Abstract
Set3 complex (Set3C) binds histone H3 dimethylated at lysine 4 (H3K4me2) to mediate deacetylation of histones in 5′ transcribed regions. To discern how Set3C affects gene expression, genome-wide transcription was analyzed in yeast undergoing a series of carbon source shifts. Deleting SET3 primarily caused changes during transition periods, as genes were induced or repressed. Surprisingly, a majority of Set3-affected genes are overlapped by non-coding RNA (ncRNA) transcription. Many Set3-repressed genes have H3K4me2 instead of me3 over promoter regions, due either to reduced H3K4me3 or ncRNA transcription from distal or anti-sense promoters. The resulting deacetylation by Set3C slows gene induction. Set3C represses internal cryptic promoters, but in different regions of genes than the Set2/Rpd3S pathway. Finally, Set3C stimulates some genes by repressing an overlapping antagonistic anti-sense transcript. These results suggest that Set3C and overlapping non-coding transcription can cooperate to fine tune the response kinetics of individual genes.
Introduction
Posttranslational modifications of histones play important roles in eukaryotic gene regulation (Kouzarides, 2007; Li et al., 2007a). Acetylation promotes efficient transcription by affecting the interaction between DNA and histones as well as recruiting downstream regulators of chromatin structure and transcription. Histone acetylation levels are controlled by the antagonistic functions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Interestingly, several of these enzymes are recruited by specific histone methylations that are linked to RNA Polymerase II (RNApII) transcription (Buratowski and Kim, 2011).
In budding yeast, Set1 and Set2 methylate H3K4 and H3K36, respectively (Krogan et al., 2003; Ng et al., 2003; Strahl et al., 2002). These methyltransferases bind specific phosphorylated forms of the RNApII subunit Rpb1 C-terminal domain (CTD), resulting in cotranscriptional methylation (Krogan et al., 2003; Ng et al., 2003). Near 5′ ends of active genes, serine 5 phosphorylated CTD helps recruit the Set1 complex (known as Set1C or COMPASS) to methylate H3 K4 (Ng et al., 2003). While Set1 is the only H3K4 methyltransferase in budding yeast, it produces different methylation states at different positions along genes. H3K4me3 is strongest near transcription start sites while H3K4me2 is enriched in 5′ transcribed regions (Kirmizis et al., 2007; Liu et al., 2005; Pokholok et al., 2005). Complementary to Set1, Set2 methyltransferase binds CTD phosphorylated at both Serine 2 and 5 to target H3 K36me2 and me3 to throughout transcribed regions but biased towards 3′ ends (Kizer et al., 2005; Krogan et al., 2003; Li et al., 2002). These methylation patterns are well conserved in all eukaryotes.
Both H3K4 and K36 methylation appear to function primarily by modulating histone acetylation (Buratowski and Kim, 2011). In 3′ transcribed regions, H3K36me by Set2 targets histone deacetylation by Rpd3 small complex, Rpd3S (Carrozza et al., 2005; Keogh et al., 2005; Li et al., 2007b). The Set2-Rpd3S pathway negatively affects transcription elongation and represses cryptic promoters within coding regions (Carrozza et al., 2005; Keogh et al., 2005; Li et al., 2007c). H3K4me3 is highly correlated with histone acetylation at promoter proximal regions, in part due to its ability to interact with PHD finger subunits of HATs and HDACs (Hung et al., 2009; Shi et al., 2006; Shi et al., 2007; Taverna et al., 2006). In higher eukaryotes, the TFIID subunit TAF3 and the chromatin remodeler Chd1 also interact with H3K4me3 (Sims et al., 2005; Vermeulen et al., 2007). The function of H3 K4me1 remains unclear.
In between the peaks of H3K4me3 and H3K36me3, H3K4me2 provides a binding site for the Set3 PHD finger protein (Kim and Buratowski, 2009). The Set3 complex (Set3C) contains two histone deacetylase (HDAC) subunits, Hos2 and Hst1 (Pijnappel et al., 2001) and deacetylates histones in 5′ transcribed regions (Kim and Buratowski, 2009). It has been proposed that Set3C is also recruited to transcribed regions through interaction with the phosphorylated CTD of RNApII, but nonetheless requires H3K4me2 to deacetylate histones (Govind et al., 2010). The function of the Set3 SET domain is still unclear and no methyltransferase activity has yet been reported. SET3 deletion genetically interacts with many chromatin factors involved in transcription regulation including Set2, SWR/Htz1 complex, SAGA, and Rpd3 large complex (Rpd3L), implicating Set3C in transcription regulation (Collins et al., 2007; Krogan et al., 2003).
The biological function of Set3C has been unclear. Although histone deacetylation typically represses transcription, Set3C has been reported to be both repressive and activating. Set3C negatively regulates meiotic genes (Pijnappel et al., 2001), yet is also required for maximal induction of the GAL1 and INO1 genes (Hang and Smith, 2011; Kim and Buratowski, 2009; Wang et al., 2002). Surprisingly, overall genome-wide transcription patterns of cells grown in synthetic complete media with glucose are largely unaffected by Set3C deletion mutants (Lenstra et al., 2011). One interesting observation is that the previously reported effects of Set3C mutations were discovered in cells undergoing metabolic changes: when media conditions were changed or upon triggering sporulation (Hang and Smith, 2011; Kim and Buratowski, 2009; Pijnappel et al., 2001; Wang et al., 2002). Here we show that Set3C is important for modulating the kinetics of many transcriptional responses during carbon source changes. Interestingly, the majority of Set3 target genes have overlapping ncRNA transcription; either Cryptic Unstable Transcripts (CUTs), Stable Uncharacterized Transcripts (SUTs), Xrn1-sensitive Unstable Transcripts (XUTs) (Lenstra et al., 2011; van Dijk et al., 2011; Xu et al., 2009) or internal cryptic initiation sites. We propose that overlapping non-coding transcription contributes to Set3C-mediated effects by altering histone methylation and acetylation patterns. Unlike activators or repressors that act as an on/off switch of transcription, Set3C instead plays an important role in the timing of transcription response during gene activation or repression.
RESULTS
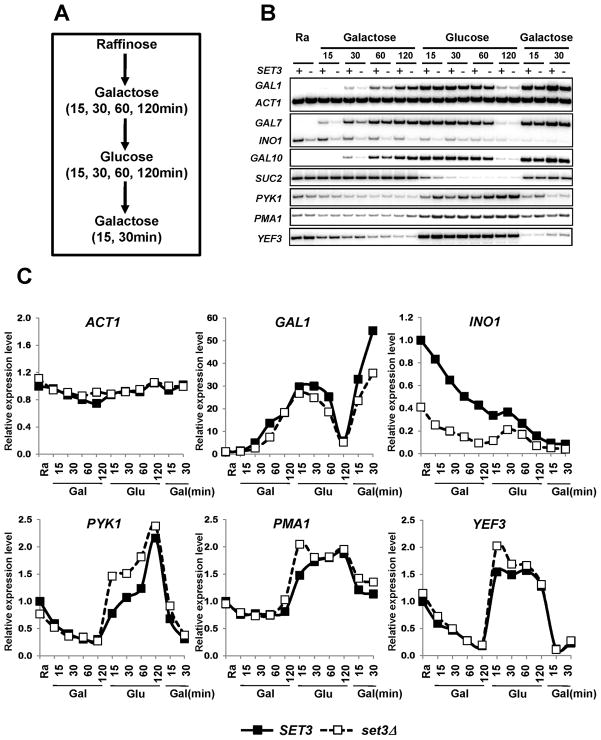
To explore the roles of Set3C, gene expression was analyzed in SET3 and set3Δ cells as they cycled through various carbon sources (Figure 1A). Cells were initially grown in Synthetic Complete medium with raffinose (SC-raffinose, 0 time point) and subsequently shifted to SC-galactose media. Samples were taken at 15, 30, 60, and 120 minutes for RNA analysis. Cells were then shifted to SC-glucose media for a similar 120 minute time course. Finally, cells were transferred back to SC-galactose for 15 and 30 minutes. Under these conditions, deletion of SET3 did not affect growth rates.
Figure 1. Set3 transiently regulates RNApII transcription.
A. Schematic representation of the time course experiments to monitor changes in mRNA levels upon carbon source shift. B. SET3 and set3Δ strains were grown in SC media containing raffinose and then sequentially shifted to SC media containing the indicated carbon sources for the times specified in the figure. + and − indicate SET3 and set3Δ, respectively. mRNA levels were determined by RT-PCR with two independent RNA samples. C. Quantitation from B. mRNA levels of SET3 cells in SC-raffinose media were set to 1.0. See also Figure S1.
To verify that the cells were responding as expected, levels of genes known to be regulated by carbon source were analyzed by RT-PCR. GAL genes were strongly induced in galactose media and repressed in glucose media (Figure 1B). GAL1, GAL7, and GAL10 mRNAs were still detected up to 60 minutes after transfer to glucose media. High level expression of SUC2 was observed in raffinose and galactose media, but this transcript was rapidly decreased in glucose media. The level of ACT1 mRNA changed very little during carbon source shifts and serves as an important normalization control (Figure 1B and 1C). As previously reported (Hang and Smith, 2011; Kim and Buratowski, 2009; Wang et al., 2002), set3Δ resulted in delayed induction of GAL1, GAL7, and GAL10 in galactose media, although levels equalized by 120 minutes (Figure 1B and 1C). INO1 level was strongly reduced by set3Δ (Figure 1B and 1C), consistent with its inositol auxotrophy (Ino-) phenotype (Villa-Garcia et al., 2011).
Set3C mutants have increased acetylation in 5′ transcribed regions of PYK1, PMA1, and YEF3, but transcription levels of these genes were unchanged under steady-state growth conditions (Kim and Buratowski, 2009). To test whether set3Δ changes the rate of activation or repression of these genes, transcript levels were monitored during carbon source shifts. In SET3 cells, these genes were down-regulated in galactose media and rapidly induced in glucose media (Figure 1B and 1C). While YEF3 mRNA levels were not strongly affected by loss of Set3, PYK1 and PMA1 transcripts were higher in set3Δ during glucose induction until they equalized at 120 minutes (Figure 1B and 1C). Therefore, as seen for GAL gene activation, the negative effect of Set3 on PYK1 and PMA1 appears transient.
To further study how the transcriptional changes upon carbon source shifts are affected by Set3C, total RNA was analyzed using high-resolution, strand-specific tiled microarrays (David et al., 2006). In wild type cells, approximately 800 genes showed significant changes relative to levels in raffinose media: 197 transcripts were increased specifically in galactose media and 94 in glucose media (Figure S1A, B). 509 transcripts were repressed in galactose relative to raffinose or glucose media (Figure S1C). In general, galactose-induced transcripts increased over the full two hour incubation, more slowly than glucose-induced genes, which were typically fully induced after 15 min (Figure S1). In all conditions, transcript levels of genes encoding transcription regulators such as GAL4, GAL3, GAL80, MIG1, and TUP1 were not affected by deletion of SET3 (Figure S2A).
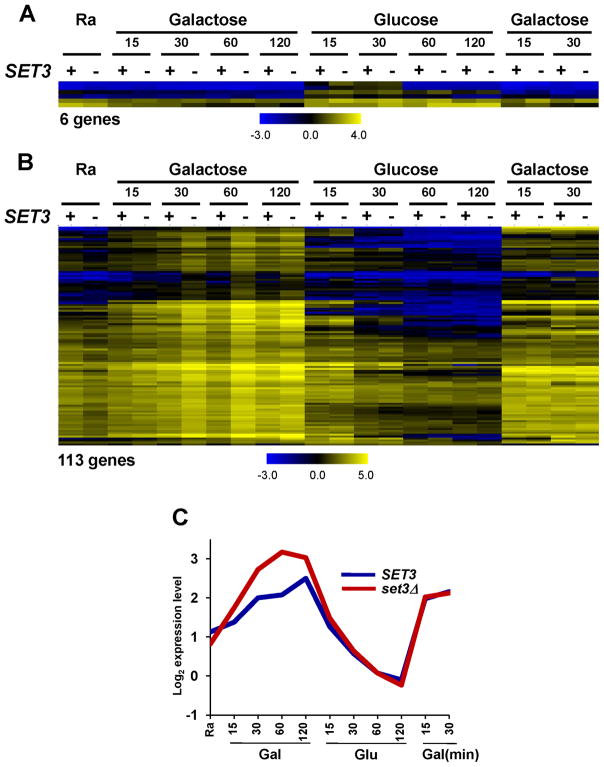
Set3-regulated genes were identified as those exhibiting at least 1.7-fold change in expression levels at one or more time points, and were further verified by visual inspection. From this analysis, 119 genes repressed by Set3C were identified. Six genes of this group were increased in glucose media (Figure 2A) and 113 genes were induced by galactose (Figure 2B). Although some genes showed differences at most or all time points, for most affected genes the greatest differences between SET3 and set3Δ occurred during the transition periods. This effect is clearly seen by calculating an averaged response pattern for the galactose-induced genes (Figure 2C).
Figure 2. Set3 negatively regulates the kinetics of transcription induction.
RNA samples from the time course in Figure 1 were analyzed by high-resolution tiling arrays. Normalized, log2-transformed mRNA expression data were visualized (color bar shows log2 scale) using the Multi Experiment Viewer program (http://www.tm4.org/). + and − indicate SET3 and set3Δ, respectively. A. Expression profiles of 6 genes upregulated in set3Δ during glucose induction. B. Expression profiles of 113 genes that are more rapidly induced in set3Δ during galactose induction. C. Averaged profile of expression signals of genes from part B. See also Figure S2.
The histone deacetylation activities of Set3C are important for its effects, since Set3-repressed genes tested were also up-regulated by hos2Δ or hst1Δ (Figure S2B). Interestingly, some genes responded more strongly to one or the other HDAC subunit, while others are sensitive to both. These gene-specific results likely reflect the different histone targets of the two HDACs (Kim and Buratowski, 2009) and show that they are non-redundant. Interaction between the Set3 PHD finger and H3K4me2 is important Set3C function, and a PHD finger mutant (W140A) that no longer binds H3K4me2 behaves similarly to set3Δ (Figure S2C). These results indicate that histone deacetylation mediates the effects of Set3C on the kinetics of transcription induction.
Remarkably, visual inspection of the array data showed that at least 61% of genes repressed by Set3 have an overlapping ncRNA (from the previously annotated CUT, SUT, or XUT classes, as well as several that are unannotated). This percentage increases to 74% for genes that at repressed 2-fold or more by Set3C. This is a minimal estimate, since there may be additional ncRNAs induced during the time course that were missed due to rapid degradation by exosome or other RNA degradation pathways. The fraction of ncRNAs seen at Set3-affected genes is much higher than in the overall genome (estimated to be about 12% by Xu et al. (2011)), suggesting that overlapping transcription may contribute to regulation by Set3C.
Upstream transcription can target Set3C to suppress gene induction
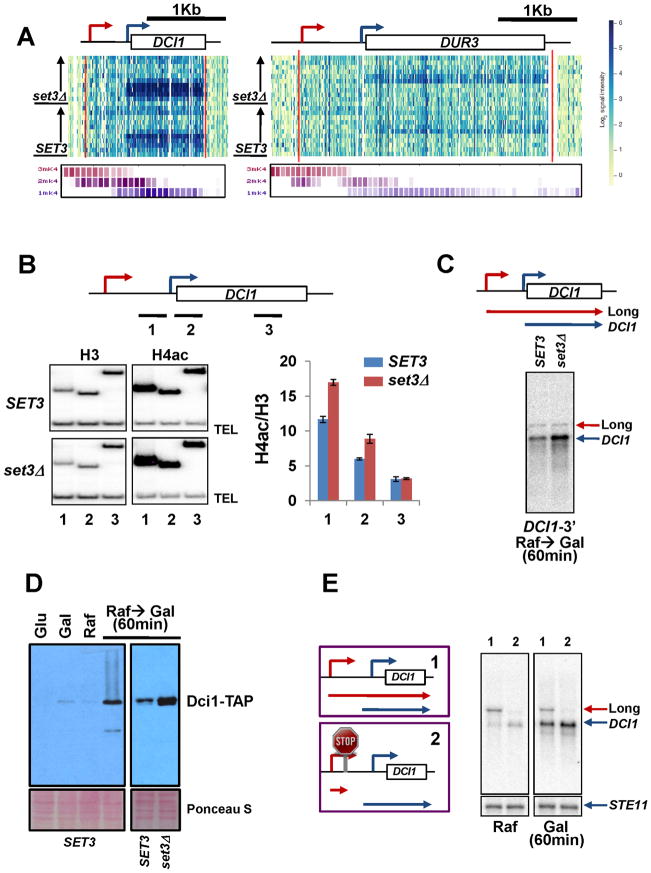
DCI1, FUN19, and ATH1 were induced in galactose media and DUR3 was transiently expressed in glucose media. These four Set3-repressed genes are annotated as having transcription start sites (TSSs) unusually far upstream (about 600 bp) of the open reading frame initiation codon (Xu et al., 2009). However, visual inspection of the array data showed that these genes have at least one additional TSS closer to the ORF (Figures 3A and S3A). The distal promoters are constitutively active, while the promoters just upstream of the ORFs are activated in specific media conditions. Interestingly, deletion of SET3 increased transcription from the proximal but not distal promoters (Figures 3A and S3A). Although H3K4me2 is typically localized in 5′ transcribed regions, genome-wide analyses of published histone methylation patterns (Kirmizis et al., 2007; Pokholok et al., 2005) show that H3K4me3 is enriched at the upstream promoter while a peak of H3K4me2 overlaps the downstream promoter (Figure 3A and Figure S3A).
Figure 3. Upstream ncRNA transcription can target H3K4me2 and Set3C repression to a downstream promoter.
A. DCI1 and DUR3 each have an ORF-distal promoter (red arrows in schematic at top) that is constitutively active, and proximal promoters (blue arrows) that are induced in specific media conditions. Transcription from the proximal promoter is increased in set3Δ. Figure shows the microarray hybridization signal for each probe arrayed at position along the gene as shown in schematic. Increased blue color designates more transcription. Red lines show the annotated start and stop of the mRNA (Xu et al., 2009), while white box shows the ORF position. The time course as in Figure 1A is arrayed from bottom to top for each strain as designated. Only the top strand signal is shown here. Histone methylation maps of DCI1 and DUR1 from cells grown in YPD are from Kirmizis et al. (Kirmizis et al., 2007) and generated by the associated website (http://www.gurdon.cam.ac.uk/~kouzarideslab/H3R2methylation.html). Each K4 methylation state is shown as a separate line of boxes representing individual probes. The intensity of color shows ChIP signal strength. The distal promoter has high levels of H3K4me3 while H3K4me2 is enriched over the proximal promoter. B. Set3 deacetylates histones in the core promoter of DCI1. Crosslinked chromatin from SET3 or set3Δ strains grown in YPD was precipitated with anti-H3 or anti-acetyl H4 as indicated. PCR analysis of the precipitated DNA was carried out on the DCI1 gene (numbered primer locations shown schematically at the top, PCR products below). TEL is from a non-transcribed region near the telomere of chromosome VI. The signals for acetyl H4 were quantitated and normalized to the total H3 signal, and the ratios were graphed (x axis shows primer number, and y axis shows ratio). Error bars show the standard deviation. C. Northern blot analysis of DCI1 transcripts with a 3′ strand-specific DNA probe. Cells were grown in SC-raffinose media and shifted to SC-galactose media for 60 minutes. Bottom panel shows two transcripts of DCI1 detected by Northern blot analysis, which are schematicized at top. D. Total proteins were extracted from cells grown in the indicated carbon sources, either for an extended time or after a 60 minute shift as indicated. Dci1-TAP protein was analyzed by immunoblot analysis. Membrane was stained with Ponceau S for loading control (bottom). E. Strains with or without the CYC1 terminator inserted between the DCI1 promoters (left panel) were grown in SC-raffinose media and shifted to SC-galactose media for 60 minutes. Right panel shows DCI1 Northern blot analysis, with transcripts schematicized in left panel. STE11 is used for a loading control (bottom). See also Figure S3.
To test whether the effect of Set3C is mediated by deacetylation of the downstream promoter, histone acetylation at DCI1 was analyzed by chromatin immunoprecipitation (ChIP) using an antibody that recognizes tetra-acetyl H4 (H4ac). Levels of acetylation were normalized to total histone content as measured by H3. Relative to SET3 cells, a set3Δ strain had increased acetylation at the downstream promoter but not the 3′ end of DCI1 (Figure 3B). This result is consistent with a model by which H3K4me2 directed by the upstream transcription unit targets Set3C deacetylation for repression of the downstream promoter.
Interestingly, although the distal promoters of these 4 genes produce transcripts in rich media, a genome-wide GFP-fusion analysis failed to detect any protein products in these growth conditions (Ghaemmaghami et al., 2003; Xu et al., 2009). Therefore, the upstream transcripts appear to be long ncRNAs. Northern blot analysis confirmed that the distal promoter of DCI1 produces a stable transcript. At 60 minutes of growth in galactose, a 3′ end DNA probe detected two transcripts. The long transcript level was unchanged but the shorter DCI1 transcript was significantly increased in set3Δ (Figure 3C). A similar increase was seen in set1Δ but not set2Δ cells, suggesting that H3K4 methylation but not H3K36 methylation is involved in this repression. At this gene, hst1Δ strongly increased the downstream DCI1 transcript but a weaker effect was seen in hos2Δ (Figure S3B).
Immunoblot analysis with a DCI1-TAP tagged strain showed that Dci1 protein is undetectable in glucose and seen at low levels in galactose or raffinose media (Figure 3D). However, it was strongly induced when cells were shifted from raffinose to galactose for 60 minutes, indicating that Dci1 protein is transiently expressed in galactose media, tracking levels of the short transcript. Also matching downstream transcript levels, Dci1 protein was increased in set3Δ (Figure 3D).
These results suggest that ncRNA transcription from an upstream distal promoter can place H3K4me2 over a downstream core promoter and that the resulting deacetylation by Set3C can inhibit or delay the expression of the downstream coding mRNA. To directly test this model, the CYC1 terminator was inserted between the two DCI1 promoters. This insertion abolished the long transcript and resulted in derepression of the shorter DCI1 transcript (Figure 3E). Also consistent with this model, H3K4me2 decreased and histone acetylation at the downstream promoter increased when upstream ncRNA transcription is blocked (Figure S3C and data not shown). These results indicate that ncRNA transcription from the DCI1 upstream promoter and Set3C cooperate to deacetylate chromatin at the downstream promoter to modulate gene induction.
Set3C represses cryptic promoters in 5′ transcribed regions
Passage of RNApII through a gene disrupts nucleosomes, which can cause cryptic transcription initiation within coding regions unless the chromatin is reassembled by elongation factors that include Spt6 and Spt16 (Cheung et al., 2008; Kaplan et al., 2003). Deacetylation of histones in 3′ transcribed regions by the Set2-Rpd3S pathway also contributes to repression of cryptic internal promoters at some genes (Carrozza et al., 2005; Li et al., 2007c). Since loss of Set3C increases acetylation in 5′ transcribed regions (Kim and Buratowski, 2009), it was pertinent to ask whether this pathway might also suppress cryptic promoters.
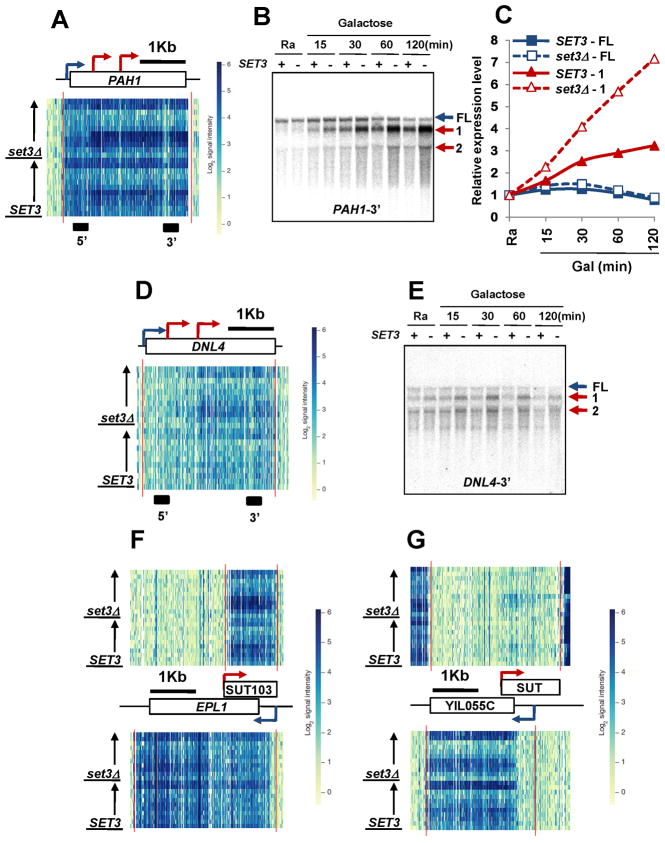
A small number of Set3-repressed genes exhibited increased sense-strand signals beginning within the open reading frame through the 3′ end, suggesting internal initiation (Figures 4A, D and S4A). For example, transcript hybridization intensities in 5′ regions of PAH1 and DNL4 were not strongly changed during the time course in either SET3 or set3Δ cells. However, during galactose incubation the signal in downstream regions increased, but even more strongly in cells lacking Set3C (Figure 4A, D). Northern blot analysis with strand-specific probes revealed several shorter transcripts that accumulated in galactose media, and these were markedly increased in set3Δ cells (Figure 4B, C, and E). Probing with 5′- or 3′-specific probes confirmed that the shorter transcripts arise from internal initiation within the coding regions (Figure S4C). As predicted, histone acetylation increased at the internal PAH1 promoter in set3Δ or hos2Δ cells (Figure S4B). Cells lacking Hos2 (Figure S4C) or carrying a Set3 PHD finger mutant (W140A, Figure S4D) also showed increased internal initiation within PAH1, indicating that recognition of H3K4me2 mediates repression of internal promoters. The PAH internal promoters were not derepressed by set2Δ (data not shown), proving that Set3 repression does not require H3K36 methylation.
Figure 4. Set3C represses cryptic promoters in 5′ transcribed regions.
A. Microarray expression profile at the PAH1 locus in SET3 and set3Δ cells. The position of the open reading frame is shown at top. Bars underneath indicate the probe positions used for Northern blotting here and Fig. S4C and D. Red and blue arrows indicate transcript start sites of short and full-length transcripts, respectively. B. Northern blot analysis of PAH1 with 3′ probe. FL indicates full-length transcript. 1 and 2 indicate short transcripts. + and − indicate SET3 and set3Δ, respectively. C. Quantitation from B. SET3 transcript levels in SC-raffinose media were set to 1.0. D. Expression data for the DNL4 locus. E. Northern blot analysis for the DNL4 gene as in B. F and G. Expression profiles for EPL1 and YIL055C loci. Red and blue arrows indicate putative transcript start sites of anti-sense transcripts and sense transcript, respectively. See also Figure S4.
Internal promoters can also be in the anti-sense direction relative to the mRNA. For example, the anti-sense transcript SUT103 overlaps the EPL1 promoter region (Figure 4F) and YIL055C has a previously uncharacterized 5′ overlapping anti-sense transcript that is expressed in galactose media (Figure 4G). The EPL1 and YIL055C transcripts were slightly increased, and the galactose-induced anti-sense transcripts were significantly higher in set3Δ cells (Figure 4F and G). Therefore, Set3C contributes to repression of cryptic promoters in 5′ transcribed regions that produce both sense and anti-sense transcripts.
Set3C-mediated repression is targeted by high H3K4me2 levels
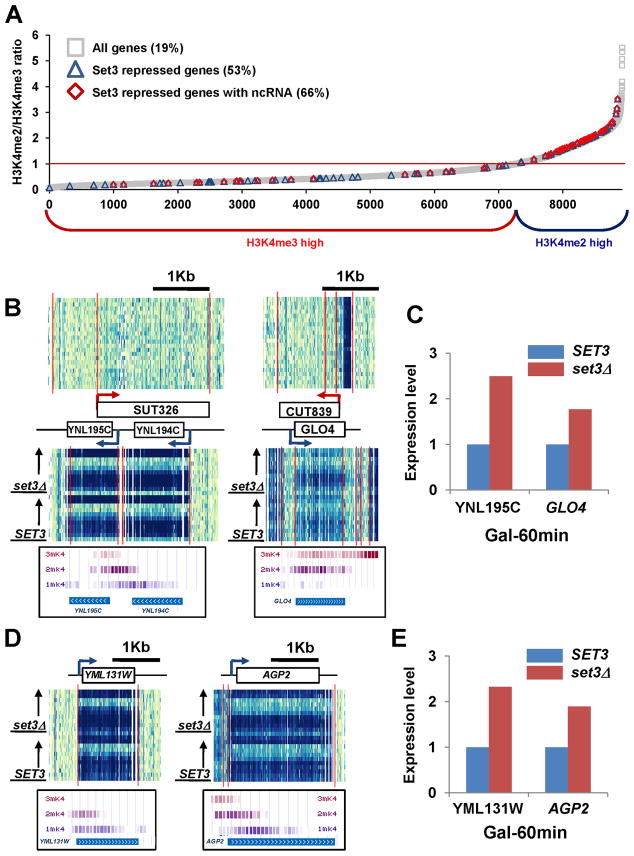
To further understand the connection between set3Δ effects and H3K4 methylation, the relative levels of H3K4me2 and me3 at 5′ ends of genes were analyzed (Kirmizis et al., 2007; Pokholok et al., 2005). Based on ChIP-chip data from Pokholok et al. (2005) for cells grown on glucose, the ratio of H3K4me2 to H3K4me3 signal at promoter regions is low at most genes, consistent with H3K4me3 enrichment at promoters. About 19% of total yeast genes show relatively higher ratios of H3K4me2 to H3K4me3. Importantly, for Set3-repressed promoters this percentage increases to 53%. Furthermore, minimally 66% of Set3-repressed promoters with overlapping ncRNA have an elevated ratio of H3K4me2 to H3K4me3. Therefore, higher H3K4me2 and ncRNA transcription correlate strongly with Set3C-mediated repression (Figure 5A).
Figure 5. H3K4me2 over promoters correlates with Set3C-mediated attenuation of transcriptional responses.
A. The ratio of H3K4me2 to H3K4me3 signal for all coding gene promoters (grey squares), Set3-repressed genes (119 genes, blue triangles), and Set3-repressed with ncRNA (73 genes, red diamonds) was plotted using data set from Pokholok et al. (2005). The percentage in parentheses indicates genes that have higher H3K4me2 relative to H3K4me3. B. Expression profiles for YNL195C and GLO4 loci. Red and blue arrows indicate putative transcript start sites of anti-sense transcripts and sense transcript, respectively. Histone methylation maps of these genes from cells grown in YPD were generated from the ChIP experiments of Kirmizis et al. (Kirmizis et al., 2007) as in Figure 3A (bottom panel). The ORF positions are shown in blue at the bottom, with white arrows showing orientation. Each K4 methylation state is shown as a separate line of boxes representing individual probes. The intensity of color shows ChIP signal strength. The promoter regions of YNL195C and GLO4 show high level of H3 K4me2. C. Expression level of these two genes at galactose 60 minute time point. mRNA levels of SET3 cells were set to 1.0. D. Expression profiles (upper panel) and histone methylation maps (bottom panel) of YML131W and AGP2 as in B. E. Expression level of these genes at galactose 60 minute time point. See also Figure S5.
Looking at some specific examples, the anti-sense transcripts SUT326 and CUT839 overlap YNL195C and GLO4 promoter regions, respectively (Figure 5B). Both mRNAs are repressed in glucose or raffinose, but are strongly induced in galactose, with even higher levels in set3Δ cells (Figure 5C). Published methylation data for cells grown in glucose reveal that the H3K4me3 and me2 peaks correspond to the overlapping ncRNA transcription (Kirmizis et al., 2007; Pokholok et al., 2005), resulting in H3K4me2 over the mRNA promoters (Figure 5B). This H3K4me2 will recruit Set3C deacetylase, explaining why induction of these two genes is stronger in set3Δ cells.
Another group of Set3C-repressed genes with high H3K4me2/me3 ratios over the promoter (Kirmizis et al., 2007; Pokholok et al., 2005) have no obvious overlapping transcripts. For example, YML131W and AGP2 had levels of H3K4me2 similar to other genes with comparable expression levels, but shifted upstream over the promoter. In contrast, H3K4me3 was significantly reduced or below the level of detection (Figure 5D). Both genes showed higher expression levels in set3Δ cells during galactose induction (Figure 5E). We hypothesized that H3K4me2 might be generated by targeted demethylation of H3K4me3. Deleting the gene for the H3K4 tri-demethylase Jhd2 (Liang et al., 2007) led to dramatically higher H3K4me3 at the Set3-repressed genes YML131W and AGP2, but not the GAL10 antisense transcript SUT013 (Figures S5A, B, and C). However, H3K4me2 levels were equally high at these genes in JHD2 and jhd2Δ cells, and both still responded to Set3 (Figure S5 and data not shown). Therefore, how H3K4me2 is targeted to these Set3 target promoters remains unclear. There may be overlapping transcription of ncRNAs that were unstable and therefore not detected, or there may be some other mechanism yet to be discovered.
Set3C positively affects some genes with anti-sense transcription
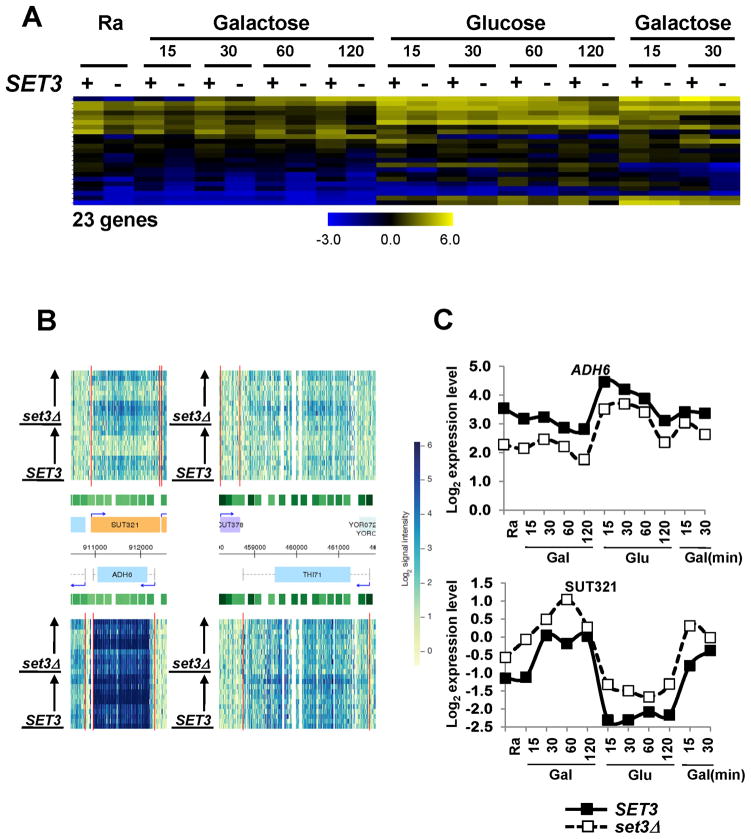
Although Set3C represses most of its target genes, it has a positive transcription effect on INO1 and several GAL genes (Figure 1B; (Pijnappel et al., 2001; Wang et al., 2002). Our microarray analysis identified 23 transcripts that require Set3 for maximal levels (Figure 6A). While the GAL genes, HXT1, and TIR3 showed delayed induction kinetics in set3Δ, most Set3-activated genes had significantly reduced transcript levels throughout the time course. In log phase cells grown in minimal media with glucose, Set3-activated genes also required the other Set3C subunits for maximal transcription (Lenstra et al., 2011). At least 74% of Set3-activated genes had an overlapping anti-sense non-coding transcript. These differed from the anti-sense pairs described above in that the ncRNA typically initiated near the 3′ end of the sense transcript, with the two promoters being outside each other’s zone of H3K4me2. These pairs were generally anti-correlated in their response to Set3. For example, sense transcript levels for ADH6, THI71, HOR2 and YKL187c were lower in set3Δ, while the overlapping anti-sense transcripts of these genes were increased in this mutant (Figures 6B, C and S6B). Similarly, GDH3 is a Set3-repressed gene, but its overlapping transcript SUT434 was decreased in set3Δ (Figure S6A).
Figure 6. Set3 represses antagonistic anti-sense transcription.
A. Hierarchical clustering of 23 genes that show reduced basal transcript levels or delayed induction in set3Δ cells. B. Expression profiles of ADH6 and THI71 loci. These genes were down-regulated while their overlapping transcripts were higher in set3Δ. C. Quantitation from B. Change in transcript level is indicated as log2 values. See also Figure S6.
Given the fact that Set3C is an HDAC, upregulation of genes by Set3 could be an indirect effect of Set3-mediated repression of an antagonistic anti-sense transcript. There are several examples of overlapping anti-sense transcription units that repress each other’s expression through the Set2/Rpd3S pathway (Hongay et al., 2006; Houseley et al., 2008; Pinskaya et al., 2009). If one of the promoters within such a pair was overlapped by a nearby third transcription unit, Set3 repression of that promoter would cause an apparent upregulation of the anti-sense transcript by Set3. Consistent with this model, we find that for many Set3-activated genes, the promoter of the overlapping anti-sense transcript is in turn overlapped by another transcript or has high H3K4me2 levels (Figures 6B and S6). In this model (Figure 7D), there is no need to invoke a separate activating function for Set3C.
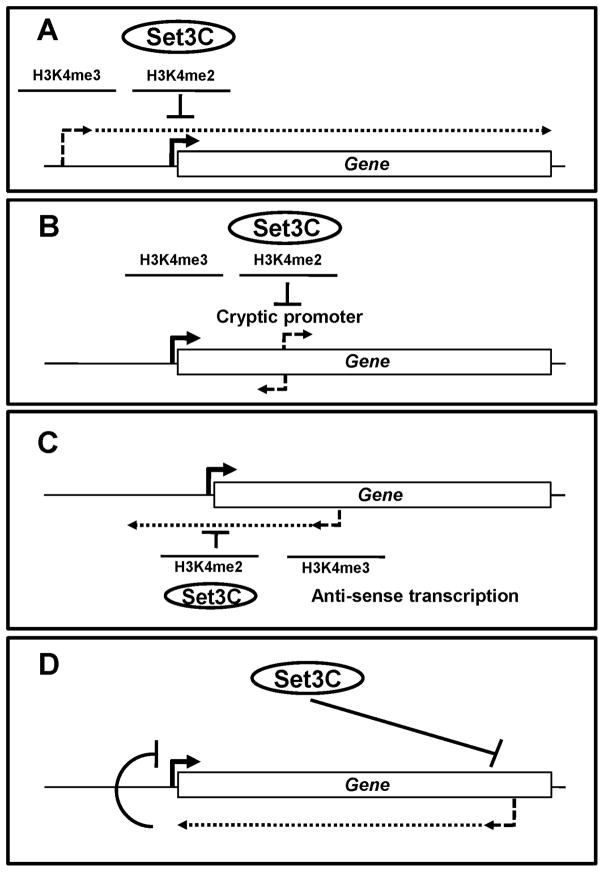
Figure 7. Models for regulation of transcription kinetics by Set3C.
A. Transcription from a distal promoter targets H3K4me2 to the proximal promoter of a downstream gene. Set3C deacetylates histones in the proximal promoter region, resulting in delayed or reduced induction. B. Deacetylation by Set3C represses cryptic promoters that produce either sense or anti-sense transcript in 5′ transcribed regions. C. Anti-sense transcription from an internal promoter or another mechanism generates H3K4me2 in the promoter region. This leads to deacetylation of this promoter by Set3C and attenuated induction. D. Set3 up-regulates genes by repressing anti-sense transcription. Repression of anti-sense transcription by Set3 leads to upregulation of overlapping sense transcription.
Discussion
In their natural environments, cells can experience rapidly changing conditions and must correspondingly change their gene expression patterns to adapt. For yeast cells, survival is dependent upon the ability to respond to different temperatures, environmental conditions, and available nutrients. Much of this gene regulation is carried out at the transcriptional level. We tend to think of gene-specific activators or repressors as on/off switches, so most gene expression analyses compare cells grown with different conditions for extended periods. However, it is also important to consider how the kinetics of transcriptional responses can affect fitness. While the immediate induction of some genes will be advantageous, for others it could be beneficial to delay expression until the inducing signal has persisted for some time. A delay mechanism can allow two genes to respond to the same activator, but with sequential expression. Here we show that the Set3C HDAC can combine with ncRNA transcription to delay or attenuate gene induction. In this model, overlapping transcription places H3K4me2 over the affected promoter, leading to deacetylation by Set3C and antagonism of gene activation (Figure 7).
Set3C does not strongly affect global transcription in steady state growth conditions (Lenstra et al., 2011), but our analysis of carbon source shifts shows that Set3C can affect the rate and magnitude of transcription transitions. Interestingly, the majority of genes affected by Set3 have an overlapping non-coding transcript. Although the configuration varies, a common theme is that a zone of H3K4me2 created by the overlapping transcription unit encompasses the Set3-repressed promoter.
At least four Set3-repressed genes have a long ncRNA produced from an upstream promoter, resulting in H3K4me2 over the downstream coding transcript promoter (Figures 3 and 7A). Other Set3-sensitive genes have internal promoters within the H3K4me2 zone (Figures 4 and 7B). Deacetylation of these promoters by Set3C inhibits their activation. In this respect, the Set1-H3K4me2-Set3C pathway bears some resemblance to the Set2-H3K36me-Rpd3S pathway (Carrozza et al., 2005; Cheung et al., 2008; Kaplan et al., 2003; Li et al., 2007c). Importantly, deletion of SET2 had no strong effect on cryptic initiation within Set3 target PAH1 and deletion of SET3 did not activate the cryptic promoters of STE11 or FLO8, targets of Set2-Rpd3S repression (Kim and Buratowski, 2009). Comparison of these and other affected genes suggests that Set3C and Rpd3S repress distinct sets of cryptic promoters, consistent with the fact that these HDACs act at different regions of genes. One interesting difference we note is that Set3-repressed internal promoters are generally active to some extent in wild-type cells, while the Set2-Rpd3S pathway appears to mediate more complete repression.
Although the transcripts produced from internal initiation sites are referred to as “cryptic”, the fact that they have persisted during evolution suggests they could have some function. In wild-type yeast, a subset of shorter transcripts is transiently induced following nutritional changes, and some of these internally initiated transcripts may produce N-terminally truncated proteins, functionally distinct from their full-length counterparts (Cheung et al., 2008; Nishizawa et al., 2008). We did not observe any truncated protein products for PAH1, but the Set3-repressed gene KCS1 has an internal internally initiated sense transcript that produces a short protein in low phosphate conditions (Nishizawa et al., 2008). Internal promoters can also create protein diversity in higher eukaryotes (Gomez-del Arco et al., 2010; Kowalczyk et al., 2012). Shorter sense-strand cryptic transcripts can also be non-coding, but instead function as “decoys” to relieve miRNA-dependent gene silencing of the longer mRNA (Leung and Sharp, 2010).
In many cases, the internal promoters of Set3-repressed genes are oriented anti-sense to the mRNA (Figures 5 and 7C). Deletion of Set3 causes these genes to be induced faster and to a greater extent than normal. Transcription from the internal promoter will generate a characteristic H3K4me3/me2 gradient in the opposite orientation to that created by sense transcription. Therefore, the overall methylation status reflects the relative strengths of the sense and anti-sense promoters, which are within each other’s zone of H3K4 dimethylation. This Set3C-mediated repression could occur at the level of promoter accessibility to initiation factors, but may also reflect inhibitory effects of deacetylation on early elongation as suggested recently (Weinberger et al, 2012).
While Set3 functions primarily as a repressor, a few genes show decreased expression in set3Δ cells (Figures 1 and 6A). Most of these genes have an overlapping transcript arising from its 3′ end that is repressed by Set3C. Genes that respond to environmental changes or stresses often have overlapping antisense transcription anti-correlated with sense transcription (Xu et al., 2011). If the promoters for the sense and anti-sense transcription units fall within each other’s zone of high H3K36 methylation, strong transcription in one direction can lead to deacetylation of the promoter driving transcription in the opposite direction. For example, under repressing conditions, anti-sense transcription from the 3′ end of GAL10 results in Rpd3S-dependent repression of the GAL10-GAL1 promoter (Houseley et al., 2008; Pinskaya et al., 2009). We suggest that Set3C indirectly promotes sense transcription at some genes by directly repressing the overlapping anti-sense transcription (Figure 7D). In many cases, a third transcription unit overlapping the Set3-repressed promoter could mediate this effect using the mechanisms described above.
Control of gene expression dynamics is very important. Responses to stimuli generally consist of “early” and “late” responding genes, often with the same transcription factor involved. “Fine-tuning” of gene induction can prevent premature responses if a signal appears only briefly, or can provide a mechanism by which genes can respond differentially to the same activator. Histone modifications can influence the kinetics as well as the magnitude of transcription changes. For example, the Rpd3L HDAC complex affects the dynamics of many stress-induced genes (Alejandro-Osorio et al., 2009; Ruiz-Roig et al., 2010).
There has been a great deal of recent attention to non-coding transcription. In many cases, the ncRNAs themselves modulate gene expression by acting as targeting or scaffolding subunits for proteins (Wang and Chang, 2011). However, we suggest that in some cases, the effects are mediated not by the ncRNA but by the co-transcriptional histone modifications that result from its expression. Bumgarner et al. (2009; 2012) recently described a set of overlapping transcripts that cause variegated expression of a yeast cell surface protein. This heterogeneous response requires the Rpd3L complex, and while it is not clear how this HDAC is targeted, this mechanism functions in cis. Similarly, the yeast SER3 gene is repressed in cis by non-coding transcription from an upstream promoter (Martens et al., 2004). Here we show that the majority of Set3C-responsive genes have overlapping non-coding transcription that places H3K4me2 over the affected promoter region. The resulting deacetylation attenuates the rate and magnitude of responses. An accompanying paper (van Werven et al., 2012) shows that Set3C and Set2 together mediate the repressive effects of long ncRNA transcription on the IME1 gene to regulate yeast sporulation. Given the conservation of chromatin modifying complexes over evolution, these types of gene circuits may help explain the prevalence of overlapping and anti-sense non-coding transcripts seen in all eukaryotes.
Experimental Procedures
Yeast Strains and Plasmids
Yeast strains and plasmids used in this study are listed in Supplemental Table S1. For time course experiments, matched SET3 and set3Δ strains were grown in 600 ml of synthetic complete media with raffinose (SC-raffinose) to an optical density at 600 nm of 0.5. For carbon source shifts, cells were collected by centrifugation at 4°C, quickly resuspended in a small volume of SC media lacking any carbon source, and then directly transferred to SC-media containing the indicated carbon source. 50 ml of cells were harvested by centrifugation at 15, 30, 60, and 120 minutes for RNA analysis.
To generate a strain that carries the CYC1 terminator between the two DCI1 promoters, the delitto perfetto strategy was used (Storici et al., 2001). Briefly, the kanMX4 and KlURA3 CORE cassette was integrated 430 bp upstream from the DCI1 ATG and the resulting strain was transformed with the 270 bp CYC1 terminator to replace the CORE cassette with the CYC1 terminator.
RT-PCR
RNA was extracted from cells with hot phenol. First-strand cDNA was prepared with 1 μg total RNA, Superscript II reverse transcriptase (Invitrogen) and gene-specific primers. One-fiftieth of the cDNA and 0.6 μCi [α-32P] dATP were used for PCR amplification, which consisted of 60 s at 94°C, followed by 23–26 cycles (determined to be in the linear range of amplification for each primer pair) of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C, followed by 2 min at 72°C). PCR signals were quantitated by phosphoimager. The sequences of oligonucleotides used in this study are listed in Supplemental Table S2.
Northern blot analysis
Total RNA was isolated from cells with hot phenol and 20 μg of total RNA were used per lane. Northern blotting was done as previously described (Marquardt et al., 2011) with oligonucleotides listed in Supplemental Table S2. Strand specific probes were generated by PCR in the presence of α-32P-dATP with only one primer. Hybridization was done in a buffer containing 1% BSA, 7 % SDS, 1mM EDTA (pH 8.0), and 300 mM Sodium phosphate buffer (pH 7.2).
Chromatin Immunoprecipitations
Chromatin immunoprecipitations were done as previously described (Kim and Buratowski, 2009) with oligonucleotides as listed in Supplemental Table S2. The following histone antibodies were used: anti-acetyl H4 (Upstate 06-598), anti-H3 K4me3 (Upstate 07-473) and anti-H3 (Abcam 1791). Anti-Rpb3 was from Neoclone.
High-resolution tiling array
Total RNA was treated with RNase-free DNaseI using Turbo DNA-free kit (Ambion). For first-strand cDNA synthesis, 20 μg of total RNA was mixed with 1.72 μg of random hexamers, 0.034 μg of oligo(dT) primer and incubated at 70°C for 10 min followed by 10 min at 25°C, then transferred on ice. The synthesis included 2,000 units of SuperScript II Reverse Transcriptase, 50 mM TrisHCl, 75 mM KCl, 3 mM MgCl2, 0.01 M DTT, dNTP + dUTP mix (0.5 mM for dCTP, dATP and dGTP; 0.4 mM for dTTP and 0.1 mM for dUTP, Invitrogen), 20 μg/mL actinomycin D in a total volume of 105 μL. The reaction was carried out in 0.2 mL tubes in a thermal cycler with the following thermal profile: 25°C for 10 min, 37°C for 30 min, 42°C for 30 min followed by 10 min at 70° for heat inactivation and 4°C on hold. Samples were then subjected to RNase treatment of 20 min at 37°C (30 units RNase H, Epicentre, 60 units of RNase Cocktail, Ambion). First-strand cDNA was purified using the MinElute PCR purification kit (Qiagen) and 5 μg were fragmented and labeled using the GeneChip WT Terminal labeling kit (Affymetrix) according to manufacturer’s protocol. The labelled cDNA samples were denatured in a volume of 300 μl containing 50 pM control oligonucleotide B2 (Affymetrix) and Hybridization mix (GeneChip Hybridization, Wash and Stain kit, Affymetrix) of which 250 μl were hybridized per array (S. cerevisiae yeast tiling array, Affymetrix, PN 520055). Hybridizations were carried out at 45°C for 16 h with 60 rpm rotation. The staining was carried out using the GeneChip Hybridization, Wash and Stain kit with fluidics protocol FS450_0001 in an Affymetrix Fluidics station. Data is plotted as in Xu et al. (2009) and can be accessed at http://steinmetzlab.embl.de/buratowski_lab/index.html. Raw data can be downloaded at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), accession number XXXXXX.
Supplementary Material
Set3C negatively regulates the kinetics of transcription induction at a some genes.
Set3C inhibits activity of cryptic internal promoters in 5′ transcribed regions.
Overlapping ncRNA transcription localizes H3K4me2 to Set3C repressed promoters.
ncRNA transcription and Set3C cooperate to fine tune the kinetics of gene induction.
Acknowledgments
We are grateful to Angelika Amon (MIT) for sharing unpublished work, Sebastian Marquardt for the Northern blot analysis protocol, and to Ollie Rando (U. Mass Worcester) and all members of the Buratowski lab for helpful advice and discussions. T.K. is a Special Fellow of the Leukemia and Lymphoma Society. This research was supported by grants GM068717 to L.M.S. and GM46498 and GM56663 to S.B. from the US National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alejandro-Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL, Gasch AP. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009;10:R57. doi: 10.1186/gb-2009-10-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-Cell Analysis Reveals that Noncoding RNAs Contribute to Clonal Heterogeneity by Modulating Transcription Factor Recruitment. Mol Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Kim T. Cold Spring Harb Symp Quant Biol. 2011. The Role of Cotranscriptional Histone Methylations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, et al. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 2010;33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang M, Smith MM. Genetic Analysis Implicates the Set3/Hos2 Histone Deacetylase in the Deposition and Remodeling of Nucleosomes Containing H2A.Z. Genetics. 2011;187:1053–1066. doi: 10.1534/genetics.110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007b;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007c;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Hazelbaker DZ, Buratowski S. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011;2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh EA. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 2008;6:2817–2830. doi: 10.1371/journal.pbio.0060326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roig C, Vieitez C, Posas F, de Nadal E. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol Microbiol. 2010;76:1049–1062. doi: 10.1111/j.1365-2958.2010.07167.x. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long non-coding RNAs mediates mating type control of gametogenesis in budding yeast. Cell. 2012 doi: 10.1016/j.cell.2012.06.049. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Villa-Garcia MJ, Choi MS, Hinz FI, Gaspar ML, Jesch SA, Henry SA. Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol Genet Genomics. 2011;285:125–149. doi: 10.1007/s00438-010-0592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Voichek Y, Tirosh I, Homung G, Amit I, Barkai N. Expression noise and acetylation profiles distinguish HDAC functions. Mol Cell. 2012;47:193–202. doi: 10.1016/j.molcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.