Abstract
The catalytic activity of L-aspartate α-decarboxylase (ADC) is essential for the growth of several micro-organisms, including Mycobacterium tuberculosis (Mtb), and has triggered efforts for the development of pharmaceutically active compounds against tuberculosis. The present study is a continuation of our recent chemoinformatics-based design approach for identifying potential drug-like inhibitors against MtbADC. We report an NMR-based protocol that allows label-free and direct monitoring of enzymatic conversion, which we have combined with a systematic testing of reported and newly identified potential inhibitors against MtbADC. Quantification of enzymatic conversion in the absence and presence of inhibitors allowed for a relative measure of the inhibitory effect (k rel). Among the newly identified compounds, D-tartrate, L-tartrate, and 2,4-dihydroxypyrimidine-5-carboxylate were found to inhibit the enzyme with k rel values of 0.36, 0.38, and 0.54, respectively. In addition to the identification of potential building blocks for the development of therapeutic agents, the current study highlights the importance of electrostatic interactions governing enzyme-inhibitor binding.
Introduction
L-aspartate α-decarboxylase (ADC, EC 4.1.1.11), encoded by the E. coli panD gene, is an enzyme responsible for the conversion of L-aspartate to β-alanine and its activity has been shown to be crucial for the growth of several microorganisms, including Mycobacterium tuberculosis (Mtb) [1]–[3]. In short, formation of β-alanine allows the synthesis of panthotenate (vitamin B5), the precursor of coenzyme A (CoA), which in turn plays an essential role in the metabolism and biosynthesis of fatty acids. The complex lipidoglycans found in the Mtb cell wall are therefore crucial for the intracellular replication, persistence, and virulence of the bacterium [4]–[6]. Impairing the pantothenate pathway by suppressing β-alanine formation has been shown to result in a significant decline in Mtb virulence [7], [8], which has motivated us to consider ADC as a potential target for therapeutic intervention against tuberculosis.
Recently, we have engaged in a chemoinformatics-based approach to identify potential drug-like inhibitors by structure-based virtual screening, which led to 28 lead molecules [9]. The purpose of our present investigation is to experimentally test a representative subset of the identified targets and contrast them with several compounds, which in part have been previously shown to be active in inhibition experiments [8], [10], [11]. Existing assays for ADC involve derivatization and separation steps [12], [13], radioactive labeling [1], [10] and laborious manometric quantification of the carbon dioxide released as a reaction by-product [14]. Extending our ongoing efforts in the design of enzyme assays [15], [16], including decarboxylase assays [17], we report a novel but simple 1H NMR protocol for monitoring ADC activity, which not only allows for direct structural information on the enzymatic reaction to be obtained and progress curves to be taken, but also serves as a convenient tool for inhibitor screening.
Materials and Methods
Inhibitors
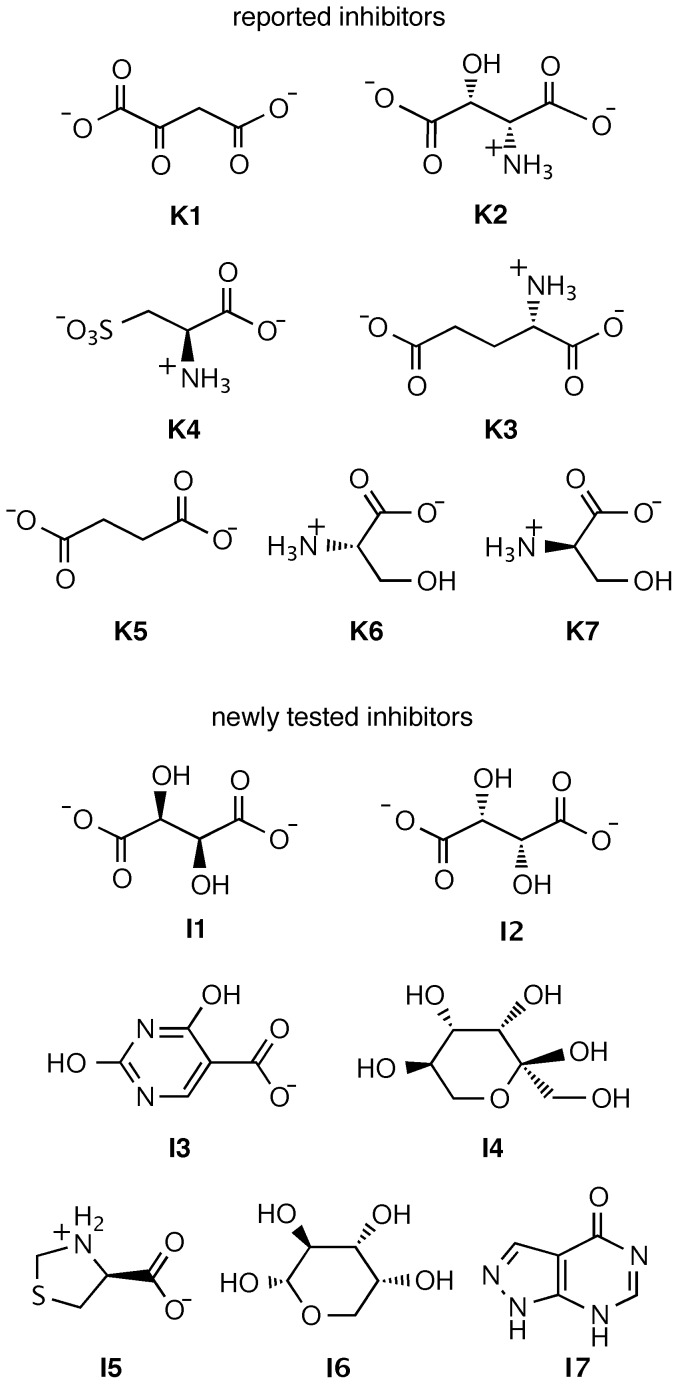
The previously reported and newly identified compounds that were tested in the present study for inhibitory effect against ADC are shown in Fig. 1. Oxaloacetate (K1), DL-threo-β-hydroxyaspartate (K2), L-glutamate (K3), L-cysteic acid (K4), succinate (K5), L-serine (K6), and D-serine (K7) had been tested previously, while D-tartrate (I1, ZINC00895296), L-tartrate (I2, ZINC00895301), 2,4-dihydroxypyrimidine-5-carboxylate (I3, ZINC00901606), D-tagatose (I4, ZINC03830878), (4S)-1,3-thiazolidin-3-ium-4-carboxylate (I5, ZINC00967474), α-D-arabinopyranose (I6, ZINC03606295), and 1,2-dihydropyrazolo[3,4-d]pyrimidin-4-one (I7, ZINC05177572) were tested for the first time, stimulated by the hits recently identified in the chemoinformatics study [9]. The compounds were purchased from Sigma-Aldrich, Germany (K1–K7 and I1–I4), or from Labotest KG, Germany (I5–I7), in the highest commercially available purity (>97%).
Figure 1. Chemical structures of known inhibitors against ADC (K1–K7) and computationally identified potential inhibitors obtained via virtual screening (I1–I7).
Protein preparation
Mycobacterium tuberculosis L-aspartate α-decarboxylase (MtbADC) was overexpressed with a C-terminal 6xHis tag in E. coli and purified as the cleaved tetrameric form by Ni2+-NTA affinity and gel filtration chromatography as described previously [2], [6]. The protein was further dialyzed against 10 mM Tris, pH 7.5.
Enzyme assays
The enzymatic reactions were carried out in D2O (Sigma-Aldrich) using 1 mM L-aspartic acid as substrate in the absence or presence of 1 mM inhibitor. The reactions were initiated by addition of enzyme (2.83 µM, as determined by using an extinction coefficient ε280 = 6400 M−1cm−1 [2] and measured on a Varian Cary 4000 UV-Vis spectrophotometer. The enzymatic activity was followed by 1H NMR using a Jeol JNM-ECX 400 spectrometer.
Results and Discussion
Enzyme assays
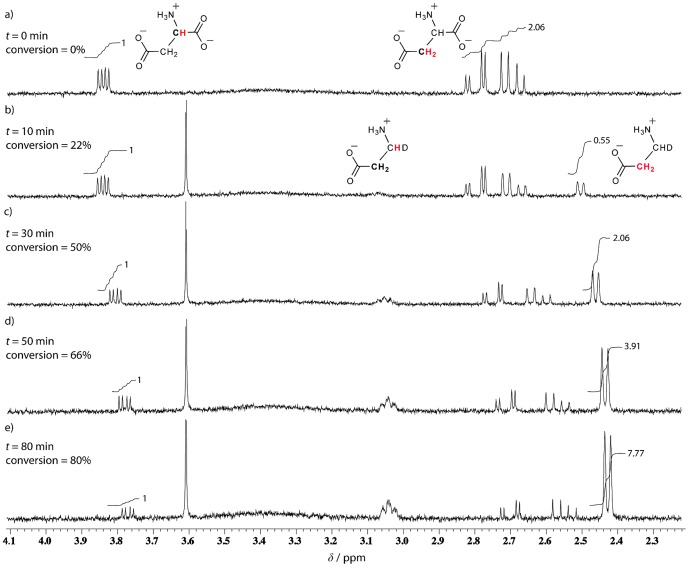
Before proceeding to inhibitor screening, a convenient assay for monitoring the ADC activity was established. Since during an enzymatic transformation, structural changes, which may be detected by differences in spectra of the substrate and product of the reaction, occur almost inevitably [18], [19], we opted for NMR spectroscopy as the technique of choice. Therefore, we validated a simple protocol using 1H NMR for monitoring the enzymatic depletion of L-aspartate and concomitant formation of β-alanine. The advantage of 1H NMR is that the technique is label-free and allows direct monitoring. To this end, the conversion of 1 mM L-aspartate in the presence of 3 µM ADC could be conveniently followed as shown in Fig. 2. L-aspartate shows two resonances, in a 1∶2 ratio (Fig. 2a). Upon addition of ADC, the signals corresponding to the product start to emerge and intensify with time while those of the substrate L-aspartate diminish and eventually completely disappear (Fig. 2 b–e). Note that, based on its structure, one would also expect two different 1H NMR peaks for β-alanine (this was confirmed by taking a spectrum for the commercial analyte). However, only the signals corresponding to the protons adjacent to the carboxylate group could be quantified in the course of the enzymatic reaction (at approximately δ = 2.44 ppm). This is a consequence of the fact that the reaction is carried out in D2O, and, therefore, the newly acquired β-alanine hydrogen is, in fact, a deuterium atom. This is also in line with both the broadness of the signal identified at approximately δ = 3.04 ppm and the splitting pattern (a doublet) of the upfield-shifted protons.The assay was optimized with respect to substrate and enzyme concentrations, such that the conversion rate was directly proportional to the enzyme concentration.
Figure 2.
Selected 1H NMR spectra of 1 mM L-aspartate a) before and b)–e) 10–80 min after addition of 3 µM ADC in D2O at 25°C. The diminishing signals of L-aspartate and the emerging ones of those corresponding to β-alanine permitted a direct monitoring of the enzymatic transformation and integration of the proton signals allowed for a kinetic profiling of the reaction (cf. Fig. 3).
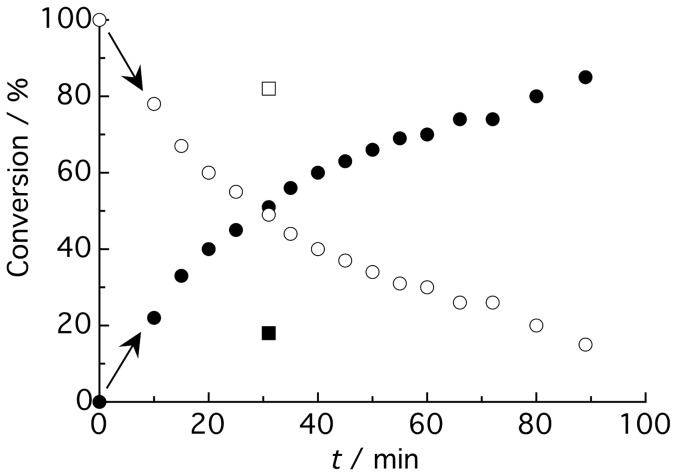
Accordingly, the depletion of the L-aspartate substrate and the concomitant formation of β-alanine could be readily detected and, upon integration, also accurately quantified. This has finally allowed us to extract a kinetic profile of the enzyme in the absence of inhibitor (empty and filled circles in Fig. 3), which does not only depict a time-resolved monitoring of β-alanine formation as a result of the enzymatic activity, but also serves as a reference for subsequent inhibition studies (for instance, D-tartrate, empty and filled squares in Fig. 3). Since it is desirable to work in the linear region of the enzyme kinetic trace, we have used in our study 50% conversion in the absence of inhibitor as a reference point, which was achieved at t = 30–40 min, depending on the enzyme batch used.
Figure 3. Kinetic monitoring of ADC activity carried out using 1 mM L-aspartate and 3 µM enzyme.
The different points correspond to conversion percentages of the individual 1H NMR spectra taken at increasing reaction times after initiation of the reaction in D2O at 25°C. Percentage of product formation and substrate depletion is represented by filled and empty circles, respectively. The percentage of product and substrate after 30 min of the reaction in the presence of D-tartrate is represented by filled and empty squares, respectively.
Inhibitors selected
Despite the relevance of ADC as a drug target, only a relatively small number of compounds have been reported to inhibit its activity [10]. Among these previously identified inhibitors, we have selected seven (K1–K7, Fig. 1), which we also tested experimentally. The inhibitory effects of these molecules have been previously quantified on E. coli ADC and inhibition constants have been determined in the mM range [10].
In our recent study [9], 336,761 molecules from three public databases have been targeted: the Maybridge (14,400 molecules), the Zinc (including the National Cancer Institute, 316,181 molecules), and the United States of America Foods and Drug Administration approved drugs database (3,180 molecules). A high-throughput virtual screening, based on the interaction of these compounds with the active site residues of the enzyme by using a stronger binding affinity as compared to that of the MtbADC:fumarate reference (−4.2 kcal/mol) as a positive criterion, led to the identification of 28 lead molecules. Herein, we tested a representative and commercially accessible subset of the positive hits (I1–I7, Fig. 1) and compared them under identical experimental conditions to the known ADC inhibitors. Apart from excluding cross-reactivity towards other pyruvoyl-dependent enzymes, all newly tested ligands were selected by ensuring a direct interaction with the well-conserved Arg54, whose role in substrate specificity has already been acknowledged [20], [21].
Inhibition studies
The enzymatic reactions for inhibitor screening were performed in the presence of 1 mM potential inhibitor, similar to the concentrations employed in previous enzymatic assays [10], [11]. The enzymatic conversion at specific reaction times in the absence and presence of inhibitor was compared and defined as k rel, which we used as a relative measure of inhibition efficiency (vide infra). The results are compiled in Table 1. As expected from literature data [10], mM inhibitor concentrations were required to induce a significant inhibitory effect for most compounds, including the newly identified ones. Since all compounds reported by Williamson and Brown act as competitive inhibitors against ADC [10], it did not come as a surprise that the most effective ones were those containing two carboxylate moieties (K1–K5), which are considered to be structural analogues to the natural substrate L-aspartate. In fact, β-hydroxyaspartate (K2), meso-diaminosuccinate (herein we tested succinate, K5), and cysteic acid (K4) have been amongst the first compounds shown to effect the pantothenate pathway [22], [23]. A recent study, where MALDI-TOF mass spectrometry has been employed as a screening methodology for identifying compounds that bind to ADC, has in fact revealed that only a few homologs of L-aspartate, such as L-glutamate (K3), can penetrate into the active site of the enzyme [11]. The only two dicarboxylate exceptions constitute the two amino acids L- and D-serine (K6 and K7), respectively, which have also been reported to inhibit β-alanine synthesis [10], [24]. Particularly, D-serine had been found to be 4.5 times more potent than its L-enantiomer (K i = 0.16 vs. 0.73 mM, respectively) [10]. In our direct label-free assays, neither K6 nor K7 showed a considerable inhibitory effect towards ADC activity. However, among all known inhibitors, the most surprising effect was exhibited by oxaloacetate K1, in the presence of which no biocatalytic activity was detected at all (k rel = 0).
Table 1. Relative inhibitory effects of selected known and newly tested compounds against ADC.[a] .
| Entry | Compound | Conversion %[b] | k rel [c] | Classification |
| Ref. | None | 50 | 1.00 | reference |
| Reported compounds [10] | ||||
| K1 | oxaloacetate | 0[d] | 0.00[d] | very strong |
| K2 | β-hydroxyaspartate | 18 | 0.36 | strong |
| K3 | L-glutamate | 20 | 0.40 | strong |
| K4 | L-cysteate | 20 | 0.40 | strong |
| K5 | succinate | 32 | 0.64 | moderate |
| K6 | L-serine | 45 | 0.90 | weak |
| K7 | D-serine | 48 | 0.96 | insignificant |
| Newly tested compounds | ||||
| I1 | D-tartrate | 18 | 0.36 | strong |
| I2 | L-tartrate | 19 | 0.38 | strong |
| I3 | 2,4-dihydroxypyrimidine-5-carboxylate | 27 | 0.54 | moderate |
| I4 | D-tagatose | 45[e] | 0.90[e] | weak |
| I5 | (4S)-1,3-thiazolidin-3-ium-4-carboxylate | 48 | 0.96 | insignificant |
| I6 | α-D-arabinopyranose | 48 | 0.96 | insignificant |
| I7 | 1,2-dihydropyrazolo[3,4-d]pyrimidin-4-one | 48 | 0.96 | insignificant |
The measurements were performed using 1 mM L-aspartate, 3 µM ADC, and 1 mM compound (potential inhibitor) in D2O at 25°C.
The conversion percentage corresponds to the product formed by integration of the 1H NMR signals corresponding to substrate and product of the enzymatic reaction after ca. 30 min upon addition of the enzyme. The time was adjusted to correspond to 50% conversion in the absence of inhibitor (reference). The absolute values were averaged from at least two independent assays.
The relative inhibitory effect, k rel, was calculated as the ratio of the conversion percentages in the presence and absence of compound.
While full inhibition was also observed when using double the enzyme concentration, i.e., 6 µM, only a small nhibitory effect could be detected (k rel = 0.9) when the assay was performed with 100 µM oxaloacetate, i.e., at a 10-fold lower inhibitor concentration.
A smaller k rel value of 0.74, suggesting moderate inhibition, was observed upon preincubation with ADC for 1 h at ambient temperature.
Complementing the chemoinformatics-based approach, our experimental data show that most molecules possessing a sizable inhibitory potential are, similar to those previously reported [10], anionic in nature. D-tartrate (I1, k rel = 0.36) was found to be a strong inhibitor, followed by the pyrimidine derivative I3 (k rel = 0.54), whose presence reduced the ADC activity by approximately 50%. Presumably, the smaller effect of I3, when compared to I1, is due to a weaker electrostatic binding with the enzyme active site. Surprisingly, D-tagatose (I4), which is a neutral compound, exhibited a slight inhibitory effect (k rel = 0.74, see footnote in Table 1), but only upon preincubation with the enzyme. It should be mentioned that all compounds in Table 1 were additionally checked under conditions of preincubation with ADC (typically 1 hour, room temperature); however, no differences with respect to the inhibition potential were noted except for D-tagatose (I4). The fact that I5, which had emerged as another hit from the chemoinformativs study, showed no detectable inhibitory effect emphasizes the need for complementary computational and experimental studies.
Based on the crystal structure [6], the affinity of molecules to the active site of ADC was postulated to be dominated by two moieties: the pyruvoyl group and the Arg54 residue. All newly tested compounds were selected upon ensuring both a direct interaction with Arg54 residue, while excluding contact with the MtbADC pyruvoyl group [9]. Our data clearly reveal that the inhibitory potential of the tested ligands is, actually, directly dependent on the number of carboxylate groups present. It transpires that in the absence of any ligand interaction with pyruvoyl, Arg54 presides over the active-site binding, which corroborates that this well conserved residue has a fundamental role in substrate specificity, namely by forming a salt bridge with the β-carboxylate of L-aspartate [21].
Apart from compounds I1 and I3–I7, which had been identified as putative inhibitors via virtual screening, we have also included L-tartrate (I2), the enantiomer of I1, in our experiments. To our surprise, an essentially identical, strong inhibitory effect as that for the D-isomer was observed (k rel = 0.38). The D-/L-tartrate example suggests that the chirality of the ligands has no large influence on their binding to the ADC active site, but that the binding is mainly driven by less specific electrostatic interactions.
Conclusions
In order to counter multi-drug resistance, attenuate Mycobacterium tuberculosis's virulence, and expedite recovery or prolong the survival of patients, additional drugs, in combination with conventional antibiotics, are continuously needed. The development of convenient and easily accessible enzymatic assays combined with drug validation tools are, thus, a compulsory requirement. In this work, we were able to monitor β-alanine formation from L-aspartate during enzymatic conversion via 1H NMR. This is the first assay reported for ADC, which allows one to accurately monitor the related biocatalytic reaction in a direct and label-free fashion. The functionality of the assay was demonstrated by its use in experimental screening for ADC inhibitors, which allowed the validation of three computationally identified inhibitors against the Mycobacterium tuberculosis L-aspartate α-decarboxylase enzyme.
Funding Statement
KS acknowledges the funding support of the Ministry of Education (Grants R154-000-439-112 and R154-000-491-112) and the Biomedical Research Council (Grant R154-000-424-305) of Singapore. The graduate research scholarship for RS is provided by National University of Singapore. MF and WMN acknowledge the support from the Fonds der Chemischen Industrie (doctoral stipend for MF) and Jacobs University Bremen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cronan JE (1980) Beta-alanine synthesis in E. coli. J Bacteriol 141: 1291–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chopra S, Pai H, Ranganathan A (2002) Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Expres Purif 25: 533–540. [DOI] [PubMed] [Google Scholar]
- 3. Spry C, Kirk K, Saliba KJ (2008) CoenzymeA biosynthesis: an antimicrobial drug target. FEMS Microbiol Rev 32: 56–106. [DOI] [PubMed] [Google Scholar]
- 4. Cox JS, Chen B, McNeil M, Jacobs WR Jr (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402: 79–83. [DOI] [PubMed] [Google Scholar]
- 5. Glickman MS, Cox JS, Jacobs WR Jr (2000) A novel mycolic acid cyclopropane synthestase is required for coding, persistence and virulence of Mycobacterium tuberculosis. Mol Cell 5: 717–727. [DOI] [PubMed] [Google Scholar]
- 6. Gopalan G, Chopra S, Ranganathan A, Swaminathan K (2006) Crystal structure of uncleaved L-aspartate-alpha-decarboxylase from Mycobacterium tuberculosis. Proteins 65: 796–802. [DOI] [PubMed] [Google Scholar]
- 7. Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, et al. (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8: 1171–1174. [DOI] [PubMed] [Google Scholar]
- 8. Webb ME, Smith AG, Abell C (2004) Biosynthesis of pantothenate. Nat Prod Rep 21: 695–721. [DOI] [PubMed] [Google Scholar]
- 9. Sharma R, Kothapalli R, van Dongen AMJ, Swaminathan K (2012) Chemoinformatic Identification of Novel Inhibitors against Mycobacterium tuberculiosis L-aspartate alpha-decarboxylase. PloS One 7: e33521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williamson JM, Brown GM (1979) Purification and properties of L-aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli. J Biol Chem 254: 8074–8082. [PubMed] [Google Scholar]
- 11. Webb ME, Stephens E, Smith AG, Abell C (2003) Rapid Screening by MALDI-TOF mass spectrometry to probe binding specificity at enzyme active sites. Chem Commun 2416–2417. [DOI] [PubMed] [Google Scholar]
- 12. Ramjee MK, Genschel U, Abell C, Smith AG (1997) Escherichia coli L-aspartate-alpha-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem J 323: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Villiers J, Koekemoer L, Strauss E (2010) 3-Fluoroaspartate and Pyruvoyl-Dependant Aspartate Decarboxylase: Exploiting the Unique Characteristics of Fluorine To Probe Reactivity and Binding. Chem Eur J 16: 10030–10041. [DOI] [PubMed] [Google Scholar]
- 14. Abell LM, O'Leary MH (1988) Isotope effect studies of the pyruvate-dependent histidine decarboxylase from Lactobacillus 30a. Biochemistry 27: 5933–5939. [DOI] [PubMed] [Google Scholar]
- 15. Sahoo H, Hennig A, Florea M, Roth D, Enderle T, et al. (2007) Single-label kinase and phosphatase assays for tyrosine phosphorylation using nanosecond time-resolved fluorescence detection. J Am Chem Soc 129: 15927–15934. [DOI] [PubMed] [Google Scholar]
- 16. Ghale G, Ramalingam V, Urbach AR, Nau WM (2011) Determining protease substrate selectivity and inhibition by label-free supramolecular tandem enzyme assays. J Am Chem Soc 133: 7528–7535. [DOI] [PubMed] [Google Scholar]
- 17. Dsouza RN, Hennig A, Nau WM (2012) Supramolecular tandem enzyme assays. Chem Eur J 18: 3444–3459. [DOI] [PubMed] [Google Scholar]
- 18. Vandenberg JI, Kuchel PW, King GF (1986) Application of progress curve analysis to in situ enzyme kinetics using 1H NMR spectroscopy. Anal Biochem 155: 38–44. [DOI] [PubMed] [Google Scholar]
- 19. Belliveau KA, Romero-Zerón LB (2010) Monitoring the enzymatic degradation of sinigrin from B. juncea meal using 1H NMR spectroscopy. Nat Prod Res 24: 24–33. [DOI] [PubMed] [Google Scholar]
- 20. Albert A, Dhanaraj V, Genschel U, Khan G, Ramjee MK, et al. (1998) Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing. Nat Struct Biol 5: 289–293. [DOI] [PubMed] [Google Scholar]
- 21. Lee BI, Suh SW (2004) Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase. J Mol Biol 340: 1–7. [DOI] [PubMed] [Google Scholar]
- 22. Shive W, Macow J (1946) Biochemical tranformations as determined by competitive analogue-metabolite growth inhibitions: I. Some transformations involving aspartic acid. J Biol Chem 162: 451–462. [PubMed] [Google Scholar]
- 23. Ravel JM, Shive W (1946) Biochemical tranformations as determined by competitive analogue-metabolite growth inhibitions: IV. prevention of pantothenic acid synthesis by cysteic acid. J Biol Chem 166: 407–415. [PubMed] [Google Scholar]
- 24. Maas WK, Davis BD (1950) Pantothenate studies. I. Interference by D-serine and L-aspartic acid with pantothenate synthesis in Escherichia coli. J Bacteriol 60: 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]