Summary
Class-I myosins are molecular motors that link cellular membranes to the actin cytoskeleton and play roles in membrane tension generation, membrane dynamics, and mechano-signal transduction [1]. The widely expressed myosin-Ic (myo1c) isoform binds tightly to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) via a pleckstrin homology domain located in the myo1c tail, which is important for its proper cellular localization [2–4]. In this study, we found that myo1c can power actin motility on fluid membranes composed of physiological concentrations of PtdIns(4,5)P2, and that this motility is inhibited by high concentrations of anionic phospholipids. Strikingly, this motility occurs along curved paths in a counterclockwise direction (i.e., the actin filaments turn in leftward circles). A biotinylated myo1c construct containing only the motor domain and the lever arm anchored via streptavidin on a membrane containing biotinylated lipid can also generate asymmetric motility, suggesting the tail domain is not required for the counterclockwise turning. We found that the ability to produce counterclockwise motility is not a universal characteristic of myosin-I motors, as membrane-bound myosin-Ia (myo1a) and myosin-Ib (myo1b) are able to power actin gliding, but the actin gliding has no substantial turning bias. This work reveals a possible mechanism for establishing asymmetry in relationship to the plasma membrane.
Experimental Procedures
Proteins and Reagents
Motility assays were performed in KMg25 (10 mM MOPS, pH 7.0, 25 mM KCl, 1 mM DTT, 1 mM EGTA, 1 mM MgCl2). Reagents and proteins, including the methods for the expression and purification of myo1c constructs, are described in Supplemental Information.
Preparation of SLBs
Small unilamellar vesicles (SUVs) for formation of supported lipid bilayers were prepared as described previously [7] with modifications. Briefly, lipids were mixed at the appropriate molar ratios, dried under vacuum, and resuspended in HNa100 (10 mM HEPES, pH 7.0, 100 mM NaCl, 1mM DTT, and 1mM EGTA) with vortexing. Suspensions were subjected to 4 freeze-thaw cycles followed by tip sonication. Samples were centrifuged for 15 min at 15,700 × g at 4°C, and supernatants were stored under N2 at 4°C for no more than four days.
Supported lipid bilayers were prepared as described [32] with modifications. Glass coverslips were soaked in 7X Cleaning Solution (MP Biomedicals) for 1 h, rinsed with copious amounts of deionized water, dried, and cleaned in a Plasma Cleaner/Sterilizer (Harrick) for 10 min. The cleaned coverslips were used immediately to construct standard motility chambers [15] using double-sided tape and vacuum grease. To make PtdIns(4,5)P2 bilayers, SUVs containing the desired mole fraction of PtdIns(4,5)P2 were incubated in chambers for 20–30 minutes. For bilayers containing 2% PtdIns(4,5)P2 and 20% DOPS, SUVs were added to chambers with 3 mM MgCl2 and incubated for 17 min. For bilayers composed of 60% DOPS, SUVs were added to chambers with CaCl2 at a final concentration of 5 mM and incubated for 40 min; the chambers were then washed with HNa100 and re-incubated with SUVs in the absence of CaCl2 for another 30 min before the final washing. All chambers were washed with 10 or more chamber volumes of HNa100 buffer before use. FRAP was performed on all SLBs to ensure the bilayers were continuous and without defect, as described in Supplemental Information.
In Vitro Motility Assays
In vitro motility assays were performed in standard motility chambers [15] at 20 and 37 °C. Briefly, for assays on nitrocellulose-coated cover slips, chambers were assembled using double stick tape and silicon vacuum grease (Beckman). Solutions were added sequentially to the chamber and incubated for 1–2 min each as follows: 0.35 mg/ml anti-myo1c monoclonal antibody in KMg25, 5 mg/ml casein made from 10 mg/ml stock in HNa100 and filtered to remove solids (Sigma), myo1c in KMg25 with 10 µM calmodulin, three washes with motility buffer, 40 nM actin filaments stabilized with Alexa Fluor 488 Phalloidin (Invitrogen) in KMg25, activation buffer (KMg25, 1mM ATP, 10 µM calmodulin, 1 mg/ml glucose, 192 U/ml glucose oxidase (Sigma), 48 µg/mL catalase (Roche)). For assays on SLBs, 2.5–7 nM actin filaments stabilized with Alexa Fluor 488nm Phalloidin were pre-mixed with the desired concentration of myosin in activation buffer before addition to the chamber. For motility on biotinylated lipid bilayers, solutions were added sequentially as follows: 0.1 mg/ml streptavidin or neutravidin (Invitrogen) in HNa100 (1–2 min), extensive washes with HNa100, 0.2–7 µM biotinylated myosin construct in the presence of 10 µM calmodulin (1–2 min), washed extensively with KMg25, 2.5–7 nM actin filaments stabilized with Alexa Fluor 488 Phalloidin in activation buffer. SLBs and actin filaments were imaged under fluorescent light illumination at 560 nm and 488 nm, respectively, on a Leica DMIRB microscope using a 100-fold magnification Leica oil objective of numerical aperture 1.4. The continuity of the SLBs was always checked before and after each time-lapse acquisition by recording the fluorescence recovery of a region bleached by constant (~1–2 min) fluorescent illumination. The rate of actin filament gliding was determined from manual tracking using MetaMorph (Universal Imaging) or the Manual Tracking plug-in for ImageJ [33]. The average gliding speed of each filament was obtained over 10 – 20 min time periods by tracking the filament position at 10 – 30 s intervals, depending of the gliding speed. Only filaments moving in a directional manner were used for analysis.
Calculation of Counterclockwise Bias
The relative angular change <Δθ> was measured between the lines connecting the leading and trailing ends of a filament after 10 min of motility. The sign of <Δθ> was defined as positive in the counterclockwise direction and negative in the clockwise direction. This measurement was then normalized by dividing by the average distance travelled <Δs> using the corresponding average velocity in each case (Table S2). Counterclockwise bias was calculated as normalized angular change (<Δθ>/<Δs>).
Results
Cell biological studies have shown that myosin-I isoforms concentrate on cell membranes, and biochemical experiments have shown that myosin-I isoforms bind directly to lipid membranes via electrostatic interactions [2, 3, 5–7]. In support of a membrane-associated function, myosin-I from the lower eukaryote, Acanthamoeba castellanii, was shown to power the gliding of actin filaments when the motor was bound to fluid, planar lipid-bilayers composed of the anionic phospholipid 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) and the neutral phospholipid 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) [8]. However, subsequent experiments found vertebrate myo1a was not able to power actin gliding while bound to membranes of similar composition that were verified to be fluid and continuous [9].
To clarify the motile properties of membrane-bound vertebrate myosin-I, we examined the motility of myosin-Ic (myo1c), the widely expressed isoform that plays roles in hearing [10], endocytosis [11], exocytosis [12], and membrane recycling [13]. Myo1c binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) via a pleckstrin homology (PH) domain in its tail domain, and membrane binding via this site is crucial for the motor’s proper cellular localization [2–4]. The affinity of myo1c for PtdIns(4,5)P2-containing membranes is further increased by binding of additional anionic phospholipids to other positively charged regions within the myo1c tail and regulatory domains [2, 14]. However, in the absence of poly-phosphoinositides, these myo1c interactions require non-physiological mole fractions of anionic phospholipids [2, 3].
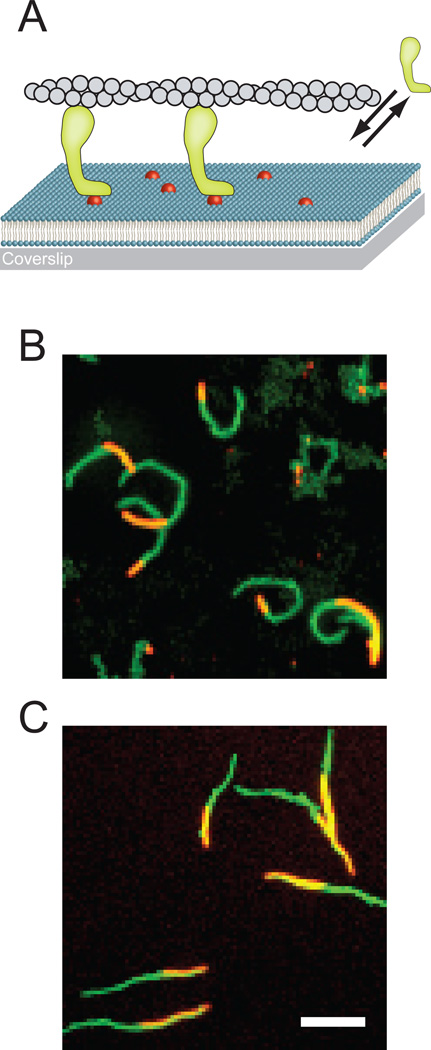
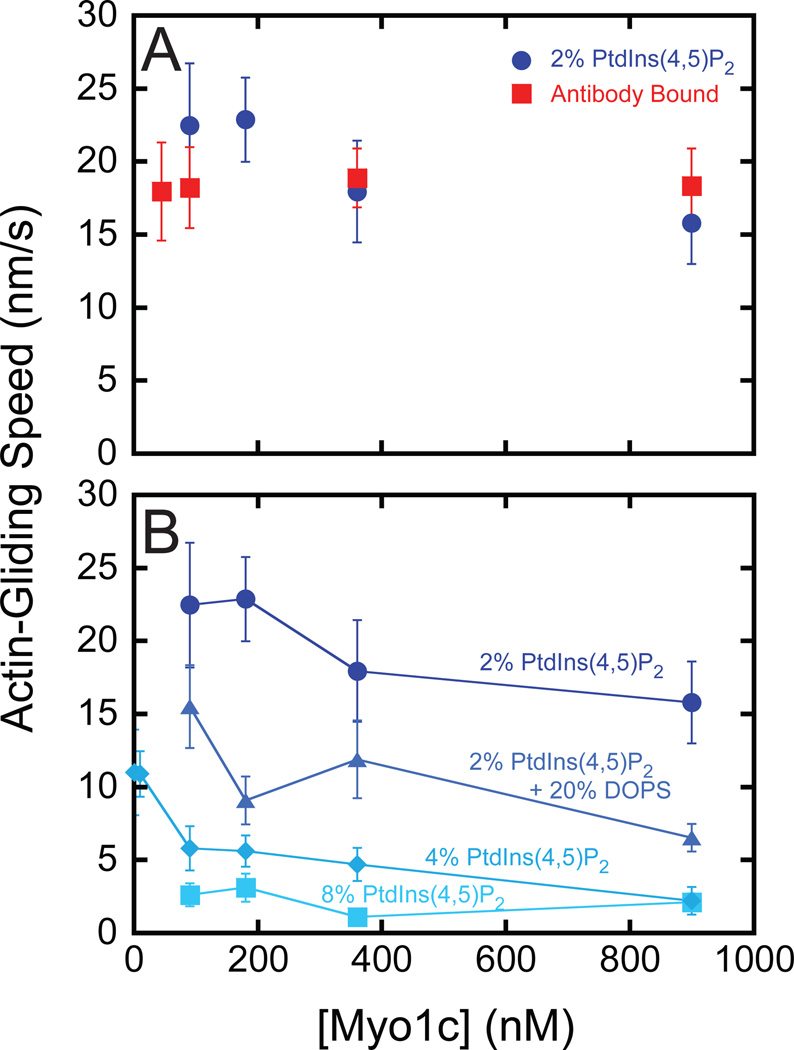
We used in vitro actin-gliding assays to measure the ability of myo1c to power motility on fluid supported lipid bilayers (SLBs) containing 2% PtdIns(4,5)P2 and 98% DOPC (see Table S1 for SLB fluidity parameters). The attachment of myo1c to PtdIns(4,5)P2-containing membranes is dynamic, i.e., it binds to and detaches from membranes [14]. Thus, a solution of premixed myosin, actin and ATP was added to the motility chamber (Figure 1A), rather than washing out free motor prior to addition of actin and ATP, as is the usual procedure in these assays [15]. Actin filaments clearly moved directionally along the surface of the SLB (Figure 1B; see also Movie S1 available online), with speeds that ranged between 16 – 22 nm/s at 22 ° C when the total myo1c concentration was varied between 90 – 900 nM (Figure 2A). Addition of myo1c at concentrations below 90 nM resulted in periods of non-directional diffusive movements of actin, as well as short periods of directed motility, consistent with a low density of myosin bound to the surface. Robust motility (70 nm/s) of 900 nM myo1c was also observed at 37 °C. Surface-attached actin filaments were not observed in the absence of myosin, and there was no binding of actin to SLBs composed only of DOPC in the presence of myo1c. Transient attachment of actin to SLBs was observed in the presence of 0.5% PtdIns(4,5)P2 and 90– 900 nM myo1c, but motility was diffusive and not directional.
Figure 1. Actin filament gliding powered by SLB-bound myosin-I.
(A) Schematic representation of actin-filament (grey) gliding powered by myosin-I (green) on supported lipid bilayers containing PtdIns(4,5)P2 (red). The lipid bilayer is spread on a glass coverslip. The double arrows indicate the dynamic interaction of myosin I with the lipid bilayer. For clarity, the representation is not drawn to scale. (B) Maximum projection from a 10 min time series of actin motility powered by myo1c at 22 °C. (C) Maximum projection from a 20 min time series of actin motility powered by myo1a at 22 °C. In both (B) and (C) the whole track of actin filaments is colored in green, while the actin filament at t = 0 s is colored in orange. Scale bar 5 µm.
Figure 2. Actin gliding speeds as a function of myo1c concentration and motility substrate.
(A) Myo1c-concentration dependence of the average actin-gliding speed when bound to (●) SLBs composed of 2% PtdIns(4,5)P2 or a (■) monoclonal antibody adsorbed to coverslips coated with nitrocellulose. (B) Myo1c-concentration dependence of actin-gliding when myo1c is bound to (●) 2% PtdIns(4,5)P2, (▲) 2% PtdIns(4,5)P2 and 20 % DOPS, (◆) 4% PtdIns(4,5)P2, or (■) 8% PtdIns(4,5)P2. Experiments were performed at 22 °C in KMg25. Error bars represent the standard deviation.
Myo1c adsorbed to a nitrocellulose coated surface using an antibody that recognizes the tail domain supported motility with speeds similar to those measured for myo1c bound to 2% PtdIns(4,5)P2 SLBs (Figure 2A). The actin gliding speed was relatively insensitive to the concentration (45 – 900 nM) of myo1c that was incubated with the antibody (Figure 2A; see Methods). Actin motility was intermittent and diffusive when 18 nM myo1c was added to the motility chamber (not shown). No actin surface attachment or motility was observed with myo1c on casein-blocked nitrocellulose in the absence of antibody.
Increasing the mole fraction of anionic phospholipids in membranes that contain 2% PtdIns(4,5)P2 has been shown to increase affinity of the membrane for myo1c [2, 3, 14]. Surprisingly, actin-gliding velocities were substantially slower when myo1c was bound to SLBs containing 2% PtdIns(4,5)P2 and 20% DOPS, 4% PtdIns(4,5)P2, or 8% PtdIns(4,5)P2 (Figure 2B; Movie S2). Actin gliding in the presence of 90–900 nM myo1c on SLBs composed of 60% DOPS (without PtdIns(4,5)P2) was too slow and irregular to determine a gliding speed, despite the high affinity of myo1c for these membranes [2]. Decreasing the myo1c concentration to 1 nM in experiments with 4% PtdIns(4,5)P2 SLBs resulted in a slight increase in the gliding speed (Figure 2B), but this increase was partly the result of rapid diffusive movements of the actin filaments due to membrane detachment. Further decreasing the myo1c concentrations resulted in complete detachment of actin from the SLBs. These results, taken together with the concentration-independent rate of antibody-bound myo1c (Figure 2A), suggest that the decreased gliding rate was not a result of increased myosin density on the surface, but was due to an effect of the increased membrane charge on the attachment of myo1c to SLBs (see below)
D-myo-inositol-1,4,5-triphosphate (Ins(1,4,5)P3), the soluble head group of PtdIns(4,5)P2, has been shown to bind tightly to the tail domain of myo1c and competitively inhibits the interaction between myo1c and PtdIns(4,5)P2 [2]. Actin motility and the attachment of filaments to SLBs was disrupted by the addition of 50 µM Ins(1,4,5)P3 to the motility chamber. Additionally, we found that myo1c was able to power acting gliding (20 ± 1.4 nm/s) when it was bound to Ins(1,4,5)P3 that was surface-attached to biotin-neutravidin via a 3-carbon linker (Movie S1). Thus, the dynamic attachment of myo1c to the inositol head group of PtdIns(4,5)P2 is sufficient to anchor myo1c at the surface to power actin motility.
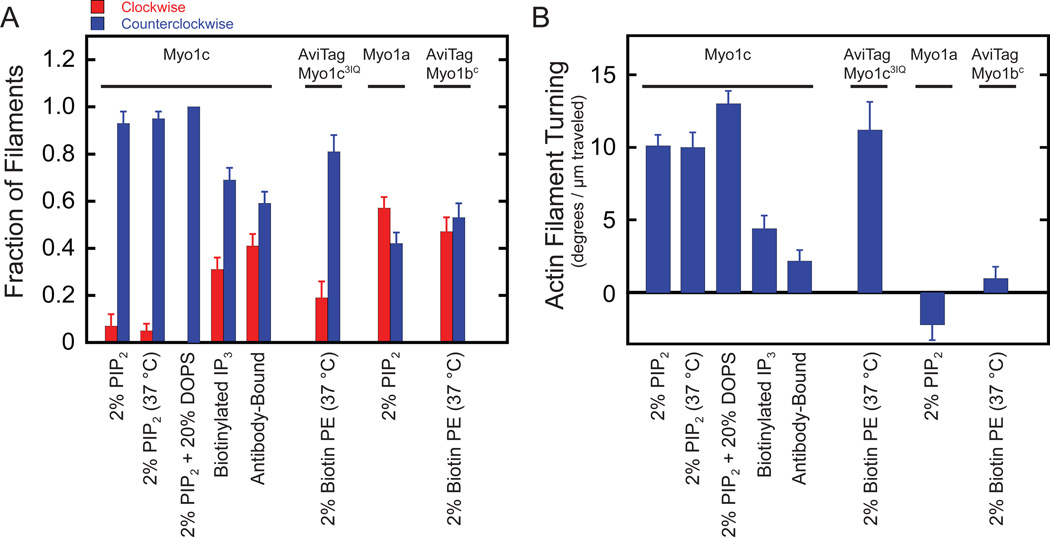
Strikingly, myo1c-powered gliding of actin filaments on SLBs containing 2% PtdIns(4,5)P2 occurred along curved paths in a counterclockwise fashion (turning left) when viewed from the objective lens side of the coverslip at both 22 °C and 37 °C (Figure 1B & 3A, Movie S1). We quantified the counterclockwise bias as an angular change, Δθ, in the actin filament, normalized to the distance traveled, Δs (<Δθ>/<Δs>; Figure 3B). We defined the angular change as positive in the counterclockwise direction and negative in the clockwise direction. The distance-normalized angular changes for motility on SLBs composed of either 2% PtdIns(4,5)P2 or 2% PtdIns(4,5)P2 and 20% DOPS were similar, 10 ± 0.73 and 13 ± 0.96 deg/µm respectively, despite the 2-fold difference in gliding velocities (Figure 2B). Myo1c anchored to nitrocellulose via an antibody supported actin gliding but not with the pronounced counterclockwise bias (2.1 ± 0.81 deg/µm) observed on SLBs (Table S2). Myo1c bound to biotinylated Ins(1,4,5)P3 had a bias of 4.6 ± 0.84 deg/µm, which is less than that of myo1c bound to PtdIns(4,5)P2-containing SLBs (Figure 3B).
Figure 3. SLB-bound myo1c powers asymmetric motility of actin filaments.
(A) Fraction of actin filaments that turn (red) clockwise and (blue) counterclockwise as a function of motility substrate and myosin-I construct or isoform. Errors were calculated by the bootstrap method (n = 100) [34] (B) Quantification of actin filament turning for the conditions listed above. Error bars are the standard errors of the mean (details in Table S2). Experiments were performed at 22 °C in KMg25, unless indicated otherwise.
In experiments performed with SLBs, myo1c dynamically attaches to and detaches from the fluid substrate. In contrast, experiments performed on a solid surface have myo1c anchored via an antibody. To test whether a fluid substrate without a dynamic interaction is sufficient for asymmetric motility, we attached a myo1c construct that contains the motor domain, the lever arm, and a biotinylated AviTag sequence at it C-terminus (myo1c3IQ) to SLBs containing 2% Biotin-PE via neutravidin. Actin gliding was too slow to quantify at 22 °C, but raising the temperature to 37 °C restored actin gliding, and asymmetric motility was observed to the same extent observed for full length myo1c on SLBs containing 2% PtdIns(4,5)P2 (Figure 3A & B). In the absence of neutravidin or myo1c3IQ, actin filaments did not attach to the bilayer.
To determine if other myosin-I family members can generate asymmetric motility, we performed actin gliding assays with full length myo1a, which has been shown to interact with anionic phospholipids, including PtdIns(4,5)P2 [5, 16], but has not been shown to be able to support motility on SLBs [9]. We found that myo1a powered actin gliding on 2% PtdIns(4,5)P2 SLBs with a velocity of 5.7 ± 0.88 nm/s at 22 °C, but the actin filaments had no significant counterclockwise bias (Figure 3A & B; Movie S1). Additionally, we anchored a construct of myosin-Ib that contains the motor domain, lever arm (4-IQ motifs), and a biotinylated AviTag sequence at its C-terminus (myo1bc) to SLBs containing 2% Biotinyl-PE via neutravidin [17, 18]. Actin motility at 37 °C had no counterclockwise bias, but exhibited a gliding speed (77 nm/s) similar to that measured previously on a solid substrate [17].
Discussion
Our results show that myo1c can power actin motility when bound to fluid lipid bilayers containing phosphoinositides with speeds similar to those found in assays where the myosin is tightly affixed to a non-fluid surface (Figure 2A). Actin gliding velocities were affected by the composition of the SLB. Gliding rates were lower on SLBs with high PtdIns(4,5)P2 content or when DOPS was added to membranes (Figure 2B). Previous binding measurements have shown that the affinity of myo1c for membranes increases with the mole fraction of PtdIns(4,5)P2 or DOPS [2], so these results indicate that actin gliding velocities decrease with increasing membrane affinity. It has been proposed that IQ motifs in the regulatory domains of myo1c [3] and myo1a [19] interact with membranes that have high mole fractions of DOPS. Interestingly, a 20 Å resolution EM structure of myo1a bound to membranes composed of 75% DOPS appears to show membrane association of the IQ motif that is distal to the motor domain [20]. Since the IQ motifs make up the force-transducing lever arm of myosin, this interaction could disrupt the working stroke, leading to slow and irregular motility. Taken together, these findings may explain why a previous study of vertebrate myo1a was not able to demonstrate motility on SLBs containing 40% DOPS [9]. We do note that one study demonstrated myo1a-supported actin gliding in the presence of anionic phospholipids [21], but it is not clear if these experiments were performed on fluid and continuous SLBs [9].
How does myosin bound to a fluid bilayer support actin gliding? Although lipid bilayers are considered fluid, both experimental measurements and simulation studies have shown that lipid bilayers are fluids of high viscosity [22]. The viscosity coefficients are in the range of 100–2000 times the viscosity coefficient of water, and subsequently the frictional load in lipid bilayers is expected to be much higher than in water. Indeed, 30 – 100 nm diameter gold nanoparticles (which are similar in size to myosin-I) attached to the head groups of single lipids have negligible effect on the diffusion of the lipid molecules in the bilayer [23]. It is therefore not surprising that myo1c molecules will experience viscous load on the tail domain during the working stroke, resulting in the displacement of the actin filament relative to the membrane. Additionally, it is likely that myo1c is able to interact with multiple anionic phospholipids [14], which may further decrease diffusion within the plane of the membrane and increase the membrane viscous load.
Our most striking result is the finding that myo1c drives asymmetric motility on lipid membranes, with actin filaments moving in a counterclockwise direction when viewed from the opposite side of the membrane. This asymmetric motility requires only the myo1c motor and regulatory domains, suggesting that the asymmetry is a property of the working stroke and not the attachment of the tail domain to the membrane. The normalized angular change (~ 10 deg/µm; Figure 3B & Table S2) is the same, within error, for different SLB compositions, different temperatures, and different myo1c constructs. This angular change could be produced if the power stroke has a counterclockwise rotational component or if the power stroke is not directly aligned with the axis of the actin filament. For example, myo1c at the leading edge of the actin filament could force the tip of the actin to have a leftward bias in its search for the next membrane-bound motor. The force required to bend an actin filament by 10 deg/µm (Table S2) is small and can be estimated by applying the equation for a cantilevered beam to the curved filaments [24]. Assuming that the flexural rigidity of actin filaments is 7.3·10−26 N·m2 [25], and that the freely diffusing end of the filament is on the order of 100 nm in length, then the force required to bend this segment is predicted to be only 0.12 pN. Interestingly, the ability to produce asymmetric motility is not a universal characteristic of myosin-I motors, since neither myo1a nor myo1b generated significant asymmetric motility on SLBs (Figure 3A & B); however, class-2 myosins are predicted to have non-axial components to their working strokes [26, 27].
While myo1c powers asymmetric motility on a variety of substrates (Figure 3A & B), a fluid substrate significantly enhances the asymmetry. The translational and rotational degrees of freedom of myo1c bound to the SLBs may allow myo1c to orient itself to interact with actin in a conformation favorable for revealing the asymmetric working stroke. On a solid substrate, the conformation and/or orientation of myo1c relative to the long axis of actin may be restricted, and the directional bias of the power stroke may be suppressed. Increasing rotational and translational flexibility may also explain the slight increase in counterclockwise bias observed when myo1c is bound to biotinylated Ins(1,4,5)P3, which is connected to surface-bound neutravidin via a 3-carbon linker rather than a surface-immobilized antibody.
Asymmetric motility powered by myo1c could play functional roles in vivo. In addition to the sliding of the actin filaments, membrane-bound myo1c could contribute a second degree of directionality relative to the membrane by applying torque to actin filaments. Several studies have shown that myosin-I family members, including a homolog of myo1c, are necessary for establishment of left-right asymmetry in invertebrate embryonic development [28, 29]. Interestingly, it has been hypothesized that cytoskeletal motors could be “F molecules” [30], which are chiral molecules that align along the anteroposterior and dorsoventral axes and convert their handedness or asymmetry into chemical or signaling differences between symmetry planes [31]. Thus, more detailed investigations of the role of myosin isoforms in symmetry breaking in vivo are warranted.
Supplementary Material
Highlights.
-
➢
Myo1c bound fluid bilayers containing PtdIns(4,5)P2 can power actin gliding.
-
➢
Membrane-bound myo1c drives actin gliding counterclockwise along curved paths.
-
➢
Myosin-Ia and myosin-Ib exhibit no counterclockwise turning bias.
Acknowledgments
We thank Tianming Lin for his excellent technical assistance and the Ostap laboratory for helpful comments. This work was supported by a NIH/NRSA fellowship (GM090551) to E.A.F. and grants from the NIH (DK075555) to M.J.T. and (GM057247) to E.M.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci U S A. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c Binds Phosphoinositides through a Putative Pleckstrin Homology Domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) J Biol Chem. 2010;285:8675–8686. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden SM, Wolenski JS, Mooseker MS. Binding of brush border myosin I to phospholipid vesicles. J Cell Biol. 1990;111:443–451. doi: 10.1083/jcb.111.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyata H, Bowers B, Korn ED. Plasma membrane association of Acanthamoeba myosin I. J Cell Biol. 1989;109:1519–1528. doi: 10.1083/jcb.109.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyrpassopoulos S, Shuman H, Ostap EM. Single-molecule adhesion forces and attachment lifetimes of myosin-I phosphoinositide interactions. Biophys J. 2010;99:3916–3922. doi: 10.1016/j.bpj.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zot HG, Doberstein SK, Pollard TD. Myosin-I moves actin filaments on a phospholipid substrate: implications for membrane targeting. J Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zot HG. Phospholipid membrane-associated brush border myosin-I activity. Cell Motil Cytoskeleton. 1995;30:26–37. doi: 10.1002/cm.970300105. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie PG. Myosin I and adaptation of mechanical transduction by the inner ear. Philos Trans R Soc Lond B Biol Sci. 2004;359:1945–1951. doi: 10.1098/rstb.2004.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokac AM, Schietroma C, Gundersen CB, Bement WM. Myosin-1c couples assembling actin to membranes to drive compensatory endocytosis. Dev Cell. 2006;11:629–640. doi: 10.1016/j.devcel.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–824. doi: 10.1038/nature01246. [DOI] [PubMed] [Google Scholar]
- 13.Brandstaetter H, Kendrick-Jones J, Buss F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J Cell Sci. 2012 doi: 10.1242/jcs.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna JM, Ostap EM. Kinetics of the interaction of myo1c with phosphoinositides. J Biol Chem. 2009;284:28650–28659. doi: 10.1074/jbc.M109.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci U S A. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazerik JN, Tyska MJ. Myosin-1A Targets to Microvilli Using Multiple Membrane Binding Motifs in the Tail Homology 1 (TH1) Domain. J Biol Chem. 2012;287:13104–13115. doi: 10.1074/jbc.M111.336313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin T, Tang N, Ostap EM. Biochemical and motile properties of Myo1b splice isoforms. J Biol Chem. 2005;280:41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 18.Laakso JM, Lewis JH, Shuman H, Ostap EM. Control of myosin-I force sensing by alternative splicing. Proc Natl Acad Sci U S A. 2010;107:698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanljung-Collins H, Collins JH. Brush border myosin I has a calmodulin/phosphatidylserine switch and tail actin-binding. Adv Exp Med Biol. 1994;358:205–213. doi: 10.1007/978-1-4615-2578-3_19. [DOI] [PubMed] [Google Scholar]
- 20.Celia H, Jontes JD, Whittaker M, Milligan RA. Two-dimensional crystallization of brush border myosin I. J Struct Biol. 1996;117:236–241. doi: 10.1006/jsbi.1996.0088. [DOI] [PubMed] [Google Scholar]
- 21.Wolenski JS, Hayden SM, Forscher P, Mooseker MS. Calcium-calmodulin and regulation of brush border myosin-I MgATPase and mechanochemistry. J Cell Biol. 1993;122:613–621. doi: 10.1083/jcb.122.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller TJ, Muller-Plathe F. Determining the local shear viscosity of a lipid bilayer system by reverse non-equilibrium molecular dynamics simulations. Chemphyschem. 2009;10:2305–2315. doi: 10.1002/cphc.200900156. [DOI] [PubMed] [Google Scholar]
- 23.Forstner MB, Kas J, Martin AF. Single Lipid Diffusion in Languir Monolayers. Langmuir. 2001;17:567–570. [Google Scholar]
- 24.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland: Sinauer Associates, Inc.; 2001. [Google Scholar]
- 25.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishizaka T, Yagi T, Tanaka Y, Ishiwata S. Right-handed rotation of an actin filament in an in vitro motile system. Nature. 1993;361:269–271. doi: 10.1038/361269a0. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins SC, Sabido-David C, Corrie JE, Irving M, Goldman YE. Fluorescence polarization transients from rhodamine isomers on the myosin regulatory light chain in skeletal muscle fibers. Biophys J. 1998;74:3093–3110. doi: 10.1016/S0006-3495(98)78016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speder P, Adam G, Noselli S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature. 2006;440:803–807. doi: 10.1038/nature04623. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi K, Maeda R, Ando T, Okumura T, Nakazawa N, Hatori R, Nakamura M, Hozumi S, Fujiwara H, Matsuno K. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science. 2011;333:339–341. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 30.Levin M. Motor protein control of ion flux is an early step in embryonic left-right asymmetry. Bioessays. 2003;25:1002–1010. doi: 10.1002/bies.10339. [DOI] [PubMed] [Google Scholar]
- 31.Brown NA, Wolpert L. The development of handedness in left/right asymmetry. Development. 1990;109:1–9. doi: 10.1242/dev.109.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 34.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Revipes in C++ New York: Cambridge University Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.