Abstract
Background
Nabidae, a family of predatory heteropterans, includes two subfamilies and five tribes. We previously reported the complete mitogenome of Alloeorhynchus bakeri, a representative of the tribe Prostemmatini in the subfamily Prostemmatinae. To gain a better understanding of architecture and evolution of mitogenome in Nabidae, mitogenomes of five species representing two tribes (Gorpini and Nabini) in the subfamily Nabinae were sequenced, and a comparative mitogenomic analysis of three nabid tribes in two subfamilies was carried out.
Methodology/Principal Findings
Nabid mitogenomes share a similar nucleotide composition and base bias, except for the control region, where differences are observed at the subfamily level. In addition, the pattern of codon usage is influenced by the GC content and consistent with the standard invertebrate mitochondrial genetic code and the preference for A+T-rich codons. The comparison among orthologous protein-coding genes shows that different genes have been subject to different rates of molecular evolution correlated with the GC content. The stems and anticodon loops of tRNAs are extremely conserved, and the nucleotide substitutions are largely restricted to TψC and DHU loops and extra arms, with insertion-deletion polymorphisms. Comparative analysis shows similar rates of substitution between the two rRNAs. Long non-coding regions are observed in most Gorpini and Nabini mtDNAs in-between trnI-trnQ and/or trnS2-nad1. The lone exception, Nabis apicalis, however, has lost three tRNAs. Overall, phylogenetic analysis using mitogenomic data is consistent with phylogenies constructed mainly form morphological traits.
Conclusions/Significance
This comparative mitogenomic analysis sheds light on the architecture and evolution of mitogenomes in the family Nabidae. Nucleotide diversity and mitogenomic traits are phylogenetically informative at subfamily level. Furthermore, inclusion of a broader range of samples representing various taxonomic levels is critical for the understanding of mitogenomic evolution in damsel bugs.
Introduction
Insect mitochondrial genome (mitogenome) typically consists a single circular molecule that is 14–20 kb long and usually contain 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs) (the large and small ribosomal subunits), and one or more non-coding (NC) regions (also referred to as the control region, CR) with essential regulatory elements for transcription and replication [1], [2]. Notable exceptions are represented by the fragmented mtDNA of some sucking lice and booklice [3]–[5]. Several advantages including simple genomic organization, high rates of evolution, and (almost) unambiguous orthology have made the mitogenomes good models for molecular evolution and abundant molecular markers for phylogeographic and phylogenetic studies at various taxonomic levels [6]–[10]. Complete mitogenomes are not only more informative than individual genes, but also provide a suite of genome level characters, such as the relative position of different genes, RNA secondary structures and models of control of replication and transcription. Mitogenomic sequencing and subsequent analysis of the 13 PCGs have increased dramatically in the past decade and the utility of mtDNAs for phylogenetic inference at various taxonomic levels has therefore been aggressively exploited [8].
Nabidae, also called damsel bugs, is a relatively small family of Heteroptera with approximately 20 genera and 500 species [11] and comprise of two subfamilies Prostemmatinae and Nabinae, and five tribes [12]. Most members of Prostemmatinae appear more stout-bodied and occasionally posses distinctive red and black color patterns. They are ground-living with enlarged and strong forelegs and appear to prey exclusively on other insects. In contrast, the Nabinae are elongate and of drab coloration, live on plant, and prey on small arthropods with simple forelegs [12], [13]. The distinctive subfamily level differences make the damsel bugs an ideal group to study the evolution of mitogenomes.
The complete mitogenome of Alloeorhynchus bakeri, a representative of the tribe Prostemmatini in the subfamily Prostemmatinae has been reported previously [14]. In this study, three complete and two nearly complete mitogenomes from two tribes, Gorpini and Nabini, in the subfamily Nabinae were sequenced. Overall, six nabid mitogenomes representing two subfamilies, three tribes, and four genera were used in the comparative analysis to: 1) assess evolutionary traits of mitogenomes from three tribes, and 2) explore the phylogenetic utility and limits of mitogenomic data at lower taxonomic levels, specifically subfamily and generic levels.
Results and Discussion
Genome Organization
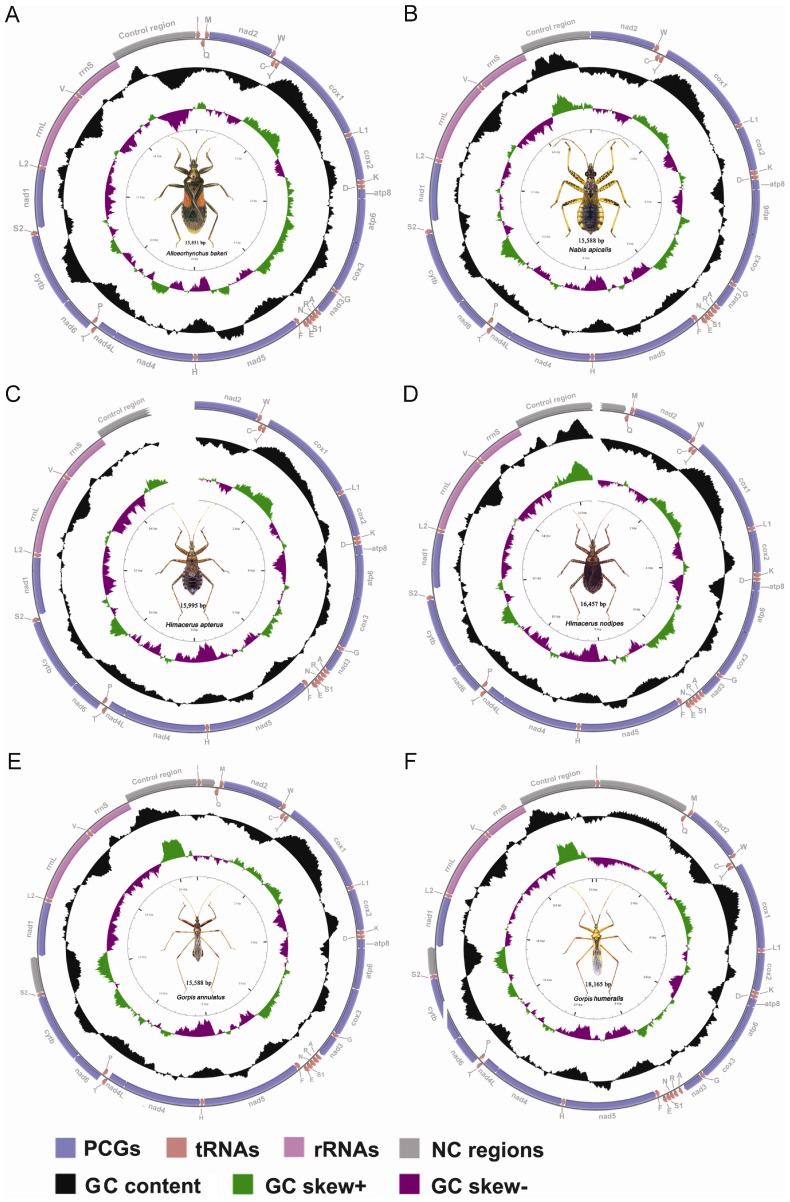
In this study, four complete and two nearly complete mitogenomes of damsel bugs representing three tribes and two subfamilies were compared. Four complete mitogenomes were from Alloeorhynchus bakeri Harris [14], Nabis apicalis Matsumura, Gorpis annulatus Paiva, and Gorpis humeralis (Distant), and two nearly complete mitogenomes were from Himacerus apterus (Fabricius) and Himacerus nodipes (Hsiao). Regions that we failed to sequence were located in or around gene nad2 and CR. Several factors including notable base bias, highly repeated regions, and stable secondary structures could result in the unsuccessful sequencing of the CR.
All six mitogenomes shared overlapping genes, typically 10 to 13 gene overlaps; the highest number (17) of gene overlaps was found in A. bakeri and the fewest number (10) was found in G. annulatus. The amount of overlap between genes was typically between 1 and 14 bp. Two pairs of overlapping genes were common to all six mitogenomes: atp8-atp6 and nad4-nad4L, and shared nearly the same 7 bp sequence (ATGATAA), with the exception of A. bakeri (ATGATAG overlap between nad4 and nad4L). This is a common feature in heteropteran mtDNAs, as well as in other arthropods [15].
Consistent with other published heteropteran mitogenomes, the six nabid mitogenomes contained all 13 PCGs and both rRNAs in the same order and direction of the hypothesized ancestral arthropod mitogenome [16] (Figure 1). Most complete mitogenomes typically had 22 tRNAs, however, only 19 tRNAs were detected in N. apicalis, with loss of the gene cluster trnI-trnQ-trnM. All detected tRNAs shared identical architectures at the tRNA loci, similar to most heteropterans.
Figure 1. Mitochondrial maps of six damsel bugs.
Direction of gene transcription is indicated by arrows. PCGs are shown as blue arrows, rRNAs as purple arrows, tRNAs as red arrows and large NC regions (>100 bp) as cyan rectangles. tRNAs are labeled according to single-letter IUPAC-IUB abbreviations (L1: UUR; L2:CUN; S1:AGN; S2:UCN). The GC content is plotted using a black sliding window, as the deviation from the average GC content of the entire sequence. GC-skew is plotted as the deviation from the average GC-skew of the entire sequence. Ticks in the inner cycle indicate the sequence length.
Most PCGs started with ATN start codons and stopped with TAA/TAG termination codons or truncated termination codons TA or T which were presumed to be completed via post-transcriptional polyadenylation [17]. Gene cox1 and nad1 has been found to employ TTG or GTG as a start codon in some species of Hemiptera, Hymenoptera, Coleoptera and Lepidoptera, thus minimizing intergenic spacing and avoiding overlaps with adjacent genes [18]–[20]. We discovered TTG start codons in cox1 gene for most sequenced damsel bugs and GTG start codon in nad1 gene for G. humeralis (Table S1).
Nucleotide Composition and Codon Usage
All analyzed nabid mitogenomes were consistently AT biased, with values from 72.6% to 76.9% and, on average, displayed positive AT- and negative GC-skews of the coding strand (Figure S1A). The J-strand of PCGs and rRNAs had a negative AT-skew in all species, that of tRNAs had a positive AT-skew (Figure S1B). The tRNAs and rRNAs had a positive GC-skew, and the 3rd codon positions had a negative GC-skew (Figure S1C). The G content was almost equal to C content in PCGs in all species. All sequences showed strong consistent base bias, except the CR showed a large variation in AT- and GC-skews.
The pattern of codon usage in all analyzed damsel bug mitogenomes was consistent with the standard invertebrate mitochondrial genetic code and the preference for A+T-rich codons. They shared the similar codon usage bias an effective number of codons (ENC) [21] equivalent to 38.45±3.58 and codon bias index (CBI) [22] equivalent to 0.65±0.07. Synonymous codons ending with A or T were clearly preferred (81.5 to 89.8% for individual species; 86.2% on average), and the most frequently used codons were AUA, UUA, AUU, UUU, UAU and AAU (Figure S2). Some G+C-rich codons were not used in some species, such as CGC in N. apicalis, UCG and GCG in H. apterus, and CUC, GCG and GGC in H. nodipes (Table S2).
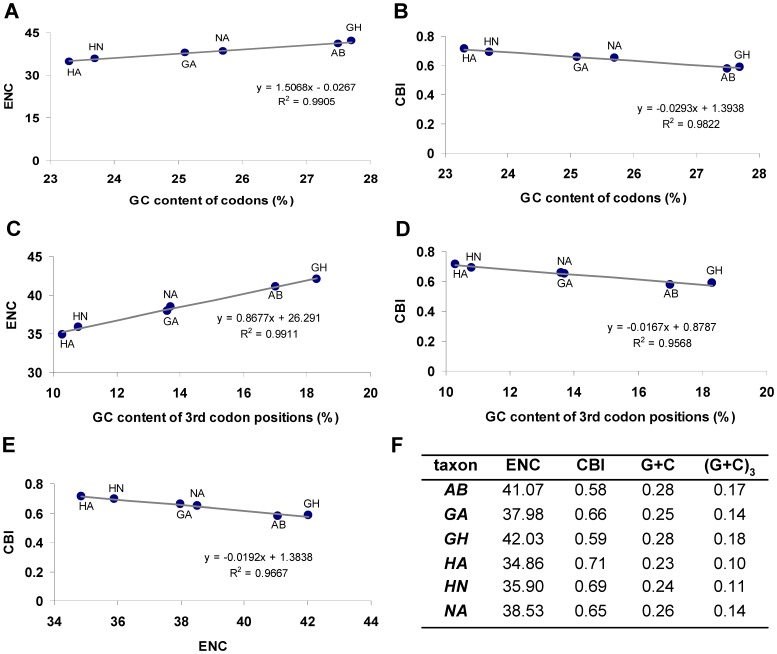
The correlations among CBI, ENC, GC content of all codons, and GC content of the 3rd codon positions in all sequenced damsel bug mitogenomes were analyzed (Figure 2). A positive correlation was observed between ENC and GC content of all codons (R2 = 0.99) (Figure 2A) and the 3rd codon positions (R2 = 0.99) (Figure 2C). Furthermore, a negative correlation was observed between CBI and GC content of all codons (R2 = 0.96) (Figure 2B), GC content of the 3rd codon positions (R2 = 0.96) (Figure 2D) and ENC (R2 = 0.97) (Figure 2E). These results are consistent with prevailing neutral mutational theories positing that genomic GC content is the most significant factor in determining codon bias among organisms [23]–[25].
Figure 2. Evaluation of codon bias across six nabid mtDNAs.
ENC, effective number of codons (out of a maximum of 61) [21]; CBI, codon bias index [22]; G+C, GC content of codons; (G+C)3, GC content of 3rd codon positions.
Evolutionary Rate
In view of the evolutionary forces acting on the mitochondrial PCGs of three nabid tribes, the average rate of non-synonymous substitutions (Ka), the average rate of synonymous substitutions (Ks), and the average ratio of Ka/Ks were calculated for each PCG, respectively [26]. The observed average Ka/Ks ratios were consistently lower than one, increasing from 0.07 for cox1 to 0.93 for atp8 (Table S3). The uniformly low values of Ka/Ks ratios for cox1–3 and cytb (Ka/Ks <1) indicate strong evolution constraints in cytochrome c oxidase [27], [28] and also suggest a strong purifying selection in the six species of nabids. The variation of GC content probably caused the different evolutionary patterns among genes, and a negative correlation was observed between the Ka/Ks and the GC content of mitochondrial PCGs (R2 = 0.85). This result is highly consistent with previous findings in true bugs [14], [29].
Transfer RNAs
The tRNAs closest to the CR (ie., trnI, trnQ and trnM), which is one of the starting points for mtDNA replication, were not detected in the complete mitogenome of N. apicalis (Figure 1). Among tRNAs, only trnS1 did not exhibit the typical clover leaf secondary structure, due to the loss of the DHU stem (Figure S3). In general, the loss of the DHU stem in trnS1 was a common feature in metazoan mtDNAs [30], and is almost always presents in Heteroptera [20], [29].
The trnA, trnR, trnN, trnQ, trnK, trnS1, trnS2, and trnV showed more mismatches in their stems (Figure S3). Mismatches were located mostly in the acceptor and anticodon stems with a single exception represented by trnV, in which the mismatch was on the TψC stem. Mismatches observed in tRNAs are corrected through the RNA-editing mechanisms that are well known for arthropod mtDNA [31]–[33].
In most nabid tRNAs, the stems and anticodon loops were extremely conserved, and the nucleotide substitutions were largely restricted to TψC and DHU loops and extra arms, with obvious insertion-deletion polymorphisms (Figures S4, S5). The presence of indels polymorphisms at different taxonomic levels underscores the potential phylogenetic value of tRNA sequences, especially when secondary structures are taken into account [30], [34], [35].
Ribosomal RNAs
Genes for the small and large subunit ribosomal RNAs (rrnS and rrnL) were adjacent on the same strand, interleaved by a single trnV, in the analyzed damsel bug mitogenomes. Both rRNAs of N. apicalis were the largest (1, 289 bp in rrnL and 820 bp in rrnS), while those in A. bakeri were the smallest (1, 252 bp in rrnL and 790 bp in rrnS) (Table S1). The size differences in both ribosomal subunits were not distinct among different species, especially in the subfamily Nabinae (1, 283±6 bp in rrnL and 812±6 bp in rrnS).
With the addition of five newly sequenced damsel bugs, the mitochondrial rRNA secondary structures were compared in three tribes. All of rRNA genes conformed to the secondary structure models proposed for A. bakeri [14] and other insects [36]–[39]. The overall structure of rrnLs, including five domains and 44 helices, were largely consistent among the analyzed damsel bugs. The multiple alignment of rrnLs extended over 1, 345 positions and contained 964 conserved (71.7%), 85 indels (6.3%) and 296 nucleotide substitutions (22.0%). Nucleotide variability was unevenly distributed among domains and helices, mainly in domains I and II.
Similarly, the secondary structures of rrnS were also largely consistent, including three domains and 27 helices. The multiple alignments spanned 849 positions and contained 608 conserved (71.6%), 59 indels (6.9%) and 182 nucleotide substitutions (21.4%). Nucleotide variability was unevenly distributed among domains and helices, mainly in domain I and the large loop between helices H577 and H673.
Comparative studies showed similar rates of substitution between two rRNAs (Figure S6). Several helices (H235, H589, H837, and H2077 in rrnL, and H39, H47, H367, H1241, and H1303 in rrnS, respectively) with high variability at the primary sequence level, showed conserved secondary structures. The phylogenetic value of structural characteristics of these helices is intriguing, suggesting the necessity of sampling at different taxonomic levels to ensure the validity of phylogenetic relationships inferred by the mitogenomic data.
Non-coding Regions
The six mitogenomes of Nabidae displayed moderate size variation, most of which could be attributed to the variety of NC regions usually caused by the presence of repetitive elements. The NC regions of six nabid mitogenomes were summarized in Table S4; the size varied from 1, 198 bp (N. apicalis, the shortest) to 3, 567 bp (G. humeralis, the largest). The proportion of NC regions appeared high in the mtDNAs of N. apicalis and G. humeralis, varying from 7.7 to 19.6%. Three distinct large NC regions were identified in the following gene pairs: trnI-trnQ, trnS2-nad1, and rrnS-trnI.
The NC region located between trnI and trnQ, appeared to be common in the Gorpis and Himacerus mitogenomes, varying in length from 221 to 1,539 bp. Tandem repeats were detected in this region of G. humeralis (1, 442 bp and six copies) and H. nodipes (540 bp and four copies) (Table S5). This region was also present in other heteropteran mtDNAs even though the nucleotide sequence could be divergent among species [29], [40]. Several arrangements at this genomic location are worth noting: (1) the gene cluster trnI- trnQ -trnM was missing in the mtDNA of N. apicalis; (2) trnI and trnQ were reversed in the flat bug Neuroctenus parus [29]; and (3) the gene order trnM-trnI-trnQ and the NC regions between trnQ and nad2 were present in almost all sequenced lepidopteran mtDNAs [37], [41]–[44]. Further investigation with a broader taxon sampling within Heteroptera is necessary to assess if this genomic location is related to the replication of mtDNA.
The NC region inserted between trnS2 and nad1, was present in all sequenced nabid mtDNAs, varying in length from 15 to 584 bp. Tandem repeats were detected in this location of G. annulatus (582 bp, 4 copies) and G. humeralis (467 bp, 3 copies) (Table S5). This intergenic spacer is also present in all partial or fully sequenced neuropterid species and other insect mtDNAs [30], and contains the binding site for the DmTFF bidirectional transcription termination factor [45].
The NC region located between rrnS and trnI, coincided with the A+T-rich region, also called the CR, including the origin of replication and promoters for transcription initiation [2], [46]–[48]. The CRs of three nabid tribes were longer than 1 kb, with high rates of nucleotide substitution and indels, and a variable number of tandem repeats.
Comparison of the CR sequences of six nabid mtDNAs revealed few relevant short blocks. The pattern of sequence conservation is in agreement with previous reports, indicating that CRs of three nabid tribes (especially the five species in the subfamily Nabinae) belong to the group 2 insect-control region [46], with the short conserved blocks, as well as the conserved secondary structures.
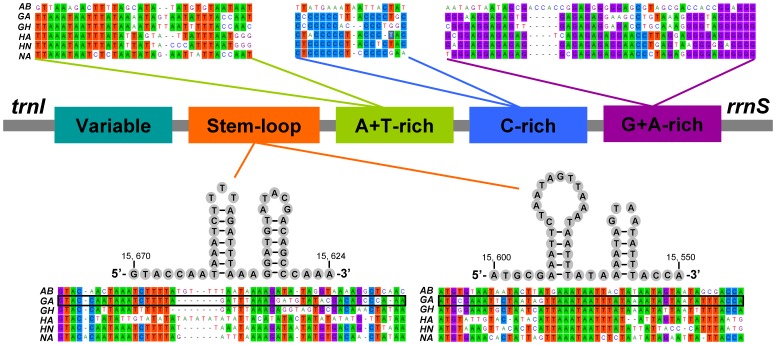
The following structural elements have been identified from comparative analysis of the CR sequences of six nabid species belonging to two subfamilies (Figure 3).
Figure 3. Organization of the control region in six nabid mtDNAs.
The colorized panes indicate the structural elements in the CR. The alignment of the primary sequences and the predicated secondary stem-loop structures are shown, respectively, above and below.
A G+A-rich sequence block was conserved at the 3′-end of the CR (near to rrnS). The overall low similarity of the primary nucleotide sequences between Nabinae and Prostemmatinae suggests that they are taxon-specific.
Upstream of the conserved G+A-rich sequence block was a conserved C-rich sequence block in Gorpini and Nabini, but not presented in Prostemmatini.
An A+T-rich sequence block was identified upstream of C-rich sequence block.
The stem-loop structures were conserved in three tribes, and are potentially associated with the second strand replication origin [47]–[51].
One variable domain concluded the rest of the region and was highly variable in both nucleotide sequence and length. Tandem repetition was observed in this region.
Comparative analysis of the structural organization of CRs among three nabid tribes revealed some highly conserved sequences, but also revealed distinct differences between the two phylogenetically-distant damsel bug subfamilies. The utility of the CR as a phylogenetic marker should be most effective at lower taxonomic ranks (subfamily level and below).
Molecular Phylogeny
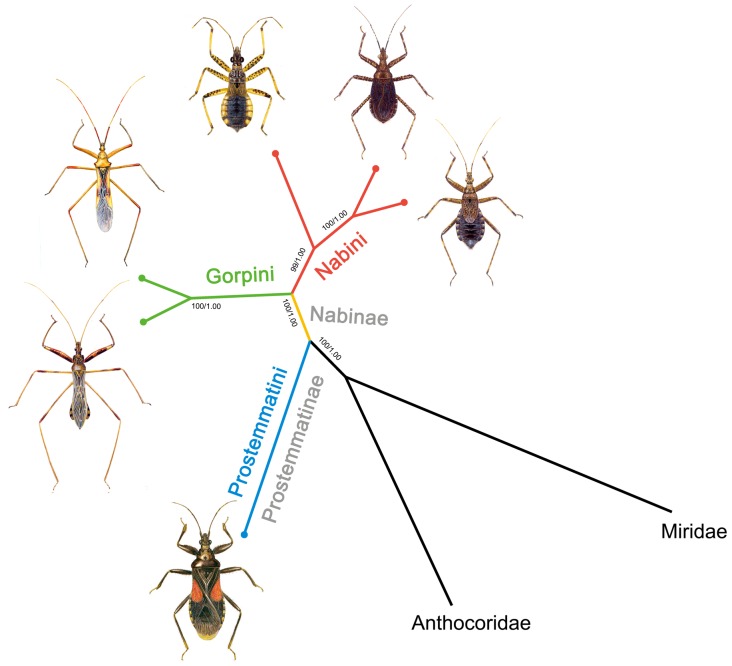
Phylogenetic analysis was performed on a concatenated nucleotide dataset of all PCGs, rRNAs and 19 tRNAs (excluding trnI, trnQ and trnM). The trees from the ML and Baysian analysis shared identical topologies and high node support values (Figure 4). The results indicated the sister-group relationships between A. bakeri (subfamily Prostemmatinae) and the other five species (subfamily Nabinae). Within the subfamily Nabinae the tribes Gorpini and Nabini were seen as sister groups. Within the tribe Nabini, the genus Nabis formed a sister group with the remaining Himacerus species.
Figure 4. Molecular phylogenetic tree of six damsel bugs.
Phylogenetic analysis was based on all mitochondrial genes (excluding trnI, trnQ and trnM). The tree was rooted with two outgroup taxa (O. niger and L. lineolaris). Numbers close to the branching points are percentages of ML bootstrap support values (left) and Bayesian posterior probabilities (right).
The relationship of three tribes from two subfamilies of Nabidae was well represented by the mitogenome data and was consistent with current phylogeny of the family Nabidae constructed mainly from morphological traits. Nabidae included two subfamilies Prostemmatinae and Nabinae, and five tribes [13]. Prostemmatinae are ground living, more stout-bodied and bright color (red and black), and display the ancestral type of karyotype (high chromosome number) [52]. This subfamily includes two tribes, Phorticini and Prostemmatini, and is considered the basal lineage in Nabidae based on their morphological and ecological traits (Figure S7). Our phylogenetic analysis based on the mitogenome sequence validates the basal position of Prostemmatini among three tribes. In addition, comparative analysis of mtDNAs also provided a genome level perspective that the architectures of Gorpini and Nabini mtDNAs were more closely related and similar, in comparison to the Prostemmatini. These similarities include having an apparently large NC region at trnI-trnQ and/or trnS2-nad, and three conserved sequence blocks (A+T-rich, C-rich and G+C-rich block) in CR (Figure S7).
Overall, nucleotide diversity and genomic traits make the mitogenomes a promising marker for resolving phylogenetic relationships at various taxonomic levels. In this comparative mitogenomic analysis, our understanding of the phylogeny of Nabidae is improved by the inclusion of representatives from three tribes and the two subfamilies.
Summary
Sequenced mitogenomes of three nabid tribes shared strong consistent nucleotide composition and base bias, except the CR showed large variation in AT- and GC-skews. The pattern of codon usage was influenced by the genomic GC content and consistent in three tribes with the standard invertebrate mitochondrial genetic code and the preference for A+T-rich codons. The comparison among orthologous PCGs showed that different genes have been subject to different rates of molecular evolution that are correlated with the GC content.
In most nabid tRNAs, stems and anticodon loops were extremely conserved, and nucleotide substitutions were largely restricted to TψC and DHU loops and extra arms, with apparent insertion-deletion polymorphisms. Comparative studies showed similar rates of substitution between the two rRNAs and some helices with high variability at the primary sequences shared good structural covariation. Architecture of six nabid mitogenomes exhibited substantial differences between the two subfamilies. Large NC regions at trnI-trnQ and/or trnS2-nad1 were present in all known nabid mtDNAs except N. apicalis in which trnI, trnQ, and trnM were missing. Comparative analysis of CRs revealed some highly conserved sequence blocks as well as distinct differences in different tribes and subfamilies. Overall, the phylogenetic analysis using mitogenomes are consistent with the current phylogeny of the family Nabidae derived mainly from morphological traits. Our study has demonstrated that nucleotide diversity and mitogenomic traits have the potential to provide insights of phylogenetic relationships at the subfamily level.
Materials and Methods
Ethics Statement
No specific permits were required for the insect collected for this study. The insect specimens were collected from road side vegetation by sweeping. The field collections did not involve endangered or protected species. The species in the family of Nabidae are common insects and are not included in the “List of Protected Animals in China”.
Samples and DNA Extraction
All species were collected in China between 2007 and 2009 (Table S6). All collections were initially preserved in 95% ethanol in the field, and transferred to −20°C for long-term storage at the China Agricultural University (CAU). For each species, the genomic DNA was extracted from one male adult’s muscle tissues of the thorax using a CTAB-based protocol [53].
PCR Amplification and Sequencing
The mitogenome was generated by amplification of overlapping PCR fragments (Table S7) using a range of universal insect mitochondrial primers [8]. Species-specific primers were designed based on the sequenced fragments to bridge gaps. PCR and sequencing reactions were conducted following [14], [20], [39].
Annotation and Bioinformatics Analysis
PCGs and rRNAs were initially identified using BLAST searches in GenBank and subsequently by alignment with genes of other true bugs. tRNAs were identified by tRNAscan-SE Search Server v.1.21 [54]. Some tRNA genes that could not be determined by tRNAscan-SE were determined in the unannotated regions by sequence similarity to tRNAs of other heteropterans. The base composition, codon usage, and nucleotide substitution were analyzed with Mega 5.0 [55]. AT-skew = (A−T)/(A+T) and GC- skew = (G−C)/(G+C) were used to measure the base-compositional difference between genes [56].
The software packages DnaSP 5.0 [57] was used to calculate the number of synonymous substitutions per synonymous site (Ks), the number of nonsynonymous substitutions per nonsynonymous site (Ka), the effective number of codons (ENC) and codon bias index (CBI) for PCGs.
Construction of Secondary Structures of RNAs and Non-coding Regions
Secondary structures of the small and large subunits of rRNAs were inferred using models predicted for A. bakeri [14] and other species of insects [36]–[39]. Stem-loops were named according to the convention of [36], [37]. Regions lacking significant homology and other non-coding regions were folded using Mfold [58].
Phylogenetic Analysis
Six nabid species with complete or nearly complete mitogenomes were used in the phylogenetic analysis, representing two subfamilies and three tribes (Table 1). Two species, Orius niger from Anthocoridae and Lygus lineolaris from Miridae, were selected for outgroup comparisons.
Table 1. List of the species included in the present study.
| Classification | Species | Accession No. | Reference |
| Anthocoridae | Orius niger | NC_012429 | [40] |
| Miridae | Lygus lineolaris | EU401991* | Direct submission |
| Nabidae | |||
| Prostemminae: Prostemmatini | Alloeorhynchus bakeri | HM235722 | [14] |
| Nabinae: Gorpini | Gorpis annulatus | JF907591 | Present study |
| Nabinae: Gorpini | Gorpis humeralis | JF927830 | Present study |
| Nabinae: Nabini | Himacerus apterus | JF927831* | Present study |
| Nabinae: Nabini | Himacerus nodipes | JF927832* | Present study |
| Nabinae: Nabini | Nabis apicalis | JF907590 | Present study |
“*”nearly complete mitogenomes. All accession numbers were retrieved from GenBank.
The complete sequences of each gene were used for phylogenetic analysis (excluding stop codons of the PCGs, trnI, trnQ and trnM). All PCGs were aligned based on amino acid sequence alignments in MEGA version 5.0 [55]. The rRNAs and tRNAs were aligned with Clustal X version 2.07 [59]. tRNA alignments were corrected according to secondary structure. Alignments of individual genes were then concatenated as a combined matrix. Model selection was based on Modeltest 3.7 [60] for nucleotide sequences. According to the Akaike information criterion, the GTR+I+G model was optimal for analysis with nucleotide alignments. MrBayes Version 3.2.1 [61] and a PHYML online web server [62], [63] were employed to reconstruct the phylogenetic trees. In Bayesian inference, two simultaneous runs of 3,000,000 generations were conducted for the matrix. Each set was sampled every 1000 generations with a burn-in of 25%. Trees inferred prior to stationarity were discarded as burn- in, and the remaining trees were used to construct a 50% majority-rule consensus tree. In ML analysis, the parameters were estimated during analysis and the node support values were assessed by bootstrap resampling (BP) [64] calculated using 100 replicates.
Supporting Information
Nucleotide composition of six nabid mitogenomes. (A) AT content; (B) AT- skew; (C) GC-skew. The values are shown for the J-strand of the whole genome, its concatenated genetic components (PCGs, tRNAs and rRNAs), 3rd codon positions in PCGs, and the control region (CR). Species are abbreviated as following: AB, A. bakeri; NA, N. apicalis; GA, G. annulatus; GH, G. humeralis; HA, H. apterus; HN, H. nodipes.
(TIF)
Codon distribution in six nabid mtDNAs. Numbers to the left refer to the total number of codon. Codon families are provided on the x axis. Most frequently used codons are highlighted in red.
(TIF)
Inferred secondary structure of tRNA families in six nabid mtDNAs. The nucleotide substitution pattern for each tRNA family was modeled using as reference the structure determined for G. annulatus. The identical nucleotides in all six nabid mtDNAs are showed by grey circles. Not conserved nucleotides are highlighted by blue circles. The tRNAs are labeled with the abbreviations of their corresponding amino acids. Inferred Watson-Crick bonds are illustrated by lines, whereas GU bonds are illustrated by dots.
(TIF)
Alignment of tRNA families ( trnA - trnL1 ) in six nabid mtDNAs. The loop regions are highlighted by the black pane.
(TIFF)
Alignment of tRNA families ( trnL2 - trnV ) in six nabid mtDNAs. The loop regions are highlighted by the black pane.
(TIFF)
Nucleotide variation in six nabid mitochondrial rRNAs.
(TIF)
Mapping various traits onto the phylogenetic tree of six damsel bugs. Ecological, morphological and mitogenomic traits are mapped onto the phylogenetic tree inferred from the mitogenomic data.
(TIF)
Structural features of six nabid mitogenomes.
(DOC)
Codon distribution in six nabid mtDNAs.
(DOC)
Evolutionary rates of six nabid mitochondrial PCGs.
(DOC)
Statistics on NC sequences in six nabid mitogenomes.
(DOC)
Occurrence of tandem repetitions in six nabid mtDNA NC regions.
(DOC)
Information of five sequenced nabid species included in the present study.
(DOC)
Primer sequences used in this study.
(DOC)
Acknowledgments
Special thanks go to John J. Obrycki (University of Kentucky) for his comments on an earlier draft.
Funding Statement
This research is supported by grants from the National Natural Science Foundation of China (Nos. 30825006, 30970394, 31061160186, 31111140015), the Special Fund for Agroscientific Research in the Public Interest (Nos. 201103012, 201103022), the Natural Science Foundation of Beijing (No. 6112013), the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences (No. O529YX5105), and College of Agriculture, University of Kentucky. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141: 173–216. [DOI] [PubMed] [Google Scholar]
- 3. Shao R, Kirkness EF, Barker SC (2009) The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus . Genome Res 19: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP (2011) Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera). BMC Genomics 12: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei DD, Shao R, Yuan ML, Dou W, Barker SC, et al. (2012) The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS ONE 7: e33973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dowton M, Castro LR, Austin AD (2002) Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates the examination of genome 'morphology'. Invertebr Syst 16: 345–356. [Google Scholar]
- 7. Boore JL (2006) The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol 21: 439–446. [DOI] [PubMed] [Google Scholar]
- 8. Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT (2006) Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst 37: 545–579. [Google Scholar]
- 9. Masta SE, Boore JL (2008) Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol Biol Evol 25: 949–959. [DOI] [PubMed] [Google Scholar]
- 10. Gissi C, Iannelli F, Pesole G (2008) Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101: 301–320. [DOI] [PubMed] [Google Scholar]
- 11.Schuh RT, Slater JA (1995) True bugs of the world (Hemiptera: Heteroptera). Classification and natural history. Ithaca: Cornell University Press.
- 12. Schuh RT, Štys P (1991) Phylogenetic analysis of cimicomorphan family relationships (Heteroptera). J N Y Entomol Soc 99: 298–350. [Google Scholar]
- 13. Lattin JD (1989) Bionomics of the Nabidae. Annu Rev Entomol 34: 383–400. [DOI] [PubMed] [Google Scholar]
- 14. Li H, Liu HY, Cao LM, Shi AM, Yang HL, et al. (2012) The complete mitochondrial genome of the damsel bug Alloeorhynchus bakeri (Hemiptera: Nabidae). Int J Bio Sci 8: 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stewart JB, Beckenbach AT (2005) Insect mitochondrial genomics: the complete mitochondrial genome sequence of the meadow spittlebug Philaenus spumarius (Hemiptera: Auchenorrhyncha: Cercopoidae). Genome 48: 46–54. [DOI] [PubMed] [Google Scholar]
- 16. Clary DO, Wolstenholme DR (1985) The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization and genetic code. J Mol Evol 22: 252–271. [DOI] [PubMed] [Google Scholar]
- 17. Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- 18. Sheffield NC, Song H, Cameron SL, Whiting MF (2008) A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol 25: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX (2010) Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to holometabolous insects. BMC Genomics 11: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H, Liu H, Shi AM, Štys P, Zhou XG, et al. (2012) The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS ONE 7: e29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morton BR (1993) Chloroplast DNA codon use: Evidence for selection at the psb A locus based on tRNA availability. J Mol Evol 37: 273–280. [DOI] [PubMed] [Google Scholar]
- 22. Wright F (1990) The “effective number of codons” used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- 23. Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH (2004) Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci USA 101: 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knight RD, Freeland SJ, Landweber LF (2001) A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol 2: RESEARCH0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herschberg R, Petrov DA (2008) Selection on codon bias. Annu Rev Genet 42: 287–299. [DOI] [PubMed] [Google Scholar]
- 26. Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- 27. Schmidt TR, Wu W, Goodman M, Grossman LI (2001) Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol Biol Evol 18: 563–569. [DOI] [PubMed] [Google Scholar]
- 28. Zsurka G, Kudina T, Peeva V, Hallmann K, Elger CE, et al. (2010) Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans. BMC Evol Biol 10: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hua JM, Li M, Dong PZ, Cui Y, Xie Q, et al. (2008) Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genomics 9: 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Negrisolo E, Babbucci M, Patarnello T (2011) The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC Genomics 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lavrov DV, Brown WM, Boore JL (2000) A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus . Proc Natl Acad Sci USA 97: 13738–13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masta SE, Boore JL (2004) The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol 21: 893. [DOI] [PubMed] [Google Scholar]
- 33. Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, et al. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fenn JD, Song H, Cameron SL, Whiting MF (2008) A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol Phylogenet Evol 49: 59–68. [DOI] [PubMed] [Google Scholar]
- 35. Miya M, Satoh TP, Nishida M (2005) The phylogenetic position of toadfishes (order Batrachoidiformes) in the higher ray-finned fish as inferred from partitioned Bayesian analysis of 102 whole mitochondrial genome sequences. Biol J Linn Soc Lond 85: 289–306. [Google Scholar]
- 36. Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR (2006) Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): Structure, organization and retrotransposable elements. Insect Mol Biol 15: 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cameron SL, Whiting MF (2008) The complete mitochondrial genome of the tobacco hornworm, Manduca sexta (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408: 112–123. [DOI] [PubMed] [Google Scholar]
- 38. Zhou ZJ, Huang Y, Shi FM (2007) The mitochondrial genome of Ruspolia dubia (Orthoptera: Conocephalidae) contains a short A+T–rich region of 70 bp in length. Genome 50: 855–866. [DOI] [PubMed] [Google Scholar]
- 39. Li H, Gao JY, Liu HY, Liu H, Liang AP, et al. (2011) The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae). Int J Bio Sci 7: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hua JM, Li M, Dong PZ, Cui Y, Xie Q, et al. (2009) Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): evidence from mitochondrial genomes. BMC Evol Biol 9: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Margam VM, Coates BS, Hellmich RL, Agunbiade T, Seufferheld MJ, et al. (2011) Mitochondrial genome sequence and expression profiling for the legume pod borer Maruca vitrata (Lepidoptera: Crambidae). PLoS ONE 6: e16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim MJ, Kang AR, Jeong HC, Kim KG, Kim I (2011) Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol Phylogenet Evol 61: 436–445. [DOI] [PubMed] [Google Scholar]
- 43. Hu J, Zhang DX, Hao JS, Huang DY, Cameron S, et al. (2010) The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): sequence, gene organization and a unique tRNA translocation event. Mol Biol Rep 37: 3431–3438. [DOI] [PubMed] [Google Scholar]
- 44. Vila M, Bjorklund M (2004) The utility of the neglected mitochondrial control region for evolutionary studies in Lepidoptera (Insecta). J Mol Evol 58: 280–290. [DOI] [PubMed] [Google Scholar]
- 45. Roberti M, Polosa PL, Bruni F, Musicco C, Gadaleta MN, et al. (2003) DmTTF, a novel mitochondrial transcription termination factor that recognizes two sequence in Drosophila melanogaster mitochondrial DNA. Nucleic Acid Res 31: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang DX, Hewitt GM (1997) Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol 25: 99–120. [Google Scholar]
- 47. Comas D, Pääbo S, Bertranpetit J (1995) Heteroplasmy in the control region of human mitochondrial DNA. Genome Res 5: 89–90. [DOI] [PubMed] [Google Scholar]
- 48. Fenn JD, Cameron SL, Whiting MF (2007) The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol Biol 16: 239–252. [DOI] [PubMed] [Google Scholar]
- 49. Dueñas JC, Gardenal CN, Llinás GA, Panzetta-Dutari GM (2006) Structural organization of the mitochondrial DNA control region in Aedes aegypti. . Genome 49: 931–937. [DOI] [PubMed] [Google Scholar]
- 50. Clary DO, Wolstenholme DR (1987) Drosophila mitochondrial DNA: Conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA. J Mol Evol 25: 116–125. [DOI] [PubMed] [Google Scholar]
- 51. Zhang DX, Szymura JM, Hewitt GM (1995) Evolution and structural conservation of the control region of insect mitochondrial DNA. J Mol Evol 40: 382–391. [DOI] [PubMed] [Google Scholar]
- 52. Nokkala C, Kuznetsova V, Grozeva S, Nokkala S (2007) Direction of karyotype evolution in the bug family Nabidae (Heteroptera): New evidence from 18S rDNA analysis. Eur J Entomol 104: 661–665. [Google Scholar]
- 53. Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 25: 4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lowe TM, Eddy SR (1997) tRNAscan–SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41: 353–358. [DOI] [PubMed] [Google Scholar]
- 57. Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 58. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 60. Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 61. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 62. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 63. Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide composition of six nabid mitogenomes. (A) AT content; (B) AT- skew; (C) GC-skew. The values are shown for the J-strand of the whole genome, its concatenated genetic components (PCGs, tRNAs and rRNAs), 3rd codon positions in PCGs, and the control region (CR). Species are abbreviated as following: AB, A. bakeri; NA, N. apicalis; GA, G. annulatus; GH, G. humeralis; HA, H. apterus; HN, H. nodipes.
(TIF)
Codon distribution in six nabid mtDNAs. Numbers to the left refer to the total number of codon. Codon families are provided on the x axis. Most frequently used codons are highlighted in red.
(TIF)
Inferred secondary structure of tRNA families in six nabid mtDNAs. The nucleotide substitution pattern for each tRNA family was modeled using as reference the structure determined for G. annulatus. The identical nucleotides in all six nabid mtDNAs are showed by grey circles. Not conserved nucleotides are highlighted by blue circles. The tRNAs are labeled with the abbreviations of their corresponding amino acids. Inferred Watson-Crick bonds are illustrated by lines, whereas GU bonds are illustrated by dots.
(TIF)
Alignment of tRNA families ( trnA - trnL1 ) in six nabid mtDNAs. The loop regions are highlighted by the black pane.
(TIFF)
Alignment of tRNA families ( trnL2 - trnV ) in six nabid mtDNAs. The loop regions are highlighted by the black pane.
(TIFF)
Nucleotide variation in six nabid mitochondrial rRNAs.
(TIF)
Mapping various traits onto the phylogenetic tree of six damsel bugs. Ecological, morphological and mitogenomic traits are mapped onto the phylogenetic tree inferred from the mitogenomic data.
(TIF)
Structural features of six nabid mitogenomes.
(DOC)
Codon distribution in six nabid mtDNAs.
(DOC)
Evolutionary rates of six nabid mitochondrial PCGs.
(DOC)
Statistics on NC sequences in six nabid mitogenomes.
(DOC)
Occurrence of tandem repetitions in six nabid mtDNA NC regions.
(DOC)
Information of five sequenced nabid species included in the present study.
(DOC)
Primer sequences used in this study.
(DOC)