Abstract
Legionellosis is mostly caused by Legionella pneumophila (Lp) and is defined by a severe respiratory illness with a case fatality rate ranging from 5 to 80%. In a previous study, we showed that a glycosaminoglycan (GAG)-binding adhesin of Lp, named Lcl, is produced during legionellosis and is unique to the L. pneumophila species. Importantly, a mutant depleted in Lcl (Δlpg2644) is impaired in adhesion to GAGs and epithelial cells and in biofilm formation. Here, we examine the molecular function(s) of Lcl and the transcriptional regulation of its encoding gene during different stages of the biofilm development. We show that the collagen repeats and the C-terminal domains of Lcl are crucial for the production of biofilm. We present evidence that Lcl is involved in the early step of surface attachment but also in intercellular interactions. Furthermore, we address the relationship between Lcl gene regulation during biofilm formation and quorum sensing (QS). In a static biofilm assay, we show that Lcl is differentially regulated during growth phases and biofilm formation. Moreover, we show that the transcriptional regulation of lpg2644, mediated by a prototype of QS signaling homoserine lactone (3OC12-HSL), may play a role during the biofilm development. Thus, transcriptional down-regulation of lpg2644 may facilitate the dispersion of Lp to reinitiate biofilm colonization on a distal surface.
Introduction
Legionellosis is a major public health concern in industrialized countries. Manifestations of this disease range from a mild respiratory illness to a severe and rapidly fatal pneumonia [1]. The case fatality rate of legionellosis ranges between 40 to 80% in untreated immuno-suppressed patients but can be reduced from 5 to 30% with appropriate case management [2], [3]. Legionellosis is acquired by inhaling airborne water droplets contaminated with Gram-negative bacilli of the Legionella genus [4]. Legionella bacteria are found worldwide and can be detected in up to 80% of freshwater sites [5]. Among more than 54 species of Legionella, L. pneumophila is the major cause of outbreaks (91.5%) and serogroup 1 (Lp1) is the predominant serotype isolated from patients (84.2%) [6], [7]. Since human-to-human transfer of the bacteria has not been reported [8], to explain the higher incidence of Lp1 compared to other Legionella species, it has been hypothesized that Lp1 is more virulent and/or more fit for survival in anthropogenic aquatic environments [9].
While it can replicate within environmental protozoa, another efficient way for Lp1 to persist and disseminate in man made water systems is to form monospecies and multispecies biofilms [10]–[14]. This mode of living acts as a protective mechanism against a variety of environmental assaults, such as antibiotics or biocide treatments [15]. Therefore, when man-made water systems are contaminated with L. pneumophila, the development of biofilm renders water disinfection strategies such as chlorination ineffective [16]. Although it remains unexplored for Legionella, in other pathogenic bacteria, the production and development of biofilms in vivo promote persistence, colonization of the airways and increases resistance to antibiotics [17]–[19].
The formation of biofilm is a developmental process with multiple steps including cell adhesion, proliferation of sessile cells forming micro-colonies which produce an extra-cellular matrix and maturation into a 3-dimensionnal shape [20]. The last stage is the detachment of bacteria from matrices to reinitiate biofilm colonization on a distal surface [20]. Biofilm formation is a highly intra/inter species regulated process that is controlled and coordinated by a wide range of factors including interconnected signalling molecules such as quorum sensing (QS) autoinducers and 3,5-cyclic diguanylic acid [21] http://www.plospathogens.org/article/info:doi/10.1371/journal.ppat.1000483 - aff1. QS is a density-dependent form of bacterial cell-cell communication that operates via secreted small molecules named autoinducers. Upon reaching a threshold concentration, autoinducers trigger induction or repression of genes involved in biofilm maturation and maintenance to functionally coordinate the behaviour of a bacterial population including the expression of virulence factors [22], [23]. A well characterized QS system is cqsAS present in Vibrio cholerae and other marine Vibrio spp. [24], [25]. The CqsAS is involved in cell density-dependent regulation of virulence and biofilm formation through the synthesis of an α-hydroxyketone autoinducer [26]. Recently, a homologous system, named lqs was described in L. pneumophila including the lqsR, lqsA and lqsS gene [27], [28]. Legionella lqsA can functionally complement a V. cholerae cqsA mutant [28]. Interestingly, the Pseudomonas aeroginosa homoserine lactone autoinducer (3OC12-HSL) was recently shown to exert a bacteriostatic activity, and decrease biofilm formation of Lp1. These effects are correlated to the transcriptional down-regulation of lqsR gene in Lp1 [29]. This finding suggests a connection between 3OC12-HSL and the uniquely identified QS system in Lp1.
Among the matrix associated proteins of biofilms, specific adhesins mediate cell to cell interactions and/or initiate surface attachment. In few cases, the same adhesins also play a role in pathogenesis by allowing the binding of planktonic bacteria to host cells or epithelium [30], [31]. In a previous study, we identified a GAG binding adhesin of Lp1 designated Legionella collagen-like protein (Lcl) [32], [33]. Lcl is produced during legionellosis and is absent from Legionella species that are rarely reported in patients and poor biofilm producers. Importantly, a mutant depleted in Lcl is dramatically impaired in biofilm formation. Here, to get insight into the molecular mechanisms used by Lp1 to produce biofilm, we investigated the molecular functions of Lcl during different stages of the biofilm development. Using recombinant variants, we show that the collagen repeats and the C-terminal domains of Lcl are crucial for the production of biofilm. We present evidence that Lcl is involved in the early step of surface attachment but also in intercellular interactions. Furthermore, we address the relationship between the regulation of Lcl gene (lpg2644) during biofilm formation and the QS system. We show that Lcl is differentially regulated during growth phases and biofilm formation. Moreover, data suggesting that the regulation of lpg2644 is QS dependent and plays a role during the late stage of biofilm development are presented. Thus, we propose that the regulation of lpg2644 gene may participate in the dispersion of bacteria to reinitiate biofilm colonization on a distal surface.
Materials and Methods
Chemical, Bacterial Strain and Cultivation
All chemicals were purchased from Sigma-Aldrich (Oakville, ON) unless otherwise noted. Restriction and cloning enzymes were purchased from New England Biolabs (Pickering, ON) and used according to manufacturer recommendations. Legionella isolates and plasmid vectors used in this study are listed in Table 1. Lp02 is a streptomycin-resistant thymidine auxotroph derived from Lp01. All mutant strains were made in the Lp02 background. Unless specified, Legionella isolates were grown on buffered charcoal yeast extract (BCYE) agar at 37°C+5% CO2 incubation for 3–4 days and Legionella pre-cultures were grown at 37°C and 100 rpm for 24 h in buffered yeast extract (BYE) broth with 100µg/ml thymidine when necessary. Escherichia coli strains and plasmids are listed in Table 1. All E. coli strains were cultured in Luria-Bertani medium, antibiotic was added to the media at concentrations of 50 µg/ml kanamycin or 100 µg/ml carbenicillin.
Table 1. Legionella species, E. coli strains and plasmids used in this study.
| MR Code | Species | Sg | Designation/Plasmid | Source/Ref |
| 23 | L. pneumophila | 1 | Lp02 | [65] |
| 36 | L. pneumophila | 1 | Lp02/pBH6119-IcmRp | [32] |
| 52 | L. pneumophila | 1 | Lp02 Δlpg2644 | [32] |
| 69 | L. pneumophila | 1 | Lp02 Δlpg2644/pBH6119 | [32] |
| 70 | L. pneumophila | 1 | Lp02 Δlpg2644/pBH6119-IcmRp | [32] |
| 73 | L. pneumophila | 1 | Lp02/pBH6119 | [32] |
| 76 | L. pneumophila | 1 | Lp02/plpg2644 | [32] |
| 77 | L. pneumophila | 1 | Lp02 Δlpg2644/plpg2644 | [32] |
| 122 | L. pneumophila | 1 | Lp02/plpg2644 Δrepeats | [32] |
| 123 | L. pneumophila | 1 | Lp02 Δlpg2644/plpg2644 Δrepeats | This study |
| 124 | L. pneumophila | 1 | Lp02 Δlpg2644 clpg2644 | [32] |
| 130 | L. pneumophila | 1 | Lp02 Δlpg2644 clpg264/pBH6119-IcmRp | [32] |
| 137 | L. pneumophila | 1 | Lp02/plpg2644 ΔC-term | This study |
| 138 | L. pneumophila | 1 | Lp02 Δlpg2644/plpg2644 ΔC-term | This study |
| 2 | E. coli TOP10 | none | Invitrogen | |
| 115 | E. coli TOP10 | pCR2.1- lpg2644 Δrepeats | This study | |
| 116 | E. coli TOP10 | pBH6119-IcmRp-lpg2644 Δrepeats | This study | |
| 131 | E. coli TOP10 | pCR2.1- lpg2644 ΔC-terminal | This study |
Sg: serogroup.
Lcl Detection by Live-cell Confocal Imaging
To detect Lcl at the surface of L.pneumophila, GFP-Lp02 and GFP-Lp02 Δ lpg2644 cells were harvested from broth cultures (OD600 = 3.0–3.5) and washed twice with phosphate-buffered saline (PBS). To reduce bacterial movement during in vivo imaging, glass coverslips were coated with poly-L lysine according to the manufacturer’s instructions (Sigma-Aldrich, Oakville, ON, Canada). Cells were centrifuged onto the coated coverslips for 5 min at RT (1000×g). Where indicated, bacteria were either incubated with anti-Lcl antibodies alone or both anti-Lcl (1∶50) and anti-Lp1 (1∶250) antibodies (Millipore) diluted in PBS simultaneously. Following 5 min incubation at RT, bacteria were washed twice and labelled with alexa–fluor conjugated secondary antibodies (Molecular probes) as indicated for 5 min on ice. The cells were then moved to a pre-cooled stage and labelled with the inner membrane impermeable FM 4–64 dye (Molecular probes) where indicated. Images were acquired using Quorum confocal spinning disc microscope with a 63×oil immersion objective (Leica DMI6000B stand with a Hamamatsu ORCA-R2 camera). Image acquisition was performed using Metamorph Wave FX software and deconvolution was performed using Volocity 4.3 software (Improvision).
General DNA Techniques
Total genomic DNAs were purified using QIAamp DNA Mini Kit (Qiagen, Mississauga, ON) and plasmid DNAs were purified using QIAprep Spin Miniprep Kit (Qiagen). DNA was quantified by UV spectrophotometry, and 5 or 10 ng was used as a template for each PCR reaction. Taq DNA polymerase, deoxynucleoside triphosphates, and buffer were used according to recommendations of the manufacturer (Invitrogen, Burlington, ON, Canada). PCR amplifications for cloning were performed using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen). A list of primers used can be found in Table 2. Sequencing reactions were performed using the BigDye® Terminator v3.1 Cycle Sequencing kit purified using the BigDye® X Terminator™ Purification kit, and run on 3130×l Genetic Analyzer (Applied Biosystems, Streetsville, ON). All clones were verified by sequencing.
Table 2. Oligonucleotides used for PCR, qRT-PCR, sequencing and cloning.
| Code | Primers/probes | Amplified DNA | Sequence (5′ to 3′) |
| 1 | lpg2644 F | lpg2644 | AGACACGTGTTGAATCCACT |
| 2 | lpg2644 R | CACCAAAAGCAATCCGGCCTCGCA | |
| 3 | lpg2644 XhoI F | AGCTCGAGCAATCCGGCCTCGCAAGCC | |
| 4 | lpg2644 EcoRI R | CGGAATTCCGGGTTGCGAGAGTTGGCTA | |
| 5 | lpg2644 Δrepeat F | GATGACGGCCAAGGTGTGCC | |
| 6 | lpg2644 Δrepeat R | TTGAGGTCCTTGAGGTCCAG | |
| 7 | lpg2644 ΔC-terminal F | TAAGCATGGCAAAACTTCAATTTTGAT | |
| 8 | lpg2644 ΔC-terminal R | TGCAGGCACACCTTGGCCGTCATC | |
| 9 | lpg2644 XbaI F | GGTCATCTAGAGAAATAAAGAATGATACATCGA | |
| 10 | lpg2644 SphI R | GTGAGCGCATGCGCAAAGCGAATTTATGAACA | |
| 11 | lpg2644 F | CTAGGTACCCATGGGCTTGTG | |
| 12 | lpg2644 R | TGCATCCCACCATGTGCTT | |
| 13 | lpg2644 probe | 6FAM- CAAACTCAGATCAAGTTAAT-MGBNFQ | |
| 14 | gyrA F | gyrA | GGCGGGCAAGGTGTTATTT |
| 15 | gyrA R | GCAAGGAGCGGACCACTTT | |
| 16 | gyrA probe | VIC-CATTTCGTTCAGTAACCTG-MGBNFQ |
SDS-PAGE and Immunoblot Analyses of Full Length and Recombinant Fragments of Lcl
Immunoblot analyses were performed with bacterial lysates and/or supernatants harvested from broth cultures at different growth phases, with or without N-(3-oxododecanoyl)-l-homoserine lactone (3 OC12-HSL) and from in 2 days and 6 days old biofilms.To obtain exponential (E, OD600 nm 0.5–2.0), post exponential (PE, OD600 nm 2–3.5), mid stationary (MS, OD600 nm 3.5–5.0) and late stationary (LS, OD600 nm >4.5) overnight pre-cultures of Lp02 were adjusted to an OD600 nm of 0.05 in BYE broth and were incubated at 37°C with constant shaking (100 rpm) respectively for 12 h, 18 h, 24 h and 42 h. Cell lysates were prepared by adjusting Legionella cultures to OD600 nm of 4.5 and washing twice with chilled 1X PBS. Equal volumes of Laemmli loading buffer containing 2-mercaptoethanol (final concentration 5%) were added to the cells. Supernatants were precipitated using 2, 2, 2-trichloroacetic acid. Pellets were washed twice with chilled acetone and resuspended in Laemmli loading buffer after normalization to the OD600 nm of their corresponding culture. Lysates and supernatants were boiled at 95°C for 10 min prior to loading onto 4–15% linear gradient Tris-HCl gels (Bio-Rad). Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to Laemmli [34]. Immunoblotting was performed as described by Towbin et al. [35]. SDS- Page Acrylamide gels were stained using Coomassie (SimplyBlue SafeStain, Invitrogen Rockville, Maryland, USA). Visual inspection of SDS-PAGE profiles with normalized secreted fractions revealed similar patterns. For immunoblot analyses, proteins were transferred onto nitrocellulose membranes (Bio-Rad). Bound Lcl (1∶20,000) and GFP (1∶5,000) (Invitrogen) antibodies were detected with peroxidase-linked anti rabbit IgG (1∶160,000).
Generation of plpg2644 Δrepeats and plpg2644 ΔC-terminal
To remove the collagen-like tandem repeats from lpg2644 gene of the pCR2.1-lpg2644 plasmid, 10 ng of plasmid were reverse PCR amplified using primers 5 and 6 (Table 2). Resulting PCR product was blunt end ligated and transformed into E. coli TOP10 (Invitrogen). Transformants with pCR2.1-lpg2644 Δrepeats were selected by carbenicillin-kanamycin resistance screening on LB agar. The lpg2644 Δrepeats fragment was next excised from the plasmid using restriction enzymes XbaI and SphI and following gel-purification was subcloned into the Legionnella expression shuttle vector pBH6119-IcmRp digested with XbaI and SphI to generate the plasmid plpg2644 Δrepeats. E. coli TOP10 transformed with plpg2644 Δrepeats were selected by carbenicillin resistance screening on LB agar and grown overnight in LB media with 50 µg/ml carbenicillin for plasmid extraction. plpg2644 Δrepeats plasmid was then transformed into Lp02 and Lp02 Δlpg2644.
To remove ΔC-terminal domain from lpg2644 gene of the pCR2.1-lpg2644 plasmid, the same procedure was used with primers 7 and 8 (table 2).
Static Biofilm Formation Assay
Biofilm assays using polystyrene 96-well plates assays (Costar polystyrene 3361, Corning Incorporated, Lowell MA) were performed according to Piao et al. with few modifications [9]. Legionella pre-cultures were grown with or without 10 µM 3OC12-HSL for 30 h in broth, adjusted to OD600 nm of 0.2 and 200 µl was used to seed each well in the 96-well microtitre plates. Following incubation at 37°C+5% CO2 for 2 or 6 days, the biomass of biofilm was quantified by crystal violet staining assay. Bacteria were stained with 40 µl 0.25% crystal violet in each well for 15 mn, washed 3 times with 200 µl sterile distilled water and were solubilised in 200 µl 95% ethanol at room temperature for 15 min. Absorbance was read at OD600 nm with a microplate reader. Each experiment was repeated three times with six technical replicates for each condition and control.
This assay was slightly modified in order to measure the effect of 3OC12-HSL (Sigma) on the regulation of lpg2644 gene on biofilm formation. Adjusted pre-cultures were seeded in 96-well microtitre plates and following a 2 h incubation at 37°C+5% CO2, with or without 10 µM 3OC12-HSL, wells were washed to remove planktonic cells. Fresh BYE media with or without 10 µM 3OC12-HSL was then added to the sessile cells for 46 h at 37°C+5% CO2 and production of biofilm was measured using the crystal violet staining assay described above.
Confocal Laser Scanning Microscopy
For confocal laser scanning microscopic examination (CLSM) of biofilms, single or mixed bacterial cultures (300 µl) were prepared in Lab-TekII chamber slides (Labtek II, VWR, Rochester, USA) according to the procedure described above. After 3 days of incubation at 37°C+5% CO2, supernatants were removed and bacteria were labelled with nucleic acid stain SYTO 62 (Invitrogen) for 30 min at RT. After two washes with sterile distilled water, 4% PFA was added for 15 min followed by one wash with sterile distilled water. The plastic wells were removed from the slide and fluoromount (DAKO north America INC, Carpinteria, USA) was added before placing a coverslip on the gasket and observed by CLSM using a Nikon Eclipse TE2000EZ inverted microscope, 100 X Plan APO oil immersion DIC N2 objective. Image acquisition and post-acquisition processing were performed using EZ-C1 Software Ver. 3.50 and the NIS-elements BR Software Ver. 3.0 for Nikon C1 Confocal Microscopy.
Attachment Assay of L. pneumophila to 96 Polystyrene Microtitre Plate using Quantitative PCR and Confocal Microscopy (CLSM)
Early stage of biofilm formation was examined by measuring the ability of cells to adhere to 96-well (flat bottom) polystyrene microtitre plate (Costar). Assays for attachment were performed using L. pneumophila strains Lp02, Lp02 harbouring an empty plasmid (Lp02 pBH6119), Lp02 deleted in lpg2644 gene (Lp02 Δlpg2644) and complemented mutant (Lp02 Δlpg2644plpg2644) (Table 1) grown on BCYE agar with ±100 µg/ml thymidine and incubated for 4 days at 37°C+5% CO2. Cell suspension was obtained by collecting colonies with a sterile swab and suspending them in 5 ml of BCYE broth. Cell suspension was adjusted to an OD600 nm of 0.1 in 25 ml of BYE broth and was incubated at 37°C with constant shaking (100 rpm). After 24 h of growth, bacteria were harvested and resuspended in BYE at OD600 nm of 2. Two hundred microliters of Legionella suspensions of each strain were aliquoted into 96-well microtitre plate (in triplicate) and then incubated for 1.5 h at 37°C+5% CO2. After 3 washes in PBS, DNA from adhering cells was purified directly from each well with a DNeasy 96 Blood and Tissue Kit according to manufacturer instructions (Qiagen). Genome copy numbers present in each well were next quantified by quantitative PCR (qPCR) using oligonucleotides and probe to gyrA (Table 2, code 14 to 16). Quantitative PCR was performed with the Universal PCR Master Mix (Applied Biosystems) using 400 nM of each specific primer and 200 nM of probe. Amplification and detection of specific products were performed with the ABI Prism 7900 detection system (Applied Biosystems) with samples first incubated at 50°C for 2 min, then at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. Attachment of stained L. pneumophila to glass cover slips was measured using confocal microscopy. Assays were performed as described above and L. pneumophila strains were labelled with nucleic acid stain SYTO 62 (Invitrogen) for 30 min at RT, washed twice and fixed with PFA (4%) for 15 min. After two washes, cover slips were mounted with DAKO and slides were analyzed by CLSM.
Flow Chamber Experiments
In order to non invasively monitor in real-time the adherence and biofilm formation of L. pneumophila under dynamic flow conditions, a continuous-flow cell system was set up (Stovall, life science, Greensboro, NC27497). Each channel (1 mm D×4 mm W×40 mm L) was equipped with bubble traps and flow breaks downstream of a peristaltic pump (IPC-N-16; Ismatec SA). The system was assembled using autoclaved silicon tubing (1 by 3 mm), sterilized with 0.5% sodium hypochlorite overnight, and rinsed excessively with sterile water. A volume of 400 µl of stationary phase grown Legionella cultures (OD600 nm, 3.0 to 3.5), adjusted to a final OD at 600 nm of 0.2 in fresh BYE was injected into flow chambers. Following 1.5 h of incubation under static condition at RT to allow attachment of cells to the chamber, cells were submitted to a constant flow of 5.5 µl/min at RT for 6 days. Biofilm structures were visualized at 2, 4, 6, 24 h, 2 and 6 days with a Nikon Eclipse TE2000EZ inverted microscope, 100 X Plan APO oil immersion DIC N2 objective, Spectra Physics 488 nm laser.
RNA Isolation and Quantitative RT-PCR Analysis
E, PE, MS and LS cultures were obtained as described above. To evaluate the impact of the LasQS 3OC12-HSL (Sigma) from P. aeruginosa on lpg2644 transcription, 10 µM 3OC12-HSL was added to MS Lp02 cell suspensions adjusted to an OD600 of 0.1 and bacteria were incubated at 37°C with constant shaking (100 rpm) for 24 hr. To prevent RNA degradation, 1 ml of each bacterial suspension was treated with RNAprotect Bacteria Reagent kit (Qiagen) and immediately stored at −20 °C.
To isolate sessile cells, biofilm assays were performed in polystyrene 6-well plates (6 ml/well) (Costar) as described above. After 2 and 6 days of incubation, culture supernatants containing planktonic cells were gently removed and suspensions cells were recovered by centrifugation (5 min, 3000 g, RT). Adhering sessile cells were next detached from the wells by pipetting using RNase free-water (Ambion Life Technologies Inc., Burlington, ON, Canada) and recovered by centrifugation (2 min, 3000 g, RT). Samples were then treated with the RNAprotect Bacteria Reagent kit (Qiagen) and immediately stored at −20 °C.
Total RNA was purified from with the RNeasy kit (Qiagen) and contaminating DNA was removed using the DNA-free kit (Ambion). Concentrations of total RNA were determined with a ND-1000 spectrophotometer (Nanodrop). Oligonucleotides and probes to lpg2644 (Table 2 code 11 to 13) and gyrA (Table 2 code 14 to 16) were designed with Primer Express 3.0 software (Applied Biosystems) and purchased from Applied Biosystems. The probes consisted of an oligonucleotide labelled at the 5' end with 6-carboxyfluorescein (6-FAM) as the reporter and at the 3' end with carboxytetramethylrhodamine (TAMRA) as the quencher. Quantitative RT-PCR analysis (qRT-PCR) was performed with the QuantiTect Multiplex RT-PCR Kit according to the manufacturer's instructions (Qiagen). The qRT-PCR mixture contained 400 nM of each gene-specific primer, 200 nM of each probe and 5 ng of RNA template. Amplification and detection of specific products were performed with the ABI Prism 7900 detection system (Applied Biosystems) with samples first incubated at 50°C for 20 min, then at 95°C for 15 min, followed by 40 cycles at 94°C for 45 s, and 60°C for 45s. Standard curves were prepared from genomic DNA of L. pneumophila Lp02 (1 to 100000 fg) for each gene to correlate the amount of DNA to cycle thresholds. An R2 of 0.99 was obtained for all of the primers pairs that were used in this study. The amount of lpg2644 RNA was normalized to the quantity of GyrA RNA present in each preparation. According to the genoscript transcriptomic database, gyrA is not significantly regulated during biofilm formation and various growth phases (http://genoscript.pasteur.fr/). Furthermore, in other Gram-negative species, transcriptomic analyses showed that the expression of gyrA is not regulated in presence of 3OC12-HSL [36]. Experiments were performed in triplicate with RNA samples isolated from three independent cultures grown at 37°C and repeated 3 times.
Results
Lcl is Expressed at the Cell Surface of L. pneumophila and Shows a Cluster Distribution
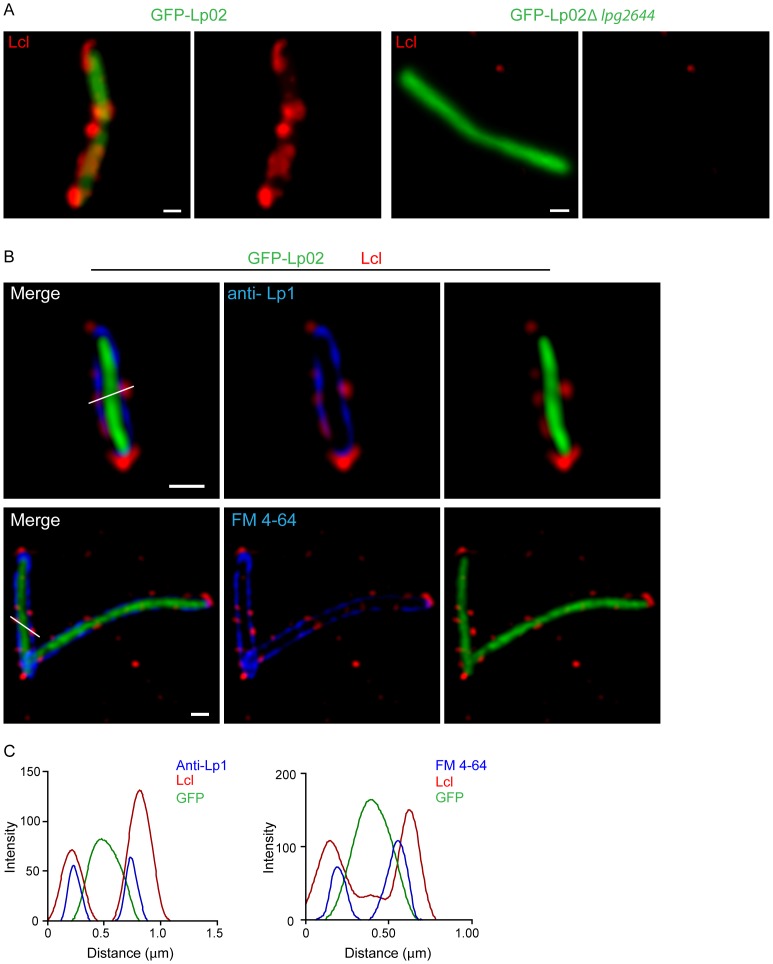
In a recent study, we discovered that Lcl promotes the formation of biofilm in L. pneumophila [32]. Deletion of Lcl encoding gene (lpg2644) dramatically decreased production of Lp1 biofilm on polystyrene matrix using a static assay. To further characterize the function of Lcl in the production of biofilms in Lp1, the cellular localization of Lcl was examined by live imaging using spinning disk confocal microscopy and live cell immuno-fluorescence assays (IFA) of Lp02 GFP expressing cells (see material and methods). Lcl anti-serum reacted with 75% of Lp02 cells. Confocal analysis revealed that Lcl is expressed at the surface of Lp02 cells, albeit not uniformly distributed (Figure 1A). No fluorescence signal was detected with Lp02 Δlpg2644 and anti-Lcl antiserum (Figure 1A). The cellular distribution of Lcl was further analysed using deconvolved confocal xy planes. To visualize the outer layers of Lp02 by live cell confocal microscopy, inner membranes and outer membrane were labelled with the dye FM4–64 and anti-Legionella pneumophila polyclonal antibodies (anti-Lp1), respectively (antibodies are not outer membrane permeable) [37] (Figure 1B). The cellular distribution of Lcl was next analysed by scanning its fluorescence intensity along cross section of the bacteria and compared with the corresponding fluorescence intensities for GFP and either anti-Lp1 or FM 4–64 (Figure 1B and C). This approach demonstrated that Lcl is expressed at the cell surface of L. pneumophila and shows a heterogeneous cluster distribution.
Figure 1. Lcl is expressed at the cell surface of L. pneumophila and shows a cluster distribution.
Localization of Lcl on the cell surface. (A) GFP-Lp02 (left) and GFP-Lp02 Δlpg2644 (right) cells were immuno-labelled using anti- Lcl antibodies followed by alexa 555 (red) conjugated secondary antibodies. Micrographs shown are merged images of 0.2 µm confocal z-stacks. Scale bars, 0.5 µm. (B) Surface localization of Lcl (red) as indicated by its co-localization with membrane labelling anti-Lp1 antibodies (top) and FM 4–64 dye (bottom). Images are confocal xy plane following deconvolution. Scale bars, 0.5 µm. (C) The localization of Lcl (red line) was determined by comparing its fluorescence intensity along the white lines from B, with cytoplasmic GFP (green line) and the outer and inner membrane markers anti-Lp1 (blue line, left) and FM 4–64 (blue line, right), respectively.
Collagen-like Tandem Repeats and C-terminal of Lcl are Required for Biofilm Formation
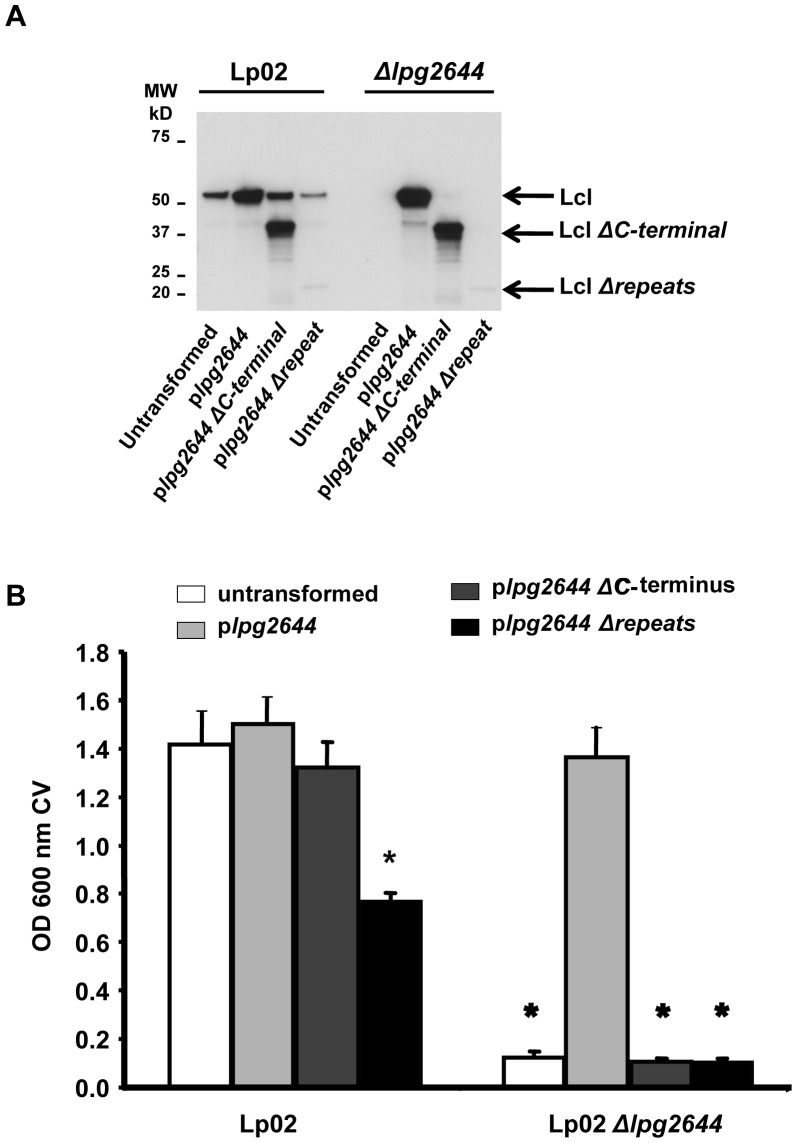
The primary sequence of Lcl is divided in 3 structural regions: the N-terminal domain including a predicted signal sequence (aa 1–68), a central polymorphic and hydrophilic region comprising collagen-like tandem repeats (aa 69–349) and a C-terminal region with no predicted function (aa 350–493). From previous report, it is know that Lcl is an immunogenic protein produced during legionellosis. This immunogenicity is partially driven by Lcl collagen-like tandem repeats [32]. To evaluate the contribution of Lcl collagenous repeats and C-terminal region of Lcl in biofilm formation, transcomplementation assays were conducted using replicative expression vectors containing lpg2644 gene without collagen-like tandem repeats (plpg2644 Δrepeats) and lpg2644 deleted in C-terminal region (plpg2644 ΔC-terminal). These plasmids were transformed into L. pneumophila Lp02 and Lp02 Δlpg2644 strains (Table 1). Immunoblotting was performed using anti-Lcl antiserum to confirm the synthesis of Lcl Δrepeats and Lcl ΔC-terminal region in Lp02 and Lp02 Δlpg2644 transformants (Figure 2A). Anti-Lcl antibodies reacted with a 50-kDa protein in control immunoblot analysis with Lp02 lysates. As expected, no anti-Lcl antibodies reactivity was observed with Lp02 Δlpg2644. Immunoblot analysis of all Lp02 transformants showed a dominant band of 50-kDa corresponding to full-length Lcl. A 25 kDa band was weakly recognized by anti-Lcl polyclonal antibodies in Lp02 plpg2644 Δrepeats and Lp02 Δlpg2644 plpg2644 Δrepeats lysates suggesting that Lcl Δrepeats was produced in these strains (Figure 2A). The weak signal observed against Lcl Δrepeats is consistent with the high immunogenicity of the tandem repeats and the use of polyclonal antibodies [32]. With Lp02 plpg2644 ΔC-terminal and Lp02 Δlpg2644 plpg2644 ΔC-terminal lysates, anti-Lcl antibodies reacted with a 32 kDa protein, which corresponds to the predicted molecular mass of Lcl ΔC-terminal.
Figure 2. The Collagen-like tandem repeat and the C-terminal domains of Lcl are essential for biofilm formation.
(A) Anti-Lcl immunoblot analyses against Lp02 and Lp02 Δlpg2644 lysates untransformed and transformed with plpg2644, plpg2644 Δrepeat and plpg2644 ΔC-terminal. The positions of the molecular size markers are indicated on the left of the blot in kDa. (B) Static biofilm assays with Lp02 and Lp02 Δlpg2644 producing recombinant variants of Lcl (plpg2644 Δrepeat and plpg2644 ΔC-terminal) and full length recombinant Lcl (plpg2644). * Two-tailed Student’s t-test P-value <0.001 versus Lp02).
Static biofilm assays were next performed with these strains using the crystal violet method. As expected, after 2 days of incubation at 37°C, Lp02 did form a biofilm while Lp02 Δlpg2644 did not (Figure 2B). In agreement with previously reported data, the biofilm defect of Lp02 Δlpg2644 could be restored at the Lp02 level by plasmid containing the full length of Lcl [32]. In contrast, the deficiency could not be rescued by plasmid expressing either a Lcl with a removed repeat region (plpg2644 Δrepeats) or a truncated C-terminal (plpg2644 ΔC-terminal). This is suggesting that these 2 domains are essential to the function of Lcl in biofilm formation. Consistent with our previous results, over-expression of Lcl in Lp02 plpg2644 did not lead to a significant increase in biofilm formation compared to Lp02 (Figure 2B). Interestingly, while Lp02 plpg2644 ΔC-terminal formed a biofilm similar to Lp02, Lp02 over-expressing Lcl without collagen-like tandem repeat domain induced a highly significant 50% decrease of Lp02 biofilm formation.
Lcl is Involved in Surface Attachment during Early Step of Biofilm Formation
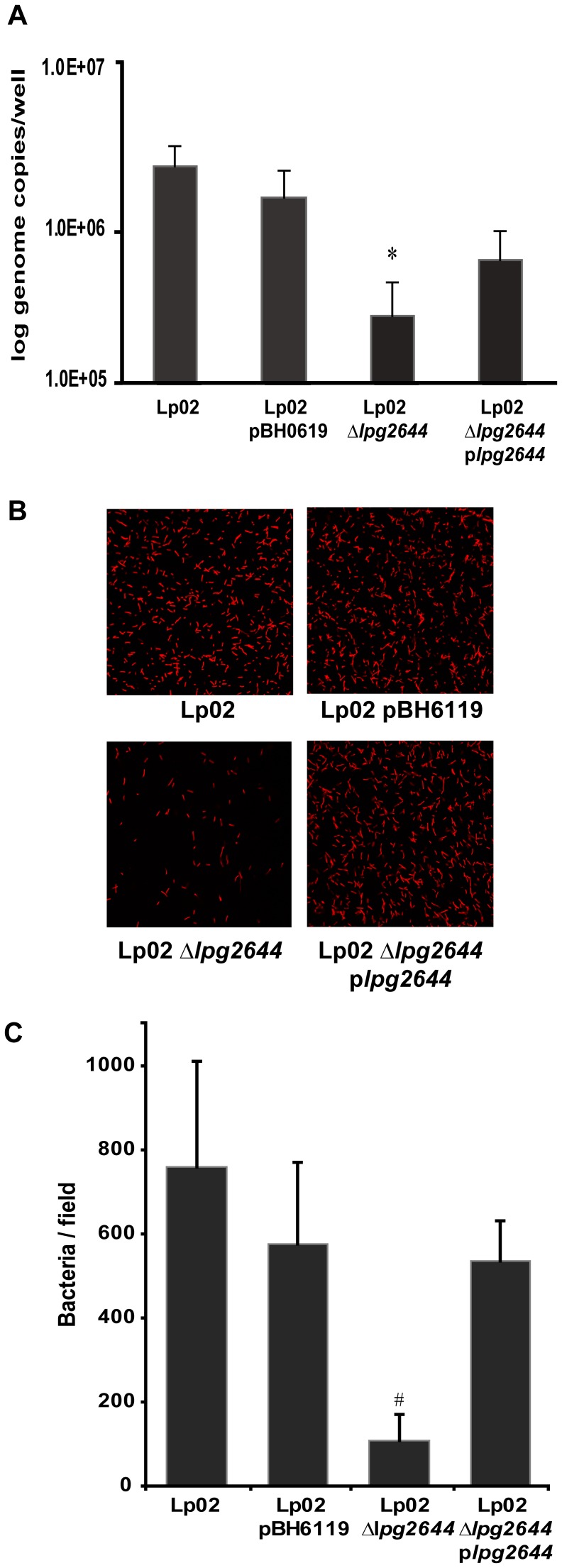
To determine whether Lcl adhesion is involved in the initial surface attachment of the biofilm formation process, we assessed the adhesion of Lp02, Lp02 pBH6119 (empty replicative plasmid), Lp02 Δlpg2644 and complemented mutant Lp02 Δlpg2644 plpg2644 to the wells of polystyrene microtitre plates by quantifying attached biomass with qPCR [32]. In control assays, the binding of Lp02 and Lp02 pBH6119 were not significantly different suggesting that the plasmid used in our experiments does not alter the binding of transformed Lp02 (Figure 3A). In contrast, the attachment of isogenic Lp02 Δlpg2644 to polystyrene was significantly reduced compared to control Lp02 strains (P≤0.001, Figure 3A). Although not fully restored, the adhesion of complemented mutant was significantly higher than Lp02 Δlpg2644 (P≤0.05) (Figure 3A). These results were further confirmed by CLSM analysis of SYTO 62 stained Legionella strains attached to glass cover slips (Figure 3B). Among 10 representative microscopy fields, the average attachment of Lp02, Lp02 pBH6119 and complemented mutant were similar whereas Lp02 Δlpg2644 showed a marked decrease attachment to surface (Figure 3C) (P<0.01). These data suggest that Lcl has a relevant role in the initial attachment of L. pneumophila to abiotic matrices.
Figure 3. Role of Lcl during the initial attachment step of biofilm formation.
The attachment of Lp02, Lp02 pBH6119, Lp02 Δlpg2644, and Lp02 Δlpg2644 plpg2644 to the wells of a microtiter plate was measured by (A) qPCR and (B and C) fluorescence microscopy. (A) The binding of Lp02 Δlpg2644 was significantly reduced compared to the other strains. Values are means+/−s.d. from three individual experiments. * Two-tailed Student’s t-test P-value <0.001 Lp02 Δlpg2644 vs. all other strains. (B) Comparative analysis of the attachment of SYTO 62 stained Legionella strains attached to glass cover slips. (C) Among 10 representative microscopy fields, the average attachment of Lp02, Lp02 pBH6119 and complemented mutant were similar whereas Lp02 Δlpg2644 showed a marked decrease attachment to surface. * Two-tailed Student’s t-test P-value <0.001.
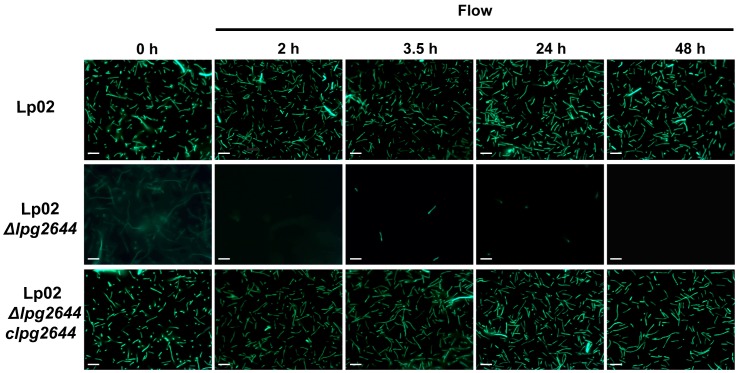
Bacteria are exposed to shear forces and in response to this stimulus, they tend to attach more firmly to matrices [38], [39]. To determine whether Lcl is likely to be a relevant adhesin in this context, flow cell biofilm assays were conducted. The presence of adhering bacteria was monitored by fluorescence microscopy using bacteria transformed with a plasmid allowing expression of recombinant green fluorescent protein (GFP) (Table 1). In control static biofilm assays, heterologous expression of GFP had no impact on bacterial growth and biofilm formation of Lp02 (data not shown). After inoculation with Lp1, flow cell chambers were incubated for 1.5 h (T = 0 h) at RT without flow to allow the cell attachment to the glass substratum. A flow of 5.5 µl/min was next applied for 6 days to allow cell growth and biofilm development. During the initial static incubation, Lp02 and complemented mutant Lp02 Δlpg2644/clpg2644 strains formed a monolayer of single dispersed cells attached to the surface (Figure 4, T = 0 h). In contrast, although cells in suspension were observed, only few Lcl isogenic mutants seem to attach to the glass surface of the flow chambers (Figure 4, T = 0 h). After the shear force was applied to the chambers, Lp02 and complemented mutant were able to form a monolayer of attached cells (Figure 4, T = 2 to 48 h). This data is consistent with observations of Mampel et al. using the same experimental model [40]. In contrast, most Lp02 Δlpg2644 cells were unable to adhere to the surface and were progressively detached by shear forces (Figure 4). This finding is consistent with our static attachment assays and it suggests that Lcl adhesin is also crucial in the initial attachment of L. pneumophila under shear flow during the early step of biofilm formation.
Figure 4. Adherence and persistence of L. pneumophila in a continuous-flow chamber system.
Fluorescence micrographs of GFP-expressing L.pneumophila strains Lp02, Lp02 Δlpg2644 and chromosomal complemented mutant (Lp02 Δlpg2644/clpg2644) in a continuous-flow chamber system operated at 5.5 µl/mn with BYE medium (magnification 100 X). During the initial attachment (0 h), Lp02 and Lp02 Δlpg2644/clpg2644 strains formed a monolayer on abiotic surface whereas Lp02 Δlpg2644 was unable to efficiently attach to the flow chambers and detached in a flow dependent manner (2 h to 48 h). Data are representative of three independent replicates. Scale bar, 5 µm.
Role of Lcl in Cell-cell and/or Cell-biofilm Matrix Components during Biofilm Formation
We next hypothesized that in addition to the initial attachment, Lcl could also play a role in cell-cell interactions and/or in the binding of Legionella to biofilm matrix components during the late stages of biofilm development. To test this hypothesis, static biofilm assays were conducted with mixed suspensions of Lp02 and Lp02 Δlpg2644 cells. Co-culture biofilms were formed on chamber slides for 3 days. To differentiate Lp02 from Lp02 Δlpg2644, wild type bacteria were transformed with pBH6119-IcmRp that allows the expression GFP and Lp02 Δlpg2644 were transformed with a promoter-less plasmid pBH6119. Plasmids were next swapped to ensure that observed phenotypes were not indirectly linked to the heterologous expression of GFP. To visualise the biofilms, bacteria were labelled with nucleic acid stain SYTO 62 and were analyzed by CLSM.
In control assays, GFP-expressing Lp02 (GFP-Lp02) and Lp02 alone developed similar mature biofilms suggesting that the expression of GFP does not alter the biofilm formation in Lp1. In assays with the GFP-expressing mutant (GFP-Lp02 Δlpg2644) and the unlabelled mutant, cells were sparsely attached to the surface and did not form micro-colonies (Figure S1).
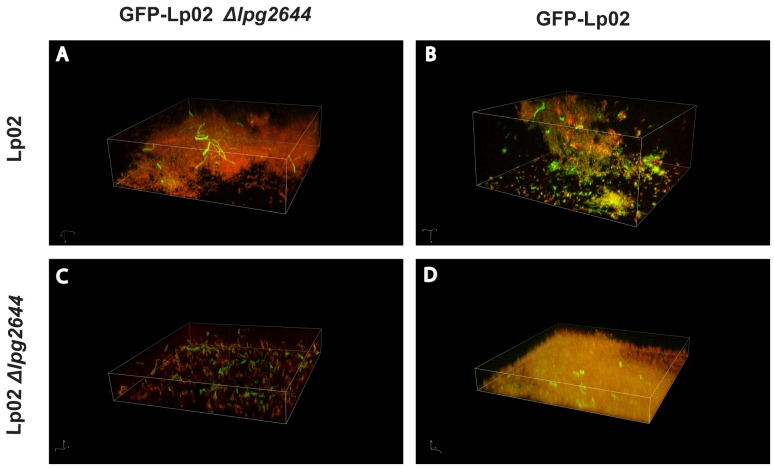
In co-culture assays with Lp02, Lp02 Δlpg2644 was unable to form micro-colonies and only few individual cells of the mutant were embedded in microcolonies of the wild type strain biofilm (Figure 5A and D). In contrast, in assays conducted with co-cultures of Lp02 and GFP-Lp02, both strains produced a three-dimensional biofilm structure in which individual green bacteria and red bacteria were interspersed homogeneously on the surface and in aggregates (Figure 5B). As expected, when Lp02 Δlpg2644 and GFP-Lp02 Δlpg2644 were used, the mutant did not form microcolonies and only sparse cells were observed (Figure 5C). Taken together these results suggest a role of Lcl in cell-cell or cell-matrix component interactions during biofilm development.
Figure 5. Representative 3D composite images of mixed culture biofilms.
Total bacteria were labelled with SYTO62 and one strain of each 1∶1 mixture was labelled with GFP. Confocal scanning laser microscopy (CSLM) micrographs show the distribution of L. pneumophila within static biofilms using (A) GFP- Lp02 Δlpg2644/Lp02, (B) GFP-Lp02/Lp02, (C) GFP- Lp02 Δlpg2644/Lp02 Δlpg2644, (D) GFP- Lp02/Lp02 Δlpg2644. (Magnification 100 X).
Regulation of Lcl Expression in different Growth Phases and Biofilm Formation
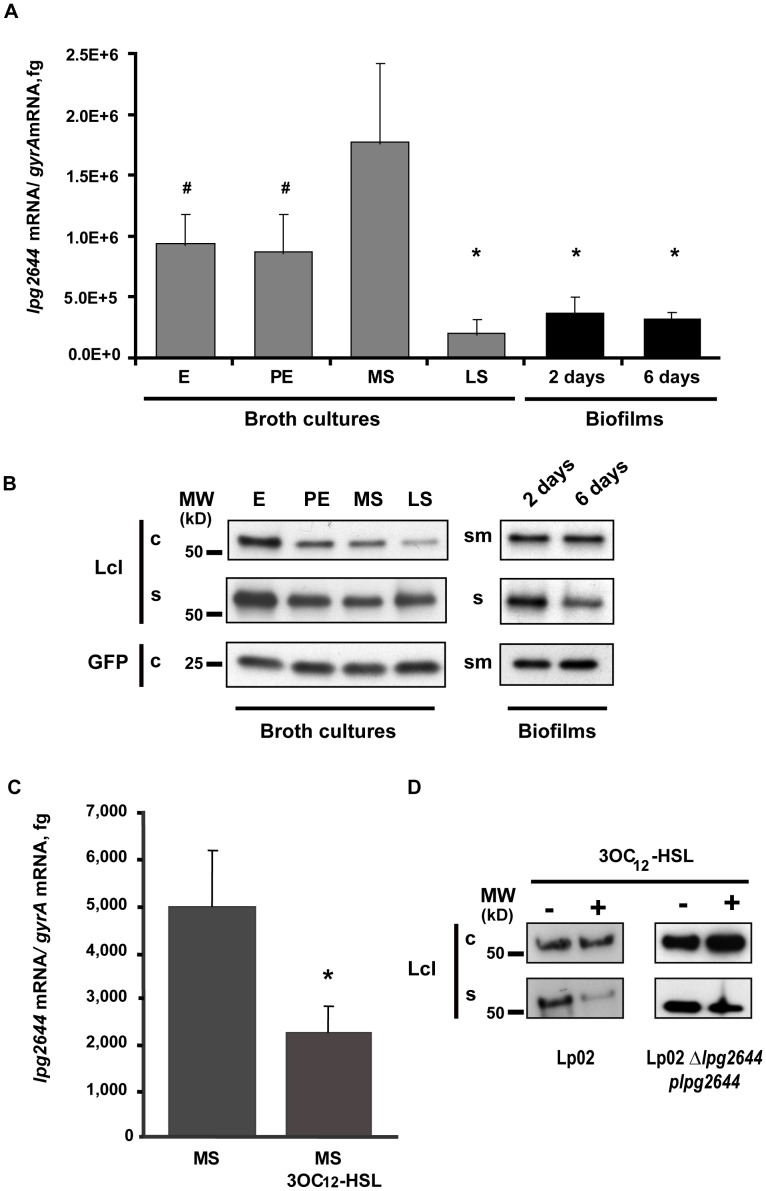
Coordination of multicellular bacterial behaviors such as biofilm formation frequently involves the transcriptional regulation of virulence and metabolic factors [41]. For Legionella bacteria, although few studies have addressed the role of transcriptional regulation and signaling in biofilm formation, there remains limited information available regarding the physiological state and gene expression within sessile cells [42], [43]. Given that Lcl is a major player in biofilm formation, we sought to investigate if Lcl expression is regulated at the transcriptional level. To verify this hypothesis, transcription levels of lcl gene were monitored by qRT-PCR using RNA from planktonic cells in broth cultures and from sessiles cells collected from static biofilms (Figure 6A). Planktonic cells were harvested during exponential (E), post exponential (PE), mid stationary (MS), and late stationary (LS) growth phases of broth cultures. RNA was also purified from sessile biofilm cells collected after 2 and 6 days of incubation (Figure 6A). In broth cultures, the relative amount of lpg2644 transcripts were 2 times lower during the E and PE phase and 8 times lower in LS phase (P≤0,001) compared to the MS phase. Interestingly, in 2 or 6 day old-biofilm assays, the amounts of lpg2644 transcripts were significantly lower than in E, PE and MS phases of the broth cultures (P≤0,001) but presented no significant (6 days) or limited differences (2 days, P≤0.05) with the LS phase. The largest differences compared to biofilms (2 and 6 days) were observed with MS growth phase bacteria which showed five times more relative amounts of lpg2644 transcripts (P≤0,001) (Figure 6A). This finding was further confirmed with the observation that the amount of lpg2644 transcripts from planktonic cells recovered from the supernatant of 2 day old biofilms was 1.7 times higher than in sessile cells (P≤0,001) (data not shown). These results indicate that the expression of lpg2644 gene differs significantly during the transition from planktonic cells to sessile cells.
Figure 6. Transcriptional regulation of lpg2644 and synthesis of Lcl in broth cultures, biofilm and in presence of 3OC12-HSL homoserine lactone.
Amounts of lpg2644 transcripts and synthesis of Lcl were respectively measured by (A) qRT-PCR and (B) anti-Lcl immunoblot analysis of cell pellet (c) supernatant (s) and sessile cells/matrix (sm) fractions in Lp02 harvested from exponential (E), post-exponential (PE), mid-stationary (MS), late stationary (LS) broth cultures and biofilms (2 and 6 days old). Quantitative RT-PCR values are means+/−s.d. from three individual experiments. # two-tailed Student’s t-test P-value ≤0.01 versus MS phase, * two-tailed Student’s t-test P-value ≤0.001 versus MS phase. (C) Effect of 3OC12-HSL homoserine lactone on lpg2644 transcription and (D) Lcl synthesis. Lp02 was grown in absence or in presence of 3OC12-HSL (10 µM) for 24 h. Transcriptional level of lpg2644 was determined by qRT-PCR analysis. Data are representative of 3 independent experiments.* Student’s t-test P-value ≤0,001 versus untreated Lp02. (D) Synthesis of Lcl was estimated by anti-Lcl immunoblot of cell pellets (c) and supernatant fractions (s) of Lp02 and Lp02 Δlpg2644 plpg2644. The positions of the molecular size markers are indicated on the left of the blot in kDa.
Next, the synthesis of Lcl protein was analysed at different growth phases and in biofilms. For broth cultures, the cells and supernatant were separated by centrifugation, and the presence of Lcl protein in each fraction was assessed by anti-Lcl immunoblot analysis. As shown in Figure 6B and 6D, secreted and cellular Lcl presented the same predicted molecular weight. Approximately 1.4 to 2.9 times less cellular Lcl was produced in LS phase than in E, PE and MS phases, as estimated by densitometry (Figure 6B). The moderate differential transcription observed between E, PE and MS phases by qRT-PCR did not translate into different amount of cellular Lcl by immunoblot analysis. Contrasting with transcriptional analysis data, the amounts of cellular and secreted Lcl in E phase were higher than any of the other growth phases. In the supernatants of PE, MS and LS broth cultures, amounts of extracellular Lcl were similar suggesting that Lcl is stable in the extracellular milieu (Figure 6 B). For biofilms assay analyses, planktonic cells and supernatants were separated from the biofilm biomass (sessile cells and matrix proteins) by collecting non-adhering material from each well. Supernatant fractions (s) were next separated from planktonic cells by centrifugation. The presence of Lcl protein in the sessile cells/matrix (sm) and the supernatant protein fractions was next assessed by anti-Lcl immunoblot analysis (Figure 6B). Although not directly comparable with broth cultures, in 2 or 6 day old-biofilm assays, the amounts of Lcl protein in the sm fractions were similar to the amounts of cellular Lcl in the E, PE and MS phases of the broth cultures (Figure 6B). This result may be explained by encasement of Lcl in the biofilm matrix of static cultures while it is released in the supernatant of broth cultures. Interestingly, the amount of Lcl found in the s fraction was reduced in 6 day old biofilms compared to 2 day old biofilms (Figure 6B). Of note, these fractions contain a mix of secreted proteins from sessile and planktonic cells and can not only be attributed to the biofilms. Taken together, these findings suggest that lpg2644 gene is moderately up-regulated during the MS phase. Once Lp1 reaches the LS phase, which is thought to mimic the behavior of bacteria from a mature biofilm, Lcl expression is down-regulated which is in agreement with the decrease transcription of Lcl gene during the late phase of biofilm maturation [44], [45].
The Transcriptional Regulation of lpg2644 Gene Plays a Role in Biofilm Development
The QS autoinducer 3OC12-HSL of P. aeruginosa which is involved in both intra and inter-species communication has recently been shown to suppress the biofilm production of Lp1 [29]. Thus, to test whether a QS system is involved in the transcriptional regulation of lpg2644 during biofilm formation, the Las QS 3OC12-HSL from P. aeruginosa was used as a model autoinducer. To rule out a possible growth-suppressive effect of LasQS 3OC12-HSL in our experimental conditions, we first determined the concentration of LasQS 3OC12-HSL that has no effect on Lp1 growth in broth cultures. A concentration range from 2.5 to 100 µM of 3OC12-HSL was tested and 10 µM was shown to have no bacteriostatic effect on Lp1 (Figure S2A and B). To evaluate the impact of this autoinducer on lpg2644 transcription, RNA was extracted from Lp02 broth cultures grown to MS phase in absence and in presence of 10 µM 3OC12-HSL. Transcription levels of lpg2644 were next determined by qRT-PCR. As shown in Figure 6C, the relative amount of lpg2644 RNA was 2.2 times lower in presence of 3OC12-HSL compared to untreated cells (P≤0.001). This data suggests that QS autoinducers such as 3OC12-HSL down-regulate the expression of Lcl which is agreement with the down-regulation of lpg2644 during the late phase of biofilm formation. The synthesis of Lcl were next examined in MS broth cultures treated with 3OC12-HSL (Figure 6D). According to densitometry analyses, the syntheses of Lcl in Lp02 were similar in cellular fractions with 3OC12-HSL and untreated cultures. Interestingly, the main differences between the 2 cultures were observed in the extracellular fractions. When treated with 3OC12-HSL, the amount of extracellular Lcl was reduced by 73% compared to an untreated culture. Of note, this reduction of extracellular Lcl in 3OC12-HSL treated Lp02 was not correlated with the accumulation of cellular proteins. A reduction of protein export by the type II secretion system is known to induce the cellular accumulation of proteins that are normally secreted [46]. As this is not the case here, our data suggest that the synthesis and the secretion of Lcl in Lp02 are both reduced in presence of 3OC12-HSL. In control assays with Lp02 Δlpg2644 plpg2644, the amount of cellular Lcl was increased in cells treated with 3OC12-HSL. In contrast, the amount of extra-cellular Lcl was decreased in 3OC12-HSL treated cells compared to untreated controls (Figure 6D).
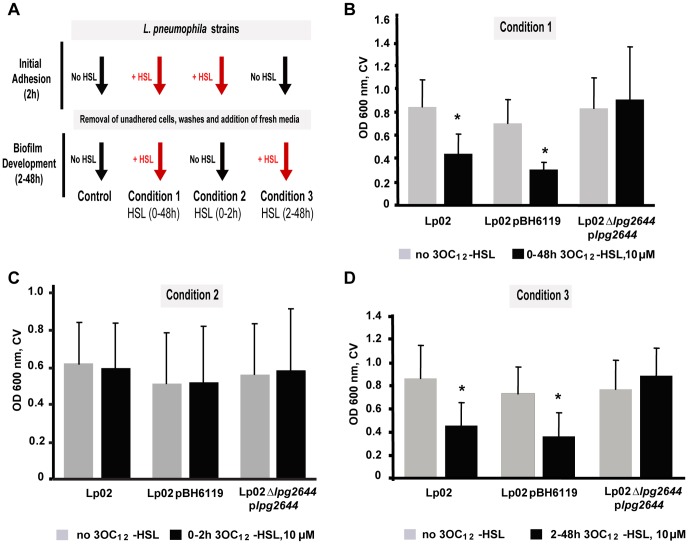
This finding raised the possibility that the transcriptional regulation of lpg2644 may have an impact on Lp1 biofilm development. Thus, static biofilm assays were conducted in presence or in absence of 3OC12-HSL (Figure 7A). As expected, in presence of 3OC12-HSL, the biofilm formation was significantly impaired in Lp02 (48% inhibition) and Lp02 pBH6119 (42% inhibition) (P≤0.001) (Figure 7A and B, Condition 1). In contrast, in trans-complementation assays with a plasmid containing lpg2644 under the control of the IcmR promotor (plpg2644) which is not regulated during biofilm formation (http://genoscript.pasteur.fr/), the biofilm production of Lp02 Δlpg2644 plpg2644 remained unchanged in presence and in absence of 3OC12-HSL (Figure 7A and B, Condition 1). Thus, when Lcl synthesis is under the control of the IcmR promoter, the constitutive expression of Lcl is sufficient to maintain biofilm integrity. These results suggest that the transcriptional regulation of Lcl gene through QS signaling plays a key role in Lp1 biofilm formation.
Figure 7. The quorum sensing dependent regulation of Lcl plays a key-role in the control of L. pneumophila biofilm development.
Flow diagrams indicating the experimental stages when Las QS 3OC12-HSL auto-inducer (HSL) was used and the corresponding static biofilm assays (A). L. pneumophila strains Lp02, Lp02 pBH6119 (empty plasmid) and Lp02 Δlpg2644 plpg2644 were exposed to Las QS 3OC12-HSL during 48 h (B), the attachment phase (0–2 h) (C) and biofilm formation (2–48 h) (D). * P value <0.001 no HSL vs+HSL.
To further investigate the impact of QS signaling on lpg2644 transcriptional regulation during the early stage (adhesion) and biofilm development, we used a modified microtitre biofilm assay. Bacterial cells were only exposed to 3OC12-HSL during the initial adhesion phase (Figure 7A, Condition 2) or during the biofilm development phase (Figure 7A, Condition 3). When 3OC12-HSL was only present during the initial adhesion phase (0–2 h), biofilms formed by Lp02, Lp02pBH6119 and Lp02 Δlpg2644 plpg2644 were comparable to controls without 3OC12-HSL (Figure 7C). This result raised the possibility that absence of effect observed on biofilms formation in condition 2 may be due to the short time exposure (2 h) to 3OC12-HSL compared to condition 3 (48 h). To control whether this is the case, similar experiments were performed with bacteria precultured (24 h) with 3OC12-HSL. With this pre-incubation, biofilms formations of Lp02pBH6119 and Lp02 Δlpg2644 plpg2644 using condition 2 remained similar (Figure S3). These results suggest that QS regulation of lpg2644 may only have a minor impact on the initial attachment phase.
In contrast, when bacteria were exclusively exposed to 3OC12-HSL during biofilm development (Figure 7A Condition 3), a marked reduction in biofilm formation was observed with Lp02 (47% inhibition) and Lp02 pBH6119 (50% inhibition) compared to their respective untreated controls (P<0.001) whereas the rescued mutant presented an unaltered biofilm (Figure 7D). These results imply that the deregulation of lpg2644 gene in the rescued mutant is sufficient to maintain the integrity of biofilms when Lp1 cells are exposed to 3OC12-HSL. Moreover, these results suggest that the transcriptional regulation of lpg2644 gene via QS bacterial auto-inducer such as 3OC12-HSL may play a role during biofilm development rather than the adhesion phase of Lp1.
Discussion
The production and colonization of biofilm is considered to play a key role in the environmental dissemination/survival and possibly the pathogenesis of Legionella pneumophila [47]. However, little is known about the components and the molecular events that mediate the production of biofilm in these pathogenic environmental bacteria [48]. Only few mediators of Legionella biofilm formation have been reported such as FliA [40], a putative twin-arginine protein transport system [49] and the c-di-GMP metabolism [43]. More recently, we reported that Lcl promotes biofilm formation by Legionella [32]. In this study, we investigated the immuno-localization, roles and regulation of this surface exposed adhesin during different stages of biofilm formation.
To evaluate the roles of the collagen-like repeats and C-terminal domains of Lcl, we transcomplemented a Lcl mutant with variants devoid of these respective domains. None of these variants could rescue the impairment in biofilm formation of the Lcl deletion mutant. Thus, both of these domains contribute to the role of Lcl in biofilm formation. Lcl was found to play a crucial role in the initial attachment of L. pneumophila to abiotic matrices during the early step of biofilm development. This was confirmed by our adhesion assays and flow cell experiments showing that attachment of an isogenic mutant Lp02 Δlpg2644 to polystyrene and glass was significantly reduced compared to Lp02. These findings are correlated with the structural differences observed by CLSM between the biofilms of Lp02 and Δlpg2644 mutant. They also support the hypothesis that Lcl may participate in cell-cell and/or cell-matrix interactions during biofilm development. In biofilm assays, the few Δlpg2644 mutant cells bound to the substratum did not produce three-dimensionnal structures in contrast to wild type strain. Moreover, in mixed cultures, the mutant did not form microcolonies and only few Lcl depleted cells were imbedded in Lp02 biofilms. Altogether, these observations suggest a role of Lcl in the anchoring of L. pneumophila bacteria to each other or to the biofilm matrix. Given that Lcl is also a secreted protein [50], this finding implies that the secreted protein on its own is not sufficient to rescue the biofilm production impairment of Δlpg2644 mutant. Thus, Lcl may need to be surface exposed to play a role in biofilm development.
L. pneumophila employs a biphasic life cycle to replicate within host cells and to spread to new niches. This process is under the control of a regulatory cascade that responds to a variety of environmental stimuli. Upon entry into stationary growth phase, L. pneumophila switches from a replicative (metabolic pathways) to a transmissive (virulent) state, which requires expression of genes involved in virulence, motility, stress resistance and cell division [44], [51]. In this report, we show that the lpg2644 gene is down regulated in two day old biofilm and during the LS phase compared to the MS growth phase. Taking into consideration that cells grown to the LS phase are known to be metabolically similar to cells of biofilms, this suggests that the pool of newly synthesized Lcl decreases during the late stage of biofilm formation [52]. Therefore, in view of the critical role of Lcl in adhesion and cell-cell or cell-matrix interactions, we propose that the observed transcriptional regulation may participate in the detachment and dispersal of matured Lp1 biofilm.
In LS phase, the transcriptional down-regulation of lpg2644 transcription correlated with a reduction of cellular Lcl protein. Interestingly, despite this reduction of cellular Lcl, the amount of extracellular Lcl in the LS phase was similar to other growth phases. This suggests that Lcl may be a relatively stable protein in the extracellular milieu of broth cultures. Moreover, while the transcription of lpg2644 was found to be down-regulated in biofilms, anti-Lcl immunoblot analyses of biofilm cell fractions revealed higher amount of Lcl compared to cell of LS phase broth cultures. To explain this difference, we are speculating that in biofilms, secreted Lcl is encased in the biofilm matrix and is co-purified with the cellular fraction. This hypothesis is in agreement with the recent discovery of secreted proteins, Bap1 and RbmC, that are selectively retained in the biofilm matrix of Vibrio cholera [53].
In Gram-negative bacteria, communication systems (QS sytems) orchestrate important temporal events including synthesis of virulence factors and biofilm formation through the use of autoinducers [54]–[56]. QS bacteria are routinely identified in man-made water systems and it is now recognized that QS systems may play a role in the regulation of environmental biofilm production [57]. In multispecies biofilms, quorum sensing also mediates interactions between bacterial species such as L. pneumophila and P. aeruginosa. Two dependent QS systems have been described in P. aeruginosa: Las and Rhl [58]–[61]. The Rhl-based QS is regulated by N-butanoyl- L-homoserine lactone (C4-HSL) produced by RhlRI whereas the Las-based QS is regulated by the autoinducer 3OC12-HSL produced by LasRI. P. aeruginosa requires the lasI gene product (3OC12-HSL) in order to develop a differentiated biofilm and 3OC12-HSL is believed to regulate the complex architecture observed in mature biofilm [55]. Las QS 3OC12-HSL is also involved in inter-species interactions and it has been shown to suppress the growth and the biofilm formation of L. pneumophila [29].
Here, we show that a non-growth-suppressive concentration of 3OC12-HSL down-regulates the transcription of lpg2644 and the secretion of Lcl in MS phase. Interestingly, the biofilm production of a complemented mutant producing Lcl under the control of a constitutive promoter is not impaired in the presence of 3OC12-HSL. This result implies that the constitutive expression of Lcl is sufficient to maintain the biofilm integrity of Lp1 and suggests that the QS regulation of Lcl may play a key role in the course of the biofilm development/maturation of this environmental pathogen. In addition, we uncover that the QS 3OC12-HSL transcriptional regulation of lpg2644 is preferentially involved after the initial adhesion stage of biofilm development. This result is agreement with data from Davies and al. showing that QS 3OC12-HSL participates exclusively in the late phase of biofilm maturation [52], [55].
In L. pneumophila, α-hydroxyketone (AHK) of the Lqs system is the only QS autoinducer that has been described to date [27], [28]. Host cell interactions are under the control of a complex virulence regulatory network including the lqs gene cluster (lqsA-lqsR-hdeD-lqsS) [27]. Although it remains unknown if this cluster regulates biofilm production in Legionella, it is homologous to the Vibrio cholerae cqsAS quorum sensing system 1 which is involved in cell density-dependent regulation of virulence and biofilm formation [24], [25], [62]. Vibrio spp. produce three distinct chemical classes of autoinducers: furanosyl borate diester AI-2, N-acyl-L-homoserine lactones (AHLs) and α-hydroxyketones (AHKs). In the Lqs system, the lqs gene cluster includes the lqsR gene that is flanked by lqsA and lqsS encoding respectively an autoinducer synthase LqsA and a sensor kinase LqsS. LqsR is a putative response regulator that promotes the induction of transmissive traits including motility and virulence (phagocytosis, intracellular replication and cytotoxicity) and inhibits entry into the replicative growth phase. Globally, this two-component system regulates the expression of a 133 kb genomic island. The auto-inducer synthase LqsA only regulates few genes whereas LqsS regulates the expression of 93 genes suggesting that LqsS responds to additional QS molecules than the α-hydroxyketone of LqsA [27], [28]. In comparative transcriptome analyses, the gene regulation patterns of ΔlqsS and ΔlqsR mutants showed a strong correlation which suggests that they share a common signal transduction pathway. Among these genes, lpg2644 was shown to be up-regulated 4.27 times in the absence of Lqs system [63], [64]. Lcl is secreted by the type II secretion system, a secretion pathway that partially relies on the Sec cytoplasmic membrane translocation system [50]. Interestingly sec genes were also found to be up-regulated in a Legionella lqs quorum sensing mutant [64]. Taking into account that the amount of secreted Lcl was reduced after treatment with HSL and in aging biofilms, these findings suggest that the synthesis and the secretion of Lcl may be co-regulated by the quorum sensing. More work is necessary to evaluate the relative contributions of the regulations of lpg2644 transcription and Lcl secretion in Lp1 biofilm formation.
The 3OC12-HSL was recently shown to regulate the transcription of lqsR [29]. Although it remains unknown if LqsS responds to homoserine lactones, considering that our study reveals that lpg2644 is also downregulated by 3OC12-HSL, it is tempting to speculate that Lp1 responds to QS signaling through at least part of this signal transduction pathway by regulating the expression of Lcl and the production of biofilm. In view of the data presented in this report, it would be interesting to further investigate the role of Lqs system on lpg2644 transcriptionnal regulation by testing the effect of spent media or purified Legionella α-hydroxyketone on Lp02 and by measuring the impact of LqS overexpression on biofilm formation and lpg2644 transcription.
In summary, we have revealed that Lcl plays a crucial role in biofilm formation during the attachment phase but also in cell-cell interactions phase and/or cell-matrix component interactions where it participates in the three-dimensional structure of the biofilm. Active detachment from the substratum is a physiologically regulated event of biofilm development and our results suggest that homoserine lactone may play a role in detachment during the dispersal phase. This is in agreement with observed down-regulation of Lcl in 2 and 6 day old-biofilms and after incubation of Lp1 with 3OC12-HSL. Although additional experiment are necessary to confirm this hypothesis, this suggests that homoserine lactones like 3OC12-HSL can stimulate the dispersion of Lp1 biofilm by down-regulating the synthesis of attachment factors such as Lcl. While few studies have addressed the transcriptional regulation/signaling of Lp1 biofilm, this report is the first to address the role and regulation of a matrix-associated protein of Lp1 biofilm. In the long term, these findings may lead to the design of alternative disinfection strategies that could more efficiently prevent colonization of man-made water systems by Legionella.
Supporting Information
Three-dimensional views of biofilm assays visualized by CLSM. Biofilm assays were performed with (A) GFP-Lp02, (B) GFP- Lp02 Δlpg2644, (C) Lp02 stained with Syto 62 and (D) Lp02 Δlpg2644 stained with SYTO62. All micrographs were taken at 3 days. Single culture of GFP-Lp02 or Lp02 stained with SYTO 62 developed a mature biofilm, whereas assays with GFP- Lp02 Δlpg2644 or SYTO62 stained Lp02 Δlpg2644 show isolated cells that did not form micro-colonies.
(TIF)
A concentration of 10 µM of 3OC12-HSL does not affect the growth of L. pneumophila in broth pre-cultures and in biofilm assays. (A) L. pneumophila was grown in BYE medium with (red square) or without (black diamond) 10 µM 3OC12-HSL at 37°C with constant shaking. Bacterial growth was determined by measuring the optical density at 600 nm. (B) Growth of L. pneumophila during biofilm formation after 48 h of incubation at 37°C with or without 3OC12-HSL (10 µM). Bacterial growth was determined by measuring the optical density at 600 nm.
(TIF)
After a 24 h pre-incubation with 3OC12-HSL, the quorum sensing dependent regulation of Lcl does not have an impact on the attachment phase (0–2 h) of biofilm development. Pre-cultures (24 h) of L. pneumophila strains Lp02, Lp02 pBH6119 (empty plasmid) and Lp02 Δlpg2644 plpg2644 with 3OC12-HSL were used in static biofilm assays under the condition 2 described in Figure 7 (attachment phase, 0–2 h).
(TIF)
Funding Statement
This work was supported by the Ontario Agency for Health protection and Promotion (http://www.oahpp.ca/) and the Canadian Institutes of Health Research (MOP-102514) (http://www.cihr-irsc.gc.ca/). AP and MT are supported by the Natural Sciences and Engineering Research Council of Canada (http://www.nserc-crsng.gc.ca/) and the Ontario Association of Medical Laboratories grants (http://www.oaml.com/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stout JE, Yu VL (1997) Eradicating Legionella from hospital water. Jama 278: 1404–1405. [PubMed] [Google Scholar]
- 2. Benin AL, Benson RF, Besser RE (2002) Trends in legionnaires disease, 1980–1998: declining mortality and new patterns of diagnosis. Clin Infect Dis 35: 1039–1046. [DOI] [PubMed] [Google Scholar]
- 3.Bartram J, Chartier Y, Lee JV, Pond K, Surman-Lee S (2007) Legionella and the prevention of legionellosis. In: Organization WH, editor. Geneva: WHO Press. 1–276.
- 4. Fields BS, Benson RF, Besser RE (2002) Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fliermans CB, Cherry WB, Orrison LH, Smith SJ, Tison DL, et al. (1981) Ecological distribution of Legionella pneumophila . Appl Environ Microbiol 41: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, et al. (2002) Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis 186: 127–128. [DOI] [PubMed] [Google Scholar]
- 7.Luck CP, Jacobs E, Roske I, Schroter-Bobsin U, Dumke R, et al. (2009) Legionella dresdenensis sp. nov. isolated from the river Elbe near Dresden in Germany. Int J Syst Evol Microbiol: Epub ahead of print. [DOI] [PubMed]
- 8. Hilbi H, Jarraud S, Hartland E, Buchrieser C (2010) Update on Legionnaires' disease: pathogenesis, epidemiology, detection and control. Mol Microbiol 76: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piao Z, Sze CC, Barysheva O, Iida K, Yoshida S (2006) Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila . Appl Environ Microbiol 72: 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vervaeren H, Temmerman R, Devos L, Boon N, Verstraete W (2006) Introduction of a boost of Legionella pneumophila into a stagnant-water model by heat treatment. FEMS Microbiol Ecol 58: 583–592. [DOI] [PubMed] [Google Scholar]
- 11. Murga R, Forster TS, Brown E, Pruckler JM, Fields BS, et al. (2001) Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147: 3121–3126. [DOI] [PubMed] [Google Scholar]
- 12. Declerck P, Behets J, van Hoef V, Ollevier F (2007) Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res 41: 3159–3167. [DOI] [PubMed] [Google Scholar]
- 13. Declerck P, Behets J, van Hoef V, Ollevier F (2007) Replication of Legionella pneumophila in floating biofilms. Curr Microbiol 55: 435–440. [DOI] [PubMed] [Google Scholar]
- 14. Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- 15. Dunne WM Jr, Pinckard JK, Hooper LV (2002) Clinical microbiology in the year 2025. J Clin Microbiol 40: 3889–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper IR, Hanlon GW (2010) Resistance of Legionella pneumophila serotype 1 biofilms to chlorine-based disinfection. J Hosp Infect 74: 152–159. [DOI] [PubMed] [Google Scholar]
- 17. Hall-Stoodley L, Stoodley P (2009) Evolving concepts in biofilm infections. Cell Microbiol 11: 1034–1043. [DOI] [PubMed] [Google Scholar]
- 18. Hoiby N, Ciofu O, Bjarnsholt T (2010) Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5: 1663–1674. [DOI] [PubMed] [Google Scholar]
- 19. Anderson GG, O'Toole GA (2008) Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol 322: 85–105. [DOI] [PubMed] [Google Scholar]
- 20. Richards JJ, Melander C (2009) Controlling bacterial biofilms. Chembiochem 10: 2287–2294. [DOI] [PubMed] [Google Scholar]
- 21. Ueda A, Attila C, Whiteley M, Wood TK (2009) Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb Biotechnol 2: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Camilli A, Bassler BL (2006) Bacterial small-molecule signaling pathways. Science 311: 1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bassler BL (2002) Small talk. Cell-to-cell communication in bacteria. Cell 109: 421–424. [DOI] [PubMed] [Google Scholar]
- 24. Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae . Cell 110: 303–314. [DOI] [PubMed] [Google Scholar]
- 25. Henke JM, Bassler BL (2004) Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus . J Bacteriol 186: 3794–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, et al. (2007) The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450: 883–886. [DOI] [PubMed] [Google Scholar]
- 27. Tiaden A, Spirig T, Weber SS, Bruggemann H, Bosshard R, et al. (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9: 2903–2920. [DOI] [PubMed] [Google Scholar]
- 28. Spirig T, Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, et al. (2008) The Legionella autoinducer synthase LqsA produces an alpha-hydroxyketone signaling molecule. J Biol Chem 283: 18113–18123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimura S, Tateda K, Ishii Y, Horikawa M, Miyairi S, et al. (2009) Pseudomonas aeruginosa Las quorum sensing autoinducer suppresses growth and biofilm production in Legionella species. Microbiology 155: 1934–1939. [DOI] [PubMed] [Google Scholar]
- 30. O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, et al. (2008) A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol 190: 3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heilmann C, Niemann S, Sinha B, Herrmann M, Kehrel BE, et al. (2004) Staphylococcus aureus fibronectin-binding protein (FnBP)-mediated adherence to platelets, and aggregation of platelets induced by FnBPA but not by FnBPB. J Infect Dis 190: 321–329. [DOI] [PubMed] [Google Scholar]
- 32. Duncan C, Prashar A, So J, Tang P, Low DE, et al. (2011) Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun 79: 2168–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vandersmissen L, De Buck E, Saels V, Coil DA, Anne J (2010) A Legionella pneumophila collagen-like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS Microbiol Lett 306: 168–176. [DOI] [PubMed] [Google Scholar]
- 34. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 35. Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bijtenhoorn P, Mayerhofer H, Muller-Dieckmann J, Utpatel C, Schipper C, et al. (2011) A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 6: e26278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fishov I, Woldringh CL (1999) Visualization of membrane domains in Escherichia coli . Mol Microbiol 32: 1166–1172. [DOI] [PubMed] [Google Scholar]
- 38. Tchesnokova V, McVeigh AL, Kidd B, Yakovenko O, Thomas WE, et al. (2010) Shear-enhanced binding of intestinal colonization factor antigen I of enterotoxigenic Escherichia coli . Mol Microbiol 76: 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I (2002) Biofilm material properties as related to shear-induced deformation and detachment phenomena. J Ind Microbiol Biotechnol 29: 361–367. [DOI] [PubMed] [Google Scholar]
- 40. Mampel J, Spirig T, Weber SS, Haagensen JA, Molin S, et al. (2006) Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol 72: 2885–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wood TK (2009) Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hindre T, Bruggemann H, Buchrieser C, Hechard Y (2008) Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154: 30–41. [DOI] [PubMed] [Google Scholar]
- 43.Carlson HK, Vance RE, Marletta MA (2010) H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol Microbiol. [DOI] [PMC free article] [PubMed]
- 44. Molofsky AB, Swanson MS (2004) Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol 53: 29–40. [DOI] [PubMed] [Google Scholar]
- 45. Byrne B, Swanson MS (1998) Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66: 3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pugsley AP (1993) The complete general secretory pathway in gram-negative bacteria. Microbiol Rev 57: 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lau HY, Ashbolt NJ (2009) The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107: 368–378. [DOI] [PubMed] [Google Scholar]
- 48. Declerck P, Behets J, Margineanu A, van Hoef V, De Keersmaecker B, et al. (2009) Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Res 164: 593–603. [DOI] [PubMed] [Google Scholar]
- 49. De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, et al. (2005) Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun 331: 1413–1420. [DOI] [PubMed] [Google Scholar]
- 50. DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP (2006) Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103: 19146–19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Molofsky AB, Swanson MS (2003) Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50: 445–461. [DOI] [PubMed] [Google Scholar]
- 52. Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56: 187–209. [DOI] [PubMed] [Google Scholar]
- 53. Absalon C, Van Dellen K, Watnick PI (2011) A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog 7: e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, et al. (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407: 762–764. [DOI] [PubMed] [Google Scholar]
- 55. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, et al. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298. [DOI] [PubMed] [Google Scholar]
- 56. Hentzer M, Eberl L, Nielsen J, Givskov M (2003) Quorum sensing : a novel target for the treatment of biofilm infections. BioDrugs 17: 241–250. [DOI] [PubMed] [Google Scholar]
- 57. Shrout JD, Nerenberg R (2012) Monitoring bacterial twitter: does quorum sensing determine the behavior of water and wastewater treatment biofilms? Environ Sci Technol 46: 1995–2005. [DOI] [PubMed] [Google Scholar]
- 58. Shiner EK, Rumbaugh KP, Williams SC (2005) Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev 29: 935–947. [DOI] [PubMed] [Google Scholar]
- 59. Parsek MR, Greenberg EP (2000) Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci U S A 97: 8789–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Juhas M, Eberl L, Tummler B (2005) Quorum sensing: the power of cooperation in the world of Pseudomonas . Environ Microbiol 7: 459–471. [DOI] [PubMed] [Google Scholar]
- 61. Fuqua C, Greenberg EP (2002) Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3: 685–695. [DOI] [PubMed] [Google Scholar]
- 62. Henke JM, Bassler BL (2004) Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi . J Bacteriol 186: 6902–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tiaden A, Spirig T, Sahr T, Walti MA, Boucke K, et al. (2010) The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila . Environ Microbiol 12: 1243–1259. [DOI] [PubMed] [Google Scholar]
- 64. Tiaden A, Spirig T, Carranza P, Bruggemann H, Riedel K, et al. (2008) Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions. J Bacteriol 190: 7532–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berger KH, Isberg RR (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila . Mol Microbiol 7: 7–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional views of biofilm assays visualized by CLSM. Biofilm assays were performed with (A) GFP-Lp02, (B) GFP- Lp02 Δlpg2644, (C) Lp02 stained with Syto 62 and (D) Lp02 Δlpg2644 stained with SYTO62. All micrographs were taken at 3 days. Single culture of GFP-Lp02 or Lp02 stained with SYTO 62 developed a mature biofilm, whereas assays with GFP- Lp02 Δlpg2644 or SYTO62 stained Lp02 Δlpg2644 show isolated cells that did not form micro-colonies.
(TIF)
A concentration of 10 µM of 3OC12-HSL does not affect the growth of L. pneumophila in broth pre-cultures and in biofilm assays. (A) L. pneumophila was grown in BYE medium with (red square) or without (black diamond) 10 µM 3OC12-HSL at 37°C with constant shaking. Bacterial growth was determined by measuring the optical density at 600 nm. (B) Growth of L. pneumophila during biofilm formation after 48 h of incubation at 37°C with or without 3OC12-HSL (10 µM). Bacterial growth was determined by measuring the optical density at 600 nm.
(TIF)
After a 24 h pre-incubation with 3OC12-HSL, the quorum sensing dependent regulation of Lcl does not have an impact on the attachment phase (0–2 h) of biofilm development. Pre-cultures (24 h) of L. pneumophila strains Lp02, Lp02 pBH6119 (empty plasmid) and Lp02 Δlpg2644 plpg2644 with 3OC12-HSL were used in static biofilm assays under the condition 2 described in Figure 7 (attachment phase, 0–2 h).
(TIF)