Abstract
Background
Despite their distinct biology, granulosa cell tumours (GCTs) are treated the same as other ovarian tumours. Intriguingly, a recurring somatic mutation in the transcription factor Forkhead Box L2 (FOXL2) 402C>G has been found in nearly all GCTs examined. This investigation aims to identify the pathogenicity of mutant FOXL2 by studying its altered transcriptional targets.
Methods
The expression of mutant FOXL2 was reduced in the GCT cell line KGN, and wildtype and mutant FOXL2 were overexpressed in the GCT cell line COV434. Total RNA was hybridised to Affymetrix U133 Plus 2 microarrays. Comparisons were made between the transcriptomes of control cells and cells altered by FOXL2 knockdown and overexpression, to detect potential transcriptional targets of mutant FOXL2.
Results
The overexpression of wildtype and mutant FOXL2 in COV434, and the silencing of mutant FOXL2 expression in KGN, has shown that mutant FOXL2 is able to differentially regulate the expression of many genes, including two well known FOXL2 targets, StAR and CYP19A. We have shown that many of the genes regulated by mutant FOXL2 are clustered into functional annotations of cell death, proliferation, and tumourigenesis. Furthermore, TGF-β signalling was found to be enriched when using the gene annotation tools GATHER and GeneSetDB. This enrichment was still significant after performing a robust permutation analysis.
Conclusion
Given that many of the transcriptional targets of mutant FOXL2 are known TGF-β signalling genes, we suggest that deregulation of this key antiproliferative pathway is one way mutant FOXL2 contributes to the pathogenesis of adult-type GCTs. We believe this pathway should be a target for future therapeutic interventions, if outcomes for women with GCTs are to improve.
Introduction
Granulosa cell tumours of the ovary (GCT) are the predominant type of ovarian sex-cord tumour, yet they comprise approximately 5% of all malignant ovarian neoplasms [1], [2]. Their rarity poses a limitation in our understanding of their aetiology and molecular pathogenesis. Divided into two distinct subtypes: adult and juvenile; the tumour cells of GCTs demonstrate several morphological, biochemical and hormonal features of normal proliferating pre-ovulatory granulosa cells. Women with GCTs tend to present with symptoms of excessive oestrogen secretion by the tumour, enabling the disease to be detected at an early stage due to the consequent symptoms of abnormal uterine bleeding, menorrhagia or cycle disturbances. However, GCTs are characterised by slow growth and a tendency to relapse, requiring patients with GCTs to undergo prolonged follow-up, as recurrences have been known to occur even forty years after the initial diagnosis [3], [4]. The overall relapse rate for women with adult-type GCTs is approximately 30%, however 70–80% of women with recurrent disease will die from GCTs [5].
In contrast to ovarian epithelial tumours, GCTs are a relatively homogenous tumour, likely to have arisen from a limited set of molecular events in specific signalling pathways [6]. Yet current GCT treatment strategies are modelled on the behaviour of ovarian epithelial tumours. Given the numerous differences between granulosa cells, and ovarian surface epithelial cells, it is likely that GCTs may require a specific treatment based on the molecular defects in the tumour itself, rather than being treated like all ovarian tumours, which only share with GCTs a common location, the ovary. With this in mind, a range of genes important in normal granulosa cell biology, as well as their relevant signalling pathways have been investigated as putative candidates involved in GCT pathogenesis (refer to Jamieson & Fuller, 2012 for a comprehensive review).
FOXL2 belongs to the large family of forkhead FOX transcription factors, and its expression is strongly maintained in granulosa cells throughout life [7], [8]. Furthermore, the overall phenotype of FOXL2 knockout mice models confirms that this gene is critical for the proper differentiation of granulosa cells [9], [10]. FOXL2 expression has also been observed in the developing eyelid, thus notably involving this gene in the pathogenesis of blepharophimosis ptosis epicanthus inversus syndrome (BPES), with or without accompanying premature ovarian failure (POF). Considering that POF is part of the phenotypic spectrum of FOXL2 mutations, FOXL2 was assumed to be a possible candidate for POF in the absence of BPES. Indeed previous work on this gene has identified two novel FOXL2 variants in two women with isolated cases of POF from New Zealand and Slovenia [11].
In 2009, the landmark study by Shah et al identified a recurring somatic mutation 402C>C in the gene FOXL2 [12]. This mutation was confirmed to be present in 97% if adult GCTs subsequently tested, and in much lower proportions (10%) in juvenile GCTs; a finding that has been replicated in independent cohorts of GCTs [13], [14], [15]. Most interestingly, this mutation was not found in any other sex-cord stromal tumours, nor in any unrelated ovarian or breast tumours [16], [17].
The 402C>G mutation results in an amino acid substitution of tryptophan for cysteine (C134W) [12], [13], which is located in the second wing on the surface of the forkhead domain. Computer modelling suggests this alteration does not disrupt the folding of the FOXL2 forkhead domain or its interactions with DNA. In addition it has been shown that mutation does not affect the localisation of the FOXL2 protein [18]. Therefore it is speculated that the pathogenicity of mutant FOXL2 occurs through changes to its interactions with other proteins. Such candidate proteins include the SMAD transcription factors and the effectors of TGF-β and BMP family signalling [19]. To date, there have been few publications exploring the pathogenicity of mutant FOXL2. The transactivation capability of mutant FOXL2 on known wildtype FOXL2 targets has been investigated, whilst one report describes the inability of mutant FOXL2 to elicit an effective apoptotic signalling cascade to be partially accountable for the pathophysiology of GCT development [18], [20]. Lastly, the aromatase gene has been identified as a direct target of mutant FOXL2 with the use of promoter-luciferase constructs [21]. Given this specific FOXL2 mutation is found in nearly all adult-type GCTs examined, it is clear this mutation must confer some survival advantage even in the heterozygous state. However further studies are required to understand the specific molecular effects of this compelling FOXL2 mutation.
The two well characterised human derived GCT lines, KGN and COV434 have each been screened by us and others for the FOXL2 mutation. The KGN line, established from a 67 year old woman with a recurrent metastatic GCT [22], was shown to be heterozygous for the 402C>G mutation [13]. However the COV434 line was shown to contain wildtype FOXL2 [13]. COV434 was also derived from a recurrent metastatic GCT, but from a much younger woman aged 27 [23]. For these reasons, KGN is considered to be representative of an adult-type GCT and COV434 a juvenile-type GCT, making both these cell lines useful models when studying the tumour properties of each subtype. Interestingly, the two cell lines also differ in their expression of FOXL2. Whilst KGN is shown to have abundant FOXL2 expression, this gene is almost absent in COV434 [13], [21]. This finding correlates to observations made by Kalfa et al, who noted FOXL2 immunochemistry to be decreased or absent in juvenile-type GCTs [24], [25].
The aim of this investigation was to identify FOXL2 transcriptional targets with relevance to GCT by analysing the effect of altering FOXL2 expression on the transcriptome of GCT cell lines. This was achieved in two ways. Mutant FOXL2 was targeted for gene silencing in KGN, whereas both wildtype and mutant FOXL2 were overexpressed in COV434. Total RNA from each perturbation experiment was then hybridised to Affymetrix U133 Plus 2 microarrays. By perturbing the expression levels of wildtype and mutant FOXL2 in these two cell lines, we wanted to see altered expression in unique suites of genes that reflect the activity of certain molecular pathways.
Methods
Cell Lines
Two human derived GCT cell lines KGN and COV434 were obtained from the Riken Cell Bank and the American Tissue Culture Collection (ATCC), respectively. Both cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine and 1% non essential amino acids, and maintained at 37°C in 5% CO2.
Plasmid Constructs
The wildtype FOXL2 plasmid (pFOXL2wt) was purchased from OriGene Techonologies (SKU SC126215, Rockville, MD). The 402 C>G mutant FOXL2 (pFOXL2m) construct was produced using site directed mutagenesis on pFOXL2wt, performed by Mutagenex http://www.mutagenex.com/01_Mutagenesis/02_Site.html (Hillsborough, NJ). The empty vector control was made by excising the FOXL2 sequence from pFOXL2wt with NotI and ligating the vector backbone. The constructs were confirmed to be correct by DNA sequence analysis.
Transient Transfections
For siRNA knockdown of mutant FOXL2, KGN cells, passage 8, were seeded at a density of 300,000 cells per well of a six well plate. Cells were transfected with FOXL2 stealth RNAi™ (cat#HSS101080 Invitrogen, NZ) or a GC matched control using Lipofectamine 2000 and Opti-MEM serum free media (Invitrogen, NZ). RNAi-lipid complexes were removed 7 h post transfection and replaced with complete media. For overexpression of wildtype or mutant FOXL2, COV434 cells, passage 9, were seeded at a density of 300,000 cells per well of a six well plate. Cells were transfected with 1 µg of control (empty vector), pFOXL2wt, or pFOXL2m using Lipofectamine 2000 and Opti-MEM serum free media. Plasmid-lipid complexes were removed 7 h post transfection and replaced with complete media. All transfections were performed in triplicate.
RNA Extraction and cDNA Synthesis
Cells were harvested 24 h post transfection and total RNA was extracted using TRIzol® reagent (Ambion, NZ). Chloroform was added to the TRIzol® to separate the phases, and the aqueous phase was combined with 70% ethanol and passed through an RNeasy column (Qiagen, Australia) according to manufacturer’s instructions. The integrity of the total RNA extracted was verified using the Experion™ automated electrophoresis system (BioRad, NZ). 0.5 µg of total RNA was reverse transcribed to cDNA using oligo(dT) primers and SuperscriptIII reverse transcriptase (Invitrogen, NZ) according to the manufacturer’s instructions. The cDNA synthesis reaction was then diluted with 100 µL of sterile water for RT-qPCR.
RT-qPCR
RT-qPCR was performed to assess the level of mutant FOXL2 knockdown in KGN, the level of overexpression of wildtype and mutant FOXL2 in COV434, and to validate both sets of microarray results. RT-qPCR primers for microarray validation were designed within the probeset region for Affymetrix U133 Plus 2 microarrays using the Primer3 software. Primer pair amplification efficiencies were calculated with the LinReg PCR applet [26]. Each reaction was performed in a final volume of 10 µL, with 1x SYBR Green master mix, 20 pmol of each primer and 2 µL of diluted cDNA. Each cDNA sample was analysed in triplicate. The expression levels of the target gene were normalised to the expression of three most stable housekeeping genes determined with the use of SLqPCR package in R. Data analysis for normalisation, relative quantification of gene expression and calculation of standard deviations was performed as outlined by Vandesompele et al [27].
Western Blotting
Western blotting was performed to assess the level of overexpression of wildtype and mutant FOXL2 in COV434. Cells were harvested 24 h post transfection in RIPA buffer and lysates were incubated with Benzonase® nuclease (Novagen, CA) to remove any residual DNA/RNA. Protein separation was performed on Mini protean TGX precast gels (BioRad, NZ) with Laemmli running buffer. Proteins were transferred onto polyvinylidene fluoride membranes. Western blotting was conducted with an anti-FOXL2 N-terminus polyclonal antibody [7] at a dilution of 1∶500 overnight at 4°C, or a monoclonal anti-vimentin antibody (Dako Corporation, CA) at 1∶1000 for 1 h at room temperature.
Microarray Labelling and Analysis
105 ng of total RNA harvested at 24 h post transfection from the knockdown and overexpression experiments was labelled using the MessageAmp™ Premier amplification according to manufacturer’s instructions (Ambion, NZ). This process involved a first and second strand synthesis, IVT labelling, and purification of the aRNA yield. Subsequently, 8.5 µg of labelled aRNA was hybridised to Affymetrix U133 Plus 2 microarrays. Hybridisation, washing and scanning of the microarrays were performed by the Centre of Proteomics and Genomics (University of Auckland, NZ) according to manufacturer’s instructions.
Bioinformatic analysis was carried out in the ‘R’ statistical environment. The.cel files from each genechip passed quality control using the ‘AffyQCReport’ package in R [28], and were subsequently normalised using the RMA algorithm with background correction [29]. Statistical analysis of different abundance between control (empty vector) and treated cells (wildtype or mutant FOXL2 overexpression, or mutant FOXL2 knockdown) was performed on log2-transformed data using the LIMMA method in R [30], to generate lists of differentially regulated genes for further functional analysis. Relationships between differentially regulated genes were explored further using the tools GATHER (Gene Annotation Tool to Help Explain Relationships) [31], GeneSetDB (http://genesetdb.auckland.ac.nz/haeremai.html, NZ) [32] and Ingenuity Pathway Analysis software (http://www.ingenuity.com, Redwood, CA).
Results
Microarray Evaluation
To identify transcriptional targets of mutant FOXL2, we have undertaken two complementary transcriptomic approaches. First, we used expression vectors containing the coding sequence of wildtype or mutant FOXL2 for overexpression in the mutation negative GCT cell line, COV434. Second, we reduced the expression of mutant FOXL2 in the GCT cell line heterozygous for the mutation, using siRNA.
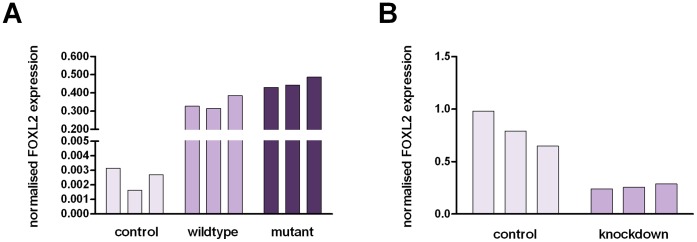
RT-qPCR was used to confirm the overexpression of wildtype and mutant FOXL2 in COV434, as well as the knockdown of mutant FOXL2 in KGN. Figure 1 depicts the changes in FOXL2 expression assessed using RT-qPCR with the data plotted as normalised expression values seen in triplicate control and treated samples. Triplicate samples were produced by performing three separate transfections on the same occasion. The RT-qPCR results show after overexpression (Figure 1A) and knockdown (Figure 1B), FOXL2 levels were greater than 120 times, and approximately 0.3 times, the levels observed in the control cells, respectively.
Figure 1. Confirmation of FOXL2 overexpression (A) and knockdown (B) assessed with RT-qPCR.
Expression values are plotted as normalised values for each control and treated sample. Errors represent technical error associated with RT-qPCR for each sample. The overexpression of wildtype and mutant FOXL2 resulted in greater than a 120 fold increase in FOXL2 expression in COV434, when compared to control cells. siRNA-targetted cells showed 0.3 times the level of FOXL2 expression observed in control cells following mutant FOXL2 knockdown in KGN.
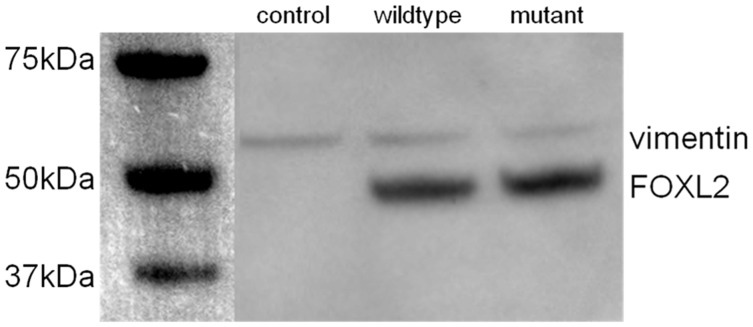
In addition, Western blotting was used to confirm the overexpression of wildtype and mutant FOXL2 expression in COV434 cells (Figure 2). A distinct band is seen in both overexpression lysates at approximately 45 kDa corresponding to the FOXL2 protein. No such band is observed in the control lysate despite the presence of similarly dense vimentin bands across all samples.
Figure 2. Western blot confirming FOXL2 overexpression in COV434.
A clear 45 kDa band in seen in lysates from wildtype and mutant FOXL2 overexpression. This band is absent in control lysates, and similar staining of vimentin across all three lysates confirms FOXL2 overexpression.
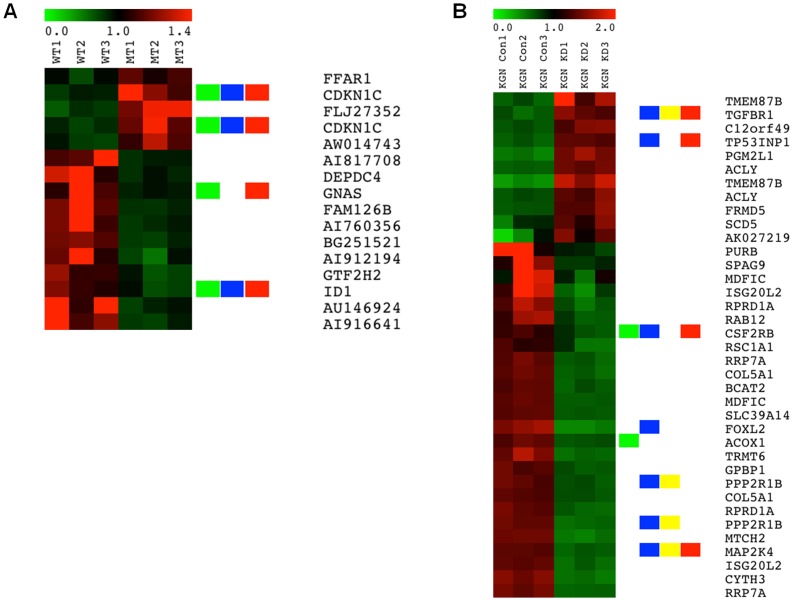
Having confirmed FOXL2 overexpression and knockdown in COV434 and KGN cells, respectively, we analysed the gene expression changes induced by altering FOXL2 expression using Affymetrix microarray gene expression analysis. The microarray results showed a mean FOXL2 expression log2 ratio of −1.77 between control cells and siRNA-targeted KGN cells, which implies a mean 3.41 fold decrease in FOXL2 expression. No microarray signals were observed for FOXL2 in either control or transfected COV434 cells. This is expected, given that FOXL2 expression is absent in this cell line, and that the probeset responsible for detecting FOXL2 expression is located in the 3′UTR of the gene, which lies outside both the wildtype and mutant FOXL2 sequences cloned into the overexpression construct. Figures 3A and B are gene expression profiles giving a snapshot of the genes that were shown to be most significantly differentially regulated following overexpression of mutant FOXL2 compared to wildtype FOXL2 in COV434 cells (A) and following mutant FOXL2 knockdown in KGN cells (B). Among these gene lists, we have highlighted genes annotated for functions of tumourigenesis, cell death, TGF-β signalling and proliferation.
Figure 3. Gene expression profiles associated with mutant FOXL2 overexpression in COV434 cells (A) and mutant FOXL2 knockdown in KGN cells (B).
Each heatmap shows signature genes of KGN with LIMMA p<0.01 and absolute fold change >2 and COV434 with LIMMA p<0.01 and absolute fold change <1.4. In the image red refers to upregulation and green is downregulation. The green, blue, yellow and red bars highlight genes annotated by tumourigenesis, cell death, TGF-β signalling and proliferation, respectively.
Mutant FOXL2 Affects the Expression of Direct Targets of Wildtype FOXL2
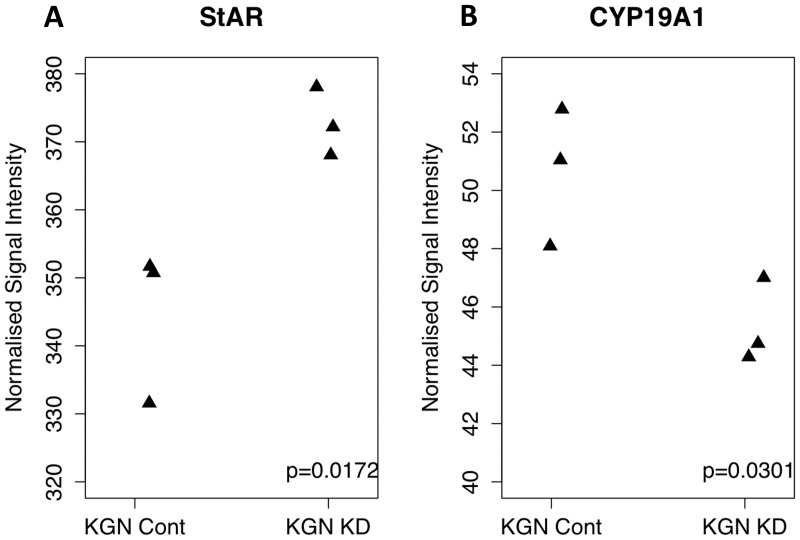
It is unclear whether the 402C>G mutation in the DNA binding domain of FOXL2 prevents FOXL2 from altering the expression of its gene targets by altering its binding capacity. To address this issue, we studied the effect of mutant FOXL2 knockdown on the abundance of the known direct FOXL2 targets, StAR and aromatase. Our hypothesis was that there would be a change in the expression of these RNAs in KGN cells after knockdown of mutant FOXL2, suggesting that mutant FOXL2 was still able to transactivate these targets. Wildtype FOXL2 normally represses StAR gene expression and upregulates the expression of aromatase (CYP19A1). Figure 4A shows in our knockdown data, there is a significant increase in the signals from StAR following FOXL2 knockdown in KGN cells (p = 0.01). Similarly, Figure 4B shows a significant decrease in the signals from the aromatase gene between the control and knockdown KGN cells (p<0.03). Together this data suggests that mutant FOXL2 is able to regulate the expression of these genes, and in its absence, the expression of these genes is altered accordingly.
Figure 4. Signal intensity plots from knockdown data for StAR (A) and CYP19A (B).
StAR, usually repressed by FOXL2 shows a significant increase (p = 0.01) in expression following the knockdown of mutant FOXL2. CYP19A, usually activated by FOXL2 shows a significant decrease (p = 0.03) in expression following the knockdown of mutant FOXL2. Signal intensities are plotted as RMA normalised data for each genechip. This data leads us to believe that mutant FOXL2 is able to regulate the expression of these FOXL2 targets.
In addition to looking at the expression of StAR and aromatase, we also screened other potential direct FOXL2 targets described by Batista et al (2007), who overexpressed wildtype FOXL2 in KGN cells. In our knockdown data, the absence of mutant FOXL2 caused an increase or decrease in the expression of genes shown to be upregulated or downregulated by Batista and colleagues, respectively, by wildtype FOXL2 (File S1). Such genes that showed consistency between our knockdown and overexpression datasets, and Batista’s gene list, included SMAD6, SOX9, SOX4 and ATF3. This would be expected if mutant FOXL2 was still able to regulate these FOXL2 targets.
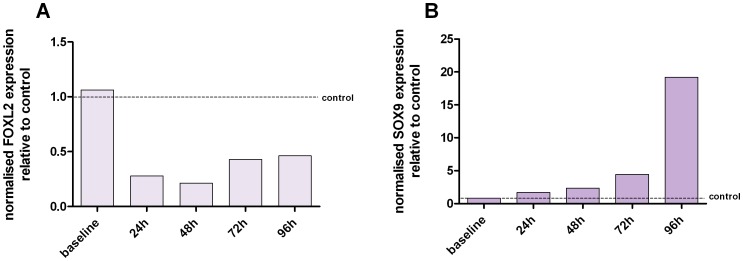
The relationship between FOXL2 and SOX9 is a well documented one (Garcia-Ortiz et al, 2009; Veitia, 2010). FOXL2 expression is important in maintaining female sex-gonads, and in its absence, a de-repression of male specific genes occurs. One of these male specific genes is SOX9. We used our microarray data to see whether this relationship was evident in the KGN cells following the knockdown of FOXL2. The microarray results showed a mean SOX9 expression log2 ratio of 0.59 between control cells and siRNA-targeted KGN cells, which implies a mean 1.55 fold increase in SOX9 expression. Given this result, we then used RT-qPCR to monitor the changes in the expression of SOX9 post FOXL2 knockdown in KGN cells over a greater time period (Figure 5). Indeed with FOXL2 levels low, the expression of SOX9 increases steadily over a 96 h time period. This inverse relationship has previously been observed in literature [33], [34], [35], [36], further adding as a form of validation for our knockdown data.
Figure 5. The expression of SOX9 post FOXL2 knockdown in KGN cells.
Figure 5A confirms the knockdown of FOXL2 expression and Figure 5B depicts SOX9 expression at each corresponding time point. Each expression value has been normalised to the expression values of three reference genes and have been plotted relative to the control cells (data not shown). Over a 96 h time period, the knockdown in FOXL2 expression resulted in a steady increase in SOX9 expression. Baseline refers to expression levels ascertained before knockdown.
Cell Line Specific Expression of FOXL2 Target Genes
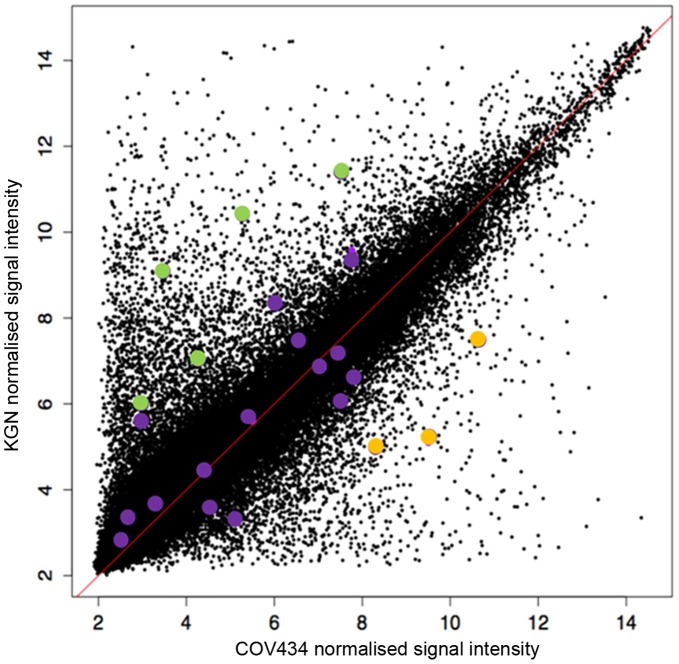
Given that the two GCT cell lines KGN and COV434 differ in their expression of FOXL2, we used our microarray data to investigate whether these cell lines also have differing expression in FOXL2 target genes. To do this, we took the mean signal intensity of each probeset across the KGN control data, and separately across the COV434 control data, and then plotted the relationship between the control data from the two cells lines (as shown in Figure 6). The points in colour represent probesets belonging to possible FOXL2 targets (outlined by Batista et al (2007) gene list). Overall Figure 6 shows the signal intensities for each probeset are mainly similar between the two cell lines. However, some genes appear to be clearly abundant in one cell line, and nearly absent in the other. This expression pattern is seen among some FOXL2 targets (purple green and yellow points, as shown in Figure 6). FOXL2 target genes that are much higher expressed in KGN than COV434 are SOX9, SOX4, FST, StAR and CYP19A1 are the green points. FOXL2 target genes that are more abundant in COV434 are the two nuclear receptors NR4A3 and NR5A2, and CDKN2A are the yellow points.
Figure 6. Cell line specific expression of FOXL2 targets.
Each point represents a single probeset. Signal intensities have been calculated for each probeset by averaging the signal intensities from the three control samples from the COV434 and KGN array data. Coloured points (purple, green and yellow) represent FOXL2 targets described by Batista et al (2007). Although most genes appear to have similar expression in both cell lines, some genes appear to have preferential expression for either cell line. FOXL2 target genes that are more abundant in KGN include SOX9, SOX4, CYP19A, StAR and FST (as shown by green points). FOXL2 target genes that are more abundant in COV434 include NR5A2, NR4A3 and CDKN2A (as shown by yellow points).
Mutant FOXL2 Regulates Genes Enriched for Functions of Tumourigenesis, Cell Death and Cell Proliferation, in Addition to TGF-β Signalling
The overexpression of wildtype and mutant FOXL2 in COV434 resulted in expression levels within 20% of each as measured by RT-qPCR (refer to Figure 1). Therefore, in order to understand the pathogenic effect of mutant FOXL2, we compared the transcriptomes of COV434 cells overexpressing wildtype and mutant FOXL2. LIMMA was used as a ranking tool and identified 340 annotated genes (p<0.01, fold changes ranging from −1.69 to 1.83) that were differentially regulated between the two treatments, that is, genes that are regulated by mutant FOXL2.
We then used the software Ingenuity Pathway Analysis (IPA) to identify relationships, functions and pathways of relevance over-represented in the list of genes differentially expressed between wildtype FOXL2 and mutant FOXL2 transfected COV434 cells. The IPA analysis revealed that the differentially expressed genes were enriched for functional annotations of tumourigenesis (p = 1.56E-5), cell death (p = 2.70E-7), and cell proliferation (p = 6.66E-7). Many of the genes were shown to belong to all three categories including SMAD3, BMPRB1, CDKN1A, CDKN2A, CDK6, BTG, JUN and INHBA (File S2).
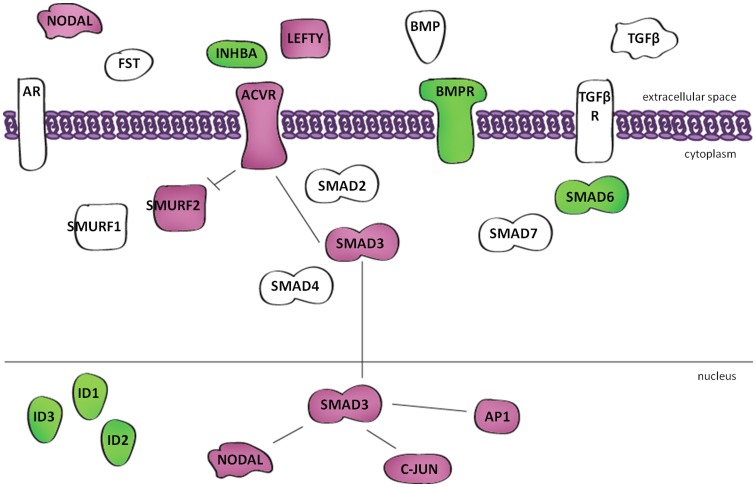
In addition, the IPA analysis revealed that many of the genes (refer to Figure 7) shown to be regulated by mutant FOXL2 mapped to the TGF-β pathway, including SMAD family members 3 and 6, the signalling ligand INHBA, and receptors belonging to BMP and activin. TGF-β signalling was also found to be enriched when using the gene annotation tools GATHER (path:hsa04350, p<0.0001, Bayes Factor 7) and GeneSetDB (p<0.001, FDR 0.19). To determine whether the relationship between mutant FOXL2 target genes and TGF-β signalling was significant, we performed a robust permutation analysis comparing the enrichment of TGF-β signalling genes in our gene list, to the enrichment of TGF-β signalling genes in 10,000 lists of randomly generated genes of equal size to our gene list (File S3). In this figure, the blue dotted lines represent the 5th and 95th percentile, respectively. The green arrow indicates that our data lies above the 95th percentile of the randomly generated data lists, indicating there is a <5% probability that the genes are enriched for TGF-β signalling by chance alone. Interestingly, TGF-β signalling was also significantly enriched when studying gene lists generated by comparing KGN cells with FOXL2 knockdown relative to the control, and also COV434 cells overexpressing mutant FOXL2 compared to the control (data not shown). However this enrichment was not seen when comparing wildtype FOXL2 overexpressed COV434 cells to control cells. Furthermore an opposite expression pattern was seen when comparing gene lists from mutant knockdown KGN cells and mutant overexpression COV434 cells in these TGF-β genes including SMAD3 and INHBA, further highlighting the involvement of the mutant gene in TGF-β signalling.
Figure 7. Mutant FOXL2 regulates the expression of genes in the TGF-β signalling pathway.
Figure schematically highlights those genes in the TGF-β pathway that are regulated by mutant FOXL2. All genes in magenta show increase in expression, genes in green show decrease in expression.
Discussion
The 402C>G mutation in FOXL2 described by Shah et al has been observed in nearly all cases of adult type GCTs that have been investigated worldwide. It is likely this mutation is pivotal in the pathogenesis of GCTs, as the FOXL2 gene is important in normal granulosa cell function. No other tumour type identified to date has the same FOXL2 mutation and it seems striking that it occurs in the same position of the gene in GCT, yet little is known about its pathogenic mechanism of action. We have adopted a transcriptomic approach to study the effect of the FOXL2 mutation by analysing how gene expression in GCT cell lines is altered after perturbing the expression of either wildtype or mutant FOXL2. The aim of this study was to identify transcriptional targets of the mutant, and thereby identify aspects of its pathogenicity.
The 402C>G mutation resides in the DNA binding domain of FOXL2 and results in an amino acid change from a cysteine to a tryptophan (C134W). However, computer modelling suggests that this amino acid substitution does not alter the conformation of the domain, suggesting the mutant protein can still bind DNA [12]. One possible explanation to describe mutant FOXL2’s behaviour, is that although able to bind DNA, the mutant is unable to bind to targets usually directly regulated by wildtype FOXL2. This would mean that the mutant FOXL2 is unable to recognise and bind the FOXL2 response element in direct wildtype FOXL2 gene targets. However, the work of Benayoun et al (2010) used reporter constructs to demonstrate the transactivation capability of mutant FOXL2 on known wildtype FOXL2 targets. With the exception of one promoter belonging to GRAS (GnRH receptor activating sequence), in which the mutant appeared to be hyperactive, the mutant behaved similarly to the wildtype protein on all other accounts [18]. We have used our microarray data to further investigate the idea that mutant FOXL2 regulates the expression of a different suite of genes compared to wildtype FOXL2.
We studied the changes in expression of known direct FOXL2 targets in our KGN knockdown data to see if mutant, compared to wildtype FOXL2, was able to differentially regulate the expression of known FOXL2 targets. Many targets of FOXL2 have been individually identified such as GnRHR, alpha-GSU, FST, FSH-beta and CYP17A1 as well as many targets obtained in several transcriptomic studies and genome-wide ChIP-on-chip experiments [33], [35], [37], [38], [39]. However we chose to focus our analysis on two transcriptional targets that are directly relevant in granulosa cell biology, namely StAR and the gene encoding aromatase (CYP19A1) [38], [40], [41]. StAR is a marker of late differentiation of granulosa cells in pre-ovulatory follicles and catalyses the translocation of cholesterol from the outer to the inner mitochondrial membrane, where it can be subsequently processed to yield steroid hormones. Also involved in steroidogenesis, aromatase is the enzyme responsible for the conversion of androgens to oestrogens in granulosa cells. We compared the normalised signal intensities of StAR and aromatase across our knockdown data in KGN cells and revealed that mutant FOXL2 was still able to alter the expression of these FOXL2 targets. Wildtype FOXL2 normally represses StAR gene expression [42] and upregulates the expression of aromatase, as consistent with our understanding of ovarian biology. In our FOXL2 knockdown data, we saw a significant increase in StAR expression (p = 0.01) and decrease in aromatase expression (p = 0.03). As we are likely to be silencing both wildtype and mutant FOXL2 alleles in the KGN line, we are suggesting that a reduction in the combined amount of FOXL2 leads to an altered regulation of the expression of these genes. However, given that we are unable to separate loss of wildtype from mutation FOXL2 function in this heterozygous cell line, we can not discount the possibility that mutant FOXL2 is unable to bind to StAR and aromatase promoters, and therefore we might not be able to measure this change in expression.
Unfortunately, in our wildtype and mutant overexpression data, the low level of expression of some FOXL2 target genes made it difficult to assess the effect of FOXL2 overexpression in COV434 cells. In this case, a more effective method of investigating mutant FOXL2s binding on wildtype FOXL2 targets may be to see if there was an enrichment of genes containing the FOXL2 response element in wildtype and mutant overexpressed COV434 cells compared to control cells. However, given the lack of specificity and discrepancy between the published FOXL2 response elements [21], [40], [43], [44], we chose to assess mutant FOXL2 binding indirectly through looking at changes in gene expression.
In addition to StAR and aromatase, we performed a similar screening method for other potential FOXL2 targets that were originally identified by Batista et al (2007), who overexpressed wildtype FOXL2 in KGN cells [38]. We looked to see whether there were consistent changes in gene expression when we altered the levels of wildtype and mutant FOXL2. In our knockdown data in KGN, the absence of mutant FOXL2 caused a significant increase or decrease in the expression of few of the genes that Batista et al (2007) showed to be downregulated or upregulated, respectively, by wildtype FOXL2 (p<0.05). This would be expected if mutant FOXL2 was still able to regulate these wildtype FOXL2 targets. File S1 details a full list of genes that showed consistency with the Batista et al (2007) gene list across both our knockdown and overexpression datasets. When comparing Batista et al’s gene list with our knockdown data, only 5% of the gene list showed consistency in direction of fold change, that is showing opposite expression patterns that would be expected when comparing gene knockdown and gene overexpression. Similarly when comparing our wildtype and mutant overexpression data with Batista et al (2007), we see only 2% of their gene list showing consistency with ours.
Such direct comparisons between gene lists can be difficult due to the differences in methodologies between the two groups. For example, Batista et al (2007) performed their experiments in KGN using a double transfection protocol, whereas we performed a single transfection protocol in both cell lines. Additionally, considering the dynamic nature of transcriptome responses, the differences in the timepoints for analysis (their 48 h post transfection versus our 24 h post transfection) may also affect the similarities, or lack thereof. As we have shown in Figure 6, the two cell lines possess differences in their global gene expression profiles, ultimately making comparisons between the two experiments problematic. Although our knockdown data and Batista’s overexpression data was performed in KGN, we are likely to be altering both mutant and wildtype alleles, where as Batista et al have only increased expression of the wildtype allele, which can also explain the differences between our gene lists. Furthermore, the expression data from each experiment was obtained from two different array platforms, different types of statistical analyses were performed to produce a working gene list and the known issue of batch effects in microarray meta-analysis could also induce spurious differences in gene activation [45]. Finally, though paradoxical, the simple comparison of knockdown to overexpression, does not always yield consistent results even when assessed completely in parallel [46]. Indeed some of these caveats discussed may also explain the lack of overlap in all of the FOXL2 transcriptome studies and between the COV434 and KGN arrays herein, yet it was reassuring to see overlap does occur among some biologically relevant genes. One gene of importance that did show such concordance was SOX9.
SOX9 is the downstream effector of the SRY gene, the Y-linked signal responsible for testis determination in male [47]. Recently, studies have shown that the ovary is constantly suppressing the expression of male-specific genes throughout life [36]. FOXL2 expression is important in maintaining the female sex gonads, even in adulthood, and in its absence a de-repression of male-specific genes such as SOX9 occurs [35]. Ulenhaut et al showed in the absence of FOXL2, the follicular structure of the ovary began to take on the structure of seminiferous tubules, and granulosa cells are reprogrammed into their male counterparts Sertoli cells. As our FOXL2 knockdown data showed a significant increase in SOX9 expression, we were interested to see the long-term effects of reduced FOXL2 expression on the expression of SOX9. As Figure 5 depicts, SOX9 expression continued to steadily rise in the absence of FOXL2 over 96 h following FOXL2 knockdown in KGN. Therefore, it was appropriate for us to undertake a microarray analysis of gene expression at a 24 h timepoint when SOX9 levels are lowest following FOXL2 knockdown. Given that we saw no other male specific genes, such as DMRT1 being significantly upregulated in our knockdown data, we are confident that the KGN granulosa cells have yet to begin their transition to Sertoli-like cells.
Given that the two GCT cell lines KGN and COV434 differ in their expression of FOXL2, we used our microarray data to investigate whether these cell lines also have differing expression in FOXL2 target genes. Figure 6 depicts the global differences in gene expression between the two cell lines, with the FOXL2 targets outlined by Batista et al (2007) highlighted in purple. Interestingly SOX9 is the top gene shown in Figure 6 to have a significantly more abundant expression in KGN cells as opposed to COV434, indicating there must be other mechanisms at play keeping SOX9 expression under control in the FOXL2 lacking cell line COV434. Other genes that are more abundantly expressed in KGN, than COV434 include the two oestrogen synthesis genes StAR and aromatase, however COV434 cells, as well as juvenile-type GCTs are both capable of oestrogen production [23], [48] and there has yet to be any data published showing oestrogen production to be higher in adult-type GCTs than juvenile-type. It appears only three FOXL2 target genes have significantly higher abundance in COV434 than KGN cells, these genes being the two steroidogenic receptors NR5A2 and NR4A3 (also known as SF-2 and NOR1 respectively), and CDKN2A. Given the preferential expression of FOXL2 target genes in either KGN or COV434 cell lines, this might suggest to us regulation of these targets may be not always be directly under FOXL2 control, and perhaps alternative mechanisms may be in place. However, it is possible the differences in the expression of FOXL2 targets is partially accounted for by the stage of granulosa cell maturation and the different genetic backgrounds of the two individuals from which the cells lines were derived, the differences in type of ‘control’ used (empty plasmid versus non-targeting siRNA), and lastly, the likelihood that both cell lines have undergone many other genomic hits after the perturbation of the FOXL2 locus. Although we are not able to use our data to provide explanations to account for the differences in FOXL2 target gene expression between COV434 and KGN, what we have done is confirm at a transcriptomic level what is already known at a pathological level; that adult and juvenile-type GCTs are quite different diseases that have arisen from the same cell type, but from likely two different mechanisms.
To gain further insights about genes regulated by mutant FOXL2, we used the software IPA, a database system for understanding how proteins work together to effect cellular change. The IPA analysis revealed that the genes shown to be regulated by mutant FOXL2 (gene list created from comparing the transcriptome of COV434 cells overexpressing wildtype or mutant FOXL2) were enriched for functional annotations of cell death, cell proliferation and tumourigenesis. It has been previously shown that FOXL2 activation tends to promote cell cycle arrest at the G1/S checkpoint through direct regulation of cyclin dependant kinases and their inhibitors [37]. It appears that mutant FOXL2, like wildtype FOXL2, upregulates the expression of the well known tumour suppressor CDKN1C (p57), in addition to CDKN1A (p21/WAF). Similarly, mutant FOXL2 caused a decrease in the expression of CDKN2A (p16/INK4a), which was shown by Arcellana et al to have reduced expression in 58% of adult GCTs [49]. So although it may appear overall that the changes in gene expression by mutant FOXL2 would be protective in the development of GCTs, it is likely that the differences in expression of a limited number of key genes are able to tip this balance in favour of tumourigenesis.
In addition, the IPA analysis revealed that many of the genes shown to be to be regulated by mutant FOXL2 mapped to the TGF-β signalling pathway. It is not surprising that this signalling pathway was enriched, as it has been previously suggested that it is likely that mutant FOXL2 has altered interactions with SMAD transcription factors and effects of the TGFβ and BMP family signalling [19]. Although Figure 7 only depicts those genes traditionally seen in TGF-β signalling diagrams, our gene list also showed the movement of other SMAD gene targets including CDKN1A, DLX3 and GADD45B. It is reassuring that we identified this pathway from our non-hypothesis driven bioinformatic analysis of transcriptomic data from different cell line studies. The TGF-β signalling pathway has been implicated in many human diseases including cancer. TGF-β signalling, originally renowned for its anti-proliferative activity, is now considered to demonstrate both tumour suppressor and oncogenic properties [50], [51]. In the current paradigm, the suppressor activities dominate in normal tissue, but during tumourigenesis, changes in TGF-β expression and cellular responses tip the balance in favour of its oncogenic activities. This process usually involves a decrease in the expression of the signalling ligand and receptor, decreased SMAD levels or activity, or a compromise in the effector function of the suppressor arm of this pathway [52]. Further supporting this idea, is the loss of expression of the antiproliferative signalling ligand INHA, and loss of BMPR1B receptor expression, as well as in increase the expression of the oncogene JUN.
It is interesting to note that the IPA functional annotations of cell death, cell proliferation, tumourigenesis and TGF-β signalling were also shown to be significantly enriched, when studying the gene lists derived from the LIMMA analysis of the mutant FOXL2 knockdown in KGN and the overexpression of mutant FOXL2 in COV434. However, when performing the same analysis with a gene list compiled from the overexpression of wildtype FOXL2 in COV434 compared to control treated cells, none of these annotations were shown to be significant. In fact, very few annotations and pathways were shown to be significantly enriched in this particular analysis. Therefore this finding demonstrates that altering expression levels of wildtype FOXL2 alone is not important for tumour development, but it is the 402C<G mutation itself in FOXL2 that is responsible for altering biological functions to contribute to GCT pathogenesis. Furthermore, it also suggests that in our gene knockdowns, it is likely that we are silencing the mutant allele as well as the wildtype allele, as if only the wildtype allele was silenced, the mutant allele expression would remain, and the gene list data would resemble the mutant overexpression data in COV434.
Conclusions
In conclusion, we have identified aspects of the pathogenicity of mutant FOXL2 through studying its transcriptional targets after perturbing its expression in two GCT cell lines COV434 and KGN. The overexpression of wildtype and mutant FOXL2 in COV434, and the silencing of mutant FOXL2 expression in KGN, has revealed that mutant FOXL2 is able to differentially regulate the expression of many genes, including two well known FOXL2 targets StAR and CYP19A. In addition, we have confirmed at a transcriptomic level, the significant difference in gene expression between adult and juvenile type GCTs, an important consideration for future therapeutic work. We have shown that many of the genes regulated by mutant FOXL2 are clustered into functional annotations of cell death, proliferation and tumourigenesis. In addition, these genes are significantly enriched for TGF-β signalling, and we suggest that deregulation of this key antiproliferative pathway is perhaps one way mutant FOXL2 contributes to the pathogenesis of adult-type GCTs.
Supporting Information
Genes shown to have similar regulation when comparing our gene list with that generated by Batista et al (2007).
(XLSX)
Outline of genes regulated by mutant FOXL2 shown to be enriched for functions of tumourigenesis, cell death and proliferation.
(XLSX)
Permutation analysis to test for enrichment of TGF-β signalling in our data. In this figure, the dotted blue lines represent the 5th and 95th percentile respectively. The green arrow indicates our data lies above the 95th percentile of randomly generated lists, thus roving the enrichment for TGF-β signalling seen in our gene lists is significant.
(TIF)
Funding Statement
This work is funded by the Sladjana M. Crosley Fund for GCT Research and the Granulosa Cell Tumour Foundation. Roseanne Rosario is supported by a University of Auckland Doctoral Scholarship and the Cancer Society of New Zealand Training Scholarship in Cancer Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Colombo N, Parma G, Zanagnolo V, Insinga A (2007) Management of ovarian stromal cell tumors. J Clin Oncol 25: 2944–2951. [DOI] [PubMed] [Google Scholar]
- 2. Jamieson S, Fuller PJ (2012) Molecular Pathogenesis of Granulosa Cell Tumors of the Ovary. Endocrine Reviews 33: 109–144. [DOI] [PubMed] [Google Scholar]
- 3. East N, Alobaid A, Goffin F, Ouallouche K, Gauthier P (2005) Granulosa cell tumour: a recurrence 40 years after initial diagnosis. J Obstet Gynaecol Can 27: 363–364. [DOI] [PubMed] [Google Scholar]
- 4. Pectasides D, Pectasides E, Psyrri A (2008) Granulosa cell tumor of the ovary. Cancer Treat Rev 34: 1–12. [DOI] [PubMed] [Google Scholar]
- 5. Fuller PJ, Chu S (2004) Signalling pathways in the molecular pathogenesis of ovarian granulosa cell tumours. Trends in Endocrinology & Metabolism 15: 122–128. [DOI] [PubMed] [Google Scholar]
- 6. Fuller PJ, Chu S, Fikret S, Burger HG (2002) Molecular pathogenesis of granulosa cell tumours. Molecular & Cellular Endocrinology 191: 89–96. [DOI] [PubMed] [Google Scholar]
- 7. Cocquet J, Pailhoux E, Jaubert F, Servel N, Xia X, et al. (2002) Evolution and expression of FOXL2. Journal of medical genetics 39: 916–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, et al. (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nature genetics 27: 159–166. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, et al. (2004) The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development (Cambridge, England) 131: 933–942. [DOI] [PubMed] [Google Scholar]
- 10. Uda M, Ottolenghi C, Crisponi L, Garcia JE, Deiana M, et al. (2004) Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Human molecular genetics 13: 1171–1181. [DOI] [PubMed] [Google Scholar]
- 11. Harris SE, Chand AL, Winship IM, Gersak K, Aittomaki K, et al. (2002) Identification of novel mutations in FOXL2 associated with premature ovarian failure. Molecular Human Reproduction 8: 729–733. [DOI] [PubMed] [Google Scholar]
- 12. Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, et al. (2009) Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med 360: 2719–2729. [DOI] [PubMed] [Google Scholar]
- 13. Jamieson S, Butzow R, Andersson N, Alexiadis M, Unkila-Kallio L, et al. (2010) The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Modern Pathology 23: 1477–1485. [DOI] [PubMed] [Google Scholar]
- 14. Kim MS, Hur SY, Yoo NJ, Lee SH (2010) Mutational analysis of FOXL2 codon 134 in granulosa cell tumour of ovary and other human cancers. J Pathol 221: 147–152. [DOI] [PubMed] [Google Scholar]
- 15. D’Angelo E, Mozos A, Nakayama D, Espinosa I, Catasus L, et al. (2011) Prognostic significance of FOXL2 mutation and mRNA expression in adult and juvenile granulosa cell tumors of the ovary. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 24: 1360–1367. [DOI] [PubMed] [Google Scholar]
- 16. Schrader KA, Gorbatcheva B, Senz J, Heravi-Moussavi A, Melnyk N, et al. (2009) The specificity of the FOXL2 c.402C>G somatic mutation: a survey of solid tumors. PloS one 4: e7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hes O, Vanecek T, Petersson F, Grossmann P, Hora M, et al. (2011) Mutational analysis (c.402C>G) of the FOXL2 gene and immunohistochemical expression of the FOXL2 protein in testicular adult type granulosa cell tumors and incompletely differentiated sex cord stromal tumors. Appl Immunohistochem Mol Morphol 19: 347–351. [DOI] [PubMed] [Google Scholar]
- 18. Benayoun BA, Caburet S, Dipietromaria A, Georges A, D’Haene B, et al. (2010) Functional exploration of the adult ovarian granulosa cell tumor-associated somatic FOXL2 mutation p.Cys134Trp (c.402C>G). PloS one 5: e8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobel M, Gilks CB, Huntsman DG (2009) Adult-type granulosa cell tumors and FOXL2 mutation. Cancer Res 69: 9160–9162. [DOI] [PubMed] [Google Scholar]
- 20. Kim JH, Yoon S, Park M, Park HO, Ko JJ, et al. (2010) Differential apoptotic activities of wild-type FOXL2 and the adult-type granulosa cell tumor-associated mutant FOXL2 (C134W). Oncogene 30: 1653–1663. [DOI] [PubMed] [Google Scholar]
- 21. Fleming NI, Knower KC, Lazarus KA, Fuller PJ, Simpson ER, et al. (2010) Aromatase Is a Direct Target of FOXL2: C134W in Granulosa Cell Tumors via a Single Highly Conserved Binding Site in the Ovarian Specific Promoter. PloS one 5: e14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishi Y, Yanase T, Mu Y, Oba K, Ichino I, et al. (2001) Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 142: 437–445. [DOI] [PubMed] [Google Scholar]
- 23. Zhang H, Vollmer M, De Geyter MD, Litzistorf Y, Ladewig A, et al. (2000) Characterization of an immortalized human granulosa cell line (COV434). Molecular Human Reproduction 6: 146–153. [DOI] [PubMed] [Google Scholar]
- 24. Kalfa N, Fellous M, Boizet-Bonhoure B, Patte C, Duvillard P, et al. (2008) Aberrant expression of ovary determining gene FOXL2 in the testis and juvenile granulosa cell tumor in children. J Urol 180: 1810–1813. [DOI] [PubMed] [Google Scholar]
- 25. Kalfa N, Philibert P, Patte C, Ecochard A, Duvillard P, et al. (2007) Extinction of FOXL2 expression in aggressive ovarian granulosa cell tumors in children. Fertil Steril 87: 896–901. [DOI] [PubMed] [Google Scholar]
- 26. Ramakers C, Ruijter J, Deprez R, Moorman A (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 27. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parman C, Halling C (2006) affyQCReports: A package to generate QC reports for affymetrix array data. [Google Scholar]
- 29. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic acids research 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 31. Chang JT, Nevins JR (2006) GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 22: 2926–2933. [DOI] [PubMed] [Google Scholar]
- 32. Araki H, Knapp C, Tsai P, Print C (2012) GeneSetDB: a comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Open Bio 2: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Ortiz JE, Pelosi E, Omari S, Nedorezov T, Piao Y, et al. (2009) Foxl2 functions in sex determination and histogenesis throughout mouse ovary development. BMC developmental biology 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schlessinger D, Garcia-Ortiz JE, Forabosco A, Uda M, Crisponi L, et al. (2010) Determination and stability of gonadal sex. Journal of andrology 31: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, et al. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139: 1130–1142. [DOI] [PubMed] [Google Scholar]
- 36. Veitia RA (2010) FOXL2 versus SOX9: a lifelong “battle of the sexes”. Bioessays 32: 375–380. [DOI] [PubMed] [Google Scholar]
- 37. Benayoun BA, Georges AB, L’Hote D, Andersson N, Dipietromaria A, et al. (2011) Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle: role of its regulation by the SIRT1 deacetylase. Human molecular genetics 20: 1673–1686. [DOI] [PubMed] [Google Scholar]
- 38. Batista F, Vaiman D, Dausset J, Fellous M, Veitia RA (2007) Potential targets of FOXL2, a transcription factor involved in craniofacial and follicular development, identified by transcriptomics. Proceedings of the National Academy of Sciences of the United States of America 104: 3330–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Escudero JM, Haller JL, Clay CM, Escudero KW (2010) Microarray analysis of Foxl2 mediated gene regulation in the mouse ovary derived KK1 granulosa cell line: Over-expression of Foxl2 leads to activation of the gonadotropin releasing hormone receptor gene promoter. J Ovarian Res 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benayoun BA, Caburet S, Dipietromaria A, Bailly-Bechet M, Batista F, et al. (2008) The identification and characterization of a FOXL2 response element provides insights into the pathogenesis of mutant alleles. Human molecular genetics 17: 3118–3127. [DOI] [PubMed] [Google Scholar]
- 41. Moumne L, Batista F, Benayoun BA, Nallathambi J, Fellous M, et al. (2008) The mutations and potential targets of the forkhead transcription factor FOXL2. Molecular and cellular endocrinology 282: 2–11. [DOI] [PubMed] [Google Scholar]
- 42. Pisarska MD, Barlow G, Kuo FT (2011) Minireview: roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology 152: 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, et al. (2003) The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Molecular and cellular endocrinology 206: 93–111. [DOI] [PubMed] [Google Scholar]
- 44. Corpuz PS, Lindaman LL, Mellon PL, Coss D (2010) FoxL2 Is required for activin induction of the mouse and human follicle-stimulating hormone beta-subunit genes. Mol Endocrinol 24: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ayroles JF, Gibson G (2006) Analysis of variance of microarray data. Methods in enzymology 411: 214–233. [DOI] [PubMed] [Google Scholar]
- 46. Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, et al. (2009) The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS genetics 5: e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schlessinger D, Garcia-Ortiz JE, Forabosco A, Uda M, Crisponi L, et al. (2009) Determination and stability of gonadal sex. Journal of andrology 31: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gittleman AM, Price AP, Coren C, Akhtar M, Donovan V, et al. (2003) Juvenile granulosa cell tumor. Clinical imaging 27: 221–224. [DOI] [PubMed] [Google Scholar]
- 49. Arcellana-Panlilio MY, Egeler RM, Ujack E, Magliocco A, Stuart GC, et al. (2002) Evidence of a role for the INK4 family of cyclin-dependent kinase inhibitors in ovarian granulosa cell tumors. Genes, chromosomes & cancer 35: 176–181. [DOI] [PubMed] [Google Scholar]
- 50. Rahimi RA, Leof EB (2007) TGF-beta signaling: a tale of two responses. Journal of cellular biochemistry 102: 593–608. [DOI] [PubMed] [Google Scholar]
- 51. Seoane J (2006) Escaping from the TGFB antiproliferative control. Carcinogenesis 27: 2148–2156. [DOI] [PubMed] [Google Scholar]
- 52. Wakefield L, Roberts A (2002) TGFb signalling: positive and negative effects on tumourigenesis. Current Opinion in Genetics & Development 12: 22–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes shown to have similar regulation when comparing our gene list with that generated by Batista et al (2007).
(XLSX)
Outline of genes regulated by mutant FOXL2 shown to be enriched for functions of tumourigenesis, cell death and proliferation.
(XLSX)
Permutation analysis to test for enrichment of TGF-β signalling in our data. In this figure, the dotted blue lines represent the 5th and 95th percentile respectively. The green arrow indicates our data lies above the 95th percentile of randomly generated lists, thus roving the enrichment for TGF-β signalling seen in our gene lists is significant.
(TIF)