Abstract
Membrane dynamics are involved in crucial processes in eukaryotic and prokaryotic cells. Membrane fusion and fission events are often catalyzed by proteins that belong to the dynamin family of large GTPases. It has recently been shown that members of the dynamin superfamily are also present in many bacterial species. Although structural information about full length bacterial dynamin-like proteins is available, their molecular role remains unclear. We have shown previously that DynA, a dynamin-like protein found in the firmicute Bacillus subtilis is able to fuse membranes in vitro. In contrast to other members of the dynamin family this membrane remodeling activity was not dependent on guanosine nucleotides, but required magnesium. DynA assemblies localize in foci that are often enriched at sites of septation and hence a potential role during bacterial cytokinesis was discussed. In order to identify potential interaction partners we constructed a bacterial-two hybrid (B2H) library and screened for DynA interacting proteins. Three potential interaction partner have been identified, YneK, RNaseY (YmdA), and YwpG. Localization of these proteins phenocopies that of DynA, supporting the potential interaction in vivo.
Keywords: Bacillus subtilis, bacterial-two hybrid library, dynamin, protein-protein interaction
Biological membranes are dynamic cellular structures that show a high degree of compartmentalization in space and time. In eukaryotic cells membrane remodeling processes are often guided by dynamins or dynamin-like (DLPs) proteins.1 Dynamin and DLPs are large GTPases that show cooperative nucleotide hydrolysis and reversible membrane-binding.1,2 Dynamin and DLPs share similar modular domain architecture with a conserved GTPase domain, an α-helical stalk region and a membrane associated domain. Classical dynamins contain a proline rich domain (PRD) domain which is involved in several protein-protein interactions.3 The stalk region is essential for dimerization4-7 of the protein and the GTPase domains undergo GTP dependent dimerization, while nucleotide hydrolysis leads to large domain movements. It is thought that these large scale domain movements drive membrane dynamics such as vesicle scission,8 mitochondrial9 and endoplasmic reticulum (ER) fusion,10 as well as fusion of mitochondrial outer,11,12 and inner membranes.13,14
Examples of membrane dynamics in bacteria are less well known since most bacteria lack internal membrane structures or organelles. However, at least during cell division the bacterial plasma membrane needs to fuse at the end of cytokinesis. Cytokinesis is driven by a complex protein machinery, termed divisome that is recruited upon assembly of FtsZ, a bacterial tubulin homologue, at the site of the future division.15 FtsZ oligomers are stabilized by bridging proteins and associated with the plasma membrane via FtsA. FtsA has an actin-like fold and harbors an amphipathic helix which mediates membrane-binding.16 Subsequently, a set of integral membrane proteins is recruited. While many of these membrane proteins (FtsL, DivIC, DivIB) seem to lack enzymatic activity, it may be speculated that they fulfill scaffolding roles within the complex protein-protein network of the divisome.17 Central components of the divisome are penicillin-binding proteins (PBPs) catalyzing cell wall synthesis.15 The dividing daughter cells are separated by inwardly growing cell wall. At the end of cytokinesis the divisome is disassembled and proteins of the Min system ensure that division is not initiated again at this site in B. subtilis.18,19 An unsolved question during this process still is how membrane fusion is accomplished. It remains elusive whether this process is actively supported by proteins.
We have recently described a bacterial dynamin-like protein, DynA that localizes in membrane associated patches in Bacillus subtilis, but is often enriched at sites of septation,20 consistent with what has been observed for a dynamin-like protein BLDP from Nostoc punctiforme.21 However, a ΔdynA strain had no division phenotype for vegetatively growing cells.20 Interestingly, B. subtilis lacking the division protein MinJ had DynA distributed all over the membrane, indicating that there might be potential protein-protein interactions that recruit DynA to the division machinery.
As a result of gene fusion, DynA is an intrinsic dimer comprising D1 and D2 regions, each of which encompasses an entire DLP. Membrane association is mediated by the D1 domain and oligomerization of DynA is homotypic, showing D1/D1 and D2/D2 contacts. DynA is an active GTPase, but requires nucleotide-binding to both GTPase domains for hydrolysis. In contrast to other DLPs that have been described so far, DynA mediates trans-tethering and membrane fusion in the absence of nucleotide. Fusion only requires Mg2+ in physiological concentrations.20 DynA was found to be dimeric in nucleotide-free solution (corresponding to DLP tetramers)20 similar to the tetrameric state of dynamin 1.22-24 Thus, GTP might be required for supramolecular assembly.
The physiological function of DynA and other bacterial DLPs remains elusive. Therefore, we aimed to identify physical interaction partners. To this end a two-hybrid strategy was chosen, because this genetic system enables sampling of a complete genome sequence independent of expression patterns of the interaction partners. N-terminally tagged DynA exhibited a complex self-interaction pattern in the BACTH system,25 indicating that it might be at least partially functional and might be a suitable bait for a two-hybrid screen. The BACTH system supports a library screening approach, because cells can be grown on nutritional selection for interaction signals. For instance the maltose metabolism is under direct control of adenylate cyclase activity.26
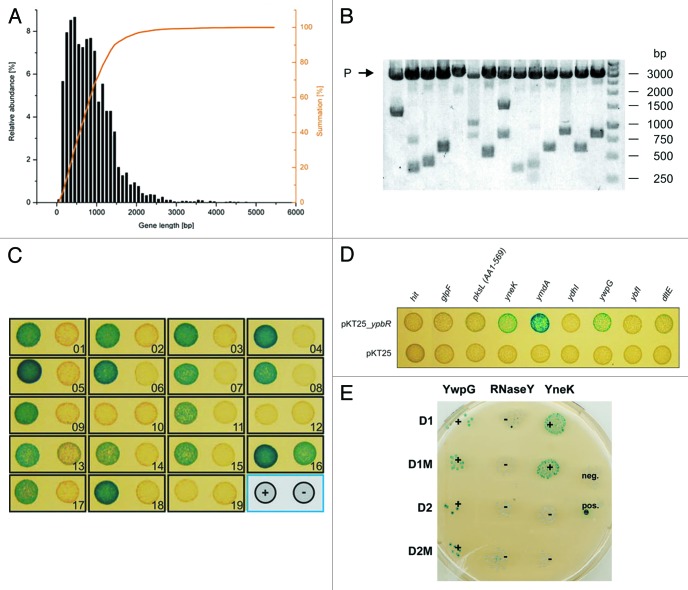
Since DynA is a membrane associated protein, it was expected that integral membrane proteins are among its interaction partners. Membrane integration of these proteins can be impaired by addition of a stably folded N-terminal domain, as was experimentally shown at least for eukaryotes.27 Therefore, a C-terminal fusion strategy was chosen for library construction to obtain a high functional coverage of the membrane proteome. The pUT18 vector was preferred to pKNT25, because it is a high copy number plasmid, which is helpful for large scale generation of recombinants and efficient plasmid recovery during the screening process. For estimation of the most appropriate insert size for the library, the gene length distribution of B. subtilis genes was determined. The average gene length of B. subtilis is 890 bp,28 with 90% of the genes having a size below 1500 bp (Fig. 1A). The library insert size was chosen to be in the range of 500 to 1500 bp, which corresponds to the majority of genes (68%) and yields an average insert size of 1000 bp. With this range of inserts an appropriate library size for the screen was estimated after Clarke and Carbon.29 The probability P for a region of length k to be present in a library of N clones is:
Figure 1. Construction of a bacterial two-hybrid library and identification of DynA (YpbR) interation partner (A) Gene length distribution of B. subtilis. (B) XbaI and SacI restriction analysis of random clones (n = 14) isolated from the pUT18 genomic library. The average insert size is 1030 bp. P, plasmid backbone. (C) Validation of plasmids found in the two-hybrid screen. Plasmids were transformed into E. coli BTH101 carrying either pKT25_dynA (+) or empty pKT25 (−) as indicated in the bottom right position. (D) Second round of validation with full length genes cloned into pUT18 vector. Note, only three full length constructs show interaction with DynA. (E) Interaction matrix of YwpG, RNaseY, and YneK with the D1 and D2 subdomains of DynA and their nucleotide-binding mutants. D1M harbors the K56A mutation and D2M the K625R mutation, respectively.
P = 1 − (1 −k/G) N
where in this case G = 4,200,000 bp is the genome size of B. subtilis. Therefore, a number of approximately 16,000 clones is sufficient to obtain a sequence coverage of 98% with the chosen average insert size of k = 1000 bp. Statistically, only one out of six plasmids contains an in-frame fusion, leading to a library size of 96,000 clones for 98% coverage. A blunt end ligation strategy with SmaI for vector digestion and AluI for fragment preparation was used. To assay the quality of the purified plasmid library, a small amount was retransformed into E. coli XL1-Blue and plasmids of isolated clones (n = 14) were subjected to restriction analysis (Fig. 1B). The plasmids showed an insert distribution over the expected range with an average insert size of 1030 bp. The library theoretically covers more than 98% of the B. subtilis genomic sequence.
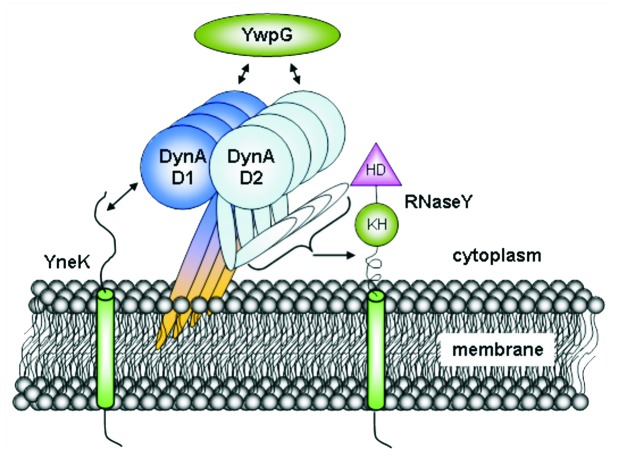
Highly competent E. coli BTH101 harboring the bait plasmid pKT25_dynA were transformed with the library aiming at 200,000 transformants and plated on M63 minimal medium with D(+)-maltose as sole carbon source supplemented with appropriate antibiotics. The screen resulted in growth of 84 clones, which were immediately patched onto fresh plates. Plasmids were isolated, retransformed into BTH101 carrying either pKT25_dynA or the empty vector and tested on BTH medium to sort out false positives. Since the initial plasmid preparations contained not only the pUT18 prey plasmids, but the pKT25_dynA bait plasmid as well, the empty vector control usually yielded a mixture of blue and white colonies due to double transformation events. 19 plasmids gave rise to mostly blue colonies in the pKT25_dynA background and predominantly white colonies in the empty vector control and were chosen for a second round of confirmation. To obtain isolated prey plasmids, the initial plasmid preparations were retransformed into BTH101 and selected for single colonies on LB with 0.5 mM IPTG, 40 mg mL-1 X-Gal supplemented with carbenicillin. Plasmids from white colonies were prepared and tested against pKT25_dynA and the empty vector as described above. Signals from 14 candidates could be confirmed and these plasmids were sequenced (Fig. 1C and Table 1). Subsequently, all genes that were identified were subcloned in pUT18 prey plasmid and tested again for interaction with the pKT25_dynA bait to ensure interaction with the full length construct. Here, three positive interactions remained, YneK, YwpG, RNaseY (YmdA), respectively (Fig. 1D). Further confirmation for an interaction of DynA with these three proteins stems from the fact that they show distinct interaction with the D1/D2 domains of DynA. DynA regions encompassing the D1 and D2 domain and their catalytically inactive mutants, D1M and D2M were introduced separately into the B2H assay and tested for interaction with YneK, YwpG and RNaseY. YneK selectively interacts with the lipid-associated D1 domain, while YwpG binds to D1 and D2. Interaction of RNaseY seems to depend on the full length protein or its oligomer, since no interaction with the isolated D1 or D2 domain was observed (Fig. 1E).
Table 1. Two-hybrid fusions encoded by the prey plasmids. Amino acid residues of the expressed fusion proteins are indicated in brackets.
| # T18-fusion | Description |
|---|---|
| 01 |
GlpF (> 65–175)-PksL(284–569) fusion glycerol permease, polyketide synthase |
| 02 |
GlpF(> 65–175)-PksL(284–569) fusion glycerol permease, polyketide synthase |
| 03 |
GlpF(> 65–175)-PksL(284–569) fusion glycerol permease, polyketide synthase |
| 04 |
YneK (1–44) hypothetical membrane protein |
| 05 |
YmdA (1–57) essential membrane protein |
| 06 |
YdhI (1–81) acetyltransferase |
| 07 |
YwpG (1–61) unknown function |
| 08 |
YbfI (1–9) HTH-type transcriptional regulator |
| 09 |
out of frame fusion |
| 10 |
out of frame fusion - |
| 11 |
DltE (1–48) involved in lipoteichoic acid biosynthesis |
| 12 |
YwpG (1–61) unknown function |
| 13 |
Hit (1–93) ubiquitous histidine triad protein |
| 14 | YdhI (1–81) acetyltransferase |
Previous data showed DynA-GFP localization to the membrane and sites of septation when expressed under its natural promoter in B. subtilis.20 Similarly DynA interacting partners YneK, RNaseY (YmdA) and YwpG tagged to GFP appeared as distinct foci on B. subtilis membrane and sites of septation (Fig. 2). Translational GFP fusions of YneK, YmdA and YwpG were also transformed into a ΔdynA background. However, no significant difference in localization of YwpG and YneK has been observed (Table 2). The only noticeable difference was a twofold decrease of RNaseY foci located at the septal region (Table 2). Since still one third of the RNaseY foci have been observed at the septum, DynA is at least not solely essential for RNaseY targeting. Since DynA depends on its septal localization on the cell division protein MinJ,30 we combined the YneK, RNaseY and YwpG GFP fusions with a minJ null allele (giving strains PSB007-009) and checked for altered localization. Indeed, in cells lacking MinJ the prominent foci seen for YneK and RNaseY were absent (Fig. 3). This observation may point to a DynA depended foci formation of YneK and RNaseY in B. subtilis, further supporting the potential interaction between these proteins. YwpG was still able to form foci at the cell pole in the absence of MinJ. Here, it seems unlikely that DynA is necessary for YwpG to form focal accumulations at the cell poles (Fig. 3).
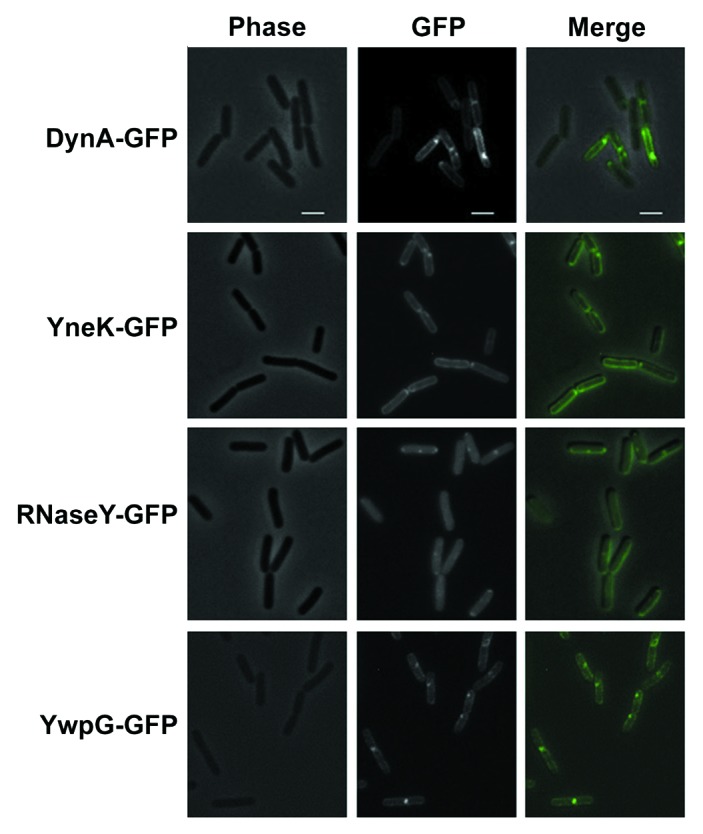
Figure 2. Localization of DynA interacting proteins. Strains (FBB018, PSB001–006) were grown in CH medium to mid-exponential phase and induced for 60 min with 0.1% xylose. YneK, RNaseY, and YwpG show similar localization compared with DynA. All proteins localize to the membrane and form foci that are often found at midcell positions. Scale bar 2 µm.
Table 2. Localization of DynA (ypbR) interaction partners into foci associated with the cell membrane.
| YneK-GFP (n = 142) |
YneK-GFP ΔdynA (n = 200) |
YwpG-GFP (n = 113) |
YwpG ΔdynA (n = 259) |
RNaseY-GFP (n = 331) |
RNaseY-ΔdynA (n = 219) |
|
|---|---|---|---|---|---|---|
| with foci |
95.1% |
97.5% |
97.3% |
95.0% |
90.6% |
91.8% |
| without foci |
4.9% |
2.5% |
2.7% |
5.0% |
9.4% |
8.2% |
| with foci at division site |
28.9% |
35.5% |
25.7% |
30.9% |
72.5% |
39.7% |
| > 1 foci/cell | 0% | 13% | 37.9% | 10.4% | 7.6% | 12.3% |
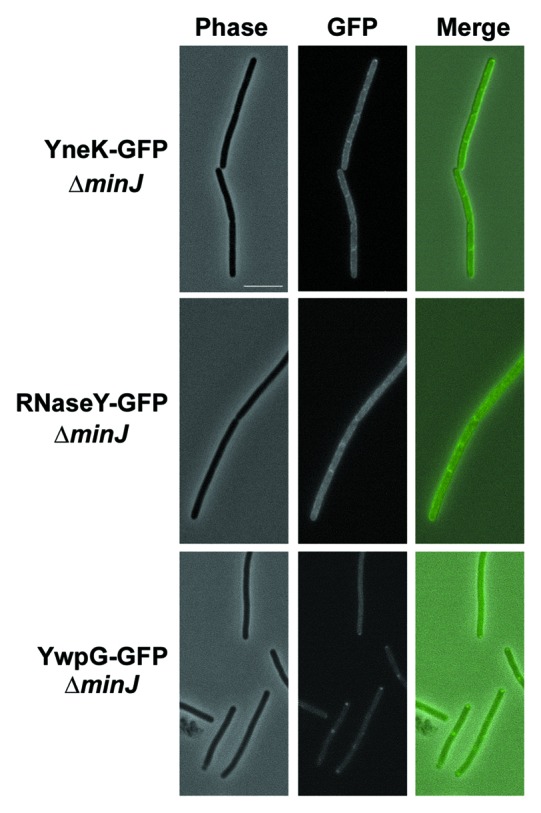
Figure 3.Localization of DynA interacting proteins in a minJ mutant background. Strains (PSB007–009) were grown in CH medium to mid-exponential phase and induced for 60 min with 0.1% xylose. YneK and RNaseY are localized to the membrane. Note that prominent foci are absent in ΔminJ strain backgrounds. YwpG localizes into discrete foci at the cell poles in absence of MinJ. Scale bar 5 µm.
In B. subtilis, DynA-GFP is enriched in focal domains or at the sites of septation whereas it is homogenously distributed across membranes in yeast.20 This indicates that in its natural host there might be additional factors which mediate clustering of DynA. Either certain lipids are required for foci formation, or the dynamin-like protein might be recruited to its focal sites by other proteins. Localization of the yeast dynamin-like protein Dnm1 on the mitochondrial outer membrane requires Fis1 and Mdv1.31-34 Fis1 is a membrane integral protein, while Mdv1 is soluble. Both proteins are required to target Dnm1 and are essential for efficient nucleation.35 The two-hybrid screen with B. subtilis DynA identified a putative transmembrane protein, YneK, which shares some limited domain similarity with Fis1. Both proteins contain a proposed bitopic domain structure. However, the putative transmembrane helices are at the N-terminal site (YneK) and towards the C-terminal end in Fis1. YwpG is a soluble protein that shares no similarity with Mdv1. Based on the different localization of YneK and YwpG (see Fig. 3) in absence of MinJ, which in turn leads to altered DynA localization, argues against a role for YneK and YwpG as putative DynA nucleation complex. Knowledge about YneK and YwpG is scarce. The gene yneK is located downstream of cotM, a gene encoding a spore coat protein, but is not expressed during sporulation.36 YwpG is upstream of the ssbB (ywpG)-glcR operon involved in DNA uptake and glucose repression.37,38 This operon is governed by the competence regulator ComK.39 However, YwpG may not be within the ssbB (ywpG)-glcR operon, which is driven by a promoter directly upstream of ywpH.39 So far no functional role has been assigned with YwpG and bioinformatic tools did not reveal any protein motif or similarity to known proteins. The obvious polar localization into foci and the chromosomal locus next to competence related genes, however, may point towards a role in DNA uptake since the competence DNA uptake machinery has been shown to be polar localized.40
The presence of RNA-binding KH domain and a highly conserved HD region suggested RNaseY to function in nucleic acid metabolism.41 RNaseY (YmdA) has been described as an essential RNase involved in processing of glycolytic mRNA,42 degradation of rpsO mRNA.43 It acts as an endonuclease by processing prematurely terminated mRNA transcripts.44 Recently, it has been shown that RNaseY is required for ribosomal RNA degradation in B. subtilis spores.45 RNaseY has a putative transmembrane region, which explains the membrane localization. RNaseY (YmdA) is localized homogenously across the membrane,46 however, occasional foci are evident that may depend on DynA (and/or MinJ; Fig. 2, Table 2). DynA-RNaseY interaction suggests that bacterial DLPs might participate in functions other than cell division. In summary we have identified three proteins that likely interact in vivo with the bacterial dynamin-like protein DynA (Fig. 4). Although, the functional role of these protein-protein interactions is not understood at present, localization studies show that all three DynA interactors show similar subcellular localization in vivo and at least two (YneK and RNaseY) may depend on DynA for correct localization. Our approach highlights the potential usefulness of a bacterial two-hybrid library. Although, caution has to be taken since false positive interactions were found in the first round of selection.
Figure 4. Interactome of DynA. B. subtilis DynA interacts specifically with YneK, YwpG and RNaseY. The YneK interaction is mediated by the D1 subdomain, while YwpG interacts with D1 and D2 domains. Interaction of RNaseY likely needs full length DynA.
Material and Methods
Strains and plasmids
All strains and plasmids used in this study are listed in Table 3. Oligonucleotides used are listed in Table 4.
Table 3. Bacterial strains and plasmids.
| Strain/plasmid |
Relevant genotype/characteristic trait |
Source |
|---|---|---|
| Bacillus subtilis | ||
| 168 |
trpC2 |
Laboratory stock |
| FBB002 |
dynA::tet trpC2 |
20 |
| FBB018 |
amyE::Pxyl-dynA-gfp spc dynA::tet trpC2 |
20 |
| PSB001 |
amyE::Pxyl-yneK-gfp spc trpC2 |
This study |
| PSB002 |
amyE::Pxyl-ywpG-gfp spc trpC2 |
This study |
| PSB003 |
amyE::Pxyl-ymdA-gfp spc trpC2 |
This study |
| PSB004 |
amyE::Pxyl-yneK-gfp spc dynA::tet trpC2 |
This study |
| PSB005 |
amyE::Pxyl-ywpG-gfp spc dynA::tet trpC2 |
This study |
| PSB006 |
amyE::Pxyl-ymdA-gfp spc dynA::tet trpC2 |
This study |
| PSB007 |
amyE::Pxyl-yneK-gfp spc minJ::tet trpC2 |
This study |
| PSB008 |
amyE::Pxyl-ywpG-gfp spc minJ::tet trpC2 |
This study |
| PSB009 |
amyE::Pxyl-ymdA-gfp spc minJ::tet trpC2 |
This study |
| RD021 | minJ::tet trpC2 | 30 |
| Escherichia coli | ||
|---|---|---|
| BTH101 |
F-, cya-99, araD139, galE15, galK16, rpsL1 (Strr), hsdR2, mcrA1, mcrB1 |
Euromedex |
| DH5α | F-endA1 hsdR17 supE44 thi-a1 λ- recA1 gyrA96 relA1 Δ(lacZYA-argf)U169 Ф80 Δ(lacZ)M15 | Invitrogen |
| Plasmids | ||
|---|---|---|
| pSG1154 |
Pxyl gfp, cat |
47 |
| pSG1154-dynA |
DynA-GFP |
20 |
| pSG1154-yneK |
YneK-GFP |
This study |
| pSG1154-ymdA |
YmdA-GFP |
This study |
| pSG1154-ywpG |
YwpG-GFP |
This study |
| pKT25 |
CyaA-T25 |
Euromedex |
| pUT18C |
CyaA-T18 |
Euromedex |
| pKNT25 |
CyaA-T25 |
Euromedex |
| pUT18 |
CyaA-T18 |
Euromedex |
| pUT18 YmdA |
CyaA-T18-YmdA |
This study |
| pUT18 YwpG |
CyaA-T18-YwpG |
This study |
| pUT18 YneK |
CyaA-T18-YneK |
This study |
| pKNT25 D1 |
CyaA-T25-DynA D1 domain |
20 |
| pKNT25 D1M |
CyaA-T25-DynA D1 K56A |
20 |
| pKNT25 D2 |
CyaA-T25-DynA D2 domain |
20 |
| pKNT25 D2M |
CyaA-T25-DynA D2 K625A |
20 |
| pKNT25 dynA | CyaA-T25-DynA | 20 |
Table 4. Oligonucleotides.
| Oligonucleotide | Describtion | Sequence | Restriction site | |
|---|---|---|---|---|
| 18C_seq_fwd |
pUT18C sequencing |
gttcgaagttctcgccggatg |
|
|
| 18C_seq_rev |
pUT18C sequencing |
cagcgggtgttggcgggtgtc |
|
|
| 18N_seq_fwd |
pUT18 sequencing |
caacgcaattaatgtgag |
|
|
| 18N_seq_rev |
pUT18 sequencing |
acgcgcctcggtgcccac |
|
|
| glpF_BTH_fwd |
glpF - > BTH |
|
cattctagagatgacagcattttggggaga |
XbaI |
| glpF_BTH_rev |
glpF - > BTH |
|
catggtacccgaatatatttagaatttgata |
KpnI |
| pksL_BTH_fwd |
pksL - > BTH |
|
cattctagagatgaggtggaggtctaacgt |
XbaI |
| pksL_BTH_rev |
pksL - > BTH |
|
catggtacccgtccgactaatgtataatcct |
KpnI |
| yneK_BTH_fwd |
yneK - > BTH |
|
catgtcgactatgctggaaggatggttttt |
SalI |
| yneK_BTH_rev |
yneK - > BTH |
|
catggtacccgtttgagaagggtctgataagg |
KpnI |
| ymdA_BTH_fwd |
ymdA - > BTH |
|
cattctagagatgaccccaattatgatggt |
XbaI |
| ymdA_BTH_rev |
ymdA - > BTH |
|
catggtacccgttttgcatactctacggctcgagtc |
KpnI |
| ydhI_BTH_fwd |
ydhI - > BTH |
|
cattctagagatgatgatcatcatcccaaacaatg |
XbaI |
| ydhI_BTH_rev |
ydhI - > BTH |
|
catggtacccgatcaattaccttcgaaaata |
KpnI |
| ywpG_BTH_fwd |
ywpG - > BTH |
|
cattctagagatgaatcaatttcgtttaaa |
XbaI |
| ywpG_BTH_rev |
ywpG - > BTH |
|
catggtacccggcctctgtttttatctttcgttttc |
KpnI |
| ybfI_BTH_fwd |
ybfI - > BTH |
|
cattctagagatgcaaaacgaaacccgcac |
XbaI |
| ybfI_BTH_rev |
ybfI - > BTH |
|
catggtacccgtctgtgaagctccttttcaa |
KpnI |
| dltE_BTH_fwd |
dltE - > BTH |
|
cattctagagatgaagatgacaaataatac |
XbaI |
| dltE_BTH_rev |
dltE - > BTH |
|
catggtacccgattctcctgcgtgttcatttgctcg |
KpnI |
| hit_BTH_fwd |
hit - > BTH |
|
cattctagagatgcattgtgcagagaattg |
XbaI |
| hit_BTH_rev |
hit - > BTH |
|
catggtacccgtgatgaggccaggcgttttgcgata |
KpnI |
| yneK_for |
yneK- gfp |
|
catctcgagatgctggaaggatggtttttatg |
XhoI |
| ynk_rev |
yneK- gfp |
|
cataagctttttgagaagggtctgataagg |
HindIII |
| ymdA_for |
ymdA- gfp |
|
catgtcgacatgaccccaattatgatgg |
SalI |
| ymdA_rev |
ymdA- gfp |
|
catgaattcttttgcatactctacggctc |
EcoRI |
| ywpG_for |
ywpG- gfp |
|
catctcgagatgaatcaatttcgtttaaaag |
XhoI |
| ywpG_rev |
ywpG- gfp |
|
cataagcttgcctctgtttttatc |
HindIII |
| Ypbr-B-2-H-F |
dynA- > BTH |
|
cattctagagacagatcaaaacag |
XbaI |
| Ypbr-B-2-H-R |
dynA- > BTH |
|
catggtaccatttttattgtattgtctg |
KpnI |
| D1_B2H_rev |
D1 - > BTH |
|
catggtaccctatcaagccttttcaccct |
KpnI |
| D2_B2H_fwd | D2 - > BTH | cattctagagatgccgaaatcagaaatcaaaa | XbaI | |
Genomic library generation
A blunt end ligation strategy with SmaI for vector digestion and AluI for fragment preparation was used. gDNA digestion was optimized as above and fragments were prepared as described with a reaction time of 40 min at 37° C using 0.5 U AluI per mg gDNA. pUT18 was digested at 40 ng ml-1 with 2 U ml-1 SmaI for 2 h at 25° C and the enzyme was inactivated for 20 min at 65° C. Subsequently, the vector was dephosphorylated with 5 U antarctic phosphatase for 1 h at 37° C to prevent self-ligation. Approximately 100,000 transformations were plated on large dishes at approximately 5,000 clones per plate and colonies were harvested by scraping each plate with 2×5 ml LB. Pooled cells were mixed for 2 h at 4° C on a rolling mixer to obtain a homogenous cell suspension. Finally, a fraction corresponding to 1 ml of a E600 = 8.0 culture was used for plasmid preparation.
Bacterial two-hybrid procedures
For two-hybrid assays, BTH101 cells were double transformed with plasmid combinations using 96-well plates and spotted on LB (50 mg ml-1 carbenicillin, 25 mg ml-1 kanamycin, 0.5 mM IPTG, 160 mg l-1 X-Gal). A double transformation of empty vectors served as negative control. Positive control was a double transformation of pUT18C_zip and pKT25_zip (Euromedex), encoding an interacting leucine zipper. For the two hybrid screen, the pUT18 library was transformed into BTH101 aiming at 200,000 transformants. Cells were regenerated in prewarmed LB for 15 min at 37° C, then washed four times in M63 and plated on M63 with 50 mg ml-1 carbenicillin, 25 mg ml-1 kanamycin, 0.5 mM IPTG, 40 mg ml-1 X-Gal and 0.2% D(+)-maltose at 100,000 clones per plate. Clones appeared after approximaltely 60 h at 30° C.
Generation of bacterial clones
C-terminal GFP fusions were constructed using the plasmid pSG1154.47 DynA interacting partners—YneK, YmdA and YwpG were PCR amplified from the genome of B. subtilis 168 with specific primers using Phusion polymerase (Thermo Scientific). DNA was digested using FastDigest® Restriction Enzymes (Fermentas). T4 DNA ligase (1 U μl-1) from Fermentas was used to ligate digested plasmid and inserts.
Null alleles of dynA (FBB002) or minJ (RD021) were transformed with chromosomal DNA of strains PSB001-003, respectively and selected on spectinomycin plates. Correct integration into the amyE locus was checked on starch (0.1%) plates that where subsequently stained with iodine.
Fluorescence microscopy
Localization of GFP tagged YneK, YmdA and YwpG in B. subtilis was studied using a Zeiss AxioImager M1 equipped with a Zeiss AxioCam HRm camera. An EC Plan-Neofluar 100x/1.3 Oil Ph3 objective was used. GFP fluorescence was monitored using filter set 38 HE eGFP. For microscopy freshly grown cells were used to inoculate 10 ml CH medium. Cells were induced with 0.1% xylose at mid-exponential phase. After 60 minutes induction 2 μl of cell culture (around OD600 1) were placed on agarose bed on a glass slide, covered with a glass slip, and observed under light microscope.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors acknowledge financial support from the Deutsche Forschungsgemeinschaft (SFB 635, project B8).
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/20215
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–47. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Pucadyil TJ, Schmid SL. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325:1217–20. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szaszák M, Gáborik Z, Turu G, McPherson PS, Clark AJ, Catt KJ, et al. Role of the proline-rich domain of dynamin-2 and its interactions with Src homology 3 domains during endocytosis of the AT1 angiotensin receptor. J Biol Chem. 2002;277:21650–6. doi: 10.1074/jbc.M200778200. [DOI] [PubMed] [Google Scholar]
- 4.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–6. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 5.Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol. 2004;11:574–5. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- 6.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, et al. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–60. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 7.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–6. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–4. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 9.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–83. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 11.Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–52. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- 12.Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–73. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–62. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 14.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–46. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 15.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–53. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 16.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–34. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 17.Wadenpohl I, Bramkamp M. DivIC stabilizes FtsL against RasP cleavage. J Bacteriol. 2010;192:5260–3. doi: 10.1128/JB.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Baarle S, Bramkamp M. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One. 2010;5:e9850. doi: 10.1371/journal.pone.0009850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bramkamp M, van Baarle S. Division site selection in rod-shaped bacteria. Curr Opin Microbiol. 2009;12:683–8. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Bürmann F, Ebert N, van Baarle S, Bramkamp M. A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol. 2011;79:1294–304. doi: 10.1111/j.1365-2958.2011.07523.x. [DOI] [PubMed] [Google Scholar]
- 21.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–9. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 22.Binns DD, Barylko B, Grichine N, Atkinson MA, Helms MK, Jameson DM, et al. Correlation between self-association modes and GTPase activation of dynamin. J Protein Chem. 1999;18:277–90. doi: 10.1023/A:1021083211267. [DOI] [PubMed] [Google Scholar]
- 23.Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–66. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–83. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–43. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–8. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 27.Denzer AJ, Nabholz CE, Spiess M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 1995;14:6311–7. doi: 10.1002/j.1460-2075.1995.tb00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–56. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 29.Clarke L, Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–9. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 30.Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol. 2008;70:1556–69. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- 31.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–20. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–52. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282:33769–75. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Chan NC, Ngo HB, Gristick H, Chan DC. Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (mdv1) coiled coil. J Biol Chem. 2012;287:9855–61. doi: 10.1074/jbc.M111.329359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–7. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henriques AO, Beall BW, Moran CP., Jr CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–97. doi: 10.1128/jb.179.6.1887-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindner C, Nijland R, van Hartskamp M, Bron S, Hamoen LW, Kuipers OP. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J Bacteriol. 2004;186:1097–105. doi: 10.1128/JB.186.4.1097-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stülke J, Martin-Verstraete I, Glaser P, Rapoport G. Characterization of glucose-repression-resistant mutants of Bacillus subtilis: identification of the glcR gene. Arch Microbiol. 2001;175:441–9. doi: 10.1007/s002030100288. [DOI] [PubMed] [Google Scholar]
- 39.Nijland R, Lindner C, van Hartskamp M, Hamoen LW, Kuipers OP. Heterologous production and secretion of Clostridium perfringens beta-toxoid in closely related Gram-positive hosts. J Biotechnol. 2007;127:361–72. doi: 10.1016/j.jbiotec.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Kidane D, Graumann PL. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell. 2005;122:73–84. doi: 10.1016/j.cell.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 41.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–72. doi: 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 42.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, et al. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–60. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao S, Bechhofer DH. Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol. 2010;192:3279–86. doi: 10.1128/JB.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–33. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segev E, Smith Y, Ben-Yehuda S. RNA dynamics in aging bacterial spores. Cell. 2012;148:139–49. doi: 10.1016/j.cell.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 46.Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology. 2006;152:2895–907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- 47.Lewis PJ, Marston AL. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene. 1999;227:101–10. doi: 10.1016/S0378-1119(98)00580-0. [DOI] [PubMed] [Google Scholar]