Abstract
Objectives
To enhance our understanding of the interstitial cystitis urine biomarker antiproliferative factor (APF), as well as interstitial cystitis biology more generally at the systems level, we reanalyzed recently published large-scale quantitative proteomics and in vivo transcriptomics data sets using an integration analysis tool that we have developed.
Materials and methods
To identify more differentially expressed genes with a lower false discovery rate from a previously published microarray data set, an integrative hypothesis-testing statistical approach was applied.
For validation experiments, expression and phosphorylation levels of select proteins were evaluated by western blotting.
Results
Integration analysis of this transcriptomics data set with our own quantitative proteomics data set identified 10 genes that are potentially regulated by APF in vivo from 4140 differentially expressed genes identified with a false discovery rate of 1%.
Of these, five (i.e. JUP, MAPKSP1, GSPT1, PTGS2/COX-2 and XPOT) were found to be prominent after network modelling of the common genes identified in the proteomics and microarray studies.
This molecular signature reflects the biological processes of cell adhesion, cell proliferation and inflammation, which is consistent with the known physiological effects of APF.
Lastly, we found the mammalian target of rapamycin pathway was down-regulated in response to APF.
Conclusions
This unbiased integration analysis of in vitro quantitative proteomics data with in vivo quantitative transcriptomics data led to the identification of potential downstream mediators of the APF signal transduction pathway.
Keywords: APF, integration analysis, interstitial cystitis, microarray, SILAC
Introduction
Interstitial cystitis (IC) is a prevalent and debilitating pelvic disorder generally accompanied by chronic pain [1,2]. This disease affects over one million Americans, especially middle-aged women, yet its aetiology remains unknown [3]. Several urinary biomarker candidates have been identified for IC [4]; among the most promising is antiproliferative factor (APF), whose biological activity is detectable in urine specimens from > 94% of patients with both ulcerative and non-ulcerative IC [5].
APF has been shown to be a nine-residue frizzled-8 protein-related sialoglycopeptide [6]. Subsequent research has gradually clarified the molecular mechanisms underlying the profound antiproliferative effect of APF. A number of proteins such as cytoskeleton-associated protein 4 [7], heparin-binding epidermal growth factor-like growth factor [8], p53 [9], E-cadherin [10] and Akt [11], as well as signal pathways, such as dual mitogen-activated protein kinase signalling pathways [12], were found to be downstream of APF. Very recently, by employing stable isotope labelling by amino acid in cell culture (SILAC)-based quantitative proteomics, we identified over 100 APF-regulated proteins in T24 human bladder cancer cells and showed that a β-catenin network containing prostaglandin G/H synthase 2/cyclooxygenase-2 (PTGS2/COX-2) is a crucial element in APF signalling [13].
Despite this progress, it remains unclear which molecules are regulated by APF in vivo because most of the findings have been established in cell culture models. Therefore, in the present study, we hypothesized that integration analysis of the in vitro proteomics data with available in vivo transcriptomics data may lead to the identification of potential APF-regulated proteins, which may be critical for the pathogenesis of IC. Recently, Gamper et al. [14] reported a quantitative transcriptomic analysis of bladder biopsies containing mucosa, lamina propria and detrusor muscle cells (at varying relative ratios and biopsy depth) from five patients with ulcerative IC and six healthy individuals as controls using Affymetrix GeneChip expression arrays (Affymetrix Corp., Santa Clara, CA, USA), with over 54 000 probe sets on the chip [14]. The study by Gamper et al. [14] is the largest published in vivo evaluation of tissues from human IC subjects without intervening cell culture steps. In an attempt to identify the potential proteins and genes regulated by APF in vivo, and to possibly expand the APF-regulated network identified by SILAC, we performed an integration analysis of our own SILAC data and the microarray data of Gamper et al. [14].
Materials and methods
Statistical analysis for the identification of differentially expressed genes (DEGs) between patients with IC and healthy controls
The microarray data set (GSE11783) of Gamper et al. [14] was retrieved from the Gene Expression Omnibus database repository (http://www.ncbi.nlm.nih.gov/geo/). To identify DEGs, we employed an integrative statistical hypothesis-testing approach that combines the adjusted P values from three methods: Student’s two-tailed t-test, the Wilcoxon rank-sum test and the median ratio test. In each of the three hypothesis tests, an empirical null probability density function was estimated for the corresponding statistic (t, rank sum and median difference) computed from 1000 random permutations of samples. Notably, 1000 is sufficiently larger than the number of all possible combinations of randomized sample groups (i.e. 11C5 = 462). An adjusted P value for the observed statistic of each probe set was then computed using the empirical probability density function [15]. The P values from the individual hypothesis tests were combined to obtain the overall P values using a meta-analysis (Stouffer’s method) as described previously [16]. Among the meta-analysis methods described [16], Stouffer’s method tends to produce small overall P values when the P values being combined are collectively small, whereas Fisher’s chi-squared method tends to produce small overall P values when one of P values is small, regardless of the other P values. In the present study, the meta-analysis was performed to select the DEGs collectively detected by the three different statistical tests (t-test, rank sum and log2-median-difference tests). Therefore, we used Stouffer’s method, rather than Fisher’s chi-squared method. False discovery rates (FDRs) were then computed from the overall P values using Storey’s method [17]. Finally, the DEGs were selected as the genes whose FDRs were < 0.01 and with fold changes > 1.4 or < 0.71. This fold change filter was used to remove potential false positives. For the same reason, we further used a fold change-based filter to avoid bias towards the t-test and rank sum test (i.e. not to remove false positive DEGs that have small fold changes but can be selected by t-test [or rank sum test] as a result of their sd).
Integration analysis of the SILAC data and the microarray data
For integration analysis, molecules that were quantitated at both protein and mRNA levels (i.e. the overlap of the SILAC data and the microarray data) were subjected to network modelling essentially as described previously [18,19]. Using the Database for Annotation, Visualization and Integrated Discovery [20], we analyzed the overlapping molecules, aiming to identify over-represented gene ontology biological processes (GOBPS) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [21], from which we selected those related to IC pathogenesis based on current knowledge. To construct a network model describing the selected GPBPs and KEGG pathways, we firstly identified the first neighbour of the overlapping molecules using the protein–protein interactome obtained from the National Center for Biotechnology Information and the KEGG databases. Cytoscape (http://www.cytoscape.org/) was then used to construct an initial network comprising the interactions among the overlapping molecules and their first neighbours. In Cytoscape, this initial network was reduced to generate a minimal subnetwork by removing the first neighbours that do not belong to the selected GOBPs and KEGG pathways. Finally, the nodes within the subnetwork were grouped into modules such that the nodes in the same modules have the same gene ontology annotations or belong to the same KEGG pathways. When a protein had multiple gene ontology annotations or belonged to multiple KEGG pathways, the protein was grouped into the most relevant module based on our understanding of biological processes or pathways related to IC pathogenesis.
Network modelling of the microarray data
DEGs that were quantitated and compared in (i) non-ulcerative tissues from patients with ulterative IC vs healthy bladder tissues and (ii) ulcerative tissues from the same patients with ulcerative IC vs healthy controls were subjected to network modelling essentially as described above.
Cell culture
Non-transformed urothelial cells (TRT-HU1) were cultured as described previously [22]. Patients with IC and age- and sex-matched healthy volunteers were recruited as described previously [13]. Primary bladder epithelial cells were propagated from transitional epithelium with submucosal bladder tissue derived from three patients with IC and three age- and sex-matched controls as described previously [13].
Western blot analysis
TRT-HU1 cells grown in six-well culture plates were treated with 1 μm synthetic asialo-APF or non-glycosylated negative control peptides (both from PolyPeptide Laboratories, San Diego, CA, USA) for 3 days after 16 h of serum starvation [6,11]. Whole cell lysates were extracted using lysis buffer purchased from Cell Signaling Technology (Beverly, MA, USA). Protein (25 μg) was subjected to SDS-PAGE separation and western blot analysis as described previously [22]. Lysates of bladder epithelial cells from patients with IC and controls were used for western blotting [13]. Antibodies against phospho-Akt (Ser473), Akt, phospho-mammalian target of rapamycin (mTOR) (Ser2448), mTOR, phospho-p70S6K (Thr389) and p70S6K were obtained from Cell Signaling Technology. Secondary antibodies and the Micro BCA protein assay kit were obtained from Pierce (Rockford, IL, USA). The ECL™ detection system was obtained from Amersham Biosciences (Little Chalfont, UK).
Results
Identification of genes differentially expressed between patients with IC and healthy controls
Gamper et al. [14] described DEGs between Hunner’s ulcer IC vs healthy control-derived mRNA. Their analysis suggested that immune and inflammatory responses are dominant in ulcerative tissue from patients with IC. However, Gamper et al. [14] used Student’s t-test for statistical analysis, which has significant limitations in the case of a small sample size. Moreover, the FDR of 8.7% for the 3618 significantly changed probe sets was relatively high compared to non-ulcer tissue in patients with Hunner’s ulcers and tissue from healthy controls (i.e. healthy) (Fig. 1A). Therefore, we decided to re-analyze the data set of Gamper et al. [14] using a more powerful statistical tool (i.e. an integrative statistical method) to calculate overall FDRs (as described in the Materials and methods).
FIG. 1.
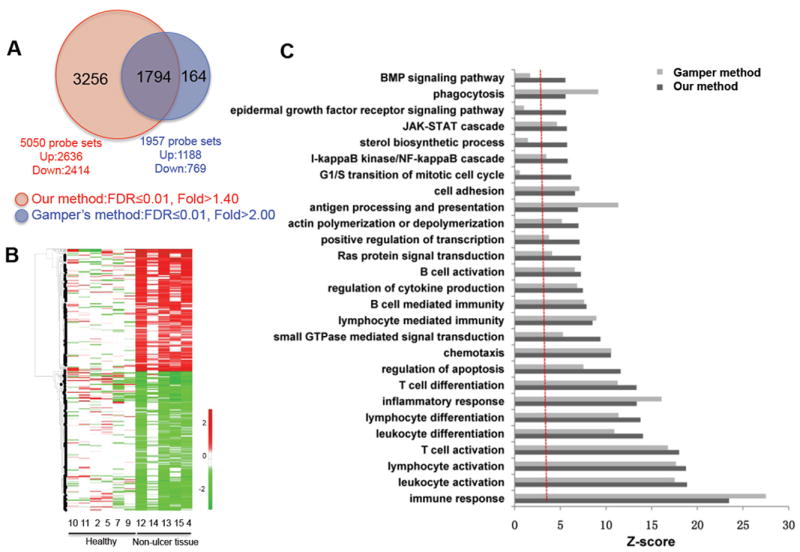
Comparison of differentially expressed genes (DEGs) of non-ulcerative tissue from patients with ulcerative interstitial cystitis (IC) vs bladder tissue from healthy controls obtained using the methods described in the present study, as well as those of Gamper et al. [14]. A, Venn diagram showing the relationships between DEGs generated using the methods described in the present study, as well as those of Gamper et al. [14]. B, Heatmap showing differential expression of the 3256 DEGs additionally identified using the methods described in the present study. The red and green colours represent the increase and the decrease of their expression levels in non-ulcerative tissue from patients with ulcerative IC compared to tissue from healthy controls, respectively. C, Functional enrichment analyses of DEGs generated using both methods. Most processes shown were commonly enriched by both DEG sets, whereas bone morphogenetic protein (BMP) and epidermal growth factor (EGF) receptor signalling pathways, sterol biosynthetic process, and G1/S transition of the mitotic cell cycle were all uniquely enriched in the DEG set of the present study. FDR, false discovery rate.
As a result of this analysis, 3034 and 3324 DEGs (FDR < 0.01 and fold change > 1.4), arising from 5050 and 5270 probe sets, were identified for comparison: (i) non-ulcerative tissue from patients with IC vs bladder tissue from healthy controls and (ii) ulcerative tissue from the same patients with IC vs bladder tissue from healthy controls, respectively, with 2218 in common. As shown in Fig. 1A, a comparison of non-ulcerative samples from patients with ulcerative IC vs healthy samples using the method described in the present study generated a more comprehensive set of DEGs with a much lower FDR than the method of Gamper et al. [14]. In addition to 1794 probe sets shared by both methods, 3256 and 163 probe sets were identified only by the method described in the present study and by the method of Gamper et al. [14], respectively (Fig. 1A).
Heatmap analysis of the 3256 probes additionally identified by the method described in the present study indicated that we were able to select DEGs showing differential expression in most (but not necessarily all) non-ulcerative tissues from patients with ulcerative IC compared to healthy tissue samples (Fig. 1B). By contrast, the method used by Gamper et al. [14] was so stringent that these probes could not be identified because no differential expression was observed in one IC sample (non-ulcer tissue 14) compared to healthy controls (Fig. 1B).
Furthermore, functional enrichment analysis of the DEG set of the present study and that of Gamper et al. [14] showed that pathways such as the immune response, immune cell activation and inflammatory response were commonly enriched (P < 0.05) in both data sets, whereas the bone morphogenetic protein and epidermal growth factor (EGF) signalling pathways, sterol biosynthetic process, and G1/S mitotic transition pathways were uniquely enriched by the DEG set of the present study (Fig. 1C). This result suggested that most of the additional 3256 probe sets are functionally redundant compared to those in the data set of Gamper et al. [14]. However, the additional DEGs were useful for integration analysis and also for the generation of a much broader network model.
Integration analysis identifies potential molecules regulated by APF in vivo
Comparison of the combined 4140 DEGs from the two DEG sets generated using our statistical method with the 102 differentially expressed proteins identified by SILAC [13] showed that the expression of 29 non-redundant genes, corresponding to 43 probe sets, was also changed in patients with IC with Hunner’s ulcer (Table 1). Among the 29 genes, 10 were consistently up- or down-regulated in both APF-treated T24 proteomics data and the human tissue gene expression data (Table 1), suggesting that they are potentially regulated by APF in vivo. Of the 10 genes, seven genes (TACSTD2, ERMP1, HSD17B4, HUWE1, MAPKSP1, GSPT1 and JUP1) were down-regulated, whereas three genes (XPOT, PTGS2/COX-2 and HIST1H4B) were up-regulated in both proteomics and transcriptomics data sets. Notably, TACSTD2, HSD17B4, HUWE1, JUP, XPOT, PTGS2/COX-2 and HIST1H4B were altered in the ulcerative bladder tissue (non-epithelium), as well as in T24 cells (epithelium-derived). This suggests that APF may regulate the expression of those genes in both epithelial and non-epithelial cells in vivo.
TABLE 1.
Overlapping differential signatures between proteomic and transcriptomic profiles
| Probe set ID | Entrez ID | Gene symbol | SILAC | GeneChip | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Fold change | Fold change | FDR | |||||
|
| |||||||
| APF/mock | N-Ul/H* | Ul/H† | N-Ul/H* | Ul/H† | |||
| 208992_s_at | 6774 | STAT3 | 0.379 | 1.690 | 2.485 | 0.004 | 0.001 |
| 208991_at | 6774 | STAT3 | 0.379 | 1.525 | 2.039 | 0.004 | 0.001 |
| 225289_at | 6774 | STAT3 | 0.379 | 1.353 | 1.906 | 0.006 | 0.005 |
| 201315_x_at | 10581 | IFITM2 | 0.425 | 1.648 | 3.090 | 0.005 | 0.001 |
| 202286_s_at | 4070 | TACSTD2 | 0.496 | 0.393 | 0.132 | 0.014 | 0.001 |
| 202422_s_at | 2182 | ACSL4 | 0.558 | 0.979 | 2.661 | 0.818 | 0.005 |
| 218342_s_at | 79956 | ERMP1 | 0.618 | 0.438 | 0.222 | 0.009 | 0.000 |
| 222603_at | 79956 | ERMP1 | 0.618 | 0.410 | 0.271 | 0.022 | 0.000 |
| 201649_at | 9246 | UBE2L6 | 0.619 | 2.906 | 1.627 | 0.005 | 0.077 |
| 209166_s_at | 4125 | MAN2B1 | 0.638 | 2.282 | 2.157 | 0.006 | 0.043 |
| 200839_s_at | 1508 | CTSB | 0.643 | 1.836 | 1.764 | 0.009 | 0.008 |
| 200838_at | 1508 | CTSB | 0.643 | 2.007 | 2.013 | 0.007 | 0.005 |
| 227961_at | 1508 | CTSB | 0.643 | 0.798 | 0.769 | 0.894 | 0.618 |
| 201413_at | 3295 | HSD17B4 | 0.646 | 0.570 | 0.521 | 0.011 | 0.006 |
| 208598_s_at | 10075 | HUWE1 | 0.646 | 0.788 | 0.662 | 0.011 | 0.009 |
| 227562_at | 8649 | MAPKSP1 | 0.659 | 0.431 | 0.396 | 0.003 | 0.002 |
| 217971_at | 8649 | MAPKSP1 | 0.659 | 0.661 | 0.628 | 0.005 | 0.003 |
| 202052_s_at | 26064 | RAI14 | 0.680 | 1.016 | 1.983 | 0.852 | 0.008 |
| 234975_at | 2935 | GSPT1 | 0.689 | 0.668 | 0.609 | 0.006 | 0.005 |
| 1554411_at | 1499 | CTNNB1 | 0.691 | 0.802 | 1.596 | 0.260 | 0.138 |
| 217933_s_at | 51056 | LAP3 | 0.695 | 3.097 | 2.180 | 0.001 | 0.002 |
| 201015_s_at | 3728 | JUP | 0.701 | 0.404 | 0.262 | 0.012 | 0.004 |
| 232666_at | 4940 | OAS3 | 0.710 | 1.264 | 1.075 | 0.013 | 0.710 |
| 202234_s_at | 6566 | SLC16A1 | 0.711 | 1.539 | 3.267 | 0.038 | 0.002 |
| 215931_s_at | 10564 | ARFGEF2 | 0.712 | 0.824 | 0.754 | 0.433 | 0.570 |
| 202562_s_at | 11161 | C14orf1 | 1.401 | 0.590 | 0.460 | 0.006 | 0.001 |
| 217188_s_at | 11161 | C14orf1 | 1.401 | 0.662 | 0.537 | 0.006 | 0.004 |
| 212600_s_at | 7385 | UQCRC2 | 1.404 | 0.772 | 0.625 | 0.018 | 0.004 |
| 200883_at | 7385 | UQCRC2 | 1.404 | 0.664 | 0.607 | 0.006 | 0.005 |
| 203634_s_at | 1374 | CPT1A | 1.407 | 0.995 | 1.047 | 0.973 | 0.373 |
| 200660_at | 6282 | S100A11 | 1.429 | 0.544 | 0.381 | 0.006 | 0.003 |
| 201250_s_at | 6513 | SLC2A1 | 1.438 | 0.396 | 0.304 | 0.020 | 0.004 |
| 209279_s_at | 50814 | NSDHL | 1.441 | 0.634 | 0.626 | 0.004 | 0.009 |
| 225359_at | 131118 | DNAJC19 | 1.455 | 0.791 | 0.667 | 0.003 | 0.006 |
| 225358_at | 131118 | DNAJC19 | 1.455 | 0.599 | 0.564 | 0.002 | 0.008 |
| 235244_at | 131076 | CCDC58 | 1.483 | 0.943 | 0.880 | 0.921 | 0.632 |
| 230097_at | 2618 | GART | 1.498 | 1.709 | 1.424 | 0.022 | 0.489 |
| 244822_at | 2618 | GART | 1.498 | 1.005 | 0.907 | 0.713 | 0.936 |
| 232103_at | 10380 | BPNT1 | 1.508 | 0.735 | 0.652 | 0.159 | 0.181 |
| 222405_at | 51495 | PTPLAD1 | 1.571 | 0.384 | 0.352 | 0.005 | 0.001 |
| 234000_s_at | 51495 | PTPLAD1 | 1.571 | 0.342 | 0.297 | 0.003 | 0.000 |
| 222404_x_at | 51495 | PTPLAD1 | 1.571 | 0.361 | 0.294 | 0.002 | 0.000 |
| 217777_s_at | 51495 | PTPLAD1 | 1.571 | 0.367 | 0.390 | 0.001 | 0.002 |
| 201596_x_at | 3875 | KRT18 | 1.596 | 0.375 | 0.183 | 0.007 | 0.001 |
| 209873_s_at | 11187 | PKP3 | 1.621 | 0.441 | 0.156 | 0.008 | 0.001 |
| 212160_at | 11260 | XPOT | 1.691 | 1.321 | 1.590 | 0.029 | 0.005 |
| 1554997_a_at | 5743 | PTGS2 | 1.790 | 1.183 | 15.836 | 0.209 | 0.006 |
| 204748_at | 5743 | PTGS2 | 1.790 | 1.102 | 6.930 | 0.228 | 0.003 |
| 218279_s_at | 8337 | HIST2H2AA3 | 1.884 | 0.988 | 1.008 | 0.285 | 0.605 |
| 210517_s_at | 9590 | AKAP12 | 1.909 | 0.450 | 0.703 | 0.008 | 0.142 |
| 227530_at | 9590 | AKAP12 | 1.909 | 0.376 | 0.544 | 0.008 | 0.017 |
| 227529_s_at | 9590 | AKAP12 | 1.909 | 0.370 | 0.490 | 0.005 | 0.014 |
| 229426_at | 9377 | COX5A | 1.983 | 0.933 | 0.830 | 0.370 | 0.137 |
| 232035_at | 8366 | HIST1H4B | 2.322 | 1.633 | 2.041 | 0.017 | 0.008 |
N-Ul/H, non-ulcerative tissue from patients with ulcerative interstitial cystitis /healthy controls.
Ul/H, ulcerative tissue/healthy controls.
APF, antiproliferative factor; FDR, false discovery rate; SILAC, stable isotope labelling by amino acids in cell culture.
To identify the molecular networks potentially regulated by APF in vivo, we performed integration analysis and network modelling. As shown in Fig. 2, 10 modules, including cell adhesion, mitogen-activated protein kinase (MAPK), GTPase regulator and cell cycle, were down-regulated, whereas nine modules, including integrin, actin regulation, transport and COX, were up-regulated. In addition, several molecules, such as PTGS2/COX-2, XPOT, mitogen-activated protein kinase scaffold protein 1 (MAPKSP1), JUP and GSPT1, are shown with similarly coloured node and node boundaries, indicating that they are up- or down-regulated at both the RNA and protein levels.
FIG. 2.
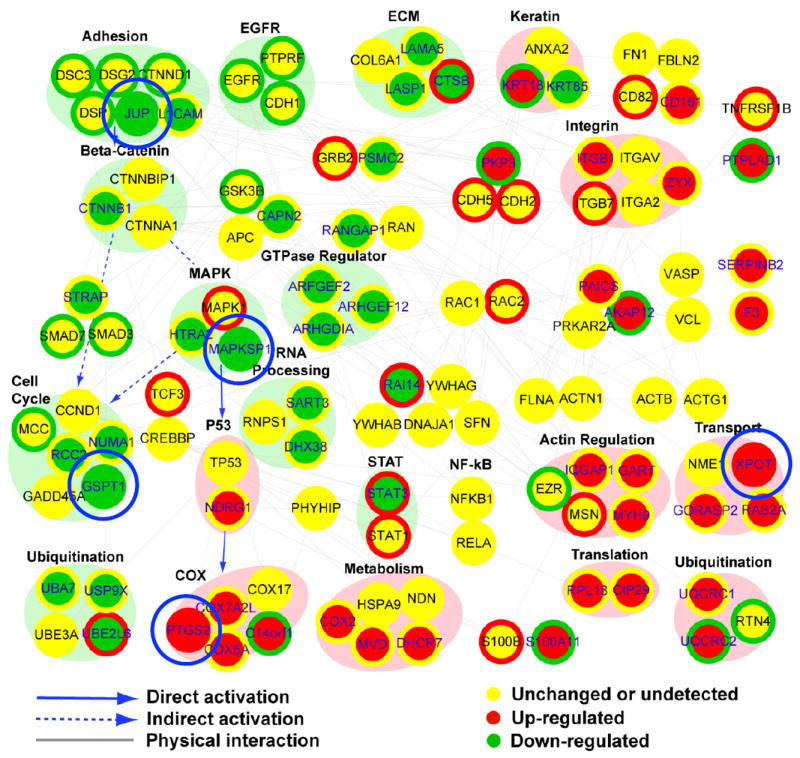
A network model generated by integrating the stable isotope labelling by amino acids in cell culture (SILAC) data of the present study and the differential RNA expression data (GSE11783) of Gamper et al. [14]. Node colour represents increases (red), no significant changes (yellows) and decreases (green) in protein abundance after antiproliferative factor (APF) treatment, as ascertained by SILAC. Changes in RNA expression levels of the corresponding nodes in patients with interstitial cystitis (IC) are shown as coloured node boundaries (donut shape) and the colour represents increases (red), no significant change (yellow) and decreases (green) in gene expression in IC patient material compared to normal specimens. Large circles (or ovals) surrounding several nodes represent modules, and colours indicate increased (pink) or decreased (light green) expression. Several molecules, such as prostaglandin G/H synthase 2/cyclooxygenase-2 (PTGS2/COX-2), mitogen-activated protein kinase scaffold protein 1 (MAPKSP1) and γ-catenin (JUP), emerged as nodes significantly perturbed in vitro and in vivo in the integrated APF network. COX; cyclooxygenase; ECM, extracellular matrix; EGFR, epidermal growth factor receptor; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor kappa B; STAT, signal transducer and activator of transcription.
Network modelling of DEGs identifies pathways changed in IC in vivo
To determine which pathways are most significantly changed in IC in vivo and whether the aforementioned APF-regulated modules are part of the significantly changed pathways, we performed network modelling analysis of the DEGs identified using the analytic method employed in the present study. As shown in Fig. 3, immune system and inflammatory pathways regulating: (i) signalling downstream of immunoreceptors, such as T cell receptor, B cell receptor and the high-affinity IgE receptor FcεRI; (iii) Toll-like receptor signalling; (iii) antigen processing and presentation; and (iv) leukocyte transendothelial migration are all up-regulated in not only ulcerative, but also non-ulcerative biopsies from patients with ulcerative IC compared to healthy bladder tissues. Previously, we found that PTGS2/COX-2, an inducible enzyme critical for the biosynthesis of pro-inflammatory prostaglandin E2, is up-regulated by APF in vitro and also overexpressed in urothelial cells explanted from IC biopsies [13]. Given that PTGS2 is also up-regulated in IC in vivo, it is probable that PTGS2/COX-2 transmits inflammatory effects of APF and that the PTGS2/COX-2 module is a critical module downstream of APF in vivo.
FIG. 3.
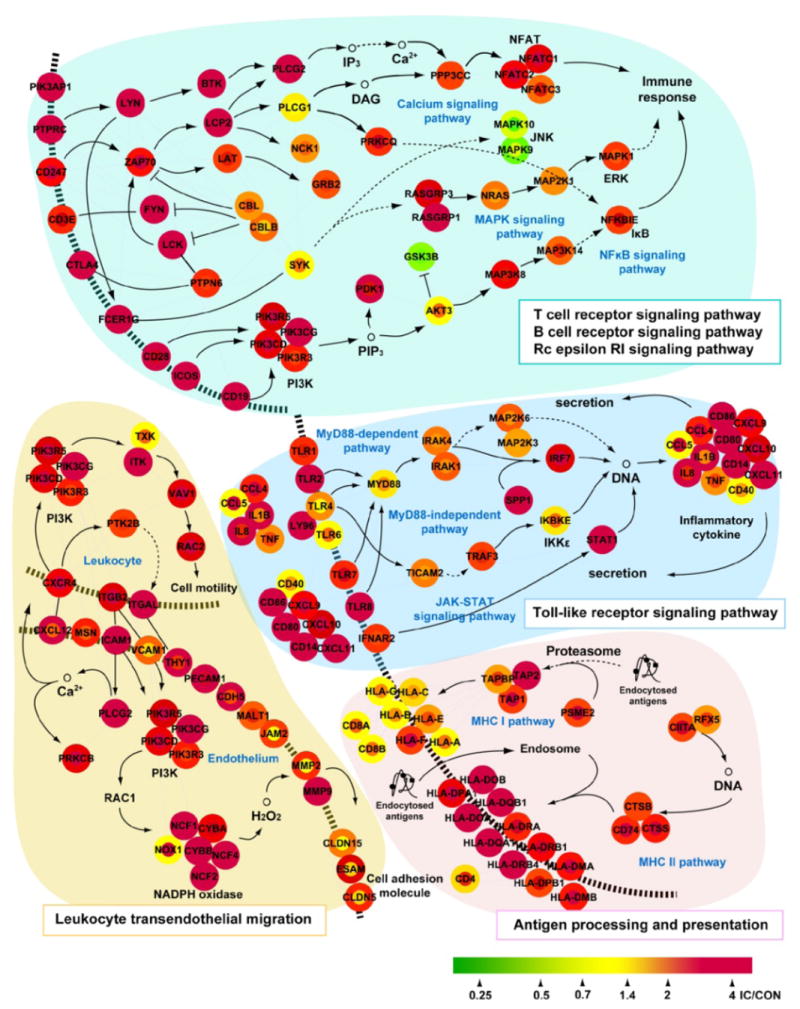
Network analysis suggests an immune response and inflammation in ulcerative tissue and non-ulcerative tissue of patients with ulcerative interstitial cystitis (IC). Microarray data from patients with IC with Hunner’s ulcer (GSE11783) predict a hypothetical network, showing an immune response and inflammation signature (P < 0.01) in both ulcerative tissue and non-ulcerative tissue. The node and node boundary colours represent changes at the mRNA level in ulcerative tissue and non-ulcerative tissue of patients with IC, respectively, compared to tissue from healthy controls. The pathway information was obtained from the Kyoto Encyclopedia of Genes and Genomes pathway database.
APF regulates the Akt/mTOR/p70S6K pathway in bladder epithelial cells
By contrast to PTGS2/COX-2, MAPKSP1 and GSPT1 are significantly down-regulated in both SILAC and microarray datasets (Fig. 2). MAPKSP1 encodes a MAPK scaffold protein that binds specifically to MEK1 but not to MEK2 [23]. It has been reported to be involved in the activation of mTORC1 [24]. GSPT1 encodes a translation factor named eukaryotic peptide chain release factor subunit 3a (eRF3a), and it has been shown that eRF3a depletion decreases translation through the inhibition of mTORC1 activity [25].
Because the phosphorylation of Akt, a crucial upstream activator of mTORC1, was previously shown to be decreased by APF [11], we further aimed to examine whether the PI3K/Akt/mTOR/p70S6K pathway signalling proteins were abnormally expressed and/or phosphorylated in APF-treated TRT-HU1 cells or cells explanted from patients with IC. When TRT-HU1 cells, which were originally explanted from normal tissue and immortalized with hTERT, were treated with APF, the phosphorylation levels of Akt-S473, mTOR-S4882 and p70S6K-T389 were down-regulated, whereas the expression levels of Akt, mTOR and p70S6K proteins were unaffected (Fig. 4, left). Moreover, a similar pattern was observed in primary bladder epithelial cells derived from patients with IC and matched controls (Fig. 4, right). These findings suggest that mTOR signalling may be important in addition to the β-catenin and MAPK pathways in the APF signal network [11-13].
FIG. 4.
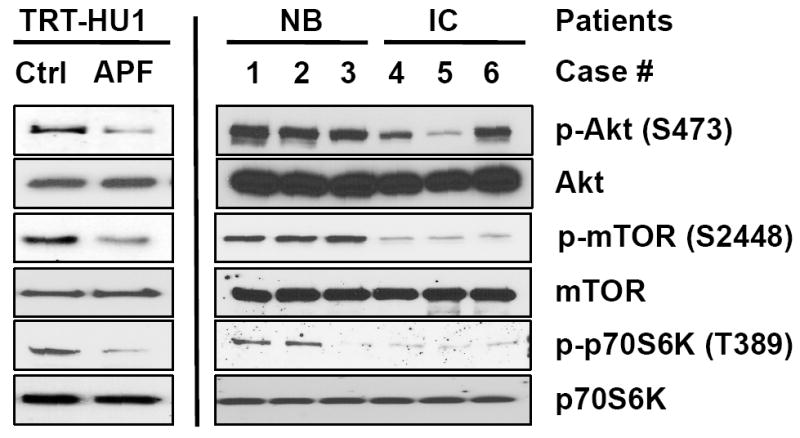
Antiproliferative factor (APF) regulates the Akt/mammalian target of rapamycin (mTOR) signalling pathway in bladder epithelial cells. Left: in TRT-HU1 cells, Akt, mTOR and p70S6K were dephosphorylated in response to APF treatment. Right: phosphorylation levels of Akt, mTOR and p70S6K were significantly reduced in primary epithelial cells explanted from interstitial cystitis (IC) bladder compared to paired normal bladder (NB)
Discussion
In the present study, the network modelling predicted that inflammatory mediators are consistently increased in both in vivo human specimens from patients with IC and in vitro APF-responsive cultured bladder cells. These findings are supportive in general terms of previous observations that mast cells (in detrusor muscle in particular), as well as cytokines and inflammatory markers (e.g. histamine, methylhistamine and interleukin-6), which often are associated with pain, hyperaemia and fibrosis, are significantly increased in the bladder tissue and urine of patients with IC [26,27]. Although the molecular networks regulated by a promising marker for IC (i.e. APF) are emerging from in vitro models using quantitative proteomics analysis [13], little is known about the molecules regulated by APF in vivo. In the present study, by using integration analysis of large-scale quantitative proteomics and transcriptomics data sets in combination with network modelling, 10 candidate genes are identified that we conclude are probably regulated by APF in vivo. Of these candidates, five (i.e. JUP, MAPKSP1, GSPT1, PTGS2/COX-2 and XPOT) are components of candidate modules that are significantly changed by APF.
Among the five prominent candidate genes identified in this analysis, JUP encodes the protein junction plakoglobin or γ-catenin. This protein is the only known constituent common to submembraneous plaques of both desmosomes and adherens junctions [28]. The down-regulation of JUP may cooperate with APF-induced down-regulation of other catenin proteins, such as α-catenin [10] and β-catenin [11,13], leading to decreased cell–cell adhesion and thus increased bladder epithelial leakiness, a common feature of IC. Similar to down-regulation of the other catenins, down-regulation of γ-catenin may also play a role in the regulation by APF of other gene expression, including E-cadherin and vimentin; decreased γ-catenin may also lead to aberrant downstream protein phosphorylation, including Akt and extracellular signal-regulated kinase (ERK) [11,12,29].
MAPKSP1 encodes a MAPK scaffold protein that binds specifically to MEK1 but not to MEK2 [23]. Recent studies showed that this protein tightly associates with p14 and that the association is important for ERK activation in response to EGF [30]. The down-regulation of MAPKSP1 in combination with APF-induced down-modulation of EGF receptor [13] would also be expected to inhibit ERK activation. This provides yet another potential mechanism by which APF could inhibit heparin-binding epidermal growth factor-like growth factor-induced activation of ERK [12]. With respect to GSPT1, which encodes eRF3a, it has been reported that eRF3a depletion causes cell cycle arrest at G1 with decreased translation through the inhibition of mTORC1 activity [25]. Notably, MAPKSP1 has also been reported to be involved in the activation of mTORC1 [24]. The findings obtained in the present study indicate that the Akt pathway is a critical downstream component of APF signalling. Another APF responsive gene, XPOT, encodes tRNA exportin, which specifically binds to mature tRNAs and exports them from the nucleus to the cytoplasm [31]. tRNAs are essential intermediates of protein synthesis. Although the physiological meaning of up-regulation of tRNA exportin in response to APF is unclear, up-regulation of tRNA exportin may comprise a compensatory mechanism for offsetting the APF-induced decrease in protein synthesis and/or APF-induced decrease in the expression of a putative tRNA synthetase-like protein [10].
Although previous studies have shown that APF plays an important role in the regulation of cell adhesion, paracellular permeability, proliferation and cell signalling, they have not addressed its possible involvement in the inflammation commonly seen in ulcerative IC. The analysis conducted in the present study showed that the increased expression of proteins involved in certain immune responses, including the activation and differentiation of leukocytes, lymphocytes and T cells, was consistently observed in the DEG set of the present study and that of Gamper et al. [14] (P < 0.05) (Fig. 1C). Recently, using SILAC-based quantitative proteomics followed by functional assays, we showed that APF can evoke an inflammatory response by inducing the overexpression of PTGS2, more commonly known as PTGS2/COX-2, in bladder T24 cancer cells [13]. It was also shown that the level of PTGS2/COX-2 is increased in urothelial cells explanted from IC biopsies compared to bladder tissue from healthy controls [13]. Analysis of the in vivo microarray data set of Gamper et al. [14] showed that PTGS2/COX-2 is overexpressed in IC. Because APF activity has been shown to be present in > 94% of patients with IC diagnosed using the same criteria [5], these findings support the conclusion that PTGS2/COX-2 may be a critical node for transmitting the inflammatory effect of APF in vivo.
In summary, integration analysis of two published large-scale data sets identified 10 potential molecules regulated by APF. Network modelling showed that inflammation is also present in non-ulcerative tissue from patients with ulcerative IC, supporting our previous findings and hypothesis suggesting that APF, whose level may greater in urine samples from patients with IC, evokes an inflammatory response by up-regulating PTGS2/COX-2 [13]. The data obtained in the present study also provide additional evidence for the role of APF in regulating the expression of proteins that in turn regulate cell adhesion, as well as Akt and Erk signalling pathway activation, corroborating information obtained by other methods [11,32]. Notably, two of these proteins (i.e. MAPKSP1 and GSPT1), which are down-regulated by APF, are also involved in the activation of mTORC1, suggesting that the mTOR pathway is potentially a critical pathway regulated by APF. Several components of the mTOR pathway are being studied as potential therapeutic targets in other diseases [33]. The analysis conducted in the present study suggests that this pathway may also be relevant in the design of diagnostic tools and medications targeting IC.
In conclusion, bioinformatics analysis of SILAC-based quantitative proteomics and transcriptomics data have predicted several candidate molecules as mediators of cellular responses evoked by APF, a sialoglycopeptide prevalent in the urine of patients with IC. Combined with earlier findings, these results suggest additional rational targets for therapeutic intervention in IC.
Acknowledgments
This research was supported by NIH grants R37 DK47556, R01 DK57691, P50 DK65298 (to M.R.F.); R01 DK52596 (to S.K.K.); Korean Ministry for Health, Welfare and Family Affairs grant A080768 and Korean MEST FPR08A1-050 (to D.H.); the WCU Project of the Korean Ministry of Education, Science and Technology (to M.R.F. and K.P.K.); and the Fishbein Family IC Research Foundation/Interstitial Cystitis Association, New York Academy of Medicine, and Children’s Hospital Boston Faculty Development (to J.K.). J.K. is an American Urological Association Foundation Research Scholar and an Eleanor and Miles Shore Scholar of Harvard Medical School. J.K. designed and coordinated this project and wrote the manuscript. W.Y. performed the proteomics data and analyses and participated in drafting the manuscript. Y.K., T.K. and D.H. performed the bioinformatics analyses. S.K. provided expertise on the biology of A.P.F. K.P.K., H.S., D.H. and M.F. participated in the design and coordination of the study. All authors were involved in writing the manuscript and have approved the final version.
Abbreviations
- APF
antiproliferative factor
- DEG
differentially expressed gene
- EGF
epidermal growth factor
- eRF3a
eukaryotic peptide chain release factor subunit 3a
- ERK
extracellular signal-regulated kinase
- FDR
false discovery rate
- IC
interstitial cystitis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen-activated protein kinase
- MAPKSP1
mitogen-activated protein kinase scaffold protein 1
- mTOR
mammalian target of rapamycin
- PGE2
prostaglandin E2
- PTGS2/COX-2
prostaglandin G/H synthase 2/cyclooxygenase-2
- SILAC
stable isotope labelling by amino acids in cell culture
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Phatak S, Foster HE., Jr The management of interstitial cystitis: an update. Nat Clin Pract Urol. 2006;3:45–53. doi: 10.1038/ncpuro0385. [DOI] [PubMed] [Google Scholar]
- 2.Bogart LM, Berry SH, Clemens JQ. Symptoms of interstitial cystitis, painful bladder syndrome and similar diseases in women: a systematic review. J Urol. 2007;177:450–6. doi: 10.1016/j.juro.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 3.Moutzouris DA, Falagas ME. Interstitial cystitis: an unsolved enigma. Clin J Am Soc Nephrol. 2009;4:1844–57. doi: 10.2215/CJN.02000309. [DOI] [PubMed] [Google Scholar]
- 4.Erickson DR, Xie SX, Bhavanandan VP, et al. A comparison of multiple urine markers for interstitial cystitis. J Urol. 2002;167:2461–9. [PubMed] [Google Scholar]
- 5.Keay SK, Zhang CO, Shoenfelt J, et al. Sensitivity and specificity of antiproliferative factor, heparin-binding epidermal growth factor-like growth factor, and epidermal growth factor as urine markers for interstitial cystitis. Urology. 2001;57:9–14. doi: 10.1016/s0090-4295(01)01127-x. [DOI] [PubMed] [Google Scholar]
- 6.Keay SK, Szekely Z, Conrads TP, et al. An antiproliferative factor from interstitial cystitis patients is a frizzled 8 protein-related sialoglycopeptide. Proc Natl Acad Sci USA. 2004;101:11803–8. doi: 10.1073/pnas.0404509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrads TP, Tocci GM, Hood BL, et al. CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J Biol Chem. 2006;281:37836–43. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 8.Keay S, Kleinberg M, Zhang CO, Hise MK, Warren JW. Bladder epithelial cells from patients with interstitial cystitis produce an inhibitor of heparin-binding epidermal growth factor-like growth factor production. J Urol. 2000;164:2112–8. [PubMed] [Google Scholar]
- 9.Kim J, Keay SK, Dimitrakov JD, Freeman MR. p53 mediates interstitial cystitis antiproliferative factor (APF)-induced growth inhibition of human urothelial cells. FEBS Lett. 2007;581:3795–9. doi: 10.1016/j.febslet.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keay S, Seillier-Moiseiwitsch F, Zhang CO, Chai TC, Zhang J. Changes in human bladder epithelial cell gene expression associated with interstitial cystitis or antiproliferative factor treatment. Physiol Genomics. 2003;14:107–15. doi: 10.1152/physiolgenomics.00055.2003. [DOI] [PubMed] [Google Scholar]
- 11.Shahjee HM, Koch KR, Guo L, Zhang CO, Keay SK. Antiproliferative factor decreases Akt phosphorylation and alters gene expression via CKAP4 in T24 bladder carcinoma cells. J Exp Clin Cancer Res. 2010;29:160. doi: 10.1186/1756-9966-29-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Keay SK, Freeman MR. Heparin-binding epidermal growth factor-like growth factor functionally antagonizes interstitial cystitis antiproliferative factor via mitogen-activated protein kinase pathway activation. BJU Int. 2009;103:541–6. doi: 10.1111/j.1464-410X.2008.08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Chung YG, Kim Y, et al. Quantitative proteomics identifies a beta-catenin network as an element of the signaling response to Frizzled-8 protein-related antiproliferative factor. Mol Cell Proteomics. 2011 Jun;10(6):M110–007492. doi: 10.1074/mcp.M110.007492. Epub 2011 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamper M, Viereck V, Geissbuhler V, et al. Gene expression profile of bladder tissue of patients with ulcerative interstitial cystitis. BMC Genomics. 2009;10:199. doi: 10.1186/1471-2164-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HJ, Suk JE, Patrick C, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285:9262–72. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang D, Rust AG, Ramsey S, et al. A data integration methodology for systems biology. Proc Natl Acad Sci USA. 2005;102:17296–301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storey JD, Tibshirani R. Technical Report No 2001-28. Stanford, CA: Department of Statistics, Stanford University; 2001. Estimating the Positive False Discovery Rate Under Dependence, with Application to DNA Microarrays. [Google Scholar]
- 18.Hwang D, Lee IY, Yoo H, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho JY, Lee M, Ahn JM, et al. Proteomic analysis of a PDEF Ets transcription factor-interacting protein complex. J Proteome Res. 2009;8:1327–37. doi: 10.1021/pr800683b. [DOI] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Ji M, Didonato JA, et al. An hTERT-immortalized human urothelial cell line that responds to anti-proliferative factor. In Vitro Cell Dev Biol Anim. 2011;47:2–9. doi: 10.1007/s11626-010-9350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–71. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 24.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chauvin C, Salhi S, Jean-Jean O. Human eukaryotic release factor 3a depletion causes cell cycle arrest at G1 phase through inhibition of the mTOR pathway. Mol Cell Biol. 2007;27:5619–29. doi: 10.1128/MCB.00035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsen D, Frye S, Trimble LA, et al. Inflammatory mediator profile in urine and bladder wash fluid of patients with interstitial cystitis. J Urol. 1994;152:355–61. doi: 10.1016/s0022-5347(17)32739-8. [DOI] [PubMed] [Google Scholar]
- 27.Saini R, Gonzalez RR, Te AE. Chronic pelvic pain syndrome and the overactive bladder: the inflammatory link. Curr Urol Rep. 2008;9:314–9. doi: 10.1007/s11934-008-0054-8. [DOI] [PubMed] [Google Scholar]
- 28.Cowin P, Kapprell HP, Franke WW, Tamkun J, Hynes RO. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. 1986;46:1063–73. doi: 10.1016/0092-8674(86)90706-3. [DOI] [PubMed] [Google Scholar]
- 29.Pan H, Gao F, Papageorgis P, Abdolmaleky HM, Faller DV, Thiagalingam S. Aberrant activation of gamma-catenin promotes genomic instability and oncogenic effects during tumor progression. Cancer Biol Ther. 2007;6:1638–43. doi: 10.4161/cbt.6.10.4904. [DOI] [PubMed] [Google Scholar]
- 30.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–14. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 31.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–73. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 32.Keay S. Cell signaling in interstitial cystitis/painful bladder syndrome. Cell Signal. 2008;20:2174–9. doi: 10.1016/j.cellsig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Tsang CK, Qi H, Liu LF, Zheng XF. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–24. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]


