Abstract
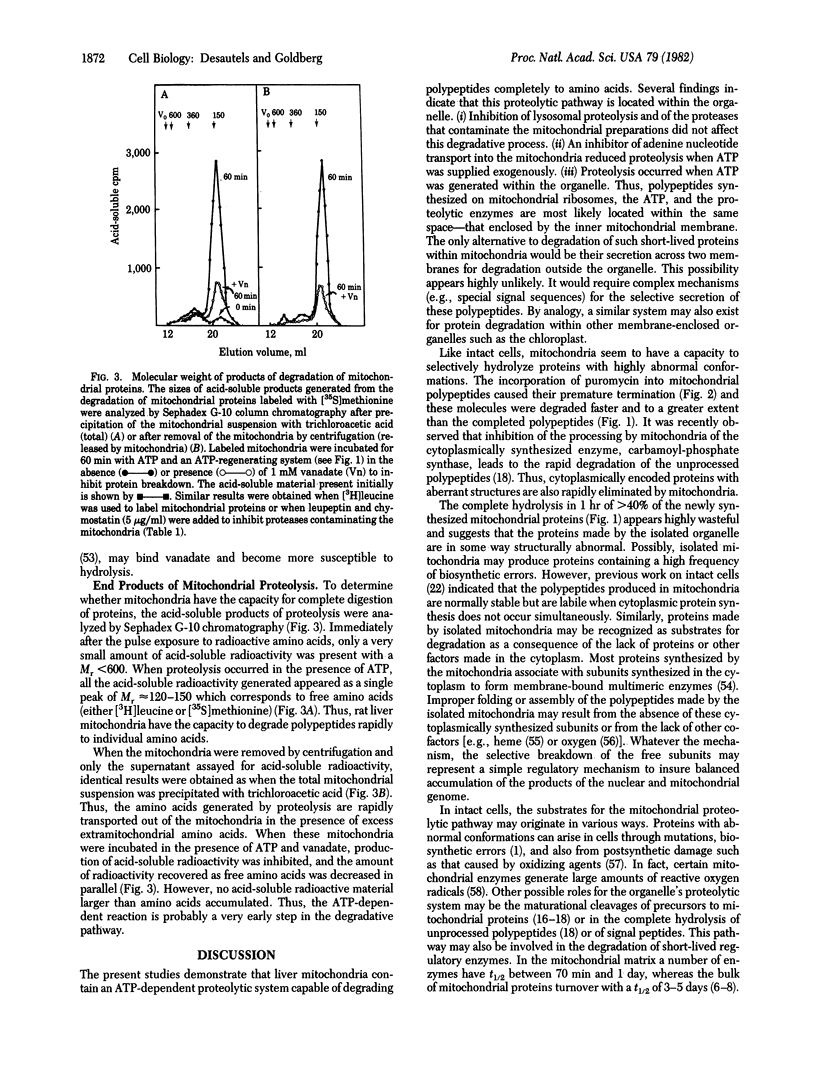
A large fraction (30-50%) of the various proteins synthesized within isolated rat liver mitochondria were degraded to amino acids within 60 min after synthesis. Incomplete mitochondrial polypeptides resulting from the incorporation of puromycin were degraded even more extensively (80% per hr). Protein breakdown was measured by the appearance of acid-soluble radioactivity and by the disappearance of labeled polypeptides detected on NaDodSO4/polyacrylamide gel electrophoresis. The amino acids generated by proteolysis were transported rapidly out of the mitochondria and no peptide intermediates accumulated in the organelle. This degradative process did not involve lysosomes or lysosomal enzymes and was markedly stimulated by ATP either generated within the mitochondria or supplied exogenously. An inhibitor of respiration (cyanide) or uncouplers of oxidative phosphorylation (oligomycin, dinitrophenol) reduced proteolysis when mitochondria were provided substrates for ATP generation. When exogenous ATP was provided, these agents did not affect proteolysis, but degradation was then sensitive to atractyloside, an inhibitor of adenine nucleotide transport. Vanadate, an inhibitor of various ATPases, blocked proteolysis even in the presence of ATP and caused a marked stabilization of nearly all polypeptide bands. Thus, mitochondria--like bacteria or the cytosol of animal cells--contain a pathway for complete degradation of proteins which seems to selectively remove polypeptides with abnormal structures. Within this organelle, ATP hydrolysis appears necessary for an initial step in this degradative process.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASHFORD T. P., PORTER K. R. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962 Jan;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Beattie D. S. Yeast versus mammalian mitochondrial protein synthesis. Methods Enzymol. 1979;56:17–29. doi: 10.1016/0076-6879(79)56005-4. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chung C. H., Goldberg A. L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Elias S., Heller H., Ferber S., Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980 Aug 25;255(16):7525–7528. [PubMed] [Google Scholar]
- Costantino P., Attardi G. Metabolic properties of the products of mitochondrial protein synthesis in HeLa cells. J Biol Chem. 1977 Mar 10;252(5):1702–1711. [PubMed] [Google Scholar]
- DeMartino G. N., Goldberg A. L. Identification and partial purification of an ATP-stimulated alkaline protease in rat liver. J Biol Chem. 1979 May 25;254(10):3712–3715. [PubMed] [Google Scholar]
- Duque-Magalhães M. C., Menezes Ferreira M. M. Cytochrome c degrading activity in rat liver mitochondria. Biochem Biophys Res Commun. 1980 Mar 13;93(1):106–112. doi: 10.1016/s0006-291x(80)80252-x. [DOI] [PubMed] [Google Scholar]
- Duque-Magalhães M. C. On a neutral proteolytic system in rat liver mitochondria. FEBS Lett. 1979 Sep 15;105(2):317–320. doi: 10.1016/0014-5793(79)80638-9. [DOI] [PubMed] [Google Scholar]
- Etlinger J. D., Goldberg A. L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977 Jan;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Fridovich I. The biology of oxygen radicals. Science. 1978 Sep 8;201(4359):875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Dice J. F. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43(0):835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Haas R., Heinrich P. C. A novel SH-type carboxypeptidase in the inner membrane of rat-liver mitochondria. Eur J Biochem. 1979 May 2;96(1):9–15. doi: 10.1111/j.1432-1033.1979.tb13007.x. [DOI] [PubMed] [Google Scholar]
- Haas R., Heinrich P. C., Sasse D. Proteolytic enzymes of rat liver mitochondria. Evidence for a mast cell origin. FEBS Lett. 1979 Jul 1;103(1):168–171. doi: 10.1016/0014-5793(79)81274-0. [DOI] [PubMed] [Google Scholar]
- Hare J. F. A novel proteinase associated with mitochondrial membranes. Biochem Biophys Res Commun. 1978 Aug 14;83(3):1206–1215. doi: 10.1016/0006-291x(78)91523-1. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt E., Trapp M., Kadenbach B. Biosynthesis of cytochrome c oxidase in isolated rat hepatocytes. FEBS Lett. 1980 Jun 16;115(1):95–99. doi: 10.1016/0014-5793(80)80734-4. [DOI] [PubMed] [Google Scholar]
- Ip M. M., Chee P. Y., Swick R. W. Turnover of hepatic mitochondrial ornithine aminotransferase and cytochrome oxidase using (14C)carbonate as tracer. Biochim Biophys Acta. 1974 Jun 20;354(1):29–38. doi: 10.1016/0304-4165(74)90049-x. [DOI] [PubMed] [Google Scholar]
- Kalnov S. L., Novikova L. A., Zubatov A. S., Luzikov V. N. Proteolysis of the products of mitochondrial protein synthesis in yeast mitochondria and submitochondrial particles. Biochem J. 1979 Jul 15;182(1):195–202. doi: 10.1042/bj1820195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunuma N., Kominami E., Kobayashi K., Banno Y., Suzuki K. Studies on new intracellular proteases in various organs of rat. 1. Purification and comparison of their properties. Eur J Biochem. 1975 Mar 3;52(1):37–50. doi: 10.1111/j.1432-1033.1975.tb03970.x. [DOI] [PubMed] [Google Scholar]
- Knecht E., Hernández-Yago J., Martinez-Ramón A., Grisolía S. Fate of proteins synthesized in mitochondria of cultured mammalian cells revealed by electron microscope radioautography. Exp Cell Res. 1980 Jan;125(1):191–199. doi: 10.1016/0014-4827(80)90203-7. [DOI] [PubMed] [Google Scholar]
- Kolarov J., Kuzela S., Wielburski A., Nelson B. D. The characterization of mitochondrial translation products in rat liver and rat hepatoma cells. FEBS Lett. 1981 Apr 6;126(1):61–65. doi: 10.1016/0014-5793(81)81033-2. [DOI] [PubMed] [Google Scholar]
- Kowit J. D., Goldberg A. L. Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J Biol Chem. 1977 Dec 10;252(23):8350–8357. [PubMed] [Google Scholar]
- Kuzela S., Luciaková K., Lakota J. Amino acid incorporation by isolated rat liver mitochondria into two protein components of mitochondrial ATPase complex. Lack of incorporation into dicyclohexylcarbodiimide binding proteolipid. FEBS Lett. 1980 Jun 2;114(2):197–201. doi: 10.1016/0014-5793(80)81113-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lipsky N. G., Pedersen P. L. Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J Biol Chem. 1981 Aug 25;256(16):8652–8657. [PubMed] [Google Scholar]
- Mori M., Miura S., Tatibana M., Cohen P. P. Characterization of a protease apparently involved in processing of pre-ornithine transcarbamylase of rat liver. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7044–7048. doi: 10.1073/pnas.77.12.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Voellmy R., Goldberg A. L. Protein degradation is stimulated by ATP in extracts of Escherichia coli. J Biol Chem. 1979 Sep 10;254(17):8194–8200. [PubMed] [Google Scholar]
- Pedersen P. L., Greenawalt J. W., Reynafarje B., Hullihen J., Decker G. L., Soper J. W., Bustamente E. Preparation and characterization of mitochondria and submitochondrial particles of rat liver and liver-derived tissues. Methods Cell Biol. 1978;20:411–481. doi: 10.1016/s0091-679x(08)62030-0. [DOI] [PubMed] [Google Scholar]
- Pestka S. Inhibitors of ribosome functions. Annu Rev Microbiol. 1971;25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- Poyton R. O., Groot G. S. Biosynthesis of polypeptides of cytochrome c oxidase by isolated mitochondria. Proc Natl Acad Sci U S A. 1975 Jan;72(1):172–176. doi: 10.1073/pnas.72.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond Y., Shore G. C. Processing of the precursor for the mitochondrial enzyme, carbamyl phosphate synthetase. Inhibition by rho-aminobenzamidine leads to very rapid degradation (clearing) of the precursor. J Biol Chem. 1981 Mar 10;256(5):2087–2090. [PubMed] [Google Scholar]
- Reijngoud D. J., Oud P. S., Kás J., Tager J. M. Relationship between medium pH and that of the lysosomal matrix as studied by two independent methods. Biochim Biophys Acta. 1976 Oct 5;448(2):290–302. doi: 10.1016/0005-2736(76)90243-1. [DOI] [PubMed] [Google Scholar]
- Russell S. M., Burgess R. J., Mayer R. J. Protein degradation in rat liver during post-natal development. Biochem J. 1980 Oct 15;192(1):321–330. doi: 10.1042/bj1920321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWIFT H., HRUBAN Z. FOCAL DEGRADATION AS A BIOLOGICAL PROCESS. Fed Proc. 1964 Sep-Oct;23:1026–1037. [PubMed] [Google Scholar]
- Saltzgaber-Müller J., Schatz G. Heme is necessary for the accumulation and assembly of cytochrome c oxidase subunits in Saccharomyces cerevisiae. J Biol Chem. 1978 Jan 10;253(1):305–310. [PubMed] [Google Scholar]
- St John A. C., Goldberg A. L. Effects of reduced energy production on protein degradation, guanosine tetraphosphate, and RNA synthesis in Escherichia coli. J Biol Chem. 1978 Apr 25;253(8):2705–2711. [PubMed] [Google Scholar]
- Stephenson G., Marzuki S., Linnane A. W. Biogenesis of mitochondria. Two-dimensional electrophoretic analysis of mitochondrial translation products in yeast. Biochim Biophys Acta. 1980 Sep 19;609(2):329–341. doi: 10.1016/0005-2787(80)90245-2. [DOI] [PubMed] [Google Scholar]
- Umezawa H. Structures and activities of protease inhibitors of microbial origin. Methods Enzymol. 1976;45:678–695. doi: 10.1016/s0076-6879(76)45058-9. [DOI] [PubMed] [Google Scholar]
- Voellmy R. W., Goldberg A. L. ATP-stimulated endoprotease is associated with the cell membrane of E. coli. Nature. 1981 Apr 2;290(5805):419–421. doi: 10.1038/290419a0. [DOI] [PubMed] [Google Scholar]
- Wheeldon L. W., Dianoux A. C., Bof M., Vignais P. V. Stable and labile products of mitochondrial protein synthesis in vitro. Eur J Biochem. 1974 Jul 1;46(1):189–199. doi: 10.1111/j.1432-1033.1974.tb03611.x. [DOI] [PubMed] [Google Scholar]
- Wheeldon L. W., Lehninger A. L. Energy-linked synthesis and decay of membrane proteins in isolated rat liver mitochondria. Biochemistry. 1966 Nov;5(11):3533–3545. doi: 10.1021/bi00875a021. [DOI] [PubMed] [Google Scholar]
- Wibo M., Poole B. Protein degradation in cultured cells. II. The uptake of chloroquine by rat fibroblasts and the inhibition of cellular protein degradation and cathepsin B1. J Cell Biol. 1974 Nov;63(2 Pt 1):430–440. doi: 10.1083/jcb.63.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow G., Schatz G. The role of oxygen in the biosynthesis of cytochrome c oxidase of yeast mitochondria. J Biol Chem. 1979 Jul 10;254(13):6088–6093. [PubMed] [Google Scholar]
- Zaman Z., Verwilghen R. L. Quantitation of proteins solubilized in sodium dodecyl sulfate-mercaptoethanol-Tris electrophoresis buffer. Anal Biochem. 1979 Nov 15;100(1):64–69. doi: 10.1016/0003-2697(79)90110-6. [DOI] [PubMed] [Google Scholar]


