Abstract
A variety of genetically encoded reporters use changes in fluorescence (or Förster) resonance energy transfer (FRET) to report on biochemical processes in living cells. The standard genetically encoded FRET pair consists of cyan and yellow fluorescent proteins (CFP and YFP), but many CFP-YFP reporters suffer from low FRET dynamic range, phototoxicity from the CFP excitation light, and complex photokinetic events such as reversible photobleaching and photoconversion. Here, we engineered two fluorescent proteins, Clover and mRuby2, which are the brightest green and red fluorescent proteins to date, and have the highest Förster radius of any ratiometric FRET pair yet described. Replacement of CFP and YFP in reporters of kinase activity, small GTPase activity, and transmembrane voltage significantly improves photostability, FRET dynamic range, and emission ratio changes. These improvements enhance detection of transient biochemical events such as neuronal action potential firing and RhoA activation in growth cones.
INTRODUCTION
FRET between fluorescent proteins is widely used to monitor biochemical processes in living cells. Most FRET-based reporters use cyan and yellow fluorescent proteins (CFPs and YFPs) as fluorophores. However, aspects of CFPs and YFPs are problematic for FRET. CFPs and YFPs can undergo rapid multi-rate and reversible photobleaching1-3, YFPs can photoconvert into cyan fluorescent species4, CFPs can photoactivate at YFP excitation wavelengths5, and the violet CFP excitation light can be phototoxic6,7. Furthermore, many CFP- and YFP-based FRET reporters produce small changes in FRET, creating detection challenges when biochemical responses are subtle or transient. Detection is especially difficult when imaged structures are small, as the response has to overcome noise or, if illumination is increased to reduce noise, fluorophore bleaching. While linker lengthening can improve responses of sensors that operate via signal-dependent interaction between domains8, sensors that operate via conformational changes in a single domain are more difficult to improve9,10. New FPs that perform well in FRET and avoid the drawbacks of CFPs and YFPs would thus be useful11,12.
Alternative FRET pairings between a green fluorescent protein (GFP) or YFP and an orange or red fluorescent protein (OFP or RFP) have been explored9,13-17, but still have major limitations. OFPs exhibit poor maturation, rapid photobleaching, and/or photoconversion to green or far-red species, all of which can lead to underestimations of FRET2,16,18. Emission from mRFP or mCherry is too weak to detect above the donor emission tail, precluding the use of ratiometric imaging to correct for differences in probe concentration12,13,16,17,19. Where comparisons can be made, pairs with previously described RFP acceptors do not improve ratio changes compared to CFP-YFP pairs9,10.
We aimed to systematically develop new FPs with properties better suited to a wide range of FRET applications. Starting from Aequoria victoria GFP and the RFP mRuby20, we engineered Clover and mRuby2, the brightest fluorescent protein and the brightest RFP characterized so far, respectively. Clover and mRuby2 conferred greater dynamic range and photostability to four existing FRET reporter designs. Among the improved reporters is a voltage sensor that allows more reliable detection of single action potentials than previous sensors. In addition, we use an improved RhoA reporter to show that RhoA activation occurs rapidly and locally in neuronal growth cones during ephrinA-stimulated retraction.
RESULTS
Evaluation of performance of existing FRET reporters
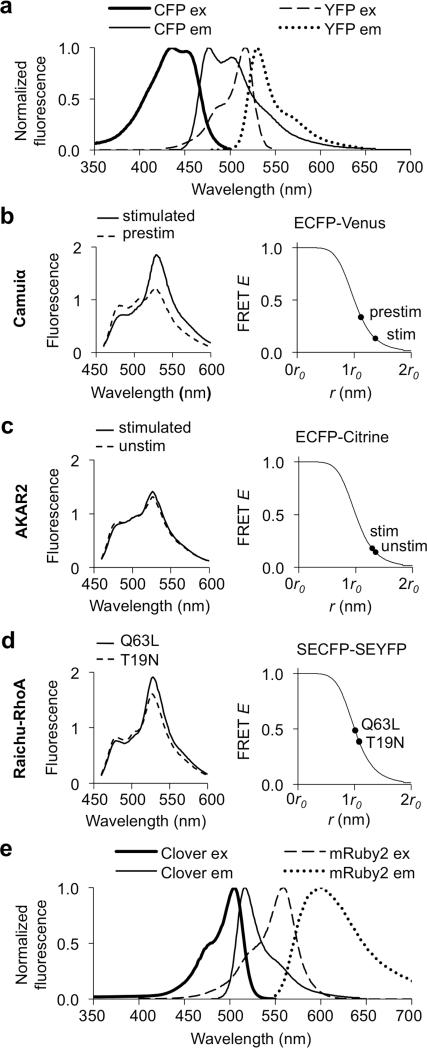
An undesirable attribute for FRET pairs is overlap between donor and acceptor emissions. Emission overlap dampens ratio changes, as an increase (or reduction) in FRET and acceptor emission is counteracted by reduction (or increase) of donor emission previously mixed in the acceptor emission channel. Current CFP-YFP pairs show extensive overlap of donor and acceptor emissions (Fig. 1a). Another important consideration is the FRET efficiency achievable by the fluorescent protein pair. FRET efficiency (E) is related to interfluorophore distance (r) by a sigmoidal curve described by the Förster equations. This curve is steepest at its midpoint, where E equals 0.5 and r equals the Förster radius r0, a parameter that depends on photophysical properties of the FRET pair and the positions of the donor emission and acceptor absorbance transition dipoles. Because of the high slope at the midpoint, reporters that operate with E near 0.5 should exhibit larger changes in E upon changes in r compared to reporters where E remains far from 0.5. To quantify the range of E over which existing reporters operate (which we refer to as FRET dynamic range), we analyzed three biosensors for which we could acquire emission spectra in basal and induced states: the Camuiα reporter of CaMKIIα activity21, the AKAR2 reporter of PKA activity22, and the Raichu-RhoA reporter of RhoA activity23. All use variants of CFP and YFP as FRET pairs. We calculated E by linear unmixing of emission spectra of each reporter in basal or induced states. We found Camuiα and AKAR2 operate with E of 0.13-0.33 (Fig. 1b) and 0.14-0.18 (Fig. 1c), while Raichu-RhoA operates closer to the midpoint of the E-r curve with a range of 0.39-0.48 (Fig. 1d). For reporters such as Camuiα and AKAR2, a FRET pair that can generate larger r0 values would be expected to improve dynamic range. Another advantage of a higher r0 is that the resulting higher FRET efficiencies lead to increased acceptor emissions, boosting the contribution of acceptor emission relative to donor bleed-through emission in the acceptor channel.
Figure 1.
Assessment of FRET reporter performance using Förster equations and emission spectra. (a) Normalized spectra of ECFP and EYFP. Variants of CFP and YFP have identical spectra. (b-d) Modeling of existing FRET reporter performance using Förster equations and emission spectra. Ranges for the FRET efficiency E were derived by fitting theoretical to experimentally-obtained emission curves (left) and were then mapped to appropriate Förster E-r curves (right) for Camuiα (b), AKAR2 (c), or Raichu-RhoA (d). (e) Clover emission and mRuby2 excitation spectra show substantial overlap while Clover and mRuby2 emission spectra are better separated than CFP and YFP emission spectra.
Given these considerations, we identified larger separation between donor and acceptor emissions and higher r0 as important design targets. To improve r0, we attempted to increase the quantum yield of the donor (ϕD), extinction coefficient of the acceptor (εA), and the integrated spectral overlap between donor emission and acceptor excitation, as the 6th power of r0 is proportional to these factors. We also favored redder wavelengths (λ), which should help increase r0 due to a λ4 term in the spectral overlap equation while reducing phototoxicity and background cellular autofluorescence24.
Evolution of Clover
Because emission of GFP overlaps substantially with absorbance of OFPs and RFPs, we first attempted to rationally improve GFP quantum yield. YFPs bear a T203Y mutation and exhibit higher quantum yields than existing GFPs. Besides creating a π–π interaction with the chromophore to cause the red-shifted spectra of YFPs25, this mutation removes a hydrogen bond between Thr203 and the chromophore. This bond may be responsible for the high vibronic shoulder in the excitation spectrum of EGFP at 470 nm, which is not observed in YFP (Supplementary Fig. 1) or in the transient I* form of wild-type GFP26 where the chromophore is not hydrogen-bonded to Thr20327. This shoulder is not mirrored in the EGFP emission spectrum, consistent with excited-state conformational changes that break the hydrogen bond prior to emission27. We hypothesized that removing this hydrogen bond in GFP may improve quantum yield by removing pathways for nonradiative decay from the excited state. To test this hypothesis, we introduced into wild-type GFP the S65G mutation to maintain a deprotonated chromophore28 and M153T and superfolder mutations to promote folding29. We then screened mutants of Thr203 for brightness in bacteria, and isolated an unusually bright T203H mutant. We subsequently mutagenesized Gln69, a position known to affect bleaching in YFPs, and found that Q69A conferred photostability superior to ECFP and Cerulean, the most commonly used FRET donors (Supplementary Fig. 1)30. Given its bright green color in the purified state, we named the resulting protein Clover.
Clover has excitation and emission maxima of 505 and 515 nm (Supplementary Fig. 1), intermediate between EGFP and EYFP. Its extinction coefficient of 111,000 M–1 cm–1 and quantum yield of 0.76 make it the brightest fluorescent protein yet characterized (Table 1). Bleaching of Clover followed first-order kinetics with less then 4% reversibility (Supplementary Fig. 1), unlike ECFP and Cerulean2,3. In confocal laser microscopy of live cells, Clover bleached similarly to YPet, previously the brightest fluorescent protein (Supplementary Fig. 1). In wild-type GFP and PA-GFP, the chromophore exists in the protonated state, but can be photoconverted to the deprotonated state by a photochemical reaction eliminating Glu22228. With a completely deprotonated chromophore at neutral pH (pKa 6.1, Supplementary Fig. 1), Clover should be incapable of photoconversion despite the presence of His203, which enhances photoconversion in PA-GFP. Indeed, we observed no photoactivation of Clover in illumination conditions that produced more than 3-fold photoactivation of PA-GFP (Supplementary Fig. 1). Clover differs from GFP by ten mutations: S30R Y39N S65G Q69A N105T Y145F M153T V163A I171V T203H (Supplementary Fig. 2). As with other GFP derivatives, Clover functions well in a variety of fusion proteins (Supplementary Figs. 3,4).
Table 1.
Properties of bright and photostable monomeric fluorescent proteins
| Protein | ref.a | λ ex b | λ em c | ε d | ϕ e | Brightnessf | pKag | Maturationh | Photostabilityi |
|---|---|---|---|---|---|---|---|---|---|
| mTurquoise2 | 30 | 434 | 474 | 30 | 0.93 | 28 | 3.1 | ND | 90j |
| mTFP1 | 45 | 462 | 488 | 64 | 0.85 | 54 | 4.3 | ND | 110 |
| EGFP | 45 | 488 | 507 | 56 | 0.60 | 34 | 6.0 | 25k | 174 |
| mWasabi | 45 | 493 | 509 | 70 | 0.80 | 56 | 6.5 | ND | 93 |
| YPet | 45 | 517 | 530 | 104 | 0.77 | 80 | 5.6 | ND | 49 |
| Clover | this work | 505 | 515 | 111 | 0.76 | 84 | 6.2 | 30 | 50 |
| mCherry | 45 | 587 | 610 | 72 | 0.22 | 16 | <4.5 | 40l | 96 |
| mKate2 | 45 | 588 | 633 | 63 | 0.40 | 25 | 6.5 | 20 | 84 |
| TagRFP-T | 2 | 555 | 584 | 81 | 0.41 | 33 | 4.6 | 100 | 337 |
| mRuby | 20 | 558 | 605 | 112 | 0.35 | 39 | 4.4 | 170 | 101j |
| mRuby2 | this work | 559 | 600 | 113 | 0.38 | 43 | 5.3 | 150 | 123 |
Source of data unless otherwise noted.
Excitation maximum in nm.
Emission maximum in nm.
Peak extinction coefficient in mM–1cm–1.
Quantum yield.
Product of ε and ϕ.
pH at which the fluorescence intensity is 50% of its maximum value.
Time in min for fluorescence to obtain half-maximal value after exposure to oxygen.
Time in s to photobleach from 1000 to 500 photons per second per molecule under arc lamp illumination.
Data from this study.
Data from ref. 46.
Data from Ref. 47.
ND=not determined.
Evolution of mRuby2
An ideal FRET partner for Clover would have a high extinction coefficient, extensive excitation overlap with Clover emission, high quantum yield, large Stokes shift, fast maturation, and high photostability. The first two characteristics would contribute to a large r0. High quantum yield and large Stokes shift are desirable to allow acceptor emission to be clearly discernable from donor emission bleedthrough in the acceptor channel, so that FRET changes can be detected by ratiometric emission imaging. Fast maturation is necessary to reduce the percentage of non-functional acceptor domains, whose presence leads to an underestimation of FRET efficiency. High photostability is important in time-lapse microscopy, because acceptor bleaching irreversibly reduces FRET dynamic range of the reporter population.
Among highly absorbing monomeric OFPs and RFPs, absorbance of mCherry and mKate derivatives exhibit poor overlap with Clover emission, while emission of mOrange and mKO derivatives are poorly separated from Clover emission. Furthermore, mKate derivatives retain substantial green fluorescence and are weak dimers31. With their brightness and large Stokes’ shifts, TagRFP and TagRFP-T are well matched optically, but exhibit complex reversible photobleaching2. The recently described mRuby20 is relatively bright and photostable (Table 2), but its photobleaching behavior had not previously been optimized.
Table 2.
Examples of r0 values of fluorescent protein FRET pairs
| FRET pair | ϕ D a | εAb (mM–1 cm–1) | r0 (nm) |
|---|---|---|---|
| ECFP-Citrine | 0.36c | 77d | 4.8 |
| ECFP-Venus | 0.36c | 92d | 5.0 |
| Cerulean-Citrine | 0.49c | 77d | 5.4 |
| Cerulean-Venus | 0.49c | 92d | 5.2 |
| SECFP-SEYFP | 0.58e | 101e | 5.4 |
| EGFP-mCherry | 0.60d | 72d | 5.4 |
| TagGFP-TagRFP | 0.59f | 100f | 5.7 |
| mTFP1-Citrine | 0.85d | 77d | 5.7 |
| mTFP1-mOrange | 0.85d | 71d | 5.7 |
| Citrine-mKate2 | 0.76d | 63d | 5.8 |
| Clover-mCherry | 0.76g | 72d | 5.8 |
| mTurquoise1-SEYFP | 0.84c | 101e | 5.8 |
| mTurquoise2-SEYFP | 0.93c | 101e | 5.9 |
| Clover-mRuby2 | 0.76g | 113g | 6.3 |
We explored the possibility of developing a FRET acceptor for Clover from mRuby. Photobleaching of mRuby on a microscope under xenon arc lamp illumination induced rapid initial loss of fluorescence (10% in 2 s, time normalized as previously described2) followed by slow bleaching (normalized time to half-initial brightness of 101 s). The precursor to mRuby, eqFP611 dimer, readily undergoes reversible photobleaching due to isomerization from a bright trans form to a dim cis form, and mutations near the chromophore affect isomerization32. We attempted to reduce fast photobleaching of mRuby due to isomerization by mutating interior residues. We screened a library of mutants for maximal FRET in tandem fusions with Clover, and performed photobleaching on individual protein solutions.
One variant showed increased FRET with Clover and improved photokinetics. This variant, which we named mRuby2, has similar spectra to mRuby (Supplementary Fig. 5) and improved brightness (ε = 113,000 versus 112,000 M–1 cm–1, ϕ = 0.38 versus 0.35, Table 1). mRuby2 differs from mRuby by four mutations: L15M I102V A119V A131P (numbering per PDB file 1UIS, Supplementary Fig. 6). Photobleaching of mRuby2 with arc lamp illumination is mono-exponential with negligible reversibility and a half-life of 121 s, compared to complex photokinetics and a 101 s half-life for mRuby (Supplementary Fig. 5). In confocal laser microscopy of live cells, mRuby2 bleached with similar kinetics to mRuby and mCherry (Supplementary Fig. 5). Finally, mRuby2 functions well in a variety of fusions (Supplementary Figs. 7,8).
The Clover-mRuby2 pair features high ϕD, εA, and overlap between donor emission and acceptor absorbance (Fig. 1e). As a result, Clover-mRuby2 has a r0 (with random interfluorophore orientation) of 6.3 nm, higher than any previous FRET pair (Table 2). To confirm that mRuby2 is an efficient FRET acceptor for Clover, we fused mRuby2 to Clover with a flexible linker and measured FRET E by spectral imaging. We used mCherry as a robustly folding RFP comparison. E in the Clover-mRuby2 fusion was 0.55 compared to 0.47 in the Clover-mCherry fusion (Supplementary Fig. 9). mRuby2 also has a larger Stokes’ shift compared to YFP or mCherry (Table 1), improving separation of acceptor and donor emissions.
Improved sensing of kinase activity
We benchmarked the performance of Clover-mRuby2 across four sensor designs differing in their sensing modality, relative position of the sensing domain, and FRET E operating range. We first used Clover-mRuby2 to retrofit Camuiα21, a reporter of CaMKIIα activity which operates at low FRET efficiencies (Fig. 1c). In Camuiα, the Venus YFP is fused to the N terminus and ECFP is fused to the C terminus of full-length CaMKIIα, and activation of CaMKIIα induces a decrease in FRET (Fig. 2a)21. We replaced YFP with Clover and ECFP with mRuby2 to create Camuiα-CR. Camuiα-CR produced a mean 29.4% peak emission ratio change in HeLa cells stimulated with ionomycin, 62% larger than original Camuiα (Fig. 2b,c). Circular permutation of Clover at two sites, after amino acid (aa) 157 and aa 173, resulted in maximum ratio changes (Fig. 2d,e) and response kinetics (Supplementary Fig. 10) similar to non-permuted Clover, suggesting that improvements were not due to more optimal fluorophore orientations. Incidentally, the intensities of Camuiα-CR in each channel did not change during baseline imaging, indicating photostability of Clover and mRuby2 (Supplementary Fig. 10). In contrast, during baseline imaging of Camuiα, the donor/acceptor ratio increased (Fig. 2c) while both fluorophore intensities decreased, indicating fast bleaching of Venus and slower bleaching of ECFP, as observed in other FRET reporters1,33.
Figure 2.
Clover-mRuby2 improves responses versus ECFP-Venus in the CaMKIIα reporter Camuiα. (a) Organization of Camuiα-CR, based on Camuiα. (b) Intensity-modulated ratiometric images of HeLa cells expressing Camuiα-CR or Camuiα before and after addition of 1 μM ionomycin at time = 0. Scale bar, 20 μm. (c) Mean donor/acceptor emission ratio changes over time. Error bars represent standard error of the mean (s.e.m.). n = 10 cells for Camuiα, n = 12 cells for Camuiα-CR. Difference in peak emission ratio change is statistically significant (P = 7.6e–5). (d,e) Similar emission ratio responses were obtained with modified Camuiα-CR reporters where Clover was replaced with circularly permuted variants. Mean peak emission ratio changes with cpClover157 (n = 4 cells) and cpClover173 (n = 10 cells) were 31.8 ± 3.5% and 32.0 ± 2.2% (mean ± s.e.m.) respectively, not significantly different from Camuiα-CR (P = 0.52 and P = 0.3 respectively).
We next replaced ECFP and Citrine in the PKA activity reporter AKAR222 with Clover and mRuby2. The resulting AKAR2-CR produced a mean 21.5% peak emission ratio change in HEK293 cells stimulated with forskolin, 117% larger than AKAR2 (Supplementary Fig. 11). Individual cellular responses also showed less fluctuation than with AKAR2. We also tested the photostability of AKAR2-CR under continuous arc lamp illumination. Mean fluorescence intensities in the individual Clover and mRuby2 channels were reduced less than 4% after 6 minutes of continual illumination (Supplementary Fig. 11 and Supplementary Video 1), while both emission signals were easily detectable over non-transfected cellular autofluorescence (signal/background ratios > 10). Although AKAR2 derivatives retaining ECFP and Citrine can be improved through extensive linker optimization34, these results further demonstrate that simple substitution with Clover-mRuby2 can improve ratio changes and enable the use of redder excitation wavelengths, and confirm suitability of Clover-mRuby2 for long-term imaging.
Improved sensing of membrane voltage
In Camuiα and AKAR2, the FPs flank a central region undergoing conformational change. We investigated whether Clover and mRuby2 could improve a FRET reporter with a different organization. In voltage-sensitive fluorescent proteins (VSFPs), FRET between tandem FPs fused to the C terminus of a transmembrane voltage sensing domain increases upon membrane depolarization (Fig. 3a)35. The existing VSFPs with the largest emission ratio changes are VSFP2.3, using Cerulean CFP and Citrine YFP36, and Mermaid14, using mUKG GFP and mKOκ OFP. For long-term neuronal imaging, CFP-YFP-based sensors like VSFP2.3 are not ideal because of phototoxicity and autofluorescence at CFP excitation wavelengths (Supplementary Fig. 12), while Mermaid exhibits fast bleaching14. Also, these VSFPs had difficulty discerning single action potentials (APs) without averaging37.
Figure 3.
Clover-mRuby2 FRET in voltage sensing. (a) Organization of VSFP-CR, based on VSFP2.3. (b) Ratiometric images of hippocampal neurons expressing VSFP-CR before and after depolarization with 50 mM KCl. Increased acceptor/donor emission was observed at the cell membranes and in neurites (arrows), but not inside the cell body (asterisk). Scale bar, 10 μm. (c) For voltage-emission ratio relationships, hippocampal neurons expressing VSFP-CR were subjected to various voltage steps from a resting potential of –70mV by patch-clamping (top panel), and emission ratio changes at each voltage were measured (bottom panel). (d) Mean emission ratio changes in response to voltage steps for VSFP2.3 (n = 39 cells) or VSFP-CR (n = 47 cells). Error bars represent s.e.m. Differences at potentials ≤ –100 mV and ≥ –40 mV are statistically significant by two-tailed t-test with Bonferroni correction for 10 repeated measures (P = 6.9e–4, 1.9e–5, 1.9e–5, 3.3e–5, 1.8e–4, 1.2e–3, 4.7e–3, and 1.5e–3 for –120, –100, –40, – 20, 0, 20, 40, and 60 mV respectively, versus required P < 0.005 for α < 0.05). (e) Ratio changes in response to a single AP were 1.03 ± 0.1% (mean ± s.e.m., n = 22 cells) for VSFP-CR and 0.82 ± 0.05% (n = 32 cells) for VSFP2.3 (P = 0.04 by two-tailed t-test). (f) VSFP-CR ratio changes (bottom) reliably detected APs (top) in a single unfiltered trace with a measured peak/noise ratio of 8.0. Measured power at the specimen plane was 1 W cm-2. At this power, the baseline ratio changed by 5% over 25 s.
We substituted Clover-mRuby2 for Cerulean-Citrine in VSFP2.3 to create VSFP-CR. VSFP-CR in HEK293 cells exhibited a basal FRET efficiency of 0.63 (Supplementary Fig. 12), similar to the Clover-mRuby2 fusion (Supplementary Fig. 9). VSFP-CR localized to the membrane of hippocampal neurons comparably to VSFP2.3 (Supplementary Fig. 12), where it responded to membrane depolarization with an increase in the mRuby2/Clover emission ratio (Fig. 3b). We related ratio changes to transmembrane voltage by patch-clamping multiple neurons expressing VSFP-CR or VSFP2.3 (Fig. 3c). Ratios responded to voltages from –100 mV to 20 mV, with the steepest response at –40 mV. With depolarization from –70 to +60 mV, VSFP-CR produced a 12.7% ratio change, 43% larger than VSFP2.3 (Fig. 3d). With hyperpolarization from –70 mV to –120 mV, VSFP-CR produced a ratio change of –2%, 67% larger than VSFP2.3 (Fig. 3d). Decreased donor and increased acceptor fluorescence both contributed to ratio increases for VSFP-CR and VSFP2.3 (Supplementary Fig. 12). Time constants of ratio changes upon activation and deactivation of VSFP-CR and VSFP2.3 were similar across voltage steps from –70 mV (Supplementary Fig. 13) and are similar to published figures for VSFP2.338. The ratio change upon depolarization from –70 to +60 mV at 20° C was best fit by a biexponential function with 5.4 ± 0.8 and 59.5 ± 5.5 ms time constants (mean ± standard error of the mean, n = 4).
We evaluated whether VSFP-CR could be used for AP detection. With evoked single APs in hippocampal neurons, membrane fluorescence of VSFP-CR (Supplemental Fig. 14) exhibited a mean emission ratio change of 1.03%, 25.6% higher than VSFP2.3 (Fig. 3e). Ratio changes reached 4% in individual neurons, allowing detection of single APs in our conditions with a peak/noise ratio of 8.0 (Fig. 3f). VSFP2.3 also responded to single APs, although the peak/noise ratio was limited to 5.9 in the best case, even with higher excitation power to compensate for its lower brightness (Supplementary Fig. 14). The emission ratio also drifted at baseline, indicating uneven bleaching of CFP and YFP, as previously observed1,33.
To determine how well VSFP-CR functions for long-term imaging compared to VSFP2.3, we first measured bleaching rates and emission intensities of donor and acceptor FPs in individual neurons expressing each sensor, and calculated the total number of photons that can be emitted. Total photonic output was 2.9-fold higher for Clover relative to Cerulean and 3.7-fold higher for mRuby2 relative to Citrine (Supplementary Fig. 14). For each neuron, we also measured the signal/noise ratio (SNR) of the ratiometric fluorescence transients induced by single APs, as defined in a separately described signal detection theoretical framework (B.A. Wilt, J.E. Fitzgerald, and M.J. Schnitzer, in press). Under conditions where AP detection is limited by optical shot noise, this measure of SNR allowed us to calculate the probability of correctly detecting a spike (PD) given a fixed false positive rate of 5%. We calculated the time over which SNR decays from 4 (PD = 99%) to 2 (PD = 64%) due to photobleaching at a fixed excitation intensity. The SNR decay time of VSFP-CR was 6.3 times longer than that of VSFP2.3 (Supplementary Fig. 14). These results indicate that VSFP-CR can be used to detect APs for a longer time than VSFP2.3 when excitation powers are adjusted for equal initial SNR, or can be used to detect APs with higher SNR than VSFP2.3 for equal measurement periods.
Detection of growth cone RhoA responses
To further determine whether Clover and mRuby2 could improve detection of biochemical events, we investigated RhoA activation during cue-dependent retraction of axonal growth cones. Ephrin stimulation induces growth cone retraction beginning within several minutes and complete by 20 minutes (Supplementary Fig. 15), a process that involves RhoA39,40 Biochemical assays in lysates detected RhoA activation at 30 minutes in central nervous system (CNS) neurons undergoing axon extension, but they cannot discern RhoA activity in neuronal growth cones, neuronal cell bodies, or astrocytes, which also express EphA receptors41. We asked whether the Raichu-RhoA reporter23 could show rapid RhoA activation specifically in growth cones in response to ephrins. Raichu-RhoA has been used to demonstrate RhoA activation during collapsin-mediated growth cone collapse of dorsal root ganglion growth cones42, but has not been used to report RhoA activation in the smaller CNS neuronal growth cones.
While Raichu-RhoA with SECFP and SEYFP already demonstrates FRET dynamic range approaching E = 0.5 (Fig. 1d), we investigated whether it could be further improved with Clover and mRuby2. Raichu-RhoA enables convenient testing because it can be encoded in either primarily activated or inactivated forms using Q63L or T19N mutations in the RhoA GTPase (Fig. 4a). The original Raichu-RhoA displayed a 30.6 ± 0.8% (mean ± standard error of the mean, n = 3 wells) peak acceptor/donor emission ratio change between Q63L and T19N mutants, corresponding to a FRET dynamic range of 0.39-0.48 (Fig. 1d). Raichu-RhoA-Clover-mRuby2 (Raichu-RhoA-CR) displayed a significantly greater ratio change of 79.5 ± 1.9% (n = 3 wells, P = 2E-5), corresponding to a wider FRET dynamic range of 0.25-0.47 (Supplementary Fig. 16). The lower FRET in the T19N form of Raichu-RhoA-CR may be due to the inability of Clover-mRuby2 to dimerize, whereas weak dimerization between CFPs and YFPs has been shown to contribute to basal and induced FRET34. We also estimated FRET E by acceptor photobleaching, obtaining 0.51 ± 0.056 and 0.10 ± 0.004 (n = 3) for active and inactive Raichu-RhoA-CR, reasonably close to the spectra-based calculations. The lower pKa of mRuby2 versus YFPs (Table 1) should render FRET between Clover and mRuby2 more pH-independent than between a CFP and a YFP. Indeed, FRET in Raichu-RhoA-CR was stable between pH 6.5 and 7.5, in contrast to the original Raichu-RhoA (Supplementary Fig. 16). mTurquoise1 and mTurquoise2 are recently described CFPs with dramatically increased quantum yields30,43, which would be expected to improve r0. We used these new CFPs (which contain the monomerizing mutation A206K) as FRET donors for the highly absorbing SEYFP (ε = 101,000 M–1 cm–1). FRET E in the inactive state was nearly unchanged (0.41 for mTurquoise1 and 0.42 for mTurquoise2, versus 0.39 for SECFP, Supplementary Fig. 16), suggesting that the opposing effects of higher r0 and increased monomerization were approximately balanced. As expected, FRET E in the active state increased with the improved CFPs (0.58 for mTurquoise1 and 0.59 for mTurquoise2, versus 0.48 for SECFP, Supplementary Fig. 16). Interestingly, emission ratio changes with the new CFP donors were still lower than with Raichu-RhoA-CR (50.0 ± 3.3% for mTurquoise1 and 46.4 ± 0.2% for mTurquoise2, versus 79.5 ± 1.9% for Clover-mRuby2, n = 3 wells each, Supplementary Fig. 16). Spectral modeling shows that Clover-mRuby2 would generate a higher ratio change if restricted to the FRET E of mTurquoise1-SEYFP or mTurquoise2-SEYFP (Supplementary Fig. 16), likely due to improved separation of donor and acceptor emissions. Similarly, mTurquoise1-SEYFP and mTurquoise2-SEYFP pairs would generate lower ratio changes than Raichu-RhoA-CR at the FRET E of Clover-mRuby2 (Supplementary Fig. 16). These comparisons confirm that the Clover-mRuby2 pair can widen FRET dynamic range, increase emission ratio changes, and improve pH stability versus CFP-YFP pairs, even with recent CFP improvements.
Figure 4.
Reporting of fast local RhoA activation in neurons with Raichu-RhoA-CR. (a) Design of Raichu-RhoA-CR, based on Raichu-RhoA. (b) Ephrin-A stimulation locally activates RhoA in a hippocampal growth cone (asterisk) from the first time point after stimulation. Scale bar is 10 μm. (c) Raichu-RhoA-CR emission ratio graphed as mean ± s.e.m. (n = 5 cells). Peak ratio change (asterisk) was significantly different from baseline by two-tailed t-test (P = 0.0187). (d) With Raichu-RhoA, peak ratio change was not significantly different from baseline (P = 0.468, n = 8 cells).
We next used Raichu-RhoA-CR to visualize RhoA activity in growth cones of hippocampal and cortical neurons from 18-day old rat embryos before and during ephrin stimulation. Bath treatment by pre-clustered ephrin-A4 and ephrin-A5 induced rapid growth cone retraction accompanied by Raichu-RhoA-CR activation (Fig. 4b). In neurons where the cell body and axonal growth cone were both visible (Fig. 4b and Supplementary Video 2), RhoA activation occurred in the growth cone from the first time point (3 min) after stimulation, when the cell body did not show RhoA activity discernable over baseline. Raichu-RhoA-CR signal then decayed between 6 and 18 min after stimulation while the growth cone shrunk. Peak Raichu-RhoA-CR activation occurred in responding growth cones at 6 min and was significantly different from baseline (Fig. 4c). In contrast, the original Raichu-RhoA could not detect changes in RhoA in responding neurons (Fig. 4d). These results with Raichu-RhoA-CR reveal that RhoA activation by ephrins can indeed occur with fast kinetics preceding growth cone retraction and with high spatial specificity.
DISCUSSION
We show in four FRET reporters – Camuiα, AKAR2, VSFP2.3, and Raichu-RhoA – that simple substitution of Clover-mRuby2 in place of the original CFP-YFP pairs confers improvements in FRET dynamic range and emission ratio changes without further optimization.
The new voltage sensor, VSFP-CR, exhibits larger ratiometric changes than its parent VSFP2.3, previously the most practically useful genetically encoded voltage indicator37. The brightness of VSFP-CR enables detection of single APs in cultured neurons with improved SNR, and its photostability, especially in the acceptor channel, extends imaging time relative to VSFP2.3. VSFP-CR also should be more practically useful for voltage sensing than the recently described archaerhodopsin ArchD95 which exhibits voltage-sensitive fluorescence44. ArchD95 fluorescence changes over baseline are larger than VSFP-CR, but its quantum yield (0.0004) is orders of magnitude lower than those of Clover (0.76) and mRuby (0.38). Imaging single APs with VSFP-CR can thus be performed at orders of magnitude lower powers than required for ArchD95 (~ 0.01 W mm–2 here vs ~10 W mm–2), making single-cell and population imaging of APs accessible using conventional arc lamps and without using high-NA oil objectives.
Experiments with the new RhoA sensor, Raichu-RhoA-CR, demonstrate that Clover-mRuby2 can improve detection of fast and localized cellular events. Our finding that localized RhoA activation in growth cones following ephrin-A stimulation occurs faster than previously detected by biochemical assays highlights the importance of using real-time optical reporters to study spatially regulated processes.
Clover-mRuby2 should allow FRET imaging with less phototoxicity and higher signal/background fluorescence than CFP and YFP, and is compatible with blue lasers and excitation filters common to most fluorescence microscopes. Clover and mRuby2 should facilitate the improvement of other existing FRET sensors and the development of new ones. Further enhancement of brightness, photostability, maturation, and Stokes’ shifts in green and red FPs should be a generalizable approach that allows for continued systematic improvement of FRET reporter dynamic range in the future.
METHODS
FRET-distance modeling
To model FRET-distance relationships, we calculated theoretical emission spectra using the equation ftotal(λ) = εD(λex)•[(1–E)•ϕD•fD(λ)+E•εA•fA(λ)]+εA(λex)•fA(λ), where ftotal is fluorescence at wavelength λ, εD(λex) is extinction coefficient of the donor at the excitation wavelength, E is FRET efficiency, ϕD is quantum yield of the donor, fD(λ) is normalized emission of the donor at wavelength λ, ϕA is quantum yield of the acceptor, fA(λ) is normalized emission of the acceptor at wavelength λ, and εA(λex) is extinction coefficient of the acceptor at the excitation wavelength. E for a measured emission spectrum was then determined as the value of E (to two significant digits) that gave the best fit between the theoretical and measured emission spectra. An apparent interfluorophore distance r was then calculated from the Förster equation E = 1/(1+(r6/r06)).
Plasmid construction
Plasmids were constructed by standard molecular biology methods, including PCR, overlap extension PCR, and restriction fragment ligation, and verified by sequencing of all cloning junctions and PCR products. Further details are described in the methods descriptions for specific reporters below. Complete plasmid sequences are available upon request and plasmids will be distributed through Addgene (http://www.addgene.org/).
Protein production and characterization
Protein mutagenesis, protein expression, and protein purification from bacterial lysates were performed as previously described31. Clover-mRuby2 tandem fusion consisted of aa 1-228 of Clover fused to aa 3-227 of mRuby2 via the linker sequence LESGGEDPMVSKGEE. Clover-mCherry consisted of aa 1-228 of Clover fused to aa 1-231 of mCherry via the linker sequence LESGGEDP. Photobleaching measurements were taken on droplets of purified protein in PBS within mineral oil using light from a 150-W xenon arc lamp (Zeiss) at 100% neutral density passed through a 480/30 nm (for Clover) or a 545/20 nm (for mRuby2) excitation filter (Chroma), focused with a 40× 1.2-numerical aperture (NA) CApochromat water-immersion lens on an Axiovert 200M inverted microscope (Zeiss). Photobleaching of CFPs was performed similarly except a 120-W mercury metal-halide arc lamp (Exfo) and a 440/20 nm excitation filter (Chroma) were used. Photobleaching times were normalized to reflect an illumination intensity initially producing 1000 photons per s per molecule as previously described2. Excitation and emission spectra on purified proteins were acquired on a SPEX Fluorolog fluorimeter (Horiba) or Safire2 (TECAN), absorption spectra were acquired on a Cary Eclipse (Varian) or Safire2 (TECAN), and extinction coefficients and quantum yields calculated as previously31.
To determine whether or not Clover would photoactivate in response to short wavelength illumination, purified PA-GFP (A206K) and Clover in PBS were illuminated with a 250W metal halide light source (Moritex, MME 250) equipped with a 395/40 nm filter. Purified protein was placed in a 96 well plate (black, flat bottom, UV transparent) and illuminated for increments of time totaling 1, 2, 4, 8, 16, 32, and 64 min. At each interval, fluorescence excitation and emission spectra were obtained with a Safire2 fluorimeter (TECAN). Excitation spectrum was obtained from 300 nm to 574 nm (emission collected at 600 nm). Emission spectrum was obtained from 480 nm to 720 nm with 450 nm excitation light. For both excitation and emission spectrum a 5 nm band pass was used for both excitation and emission.
HeLa and HEK293 cell culture and transfection
Unless otherwise noted, cell culture and transfection was carried out as described below. Cells were maintained in high glucose Dulbecco's Modified Eagle Medium (DMEM, HyClone) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 2 mM glutamine (Sigma) at 37° C in air with 5% CO2. Cells were transfected at 75-90% confluency with Lipofectamine 2000 (Invitrogen) in 35 mm coverglass-bottom dishes (In Vitro Scientific) or LabTek 8-chamber slides (Nunc). Transfections were carried out according to manufacturer's instructions, except that amounts of DNA and transfection reagent were halved to reduce cell toxicity. The medium was refreshed 4-6 h after transfection. Cells were cultured for another 12-36 h before experiments were performed.
Live cell fluorescent protein photobleaching
Spinning disk confocal and widefield microscopy photobleaching experiments were performed with fusions of Clover or mRuby2 to human histone H2B to confine fluorescence to the nucleus. HeLa cells were cultured in DMEM supplemented with 12.5% calf serum on 35 mm Delta-T (Bioptechs) imaging dishes and transfected 1 day before imaging. Widefield photobleaching was performed on a Nikon TE2000 inverted microscope equipped with a Nikon Plan Fluorite 40x dry objective (NA = 0.85) and an X-Cite Exacte metal halide lamp (Lumen Dynamics). Photobleaching was conducted using BrightLine GFP-4050A and TRITC-B-000 filter cubes (Semrock) and a Newport (Irvine, CA, U.S.A.) 1918-C optical power meter was used to verify that the illumination power at the objective was 7.3 mW cm–2. Power moderation was achieved by using an adjustable iris diaphragm contained within the lamp. With the aperture opening set and neutral density filters in the illuminator inserted (to avoid photobleaching), a region containing 10-20 evenly bright nuclei was located. The neutral density was then removed from the light path and the region was photobleached continuously with a set exposure time (75 ms for Clover and 100 ms for mRuby2) for 6 minutes (mRuby2) or 9 minutes (Clover) for a total of 1800-3000 frames. Images were collected with a QImaging Retiga EXi camera (Photometrics). Multiple regions were photobleached to ensure that data for 30 nuclei could be averaged. The raw data was collected and analyzed using NIS-Elements software (Nikon). Spinning disk confocal photobleaching measurements were collected on a CSU spinning disk confocal microscope with a 63× 1.4-NA Plan-Apo oil-immersion objective (Zeiss) and FITC and TRITC filter sets (Chroma). Lasers (488 nm diode for Clover and 561 nm diode for mRuby2) were tuned to an output power of 6.2 mW cm–2 (Clover) or 3.43 mW cm–2 (mRuby2), at the objective, with a FieldMax II-TO power meter (Coherent). Images were captured using a Evolve electron-multiplied CCD camera (Photometrics) with the gain multiplier turned off. For each image set, using minimum laser power, a region of evenly bright nuclei was located. The laser power was raised back to the bleach level and each region was photobleached continuously for 5-8 minutes for a total of 1000-2000 frames, with multiple regions being bleached to ensure data for 30 nuclei. Raw data was collected with the AxioVision software (Zeiss) and then analyzed with NIS-Elements software (Nikon).
Ratiometric image analysis
FRET measurements were quantified using ImageJ (NIH). Raw 16-bit TIFF files were imported into ImageJ, then regions were drawn on random transfected cells for reporter responses and on untransfected cells for background measurements. Emission ratios were obtained by calculating background-subtracted FRET intensities divided by background-subtracted donor intensities, except for Camuiα and Camuiα-CR, for which net donor over net FRET ratios were calculated. Time-course ratio measurements were normalized to baseline pre-stimulation values. Additional analyses were performed for voltage sensors as described below. Intensity-modulated displays were generated using a full-spectrum look-up table with minimum values in blue and maximum values in red and with intensity modulation by the acceptor channel.
Camuiα characterization and improvement
Camuiα was generously provided by Yasunori Hayashi. Camuiα was subcloned from the pEGFP backbone into pcDNA 3.1. For determination of the operating FRET efficiency range of Camuiα with CFP-YFP, FRET was determined in HEK293FT cell lysates. Two days following transfection, cells were homogenized in CaMKII assay buffer composed of 40 mM HEPES-NA (pH 8.0), 0.1 mM EGTA, 5 mM magnesium acetate, 0.01% Tween-20, 1 mM DTT, and protease inhibitor cocktail (Roche). After centrifugation, supernatant was transferred to a 96-well Special Optics flat-bottom tissue culture-treated microplate (Corning). Fluorescence spectra pre-stimulation were obtained on a Safire2 (TECAN) fluorimeter using 430 nm excitation light with 5 nm bandwidth. Emission was collected from 460 to 650 nm in 1 nm steps with 5 nm bandwidth. To activate the reporter, 1 mM calcium was added in the presence of 1 μM calmodulin (Calbiochem) and 50 μM ATP (Calbiochem) at room temperature. Spectra was measured after activation.
To construct Camuiα-CR, Clover (aa 1-228) was amplified with a N-terminal NheI site and a C-terminal extension encoding the linker between Venus and the CaMKIIα domain. The CaMKIIα domain flanked by linkers on either end was PCR amplified. mRuby2 (aa 1-237) was PCR amplified with a N-terminal extension encoding the linker following the CaMKIIα domain and with an ApaI restriction site at the C terminus. The full insert was assembled by overlap PCR and cloned into the pcDNA3.1 backbone using restrictions site NheI and ApaI. Circular permutants of Clover were constructed by amplifying two pieces: Clover (aa 157-228 or aa 173-228) with a NheI restriction site and a start codon at the N terminus and a five amino acid linker “GGSGG” at the C terminus, and Clover (aa 1-156, aa 1-172) with the “GGSGG” linker at the N terminus plus overlap with the linker preceding the CaMKIIα domain. The full circularly permuted protein sequence was assembled by overlap PCR and then used to assemble the full reporter sequence in a second overlap PCR step.
For microscopy experiments, cells were cultured and transfected in chamber slides and imaged two days following transfection. Cells were serum starved in serum-free DMEM for 4-6 hours, then washed two times with Hank's Balanced Salt Solution (HBSS, HyClone) and maintained in HBSS with 2 mM calcium. Cells were imaged using a cooled CCD camera (SPOT Pursuit USB2.0) and a 40× 1.2-NA C-Apochromat water-immersion objective on an Axiovert 200M inverted microscope (Zeiss) controlled by Micro-manager 1.4 software49 on a 17-inch 2.5-GHz Core 2 Duo MacBook Pro running Mac OS 10.6.8 (Apple). Illumination was provided by an Exfo metal-halide light source attenuated with a 10% neutral-density filter. Consecutive FRET and donor emission images were acquired with the following filters (ex = excitation, em = emission): ex 440/20 nm and em 480/40 nm for CFP, ex 440/20 nm and em 535/35 nm for CFPYFP FRET, ex 485/30 nm and em 530/40 nm for Clover, and ex 485/30 nm and em 595/70 nm for Clover-mRuby2 FRET). After baseline acquisition for 6 min, cells were stimulated with 1 μM of the calcium ionophore ionomycin and images were acquired for 10 min.
AKAR2 characterization and improvement
To construct AKAR2-CR, a PCR fragment encoding Clover (aa 1-228) was cloned between restriction sites BamHI and SphI to replace CFP, and a PCR fragment encoding mRuby2 (aa 1-237) was cloned between SacI and EcoRI to replace YFP in AKAR2. For determination of the operating FRET efficiency range of AKAR2 with CFP-YFP, transfected HEK293 cells expressing AKAR were trypsinized and transferred to 96-well Special Optics flat-bottom tissue culture-treated microplates two days after transfection. Cells were allowed to adhere for 3 h at 37° C and 5% CO2. in DMEM with 10% FBS, then incubated in serum free DMEM for 3 h. Cells were then washed twice with PBS and transferred to Dulbecco's Phosphate Buffered Saline (DPBS, Invitrogen). Emission spectra before and after stimulation with 50 μM forskolin (Cell Signaling Technology) and 3-siobutyl-1-methylxanthine (100 μM IBMX; Cell Signaling Technology) were then obtained using a Safire2 fluorimeter (TECAN) with settings as above for Camuiα.
For time-lapse imaging, HeLa cells were transfected and cultured for two days on glass-bottom dishes in DMEM with 10% FBS, then incubated in serum free DMEM for 3 h. Cells were imaged with a C-Apochromat 40× 1.2NA water-immersion objective on a Zeiss Axiovert 200M controlled by a custom-built computer running Windows XP (Microsoft) and Meta-Fluor software (Molecular Devices). Excitation light from a 150 W xenon arc lamp was passed through a 10% neutral density filter. Consecutive FRET and donor emission images were acquired with 500ms exposures each every 15s with the following filters (ex = excitation, em = emission): ex 420/20 nm and em 475/40 nm for CFP, ex 420/20 nm and em 535/25 nm for CFP-YFP FRET, ex 460/20 nm and em 535/45 nm for Clover, and ex 460/20 nm and em 595/50 nm for Clover-mRuby2 FRET). Signal/noise ratio was calculated as mean ratio change divided by standard variation of the 8 time points from 4 to 6 min after stimulation. Conditions for continual imaging of AKAR-CR were similar, except illumination was continual and emission filters were cycled between 535/45 nm for Clover and 595/50 nm for mRuby2 as quickly as possible. Under these conditions, we estimate the excitation rate per molecule of Clover to be 4.3% of that used in the normalized in vitro photobleaching conditions.
Quantification of cellular autofluorescence
Rat hippocampal cells were dissected from postnatal day 0 Sprague-Dawley pups (Charles River Labs), dissociated with papain, and cultured in Neurobasal supplemented with glutamine and B27 (Invitrogen) as previously described40. At 9 days in vitro, cells were fixed with 4% paraformaldehyde at room temperature for 20 minutes, washed in PBS, then imaged on an FV1000 laser scanning confocal microscope (Olympus) through a 40x N.A. 1.3 oil immersion objective. Powers of 440nm and 488nm laser lines were quantified using an X-Cite XP750 power sensor connected to an XR2100 power meter (Exfo). Autofluorescence in the CFP channel was imaged using the 440 nm laser at 25μW and a 480/40 nm emission filter. Autofluorescence in the GFP channel was imaged using the 488nm laser at 25μW and a 600/60 emission filter. ImageJ was used to quantify background-subtracted whole cell autofluorescence.
Voltage sensor characterization and improvement
pcDNA3.1(–)VSFP2.3 was constructed from pcDNA3.1(–)VSFP2.1 (Addgene plasmid 16255) using site-directed mutagenesis and subcloned in the NheI-HindIII sites of pcDNA3.1/Puro-CAG, a pcDNA3.1 derivative in which a cytomegalovirus enhancer and chicken beta-actin promoter replace the cytomegalovirus enhancer-promoter. We constructed VSFP-CR by PCR overlap assembly to fuse PCR fragments encoding the voltage-sensitive domain and 15-aa phosphobinding motif from VSFP2.3, Clover aa 1-228, the linker “LESGGE”, and mRuby2 aa 1-237.
For quantification of voltage sensor membrane localization, dissociated rat hippocampal neurons were transfected 7 days post-plating using Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. At 9 days in vitro, cells were fixed with 4% paraformaldehyde at room temperature for 20 minutes, washed in PBS, then imaged on an FV1000 laser scanning confocal microscope (Olympus) through a 40× 1.3-NA oil immersion objective. CFPs were imaged using a 440 nm laser and a 480/40 nm emission filter, YFPs with a 515 nm laser and a 580/100 nm emission filter, GFPs with a 488 nm laser and a 530/40 nm emission filter and OFPs with a 559 nm laser and a 600/60 nm emission filter. Neurons with intermediate levels of sensor expression were chosen for imaging. Neurons with very low expression levels were discarded due to signal insufficiency, while overexpressors usually showed abnormal morphology. A single slice located approximately mid-way between the top and bottom of each neuron was imaged using an 1-Airy pinhole setting. Data analysis was performed blind. ImageJ (NIH) was used for manual tracing of the plasma membrane, and for quantification of background-subtracted total and membrane-only fluorescence.
For depolarization experiments, neurons were electroporated after dissection before plating, and stimulated at 3 days in vitro by addition of 150 mM KCl to a final concentration of 50 mM while imaging was performed at 1 min intervals at room temperature Cells were imaged using a cooled CCD camera (Hamamatsu Orca AG) and a 40× 1.2-NA C-Apochromat water-immersion objective on an Axiovert 200M inverted microscope (Zeiss), controlled by a 17-inch 2.5-GHz Core 2 Duo MacBook Pro running Mac OS 10.6.8 (Apple) and Micro-manager 1.4. For patch-clamp experiments, cultured neurons were transfected after 4-7 days in vitro using calcium phosphate. Two days after transfection, cells were patch-clamped at 20º C using an Axopatch 700B amplifier (Axon Instruments) with borosilicate glass electrodes with resistances of 3-6 MOhm. Cells were superfused in a chamber mounted on the stage of a BX51WI epifluorescent microscope with a 40x 0.8-NA water immersion objective (Olympus). The extracellular solution contained 150 mM NaCl, 4 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM CaCl2, and 2 mM MgCl2. The intracellular solution contained 129 mM K-gluconate, 10 mM KCl, 10 mM HEPES, and 4 mM ATP.
Fluorescence traces were measured while cells were voltage-clamped or current-clamped in whole-cell mode. Access resistances were 5-25 MOhm and monitored throughout the recording. Step voltage depolarizations were applied to change the membrane potential from a holding voltage of –70 mV to voltages ranging from –120 mV to 100 mV in 20 mV steps for 1 s. To generate action potentials, 1 nA of current was injected for 2 ms. Electrode voltages and currents were recorded using pClamp (Axon Instruments) and analyzed using custom software written in MATLAB. Voltage traces were corrected for the junction potential post hoc. To measure the fast on kinetics of Clover, emission was passed through a 520/40 bandpass filter to a Hamamatsu R6357 PMT and sampled at 5-10 kHz.
Fluorescence excitation was delivered using a mercury arc lamp passed through either 435/20 or 475/25 nm excitation filters (for CFP-YFP and Clover-mRuby2 FRET pairs, respectively). Fluorescence emission was passed through a 455 nm dichroic or 495 nm dichroic and long pass emission filter (for CFP-YFP and Clover-mRuby2, respectively) to a DualView emission splitter (Optical Insights). Emission was then split by a 505 nm dichroic and passed through 465/30 nm and 535/30 nm emission filters (CFP-YFP) or a 565 nm dichroic and 520/40 nm and 630/50 nm emission filters (Clover-mRuby2). Emission was recorded at 100 Hz by an Andor iXon 897 electron-multiplied charge-coupled device camera cooled to –80° C. To avoid intracellular aggregates, pixels were ranked by their signal/noise ratio and the top 5% (patch-clamp analysis) or 15% (evoked AP analysis) of all image pixels (including background) were summed to produce fluorescence time traces for further analysis. Peak-to-noise ratios were defined as the ratio of baseline-subtracted peak emission ratio changes and the standard deviation of the baseline trace in 100 ms windows before and after the AP train. Noise in fluorescence traces was found to approach the theoretical limit set by electron multiplication amplified shot noise for the individual channels and for their ratio.
Integrated photon output and signal/noise ratio decay measurements
Photobleaching curves of donor and acceptor channels were acquired on multiple neurons patch-clamped in current clamp mode with 488 nm (for VSFP-CR) and 445 nm (for VSFP2.3) lasers (OBIS, Coherent) at 10 W cm–2. Cellular emission intensities F in photons/s were calculated using the known gain settings of our camera. Photobleaching times τ in the donor and acceptor channels were derived from monoexponential fits to baseline fluorescence at the cellular resting potential. The number of emitted photons was calculated as:
The SNR of an AP is defined in a separate manuscript (B.A. Wilt, J.E. Fitzgerald, and M.J. Schnitzer. Photon shot-noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys. J., in press), where the SNR describes the tradeoff between true and false detections of APs by the relation PD = 1–Φ(Φ–1(1–PF)–SNR), where PD is the detection probability, PF the false positive rate and Φ is the cumulative distributive function of a standard normal distribution. The fluorescence transient evoked by an AP was modeled to be a step function followed by a monoexponential decay equal to the off time of the probe τoff. In the limits of high photon counts and small ratiometric changes ΔR/R << 1, the SNR of an AP is (ΔR/R)•(F1F2τoff /2(F1+F2))1/2, where F1 and F2 are the intensities of the donor and acceptor. Assuming a reciprocal relation between excitation intensity and photobleaching time, the time to decay from an AP SNR of θ to θ/2 was found to equal T1/2 = ln(4) SNRC2τoff/2θ2, where SNRC, the signal-to-noise capacity, is a dimensionless measure of a probe's ability to report activity and is defined as the instantaneous peak-to-noise ratio of a probe integrated over all time. For a ratiometric indicator,
Raichu-RhoA characterization and improvement
pCAGGS-Raichu-RhoA was kindly provided by Michiyuki Matsuda. Raichu-RhoA-CR was constructed by replacing SEYFP with Clover (aa 1-227) at restriction sites EcoRI and XhoI and SECFP with mRuby2 (aa 1-238) at restriction sites NotI and XbaI in pCAGGS-Raichu-RhoA. Raichu-RhoA-mTurquoise1 and Raichu-RhoA-mTurquoise2 were constructed similarly by replacing SECFP with mTurquoise1 and mTurquoise2 at restriction sites NotI and XbaI. To determine operating FRET efficiency, HEK293FT cells expressing dominant-negative (T19N) or constitutive-active (Q63L) variants of each reporter were trypsinized and transferred to a 96 well plate. Emission spectra were obtained with a Safire2 (TECAN) with the same measurement parameters as used for Camuiα. To determine the pH dependency of FRET efficiency, HeLa cells transfected with Raichu-RhoA and Raichu-RhoA-CR (Q63L and T19N for each construct) were lysed two days after transfection in lysis buffer at a range of pHs. The buffer consisted of 20 mM Tris-HCl (pH – 6.5, 6.75, 7.0, 7.25, 7.5), 100 mM NaCl, 0.5% Triton X-100, and 5mM MgCl2. Following lysis, fluorescence spectra was obtained with the Safire2 fluorimeter.
To measure FRET efficiency through acceptor photobleaching, HEK293FT cells transfected with Raichu-RhoA-CR were collected and transferred to a 96 well plate with UV transparent, flat bottom and black walls. Cells were imaged and bleached with the 40× 1.2-NA C-Apochromat water-immersion objective on an Axiovert 200M (Zeiss). Illumination was provided by an Exfo metal-halide light source. FRET and donor images were acquired before acceptor photobleaching. mRuby2 was bleached at 100% neutral density using 545/10 nm light until < 5% initial fluorescence intensity remained. Donor and FRET emission images were acquired after and before bleaching with the same settings. ImageJ was used for analysis. Whole cells were selected as regions to measure donor fluorescence before and after acceptor photobleaching. FRET efficiency was determined as 1 minus the fraction of remaining Clover signal. The same process was used with Raichu-RhoA but bleaching of SECFP precluded the determination of FRET efficiency.
For growth cone retraction experiments, primary hippocampal or cortical neurons were dissected from embryonic day 18 rats (Charles River Labs) and dissociated with papain as previously described40. Neurons were transfected by electroporation with a rat neuron Nucleofector kit (Lonza) and plated in chamber slides. Prior to plating, slides were coated by incubating with 0.25 mg/mL poly-D-lysine (Sigma) in borate buffer for 12-24 h, washing three times with water for 15 min, incubating with 18 μg/mL laminin (BD Biosciences) in Neurobasal for 12-16 h, and washing. Neurons were plated in Neurobasal with Glutamax, penicillin and streptomycin, and 5% FBS (all purchased from Invitrogen). Plating media was replaced 12 hours later with phenol red-free Neurobasal media with Glutamax and B27. Neurons were imaged 1-2 d after transfection in HBSS with the 40x 1.2NA C-Apochromat water-immersion objective on the Zeiss Axiovert 200M. 10 μg/mL of ephrin-A4-Fc and ephrin-A5-Fc (R&D Systems) were separately preclustered with 50 μg/mL of goat anti-human IgG (H+L) (Jackson ImmunoResearch) for 1 h, then mixed. Baseline measurements were taken in FRET donor and acceptor channels every 3 min for 21 min. Pre-clustered ephrin-A was added to chambers at a final concentration of 5 μg/mL of ephrin-A and 25 μg/mL of IgG. FRET imaging was continued for another 21 min.
Statistical methods
Statistical comparisons between single pairs of pre-identified measures of interest were performed with a two-tailed Student t-test with significance level set at α = 0.05 unless otherwise stated. F-tests were used to determine the equivalence of variance for each comparison, and t-tests assuming equal or unequal variances were selected accordingly. For statistical comparisons between voltage reporters for responses at a series of voltages above and below the holding potential, a two-tailed Student t-test was performed where the P-value required for statistical significance at the α < 0.05 level was adjusted to 0.005 by the Bonferroni method. Statistical tests were performed in Excel (Microsoft) and MatLab (Mathworks).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y. Hayashi (RIKEN Brain Science Institute) for the Camuiα plasmid, J. Zhang (John Hopkins Medicine) for the AKAR2 plasmid, M. Matsuda (Kyoto University) for the Raichu-RhoA plasmid, and Paulmurugan Ramasamy (Stanford University) for the pcDNA3.1/Puro-CAG plasmid. We thank N. Desai for help with cloning of the voltage sensors, members of the Lin laboratory for helpful discussion, and M. E. Greenberg (Department of Neurobiology, Harvard Medical School) for advice and resources during axon guidance experiments. This work was supported by the Burroughs Wellcome Fund (M.Z.L.), a Stanford University Bio-X Interdisciplinary Initiatives Project grant (M.Z.L. and M.J.S.), a Siebel Foundation Scholarship (A.L.), the Stanford CNC Program (Y.G., J.D.M., M.J.S.), the National Academy of Sciences Keck Futures Initiative (Y.G., J.D.M., M.J.S.), National Science Foundation grant 1134416 (M.Z.L.), and National Institutes of Health grants R01NS076860 (M.Z.L.) and 4R37NS027177-23 (R.Y.T.). M.Z.L. is a Rita Allen Foundation Scholar.
Footnotes
AUTHOR CONTRIBUTIONS
M.Z.L. conceived the study. A.L., F.S.-P, M.Z.L., Y.G., and J.D.M. designed and performed FRET experiments and analyzed data. M.R.M. and M.Z.L. created and characterized fluorescent protein variants. M.A.B. and M.W.D. created and characterized fluorescent protein targeting fusions. P.J.C. and M.W.D. performed live-cell photobleaching experiments. J.W. provided unique reagents. M.S. and R.Y.T. provided ideas and advice. A.L., F.S.-P., and M.Z.L. wrote the manuscript.
REFERENCES
- 1.Reiff DF, et al. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 4.Raarup MK, et al. Enhanced yellow fluorescent protein photoconversion to a cyan fluorescent protein-like species is sensitive to thermal and diffusion conditions. J Biomed Opt. 2009;14:034039. doi: 10.1117/1.3103338. [DOI] [PubMed] [Google Scholar]
- 5.Malkani N, Schmid JA. Some secrets of fluorescent proteins: distinct bleaching in various mounting fluids and photoactivation of cyan fluorescent proteins at YFP-excitation. PLoS One. 2011;6:e18586. doi: 10.1371/journal.pone.0018586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 2003;36:280–290. doi: 10.1046/j.1365-313x.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- 7.Hockberger PE, et al. Activation of flavin-containing oxidases underlies light-induced production of H2O2 in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:6255–6260. doi: 10.1073/pnas.96.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwok S, et al. Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem Biophys Res Commun. 2008;369:519–525. doi: 10.1016/j.bbrc.2008.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mutoh H, et al. Spectrally-resolved response properties of the three most advanced FRET based fluorescent protein voltage probes. PLoS One. 2009;4:e4555. doi: 10.1371/journal.pone.0004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.van der Krogt GN, Ogink J, Ponsioen B, Jalink K. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS One. 2008;3:e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 14.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbo D, et al. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 2009;9:24. doi: 10.1186/1472-6750-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TWJ. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS One. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 18.Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Photoconversion in orange and red fluorescent proteins. Nat Methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey CD, et al. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci U S A. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kredel S, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takao K, et al. Visualization of synaptic Ca2+ /calmodulin-dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–3112. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 23.Yoshizaki H, et al. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol Annu Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 25.Ormo M, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 26.Kennis JT, et al. Uncovering the hidden ground state of green fluorescent protein. Proc Natl Acad Sci U S A. 2004;101:17988–17993. doi: 10.1073/pnas.0404262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brejc K, et al. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci U S A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson JN, et al. Structure and mechanism of the photoactivatable green fluorescent protein. J Am Chem Soc. 2009;131:4176–4177. doi: 10.1021/ja808851n. [DOI] [PubMed] [Google Scholar]
- 29.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 30.Goedhart J, et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods. 2010;7:137–139. doi: 10.1038/nmeth.1415. [DOI] [PubMed] [Google Scholar]
- 31.Lin MZ, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kredel S, et al. Optimized and far-red-emitting variants of fluorescent protein eqFP611. Chem Biol. 2008;15:224–233. doi: 10.1016/j.chembiol.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat Protoc. 2009;4:1623–1631. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- 34.Kotera I, Iwasaki T, Imamura H, Noji H, Nagai T. Reversible dimerization of Aequorea victoria fluorescent proteins increases the dynamic range of FRET-based indicators. ACS Chem Biol. 2010;5:215–222. doi: 10.1021/cb900263z. [DOI] [PubMed] [Google Scholar]
- 35.Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- 36.Lundby A, Mutoh H, Dimitrov D, Akemann W, Knopfel T. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS One. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akemann W, Middleton SJ, Knopfel T. Optical imaging as a link between cellular neurophysiology and circuit modeling. Front Cell Neurosci. 2009;3:5. doi: 10.3389/neuro.03.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundby A, Akemann W, Knopfel T. Biophysical characterization of the fluorescent protein voltage probe VSFP2.3 based on the voltage-sensing domain of Ci-VSP. Eur Biophys J. 2010;39:1625–1635. doi: 10.1007/s00249-010-0620-0. [DOI] [PubMed] [Google Scholar]
- 39.Wahl S, Barth H, Ciossek T, Aktories K, Mueller BK. Ephrin-A5 induces collapse of growth cones by activating Rho and Rho kinase. J Cell Biol. 2000;149:263–270. doi: 10.1083/jcb.149.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shamah SM, et al. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- 41.Nestor MW, Mok LP, Tulapurkar ME, Thompson SM. Plasticity of neuron-glial interactions mediated by astrocytic EphARs. J Neurosci. 2007;27:12817–12828. doi: 10.1523/JNEUROSCI.2442-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura T, Aoki K, Matsuda M. Monitoring spatio-temporal regulation of Ras and Rho GTPase with GFP-based FRET probes. Methods. 2005;37:146–153. doi: 10.1016/j.ymeth.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 43.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 47.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 48.Mori MX, Imai Y, Itsuki K, Inoue R. Quantitative measurement of Ca(2+)-dependent calmodulin-target binding by Fura-2 and CFP and YFP FRET imaging in living cells. Biochemistry. 2011;50:4685–4696. doi: 10.1021/bi200287x. [DOI] [PubMed] [Google Scholar]
- 49.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.