Abstract
Human soluble calcium-activated nucleotidase 1 (hSCAN-1) represents a new family of apyrase enzymes that catalyze the hydrolysis of nucleotide di- and tri-phosphates, thereby modulating extra-cellular purinergic and pyrimidinergic signaling. Among well-characterized phosphoryl transfer enzymes, hSCAN-1 is unique not only in its unusual calcium-dependent activation, but also in its novel phosphate-binding motif. Its catalytic site does not utilize backbone amide groups to bind phosphate, as in the common P-loop, but contains a large cluster of acidic ionizable side chains. By employing a state-of-the-art computational approach, we have revealed a previously uncharacterized catalytic calcium-binding site in hSCAN-1, which elucidates the unusual calcium-dependence of its apyrase activity. In a high-order coordination shell, the newly identified calcium ion organizes the active site residues to mediate nucleotide binding, to orient the nucleophilic water, and to facilitate the phosphoryl transfer reaction. From ab initio QM/MM molecular dynamics simulations with umbrella sampling, we have characterized a reverse protonation catalytic mechanism for hSCAN-1 and determined its free energy reaction profile. Our results are consistent with available experimental studies and provide new detailed insight into the structure-function relationship of this novel calcium-activated phosphoryl transfer enzyme.
1. Introduction
The apyrases are a class of extracellular enzymes responsible for modulating levels of extracellular signals.1–6 Many biochemical signals that originate outside the cell, as in immune response, as well as intercellular signals, which dictate the cell cycle and cell proliferation, are modulated by the concentration of nucleotide di- and tri-phosphates (e.g., ATP, GDP).7,8 Apyrases act to monitor the appropriate balance of these purinergic and pyrimidinergic signals by hydrolyzing, for example, ATP to ADP or GDP to GMP. Two functionally similar but evolutionarily unrelated families of apyrases have been identified: nucleotide triphosphate diphosphohydrolases (named NTPDases)1–3 and calcium-activated nucleotidases (CANs).4–6
The first characterized CAN was isolated from the saliva of blood-sucking insects, and it promotes blood flow in their subjects during feeding. In mammals, high levels of ADP are normally released as a purinergic signal in the event of tissue damage. ADP can bind to platelets collecting at the site of an insect bite; this triggers the recruitment and activation of additional platelets from the blood stream, which cascades into blood coagulation. The insect apyrase is excreted to interfere with this signal through rapid hydrolysis of ADP to AMP.9 Interest in the CAN family was broadened once a human homologue of the insect apyrase was identified.5,6 It had been proposed that a native human protein with the ability to rapidly oppose blood coagulation could be a valuable therapeutic agent in treating unwanted blood clotting: myocardial infarction or ischemic stroke.10 However, while this human soluble CAN, hSCAN-1, is evolutionarily homologous to the insect CAN, its substrate specificity is inverted, selecting GDP hydrolysis over ADP hydrolysis.6 In a subsequent effort to re-engineer hSCAN-1 from a GDPase into a highly active ADPase, significant progress has been made, augmenting ADPase activity more than 100-fold and shifting the ADPase/GDPase activity ratio from 0.005 to 41.4.4 Still, despite these successes in enzyme redesign, it should be noted that the overall ADPase activity of these mutants does not match the original GDPase activity of wild-type hSCAN-1. This low enzymatic activity limits the potential of using mutated hSCAN-1 as a therapeutic compound. At this point, a thorough mechanistic understanding could facilitate continued enzyme engineering efforts.
In comparison with other well-characterized phosphoryl transfer enzymes, hSCAN-1 is novel in terms of its sequence and three-dimensional structure, most interestingly in its phosphate–binding motif.4,11 It has previously been confirmed that none of the nucleotide-binding motifs indexed in the PROSITE12 or Pfam13 sequence databases can be found within the hSCAN-1 sequence. Furthermore, Pfinder,14 a program developed to predict phosphate-binding sites in proteins based on structural comparison to known phosphate-binding motifs, failed to identify the hSCAN-1 binding site. Nearly all well-characterized nucleotide-binding enzymes (including kinases, motor proteins, G proteins, and ATP synthase) utilize the common phosphate binding P-loop/Walker A motif,11 a structural motif of consecutive backbone NH groups oriented in an inward-facing loop to stabilize the negatively charged phosphates.15 The phosphate-binding families that lack a complete P-loop (including the NTPDase apyrases and the related Actin/HSP70 family) still rely on some backbone amide-phosphate interaction for substrate binding.16 hSCAN-1 does not contain a P-loop and lacks backbone amide-phosphate interaction altogether; it presents a unique phosphate-binding mechanism.
Another distinct feature of the CANs is the nature of their divalent cation dependence. The NTPDase family of apyrases is also cation dependent, but displays activity in the presence of Mg2+, Ca2+, or Mn2+,17–19 and the kinetic relationship expressed between cationic concentration and enzymatic activity is linear, indicating a traditional metal ion cosubstrate.20 Thus it came as a surprise that hSCAN-1 is exclusively activated by calcium, and kinetics experiments show an unusual and unexplained sigmoidal relationship between enzymatic rate and calcium concentration.20 Crystal structures of hSCAN-1, the apo structure as well as in complex with a substrate analog, have identified a single cation binding site in which the metal clearly plays a tertiary structural role, coordinating five distant residues and bringing together the five blades of its beta propeller tertiary structure4,21 (see Figure 1a), but this structural dependence alone does not clarify the kinetics results. In order to justify the sigmoidal relationship between enzymatic rate and calcium concentration, it has been proposed that a second calcium may be required to act in binding and/or hydrolyzing the nucleotide.22 This hypothesis is consistent with the kinetics results, but uncorroborated by any crystal structure of hSCAN-1,4 since none contains such a second cation.
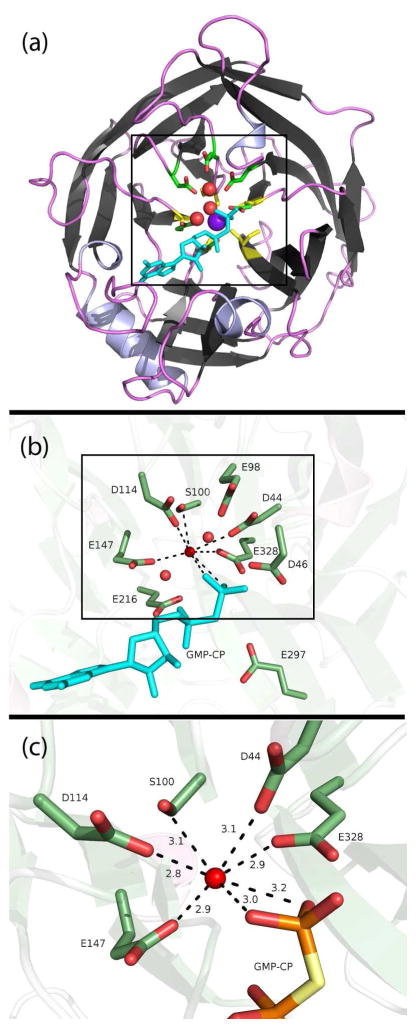
Figure 1.
(a) Cartoon depiction of the hSCAN-1 5-bladed beta-propellar crystal structure (PDB code: 1S1D).4 The crystalized structural calcium is centered in purple, and the five calcium-coordinated carbonyl groups (one from each beta sheet) are highlighted in yellow. Three crystalized waters in the active site are shown as red spheres, proximal acidic ionizable residues are shown as sticks, and the substrate analog, GMP-CP, is depicted in cyan. (b) A closer view highlighting the eight ionizable aspartates and glutamates and a serine, all within 5.5 Å of the negatively charged phosphates. (c) A close-up of the apparent hepta-coordination around a crystal-assigned water, or potentially misassigned cation. The distances are shown in Å.
In the current study, through molecular dynamics simulations, we have revealed a second calcium-binding site and characterized the metal’s critical role in organizing a new type of phosphate-binding motif. This catalytic calcium is structurally essential to the stability of the highly organized cluster of acidic ionizable residues comprising the novel hSCAN-1 active site. Through Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, a state-of-the-art approach to simulating enzymatic reactions, we have identified a well-positioned nucleophilic water, elucidated the catalytic role of calcium’s flexible high-order coordination chemistry, and suggest a reverse protonation mechanism for the hydrolysis of GDP.
2. Method
The initial model of the enzyme-substrate complex was prepared on the basis of the crystal structure of hSCAN-1 in complex with a substrate analog GMP-CP, subunit B (PDB code: 1S1D).4 (It should be noted that the residue numbering scheme used in this paper is the same as that in the original reference,4 which is different from the numbering in the 1S1D pdb file by 2 units.) The ligand GMP-CP was replaced with the substrate GDP. The protonation states of ionizable residues under physiological conditions were determined via the program H++,23 a web-based tool that uses Poisson-Boltzmann continuum electrostatics calculations to calculate the pKa shift of each ionizable residue due to its surrounding environment and allows for the inclusion of the substrate and ions in the calculation. The tleap Amber tool was used to add protons. The prepared protein system was solvated into a rectangular TIP3P water box with a 10 Å buffer distance on each side, and neutralized by adding Na+ ions. The resulting simulation system has about 40,000 atoms. After a series of minimizations and equilibrations, the standard molecular dynamics simulation with periodic boundary condition at 300K and 1 atm was carried out for up to 30 ns. Long-range electrostatic interactions were treated with particle mesh Ewald (PME) method. All minimizations and MD simulations were carried out with AMBER10 molecule dynamics package,24 employing the Amber99SB force field25–27 for the protein and the TIP3P model28 for the water. The force field parameters for the GDP substrate were parameterized by Carlson et al.29
With a snapshot taken from the productive MD trajectory, the QM/MM system was prepared by removing all water molecules beyond a 27 Å radius from the β-phosphate of the substrate. The QM subsystem, as illustrated in Figure S1, which includes the side chains of Asp44, Asp46, Glu98, and Asp114, the phosphate portion of the GDP, the nucleophilic water, and the catalytic calcium ion, was described at the B3LYP/6-31G* level. This level of QM treatment has been tested and employed successfully in other contemporary ab initio QM/MM studies of phosphoryl transfer enzymes.30–36 The MM subsystem was treated by the same force field as in the previous classical MD simulations. The QM/MM boundary was described by the pseudobond approach with the improved parameters.37–39 The spherical boundary condition was employed, in which only atoms within a 20 Å radius from the beta phosphate were allowed to move. Cutoffs of 18 and 12 Å were specified for electrostatic and van der Waals interactions within the MM subsystem, but no cutoff for electrostatic interactions between the QM and MM subsystems was used. All ab initio QM/MM calculations were performed with a combination of modified Q-Chem3.040 and Tinker41 programs.
Following initial unrestrained QM/MM MD equilibration, the QM/MM system was minimized with an iterative minimization procedure.42 Given a proposed reaction scheme, a minimum reaction energy path was then determined with the reaction coordinate driving method.42 Given a promising reaction profile, the MM sub-system for each structure determined along the path was subsequently equilibrated with 500 ps MD simulations, with the QM sub-system fixed. The resulting snapshots were used as the starting structures for ab initio QM/MM MD simulations with umbrella sampling. For each window, a 25 ps B3LYP(6-31G*) QM/MM MD simulation was carried out, in which the Beeman algorithm43 was used with a time step of 1 fs, and the Berendsen thermostat44 was employed to control the temperature around 300 K. From these biased simulations, the free energy profile for each reaction was obtained with the weighted histogram analysis method (WHAM).45–48 This computational protocol has been successful in studying various enzyme systems and their catalytic mechanisms.34,49–57
3. Results
3.1. Revelation and Characterization of the Catalytic Calcium-Binding Site
The CAN family of apyrases is hallmarked by its exclusive and unusual calcium activation. In the crystallization of hSCAN-1, a single calcium cation was identified in its structure, centrally positioned to coordinate the five blades of its beta-propeller tertiary structure. However, while this calcium-coordinated beta-propeller defines the global structure of hSCAN-1, this organization alone does not confer its catalytic activity.22 The active site of hSCAN-1 is quite distinct from other well-characterized phosphate binding enzymes. It comprises a large cluster of acidic ionizable residues whose structural organization appears to rely on a complex hydrogen-bonding network, and the uncertain nature of the multiple protonation assignments presents the initial challenge in modeling the reactant complex of hSCAN-1. We prepared many competing models representing plausible protonation states and proton positions and carried out molecular dynamics simulations for each. Trajectories were evaluated for active site stability, consistency with the crystal structure, and the proper positioning of a water for nucleophilic attack. For promising models, we continued the examination using the QM/MM-MD method to calculate and compare free energy profiles for various catalytic mechanisms. As the outcome of this extensive computational investigation, we have revealed that a second calcium cation is required to organize active site residues into the catalytically active reactant state conformation, for which a single model demonstrated remarkable active site stability. The catalytic calcium we have identified utilizes a high-order coordination shell to coordinate the cluster of ionizable residues to mediate nucleotide binding, to orient the nucleophilic water, and to facilitate the phosphoryl transfer.
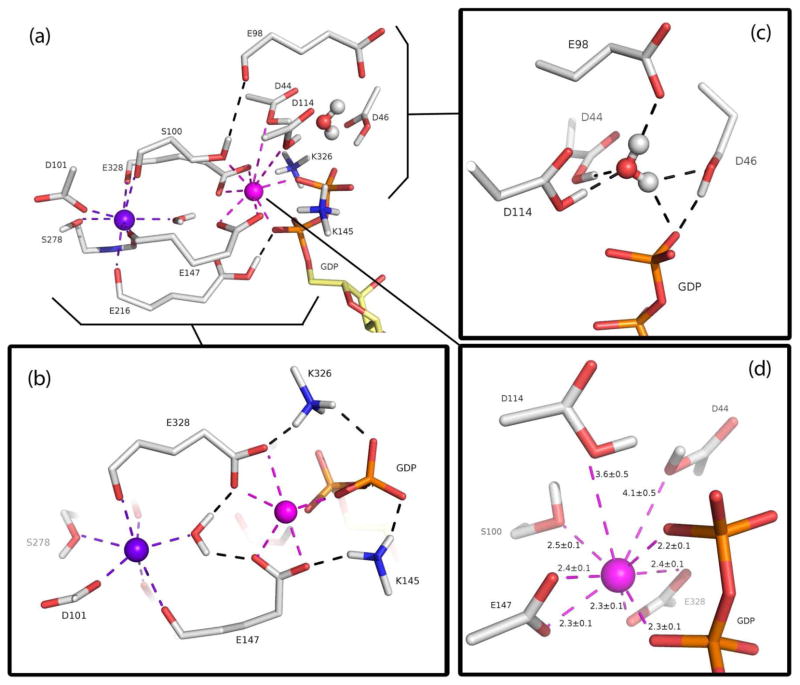
Figure 2a is an overall depiction of the novel dual-calcium phosphate-binding active site structure employed by hSCAN-1. The two calcium cations are in close proximity with their electrostatic repulsion buffered by a single water molecule. The previously identified structural calcium, displayed in purple, has an eight-fold coordination structure that includes the five backbone carbonyl groups, from remotely distributed amino acids, that bring together the global beta-propeller structure. The side chains of three of these residues—Ser100, Glu147, and Glu328—extend in parallel to make direct contact with the newly identified catalytic calcium, displayed in magenta. In addition to directly binding phosphate, the catalytic calcium is responsible for structurally organizing the cluster of acidic ionizable residues that distinguishes the active site of hSCAN-1 from previously characterized phosphate-binding structural motifs (see Figure 2d). There are two pairs of “mirrored” residues directly coordinated to the catalytic calcium: Glu147/Glu328 are deprotonated and form bidentate interactions, Asp44/Asp114 are protonated and form specific monodentate interactions through their hydroxyl oxygens. The side chain of Ser100 completes the catalytic calcium coordination shell.
Figure 2.
(a) Representation of the characterized hSCAN-1 active site at the reactant state, highlighting the spatial relationship between the structural calcium (purple) and the newly identified catalytic calcium (magenta). All calcium-coordinated groups, catalytically important residues, and a central water are shown as sticks. The nucleophilic water is shown as ball-and-stick. (b) A bird’s-eye view revealing the novel, symmetric calcium-mediated salt bridge phosphate-binding motif. (c) A close-up of the precise coordination and positioning of the nucleophilic water through hydrogen bonds. The three aspartates are in their protonated states. (d) The nine-fold coordination distances around the catalytic calcium: 7 tighter interactions including the 2 bidentate glutamates and 2 looser coordinations with the protonated aspartates. The distances shown are averages from the 30 ns MD simulation of the reactant state, in Å.
The proper positioning and orientation of a nucleophilic water, into a reactive position proximal to the terminal phosphate, is considered the key to enzymatic phosphoryl transfer by hydrolysis.58 In hSCAN-1, this is achieved through hydrogen bonding interactions with four residues: Asp44, Asp46, Glu98, and Asp114 (see Figure 2c). The direct coordination of the protonated Asp44/Asp114 pair to the catalytic calcium positions their respective hydroxyl groups to hydrogen bond the nucleophilic oxygen in a near perfect tetrahedral arrangement. Simultaneously, the protons of the nucleophilic water are tethered through hydrogen bonds to the deprotonated Glu98 and the protonated Asp46, which is stabilized through its own hydrogen bond to the terminal phosphate. This four-point coordination secures the nucleophilic water in its pre-attack position adjacent to the phosphate for the entirety of our 30 ns molecular dynamics simulation of the hSCAN-1 reactant state.
The bidentate coordination of the Glu147/Glu328 pair to the catalytic calcium is indirectly important to the binding and coordination of the phosphate group itself. Deprotonated under physiological conditions, the negative charges on these glutamates prompt interactions with Lys145/Lys326, and these positively charged residues flank the phosphate moiety and coordinate its positioning directly (see Figure 2b). The side chain of Ser100, whose backbone is also tethered by the structural calcium, forms a loose hydrogen bond to the carbonyl group on the backbone of Glu98, serving to bind the active site together (see Figure 2a).
Arriving at the reactant state of hSCAN-1 was not a direct path. Eight glutamates and aspartates are organized within 5.5 Å of the phosphate moiety of the bound substrate (see Figure 1b). According to the initial Poisson-Boltzmann based pKa-shift calculations, six of these eight residues were to be protonated/neutralized in this complex (see Table S1). Since each residue could be protonated at one of two oxygens, there were 64 possible permutations of this protonation state. Some of the likely protonation positions could be surmised from the relevant inter-atomic distances within the crystal structure, but several protonation assignments remained uncertain, and we built and simulated models for each plausible state.
Of all the models simulated, only two resulted in trajectories in which the active site organization seen in the crystal structure was well preserved. In the first of these simulations, an intricate network of interactions between the active site residues, waters, and the GDP did form, but this adherence to the initial structure is short-lived. The RMSD of the active site residues begins to drift following the initial two nanoseconds of simulation (see Figure S2). Furthermore, calculations exploring the energetics of phosphoryl transfer from this state resulted in exceedingly high activation energies (see Figure S9). In the second of the promising simulations, a Na+ counter-ion was observed to spontaneously migrate from the bulk solution to within the active site. This cation became coordinated to the β-phosphate as well as to the side chains of Asp44, Ser100, Asp114, Glu147, and Glu328. We inspected the corresponding position in the crystal structure and found that it matched the exact position of an assigned crystal water molecule. In the crystal structure, the corresponding oxygen atom appeared to have around it a hepta-coordinated arrangement of the same set of electron rich oxygen atoms, as shown in Figure 1c. While the resolution of the hSCAN-1 crystal structure is fairly high, at 1.6 Å, it is generally accepted that, beyond a resolution of 1.5 Å, it can be difficult to distinguish between a water molecule and various cations, such as sodium or calcium.59 We began to consider the likelihood that an important metal cation, responsible for stabilizing the structural coordination of these ionizable residues, had been misassigned in the solution of the crystal structure.
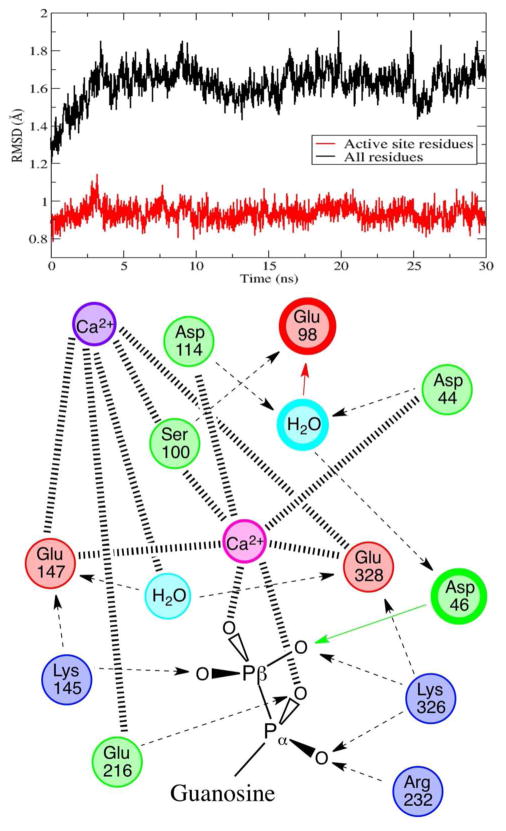
Exploring the possibility of a second calcium-binding site, we replaced the questionable crystal water molecule with a second calcium cation, but we also needed to reconsider how this divalent cation would alter the protonation state of the ionizable binding site. According to a recalculation of the residue pKa-shifts, with this second calcium in place, Glu147 and Glu328 were to be deprotonated now, and Asp114 was relatively ambiguous, with a predicted pKa of 5.9. After building and simulating models respective of these protonation states, we determined a single reactant state and conformation that demonstrated exceptional active site stability throughout the entire 30 ns molecular dynamics simulation and presented a viable reactant state with a properly coordinated substrate and potential water nucleophile (see Figure 3 and Figure S3).
Figure 3.
Top: heavy atom RMSD from the crystal structure for all residues within 8 Å of the terminal phosphate (in red), highlighting active site stability for 30 ns of molecular dynamics, and for the entire enzyme (in black). Bottom: schematic of the structural interactions between the active site residues, the structural calcium, the catalytic calcium, the nucleophilic water, an active site water, and the GDP substrate. Hashed lines represent coordination to calcium, dashed lines represent hydrogen bonding with arrows pointing to H-bond acceptors, colored arrows and bold circles depict proton transfers and residues involved in the catalytic chemistry.
3.2. Reverse Protonation Phosphoryl Transfer Mechanism
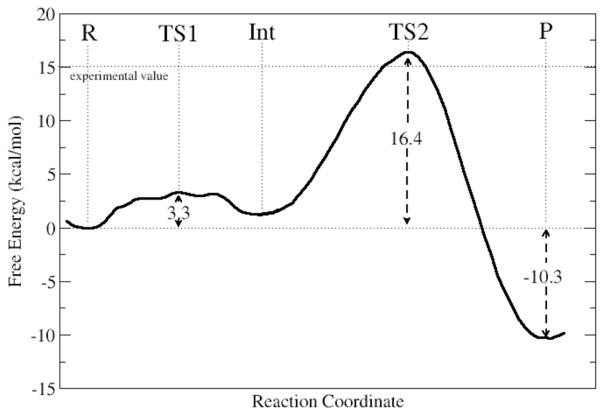
In the pursuit of the catalytic mechanism of hSCAN-1, we are exploring the free energy associated with the hydrolysis of GDP to GMP and inorganic phosphate. The energetic barrier to this reaction is primarily brought down by the pre-positioning of a single water molecule for nucleophilic attack of the β-phosphate. In the reactant structure we have presented, we have revealed a well-designed water-coordinating site, with four hydrogen bond contact points (see Figure 2c), clearly indicating the nucleophile for the reaction. We have employed the QM/MM-MD method, coupled with umbrella sampling, to explore various mechanistic schemes for hSCAN-1. We have investigated plausible mechanisms starting from our proposed reactant state as well as from alternative, less stable systems (see Figures S9–S12 for examples). Ultimately, the most promising mechanism did derive from our proposed reactant, and our calculations suggest a novel, two-step, reverse protonation phosphoryl transfer. The overall activation energy from the full free energy profile was calculated to be 16.4 kcal/mol (see Figure 4), which is in good agreement with the 15.0 kcal/mol barrier predicted from experimental kinetics.4
Figure 4.
Free energy profile for the complete two-step phosphoryl transfer reaction. The activation free energy calculated from experiment, 15.04 kcal/mol, is shown by the dotted line. Activation energies for TS1/TS2 and the overall change in free energy are shown in kcal/mol.
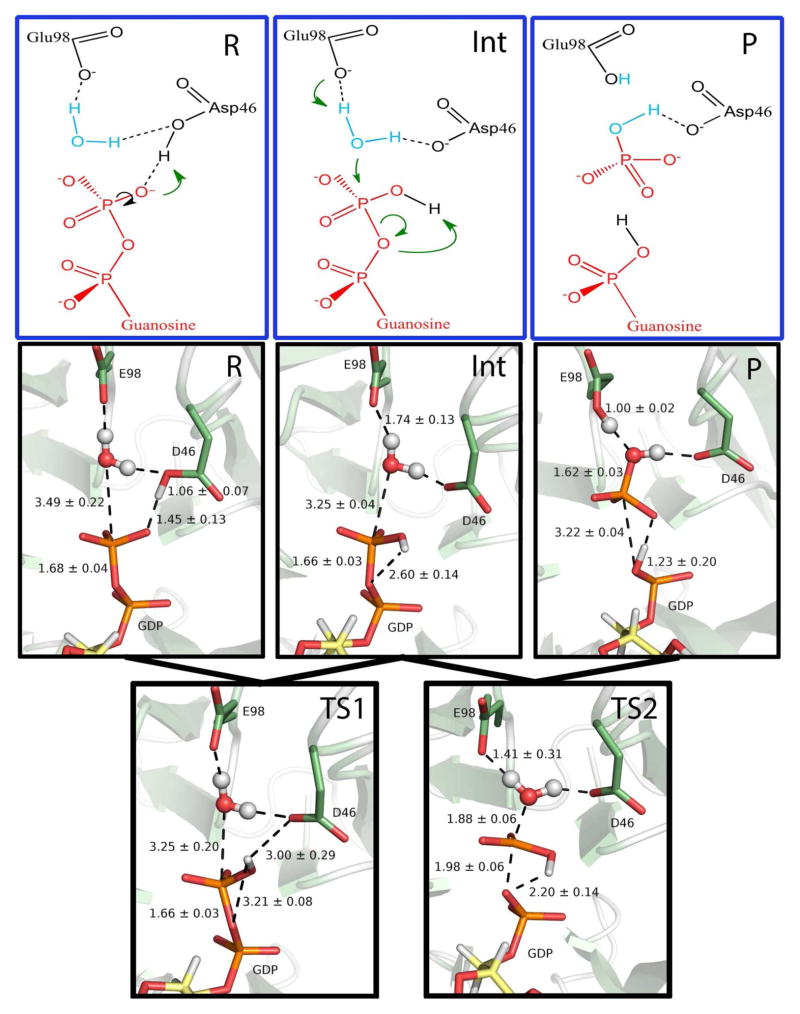
In the first step of the reverse protonation mechanism, the proton shared in a low-barrier hydrogen bond between Asp46 and the β-phosphate (see Figure S4) transfers to the phosphate and rotates away from Asp46 and toward the bridging-oxygen leaving group (see Figure 5, Figure S5). This initial step requires 3.3 kcal/mol and results in a semi-stable intermediate (see Figure S6). From this intermediate, the β-phosphate is transferred directly to the nucleophilic water, in the rate-limiting step, with a barrier of 15.1 kcal/mol (see Figure S7). For the second proton transfer of the reverse protonation mechanism, the basic Glu98 extracts a proton from the nucleophilic water near the transition state of the second step. Near the product state, the phosphorylproton, originally from Asp46, transfers spontaneously to neutralize the leaving group oxygen. The overall hydrolysis of this high-energy phosphate bond is exothermic, with a total ΔG = −11.6 kcal/mol. The phosphoryl transfer is concerted and proceeds through a near synchronous mechanism, with the breaking O-P bond at 1.98 ± 0.06 Å and the forming P-O(wat) bond at 1.88 ± 0.06 Å at the transition state (see Figure 5). Once the products GMP and Pi are released from the active site, pKa calculations of the apo protein indicate that each of the acidic ionizable residues within the active site will become deprotonated. Then as a new GDP substrate binds, its negative charges prompt the protonation of Asp44, Asp46, Asp114, and Glu216, resetting the system to the reactive protonation states for the subsequent enzymatic turnover.
Figure 5.
Line drawings and structures along the hSCAN-1 reverse protonation reaction path, with average distances and standard deviations from the QM/MM-MD sampling shown for the phosphoryl transfer and the proton transfers, in Å. From the reactant state (R) to the intermediate (Int), a proton transfers from D46 to the β-phosphate and rotates away from D46 and toward the leaving group oxygen. From the intermediate to the product (P), the nucleophilic attack takes place, with a proton abstraction by the general base E98 and a proton transfer to the leaving group.
In order to assess the validity of the determined transition state, we have run a series of unrestrained ab initio QM/MM MD simulations initiated from 25 snapshots around the characterized transition state region, RC = 0.1 Å. Two 500 fs simulations have been run for each snapshot, one with Boltzmann sampled velocities and the other with the reverse of these velocities. Among all 25 pairs, 6 resulted in both trajectories relaxing to the reactant; 7 resulted in both trajectories relaxing to the product; and the remaining 12 have one trajectory falling to the reactant and the other falling to the product. These results indicate that the determined transition state is meaningful.
4. Discussion
Enzymes involved in nucleotide binding and phosphoryl transfer are among the most studied and therapeutically targeted biomolecular systems (e.g. G-proteins, Kinases), and extensive research has been devoted to understanding and characterizing the structural mechanisms by which these enzymes achieve phosphate recognition.11–13,60–62 The structural P-loop, otherwise known as the Walker A motif,15 has been recognized as the most common phosphate-binding structural motif, shared by 13 out of 27 nucleotide-binding superfamilies.11 The motif is characterized by the coordination of multiple consecutive backbone amide groups oriented into an inward facing ring to encircle and electrostatically stabilize the binding of the highly negative phosphate groups. (see Figure S8 for a representative P-loop from the Yersinia tyrosine phosphatase).63 Therefore, the crystalized phosphate-binding catalytic site of hSCAN-1 is fundamentally surprising, not only for the absence of backbone amide incorporation, but for the seemingly paradoxical abundance of acidic ionizable side chains, which impose additional electron density to the phosphate-binding environment (see Figure 1b).
Our theoretical revelation of a previously uncharacterized catalytic calcium-binding site reconciles the incongruous connection between acidic side chains and phosphate binding. In a symmetric fashion, as illustrated in Figure 2b, the newly identified calcium ion simultaneously binds to the phosphate groups and stabilizes the structure and deprotonated states of two negatively charged glutamates. Each of these glutamates brings in a positively charged lysine side chain to form a novel calcium-mediated salt bridge phosphate-binding motif. The importance of this specific charge scheme and the bidentate coordination of these glutamates is corroborated by documented mutagenesis results; both the E147Q and the E328Q mutations reduce kcat by 2–3 orders of magnitude.
The identification of this second calcium ion also explains the previously puzzling sigmoidal relationship between enzymatic rate and calcium concentration. And further, our results provide a rich basis for understanding why hSCAN-1 is exclusively dependent on calcium and, in general, how catalytic calcium-activation is achieved. Divalent cations are typically associated with nucleotide di- and tri-phosphates in solution, and it is not uncommon for these ions to accompany a nucleotide substrate into an enzymatic active site as a requisite co-substrate. However, in these cases, as with the NTPDase family of apyrases, there is no exclusive selectivity of calcium over magnesium or manganese as the cosubstrate.17–19 While magnesium and manganese are comparatively inflexible and relatively strict hexa-coordinators, and Zinc, while similar to calcium in its coordination flexibility, favors lower coordination numbers as well, primarily 4 and 6, calcium can support coordination numbers from 6 to 10 (see Table S2).64 Our proposed hSCAN-1 reactant state demonstrates the unique nature of calcium’s nine-fold coordination, in which, specifically, 7 interactions are tightly coordinated and 2 interactions are looser (see Figure 2d). The 2 loose interactions involve the hydroxyl oxygens of Asp44 and Asp114, and this is the key to the proper orientation of the nucleophilic water (see Figure 2c). This result too is supported by experimental mutagenesis data, which shows significant reduction of kcat for the D44N and the D114N mutations, over two and three orders of magnitude respectively.4 When mutated to asparagine, an amide proton replaces the analogous position of the hydroxyl lone-pair. This mutation would impede the proposed calcium coordination and thus hinder the essential nucleophilic orientation.
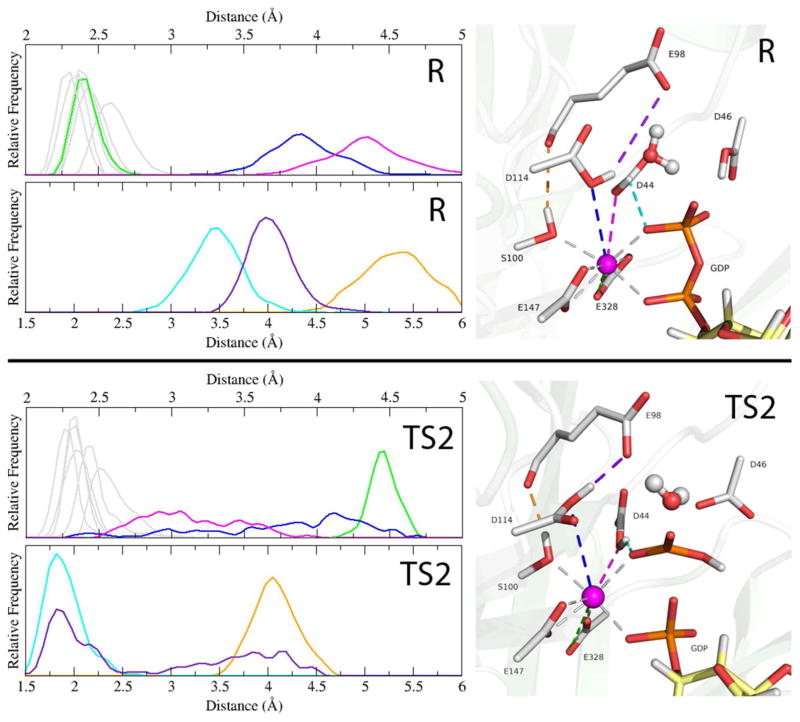
Mechanistically, from the reactant to the transition state 2, the coordination around the catalytic calcium undergoes an important shift. Presented as histograms taken from our QM/MM MD trajectories, Figure 6 shows the distances of the calcium-coordinated interactions at the reactant and TS2, and also shows key hydrogen bonds that form as a result of the calcium coordination rearrangement. In the rotation of the phosphoryl proton away from Asp46 and toward the leaving group oxygen, Glu328 has shifted from bidentate to monodentate (the lost coordination is monitored in the bottom panel of Figure S6). This coordination change resembles the carboxylate shift mechanism proposed for other metal binding enzymes to some extent,65,66 but serves a distinct catalytic role in hSCAN-1. Our simulations indicate that the bidentate coordination of Glu328 is stable in the reactant enzyme-substrate complex, and the shift to monodentate is associated with a subtle structural rearrangement of the remaining coordinated interactions. This carboxylate shift promotes the formation of the transition state 2, most importantly, by allowing space for a stronger interaction between the calcium and Asp44. The loose-coordination symmetry of Asp44/Asp114 that oriented the nucleophile in the reactant state is lost, and these residues reorient and form hydrogen bonds that facilitate nucleophilic attack and the phosphoryl transfer; Asp114 loosens its coordination and shifts up to interact with Glu98, and Asp44, now more strongly coordinated, shifts down to interact with the transferring phosphate. Also depicted is the TS2 stabilizing role of Ser100 in forming a stronger hydrogen bond with the carbonyl of Glu98 as the active site gets generally compressed toward the transition state. This role may justify the significant attenuation of kcat, nearly two orders of magnitude, for the S100A mutation.
Figure 6.
Shown for the reactant state (R) and the rate determining transition state 2 (TS2) are histograms of the coordination distances around the catalytic calcium (upper plots) and 3 selected atom-atom distances (lower plots), all from the QM/MM-MD simulations. In the structural depictions on the right, the corresponding distances are shown as dashes and color coordinated to the histogram plots. Calcium-coordinated interactions that remain relatively constant from R to TS2 are greyed out. Highlighted is the flexibility of the calcium coordination as one of the E328 interactions (green) shifts out of the coordination shell as D44 (magenta) becomes more tightly coordinated in order to form a hydrogen bond with the transferring phosphate (cyan), and D114 is slightly more loosely coordinated (blue) as it forms a hydrogen bond with E98 (purple). These two new hydrogen bonds replace the nucleophile-coordinating interactions lost during the attack and stabilize the transition state. In orange is the TS2 stabilizing interaction between S100 and the carbonyl of E98.
Our characterized reverse protonation mechanism contains elements of both the general base mechanism and the substrate-assisted mechanism, the two existing mechanisms generally considered for the hydrolysis of di- and tri-nucleotide phosphates.67–75 In a classical substrate-assisted mechanism, the substrate serves to abstract the proton from the attacking nucleophilic water. Our mechanism resembles the general base mechanism in the abstraction of this proton—Asp114 acts as the general base—but the substrate is involved in proton abstraction from the neutralized Asp46. This novel combination of mechanistic features allows for the thermodynamically important protonation of the leaving group through the substrate, but avoids the sterically unfavorable geometry that results from a simultaneous nucleophilic attack and substrate-assisted nucleophilic proton abstraction. Our characterized transition state for the phosphoral transfer is 56% associative, based on Pauling’s formulation,76,77 with a P—O forming distances of 1.88 Å. The calculated free energy barrier of 16.4 kcal/mol is the lowest of the various enzyme-reactant complexes and potential mechanisms we have modeled (see Figures S10–S12 for results from an alternate protonation state and alternate mechanisms) and is in good agreement with the estimated 15.0 kcal/mol, based on the experimental kcat of 1.0 × 104 min−1 and calculated from transition state theory.4
5. Conclusion
Human soluble calcium-activated nucleotidase 1 (hSCAN-1) is the only known human CAN, a relatively unstudied protein family that is uniquely calcium dependent. It possesses a nucleotide binding mechanism distinct from all other characterized phosphate binding motifs. Our computational work on hSCAN-1 complements the experimental structural and biochemical studies to help elucidate the detailed atomistic structure, reactivity, and mechanism of this novel phosphoryl transfer enzyme.
The unusual nature of the calcium dependence in the CAN family is due to a dual calcium-binding active site. One calcium, identified in the crystal structure, is responsible for coordination of the general tertiary structure of the enzyme, the 5-bladed beta propeller. The second calcium, which is essential for catalytic activity, interacts directly with the nucleotide substrate and serves to coordinate, directly, the residues responsible for orienting the nucleophilic water and, indirectly, the residues important to substrate binding and transition state stabilization. The highly specific nature of the CAN calcium dependence, which cannot be met by other divalent cations, is due to the active site design, which requires a high-order coordination to organize the reactant structure and then relies on a flexibility to shift to an alternately-coordinated intermediate that promotes transition state stabilization.
In general, purinergic and pyrimidinergic signaling is an emerging topic, related to neurotransmission, cellular function, and intercellular communication. The biological and biomedical significance of the CANs in particular is only beginning to be understood. Expression of hSCAN-1 and its cross-species homologues has recently been implicated in various disease states (e.g., Desbuquois dysplasia78 and prostate cancer79) and associated with certain cellular processes (e.g. unfolded protein stress response80 and embryogenesis81). Moreover, the previous experimental redesign of hSCAN-1 has established this enzyme as a viable but still challenging subject for protein engineering research. Our computational study should facilitate the further investigation into the structure/function relationship of this important and novel extracellular signaling enzyme, stimulate ongoing protein design progress with respect to hSCAN-1, and contribute to the known diversity of enzymatic phosphoryl transfer in both phosphate binding motif and catalytic mechanism.
Supplementary Material
Acknowledgments
This work was supported by NSF-IGERT and NIH (R01-GM079223, R21-GM097530 and R01-AI94599). We thank TeraGrid and NYU-ITS for providing computational resources.
Footnotes
Supporting Information: Tables S1-S2, Figures S1-S12, and the complete citation for references 10, 13, 24, 40, 78, and 79. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zimmermann H. Nat Med. 1999;5:987. doi: 10.1038/12419. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann H. Drug Dev Res. 2001;52:44. [Google Scholar]
- 3.Zimmermann H. Trends Pharmacol Sci. 1999;20:231. doi: 10.1016/s0165-6147(99)01293-6. [DOI] [PubMed] [Google Scholar]
- 4.Dai J, Liu J, Deng Y, Smith TM, Lu M. Cell. 2004;116:649. doi: 10.1016/s0092-8674(04)00172-2. [DOI] [PubMed] [Google Scholar]
- 5.Valenzuela JG, Charlab R, Galperin MY, Ribeiro JMC. J Biol Chem. 1998;273:30583. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- 6.Smith TM, Hicks-Berger CA, Kim S, Kirley TL. Arch Biochem Biophys. 2002;406:105. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- 7.Di Virgilio F, Chiozzi P, Ferrari D, Falzoni S, Sanz JM, Morelli A, Torboli M, Bolognesi G, Baricordi OR. Blood. 2001;97:587. doi: 10.1182/blood.v97.3.587. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann H. Pflügers Arch. 2006;452:573. doi: 10.1007/s00424-006-0067-4. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro JM. Annu Rev Entomol. 1987;32:463. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 10.Gayle RB, et al. J Clin Invest. 1998;101:1851. doi: 10.1172/JCI1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinoshita K, Sadanami K, Kidera A, Go N. Protein Eng. 1999;12:11. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. Nucleic Acids Res. 2010;38:D161. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, et al. Nucleic Acids Res. 2010;38:D211. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parca L, Gherardini PF, Helmer-Citterich M, Ausiello G. Nucleic Acids Res. 2011;39:1231. doi: 10.1093/nar/gkq987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker JE, Saraste M, Runswick MJ, Gay NJ. EMBO J. 1982;1:945. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zebisch M, Sträter N. Proc Natl Acad Sci USA. 2008;105:6882. doi: 10.1073/pnas.0802535105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asai T, O’Sullivan WJ, Tatibana M. J Biol Chem. 1983;258:6816. [PubMed] [Google Scholar]
- 18.Cheeseman MT. Insect Biochem Mol Biol. 1998;28:1025. doi: 10.1016/s0965-1748(98)00093-9. [DOI] [PubMed] [Google Scholar]
- 19.Ostuni MA, Egido P, Peranzi G, Alonso GL, Lacapere JJ, Gonzalez DA. Physiological research/Academia Scientiarum Bohemoslovaca. 2009;58:843. doi: 10.33549/physiolres.931682. [DOI] [PubMed] [Google Scholar]
- 20.Murphy DM, Ivanenkov VV, Kirley TL. Biochemistry. 2003;42:2412. doi: 10.1021/bi026763b. [DOI] [PubMed] [Google Scholar]
- 21.Yang M, Horii K, Herr AB, Kirley TL. J Biol Chem. 2006;281:28307. doi: 10.1074/jbc.M604413200. [DOI] [PubMed] [Google Scholar]
- 22.Yang M, Kirley TL. Biochemistry. 2004;43:9185. doi: 10.1021/bi049565o. [DOI] [PubMed] [Google Scholar]
- 23.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic Acids Res. 2005;33:W368. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Case DA, et al. AMBER 10. University of California; San Francisco: [Google Scholar]
- 25.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179. [Google Scholar]
- 26.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049. [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 29.Meagher KL, Redman LT, Carlson HA. J Comput Chem. 2003;24:1016. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Zhang Y, McCammon JA. J Am Chem Soc. 2005;127:1553. doi: 10.1021/ja0464084. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Broyde S, Zhang Y. J Mol Biol. 2009;389:787. doi: 10.1016/j.jmb.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks JM, Hu H, Rudolph J, Yang W. J Phys Chem B. 2009;113:5217. doi: 10.1021/jp805137x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith GK, Ke Z, Guo H, Hengge AC. J Phys Chem B. 2011;115:13713. doi: 10.1021/jp207532s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lior-Hoffmann L, Wang L, Wang S, Geacintov NE, Broyde S, Zhang Y. Nucleic Acids Res. first published online July 5, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang LH, Yu XY, Hu P, Broyde S, Zhang YK. J Am Chem Soc. 2007;129:4731. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosta E, Nowotny M, Yang W, Hummer G. J Am Chem Soc. 2011;133:8934. doi: 10.1021/ja200173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y. J Chem Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YK. Theor Chem Acc. 2006;116:43. [Google Scholar]
- 39.Zhang YK, Lee TS, Yang WT. J Chem Phys. 1999;110:46. [Google Scholar]
- 40.Shao Y, et al. Q-Chem, , version 3.0; Q. [Google Scholar]
- 41.Ponder JW. TINKER, Software Tools for Molecular Design, Version 4.2. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Liu H, Yang W. J Chem Phys. 2000;112:3483. [Google Scholar]
- 43.Beeman D. J Comput Phys. 1976;20:9. [Google Scholar]
- 44.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684. [Google Scholar]
- 45.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J Comput Chem. 1992;13:1011. [Google Scholar]
- 46.Souaille M, Roux B. Comput Phys Commun. 2001;135:40. [Google Scholar]
- 47.Ferrenberg AM, Swendsen RH. Phys Rev Lett. 1988;61:2635. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- 48.Grossfield A. WHAM: the weighted histogram analysis method, Version 206. 2012
- 49.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:3806. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:16721. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ke Z, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:16705. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:12750. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Hu P, Zhang Y. J Phys Chem B. 2007;111:3758. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu R, Wang S, Zhou N, Cao Z, Zhang Y. J Am Chem Soc. 2010;132:9471. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Wang S, Zhang Y. J Phys Chem B. 2010;114:8817. doi: 10.1021/jp104258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ke Z, Smith GK, Zhang Y, Guo H. J Am Chem Soc. 2011;133:11103. doi: 10.1021/ja204378q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou YZ, Zhang YK. Chem Commun. 2011;47:1577. doi: 10.1039/c0cc04112b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cleland WW, Hengge AC. Chem Rev. 2006;106:3252. doi: 10.1021/cr050287o. [DOI] [PubMed] [Google Scholar]
- 59.Wlodawer A, Minor W, Dauter Z, Jaskolski M. FEBS J. 2008;275:1. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parca L, Gherardini PF, Helmer-Citterich M, Ausiello G. Nucleic Acids Res. 2011;39:1231. doi: 10.1093/nar/gkq987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirsch AK, Fischer FR, Diederich F. Angew Chem. 2007;46:338. doi: 10.1002/anie.200603420. [DOI] [PubMed] [Google Scholar]
- 62.Brakoulias A, Jackson RM. Proteins. 2004;56:250. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 63.Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA. Nature. 1994;370:571. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 64.Glusker JP, Katz AK, Bock CW. The Rigaku Journal. 1999;16:8. [Google Scholar]
- 65.Sousa SF, Fernandes PA, Ramos MJ. Biophys J. 2005;88:483. doi: 10.1529/biophysj.104.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sousa SF, Fernandes PA, Ramos MJ. J Am Chem Soc. 2006;129:1378. doi: 10.1021/ja067103n. [DOI] [PubMed] [Google Scholar]
- 67.Liljas A, Ehrenberg M, Åqvist J. Science. 2011;333:37. doi: 10.1126/science.1202532. [DOI] [PubMed] [Google Scholar]
- 68.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. Science. 2010;330:835. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wittinghofer A. Trends Biochem Sci. 2006;31:20. doi: 10.1016/j.tibs.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 70.Martin-Garcia F, Mendieta-Moreno JI, Lopez-Vinas E, Gomez-Puertas P, Mendieta J. Biophys J. 2012;102:152. doi: 10.1016/j.bpj.2011.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parke CL, Wojcik EJ, Kim S, Worthylake DK. TJ Biol Chem. 2010;285:5859. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grigorenko BL, Shadrina MS, Topol IA, Collins JR, Nemukhin AV. Biochim Biophys Acta. 2008;1784:1908. doi: 10.1016/j.bbapap.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasqualato S, Cherfils J. Structure. 2005;13:533. doi: 10.1016/j.str.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 74.Schweins T, Geyer M, Scheffzek K, Warshel A, Kalbitzer HR, Wittinghofer A. Nat Struct Biol. 1995;2:36. doi: 10.1038/nsb0195-36. [DOI] [PubMed] [Google Scholar]
- 75.Schweins T, Langen R, Warshel A. Nat Struct Biol. 1994;1:476. doi: 10.1038/nsb0794-476. [DOI] [PubMed] [Google Scholar]
- 76.Pauling L. The nature of the chemical bond and the structure of molecules and crystals; an introduction to modern structural chemistry. 3. Cornell University Press; Ithaca, N.Y: 1960. [Google Scholar]
- 77.Mildvan AS. Proteins. 1997;29:401. [PubMed] [Google Scholar]
- 78.Huber C, et al. Am J Hum Genet. 2009;85:706. doi: 10.1016/j.ajhg.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerhardt J, et al. Am J Pathol. 2011;178:1847. doi: 10.1016/j.ajpath.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uccelletti D, Pascoli A, Farina F, Alberti A, Mancini P, Hirschberg CB, Palleschi C. Mol Biol Cell. 2008;19:1337. doi: 10.1091/mbc.E07-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devader C, Webb RJ, Thomas GM, Dale L. Gene. 2006;367:135. doi: 10.1016/j.gene.2005.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.