Abstract
l(2)01810 causes glutamine-dependent megamitochondrial formation when it is overexpressed in Drosophila cells. In the present study, we elucidated the function of l(2)01810 during megamitochondrial formation. The overexpression of l(2)01810 and the inhibition of glutamine synthesis showed that l(2)01810 is involved in the accumulation of glutamate. l(2)01810 was predicted to contain transmembrane domains and was found to be localized to the plasma membrane. By using 14C-labelled glutamate, l(2)01810 was confirmed to uptake glutamate into Drosophila cells with high affinity (Km =69.4 µM). Also, l(2)01810 uptakes glutamate in a Na+ -independent manner. Interestingly, however, this uptake was not inhibited by cystine, which is a competitive inhibitor of Na+ -independent glutamate transporters, but by aspartate. A signal peptide consisting of 34 amino acid residues targeting to endoplasmic reticulum was predicted at the N-terminus of l(2)01810 and this signal peptide is essential for the protein’s localization to the plasma membrane. In addition, l(2)01810 has a conserved functional domain of a vesicular-type glutamate transporter, and Arg146 in this domain was found to play a key role in glutamate transport and megamitochondrial formation. These results indicate that l(2)01810 is a novel type of glutamate transporter and that glutamate uptake is a rate-limiting step for megamitochondrial formation.
Keywords: glutamate transporter, l(2)01810, megamitochondrion, nutritional stress, signal peptide
INTRODUCTION
Megamitochondria are enlarged round mitochondria which are formed either by fusion of normal mitochondria or by inhibition of cleavage during mitochondrial division [1,2]. When compared with normal mitochondria, megamitochondria manifest unique characteristics in addition to their enlarged size, such as unfolded and shortened cristae that are confined to the membrane periphery and dense granules that are scattered throughout the enlarged matrix [3].
Formation of megamitochondria is induced by various conditions such as treatment with drugs, hormonal disturbance, malnutrition, high protein diet, ethanol intoxication, virus infection, and even by exercise [2]. Interestingly, megamitochondria were also found in the brown adipose cells of hibernating animals [4].
Recently, it was found that the increase of l(2)01810 expression by either the knockdown of SPS1 (selenophosphate synthetase 1) or by the overexpression of l(2)01810 in Drosophila cells resulted in glutamine-dependent megamitochondrial formation [5]. SPS1 knockdown induced the increase of both l(2)01810 and GS1 (glutamine synthetase 1), which led to the accumulation of intracellular glutamine. GS1 is responsible for the conversion of glutamate into glutamine [6]. However, the function of l(2)01810 with regard to megamitochondrial formation has not been determined. l(2)01810 was originally identified during genome-wide P element insertion mutagenesis. This gene is located on Drosophila chromosome 2. When it was knocked out by P element insertion, the mutant showed lethality during embryogenesis, suggesting that it is an essential gene [7].
Most intracellular glutamate residues are supplied from the diet and are taken up from the outside of cells by GLUTs (glutamate transporters). GLUTs can be classified into two types according to their subcellular localization: plasma-membrane-type and intracellular-type [8]. GLUTs of the plasma-membrane-type take up extracellular glutamate into the cytoplasm of a cell. EAATs (excitatory amino acid transporters) and cystine/glutamate transporters are typical plasma-membrane-type GLUTs. EAATs have a high affinity for glutamate and their activities are Na+ -dependent. Cystine/glutamate transporters are antiporters that normally take up cystine and release glutamate, but when the concentration of extracellular glutamate is high, glutamate can be taken up with a high affinity in a Na+ -independent manner [9,10]. There are two types of intracellular GLUTs, vesicular and mitochondrial GLUTs. These proteins are localized in the membrane of vesicles or mitochondria and play a role in the redistribution of glutamate residues present in the cytoplasm [8]. VGLUTs (vesicular GLUTs) have a low affinity to glutamate and their functions are Na+ -independent [8]. The MGLUTs (mitochondrial glutamate transporters) are a subfamily of MCs (mitochondrial carriers) and transport cytoplasmic glutamate residues into mitochondria [11].
In the present study, we examined the molecular and biochemical properties of l(2)01810 and found that it is a novel type of GLUT. We also found that the increase of intracellular glutamate levels led to glutamine-dependent megamitochondrial formation. Our results suggest that the high glutamate level can serve as a signal for megamitochondrial formation.
EXPERIMENTAL
Materials
Materials were purchased from the following sources: Drosophila SL2 (Schneider cell line 2), antibiotics and antimycotics from Invitrogen; HyQ SFX-Insect medium from Hyclone; MitoTracker Red from Molecular Probes; dimethyldioctadecyl ammonium bromide, methionine sulfoximine, concanavalin A, glutamine/glutamate determination kit (GLN-1), mouse anti-tubulin, mouse anti-HA (haemagglutinin) and goat anti-mouse IgG from Sigma; Hybond-ECL nitrocellulose membrane from Amersham Biosciences; restriction enzyme from Beams-Bio; TRIzol reagent, Pfx DNA polymerase and protein size marker from Invitrogen; L-[U-14C] glutamate and 32Pi from PerkinElmer; Lab-Tek chambered coverglass from Nunc; and oligonucleotides from Cosmo Genetech.
Vector construction
To express l(2)01810–GFP (green fluorescent protein), dVGLUT–GFP and dEAAT1–GFP fusion proteins, in which GFP is tagged at their C-terminus, the HA portion of pAcl(2)01810-HA, which contains the l(2)01810 sequence originating from the SL2 cell cDNA sequence (GenBank® accession number JF411011) [5], was replaced with the GFP ORF (open reading frame) using BamHI and NotI sites. The vector was designated pAcl(2)01810-GFP. pAcdVGLUT-GFP and pAcdEAAT1-GFP were constructed by replacing the l(2)01810 ORF in pAc1(2)01810-GFP with dVGLUT and dEAAT1 ORF respectively, using the following specific primers: dEAAT1, 5′-aagcttgctagccaccatgacgcgacccaaacagga-3′ and 5′-aattccggatcctccttcatctcgtggccat-3′; and for dVGLUT, 5′-aagcttgctagccaccatgaagggtctgacggcgtt-3′ and 5′-aagcttagatcttgctgctggtatccctgcgg-3′.
To express the l(2)01810 signal peptide and GFP fusion protein, the l(2)01810 ORF of pAcl(2)01810-GFP was replaced with the putative signal peptide of l(2)01810 prepared by PCR using Pfx DNA polymerase and the primers 5′-aagcttgctagccaccatgacgcaacagccacaatg-3′ and 5′-aagcttggatccgtgtaggcattcaggatgg-3′, at the NheI and BamHI sites. The vector was designated pAcl(2)01810SP-GFP (where SP is signal peptide). To express the l(2)01810 SP deletion mutant, DNA fragments containing amino acids 34–496 of l(2)01810 and the start codon at the 5′ end were prepared by PCR using the primers 5′-aagcttgctagccaccatgtacacgatgcgcgtctgtct-3′ and 5′-aagcttggatccgagctctgctccagaccac-3′ and then the ORFs of l(2)01810 in pAcl(2)01810-HA and pAcl(2)01810-GFP were replaced by the truncated fragment using NheI and BamHI sites. The vectors were designated pAcΔSPl(2)01810-HA and pAcΔSPl(2)01810-GFP respectively.
Site-directed mutagenesis was performed at Arg146 and Glu153, which are located in the putative domain of l(2)01810, by employing PCR mutagenesis methods using the following primers: for R146A, 5′-tgtgaccgcagttctgatgg-3′ and 5′-ccatcagaactgcggtcaca-3′, and for E153A, 5′-ggtctgggcgcgggaaccac-3′ and 5′-gtggttcccgcgcccagacc-3′. The resulting fragments were inserted into pAcl(2)01810-HA by replacing the l(2)01810 ORF with the DNA fragment and the sequences were confirmed by nucleotide sequencing. The resulting vectors were designated pAcl(2)01810R146A-HA and pAcl(2)01810E153A-HA respectively.
SL2 cell culture, DNA transfection and reagent treatment
SL2 cell culture and DNA transfection of SL2 cells were performed as described previously [5,12] with minor modifications. Briefly, in a single transfection experiment, 2 µg of vector was mixed with 100 µl of dimethyldioctadecyl ammonium bromide (125 µg/ml) and 200 µl of HyQ SFX-Insect medium, incubated for 30 min at room temperature (20°C), and then added to a well of a six-well plate containing 6×106 cells and the cells were incubated for 48–72h at 25°C. In co-transfection experiments, 4 µg of PDI (protein disulfide-isomerase)–DsRed {ER (endoplasmic reticulum) marker protein [13]} and 2 µg of pAcGFP or pAcl(2)01810SP-GFP were used. For inhibition of glutamine synthetase activity in SL2 cells, cells were incubated with medium containing 3 mM MSO (methionine sulfoximine) for 3 days.
Mitochondrial staining and confocal microscopy
Mitochondrial staining and confocal microscopy were carried out as described previously [5] with minor modifications. Briefly, 0.5×106 cells were plated on to a chambered coverglass 1 day before staining. SL2 cells were incubated with 1 µM MitoTracker Red for 30 min at 25°C and then washed three times with HyQ SFX-Insect medium. To determine subcellular localization, 1 day after transfection of SL2 cells with vectors that can express GFP fusion proteins, the cells were transferred on to a chambered coverglass. For observation of ER localization, co-transfected cells with a vector that can express DsRed or GFP fusion protein were transferred on to concanavalin A-coated chambered coverglasses. Cells were then observed with a LSM510 confocal microscope (Carl Zeiss) at a 512×512 pixel resolution through an ×63 C-Apochromat objective. Excitation wavelengths were 543 nm for MitoTracker Red and DsRed, and 488 nm for GFP.
Intracellular glutamine and glutamate measurement
Glutamine and glutamate measurements were carried out as described previously [5] with minor modifications. At 3 days after transfection, 3–5×107 cells were harvested, washed with ice-cold PBS and extracted with 10% perchloric acid, followed by centrifugation at 3000 g for 3 min. After neutralizing the supernatants with 5 M KOH, extracts were incubated for 10 min on ice followed by centrifugation at 15000 g for 3 min. Glutamine and glutamate levels were determined in the final supernatants using the glutamine/glutamate determination kit (Sigma) according to the manufacturer’s instructions. Glutamine and glutamate levels were expressed in nmoles/107 cells.
Glutamate- and phosphate-uptake assay
The glutamate-uptake assay was performed as described previously [14] with minor modifications. At 2days after transfection with 2 µg of an appropriate vector, cells were gently washed three times with Hanks balanced salt solution containing 137.9 mM NaCl, 5.33 mM KCl, 0.37 mM Na2HPO4, 0.44 mM KH2PO4 and 5.6 mM glucose that had been adjusted to pH 6.8 and incubated with 1 ml of the same medium. l-glutamate (0.1 mM) and 0.165 µCi/ml of l-[U-14C] glutamate were added to the medium. Incubation was stopped after 10 min by removing the medium and rinsing the cells three times with ice-cold Hanks balanced salt solution. For kinetic studies, glutamate-uptake rates were assessed over a concentration range of 6.25 µM to 1.6 mM unlabelled glutamate plus 0.165 µCi/ml l-[U-14C] glutamate. To determine Na+ -dependency, glutamate uptake was measured in either standard Hanks balanced salt solution, which contains Na+, or Na+ -free medium prepared by replacing NaCl with choline chloride in Hanks balanced salt solution. To measure the inhibitory effect of competitors on glutamate uptake, the uptake assay was performed in medium containing 2 µM l-glutamate and 0.5 mM of competitors such as l-aspartate, l-cystine or l-glutamine.
The phosphate uptake assay was performed as described previously [15] with minor modifications. Cells were gently washed three times with prewarmed (25°C) phosphate uptake solution (137 mM NaCl, 5.4 mM KCl, 2.8 mM CaCl2, 1.2 mM MgCl2 and 10 mM Hepes/Tris, pH 6.8) and incubated with 1 ml of the solution. KH2PO4 (0.1 mM) and 1 µCi/ml 32Pi were added to the medium. After 10 min, the solution was removed and the cells rinsed three times with ice-cold phosphate uptake solution. Cells were harvested and lysed with 0.5 M NaOH. The radioactivities of lysates were measured with a Tri-Carb 2900TR liquid scintillation analyser (PerkinElmer) and protein concentrations of the lysates were measured using the Bradford method [16]. The uptake of glutamate and phosphate are expressed in nmol/mg of protein per min.
Western blotting
Western blot analysis was performed as described previously [17] with minor modifications. Briefly, 2days after transfection, cells were harvested and lysed for 30 min on ice with RIPA lysis buffer [50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1%(v/v) Nonidet P40, 0.5% sodium deoxycholate and 0.1% SDS, containing 0.1% protease inhibitor mixture (Sigma; P2714)]. The lysates were centrifuged at 15000 g for 15 min and the protein amounts in the supernatants were measured as described above. Proteins (20 µg) were separated by SDS/PAGE (10%gels) and transferred to a Hybond-ECL nitrocellulose membrane. The membrane was blocked with TBST (50 mM Tris/HCl, pH 7.5, 150 mM NaCl and 0.1% Tween 20) containing 5% (w/v) non-fat dried skimmed milk powder for 1 h at room temperature and then incubated with an anti-HA antibody (1:20 000) or anti-tubulin antibody (1: 200000) diluted with 5% (w/v) non-fat dried skimmed milk powder in TBST for 1 h at room temperature. After washing with TBST, the membrane was incubated with goat anti-mouse IgG (1:10 000) for 1 h at room temperature.
Bioinformatical analyses
To predict the localization and membrane topology, TMHMM, HMMTOP and PSORT II software were used as described previously [13]. The prediction of conserved domains was performed by searching NCBI’s CDD (conserved domain database; [18]) and using the Motif scan program [19]. SP or signal anchor were predicted by using Signal P 3.0 software [20]. To compare the characteristics among different types of GLUTs or their homologues in different model organisms [Dmel (Drosophila melanogaster), Cele (Caenorhabditis elegans), Drer (Danio rerio), Mmus (Mus musculus), Rnor (Rattus norvegicus) and Hsap (Homo sapiens)], the protein sequences were retrieved from the NCBI database; Dmel Eaat1 (NP_723454), Cele glt-5 (NP_496094), Drer SLC1a3 (solute carrier 1a3; NP_997805), Mmus Eaat1 (NP_683740), Hsap Eaat1 (NP_004163), Cele glt-1 (NP_001024393), Drer Eaat2 (NP_956273), Mmus Eaat2 (NP_035523), Hsap Eaat2 (NP_004162), Dmel Eaat2 (NP_001162844), Dmel genderblind (NP_651536), Cele aat-1 (NP_501707), Drer SLC7a11-like (XP_001919426), Mmus xCT (NP_036120), Hsap xCT (NP_055146), Dmel l(2)01810 (NP_620115), Dmel VGLUT (NP_608681), Cele eat-4 (NP_499023), Drer SLC17a7 (NP_001092225), Hsap VGLUT1 (NP_064705), Rnor VGLUT1 (NP_446311), Dmel arala1 (NP_733365), Cele Q21153 (NP_497274), Drer SLC25a12 (NP_997947), Mmus aralar1 (NP_766024) and Hsap aralar1 (NP_003696). All amino acid sequences in FASTA format were aligned using ClustalW v.1.8 and the alignment was bootstrapped (1000 replicates; seed = 1000) and phylogenic trees were constructed by UPGMA method using MEGA v.4.0 software [21].
Statistical analysis
Experiments for the measurements of glutamine and glutamate levels, and glutamate uptake were performed in triplicate. Statistical analyses were performed with the aid of the statistical package program GraphPad Prism 4. To determine the statistical significance of the observed changes in the groups of data, unpaired Student’s t test or one-way ANOVA test [significance level (α) = 0.05] followed by Tukey’s multiple comparison test were performed.
RESULTS
l(2)01810 participates in the increase of the intracellular glutamate level
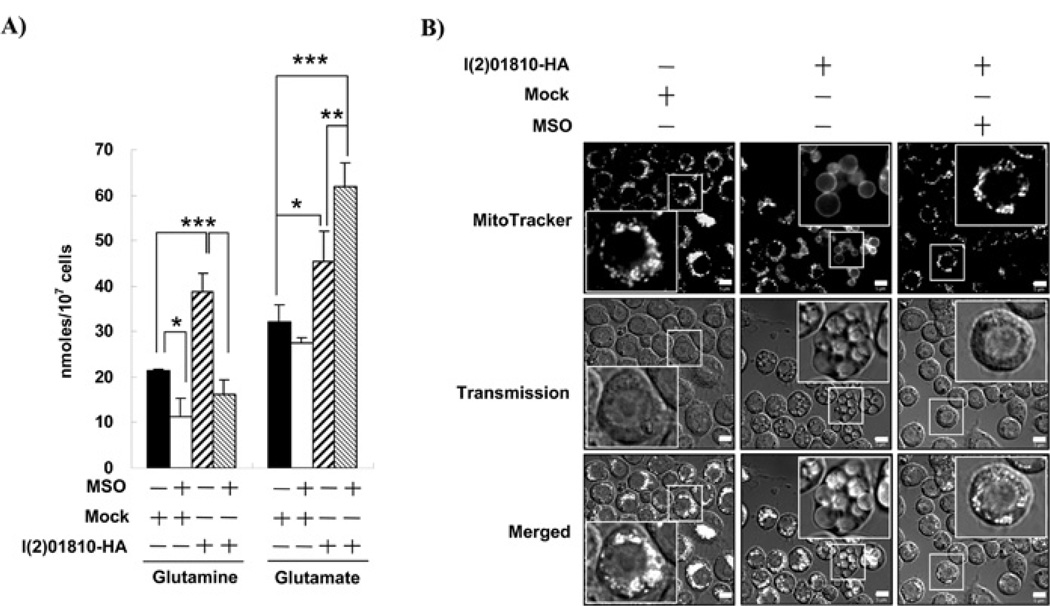
To assess the role of l(2)01810 in the accumulation of glutamine, l(2)01810 was overexpressed in SL2 cells and the levels of glutamine and glutamate were measured. As shown in Figure 1(A), the glutamine level in the l(2)01810-overexpressing cells was approximately 2-fold higher than that in mock-transfected control cells. However, treatment with MSO, an inhibitor of GS1, significantly decreased the level of glutamine in l(2)01810-overexpressing cells (more than 2-fold, P < 0.001). In addition, the glutamine levels in mock-transfected control cells also decreased approximately 2-fold by MSO treatment (P < 0.05). These results suggest that the increase in the intracellular glutamine level was dependent on GS1 activity. The overexpression of l(2)01810 increased glutamate levels in SL2 cells by approximately 1.5-fold compared with that in the mock-transfected control cells (P < 0.05). Interestingly, MSO treatment did not decrease the glutamate levels, but rather increased it by 1.3-fold in the l(2)01810-overexpressing cells (P < 0.01). These results clearly indicate that l(2)01810 is involved in the process of glutamate accumulation. Since glutamate is used as a precursor during glutamine synthesis, the inhibition of GS1 activity should result in accumulation of its substrate in the cell. It should be noted that MSO treatment in control cells transfected with the backbone vector did not increase glutamate levels (see Figure 1A). Therefore it can be concluded that the increase of glutamate levels in the MSO-treated cells where l(2)01810 was overexpressed was solely dependent on the increase of l(2)01810 expression. These results strongly suggest that l(2)01810 is involved in the accumulation of glutamate in the cell.
Figure 1. l(2)01810 regulates intracellular glutamate levels.
(A) Effect of l(2)01810 overexpression on intracellular glutamine and glutamate levels. After transfection with a vector that can express l(2)01810, cells were incubated with or without MSO and the levels of glutamine and glutamate were measured (see the Experimental section). The vectors and MSO are shown on the x axis. Mock represents the cells transfected with backbone vector and used as a control. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison test. *, ** and *** indicate significance at P < 0.05, 0.01 and 0.001 respectively. (B)Megamitochondrial formation induced by l(2)01810 overexpression is dependent only on the glutamine level. Cells were transfected with backbone (designatedmock) or l(2)01810-overexpressing vector and incubated in the absence or presence of MSO for 3 days. Confocal microscopy was carried out following MitoTracker Red staining. Scale bars represent 5 µm. Examples of normal mitochondria and megamitochordia are boxed within the respective panels and are then magnified 6-fold in the larger boxes to clearly show normal mitochondria or megamitochondria.
As reported previously, the intracellular glutamine level is closely related to megamitochondrial formation and the overexpression of l(2)01810 induced megamitochondrial formation [5]. We also examined whether there is any relationship between glutamate levels and megamitochondrial formation. Interestingly, megamitochondrial formation induced by l(2)01810 overexpression was inhibited by MSO treatment (Figure 1B). This result indicates that megamitochondrial formation is dependent only on the intracellular glutamine level, but the high level of glutamate in the cell can induce the conversion of glutamate into glutamine byGS1 activity leading to the increase of glutamine levels.
l(2)01810 is a high-affinity GLUT
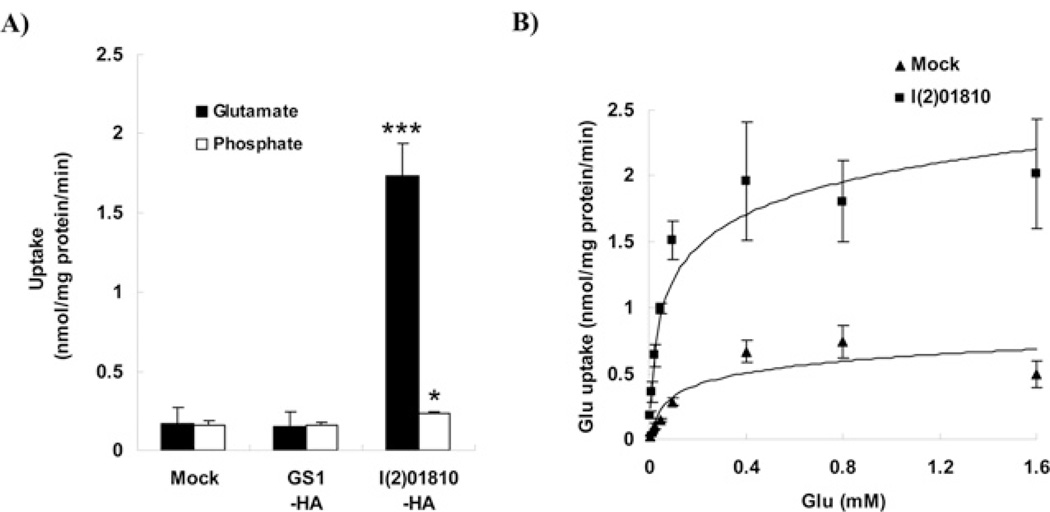
Glutamate can accumulate in the cell either by increasing its synthesis or by increasing its uptake. Therefore it is necessary to determine whether l(2)01810 is responsible for glutamate synthesis or for transport. To answer this question, we searched for proteins that have high sequence homology with l(2)01810. After a BLAST search of the NCBI database, a phylogenic tree against the homologous proteins was constructed. As shown in Supplementary Figure S1 (at http://www.BiochemJ.org/bj/439/bj4390277add.htm), the proteins annotated as ‘sodium-dependent phosphate transporter’ showed the highest homology (70–80% in similarity) with l(2)01810. Considering that l(2)01810 is involved in glutamate accumulation and that it has high sequence homology with phosphate transporters, we hypothesized that the function of l(2)01810 is to transport glutamate into a cell. To test this hypothesis, we measured glutamate-uptake activity in cells that overexpress l(2)01810. After transfection of SL2 cells with a vector expressing l(2)01810, [14C]glutamate was added to the medium and the radioactivity transported into the cell was measured (see the Experimental section).As shown in Figure 2(A), the glutamate uptake in the l(2)01810-overexpressing cells (1.73 nmol/mg of protein per min) was increased 11-fold compared with that in control cells (0.16 nmol/mg of protein per min). Since l(2)01810 has high sequence homology with Na+ -dependent phosphate transporters, we examined whether it has phosphate transporter activity. However, the phosphate uptake in l(2)01810-overexpressing cells (0.23 nmol/mg of protein per min) did not change significantly compared with that in control cells (0.16 nmol/mg of protein permin), even in theNa+ -containing medium. These results indicate that l(2)01810 functions as a GLUT, not as a phosphate transporter, even though l(2)01810 has high sequence homology with phosphate transporters.
Figure 2. l(2)01810 is a high-affinity GLUT.
(A) l(2)01810 has glutamate, but not Pi, uptake activity. At 2 days after transfection with the plasmids indicated on the x axis, glutamate and phosphate uptake were measured as described in the Experimental section. Closed and open bars represent uptake levels of glutamate and phosphate respectively. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison test. * and *** indicate significance at P < 0.05 and 0.001 respectively. (B) Dose–response curves for glutamate uptake. ▲ and ■ designate mock (backbone vector) and pAcl(2)01810-transfected SL2 cells respectively. The curves were obtained by fitting the Michaelis–Menten function to the data.
Since GLUTs can be classified into two types according to their affinity to glutamate (low-affinity and high-affinity GLUTs), we examined the affinity of l(2)01810 to glutamate. Kinetic parameters such as Km and Vmax were measured by performing dose–response experiments in l(2)01810-overexpressing cells. As shown in Figure 2(B), the glutamate uptake occurred in a dose-dependent manner in both control and l(2)01810-overexpressing cells and glutamate uptake was saturated at a high concentration (400 µM). Lineweaver–Burk analysis was used to determine Vmax and Km values. The Vmax in l(2)01810-overexpressing cells and in the control cells were 2.64 nmol/mg of protein per min and 0.8 nmol/mg of protein per min respectively. The Km value in l(2)01810-overexpressing cells was 69.4 µM. GLUTs such as EAATs are known as high-affinity GLUTs and their Km values are 48–97 µM in EAAT-overexpressing cells [22], but those of low-affinity GLUTs are greater than 0.5 mM [23]. Since the Km value in l(2)01810-overexpressing cells is similar to that in EAAT-overexpressing cells, it can be concluded that l(2)01810 is a high-affinity GLUT.
l(2)01810 is localized to the plasma membrane
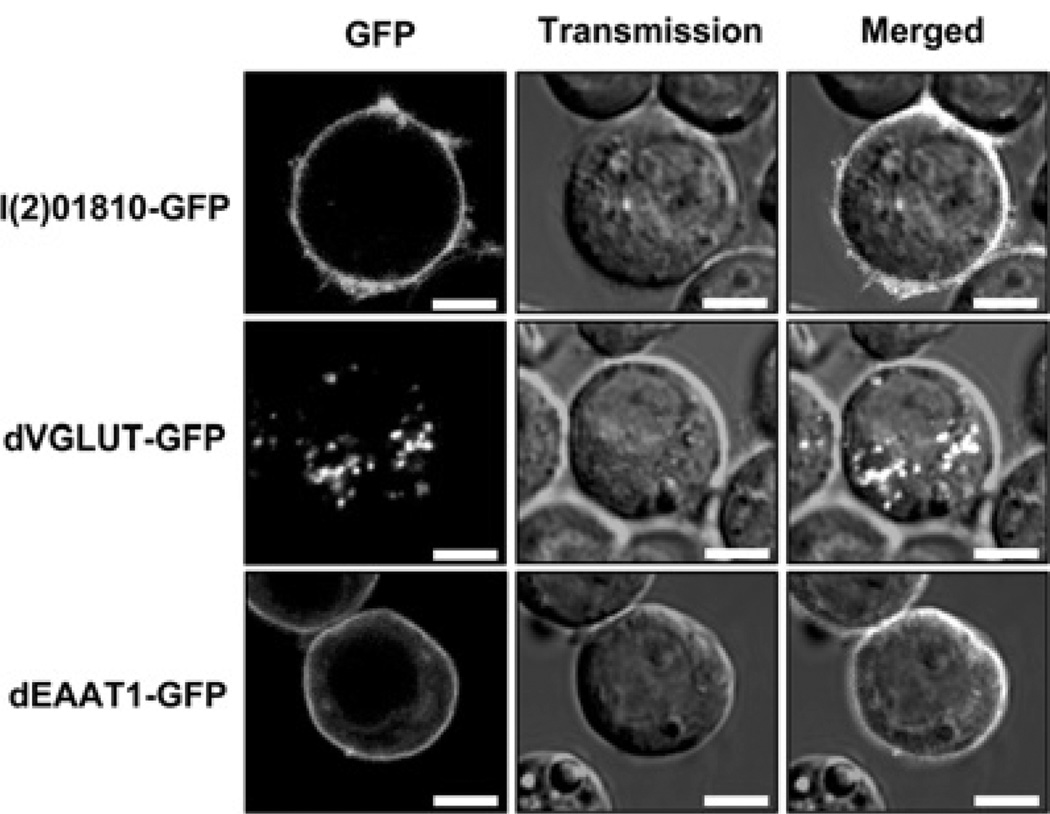
As described in the Introduction, GLUTs can be classified into either plasma-membrane-types or intracellular-types according to their subcellular localization [8]. To determine the subcellular localization of l(2)01810, we first analysed the amino acid sequence of l(2)01810 by employing bioinformatical methods. Membrane topology analysis using PSORT II, TMHMM and HMMTOP software predicted with high probability that l(2)01810 contains 9–11 transmembrane domains, suggesting it is a plasma membrane protein. To confirm this prediction, SL2 cells were transfected with a vector that can express the l(2)01810–GFP fusion protein and the subcellular localization of l(2)01810 was examined under a confocal microscope. As shown in Figure 3, l(2)01810 was found to be localized on the plasma membrane and this pattern is similar to that of dEAAT, which has been known to be localized on the plasma membrane. A minor difference was found between l(2)01810 and dEAAT. Although dEAAT forms smooth ring structures along with the plasma membrane, l(2)01810 forms fuzzy ring structures on the cell surface. In contrast with dEAAT, dVGLUTs were localized to the intracellular vesicles. The dVGLUT result is consistent with observations made by another group [24]. It should be noted that subcellular localization of l(2)01810 is more similar to that of dEAAT than that of dVGLUT, which has a higher sequence homology with l(2)01810 than dEAAT (see the Discussion).
Figure 3. Subcellular localization of l(2)01810.
SL2 cells were transfected for 2 days with l(2)01810, dVGLUT and dEAAT1 expression vectors with GFP tagged at the C-terminus of each protein and the cells were examined under a confocal microscope. Scale bar = 5 µm.
SP of l(2)01810 is important for its localization and function
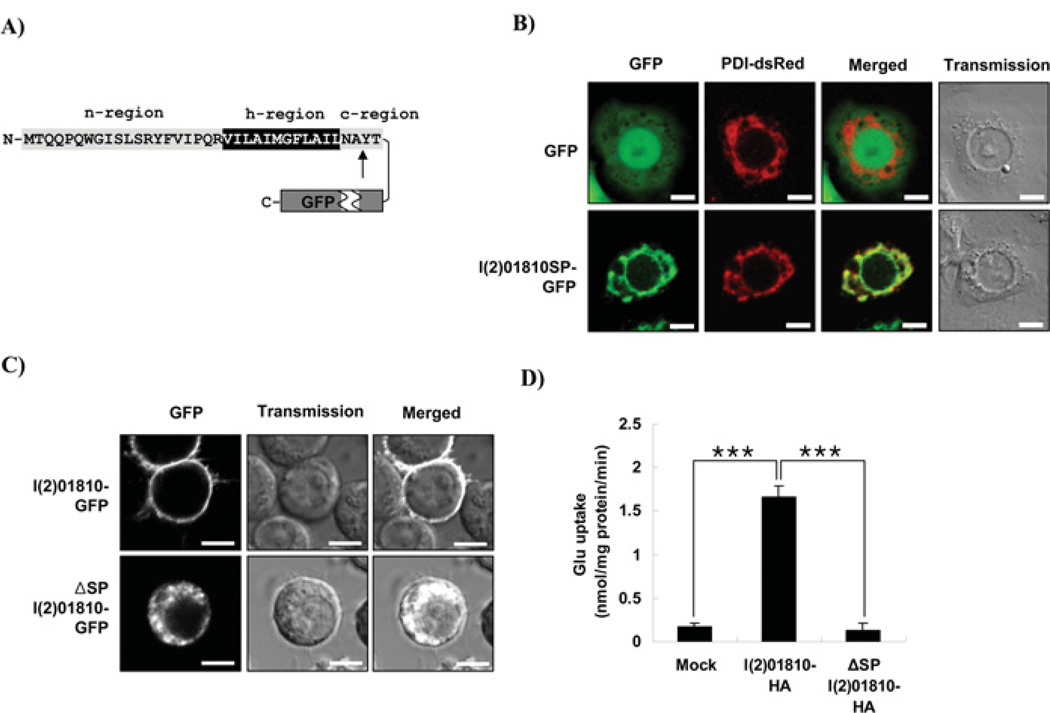
Since l(2)01810 is localized on the plasma membrane, there is a high possibility that it contains a SP (or signal sequence) for its accurate targeting and membrane insertion [25,26]. By using Signal P software, we predicted that l(2)01810 has a SP at its N-terminus (amino acid position 1–34). To test whether the predicted SP has the capability to move its cargo protein to the ER, a vector that can express l(2)01810 SP–GFP fusion protein [l(2)01810SP–GFP, see Figure 4A] was transfected into SL2 cells and the subcellular localization of this protein was observed under a confocal microscope. As shown in Figure 4(B), l(2)01810SP–GFP fusion proteins were observed in the structures which were completely overlapped with those stained with an ER marker (PDI–DsRed), suggesting that the SP of l(2)01810 facilitates GFP to be expressed on the membrane of the ER and the GFP is localized in the ER (lower panel). However, the control GFP is localized throughout the cell (upper panel). The SP-deleted mutant [ΔSP l(2)01810–GFP] lost its ability to localize in the plasma membrane (Figure 4C). The GLUT activity of the mutant where the signal peptide was deleted [ΔSP l(2)01810-HA] was lost completely (Figure 4D). These results suggest that the SP is required for trafficking and that the localization of l(2)01810 to the plasma membrane is essential for its activity.
Figure 4. Effect of putative SP of l(2)01810 on its localization and function.
(A) Schematic illustration of the l(2)01810 SP–GFP fusion protein. The tripartite structure (n-, h- and c-region) of the l(2)01810 signal peptide predicted by the Signal P program is represented. The predicted cleavage site by signal peptidase 1 at the position between amino acids 34 and 35 is indicated by an arrow. N- and -C indicated the N- and C-terminus respectively. (B) Subcellular localization of l(2)01810SP–GFP. After 2 days of co-transfection of GFP or l(2)01810SP–GFP with PDI–DsRed into SL2 cells, the cells were transferred on to a concanavalin A-coated chamber slide and observed under a confocal microscope. Scale bars represent 5 µm. (C) Subcellular localization of l(2)01810 SP deletion mutants [ΔSP l(2)01810]. After 2 days of transfection, GFP fluorescence was observed using a confocal microscope. Scale bars = 5 µm. (D) Glutamate uptake by l(2)01810 SP deletion mutant. After 2 days of transfection, glutamate-uptake rates were measured. Mock represents the cells transfected with backbone vector and used as a control. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison test. ***P < 0.001.
l(2)01810 is a novel type of Na+ -independent GLUT
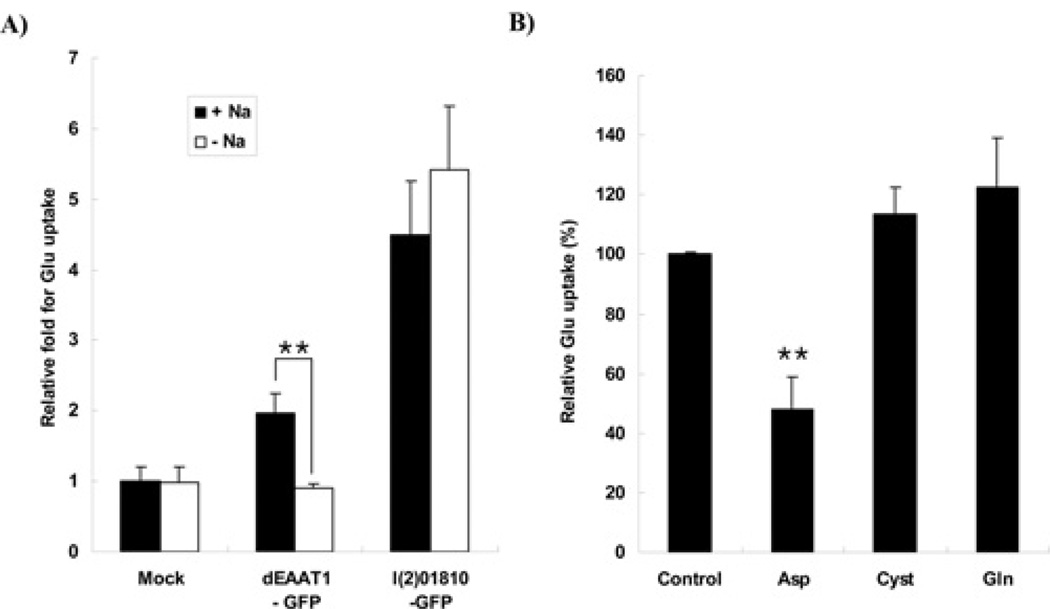
Since the plasma-membrane-type GLUTs are either Na+ -dependent or independent for their activity [27], we examined the effect of Na+ on the activity of l(2)01810. As described in the Experimental section, the cells overexpressing l(2)01810 were measured for their glutamate-uptake activities in Na+ -free medium or standard uptake medium. Glutamate uptake by l(2)01810 in Na+ -free medium was similar to that in standard uptake medium suggesting that glutamate transport by l(2)01810 is Na+ -independent (Figure 5A). It should be noted that the HA-tagged l(2)01810 also showed Na+ -independent GLUT activity (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/439/bj4390277add.htm). However, this property is reversed in dEAAT1, which is a plasma-membrane-type of GLUT. Glutamate uptake was decreased to background when measured in Na+ -free medium indicating that dEAAT1 is a Na+ -dependent enzyme, and this result is consistent with that reported by a previous study [28].
Figure 5. l(2)01810 is a Na+ -independent GLUT which can be inhibited by aspartate.
(A) l(2)01810 is a Na+ -independent GLUT. After 2 days of transfection, glutamate-uptake rates were measured as described in the Experimental section. Closed and open bars indicate Na+ -containing and Na+ -free medium respectively. The y axis represents the relative glutamate uptake of each sample compared with the mock-transfected cells in Na+ -containing medium. Glutamate uptake by the mock-transfected cells was set to 1. The vectors used are shown on the x axis. Mock represents the cells transfected with backbone vector and used as a control. Statistical significance was tested by unpaired Student’s t test. **P < 0.01. (B) Inhibitory effect of potential competitors on glutamate uptake. After 2 days of transfection with l(2)01810-expressing vector, the glutamate-uptake assay was performed. The glutamate uptake of control cells (no competitor) was set to 100%. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison test. **P < 0.01.
Since the cystine/glutamate transporter, which is a plasma-membrane-type GLUT, has been known to be Na+ -independent and its activity is inhibited by l-cystine (see [8] and references therein), we also examined whether glutamate-transport activity was inhibited by l-cystine. After pAcl(2)01810-HA was transfected, a competitive inhibition assay for glutamate uptake was performed by treating the cells with various potential competitive inhibitors (see the Experimental section for details). Surprisingly, l-cystine and l-glutamine did not inhibit glutamate uptake by l(2)01810 (Figure 5B). On the other hand, l-aspartate (0.5 mM) inhibited glutamate uptake to 52% (P < 0.01), and this characteristic is common with that of EAATs. These results show that l(2)01810 has unique biochemical characteristics: Na+ -dependency of l(2)01810 is similar to that of cystine/glutamate transporters, but different from that of EAATs; both l(2)01810 and EAATs can be competitively inhibited by l-aspartate, but cystine/glutamate transporters can be inhibited by l-cystine. Therefore it can be concluded that l(2)01810 is a novel type of GLUT.
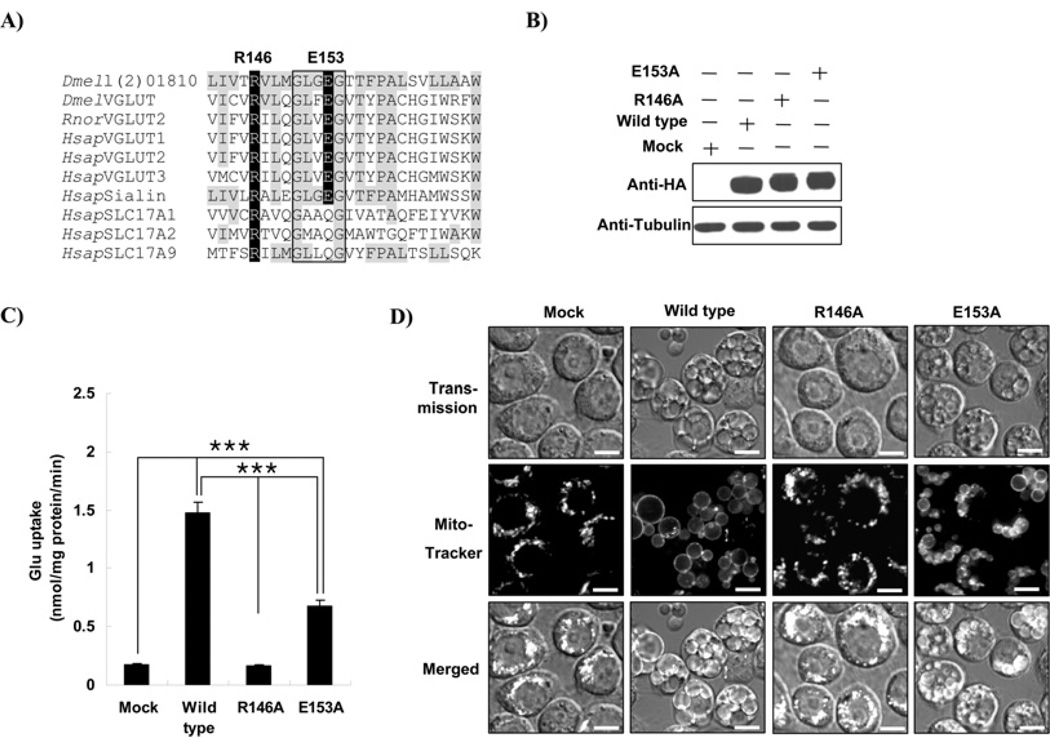
TM4 (transmembrane domain 4) is required for the activity of l(2)01810
To determine the functional domain of l(2)01810, domain search analysis was performed as described in the Experimental section. As a result, we found that this protein has conserved transmembrane domains found within the MFS (major facilitator superfamily), the largest membrane transporter family. Among the MFS members, the solute carrier 17A (SLC17A) transporter family proteins showed the highest sequence homology with l(2)01810, especially in TM4 (see Figure 6A). Since Arg146 and Glu153 were found to play key roles in the rat VGLUT system [29], we examined the effect of these amino acid residues of l(2)01810 on glutamate-uptake activity by substituting these amino acids with alanine residues. When the vectors that express mutant l(2)01810 were transfected into SL2 cells, the expression levels of both wild-type and mutants were significantly elevated from the control cells (Figure 6B). Glutamate-uptake activity was almost completely lost in the Arg146 substitution mutant (R146A), whereas the Glu153 mutant (E153A) showed a decrease in glutamate-uptake activity by half compared with wild-type (P < 0.001; see Figure 6C). These results indicate that Arg146 and Glu153 in TM4 play essential roles during glutamate uptake and TM4 is the functional domain of l(2)01810. We also investigated whether the mutants have the capability of making megamitochondria. As shown in Figure 6(D), the mutant R146A, which lost GLUT activity, showed no megamitochondrial formation, whereas the mutant E153A, which retained GLUT activity in approximately 50% of that of wild-type, formed megamitochondria, but the sizes of megamitochondria were smaller than those found in wild-type. These results suggest that megamitochondrial formation correlated positively with glutamate-transport activity of the cell and that l(2)01810 is responsible for this activity.
Figure 6. TM4 is required for glutamate-transport activity.
(A) Amino acid sequences in the putative TM4 of the SLC17A family. Amino acid sequences were aligned using ClustalW. Identical residues between l(2)01810 and other proteins are shown in grey boxes. Charged residues conserved in the TM4 of VGLUTs are shown in dark boxes. The conserved GxxxG motif among the SLC17A family is shown in the rectangular box. (B) Western blot analysis of transfected cells. After 2 days of transfection, expression levels were determined by Western blot analysis using an anti-HA antibody. α-Tubulin was used as a loading control. (C) Glutamate uptake assay. Glutamate-uptake assays on the mutants were performed as described in the Experimental section. The proteins used are shown on the x axis. Statistical significance was tested by one-way ANOVA followed by Tukey’s multiple comparison test. ***P < 0.001. (D) Loss of glutamate-transport activity in the l(2)01810 mutant does not induce megamitochondrial formation. Cells were transfected with backbone (Mock) or l(2)01810 mutants overexpressing vector and incubated for 3 days. Confocal microscopy was carried out following MitoTracker Red staining. Scale bars represent 5 µm.
DISCUSSION
In a previous study, l(2)01810 was found to play a major role in the formation of megamitochondria [5]. However, its function has not been elucidated other than demonstrating that it is an essential protein in flies [7]. In the present study, we further elucidated the function of l(2)01810 in Drosophila by employing bioinformatical, molecular and biochemical methods.
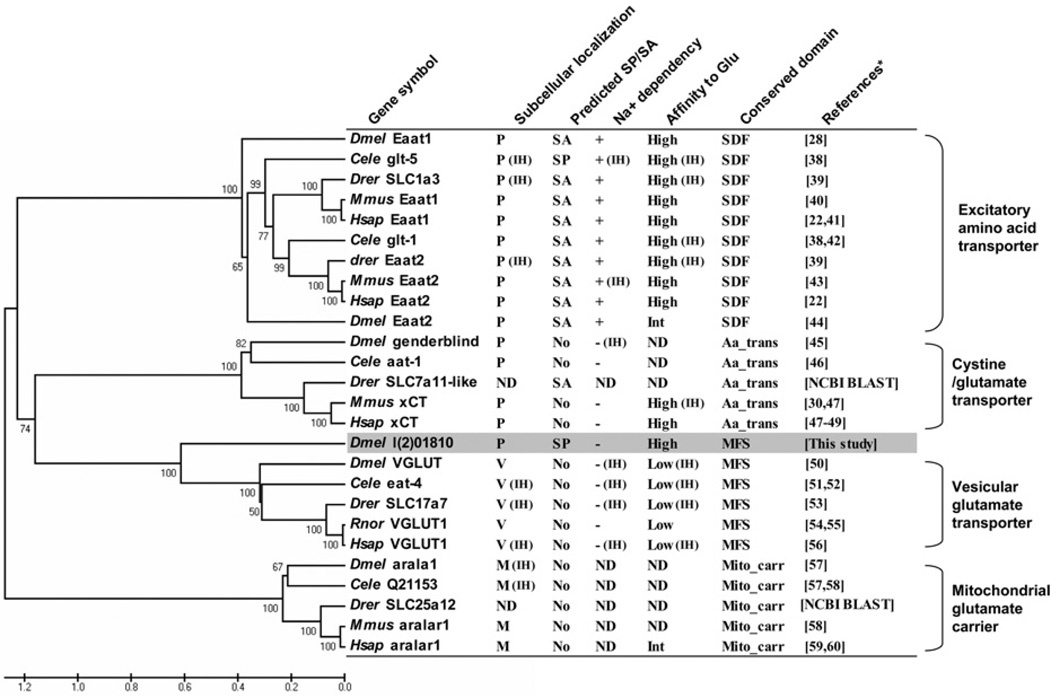
The phylogenetic relationship and physicochemical properties between l(2)01810 and its sequence homologues are summarized in Figure 7. In terms of amino acid sequence, l(2)01810 has the highest homology with VGLUTs (~27% in identity) and it also has a conserved domain of VGLUTs (i.e. MFS domain). The major difference between l(2)01810 and VGLUTs is that l(2)01810 is localized in the plasma membrane, whereas VGLUTs are localized in the membranes of vesicles. Although EAATs and xCTs are also localized in the plasma membrane, their sequence homologies with l(2)01810 are much lower than VGLUTs [the amino acid sequence homology between l(2)01810 and EAATs is approximately 3% and that between l(2)01810 and xCTs is approximately 7%]. In addition, EAATs are Na+ -dependent. xCTs are localized on the plasma membrane, Na+ -independent, and have a high affinity to glutamate. These properties are also shared with l(2)01810. However, there are two characteristics of xCTs which are distinct from those of l(2)01810, and they are membrane trafficking mechanism and cystine competition. xCTs do not contain a SP and require binding with the 4F2hc (heavy chain of 4F2 cell-surface antigen) for movement to the plasma membrane [30]. Also, glutamate uptake by xCTs can be competitively inhibited by l-cystine (see also [31,32]). However, l(2)01810 is not inhibited by l-cystine, but rather by l-aspartate. In addition, although xCTs contain a transmembrane amino acid transporter protein domain, l(2)01810 has an MFS domain. As shown in Figure 7, l(2)01810 takes a position between VGLUTs and xCTs in the phylogenic tree. Therefore it can be concluded that l(2)01810 has different properties from other known GLUTs. It should be noted that l(2)01810 is annotated as a sodium:phosphate symporter according to FlyBase and has a high homology with the insect proteins annotated as sodium:phosphate symporters, especially CG9254 of Dmel (85% similarity; Supplementary Figure S1). However, l(2)01810 did not show any phosphate transport activity, but showed glutamate-transport activity. Therefore there is a possibility that CG9254 also has glutamate-transport activity. Although orthologues of l(2)01810 are found in other insect species, it remains to be determined whether they have the same properties as l(2)01810. l(2)01810 showed the highest homology (approximately 40% similarity) with mouse and human SLC17A5 (also named as sialin) proteins, when searched with NCBI BLAST. However, all SLC17A5 proteins contain a signal anchor instead of a SP (predicted by Signal P software) and they are localized in the late endosome and the lysosome [33]. In addition, both CG4330 and CG4288 were predicted as a Drosophila homologue of SLC17A5 using NCBI HomoloGene. Therefore it appears that there is no homologue of l(2)01810 in mammals.
Figure 7. Comparison of characteristics between l(2)01810 and other GLUTs.
A phylogenic tree was constructed using the amino acid sequences of various GLUTs. Numbers at each node represent the percentage bootstrap value of 1000 replicates. Physicochemical properties marked on the top were obtained by searching the NCBI database for the amino acid sequence; and the conserved domain was observed by running Signal P 3.0 software for SP prediction, and from the references cited for subcellular localization, Na+ -dependence and affinity to glutamate. Aa_Trans, transmembrane amino acid transporter protein; IH, inferred from homologues; M, mitochondrial membrane; Mito_carr, mitochondrial carrier protein; ND, not determined; P, plasma membrane; SA, signal anchor; SDF, sodium:dicarboxylate symporter family; V, vesicular membrane. ‘High’, ‘Int’ and ‘Low’ affinity to glutamate were defined as the Km < 100 µM, Km = 100–250 µM and Km > 500 µM respectively. Data for the gene symbols were taken from [22,28,30,38–60], as shown in the Figure.
Unlike any other known GLUTs, which are expressed primarily in the CNS (central nervous system) such as brain, and thoracic and abdominal ganglion, l(2)01810 is not expressed in the CNS, but instead is mainly expressed in the digestive system such as the gut, Malpighian tubule, salivary gland and the reproductive system such as ovaries (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/439/bj4390277add.htm). Since l(2)01810 is expressed mostly in the digestive system and has glutamate-transport activity, it can be assumed that l(2)01810 is required for the transport of dietary glutamate. Generally, the dietary proteins are degraded by digestive enzymes secreted from the stomach, pancreas and small intestine, and the resulting free amino acids are absorbed into enterocytes in the small intestine by various amino acid transporters [34,35]. When amino acids are used for energy production, toxic ammonia is produced. Ammonia is often converted into glutamate to detoxify ammonia, transported into the liver and finally converted into urea. In insects, the gut and Malpighian tubule belong to the digestive system. The Malpighian tubule is involved primarily in the extraction of metabolic waste and urine synthesis [36]. However, insects do not produce urea to detoxify ammonia, but convert ammonia into uric acid. In this process, glutamate is also used as an ammonium donor [37].
High levels of glutamate in a cell cause an adverse effect and the excess glutamate is removed by converting it into glutamine. The resulting increased level of glutamine will lead to the formation of megamitochondria. Since the overexpression of l(2)01810 led to megamitochondrial formation, it appears that the glutamate transport or glutamate accumulation is a rate-limiting step for megamitochondrial formation. GS1 seems to serve as a sensor for high levels of glutamate in the cell. The reason why l(2)01810 is expressed abundantly in ovaries, but not in testes, is not clear. One possibility is that l(2)01810 is a maternal gene and, therefore, plays an essential role during early embryogenesis.
The present study shows that the function of l(2)01810 during megamitochondrial formation is to uptake extracellular glutamate into a cell. In addition, this protein is a novel type of GLUT which takes up dietary and metabolic glutamate mainly into non-neuronal cells such as the gut, the Malpighian tubule and even the ovaries. We therefore propose to rename l(2)01810 dietary and metabolic glutamate transporter (DMGLUT).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Jeongsil Kim-Ha (Department of Molecular Biology, Sejong University, Seoul, Korea) and Professor Jeongbin Yim (Department of Biological Sciences, Seoul National University, Seoul, Korea) for providing Drosophila cell lines and cDNAs isolated from adult flies respectively. Sequencing experiments were performed in The Genome Research Facility of the Department of Biological Sciences, Seoul National University.
FUNDING
This work was supported by the Priority Research Centers Program and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [grant numbers 2009-0094020 and 2011-0012947 (to B.J.L.)] and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. M.S.S., J.Y.K., K.H.L. and H.K.J. were supported by a Brain Korea 21 Research Fellowship from the Korea Ministry of Education and Human Resources Development.
Abbreviations used
- Cele
Caenorhabditis elegans
- CNS
central nervous system
- Dmel
Drosophila melanogaster
- Drer
Danio rerio
- EAAT
excitatory amino acid transporter
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GLUT
glutamate transporter
- GS1
glutamine synthetase 1
- HA
haemagglutinin
- Hsap
Homo sapiens
- MFS
major facilitator superfamily
- Mmus
Mus musculus
- MSO
methionine sulfoximine
- ORF
open reading frame
- PDI
protein disulfide-isomerase
- Rnor
Rattus norvegicus
- SL2
Schneider cell line 2
- SLC
solute carrier
- SP
signal peptide
- SPS1
selenophosphate synthetase 1
- TM4
transmembrane domain 4
- VGLUT
vesicular GLUT
Footnotes
AUTHOR CONTRIBUTION
Myoung Sup Shim, Bradley Carlson and Byeong Jae Lee contributed to the study design and concept. Myoung Sup Shim, Jin Young Kim, Kwang Hee Lee and Hee Kyoung Jung performed experiments. Myoung Sup Shim, Bradley Carlson, Xue-Ming Xu, Dolph Hatfield and Byeong Jae Lee analysed data. Myoung Sup Shim, Dolph Hatfield and Byeong Jae Lee wrote the paper. Byeong Jae Lee directed the project.
REFERENCES
- 1.Tandler B, Erlandson RA, Wynder EL. Riboflavin and mouse hepatic cell structure and function. I. Ultrastructural alterations in simple deficiency. Am. J. Pathol. 1968;52:69–95. [PMC free article] [PubMed] [Google Scholar]
- 2.Wakabayashi T. Megamitochondria formation – physiology and pathology. J. Cell. Mol. Med. 2002;6:497–538. doi: 10.1111/j.1582-4934.2002.tb00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tandler B, Hoppel CL. Studies on giant mitochondria. Ann. N.Y. Acad. Sci. 1986;488:65–81. doi: 10.1111/j.1749-6632.1986.tb46548.x. [DOI] [PubMed] [Google Scholar]
- 4.Grodums EI. Ultrastructural changes in the mitochondria of brown adipose cells during the hibernation cycle of Citellus lateralis. Cell Tissue Res. 1977;185:231–237. doi: 10.1007/BF00220667. [DOI] [PubMed] [Google Scholar]
- 5.Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, Lee BJ. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J. Biol. Chem. 2009;284:32881–32894. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caizzi R, Bozzetti MP, Caggese C, Ritossa F. Homologous nuclear genes encode cytoplasmic and mitochondrial glutamine synthetase in Drosophila melanogaster. J. Mol. Biol. 1990;212:17–26. doi: 10.1016/0022-2836(90)90301-2. [DOI] [PubMed] [Google Scholar]
- 7.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danbolt NC. Glutamate uptake. Prog. Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 9.Patel SA, Warren BA, Rhoderick JF, Bridges RJ. Differentiation of substrate and non-substrate inhibitors of transport system xc− : an obligate exchanger of L-glutamate and L-cystine. Neuropharmacology. 2004;46:273–284. doi: 10.1016/j.neuropharm.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Bringmann A, Pannicke T, Biedermann B, Francke M, Iandiev I, Grosche J, Wiedemann P, Albrecht J, Reichenbach A. Role of retinal glial cells in neurotransmitter uptake and metabolism. Neurochem. Int. 2009;54:143–160. doi: 10.1016/j.neuint.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflügers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 12.Jin JS, Baek S, Lee H, Oh MY, Koo YE, Shim MS, Kwon SY, Jeon I, Park SY, Baek K, et al. A DNA replication-related element downstream from the initiation site of Drosophila selenophosphate synthetase 2 gene is essential for its transcription. Nucleic Acids Res. 2004;32:2482–2493. doi: 10.1093/nar/gkh569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CL, Shim MS, Chung J, Yoo HS, Ha JM, Kim JY, Choi J, Zang HL, Hou X, Carlson BA, et al. G-rich, a Drosophila selenoprotein, is a Golgi-resident type III membrane protein. Biochem. Biophys. Res. Commun. 2006;348:1296–1301. doi: 10.1016/j.bbrc.2006.07.203. [DOI] [PubMed] [Google Scholar]
- 14.dos Santos AQ, Nardin P, Funchal C, de Almeida LM, Jacques-Silva MC, Wofchuk ST, Goncçalves CA, Gottfried C. Resveratrol increases glutamate uptake and glutamine synthetase activity in C6 glioma cells. Arch. Biochem. Biophys. 2006;453:161–167. doi: 10.1016/j.abb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, Haito S, Furumoto M, Uehata Y, Sakurai A, Segawa H, Tatsumi S, Kuwahata M, Miyamoto K. Unique uptake and efflux systems of inorganic phosphate in osteoclast-like cells. Am. J. Physiol. Cell Physiol. 2007;292:C526–C534. doi: 10.1152/ajpcell.00357.2006. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim JY, Lee KH, Shim MS, Shin H, Xu XM, Carlson BA, Hatfield DL, Lee BJ. Human selenophosphate synthetase 1 has five splice variants with unique interactions, subcellular localizations and expression patterns. Biochem. Biophys. Res. Commun. 2010;397:53–58. doi: 10.1016/j.bbrc.2010.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer AM, Anderson JB, Derbyshire MK, Scott CD, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserve domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J. Neurosci. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston GAR. Glutamate uptake and its possible role in neurotransmitter inactivation. In: Roberts PJ, Storm-Mathisen J, Johnston GAR, editors. Glutamate: Transmitter in the Central Nervous System. Chichester: Wiley; 1981. pp. 77–87. [Google Scholar]
- 24.Fei H, Karnezis T, Reimer RJ, Krantz DE. Membrane topology of the Drosophila vesicular glutamate transporter. J. Neurochem. 2007;101:1662–1671. doi: 10.1111/j.1471-4159.2007.04518.x. [DOI] [PubMed] [Google Scholar]
- 25.Gierasch LM. Signal sequences. Biochemistry. 1989;28:923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- 26.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 27.Erecińska M, Silver IA. Metabolism and role of glutamate in mammalian brain. Prog. Neurobiol. 1990;35:245–296. doi: 10.1016/0301-0082(90)90013-7. [DOI] [PubMed] [Google Scholar]
- 28.Seal RP, Daniels GM, Wolfgang WJ, Forte MA, Amara SG. Identification and characterization of a cDNA encoding a neuronal glutamate transporter from Drosophila melanogaster. Recept. Channels. 1998;6:51–64. [PubMed] [Google Scholar]
- 29.Juge N, Yoshida Y, Yatsushiro S, Omote H, Moriyama Y. Vesicular glutamate transporter contains two independent transport machineries. J. Biol. Chem. 2006;281:39499–39506. doi: 10.1074/jbc.M607670200. [DOI] [PubMed] [Google Scholar]
- 30.Sato H, Tamba M, Ishii T, Bannai S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 1999;274:11455–11458. doi: 10.1074/jbc.274.17.11455. [DOI] [PubMed] [Google Scholar]
- 31.Bannai S. Exchange of cystine and glutamate across plasma membrane of human fibroblasts. J. Biol. Chem. 1986;261:2256–2263. [PubMed] [Google Scholar]
- 32.Cho Y, Bannai S. Uptake of glutamate and cysteine in C-6 glioma cells and in cultured astrocytes. J. Neurochem. 1990;55:2091–2097. doi: 10.1111/j.1471-4159.1990.tb05800.x. [DOI] [PubMed] [Google Scholar]
- 33.Morin P, Sagné C, Gasnier B. Functional characterization of wild-type and mutant human sialin. EMBO J. 2004;23:4560–4570. doi: 10.1038/sj.emboj.7600464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adibi SA, Gray SJ, Menden E. The kinetics of amino acid absorption and alteration of plasma composition of free amino acids after intestinal perfusion of amino acid mixtures. Am. J. Clin. Nutr. 1967;20:24–33. doi: 10.1093/ajcn/20.1.24. [DOI] [PubMed] [Google Scholar]
- 35.Bröer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 2008;88:249–286. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- 36.McGavin GC. Essential Entomology; An Order By Order Introduction. New York: Oxford University Press; 2001. [Google Scholar]
- 37.Vorhaben JE, Campbell JW. Glutamine synthetase. A mitochondrial enzyme in uricotelic species. J. Biol. Chem. 1972;247:2763–2767. [PubMed] [Google Scholar]
- 38.Mano I, Straud S, Driscoll M. Caenorhabditis elegans glutamate transporters influence synaptic function and behavior at sites distant from the synapse. J. Biol. Chem. 2007;282:34412–34419. doi: 10.1074/jbc.M704134200. [DOI] [PubMed] [Google Scholar]
- 39.Rico EP, de Oliveira DL, Rosemberg DB, Mussulini BH, Bonan CD, Dias RD, Wofchuk S, Souza DO, Bogo MR. Expression and functional analysis of Na+ -dependent glutamate transporters from zebrafish brain. Brain Res. Bull. 2010;81:517–523. doi: 10.1016/j.brainresbull.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka K. Cloning and expression of a glutamate transporter from mouse brain. Neurosci. Lett. 1993;159:183–186. doi: 10.1016/0304-3940(93)90829-a. [DOI] [PubMed] [Google Scholar]
- 41.Kawakami H, Tanaka K, Nakayama T, Inoue K, Nakamura S. Cloning and expression of a human glutamate transporter. Biochem. Biophys. Res. Commun. 1994;199:171–176. doi: 10.1006/bbrc.1994.1210. [DOI] [PubMed] [Google Scholar]
- 42.Kawano T, Takuwa K, Nakajima T. Structure and activity of a new form of the glutamate transporter of the nematode Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 1997;61:927–929. doi: 10.1271/bbb.61.927. [DOI] [PubMed] [Google Scholar]
- 43.Kirschner MA, Copeland NG, Gilbert DJ, Jenkins NA, Amara SG. Mouse excitatory amino acid transporter EAAT2: isolation, characterization, and proximity to neuroexcitability loci on mouse chromosome 2. Genomics. 1994;24:218–224. doi: 10.1006/geno.1994.1609. [DOI] [PubMed] [Google Scholar]
- 44.Besson MT, Soustelle L, Birman S. Selective high-affinity transport of aspartate by a Drosophila homologue of the excitatory amino-acid transporters. Curr. Biol. 2000;10:207–210. doi: 10.1016/s0960-9822(00)00339-0. [DOI] [PubMed] [Google Scholar]
- 45.Augustin H, Grosjean Y, Chen K, Sheng Q, Featherstone DE. Nonvesicular release of glutamate by glial xCT transporters suppresses glutamate receptor clustering in vivo. J. Neurosci. 2007;27:111–123. doi: 10.1523/JNEUROSCI.4770-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veljkovic E, Stasiuk S, Skelly PJ, Shoemaker CB, Verrey F. Functional characterization of Caenorhabditis elegans heteromeric amino acid transporters. J. Biol. Chem. 2004;279:7655–7662. doi: 10.1074/jbc.M309528200. [DOI] [PubMed] [Google Scholar]
- 47.Bridges CC, Kekuda R, Wang H, Prasad PD, Mehta P, Huang W, Smith SB, Ganapathy V. Structure, function, and regulation of human cystine/glutamate transporter in retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 2001;42:47–54. [PubMed] [Google Scholar]
- 48.Sato H, Tamba M, Kuriyama-Matsumura K, Okuno S, Bannai S. Molecular cloning and expression of human xCT, the light chain of amino acid transport system xc−. Antioxid. Redox Signaling. 2000;2:665–671. doi: 10.1089/ars.2000.2.4-665. [DOI] [PubMed] [Google Scholar]
- 49.Kim JY, Kanai Y, Chairoungdua A, Cha SH, Matsuo H, Kim DK, Inatomi J, Sawa H, Ida Y, Endou H. Human cystine/glutamate transporter: cDNA cloning and upregulation by oxidative stress in glioma cells. Biochim. Biophys. Acta. 2001;1512:335–344. doi: 10.1016/s0005-2736(01)00338-8. [DOI] [PubMed] [Google Scholar]
- 50.Daniels RW, Collins CA, Gelfand MV, Dant J, Brooks ES, Krantz DE, DiAntonio A. Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J. Neurosci. 2004;24:10466–10474. doi: 10.1523/JNEUROSCI.3001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee RY, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans. J. Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rand JB, Duerr JS, Frisby DL. Neurogenetics of vesicular transporters in C. elegans. FASEB J. 2000;14:2414–2422. doi: 10.1096/fj.00-0313rev. [DOI] [PubMed] [Google Scholar]
- 53.Higashijima S, Mandel G, Fetcho JR. Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J. Comp. Neurol. 2004;480:1–18. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- 54.Bellocchio EE, Reimer RJ, Fremeau RT, Jr, Edwards RH. Uptake of glutamate into synaptic vesicles by an inorganic phosphate transporter. Science. 2000;289:957–960. doi: 10.1126/science.289.5481.957. [DOI] [PubMed] [Google Scholar]
- 55.Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- 56.Ni B, Du Y, Wu X, DeHoff BS, Rosteck PR, Jr, Paul SM. Molecular cloning, expression, and chromosomal localization of a human brain-specific Na+ -dependent inorganic phosphate cotransporter. J. Neurochem. 1996;66:2227–2238. doi: 10.1046/j.1471-4159.1996.66062227.x. [DOI] [PubMed] [Google Scholar]
- 57.Del Arco A, Agudo M, Satrústegui J. Characterization of a second member of the subfamily of calcium-binding mitochondrial carriers expressed in human non-excitable tissues. Biochem. J. 2000;345:725–732. doi: 10.1042/0264-6021:3450725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Arco A, Morcillo J, Martínez-Morales JR, Galián C, Martos V, Bovolenta P, Satrústegui J. Expression of the aspartate/glutamate mitochondrial carriers aralar1 and citrin during development and in adult rat tissues. Eur. J. Biochem. 2002;269:3313–3320. doi: 10.1046/j.1432-1033.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- 59.Del Arco A, Satrústegui J. Molecular cloning of Aralar, a new member of the mitochondrial carrier superfamily that binds calcium and is present in human muscle and brain. J. Biol. Chem. 1998;273:23327–23334. doi: 10.1074/jbc.273.36.23327. [DOI] [PubMed] [Google Scholar]
- 60.Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrústegui J, Palmieri F. Citrin and aralar1 are Ca2+ -stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.