
0.322 |
|
| Onset of lung symptons |
W:0.021 |
0.885 |
W:0.039 |
0.843 |
W:0.401 |
0.526 |
W:1.302 |
0.31 |
|
| BMI |
W:1.169 |
0.28 |
W:0.687 |
0.407 |
W:0.436 |
0.509 |
W:2.498 |
0.114 |
|
| Nasal poliposys |
W:0.62 |
0.431 |
W:0.984 |
0.321 |
W:0.419 |
0.517 |
W:1.26 |
0.262 |
|
| Diabetes |
W:0.358 |
0.55 |
W:0.016 |
0.901 |
W:0.174 |
0.676 |
W:0.184 |
0.668 |
|
| Osteoporosis |
W:0.877 |
0.349 |
W:1.056 |
0.0304 |
W:0.561 |
0.454 |
W:0.083 |
0.773 |
|
| Pancreatic insuficience |
W:1.6 |
0.206 |
W:0.693 |
0.406 |
W:1.063 |
0.302 |
W:0.182 |
0.669 |
|
| Meconium ileus |
W-0.252 |
0.616 |
W:3.813 |
0.051 |
W:1.109 |
0.292 |
W:1.498 |
0.221 |
|
| SaO2 |
F:2.131 |
0.142 |
F:0.022 |
0.884 |
F:1.868 |
0.178 |
F:1.344 |
0.25 |
|
| Scores |
Bhalla |
F:6.526 |
0.012 |
F:0.2 |
0.689 |
F:4.942 |
0.032 |
F:4.013 |
0.049 |
| Kanga |
F:1.3 |
0.256 |
F:0.486 |
0.492 |
F:0.027 |
0.871 |
F:3.765 |
0.057 |
|
| SK |
F:2.361 |
0.127 |
F:0.286 |
0.597 |
F:1.042 |
0.312 |
F:1.243 |
0.269 |
|
| FVC |
F:0.139 |
0.71 |
F:0.829 |
0.37 |
F:0.918 |
0.345 |
F:0.93 |
0.339 |
|
| FEV1 |
F:0.785 |
0.377 |
F:0.622 |
0.436 |
F:0.907 |
0.348 |
F:2.797 |
0.099 |
|
| FEV1/FVC |
F:0.891 |
0.347 |
F:0.005 |
0.943 |
F:2.212 |
0.146 |
F:0.156 |
0.694 |
|
| FEF25-75% |
F:0.42 |
0.518 |
F:0.112 |
0.735 |
F:0.02 |
0.887 |
F:1.048 |
0.31 |
|
| 1aP. aeruginosa isolated |
W:0.962 |
0.327 |
W:0.702 |
0.402 |
W:0.16 |
0.69 |
W:0.099 |
0.753 |
|
| Isolated Bacteria | PAM |
W:0.92 |
0.338 |
W:0.141 |
0.708 |
W:2.016 |
0.156 |
W:0.165 |
0.684 |
| PANM |
W:1.21 |
0.272 |
W:1.149 |
0.284 |
W:0.987 |
0.753 |
W:0.262 |
0.609 |
|
| AX |
W:3.2 |
0.074 |
W:0.038 |
0.845 |
W:0.642 |
0.423 |
W:2.911 |
0.088 |
|
| BC |
W:4.290 |
0.038 |
W:0.1 |
0.753 |
W:3.681 |
0.055 |
W:0.341 |
0.559 |
|
| SA | W:0.209 | 0.65 | W:1.151 | 0.283 | W:0.191 | 0.662 | W:1.102 | 0.294 | |
Analysis by linear regression (F) and logistic regression (W) test . Values below of 0.05 for p denote a clinical correlation between variables. E - Statistical,% - percentage, SaO2 - transcutaneous oxygen saturation, FEV1 – forced expiratory volume in 1 second, FVC – forced expiratory capacity, FEV1/FVC – ratio between two variables, forced FEF25-75, SC - expiratory flow at 25-75% of the pulmonary volume, SK - Shwachman-Kulczycki, PAM – P. aeruginosa mucoid, PAM – P. aeruginosa non mucoid, AX – A. xylosoxidans, BC – B. cepacia, SA – S. aureus, BMI – body mass index, CF - cystic fibrosis, ACE – angiotensin converting enzyme. No identified mutation (44 patients) – patients without of identified mutation in classes I, II or III. One identified mutation (51 patients) – patient with one identified mutation in class I, II, or III. Two identified mutation (85 patients) – patient with two mutations in class I, II and/or III. Others identified mutations as class IV (P205S e R334W) was included in the statistical analysis in the not identified mutation subgroup, to minimize the associated factor with the mutation classes in the CFTR gene.
The ACE gene D/I polymorphism was associated with the onset of clinical manifestations (Table 3), in the subgroup of patients with one identified CFTR mutation. We observed that patients with I/I genotype had OR: 0.297 (0.084 – 0.995), as protection factor, and the ones with D/D genotype had OR: 1.519 (1.074 to 2.146), as a severity factor.
Table 3.
Association ofACEgene D/I polymorphism with onset of clinical symptoms of patients in months considering the cohorts toCFTRmutation
| Groups | ACEgenotype | ≤ 3 months | > 3 months | Total | X2 | p | X2 | p | OR (CI 5-95%) |
|---|---|---|---|---|---|---|---|---|---|
| Without taking CFTR mutation into account |
I/I |
38 |
28 |
66 |
0,880 |
0,644 |
0.012 |
0.9136 |
1.035 (0.555 - 1.931) |
| I/D |
44 |
37 |
81 |
0.473 |
0.492 |
0.808 (0.4396 -1.484) |
|||
| D/D |
15 |
8 |
23 |
0.723 |
0.395 |
1.486 (0.5937, 3.721) |
|||
| No identified CFTR mutation |
I/I |
13 |
6 |
19 |
1,685 |
0,431 |
0.825 |
0.364 |
1.806 (0.5017 -6.498) |
| I/D |
9 |
9 |
18 |
1.624 |
0.203 |
0.438 (0.122 - 1.576) |
|||
| D/D |
3 |
1 |
4 |
0.357 |
0.977 |
2.045 (0.194 - 21.58) |
|||
| One CFTR mutation identified class I, II or III |
I/I |
10 |
11 |
21 |
5,564 |
0,062 |
4.217 |
0.049 |
0.297 (0.084 – 0.995) |
| I/D |
14 |
6 |
20 |
0.521 |
0.471 |
1.167 (0.775 – 1.757) |
|||
| D/D |
8 |
1 |
9 |
2.951 |
0.097 |
5,667 (0,647 – 49,61) |
|||
| Two CFTR mutation identified class I, II and/or III | I/I |
15 |
11 |
26 |
0,599 | 1,026 | 0.773 |
0.379 |
1.527 (0.593 - 3.936) |
| I/D |
21 |
22 |
43 |
0.122 |
0.727 |
0.854 (0.352 - 2.072) |
|||
| D/D | 4 | 6 | 10 | 0.511 | 0.704 | 0.611 (0.158 - 2.358) |
Values below 0.05 for p denote clinical correlation between variables. OR - odds ratio, CI- confidence interval, ≤ − less than or equal to, > − greater, D - deleted allele, I - insertion allele. No identified mutation (44 patients) – patients without of identified mutation in classes I, II or III. One identified mutation (51 patients) – patient with one identified mutation in class I, II, or III. Two identified mutation (85 patients) – patient with two mutations in class I, II or III.
The ACE*D is associated with a higher gene expression and, consequently, promotes a greater inflammatory response in the body, leading to early symptoms [8,10,12,14,24]. The earliest onset of signs and symptoms are accompanied by early onset of inflammation and deterioration of lung and pancreatic functions. These symptoms are characteristic of severe patients.
An association of the infection/colonization by B. cepacia with ACE gene D/I polymorphism was identified for patients without taking the CFTR mutation into account, OR: 4.509 (1.513 - 10.89), and for patients with one CFTR mutation identified to class I, II or III, OR: (1.43 - 40.38), for the D/D genotype (Table 4).
Table 4.
Association of theACEgene D/I polymorphism, withoutCFTRgenotype distribution and presence ofB. cepacia(BC)
| Group | Ace genotype | Presence | Absence | Total | X2 | p | X2 | p | OR (CI 5-95%) |
|---|---|---|---|---|---|---|---|---|---|
| Without taking CFTR mutation into account |
I/I |
8 |
64 |
72 |
8.654 |
0.013 |
0.468 |
0.498 |
0.699 (0.319 – 1.534) |
| I/D |
9 |
74 |
82 |
0.814 |
0.182 |
0.651 (0.304 – 1.394) |
|||
| D/D |
8 |
16 |
24 |
8,653 |
0.003 |
4.509 (1.513 – 10.89) |
|||
| No identified CFTR mutation |
I/I |
3 |
18 |
21 |
0.530 |
0.767 |
0.003 |
1.29 |
1.056 (0.188 - 5.925) |
| I/D |
2 |
16 |
18 |
0.204 |
1.00 |
0.656 (0.107 - 4.041) |
|||
| D/D |
1 |
3 |
4 |
0.438 |
0.93 |
2.267 (0.196 - 26.27) |
|||
| One CFTR mutation identified class I, II or III |
I/I |
2 |
20 |
22 |
5.539 |
0.063 |
1.248 |
0.466 |
0.383 (0.069 - 2.117) |
| I/D |
2 |
18 |
20 |
0.787 |
0.629 |
0.463 (0.084 - 2.562) |
|||
| D/D |
4 |
5 |
9 |
6.834 |
0.027 |
7.6 (1.43 - 40.38) |
|||
| Two CFTR mutation identified class I, II or III | I/I |
3 |
26 |
29 |
0.511 | 0.774 | 0.084 |
1.07 |
0.8077 (0.193 - 3.387) |
| I/D |
5 |
40 |
45 |
0.039 |
1.10 |
0.875 (0.234 - 3.275) |
|||
| D/D | 2 | 9 | 11 | 0.495 | 0.764 | 1.833 (0.335 - 10.02) |
Values below 0.05 for p denote clinical correlation between variables. OR - odds ratio, CI- confidence interval, D - deleted allele, I - insertion allele. No identified mutation (44 patients) – patients without of identified mutation in classes I, II or III. One identified mutation (51 patients) – patient with one identified mutation in class I, II, or III. Two identified mutation (85 patients) – patient with two mutations in class I, II or III.
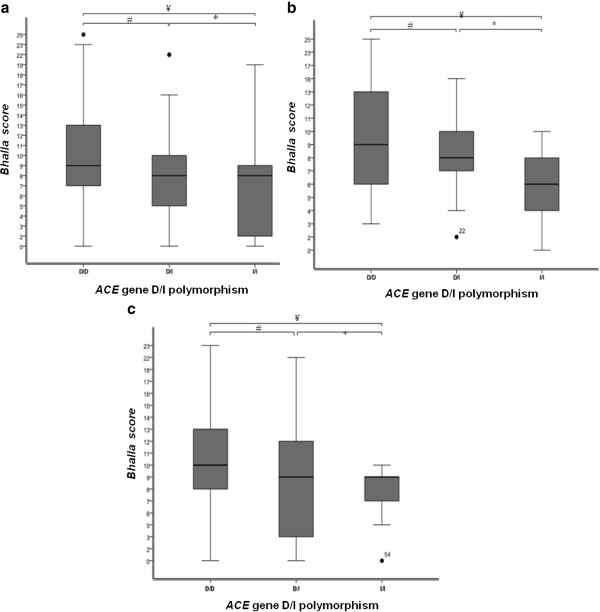
In the analysis of the BS and ACE gene D/I polymorphism, an association was found when no grouping by CFTR genotype occurred (p = 0.015), as well in the subgroup of patients for whom one class I, II and/or III mutation have been identified (p = 0.038), and in the subgroup of patients for whom two class I, II and/or III mutation have been identified (p = 0.042) (Figure 1). There was no difference between BS and the age of patients after categorization. Younger patients (≤ 154 months) had the same distribution of BS as older patients (> 154 months) (p = 0.761). Age is not a variable that contributes to the association between the ACE gene D/I polymorphism and BS. The analysis of an association between the BS and the age of patients with CF was performed in order to show that age had no influence on the score value analysis. We can conclude that the ACE gene D/I polymorphism acts in genetic modulation by association with BS. The BS is a computed tomography score, which measures pulmonary involvement, therapeutic effects and selection of patients for transplantation, which detects anatomical changes of the lung parenchyma [15,25]. The BS has low variation between examiners, good reproducibility, high sensitivity and specificity, and high correlation with pulmonary function test [15]. The values obtained in the score can predict severity associated with deterioration of the structure of the lung parenchyma, which later in clinical evolution can be observed by other variables such as BMI and lung function.
Figure 1.
Association of clinical data, with numerical distribution, withACEgene D/I polymorphism and subgroups ofCFTRmutations. a Bloxplot denoting the association of the ACE gene D/I polymorphism in patients without taking CFTR gene into account. There were significant differences between groups of patients with the genotypes D/D and I/I. # p: 0.092 * p: 0.768, ¥ p: 0.045. b. Bloxplot denoting the association of the ACE gene D/I polymorphism in the subgroup of patients for whom one class I, II and III mutation have been identified. There were significant differences between groups of patients with genotypes D/D and I/I. # p: 0.478, * p: 0.183; ¥ p: 0.043. c. Bloxplot denoting the association of the ACE gene D/I polymorphism in the subgroup of patients for whom two class I, II and/or III mutation have been identified. There were significant differences between groups of patients with genotype D/D and I/I. # p: 0.789 * p: 0.505; ¥ p: 0.05. Analysis was performed by Mann–Whitney test considering a p-value of 0.05 as statistically significant
Evolution of CF is secondary to mutation class in the CFTR gene and environment factors. Many studies have correlated mutations, polymorphisms and clinical variables to CF [5,26]. Association studies commonly face the problem of having insufficient sample size for the number of mutations in the CFTR gene to achieve a homogeneous population and characterize the follow-up of chronic and persistent lung disease [27].
Unlike other genetic diseases such as asthma, CF is monogenic. It was expected that mutations in the CFTR gene would determine the CF severity. Patients with mutations of classes I, II and III have more severe clinical forms than those with mutations IV, V and VI. However, we can observe changes in severity of CF in patients with identical mutations in the CFTR gene [28]. Our study allowed us to characterize the association between the CFTR gene, the environment and one possible CF modifier gene in patients of a Reference University Center, using a statistical method of gene association versus clinical markers.
The main environmental factor in the clinical variability of CF is the patients ´ access to treatment [28]. At our center, treatment is guaranteed by the public health system, which allows equal access for all patients included in the study, and it is not an additional factor in the analysis of data, which is not true in all CF centers in Brazil. Unlike the U.S. where the private system ensures better treatment in CF [29], in Brazil, the public health system is the reference.
Some review articles have suggested a possible modulation of the CF severity by the ACE gene. This fact is based on the proinflammatory property of the ACE protein [2,3,30]. To the best of our knowledge, few studies had characterized the ACE gene as a potential factor in the clinical CF severity [8,30,31]. Bartlett et al. (2009) [31], in a multicenter study, studied the same polymorphism in relation to the propensity for liver disease in patients with CF. They genotyped 124 patients with CF and liver disease and 843 patients with CF and no liver disease. In addition to this polymorphism, four other genes, and their polymorphisms, were analyzed. The polymorphism D/I in the ACE gene was not associated with the presence of liver disease in CF patients, OR: 1.11 (0.85 to 1.44).
Finally, the presence of B. cepacia complex increases inflammation, favoring the exacerbation of immune response, further deterioration of the bronchopulmonary structure and causing rapid deterioration of lung function [32]. More studies to determine whether the presence of the D/D genotype causes increased gene expression and, therefore, facilitates chronic infection by different bacteria are needed. The D/D genotype of the ACE gene D/I polymorphism was significantly associated with higher values of BS. Higher values on the BS are associated with greater clinical severity [15]. Patients with the D/D genotype had higher severity, when compared to patients with the I/I genotype. This data confirms that higher gene expression, given by the D allele, leads to a change in the structure of the lung parenchyma, with subsequent increases in the value of the score.
Our data suggest that ACE gene should be studied in other populations, principally in populations with high prevalence of chronic pulmonary infection by B. cepacia, early onset of clinical manifestations and early onset of severe lung disease (showed by BS).
Patient’s subgroups that were defined on the basis of CFTR mutation analysis may also to be different in comorbidities which may unmask the role of the ACE gene as modifier that was studied in this study, being a limitation of our work.
Conclusion
CF patients with the D/D genotype for the ACE gene D/I polymorphism have a higher risk for chronic infection with BC and deterioration of lung function, characterized by a high BS. There was an association between the presence of the D allele ACE gene and the severity of CF. Further studies are needed to verify the pro-inflammatory activity of this gene in CF, along with a larger CF population with homogeneous CFTR mutation; suggesting that in this case, a multicenter study is necessary.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FALM: made substantial contributions to conception and design, acquisition of data, and analysis and interpretation of data; involved in drafting the manuscript and revising it for critically important intellectual content. TDRH: participated in the design of the study and in the collection of clinical markers. CSB: carried out the molecular genetic studies and drafted the manuscript. AFR: has been involved in drafting the manuscript and revising it critically for important intellectual content. LCB: performed genotyping for CFTR mutation. JDR: has given final approval for the publishing of this version. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Fernando A L Marson, Email: fernandolimamarson@hotmail.com.
Carmen S Bertuzzo, Email: bertuzzo@fcm.unicamp.br.
Taís D R Hortencio, Email: taisdaiene@hotmail.com.
José D Ribeiro, Email: jdirceuribeiro@gmail.com.
Luciana C Bonadia, Email: luciana.bonadia@gmail.com.
Antônio F Ribeiro, Email: anferi@fcm.unicamp.br.
Acknowledgments
Kátia Cristina Alberto Aguiar, Aline Gonçalves and Simoni Avansini – assistance in data collection and organization of ideas. Rodrigo Secolin – Enghish review. Maria Angela Ribeiro – spirometry analysis. We thank Frauke Stanke, Rossella Tomaiuolo and Salmo Raskin the excellent contributions with suggestions, corrections and criticisms that have greatly improved our work.
References
- Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- Davies JC, Griesenbach U, Alton E. Modifier genes in cystic fibrosis. Pediatr Pulmonol. 2005;39:383–391. doi: 10.1002/ppul.20198. [DOI] [PubMed] [Google Scholar]
- Slieker MG, Sanders EA, Rijkers GT, Ruven HJ, van der Ent CK. Disease modifying genes in cystic fibrosis. J Cyst Fibros. 2005;4:7–13. doi: 10.1016/j.jcf.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Belcher CN, Vij N. Protein Processing and Inflammatory Signaling in Cystic Fibrosis: Challenges and Therapeutic Strategies. Current Molecular Medicine. 2010;10:82–94. doi: 10.2174/156652410791065408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culling B, Ogle R. Genetic Counselling Issues in Cystic Fibrosis. Paediatr Respir Rev. 2010;11:75–79. doi: 10.1016/j.prrv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Stanke F, Becker T, Kumar V, Hedtfeld S, Becker C, Cuppens H, Tamm S, Yarden J, Laabs U, Siebert B, Fernandez L, Macek M, Jr, Radojkovic D, Ballmann M, Greipel J, Cassiman JJ, Wienker TF, Tümmler B. Genes that determine immunology and inflammation modify the basic defect of impaired ion conductance in cystic fibrosis epithelia. J Med Genet. 2010. pp. 1–8. [DOI] [PMC free article] [PubMed]
- Arkwright PD, Pravica V, Geraghty PJ, Super M, Webb AK, Schwarz M, Hutchinson IV. End-organ dysfunction in cystic fibrosis: association with angiotensin I converting enzyme and cytokine gene polymorphisms. Am J Respir Crit Care Med. 2003;167:384–389. doi: 10.1164/rccm.200204-364OC. [DOI] [PubMed] [Google Scholar]
- K-Raman P, Krishnan P, Ruiz F, Purushothaman M, Wiley J, Zubatov Y, Kini AS, Sharma SK, Fallon JT, Fuster V, Moreno PR. Increased angiotensin converting enzyme is associated with increased inflammation and neovascularization in peripheral vascular disease: mechanistic role of angiotensin II type I receptor, interleukin-6, and tumor necrosis factor alpha in diabetic atherosclerosis. J Am Coll Cardio. 2010;55:156–163. doi: 10.1016/j.jacc.2009.11.004. [DOI] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-Converting enzyme gene accounting for half the variance of serum levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information – NCBI. Available in: http://www.ncbi.nlm.nih.gov/. Acess in: 04/26/2012.
- Messadi E, Vincent MP, Griol-Charhbili V, Mandet C, Colucci J, Krege JH, Bruneval P, Bouby N, Smithies O, Alhenc-Gelas F, Richer C. Genetically determined angiotensin converting enzyme level and myocardial tolerance to ischemia. FASEB J. 2010;24:4691–4700. doi: 10.1096/fj.10-165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadia LC. Correlação entre aspectos clínicos, moleculares e fisiológicos de pacientes adultos com hipótese diagnóstica de fibrose cística de um centro de referência no Brasil. Phd thesis, University of Campinas, Departement of Genetics; 2011. [Google Scholar]
- Ogus C, Ket S, Bilgen T, Keser I, Cilli A, Gocmen AY, Tosun O, Gumuslu S. Insertion/deletion polymorphism and serum activity of the angiotensin-converting enzyme in Turkish patients with obstructive sleep apnea syndrome. Biochem Genet. 2010;48:516–523. doi: 10.1007/s10528-010-9335-2. [DOI] [PubMed] [Google Scholar]
- Santos CIS, Ribeiro JD, Ribeiro AF, Hessel G. Critical analysis of scoring systems used in the assessment of Cystic Fibrosis severity: state of the art. J Bras Pneumol. 2004;30(3):286–298. [Google Scholar]
- WHO Antro. computer program]. Version 3.0.1. WORLD HEALTH ORGANIZATION, Geneva; 2006. [Google Scholar]
- WHO Antro PLUS. computer program]. Version 1.0.2. WORLD HEALTH ORGANIZATION, Geneva; 2007. [Google Scholar]
- Manual of nutrition. Avaliação nutricional da criança e do adolescente: manual de orientação. Sociedade Brasileira de Pediatria, Departamento de Nutrologia. 2009. pp. 1–107.
- American Thoracic Society (ATS) Disponible in: http://www.thoracic.org/. Acess: 05/11/2011.
- SPSS 17.0 for Windows (computer program) Statistical Package for Social Science (SPSS). Release Version 17.0.1. Incorporation, Chicago (IL): SPSS; 2011. Available from: http:// www.spss.com. [Google Scholar]
- Drăghici S. Data analysis tools for DNA microarrays. Chapman & Hall/CRC, New York; 2003. [Google Scholar]
- Faul F, Erdfelde E, Lang AG, Buchner A. G*Power 3: A flexible sta-tistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Research Methods. 2007;39:175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
- Yugar-Toledo JC, Martin JF, Krieger JE, Pereira AC, Demacq C, Coelho OR, Pimenta E, Calhoun DA, Júnior HM. Gene variation in resistant hypertension: multilocus analysis of the angiotensin 1-converting enzyme, angiotensinogen, and endothelial nitric oxide synthase genes. DNA Cell Biol. 2011;30(8):555–564. doi: 10.1089/dna.2010.1156. [DOI] [PubMed] [Google Scholar]
- Mehri S, Baudin B, Mahjoub S, Zaroui A, Bénéteau-Burnat B, Mechmeche R, Hammami M, Ben Arab S. Angiotensin-converting enzyme insertion/deletion gene polymorphism in a Tunisian healthy and acute myocardial infarction population. Genetic Testing and Molecular Biomarkers. 2010;14(1):85–91. doi: 10.1089/gtmb.2009.0105. [DOI] [PubMed] [Google Scholar]
- Albi G, Rayón-Aledo JC, Caballero P, Rosado P, García-Esparza E. Cystic fibrosis in images: the Bhalla scoring system for computed tomography in paediatric patients. Radiologia. 2011. in print. [DOI] [PubMed]
- Drumm ML, Ziady AG, Davis PB. Genetic variation and clinical heterogeneity in cystic fibrosis. Annu Rev Pathol. 2012;7:267–82. doi: 10.1146/annurev-pathol-011811-120900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston RS, Tomlinson IP. Modifier genes in humans: strategies for identification. Eur J Hum Gen. 1998;6:80–88. doi: 10.1038/sj.ejhg.5200156. [DOI] [PubMed] [Google Scholar]
- Bush A. Genes in their environment: how can we read the riddles? J Pediatr. 2008;84(3):185–188. doi: 10.2223/JPED.1789. [DOI] [PubMed] [Google Scholar]
- Schechter MS. Non-genetic influences on CF lung disease: the role of sociodemographic characteristics, environmental exposuresand healthcare interventions. Pediatr Pulmonol Suppl. 2004;26:82–85. doi: 10.1002/ppul.70061. [DOI] [PubMed] [Google Scholar]
- Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM, Darrah RJ, Dorfman R, Sandford AJ, Corey M, Zielenski J, Durie P, Goddard K, Yankaskas JR, Wright FA, Knowles MR. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med. 2005;353(14):1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- Bartlett JR, Friedman KJ, Ling SC, Pace RG, Bell SC, Bourke B, Castaldo G, Castellani C, Cipolli M, Colombo C, Colombo JL, Debray D, Fernandez A, Lacaille F, Macek M Jr, Rowland M, Salvatore F, Taylor CJ, Wainwright C, Wilschanski M, Zemková D, Hannah WB, Phillips MJ, Corey M, Zielenski J, Dorfman R, Wang Y, Zou F, Silverman LM, Drumm ML, Wright FA, Lange EM, Durie PR, Knowles MR. Genetic modifiers of liver disease in cystic fibrosis. JAMA. 2009;302:1076–1083. doi: 10.1001/jama.2009.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Jang TN, Liu CY, Fung CP, Yu KW, Wong WW. Characteristics of patients with Burkholderia cepacia bacteremia. J Microbiol Immunol Infec. 2001;34:215–219. [PubMed] [Google Scholar]