Abstract
Tel2, a protein conserved from yeast to vertebrates, is an essential regulator of diverse cellular processes including telomere maintenance, DNA damage checkpoints, DNA repair, biological clocks, and cell signaling. The Drosophila Tel2 protein is produced as a translational fusion with EpsinR, a Clathrin adapter that facilitates vesicle trafficking between the Golgi and endosomes. EpsinR and Tel2 are encoded by a Drosophila gene called lqfR. lqfR is required for viability, and its specific roles include cell growth, proliferation, and planar cell polarity. We find that all of these functions of lqfR are attributed entirely to Tel2, not EpsinR. In addition, we find that Drosophila LqfR/Tel2 is a component of one or more protein complexes that contain E-cadherin and Armadillo. Moreover, Tel2 modulates E-cadherin and Armadillo cellular dynamics. We propose that at least one of the functions of Drosophila Tel2 is regulation of Wingless signaling.
Introduction
Tel2 is a protein shown to be essential in yeast, nematodes, and vertebrates, that functions in diverse pathways for reacting to a variety of cellular stresses and cues including DNA damage, abnormal mRNAs, nutrient availability, mitogens, and cell cycle progression [1]. Tel2 functions as a co-chaperone with Hsp90 in PIKK complex assembly [2]–[4]. The role of Tel2 in PIKK assembly has been proposed to explain all of its functions, but this point is highly controversial [5]–[7].
The tel2 gene was identified originally as an essential gene in budding yeast S. cerevisiae in a screen for mutants with short telomeres [8]. Genes homologous to tel2 were found to be essential also in S. pombe, C. elegans, and mice, but the phenotypes of the mutants and subsequent biochemical studies indicated that Tel2 function is not limited to telomere dynamics [2], [6], [7], [9]–[17].
In the course of a study of the Drosophila gene encoding Golgi Epsin or Epsin-Related (EpsinR), we and others [18] discovered that one isoform of Drosophila EpsinR is a translational fusion with the only Tel2 coding sequences in Drosophila. EpsinR is multi-modular protein conserved from yeast to vertebrates that promotes Clathrin-coated vesicle formation at the trans-Golgi network and endosomes and thereby modulates Golgi-endosome trafficking [19]–[26]. A similar protein conserved in yeast through vertebrates, endocytic Epsin, promotes Clathrin-coated vesicle formation at the plasma membrane [27], [28]. Endocytic Epsin is an essential component of the Notch signaling pathway [29], [30]. As endocytosis and endosomal trafficking play key roles in a variety of signaling mechanisms [31], we were curious whether like endocytic Epsin, Golgi Epsin might be crucial to a particular signaling pathway. To this end, we generated Drosophila with loss-of-function mutations in the single EpsinR gene, called liquid facets-Related (lqfR) [32]. The lqfR mutant phenotype is complex; there are defects in planar cell polarity and cell size, proliferation, and patterning [32]. Here we show that these morphological defects of lqfR mutants are due entirely to the loss of Tel2 activity. Moreover, we show that the essential Tel2 function in Drosophila is at least in part direct regulation of the Wingless signaling pathway.
Results and Discussion
Exon 6 of lqfRa encodes the Drosophila Tel2 homolog
The lqfR gene pre-mRNA is alternatively spliced to generate mRNAs with different C-terminal exons and thus two different proteins, LqfRa (1415 aa) and LqfRb (649 aa) (Fig. 1) [18], [32]. Both LqfRa and LqfRb have structural elements characteristic of Golgi Epsin: the ENTH domain and binding motifs for AP-1 and Clathrin. The larger protein also contains a domain encoded by its LqfRa-specific C-terminal exon 6 (921 aa) that is homologous to Tel2. Tel2 is a Y-shaped protein in the HEAT repeat family of superhelical proteins, in which 32 interacting α-helices are packed to generate two α-solenoids that form the long (21 α-helices) and short (11 α-helices) lines of the Y. Human Tel2 and LqfRa exon6 are 19% identical and 13% similar in amino acid sequence throughout the length of their polypeptide chains (Fig. S1).
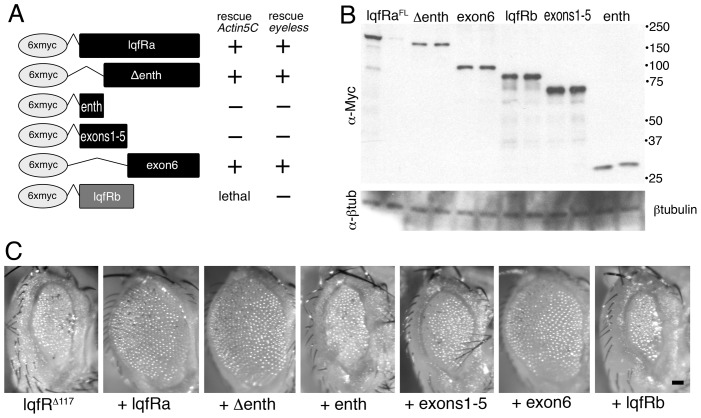
Figure 1. lqfR gene products.
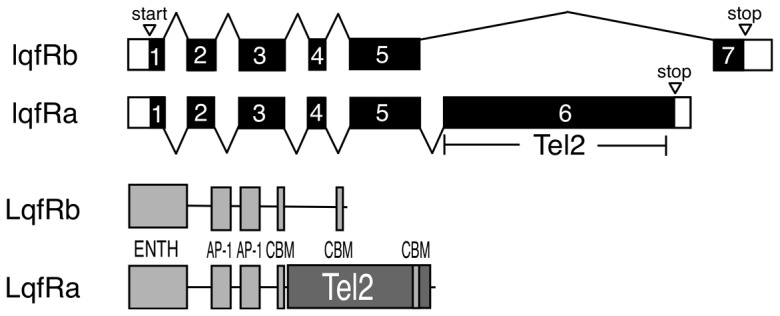
At top is a diagram of the two lqfR mRNAs formed by alternative pre-mRNA splicing. Exons 1–7 are indicated by bars and introns by bent lines. The black region in each transcript is the open reading frame. The larger transcript, lqfRa, contains exon6 which encodes Tel2. At bottom are the protein products of each mRNA. ENTH = Epsin N-terminal homology domain, AP-1 = binding motif for the Clathrin adapter AP-1, CBM = Clathrin binding motif.
The evidence that exon 6 is not a separate gene - that the lqfRa splice form truly exists - is convincing. First, there is compelling evidence that exons 5 and 6 are joined in an mRNA; an RT-PCR amplification product containing exon 5 spliced to exon 6 has been generated [32]. Moreover, an exon 3–5 probe hybridizes not only to a species the size of lqfRb, but also to a larger mRNA corresponding in size to lqfRb that also hybridizes to an exon 6 probe. Second, there is strong evidence that LqfRa and LqfRb proteins both exist; antibodies generated to parts of LqfR that exclude the region encoded by exon 6 hybridize to two bands on Western blots, one corresponding in size roughly to LqfRa, and the other to LqfRb [18], [32].
We were curious to determine whether or not the fusion of the genes for Golgi Epsin and Tel2 was specific to Drosophila. We performed two BLAST searches (at www.uniprot.org), one using as a query the amino acid sequence of LqfR exons 1–5, and the other using exon 6. In some species that had clear homologs of both genes, both queries identified the same gene or adjacent genes, indicating that lqfR and tel2 are likely fused in that species. In other species each query identified distinct, non-adjacent genes. Although our analysis was not exhaustive, we did find that the lqfR and tel2 genes were likely fused in all queried Drosophila species and also in other insects in the database, but not in yeast, nematodes, nor any vertebrates (data not shown).
Exon 6 of lqfRa is necessary and sufficient for all lqfR/Tel2 gene functions tested
We found previously [32] that either full-length LqfRa fused at its C-terminus to GFP (LqfRaFL-GFP) or a version of the fusion protein that lacks the ENTH domain (LqfRaΔENTH -GFP), when expressed using Gal4/UAS and the ubiquitous Actin5C-gal4 driver, is sufficient to rescue all of the obvious defects due to loss of lqfR+ gene activity: these include larval lethality and the absence of imaginal discs. The dispensability of the ENTH domain was not entirely surprising, as endocytic Epsin also functions well without its ENTH domain [33], [34]. However, further structure/function experiments did yield results that were completely unexpected.
First, we generated five UAS transgenes in P element vectors, in which full-length (FL) lqfRa or four deletion derivatives were tagged with 6xmyc epitope coding sequences at their 5′ ends (Fig. 2A) and used them to transform Drosophila. Each transgene was tested for its ability when expressed with Actin5C-gal4 to substitute for the endogenous lqfR gene. The results obtained by expressing LqfRaFL-GFP or LqfRaΔENTH-GFP described above were recapitulated by 6xmyc-LqfRaFL and 6xmyc-LqfRaΔENTH: expression of either protein rescued lqfR null mutants to wild-type (Fig. 2A). In contrast, neither the ENTH domain alone (6xmyc-LqfRENTH) nor exons 1–5 alone (6xmyc-LqfRex1-5) had any rescuing activity (Fig. 2A). This was not due to a failure of transgene expression as the 6xmyc-LqfRENTH and 6xmyc-LqfRex1-5 proteins accumulated in the flies to levels at least as high as 6xmyc-LqfRaΔENTH (Fig. 2B). The most remarkable result was that exon 6 alone (6xmyc-LqfRexon6) rescued lqfR null mutants to wild-type (Fig. 2A). In summary, we found that expression of exon 6, which contains only the Tel2-like region of LqfRa, was sufficient to rescue the imaginal disc proliferation and patterning defects of lqfR null mutants and no other portions of LqfRa were able to provide any rescuing activity independently.
Figure 2. Rescue of lqfR null mutant phenotype by lqfRa exon 6.
(A) At left, the table shows six epitope-tagged proteins expressed in Drosophila by a UAS transgene. The columns at right show the results when each transgene was expressed in a lqfRΔ117 or lqfRΔ117/Df(3R)Exel6191 background with either an Actin5C-gal4 or an eyeless-gal4 driver. +: lethality and externally obvious morphological defects were rescued, − : no rescue. (B) A blot of electrophoresed adult fly protein extracts probed first with antibodies to the Myc tag (α-Myc) and reprobed with antibodies to β-tubulin (α-βtub) as a loading control. The flies contain the UAS construct indicated and an eyeless-gal4 driver. The genotypes of the flies used were: EGUF/UAS; FRT82B lqfRΔ117/TM6B. For each UAS construct, two different P element transformant lines were tested. Note that one of the UAS-lqfRaFL lines expressed little or no protein and this line also failed to rescue the lqfRΔ117 mutant phenotype. The numbers at the right of the blot indicate the positions of corresponding size markers (kD). (C) Light microscope images of the eyes of adult flies. The flies are lqfRΔ117/lqfR+ and their eyes are lqfRΔ117 homozygous clones. The fly at the very left has no UAS transgene and the others contain a copy of the UAS transgene indicated, expressed by eyeless-gal4. The genotypes of the flies were: EGUF/UAS; FRT82B lqfRΔ117/FRT 82B GMR-hid. scale bar: ∼50 µm.
The results so far predict that LqfRb, which does not contain exon 6, would not have any rescuing activity. We could not test LqfRb in the assay described above because unexpectedly, expression of UAS-6xmyc-lqfRb with Actin5C-gal4 was lethal. To overcome this obstacle, we expressed 6xmyc-lqfRb in the eye only, and asked if the eye morphology defects in eyes with no LqfR protein in otherwise normal flies (lqfRΔ117/lqfR+ flies with homozygous lqfRΔ117 eyes generated using FRT/GMR-hid) were rescued. lqfRΔ117 eyes are tiny and rough compared to wild-type (Fig. 2C). Flies expressing 6xmyc-LqfRb with ey-gal4 were viable, and we found that 6xmyc-LqfRb had no rescue activity in the eye (Fig. 2A,C). As controls, we tested the four transgenes described above and we found the same results with those as we did with the Act5C-gal4 driver: 6xmyc-LqfRaFL, 6xmyc-LqfRaΔENTH, or 6xmyc-LqfRexon6 rescued lqfRΔ117 eyes to wild-type and 6xmyc-LqfRex1-5 did not rescue (Fig. 2A,C). The failure of 6xmyc-LqfRb to rescue was not due to failure of protein expression, as LqfRb protein accumulated to levels similar to those of LqfRexon6 (Fig. 2B). The inability of lqfRb to complement the lqfR mutant phenotype is consistent with the finding that exon 6 alone of lqfRa is sufficient to do so.
We conclude that Golgi Epsin and Tel2, although fused in LqfRa, are independent protein functions. Moreover, the external morphology and lethality aspects of the mutant phenotype described for lqfR null mutants reflects only the loss of Tel2 activity, and not the loss of Golgi Epsin. We therefore propose renaming the lqfR gene lqfR/tel2.
The Tel2-like portion of LqfRa encoded by exon 6 expressed alone is mainly nuclear
Using either of two different polyclonal antibodies, one to LqfR exons 1–5 and the other to an ENTH-less LqfRb, LqfR was shown to colocalize with Golgi markers in the eye and elsewhere [18], [32]. We were curious to know where the truncated protein consisting of LqfRa exon 6 alone (6xmyc-LqfRexon6) accumulates in the cell. Full length 6xmyc-LqfRaFL monitored with anti-Myc had a cytoplasmic localization pattern similar to that of endogenous LqfR and other Golgi markers (Fig. 3) [18], [32]. By contrast, 6xmyc-LqfRexon6 was mainly nuclear (Fig. 3). Co-labeling with TOPRO3 suggests that exon6 is at the nuclear envelope because it does not colocalize with DNA, but surrounds it (Fig. 3). Further experiments are required to determine whether Tel2 is localized to the nuclear side or the cytoplasmic side of the nuclear envelope. Nevertheless, the majority of the Tel2-like portion of LqfRa does not localize to the Golgi as the full length LqfR protein does, and yet it is sufficient to rescue the lqfR/Tel2 mutant phenotype. The implication is that the essential lqfRa/tel2 gene function may not be at the Golgi.
Figure 3. Subcellular localization of Myc-tagged LqfR proteins.
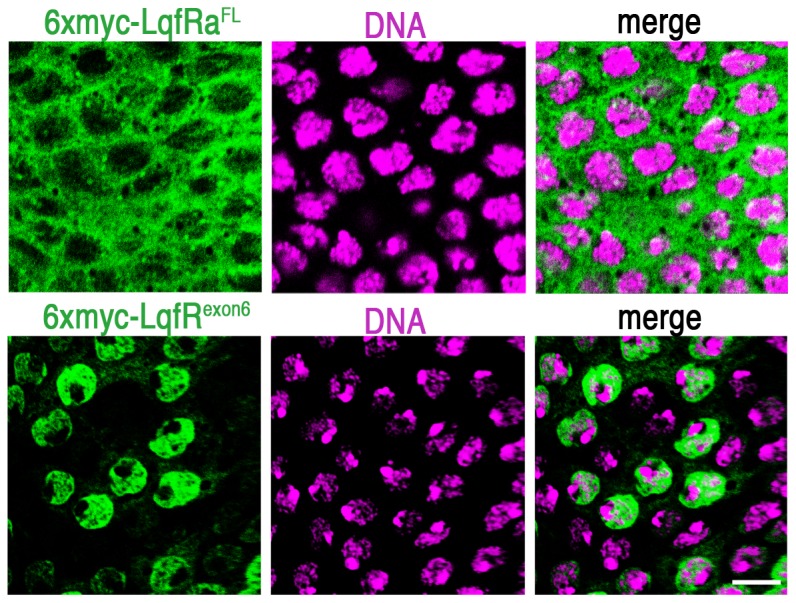
Confocal microscope images of third instar larval eye disc tissue from two different discs (each row is a single disc) are shown. The portion of the eye disc shown is the peripodial epithelium, a layer of cells that lies atop the cell layer that forms the retina. The peripodial cells are large and flat the nuclei and cytoplasm are distinguished more easily than in the retinal cells. The discs were immunostained with antibodies to the Myc epitope (green) and the DNA stain TOPRO3 (purple). The Myc-tagged proteins indicated were expressed by UAS transgenes using an Actin5C-gal4 driver. scale bar: ∼10 µm.
These results raise a question: as LqfRa/Tel2 contains the amino acids encoded by exons 1–5 and exon 6, why does the antibody to LqfR exons 1–5 not include the labeling pattern of 6xmyc-LqfRexon6 – that is the nuclear envelope? One explanation may lie in the observation that LqfRb, which lacks the exon 6-encoded amino acids, is the majority of the LqfR protein present in eye discs [32]. Thus, the antibody to exons 1–5 may be detecting LqfRb only. Alternatively, it is possible that the exon 6-encoded Tel2 region of LqfRa is cleaved post-translationally from the N-terminus of the protein, so that the antibody to exons 1–5 does not detect exon 6-encoded protein. Yet another possibility is that is LqfRa/Tel2 normally shuttles between the cytoplasm and the nucleus and the 6xmyc-Tel2 protein fusion is retained at the nuclear envelope abnormally. The generation of an antibody specific to the Tel2-like region of LqfRa might help to distinguish among these alternatives.
Wingless pathway genes interact strongly with lqfR/tel2
The specific cell growth and patterning defects in lqfR/Tel2 mutants are suggestive of defects in a variety of different signaling pathways [32]. Wingless signaling, for example, regulates both cell proliferation and patterning in the eye [35]. Wingless regulates initiation of the wave front of eye morphogenesis called the morphogenetic furrow. In addition, Wingless expressed at the lateral margins of the eye disc forms a gradient that results in formation of a dorsal/ventral midline called the equator about which the facets, or ommatidia, are mirror-image symmetrical. Separation of eye and head cuticle tissue also requires Wingless. As the lqfR/tel2 mutant phenotype includes defects in morphogenetic furrow movement and planar cell polarity in both the eye and wing [32], it seemed reasonable that the function of lqfR/tel2 could somehow relate to the Wingless pathway.
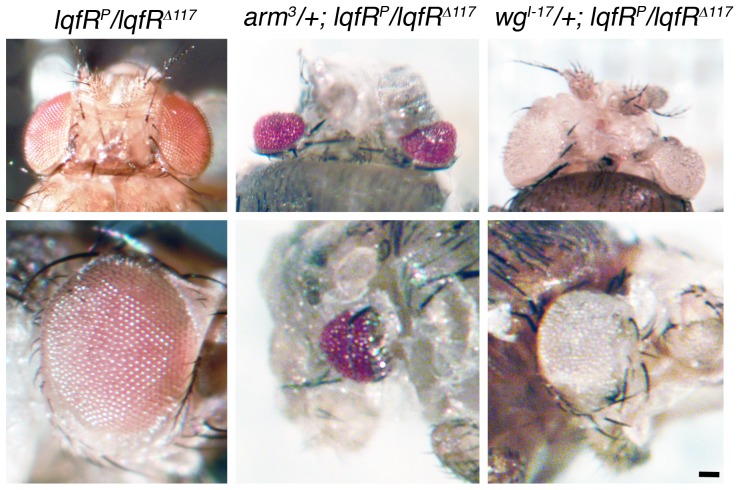
We tested two genes encoding core components of the Wingless pathway, wingless and armadillo, for interactions with lqfR/Tel2. Wingless ligand binds its receptor Frizzled which results in accumulation in the nucleus of the transcriptional regulator Armadillo [36]. We found strong genetic interactions between lqfR/tel2 and each of the two Wingless pathway genes. Heterozygotes for a hypomorphic allele and a null allele of lqfR (lqfRP/lqfRΔ117) are semi-viable and the adult escapers may have normal, slightly roughened, or kidney-shaped eyes [32] (Fig. 4). Flies that were lqfRP/lqfRΔ117 and also heterozygous for a loss-of-function allele of armadillo (arm3 or arm8) died in their pupal cases. (arm-/arm+ flies appear wild-type.) The pupae had small or absent eyes, small head capsules, and also had morphological defects in the head cuticle, wings, and legs (Fig. 4). wingless loss-of-function mutations (wgI-17 or wgI-8) had similar but weaker effects when heterozygous in a lqfRP/lqfRΔ117 background; there were viable adult escapers with severely defective eyes varying from kidney-shaped to nearly absent, and also with defects in the head cuticle (Fig. 4). (wg−/wg+ animals appear wild-type.) These strong genetic interactions suggest that lqfR/tel2 may function in the Wingless signaling pathway.
Figure 4. Genetic interactions between lqfR, armadillo, and wingless.
Shown are light microscope images of adult fly heads (dorsal view, top row), and eyes (bottom row). The genotypes of each column are indicated at top. The same fly is shown in the top and bottom rows. scale bar: ∼50 µm.
Wingless target gene expression depends to some extent on lqfR/tel2 function
The dominant enhancement of lqfR/tel2 mutant phenotypes by loss of function mutations in Wingless pathway genes suggests that lqfR/tel2 facilitates Wingless pathway activation. To test this idea, we generated lqfR/tel2 null clones in eye discs and monitored expression of the Wingless target genes dachsous (ds) [36], [37] and optomotor blind (omb) [38]. Weak effects on target gene expression were apparent in both cases. As the effects on ds expression was stronger, this data is shown below.
The dachsous gene encodes an atypical cadherin adhesion protein involved in cell polarity and cell growth and is a transcriptional target of Arm [37], [38]. ds-lacZ enhancer trap lines express β-galactosidase in response to Wg pathway activation [38]. Wingless ligand is expressed in the dorsal- and ventral-most margins of the eye disc and the protein forms a gradient with its lowest point at the dorsal/ventral axis (the equator) [40]. β-galactosidase expression by ds-lacZ reflects the Wg gradient [39]. We found that ds-lacZ expression was reduced in lqfR/tel2 null clones (Fig. 5). Moreover, we found that a dachsous loss-of-function mutation, ds38K, is as strong a dominant enhancer of the lqfRP/lqfRΔ117 mutant phenotype as are armadillo mutations (Fig. 5). These results suggest that in the absence of LqfR/Tel2, Wg signaling is less efficient than it is normally.
Figure 5. Genetic interactions between lqfR and dachsous.
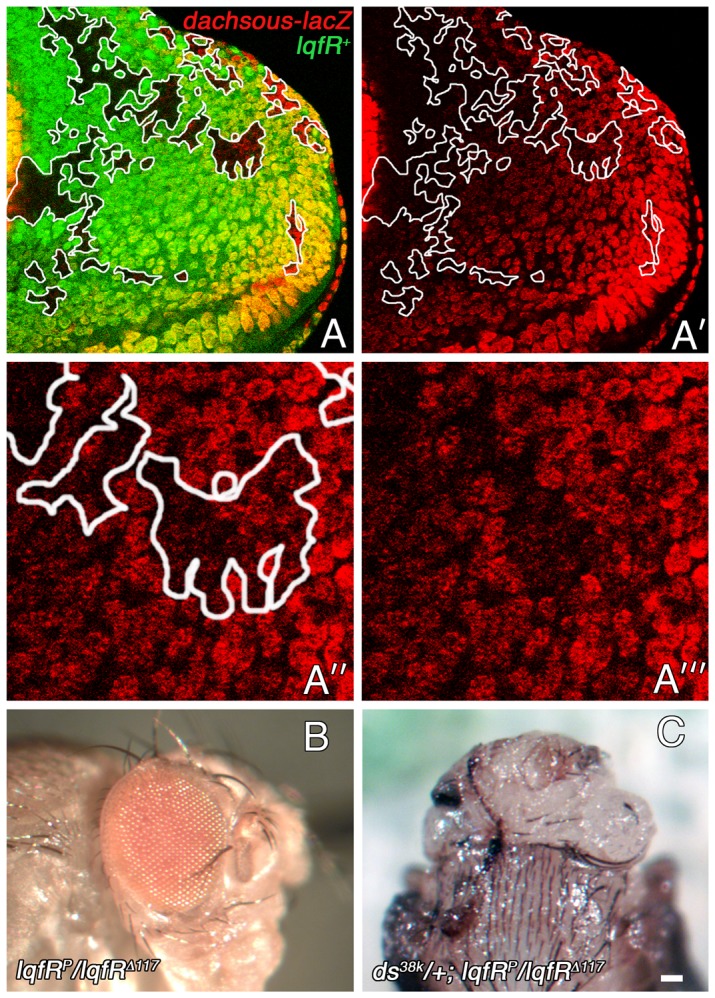
(A-A′″) Confocal microscope images of an eye disc immunostained with antibodies to β-galactosidase are shown. The disc expresses GFP in all cells except for lqfRΔ117 homozygous clones. The genotype is ds-lacZ/+; FRT82B lqfRΔ117/FRT82B ubi-gfp. (A′) Clones are outlined. (A″,A″′) Enlargements of part of A′ which shows that ds-lacZ expression levels are lower in lqfRΔ117 clones than in adjacent wild-type tissue. (B) A light microscope image of an eye from an adult fly hypomorphic for lqfR is shown. (C) The head (dorsal view) of a pupa that will not eclose dissected from its pupal case. ds38k/ds+ animals (not shown) appear wild-type. scale bar: ∼10 µm in A,A′; ∼5 µm in A″, A″′; ∼100 µm in B,C.
The effects of lqfR/tel2 loss of function on Wingless target gene expression are weaker than expected based on the dramatic genetic interactions between lqfR/tel2 mutations and mutations in arm, wg, or ds. (We also monitored expression of the Wingless target gene senseless (sens) [41] in wing disc lqfR/tel2 null clones but could see no effect). One possible explanation is that the effects of a modest decrease in Wingless signaling are amplified by downstream effects on other signaling pathways. Therefore, the combined effect of losses in several signaling pathways in lqfR/tel2 mutants could account for the striking genetic interactions.
Plasma membrane levels of E-cadherin and Armadillo increase in the absence of lqfR/Tel2 activity
In a mutagenesis screen for dominant enhancers of the lqfR/tel2 mutant eye phenotype (Lee et al., manuscript preparation), we identified loss-of-function alleles of polychaetoid, which encodes the Drosophila homolog of vertebrate ZO-1 [42]. in ZO-1/Polychaetoid is present at tight junctions and adherens junctions, where it connects other proteins present there to the actin cytoskeleton [42]–[44]. Although the mechanism is unclear, loss of Polychaetoid in Drosophila results in increased accumulation of the cell adhesion protein E-cadherin at the plasma membrane [43].
The transmembrane protein E-cadherin is a central component of adherens junctions through homotypic interactions between E-cadherin extracellular domains on adjacent cells [45]. The intracellular domain of E-cadherin binds proteins, including Armadillo and α-catenin, which are essential for E-cadherin's function as a cell adhesion protein [46], [47] Because E-cadherin binds Armadillo, E-cadherin function effects Wingless signaling [48]. Notably, E-cadherin overexpression antagonizes Wingless signaling, presumably by preventing Armadillo from entering the nucleus [49], [50].
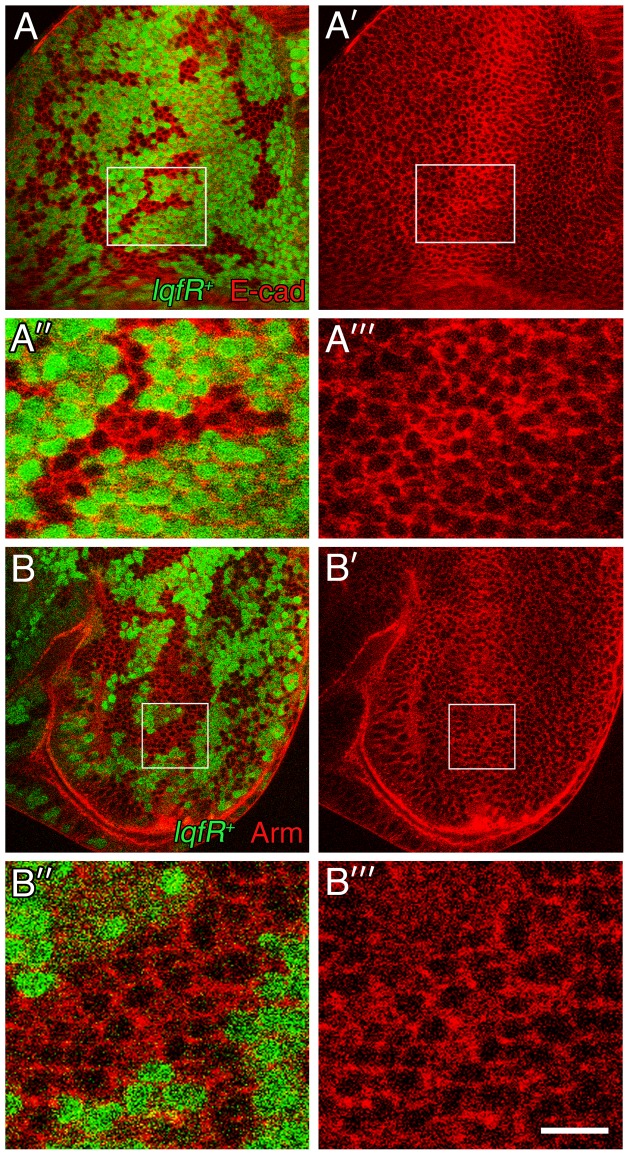
We wondered whether the genetic interaction between polychaetoid and lqfR/tel2, which suggests that both genes facilitate Wingless signaling, could be explained by the effect of Polychaetoid on E-cadherin levels. To test this hypothesis, first we asked whether E-cadherin levels, which increase in the absence of Polychaetoid, were also elevated in eye disc clones lacking LqfR/Tel2. We found that E-cadherin levels do indeed increase in lqfR/tel2 null clones; this effect appears most dramatic near the morphogenetic furrow where the highest levels of E-cadherin accumulate normally (Fig. 6). In discs where either 6xmyc-LqfRaFL or 6xmyc-LqfRexon6 were overexpressed, lqfR/tel2 null clones had the same levels of E-cadherin as surrounding wild-type tissue, confirming that the effect on Cadherin is mediated by Tel2 (Fig. S2). Like E-cadherin levels, Armadillo levels at the plasma membrane also are higher than usual in the absence of LqfR/Tel2 (Fig. 6). We conclude that the Tel2-like portion of LqfRa modulates Wingless signaling through an effect, either direct or indirect, on E-Cadherin levels and Armadillo localization.
Figure 6. E-cadherin and Armadillo protein accumulation in lqfR- clones.
(A,A′) Confocal microscope images of an eye disc immunostained with E-cadherin antibodies (red). lqfR- clones are marked by the absence of GFP (green). (A″,A″′) Enlargements of the boxed regions in A and A′. (B,B′) Confocal microscope images of an eye disc immunostained with Armadillo antibodies (red). lqfR- clones are marked by the absence of GFP (green). The genotype for both experiments is ey-flp; FRT82B lqfRΔ117/FRT82B ubi-gfp. scale bar: ∼40 µm in A,A′,B,B′; ∼10 µm in A″, A″′,B″,B″′.
LqfR/Tel2 interacts physically with E-cadherin, Armadillo and α-catenin
To determine whether or not the effect of LqfR/Tel2 on E-cadherin and Armadillo is direct, we asked whether LqfR/Tel2 is present in a complex with either protein. We also tested for physical interactions between LqfR/Tel2 and the adherens junction protein α-catenin, which binds Armadillo. First, we used antibodies to GFP to immunoprecipitate LqfRa-GFP from fly embryos that overexpress it (Actin5C>lqfRa-gfp). Next, using antibodies to each of the three proteins on blots, we determined whether E-cadherin, Armadillo, or α-catenin were also present in the precipitate. We found that each of the three proteins co-immunoprecipitated with LqfRa-GFP (Fig. 7). The adherens junction proteins are not binding to GFP because we did the same experiment with embryos that overexpress the ENTH domain only fused to GFP (LqfRENTH-GFP) and we found that LqfRENTH-GFP did not coimmunoprecipate with any of the three proteins (Fig. 7). Moreover, the correlation between rescue of the lqfR/tel2 mutant phenotype and binding to the adherens junction proteins (LqfRa-GFP both rescues and binds and LqfRENTH-GFP does neither) suggests that the interaction between LqfR/Tel2 and E-cadherin, Armadillo and α-catenin may be relevant to the lqfR/tel2 mutant phenotype and that the effect of LqfR/Tel2 on adherens junctions is direct. Further experiments are required to determine whether or not all four proteins are present in a single complex.
Figure 7. Coimmunoprecipitation of LqfRa and Wingless pathway proteins.
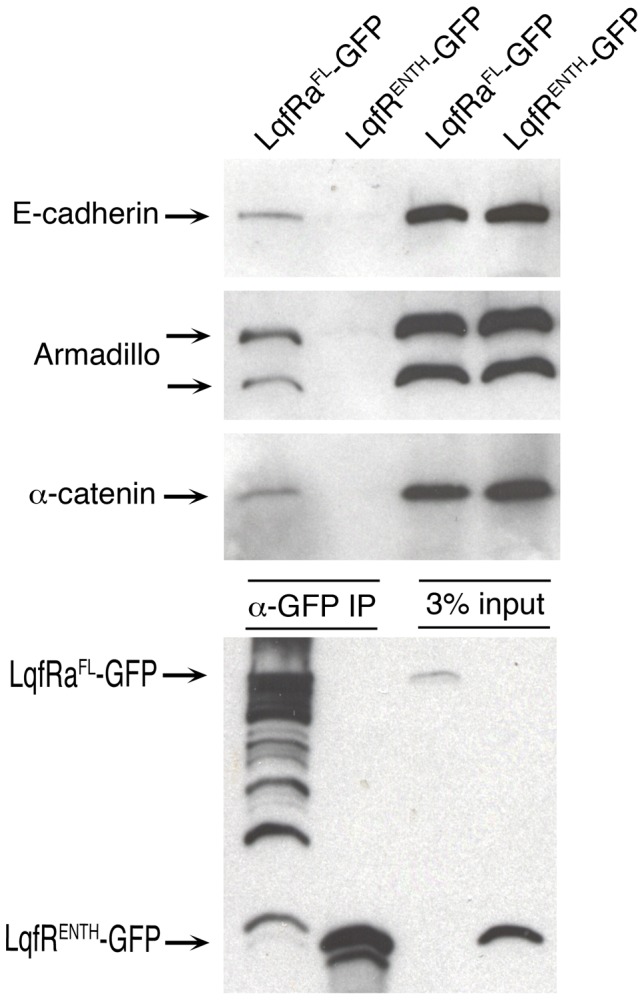
Shown is a blot of protein extracts, before and after immunoprecipitation, from embryos expressing either LqfRaFL-GFP or LqfRaENTH-GFP as a negative control. The LqfR protein fusions were expressed from UAS transgenes using an Actin5C-gal4 driver. The two leftmost lanes (α-GFP IP) are immunoprecipitates using GFP antibodies, and the rightmost lanes (3% input) are aliquots of the protein extracts used, loaded to show that equivalent amounts of protein were present in each extract subjected to immunoprecipitation.
LqfR/Tel2 is not required for Wntless-mediated Wingless secretion
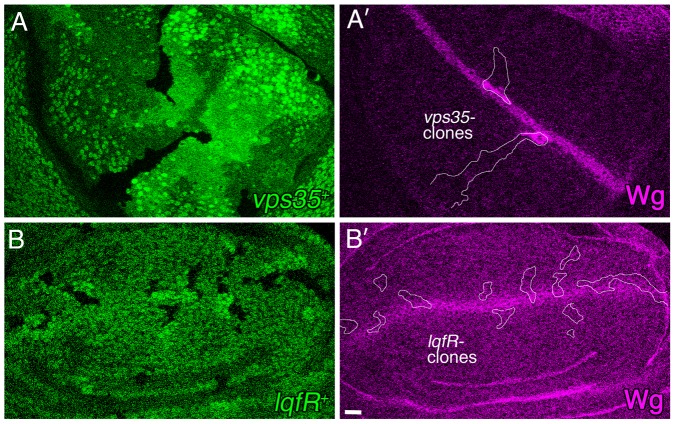
Wingless secretion requires the transmembrane protein Wntless, which binds to Wingless at the Golgi and guides it to the plasma membrane. After releasing Wingless, Wntless is endocytosed and trafficked back to the Golgi. Retrograde trafficking of Wntless from endosomes to Golgi is essential for Wingless secretion and requires the retromer complex. Cell clones lacking retromer complex proteins cannot secrete Wingless and instead Wingless accumulates inside the cells [51]–[53]. In human cultured cells, EpsinR is required for retromer complex function [54], [55]. We have shown that loss of Tel2 activity, and not loss of Golgi Epsin, is the reason why Wingless signaling falters in lqfR/tel2 mutants. Nevertheless, as Golgi Epsin is expected to be required for Wingless secretion, we tested whether or not lqfR/tel2 activity is required. Port et al., 2008 showed that cell clones in the wing disc lacking the retromer protein Dvps35 accumulate Wingless and we were able to replicate this result (Fig. 8A,A′). In contrast, lqfR/tel2 null clones have wild-type Wingless levels (Fig. 8B,B′) and we infer that Wingless is secreted normally from the mutant cells. This result is consistent with the observation that Tel2 expression rescues the lqfR/tel2 null mutant phenotype. Moreover, we conclude that in Drosophila, Golgi Epsin is not always required for retromer complex function.
Figure 8. Wg protein secretion in lqfR- clones.
Shown are confocal microscope images of two third instar larval wing discs immunostained with Wg antibodies (purple). Wingless is expressed and secreted by a stripe of cells at the dorsal/ventral boundary. Homozygous mutant clones are marked by the absence of GFP (green). (A,A′) A wing disc with vps35E42 homozygous clones, outlined in white in A′. The genotype is hs-flp; FRT42D vps35E42/FRT42D ubi-gfp. (B,B′) A wing disc with lqfRΔ117 mutant clones, outlined in white in B′. The genotype is hs-flp; FRT82B lqfRΔ117/FRT82B ubi-gfp. scale bar: ∼10 µm.
Conclusions
Many of the results presented here were unexpected and raise questions that remain to be answered. First, we were surprised to find that in Drosophila, EpsinR and Tel2 proteins are fused. The gene fusion appears to have no consequence for the essential function of the locus in Drosophila. Whether or not the two parts of the protein function together in other contexts, for example during oogenesis or in redundant functions undetectable in our experiments, is unknown. It was also unexpected that only the Tel2 function is essential in Drosophila; EpsinR is not essential for viability in flies and EpsinR is not required for the function of the retromer complex in Wntless recycling. Whether retrograde trafficking of Wntless is the exception, or whether retromer complex function is generally EpsinR-independent in Drosophila remains to be determined.
At least one aspect of Tel2's essential function in Drosophila is to promote Wingless signaling through modulation of adherens junction proteins. Whether or not the lqfR/tel2 mutant phenotype in its entirety reflects a failure of Wingless signaling is yet unclear. There are mutant phenotypes of lqfR mutant flies that are not easily explained by a loss of Wingless signaling [32]. Moreover, recent results link the function of LqfR/Tel2 to a PIKK complex [56]. Nevertheless, the results presented suggest that the association of Tel2 with adherens junction proteins prevents the accumulation of excess E-cadherin at the plasma membrane which would otherwise sequester Armadillo and prevent efficient Wingless signaling (Fig. 9). Further experiments are required to determine precisely how Tel2 activity affects E-cadherin levels. It is interesting that loss of Tel2 activity in C.elegans phenocopies, at least in part, loss of Wnt signaling proteins rather than loss of PIKK activities [7]. The results presented here pave the way for genetic experiments to determine whether or not Tel2 activity in Drosophila always involves PIKK complexes.
Figure 9. The effect of Tel2 on Wingless signaling.
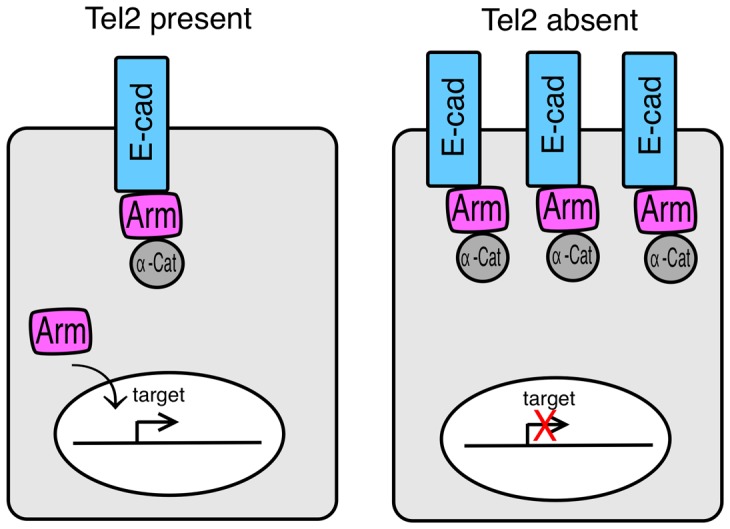
A model for how Wingless signaling is compromised in the absence of Tel2 is illustrated. We speculate that in the absence of Tel2, increased E-cadherin at the plasma membrane sequesters Armadillo (Arm) so that little remains free in the cytoplasm to enter the nucleus in response to Wingless signaling.
Materials and Methods
Drosophila strains
The following mutations were used: lqfRP (FBal0230310), lqfRΔ117 (FBal0191038), Df(3R)Exel6191 (Fab0038246), ds38k (FBal0028156), arm3 (FBal0000712), arm8 (FBal0000716), vps35E42 (FBal0221801), wgI-17 (FBal0018509). The following transgenic lines were used: Actin5C-gal4 (FBti0012293), ey-gal4 (FBti0012711), EGUF (FBti0012712, FBti0012284), hs-flp (FBti0015982), ey-flp (FBti0015982), FRT82B (FBti0002074), FRT42D (FBti141188), ubi-gfp (FBti0012695, FBti0015577), ds-lacZ (FBal0045522), GMR-hid (FBti0012710), UASt-lqfRaFL-gfp [32], UASt-lqfRaENTH-gfp [57]. The following transgenic lines (on chromosome 2) were generated: UASt-6xmyc-lqfRaFL, UASt-6xmyc-lqfRaΔENTH, UASt-6xmyc-lqfRaexons1-5, UASt-6xmyc-lqfRaexon6, UASt-6xmyc-lqfRb. Chromosomes used are indicated in the figure legends.
Transgene construction and transformation
DNA fragments for four of the lqfR P element constructs were generated as follows. First, the template pUASt-lqfRa-gfp [32] was used with the following primer pairs to amplify four different products:
lqfRaFL:
F: 5′- CACCGTGGATAAATTCATCAGCATGTGGAAAG
R: 5′- TTAGGCAGCCTGTTCCATGGCG
lqfRaΔENTH:
F: 5′- CACCGTGGATAAATTCATCAGCATGTGGAAAG
R: 5′-TTAGGCAGCCTGTTCCATGGCG
lqfRaexon6:
F: 5′-CACCGCTGTTGAAGAGCAGTTGGCATCC
R: 5′-TTAGGCAGCCTGTTCCATGGCG
lqfRb:
F: 5′- CACCATGCACGTGGTGGATAAATTCATCAG
R: 5′- TTATCATTGAAACAAGTCGAATGCCG
The PCR products were ligated into pENTR/D-TOPO (Invitrogen). A DNA fragment containing lqfRaexons1-5 was generated by modifiying pENTR-lqfRaFL. The plasmid was restricted with Stu I and Sna BI and then religated, thus removing the final 38 bp of exon 5 and most of exon 6, except for 402 bp at the 3′ end. The lqfR fragments in pENTR-lqfR were transferred to the pTMW vector (Drosophila Genomics Resource Center, #1107) by using site-specific recombination (http://emb.carnegiescience.edu/labs/murphy/Gateway%20vectors.html). The sequence of each pTMW-lqfR plasmid was verified. P element-mediated transformation of yw was performed by Genetivision (Houston, TX).
Analysis of eyes
For immunofluorescence, eye discs were fixed in PEMS and antibody incubations and washes were in PBST as described [58]. Primary antibodies used (DSHB = Developmental Studies Hybridoma Bank): rat anti-E-cadherin (DSHB∶DCAD2, used 1∶100), mouse anti-Armadillo (DSHB∶N27A1, used 1∶100), mouse anti-β-balactosidase (DSHB∶40-1a, used 1∶50), mouse anti-Myc (Santa Cruz Biotechnology∶sc-40, used 1∶20), mouse anti-Wingless (DSHB∶4D4, used 1∶100). Secondary antibodies were as in Lee et al., 2009. Confocal microscopy of eye discs, light microscopy of adult eyes, and image processing was as described [32].
Protein blot in Figure 2
Protein extracts of 2 adult flies containing one copy each of the transgene indicated and the ey-gal4 driver were made by homogenizing and boiling in 2× Laemmli Buffer. After SDS-PAGE, Western blotting and probing was performed as described [59]. Primary antibodies were mouse anti-β-tubulin (DSHB∶E7, 1∶100), and anti-Myc (Santa Cruz Biotechnology∶sc-40, used 1∶500) [32].
Immunoprecipitation
Protein extracts were prepared from Act>lqfRa-gfp and Act>lqfRENTH-gfp embryos: GFP-positive embryos were homogenized in 100 µl lysis buffer (1% NP40, 0.5% deoxycholate, 1 mM DTT, 150 mM NaCl, 50 mM Tris pH 8.0 with protease inhibitor cocktail [Roche, complete-mini, EDTA-free] and 2 mM PMSF). Lysis buffer (300 µl) was added followed by centrifugation at 12,000 rpm at 4°C. A 300 µl aliquot was removed and mixed with 20 µl of a 50% slurry of GFP-trapA (Chromotek) and a 10 µl aliquot was mixed with 2× SDS loading buffer as a loading control. After incubating 2 hrs. with mild shaking at 4°C, the 300 µl aliquot was spun down, the pellet collected and washed for 5 min. with shaking in 1 ml lysis buffer, and then washed again for 10 min. with shaking in 1 ml of 500 mM NaCl. The pellet was washed 4 times more in 1 ml of 500 mM NaCl and then mixed with 20 µl of 2× Laemmli Buffer. Each sample was boiled for 5 min, microfuged, and the supernatant subjected to SDS-PAGE in a 7.5% gel. Western blotting was performed as described (Chen et al., 2002). Primary antibodies were: rat anti-E-cadherin (DSHB∶DCAD2, used 1∶1000), mouse anti-Armadillo (DSHB∶N27A1, used 1∶500), rat anti-α-catenin (DSHB∶DCAT-1, used 1∶100), rat anti-GFP (Chromotek∶3H9, used 1∶1000). Secondary antibodies were from Santa Cruz Biotechnology and used at 1∶5000: goat anti-rat HRP , goat anti-mouse HRP, goat anti-rat HRP.
Supporting Information
Amino acid sequence alignment of human and yeast Tel2 and Drosophila LqfR-exon 6. The amino acid sequences of H. sapiens Tel2, D. melanogaster LqfR exon 6, and S. cerevisiae Tel2 were aligned using MacVector and the results are shown. H. sapiens vs. S. cerevisiae: aligned length = 850, gaps = 23, identities = 116 (13%), similarities = 102 (12%). H. sapiens vs. D. melanogaster: aligned length = 929, gaps = 15, identities = 181 (19%), similarities – 158 (17%). D. melanogaster vs. S. cerevisiae: aligned length = 924, gaps = 18, identities = 110 (11%), similarities = 121 (13%).
(TIF)
Rescue of E-cadherin accumulation abnormality in lqfR - clones by transgene expression. Confocal microscope images of three third instar larval eye discs immunostained with antibodies to E-cadherin (red). lqfR- clones are marked by the absence of GFP (green). The images at bottom are identical to the ones at the top except only the red layer is shown and the clone is outlined. (A–C′) The discs express the transgenes indicated. The genotype is ey-flp; FRT82B lqfRΔ117/FRT82B ubi-gfp in all panels, with the addition of Act5C-gal4, UAS-lqfRa/ + (B,B′) and Act5C-gal4, UAS-lqfRaexon6/ + (C,C′) on chromosome 2. scale bar: ∼10 µm in A–B′; ∼25 µm in C,C′
(TIF)
Acknowledgments
We are grateful to Konrad Basler, Xinhua Lin, and the Bloomington Drosophila Stock Center for flies. We acknowledge the DNA sequencing and confocal microscope facilities of the ICMB at UT Austin, and we thank Paul Macdonald for the use of his confocal microscope.
Funding Statement
This work was supported by the National Institute of Child Health and Human Development (NICHD) Grant RO1-HD30680. The funders had no role in study design, data collection and alaysis, decision to publish, or preparation of the manuscript.
References
- 1. Chang M, Lingner J (2008) Tel2 finally tells one story. Science 320: 60–61. [DOI] [PubMed] [Google Scholar]
- 2. Takai H, Wang RC, Takai KK, Yang H, de Lange T (2007) Tel2 regulates the stability of PI3K-related protein kinases. Cell 131: 1248–1259. [DOI] [PubMed] [Google Scholar]
- 3. Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, et al. (2010) CK2 phospho-dependent binding of R2TP complex to Tel2 is essential for mTOR and SMG1 stability. Mol Cell 39: 839–850. [DOI] [PubMed] [Google Scholar]
- 4. Takai H, Xie Y, de Lange T, Paveletich NP (2010) Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 24: 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson CM, Blackburn EH (2008) Mec1 function in the DNA damage response does not require its interaction with Tel2. Cell Cycle 7: 3695–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson CM, Korkin D, Smith DL, Makovets S, Seidel JJ, et al. (2008) Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev 22: 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moser SC, von Elsner S, Bussing I, Alpi A, Schnabel R, et al. (2009) Functional dissection of Caenorhabditis elegans CLK-2/TEL2 cell cycle defects during embryogenesis and germline development. PLoS Gen 5: e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lustig AJ, Petes TD (1986) Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA 83: 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lakowski B, Hekimi S (1996) Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013. [DOI] [PubMed] [Google Scholar]
- 10. Shikata M, Ishikawa F, Kanoh J (2007) Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J Biol Chem 282: 5346–5355. [DOI] [PubMed] [Google Scholar]
- 11. Jiang N, Benard CY, Kebir H, Shoubridge EA, Hekimi S (2003) Human CLK2 links cell cycle progression, apoptosis, and telomere length. J Biol Chem 278: 21678–21684. [DOI] [PubMed] [Google Scholar]
- 12. Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, et al. (2007) HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol 9: 391–401. [DOI] [PubMed] [Google Scholar]
- 13. Ahmed S, Alpi A, Hengartner MO, Gartner A (2001) C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr Biol 11: 1934–1944. [DOI] [PubMed] [Google Scholar]
- 14. Bénard C, McCright B, Zhang Y, Felkai S, Lakowski B, et al. (2001) The C. elegans maternal-effect gene clk-2 is essential for embryonic development, encodes a protein homologous to yeast Tel2p and affects telomere length. Development 128: 4045–4055. [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Muse T, Boulton SJ (2005) Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans . EMBO J 24: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hurov KE, Cotta-Ramusino C, Elledge SJ (2010) A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24: 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, et al. (2010) Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem 285: 20109–20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leventis PA, Da Sylva TR, Rajwans N, Wasiak S, McPherson PS, et al. (2011) Liquid facets-Related (LqfR) is required for egg chamber morphogenesis during Drosophila oogenesis. PLoS ONE 6: e25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duncan MC, Castaguta G, Payne GS (2003) Yeast epsin-related proteins required fro Golgi-endosome traffic define a γ-adaptin ear-binding motif. Nat Cell Biol 5: 77–81. [DOI] [PubMed] [Google Scholar]
- 20. Kalthoff C, Groos S, Kohl R, Mahrhold S, Ungewickell EJ (2002) Clint: A novel clathrin-binding ENTH-domain protein at the Golgi. Mol Biol Cell 13: 4060–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wasiak S, Legendre-Guillemin V, Puertollano R, Blondeau F, Girard M, et al. (2002) Enthoprotein: a novel clathrin-associated protein identified through subcellular proteomics. J Cell Biol 158: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirst J, Motley A, Harasaki K, Chew SYP, Robinson MS (2003) EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol Biol Cell 14: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mills IG, Praefcke GJK, Vallis Y, Peter BJ, Olesen LE, et al. (2003) EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol 160: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duncan MC, Payne GS (2003) ENTH/ANTH domains expand to the Golgi. Trends Cell Biol 13: 211–215. [DOI] [PubMed] [Google Scholar]
- 25. Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS (2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117: 9–18. [DOI] [PubMed] [Google Scholar]
- 26. Dodd ME, Hatzold J, Mathias JR, Waters KB, Bennin DA, et al. (2009) The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development 136: 2591–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wendland B (2002) Epsins: adaptors in endocytosis? Nat Rev Mol Cell Biol 3: 971–977. [DOI] [PubMed] [Google Scholar]
- 28. Aguilar RC, Wendland B (2005) Endocytosis of membrane receptors: two pathways are better than one. Proc Natl Acad Sci USA 103: 4116–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Overstreet E, Fitch E, Fischer JA (2004) Fat facets and Liquid facets promote Delta endocytosis and signaling in the signaling cells. Development 131: 5355–5366. [DOI] [PubMed] [Google Scholar]
- 30. Wang W, Struhl G (2004) Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development 131: 5367–5380. [DOI] [PubMed] [Google Scholar]
- 31. Platta HW, Stenmark H (2011) Endocytosis and signaling. Curr Opin Cell Biol 23: 393–403. [DOI] [PubMed] [Google Scholar]
- 32. Lee J-H, Overstreet E, Fitch E, Fleenor S, Fischer JA (2009) Drosophila liquid facets-Related encodes Golgi Epsin and is an essential gene required for cell proliferation, growth, and patterning. Dev Biol 331: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Overstreet E, Chen X, Wendland B, Fischer JA (2003) Either part of a Drosophila epsin (Liquid facets) protein, divided after the ENTH domain, functions in the internalization of Delta in the developing eye. Curr Biol 13: 854–860. [DOI] [PubMed] [Google Scholar]
- 34. Xie X, Cho B, Fischer JA (2012) Drosophila Epsin's role in Notch ligand cells requires three Epsin protein functions: The lipid binding function of the ENTH domain, a single Ubiquitin interaction motif, and a subset of the C-terminal protein binding modules. Dev Biol 363: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenny A (2010) Planar cell polarity in the Drosophila eye. In: Cagan RL, Reh TA, eds. Invertebrate and Veterbrate Eye Development. Curr Top Dev Biol 93: , 190–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clevers H (2006) Wnt/β-catenin signaling in development and disease. Cell 127: 469–480. [DOI] [PubMed] [Google Scholar]
- 37. Clark HF, Brentrup D, Schneitz K, Bieber A, Goodman C, et al. (1995) dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila . Genes Dev 9: 1530–1542. [DOI] [PubMed] [Google Scholar]
- 38. Yang CH, Axelrod JD, Simon MA (2002) Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108: 675–688. [DOI] [PubMed] [Google Scholar]
- 39. Zecca M, Basler K, Struhl G (1996) Cell Direct and long-range action of a Wingless morphogen gradient. Cell 87: 833–844. [DOI] [PubMed] [Google Scholar]
- 40. Treisman JE, Rubin GM (1995) Wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121: 3519–3527. [DOI] [PubMed] [Google Scholar]
- 41. Parker DS, Jemison J, Cadigan KM (2002) Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila . Development 129: 2565–2576. [DOI] [PubMed] [Google Scholar]
- 42. Takahisa M, Togashi S, Suzuki T, Kobayashi M, Murayama A, et al. (1996) The Drosophila tamou gene, a component of the activating pathway of extramacrochaetae expression, encodes a protein homologous to mammalian cell-cell junction-associated protein ZO-1. Genes Dev 10: 1783–1795. [DOI] [PubMed] [Google Scholar]
- 43. Seppa MJ, Johnson RI, Bao S, Cagan RL (2008) Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev Biol 318: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gonzalez-Mariscal L, Betanzos A, Avila-Flores A (2000) MAGUK proteins: structure and role in the tight junction. Semin Cell Dev Biol 11: 315–324. [DOI] [PubMed] [Google Scholar]
- 45. Kemler R (1992) Classical cadherins. Semin Cell Biol 3: 149–133. [DOI] [PubMed] [Google Scholar]
- 46. Chen YT, Stewart DB, Nelson WJ (1999) Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol 144: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huber AJ, Stewart DB, Laurents DV, Nelson WJ, Weis WI (2001) The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin: A possible mechanism for regulating cadherin turnover. J Biol Chem 276: 12301–12309. [DOI] [PubMed] [Google Scholar]
- 48. Heuberger J, Birchmeier W (2010) Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spr Harb Perspect Biol 2: a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanson B, White P, Vincent JP (1996) Uncoupling cadherin-based adhesion from Wingless signaling in Drosophila . Nature 383: 627–630. [DOI] [PubMed] [Google Scholar]
- 50. Gottardi CJ, Wong E, Gumbiner BM (2001) E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol 153: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, et al. (2008) The retromer complex influences Wnt secretion by recycling Wntless from endosomes to the trans-Golgi network. Dev Cell 14: 120–131. [DOI] [PubMed] [Google Scholar]
- 52. Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, et al. (2008) Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol 10: 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Port F, Kuster M, Herr P, Furger E, Banziger C, et al. (2008) Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat Cell Biol 10: 178–185. [DOI] [PubMed] [Google Scholar]
- 54. Saint-Pol A, Yelamos B, Amessou M, Mills IG, Dugast M, et al. (2004) Clathrin adapter EpsinR is required for retrograde sorting on early endosomal membranes. Dev Cell 6: 525–538. [DOI] [PubMed] [Google Scholar]
- 55. Popoff V, Mardones GA, Tenza D, Rojas R, Lamaze C (2007) The retromer complex and clathrin define an early endosomal retrograde exit site. J Cell Sci 120: 2022–2031. [DOI] [PubMed] [Google Scholar]
- 56. Glatter T, Schittenhelm RB, Rinner O, Roguska K, Wepf A, et al. (2011) Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Sys Biol 7: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overstreet E (2005) PhD thesis, The University of Texas at Austin.
- 58. Fischer-Vize JA, Rubin GM, Lehmann R (1992) The fat facets gene is required for Drosophila eye and embryo development. Development 116: 985–1000. [DOI] [PubMed] [Google Scholar]
- 59. Chen X, Zhang B, Fischer JA (2002) A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Faf facets. Genes Dev 16: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of human and yeast Tel2 and Drosophila LqfR-exon 6. The amino acid sequences of H. sapiens Tel2, D. melanogaster LqfR exon 6, and S. cerevisiae Tel2 were aligned using MacVector and the results are shown. H. sapiens vs. S. cerevisiae: aligned length = 850, gaps = 23, identities = 116 (13%), similarities = 102 (12%). H. sapiens vs. D. melanogaster: aligned length = 929, gaps = 15, identities = 181 (19%), similarities – 158 (17%). D. melanogaster vs. S. cerevisiae: aligned length = 924, gaps = 18, identities = 110 (11%), similarities = 121 (13%).
(TIF)
Rescue of E-cadherin accumulation abnormality in lqfR - clones by transgene expression. Confocal microscope images of three third instar larval eye discs immunostained with antibodies to E-cadherin (red). lqfR- clones are marked by the absence of GFP (green). The images at bottom are identical to the ones at the top except only the red layer is shown and the clone is outlined. (A–C′) The discs express the transgenes indicated. The genotype is ey-flp; FRT82B lqfRΔ117/FRT82B ubi-gfp in all panels, with the addition of Act5C-gal4, UAS-lqfRa/ + (B,B′) and Act5C-gal4, UAS-lqfRaexon6/ + (C,C′) on chromosome 2. scale bar: ∼10 µm in A–B′; ∼25 µm in C,C′
(TIF)