Tachedjian et al. (1) report in this issue of PNAS that one class of inhibitors of the reverse transcriptase of HIV type-1 (HIV-1) can enhance the dimerization of the enzyme. This unexpected finding provides insight into the actions of the inhibitors and into the dimerization process, neither of which is well understood.
Therapies involving three-drug combinations are now the standard treatment for HIV-1 infections. Despite the successes with such treatments, there are still considerable problems with drug toxicity and the development of viral resistance. The approved drugs inhibit two viral enzymes: reverse transcriptase (RT) and protease. All of the protease inhibitors are substrate analogs; however, there are two types of RT inhibitors, nucleoside analogs and nonnucleoside inhibitors (NNRTIs). The mode of action of nucleoside analogs is straightforward. Nucleoside analogs lack a 3′OH and, when incorporated into viral DNA by HIV-1 RT, act as chain terminators, blocking viral DNA synthesis. NNRTIs are noncompetitive inhibitors, binding to a hydrophobic pocket in HIV-1 RT near the polymerase active site (Fig. 1), which somehow prevents RT from carrying out the polymerization reaction. NNRTIs do not interfere with substrate binding. Kinetic analysis shows that NNRTIs inhibit the chemistry step (2, 3); however, we do not yet understand how the binding of an NNRTI blocks dNTP incorporation. Clearly, we need to know more about the interactions of HIV-1 RT and nonnucleoside inhibitors. This is part of what makes the report by Tachedjian et al. (1) so interesting; it provides insight into the interactions of HIV-1 RT and NNRTIs.
Figure 1.
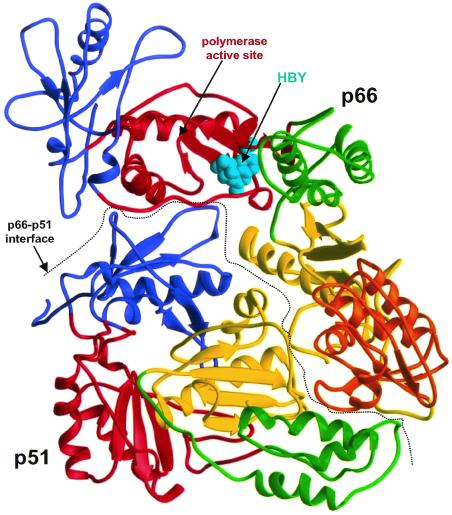
The structure of HIV-1 RT with a bound NNRTI. The p66 subunit is on the upper right; the p51 subunit is on the lower left. The interface between the subunits is shown by a dotted line. The individual subdomains of the two subunits are color coded: blue, fingers; red, palms; green, thumbs; yellow, connection domains. The RNaseH domain is gold and the NNRTI (HBY) is cyan. The approximate position of the polymerase active site is indicated by an arrow. The figure was prepared from the coordinates of Hsiou et al. (ref. 4; Protein Data Bank ID code 1bqm).
Retroviral RTs have two separate catalytic activities—a polymerase that can copy either an RNA or a DNA substrate into DNA and an RNaseH that cleaves RNA if, and only if, it is part of an RNA/DNA duplex. HIV-1 RT is a heterodimer; the larger subunit, p66, is 560-aa in length and contains both a polymerase and an RNaseH domain. The smaller subunit, p51, contains the first 440 amino acids of p66 that correspond closely, but not exactly, to the polymerase domain. The structure of the polymerase domain of p66 has been likened to the human right hand, composed of fingers, palm, and thumb subdomains, and a connection subdomain linking the polymerase and RNaseH domains (Fig. 1). Although the p51 subunit contains the same four subdomains that are found in the polymerase domain of p66 (fingers, palm, thumb, and connection), the physical relationships of the subdomains differ in p66 and p51 (5, 6). Because of these differences, the same amino acid sequence adopts somewhat different structures in the two subunits and in some cases, the same amino acid sequences play distinct roles in the two subunits of the protein. For example, the polymerase active site lies in the palm of p66. Although p51 contains the same amino acids that form the polymerase active site in p66, there is no functional polymerase active site in p51, and the p51 subunit plays no direct role in polymerization (7, 8).
Only dimeric forms of the enzyme are active. Presumably, the role of p51 is primarily structural. It seems to provide a scaffold that is essential for the ability of p66 to function as either a polymerase or an RNaseH. Moreover, at least in vitro, the second (structural) component does not have to be p51 nor does the catalytic subunit need to be p66. Homodimers p66/p66 and p51/p51, although less stable than p66/p51 heterodimers, have polymerase activity (p66/p66 also has RNaseH activity; p51/p51 lacks an RNaseH domain; refs. 9 and 10). All of this makes the folding and dimerization of HIV-1 RT an intriguing and difficult problem. This problem provides the second important reason to read the work of Tachedjian et al. (1); they have shown that several NNRTIs can enhance the dimerization of HIV-1 RT. Several things are already clear. (i) The ability of the NNRTIs to influence dimerization requires that the drugs bind to the enzyme; the NNRTIs have a greatly reduced effect on the dimerization of drug-resistant RTs. (ii) The NNRTIs can act posttranslationally, and dimerization is enhanced in extracts after the subunits have been synthesized. (iii) Prebinding the drugs (and washing out unbound drug) promotes dimerization if the drug is prebound to p66, but does not promote dimerization if the drug is prebound to p51. (iv) The drugs can enhance the dimerization of wild-type HIV-1 RT and of RTs with mutations that interfere with dimerization. (v) The position of the drug-binding pocket, which lies almost entirely within p66, argues that the effect is indirect. NNRTIs do not bind along the p66/p51 interface and do not simply act as a “glue” to bind the subunits together (Fig. 1).
Part of the reason that the paper is so interesting is not the questions it answers, but the questions it raises. What is the mechanism by which the NNRTIs stimulate dimerization? As the authors point out, there are some intriguing possibilities. As has been mentioned already, the data show that enhanced dimerization occurs if the drug is prebound to p66 but not to p51. Both p66 and p51 can form homodimers, although both homodimers are less stable than the heterodimer (the p51/p51 homodimer is so weak that it is barely detectable). However, the fact that these homodimers form (and are enzymatically active) strongly suggests that both p66 and p51 can, under some circumstances, adopt either the configuration of p66 or p51 in the heterodimer. Does the drug bind to monomeric p66, locking it into the configuration suitable for dimerization with p51? If this is the mechanism, why does the drug not bind also to p51 and stabilize it in a similar configuration? Clearly, there is a preference for p66 and p51 to adopt the configurations they have in the heterodimer; this helps explain the hierarchical stability of the various HIV-1 RT dimers (p66/p51 > p66/p66 > p51/p51). One possibility is the drug does bind to p51, but binds so weakly that it is not detected in the relatively stringent assay described in the paper.
Alternatively, the NNRTIs might bind only to dimeric forms of RT. The p66/p66 homodimer is sufficiently stable that considerable NNRTI could be bound; the small amount of p51/p51 homodimer would bind only a negligible amount of drug. In this model, the drug would somehow enhance the amount of dimer and/or the exchange of subunits. It is also possible that, at least in some cases, an NNRTI might provide a hydrophobic core and enhance proper folding of the RT. There is also a question of whether NNRTIs can, under certain circumstances, enhance the activity of HIV-1 RT (either wild-type or mutants) in the context of viral replication. No matter what answers are obtained to these questions, there is much to learn and the work of Tachedjian et al. (1) has provided pioneering information, insights, and ways to explore the problem. We can all look forward to the next installment in this developing story.
Acknowledgments
I am grateful to John Coffin for helpful discussions, to Hilda Marusiodis for help in preparing the manuscript, and to Stefan Sarafianos and Eddy Arnold for preparing the figure.
Footnotes
See companion article on page 7188.
References
- 1.Tachedjian G, Orlova M, Sarafianos S G, Arnold E, Goff S P. Proc Natl Acad Sci USA. 2001;98:7188–7193. doi: 10.1073/pnas.121055998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rittinger K, Divita G, Goody R S. Proc Natl Acad Sci USA. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spence R A, Kati W M, Anderson K S, Johnson K A. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsiou Y, Das K, Ding J, Clark A J, Jr, Kleim J P, Rosner M, Winkler I, Reiss G, Hughes S H, Arnold E. J Mol Biol. 1998;284:313–323. doi: 10.1006/jmbi.1998.2171. [DOI] [PubMed] [Google Scholar]
- 5.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 6.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P K, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hostomsky Z, Hostomska Z, Fu T B, Taylor J. J Virol. 1992;66:3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Restle T, Muller B, Goody R S. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 10.Bavand M R, Wagner R, Richmond T J. Biochemistry. 1993;32:10543–10552. doi: 10.1021/bi00091a003. [DOI] [PubMed] [Google Scholar]