Abstract
Biological indicators have numerous and widespread utility in personalized medicine, but the measurement of these indicators also pose many technological and practical challenges. Blood/plasma has typically been used as the sample source with which to measure these indicators, but the invasiveness associated with procurement of samples has led to increased interest in saliva as an attractive alternative. However, there are unique issues associated with the measurement of saliva biomarkers. These issues are compounded by the imperfect correlation between saliva and plasma with respect to biomarker profiles. In this manuscript, we address the technical challenges associated with saliva biomarker quantification describe a high-content microarray assay that employs both grating-coupled surface plasmon resonance imaging surface plasmon coupled emission modalities in a highly sensitive assay that has a large dynamic range. This powerful approach provides the tools to map the proteome of saliva, which in turn should greatly enhance the utility of salivary biomarker profiles in personalized medicine.
Keywords: saliva, biomarker, proteomics, surface plasmon resonance, emission microarray, SPRI, SPCE, personalized medicine, biofluid, biomolecule
Unit Introduction
The transformational promise of personalized medicine depends largely on technological advances that will allow multiplexed measurement and characterization of bioindicators of health and disease. An ability to simultaneously profile multiple objective indicators of a biological state (biomarkers) will facilitate screening, diagnosis and risk-stratification, guide targeted therapy, and monitor response to treatment across a wide spectrum of disease. Traditionally, blood has been the dominant choice as biofluid because it is exposed to every organ system as it flows through the body and mingles with proteins and biomolecules reflective of different biological pathways implicated in disease. Limitations to the use of blood, however, include the invasiveness of the sample procurement, the need for trained personnel, the need to physically isolate cells from the liquid component, and the limited quantity available from infants and children. Additionally, the stress and pain associated with a blood draw can alter the biomarker profile and confound interpretation of the results. Saliva offers an attractive alternate to conventional biofluids such as blood and urine because collecting saliva is much easier and better accepted by the patient. Beyond eliminating the risk of disease transmission due to accidental needle-sticks, the self-collection possible with saliva allows multiple, repeat samples at close intervals.
One of the compelling features of saliva as biofluid is the presence of many of the biomarkers that exist in plasma, although at much lower levels ; for review see (Lee and Wong, 2009) (Kawas et al.; Parahitiyawa et al.). As with blood, saliva contains a range of nucleic acids, drug metabolites, enzymes, hormones, antibodies, antimicrobial constituents, cytokines, microbes and other peptides (Rehak et al., 2000; Zelles et al., 1995) (Table 1). Compared to the 2698 proteins found in plasma, the salivary proteome is estimated to contain more than 2290 measurable proteins (Loo et al.). The finding that over 40% of candidate markers for diseases such as cancer, cardiovascular disease, and stroke can be found in whole saliva, underscores the potential of saliva to reflect a range of physiologic states including emotional, endocrinal, nutritional and metabolic variations.
Table 1.
Potential Salivary Biomarkers
| Class of Biomarker |
Biomarker | Associated Signaling Pathway |
Range Measured in Saliva |
Method(s) Used for Detection |
Reference(s) |
|---|---|---|---|---|---|
| Enzyme | α-amylase | Symptho-adrenomedullary | 100–900 µg/ml, follows diurnal rhythm | Kinetic colorimetric assay | (Noto et al., 2005) |
| Lysozyme | Immune | 20–68.2 µg / min | ELISA | (Yang et al., 2002) | |
| Hormone | Chromogranin A | Symptho-adrenomedullary | 0.3–0.45 pmol/mg protein, follows diurnal rhythm | ELISA | (Nakane et al., 2002) (Giampaolo et al., 2002) (Soo-Quee Koh and Choon-Huat Koh, 2007) |
| Cortisol | HPA | 3.5–27 mg/dl | Competition ELISA | (Ng et al., 2003) (Sluiter et al., 2003) | |
| DHEA | HPA | 0.5 – 1.0 nmol, 40–550 pg/ml | ELISA, RIA | (Granger et al., 1999; Shirotsuki et al., 2009) | |
| Leptin | 3–6 pmol/L | RIA | (Randeva et al., 2003) | ||
| Immunoglobulin | IgA | HPA | 100–900 µg/ml | ELISA | (Yang et al., 2002) (Ng et al., 2004) |
| Neurotrophins | Nerve Growth Factor (NGF) | HPA | (Borelli et al.) (Jang et al.) (Saruta et al.) | ||
| Brain-derived Neurotrophic Factor (Saruta et al.) | HPA | (Saruta et al.) | |||
| Neuropeptides | Beta-endorphin | HPA | 1300–2800 pg/ml | ELISA | (Rai et al.; Takai et al., 2007) |
| melatonin | HPA | 2–50 pg/ml | ELISA | (McVicar et al., 2007; Shinohara and Kodama) | |
| Oxytocin | 2–10 pg/ml | EIA | (Carter et al., 2007) | ||
| Substance P | 10–130 pg/ml | ELISA | (Borelli et al.) (Jang et al.) | ||
| Cytokines | IL-1 beta | 200–800 pg/ml | ELISA, Luminex | (He et al.; Teles et al., 2009) | |
| TNF-alpha | 40–90 pg/ml | RIA, Luminex | (He et al.; Teles et al., 2009) | ||
| IL-2 | 3–75 pg/ml | ELISA, Luminex | (Simcic et al., 2006) (Teles et al., 2009) (Leigh et al., 2002) | ||
| IL-4 | 17–110 pg/ml | ELISA, Luminex | (Liu et al., 2009) (Teles et al., 2009) (Leigh et al., 2002) | ||
| IL-5 | 30–200 pg/ml | Luminex | (Teles et al., 2009) | ||
| IL-6 | 25–50 pg/ml | Luminex | (Teles et al., 2009){ | ||
| IL-8 | 1500–2300 pg/ml | Luminex | (Teles et al., 2009) | ||
| IL-10 | 100–450 pg/ml | Luminex | (Teles et al., 2009) (Leigh et al., 2002) | ||
| IL-12 | 12–28 pg/ mg protein | ELISA | (Leigh et al., 2002) | ||
| IFN-gamma | 6–150 pg/ml | ELISA, Luminex | (Liu et al., 2009) (Teles et al., 2009) (Leigh et al., 2002) | ||
| IL-6 | 2–37 pg/ml | ELISA, Luminex | (Simcic et al., 2006) | ||
| GM-CSF | 30–90 pg/ml | Luminex | (Teles et al., 2009) | ||
| Drug Metabolites | Cotinine | 0–1000+ ng/ml | Colorimetric immunoassay | (Marrone et al.) | |
| Cannabinoid | 2–293 ng/ml | ELISA | (Schwope et al.) | ||
| Cocaine | 0–2282 ng/ml | GC-MS | (Bosker and Huestis, 2009) | ||
| Amphetamines | 3–1654 ng/ml | EIA | (Cooper et al., 2006) | ||
| Opiates | 23.6–191 ng/ml | EIA | (Barnes et al., 2003) | ||
| Phencyclidine | 2–10 ng/ml | EIA | (Cone et al., 2002) | ||
| Cellular Biomarkers | T-lymphocyte/ NK cell ratio | Negative correlation associated with stress | Flow cytometry | (Dos-Santos et al., 2009) | |
| B-lymphocyte percentage | Negative correlation associated with stress | Flow cytometry | (Dos-Santos et al., 2009) |
However, using saliva as a diagnostic biofluid also presents its own unique set of challenges, which have been the subject of recent reviews. These challenges include the wide dynamic range of the salivary proteome, confounding effects of diurnal variation (Harden et al.), the presence of proteolytic enzymes that decrease biomarker stability (Thomadaki et al.), high abundance glycoproteins, and the need to control for salivary flow rate, protein concentration, and method of collection. Additionally, salivary and serum biomarker levels do not correlate well when the route of transport across biological membranes requires more than passive diffusion, which limits the size and type of systemic disease biomarkers that can be assayed in saliva.
Many of the current proteomic technologies rely on mass spectrometry for interrogating the complex plasma and saliva proteomes. The substantive improvements in sensitivity and mass accuracy notwithstanding, the complexities of the saliva matrix can exceed the capacity of current mass spectrometry platforms to resolve the individual protein and peptide componenst fully in a single analysis. Often, in-depth analysis of the biofluid requires extensive sample preparation or fractionation. Promising developments in analytical technologies that utilize Surface plasmon resonance (SPR) are opening up new opportunities for multiplexed measurements of salivary analytes with a wide dynamic range. Surface plasmon resonance (SPR) based technologies have been used to identify cellular and protein biomarkers in complex fluids. (Jin et al., 2006) (Rice et al.). One distinctive advantage of SPR-based sensors is the capacity to carry out real-time measurement without labeling (Liedberg et al., 1983). In a grating-coupled imaging system (GCSPRI), a grating, printed on the chip surface, allows light to be coupled into the surface plasmon at a specific resonance angle. Only molecular interactions close to the surface will influence the SPR measurement because the penetration depth of the surface plasmon is only about 200 nm from the surface (Liedberg et al., 1993). Non-specific binding, however, can limit the sensitivity of this approach for complex sample analysis.
To overcome sensitivity limitations, a fluorescence measurement can be made that takes advantage of surface plasmon coupled emission (SPCE). In an SPCE system, the fluorescence of molecules within the 200 nm evanescent field penetration depth couples to the surface plasmons in the metal. The emission is then re-radiated with high efficiency and with a high degree of polarization and directionality at a resonance angle that corresponds to a wave vector matching the emission wavelength (Reilly et al., 2006). The increase in fluorophore excitation and emission collection efficiency improves the sensitivity of SPCE microarrays to picomolar sensitivity for most analytes.
Strategic Planning
The most commonly used biomarkers are macromolecules including disease-relevant proteins or nucleic acids. Both soluble and particulate biomarkers, including cells, viral particles or bacteria, are highly amenable to detection by SPR/SPCE using antibodies. Alternatively, enzymatic matrix metalloproteinase 9 (MMP-9) activity, for example, can be measured using a SPCE assay when fluorescently labeled gelatin is printed on the sensor chip. A decrease in the fluorescence following exposure to a sample of saliva correlates with increased MMP-9 activity. Alternatively, a panel of cognate antigens can be immobilized on the sensor chip to quantify antibody-binding activity or epitope mapping.
The presence of specific patient cell populations can also function as disease biomarkers. If a given cell subpopulation is known to be differentially regulated and can be captured with a specific antibody, then surveying the levels of this cell population within a biofluid may provide biological insight (Dos-Santos et al., 2009). Alternatively, cells captured on a sensor chip can be exposed to labeled secondary antibodies specific to various cytokines or cell specific membrane markers in order to phenotype cells (Rice et al.).
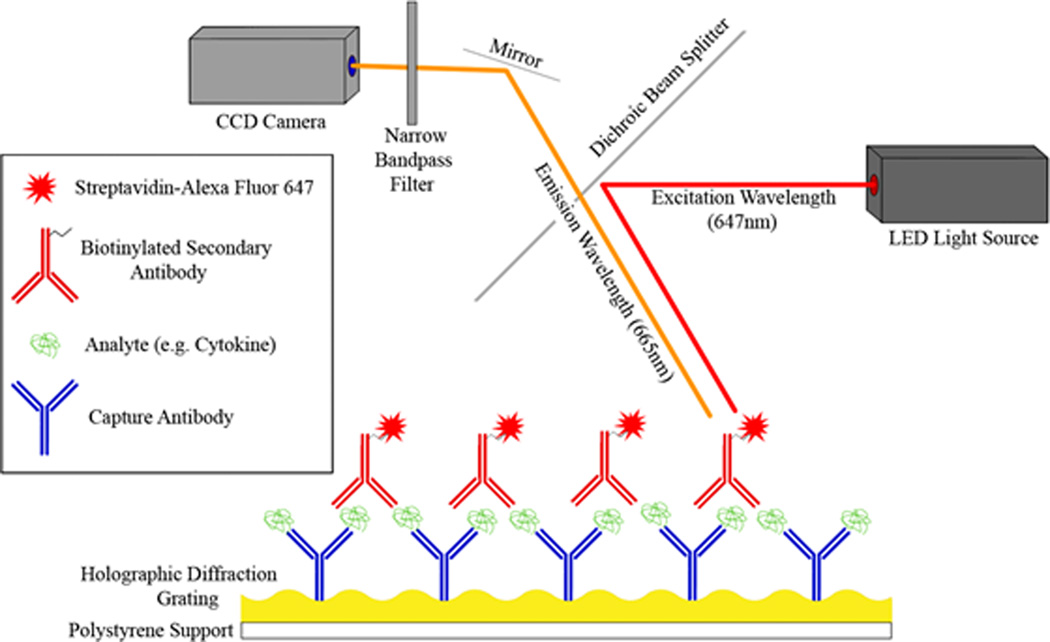
Samples can be assayed by SPR/SPCE using a direct or indirect format assays. In a direct approach, protein in a sample is labeled with a fluorescent dye, separated, recirculated over capture antibody microarray, followed by fluorescent imaging (Sanchez-Carbayo, 2006). Indirect sandwich immunoassays also consist of a capture antibody microarray. Unlabeled analytes in the sample are exposed to the chip surface, followed by a cocktail of secondary antibodies. Fluorescent labels can be directly conjugated to secondary antibodies, or bound using streptavidin-biotin (Fig 1).
Figure 1. Indirect Fluorescent Immunoassay using Surface Plasmon coupled emission (SPCE).
In this SPCE assay diagram, capture antibodies have been printed on a gold sensor chip which has a holographic diffraction grating. Analyte capture was detected by recirculating an analyte-specific biotinylated secondary antibody and streptavidin-Alexa Fluor 647 over the chip. Fluorophores on the chip were then excited using an LED light source, and a CCD camera detected the resulting fluorescence. In addition to the standard fluorescent excitation shown above, the interaction of the surface plasmon increases the directional fluorescent emission which amplifies this fluorescent signal ~80-fold.
The primary advantages of the indirect immunoassay technique are its increased sensitivity due to signal amplification using polyclonal secondary antibodies with multiple biotin tags. These use quantum dots and other reagents which amplify the signal to noise ratio (Li et al., 2003) (Brown et al., 2004) (Goldman et al., 2002). These assays are limited, however, by the increased time required for the multiple incubation and wash steps, the limited availability of matched-pair antibodies for target biomarkers, molecular size requirements, and the potential for antibody cross-reactivity (Sanchez-Carbayo, 2006). This potential for cross-reactivity results in the need for a large number of controls and limits the multiplexing capacity of immunoassay microarrays (Brown et al., 2004; Sanchez-Carbayo, 2006).
The direct labeling approach has the advantage of significantly reduced time per assay and fewer controls for nonspecific binding, thereby reducing the cost-per-assay (Li et al., 2003) (Brown et al., 2004). The primary disadvantage of this technique is the potential reduction in sensitivity. Li et al. compared relative background-subtracted fluorescence produced by multiple cytokine immunoassays in both direct and indirect formats and discovered an inconsistent signal between cytokines in the direct but not indirect labeling experiments, despite presenting equal concentrations of all 5 cytokines in all assays. They posit that this inconsistency is due to differential labeling of cytokines by the fluorescent dye, a result that they confirm with standard SPR techniques (Li et al., 2003). Thus, while direct labeling makes samples readily usable in the SPCE format, fluorescence does not always correlate well with relative protein concentration depending on the biofluid and the analyte panel of interest. The relative advantages of each approach must be considered when planning biomarker signature identification experiments.
Proteins can be immobilized onto gold surface by physical adsorption, or covalent attachment that can be orientation specific. Physical adsorption has been widely used because of the advantages including simplicity and high densities of surface capture ligand deposition. Covalent attachment provides a more reproducible and robust protein immobilization strategy, usually through amine or sulfhydryl reaction chemistry. Oriented immobilization is a preferred method to increase protein activity by orienting the binding site of the capture ligand towards the sample interface, although the density of capture ligand bound is usually lower. When immobilizing a capture ligand it is important to remember that binding properties can be changed by contact with sensor surfaces. Proteins are affected at the sensor surfaces by van der Waals hydrophobic and electrostatic interactions, interfacial perturbations by multipoint attachments to the surface, pH environment, surface charge, co-adsorption of low-molecular-weight ions, and isoelectric points of proteins (Moulin et al., 1999) which must all be considered.
The appropriate choice of immobilization strategies is important to maximize binding activity of the capture ligand and to minimize non-specific binding of proteins and non-specific adsorption of cells. Examples of surface modification that link the capture ligand to the gold surface include monolayers of alkane dithiols or polyethylene glycol, or a modified dextran hydrogel. This crosslinking layer is covalently attached to the gold through a thiolate bond mediated by a free sulfhydryl group. The other end of the crosslinker includes include a functional group that is reactive with a non-binding site on the capture ligand. A N-hydroxysuccinimide (NHS)/N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC) reaction is often used for attachment of proteins or peptides because of the availability of free amine groups (Sehgal and Vijay, 1994). Alternatively, streptavidin or protein G can be attached to the crosslinker for the attachment of biotinylated capture ligands or immunoglobulins, respectively.
For SPR and SPCE applications it is necessary to block printed sensor chips to prevent nonspecific binding. A number of common protein blocking reagents are commercially available, with some of the most common listed in Table 2. Bovine serum albumin (BSA) is a common blocking reagent due to its relatively low cost and the abundance of albumin in serum. It is important to consider, however, that BSA may be immunoreactive with polyvalent sera due to the presence of BSA in cell culture media. Cold water Telostean fish gelatin is another common blocking reagent which is immunologically distinct from proteins likely to be used in SPR/SPCE applications, making it a preferred reagent in situations where the use of BSA may confound experimental results. It may also be necessary to specifically quench free NHS, protein A/G, or avidin groups using an excess of ethanolamine, IgG, or biotin respectively.
Table 2.
Blocking Reagents
| Blocking Agent | Suppliers | Commonly used concentration range |
|---|---|---|
| Bovine Serum Albumin (BSA) | Jackson ImmunoResearch Laboratories; Sigma-Aldrich | 1%–10% |
| Casein | Sigma-Aldrich | 5%–10% |
| 45% Tclostcan fish gclatin in water | Sigma-Aldrich | 1%–5% dilution of stock solution |
| Non-fat dried milk (NFDM) | Local groccry store; Sigma Chemical | 2%–5% |
| Goat serium | Sigma-Aldrich | 1%–10% |
Multiplexed assays have the advantage of high throughput content analysis that allows for thousands of independent measurements to be made simultaneously. It is important therefore to assimilate proper controls to accurately measure the contribution of background using isotype control antibodies or scrambled peptides, for example. Additionally, when analyzing for differences in biomarker expression between groups, it is important to adequately control for type 1 error rate. Additionally, the use of at least three ROI replicates to establish a mean (μ) and intra-assay variance (σ) for individual biomarker measurements is important. Statistical analysis programs including Significance Analysis of Microarrays (SAM) can help to identify differences between groups while adequately controlling for type 1 error in large data sets.
Basic Protocol I - Saliva collection and storage
Proper collection of patient salivary samples for microarray analysis allows for improved consistency between subjects. Passive drool that collects in the mouth can be collected using a syringe or a tube. Alternatively, subjects can use a Salivette® (Sarstedt, Newton, NC) and samples should be collected according to manufacturer’s instructions. It is important to rapidly chill samples on ice after collection in order to ensure minimal degradation of cytokines and other biomarkers of interest. The collection procedure generally takes 5–15 minutes per patient, and is completely non-invasive and painless.
Materials
15 mL Tubes
Ice Bucket or Cold Storage Container
−80°C Freezer
1.5 mL Micro-centrifuge Tubes
1 mL syringe
0.22 µM syringe filter (Fisher Scientific)
Complete Protease Inhibitor Cocktail (Roche)
Collection and Storage
Rinse out mouth with water one or two times to remove contaminants insofar as possible.
- Place the collection tube provided at the lower lip and let saliva flow into the tube until 5 mL of fluid has been collected.While passive drool is the standard collection protocol, a syringe may be used to facilitate collection when necessary.
Seal the tube and place into an ice bucket or other cold storage container
Centrifuge the sample at 2600 rpm for 15 minutes at −4°C.
Create 1 mL aliquots of the supernatant from the centrifugation tube.
Using a 1 mL syringe, pass each aliquot through a 0.22 µM filter and collect in a 1.5 mL micro-centrifuge tube.
Add an appropriate volume of protease inhibitor cocktail (~100 µl) and briefly vortex.
Log samples and store at −80°C.
Thaw samples on ice before use in an assay.
Basic Protocol II –GCSPRI/SPCE Microarray Sensor Chip Preparation
GCSPR and SPCE assays make use of gold sensor chips that contain a diffraction grating. These chips can be treated with a chemical crosslinker to improve the uniformity and reproducibility of protein deposition or left as an unmodified gold surface. The following protocol describes surface treatment with DSP for covalent attachment of proteins through a reactive NHS group. Antibodies (or other proteins of interest) are deposited onto the chip in a microarray format using a robotic spotting system (Spotbot II, ArrayIt Microarray Technology) and microspotting pins. Spotting may also be performed manually using a micropipette, however the use of an automated spotting system allows for more precise control over the size and spacing of spots on the sensor chip. When spotted using a robotic spotter, 1 cm2 of active sensor surface can contain over one thousand discrete spots.
Materials
95% Ethanol
Ultrapure H2O (18 mΩ)
DMSO, commercial grade
DSP (Dithiobis(succinimidyl) propionate) (Thermo Scientific)
Spotbot II Automated Spotting System (ArrayIt Microarray Technology)
Gold GCSPRI/SPCE dual mode sensor chips (Ciencia)
384 well low-protein binding V bottom Microplate (Thermo scientific)
Capture Antibody or Protein (in PBS with no carrier protein)
Pre-printing and Sensor Chip Surface Modification with DSP
-
1Add 10 µL aliquots of capture and control antibodies to sequential wells (A1–24, B1–24, etc.) of a 384 well microplate at a final concentration of 0.25 – 0.5 mg/mL. Aliquot only one antibody or protein per well. Also include wells with each buffer without protein as negative control (blocking buffer, sample buffer, etc.). A biotinylated protein should be added to the last well as a positive control for assays employing a biotin-streptavidin reaction.When preparing the 384 well plate, keep in mind that antibodies will be spotted onto the sensor chip in the order that they are present on the plate. It is beneficial to include 10 µL aliquots of any reagent buffers in wells at both the beginning and end of your microarray. These can serve as internal controls for auto-fluorescence on your printed sensor chips and to ensure there is no protein carryover between printed spots.
-
2Cover the 384 well microplate with an adherent foil cover, and spin the plate at 500 rpm for 3 minutes at 4°C to make sure all liquid is in the bottom of the wells.Be careful when moving the plate following this spin to make sure liquid remains at the bottom of the well before printing.
-
3
Rinse gold sensor chips sequentially with 95% ethanol and H2O (18 mΩ) for 10 seconds each. Dry chips using pressurized filtered air, and place chips in a plastic petri dish with the active gold surface facing up.
-
4Make a solution of DSP by adding 4 mg of DSP powder for every 1 mL of DMSO and gently vortex to dissolve the powder. Carefully pipette 300 uL of this solution onto the center of each sensor chip. The resulting drop of liquid should completely cover the 1 cm2 active surface at the center of the chip. Close the petri dish cover and allow chips to incubate for 30 minutes in the dark at room temperature. While chips are incubating, proceed with steps 5–10.DSP should be equilibrated to room temperature prior to opening in order to prevent condensation following use.
Preparing the Robotic Spotter
-
5Prepare automated spotting apparatus by turning on the humidity, wash buffer, vacuum, and sonication elements (optional) of the machine.If you are using a manual spotting machine, see Alternate protocol 1 for a modified procedure.
-
6Create a new spotting procedure program in silico to print the number of spot replicates from the wells of a 384 well plate onto the sensor chip(s).A number of settings in the spotting program can be modified where experimentally appropriate. The size of the pin and resulting spot should be considered when deciding on the spacing between spots.
-
7
Using gloves, remove the print head from the spotting machine, wash with 95% ethanol, carefully scrub the pinholes with a small brush, dry with pressurized filtered air, and return the print head to the spotting machine.
-
8
Using gloves, briefly sonicate the printing pin in H2O (18 mΩ), dry with filtered pressurized air, and examine the pin under 10× magnification to confirm that the hollow tip it is free of debris. Insert the pin into the print head, taking care not to touch the tip of the pin.
-
9
Using gloves, insert a clean slide into the preprinting area of the spotting apparatus.
-
10Uncover and insert the 384 well plate into the spotting machine in the appropriate orientation.If the plate is unintentionally shaken or otherwise disturbed, repeat step 2 prior to inserting the plate into the spotting machine.
Sensor Chip Printing
-
11
Once sensor chips have incubated with DSP for 30 minutes (Step 4), wash each chip with 5 mL of DMSO, then wash each chip with H2O (18 mΩ) for 10 seconds, and dry each chip with pressurized filtered air.
-
12Once dry, immediately insert the chips into the spotting apparatus and begin the spotting program.The protein binding activity of DSP rapidly decreases over time, so it is important to begin the printing of chips as quickly as possible following the completion of Step 11.
-
13
After spotting is complete, remove and store the 384 well plate. Remove the pin and repeat step 8 before storing. Turn off the wash, vacuum, and sonication elements of the spotting machine.
-
14
Allow the chips to incubate in the humid environment (80% humidity) for 1 hour.
-
15
Turn off the humidity control and store the chips in a desiccated environment at 4°C in the dark for up to two weeks until use.
Basic Protocol III –Saliva Indirect Immunoassay Microarray Using SPCE
This SPCE assay employs a sandwich immunoassay (indirect) design. Printed chips are exposed to alternating wash steps and blocking buffer, sample recirculation, biotinylated detection antibody cocktail recirculation, and finally streptavidin-alexafluor647 recirculation.
Materials
Printed Gold Sensor Chip [see Basic Protocol II] (Ciencia)
Glass Coverslip Window with Inlet/Outlet Ports (Ciencia)
Double-sided adhesive gasket (Ciencia)
GCSPR/SPCE Instrument (Ciencia)
Matched-pair biotinylated Detection Antibodies
Saliva Sample [see Basic Protocol I]
Streptavidin-Alexa Fluor 647 (Invitrogen)
Blocking Buffer
Running Buffer
PBS-T (Wash Buffer)
Sensor Chip Flow cell Assembly and Reagent Preparation
-
1
Create a cocktail of matched-pair secondary detection antibodies corresponding to the primary capture antibodies printed on the sensor chip. Dilute each antibody in reagent buffer to a working concentration. This concentration can be determined empirically and is usually similar to the suggested working concentration for an ELISA. The final volume of the secondary antibody cocktail should include 1.5 mL for each sensor chip. Make this cocktail in one batch and aliquot 1.5 mL per assay to minimize inter-assay variation.
-
2Prepare streptavidin-alexafluor647 in reagent buffer at 1 µg/mL. Make this reagent in one batch and aliquot 10 mL per assay to minimize inter-assay variation.This reagent is light sensitive. Cover tube with aluminum foil to prevent loss of fluorescence.
-
3
Prepare 50 mL reservoirs of PBST and Blocking buffer.
-
4
Thaw saliva sample on ice and dilute sample 1:1 with PBS to a final volume of 1.5 mL.
-
5Allow the previously printed sensor chips to warm to room temperature. Then assemble a flow cell on each chip by attaching a glass window to the chip surface using a double-sided adhesive gasket. Apply a small amount of pressure to the window in order to ensure the gasket is completely sealed to the chip and to the window.While attaching the window, do not come into contact with the ROI-containing areas of your chip.
-
6
Insert the assembled chip into the chip holder of the GCSPR/SPCE instrument. Using the instrument software, capture an image of the chip in SPR mode. Using this image, create and save an ROI template over the image of your microarray.
SPCE Assay
-
7
Insert a previously used “dummy” chip into the chip holder of the GCSPR/SPCE instrument. Run H2O (18 mΩ) over these chips for 5 minutes through each of the fluidic channels (1–5) at a flow rate of 0.5 mL/min in order to wash out the fluidics.
-
8
Place the inlet for each fluidic line into one of the reagent tubes (1. PBST 2. Blocking Buffer 3. Saliva sample 4. Secondary Antibody Cocktail 5. Streptavidin-alexafluor647 cocktail). Prime each line for 20 seconds at 0.5 mL/min. As a last step, prime all of the common line fluidics with blocking buffer at 0.5 mL/min for 3 min.
-
9
Remove the previously used sensor chips from the fluidics systems and insert the new chip assembled in step 3. Block these chips with the blocking reagent from step 6 for 30 minutes at a flow rate of 0.5 mL/min.
-
10
Wash chips with PBST for 10 minutes at a flow rate of 0.5 mL/min. Begin recording SPR angle data. Use this first wash step to generate a baseline SPR angle measurement for each individual ROI.
-
11
Recirculate the saliva sample over the chip for 1 hour at a flow rate of 0.5 mL/min.
-
12
Wash chip with PBST for 5 minutes at a flow rate of 0.5 mL/min.
-
13
Recirculate the detection antibody cocktail prepared in step 1 over the chip for 1 hour at 0.5 mL/min.
-
14
Wash chip with PBST for 5 minutes at a flow rate of 0.5 mL/min.
-
15
Flow the streptavidin-alexafluor647 solution over the chip for 30 minutes at 0.5 mL/min.
-
16
Wash chips with PBST for 10 minutes at a flow rate of 0.5 mL/min. Stop recording SPR data and save the SPR data file.
-
17Switch the laser to SPCE wavelength. Record an angle scan and determine the angle of maximum fluorescence for the chip. Set the camera angle to the maximum SPCE angle and record the fluorescence at each ROI. Save the fluorescent data file.Increasing the fluorescence exposure time will increase the fluorescence at each ROI. Set the exposure length so that the range of fluorescence values for the entire microarray falls between 0 and 1800 arbitrary fluorescence units.
Alternate Protocol I – Manual Chip Printing
If a robotic spotter is not available, a manually operated printer or micropipette may be used instead. An example is MicroCASTer™ Pin System (Schleicher & Schuell, Inc.). Alternatively, a 0.1 – 2 µl micropipettor may be used.
Additional Materials
Chip Printing Reagents [see Basic Protocol II]
MicroCaster System 8 pin manual arrayer (Schleicher and Schuell Bioscience, or other manual spotting system)
-
1
Add 100 µL aliquots of capture and control antibodies or other reagents to sequential wells (A1–12, B1–12, etc.) of a 96 well low-protein binding B0-bottom microplate at a final concentration of 0.25 – 0.5 mg/mL.
-
2
Complete Step 3–4 of Basic Protocol II.
-
3
Sonicate and rinse the printing pins in ethanol followed by dH2O, and dry using filtered pressurized air.
-
4
Complete Step 11 of Basic Protocol II.
-
5
Insert chips into the spotting apparatus in the correct orientation to the grating.
-
6
Use the pins to transfer antibodies from the 96 well plate to the chip, moving the chip horizontally using the MicroCaster indexing system in order to produce the desired number of spots.
-
7
Once spotting has completed, store the chips in a humid environment (>80% humidity) for 1 hour.
-
8
Store chips at 4°C in a desiccated environment in the dark for up to two weeks.
Alternate Protocol II –Direct Fluorescent Immunoassay Microarray Using SPCE
For a direct fluorescent immunoassay in a microarray format, all proteins in the sample (saliva, whole cell lysate, etc.) are labeled through free amines with a fluorescent molecule. This fluorescently labeled protein solution is then recirculated over the antibody microarray printed on a sensor chip, and the resulting fluorescence of each ROI indicates the presence of that antigen in the sample solution. For insight into the advantages and disadvantages of each approach, consult the Strategic Planning section.
Materials
Printed Gold Sensor Chip [see Basic Protocol II] (Ciencia)
Glass Coverslip Window with Inlet/Outlet Ports (Ciencia)
Double-sided adhesive gasket (Ciencia)
GCSPR/SPCE Instrument (Ciencia)
SpectraMax M2 Microplate Reader (Molecular Devices, or other fluorescent plate reader)
M-PER Mammalian Protein Extraction Reagent (Thermo Scientific)
Complete Protease Inhibitor Cocktail (Roche)
Alexafluor647-NHS (Invitrogen)
1 M Sodium Bicarbonate in ultrapure H2O (18 mΩ)
Sephadex G-100 Size Exclusion Resin (Pharmacia Fine Chemicals)
15 mL Column
PBS
PBST
Blocking Buffer
Fluorescent Labeling of Saliva or Cell Lysate Proteins
-
1
Resuspend 1 mg of NHS reactive Alexa Fluor 647 dye in 250 µL DMSO and separate the sample into 10 µL aliquots. Store aliquots at −20°C.
-
2
For saliva samples, thaw saliva samples on ice. Mix saliva 1:1 with ultrapure H2O (18 mΩ) to a volume of 0.5 mL. Add 3.3 µL of Alexa Fluor 647 (prepared in Step 1) and 72.2 µL of 1 M sodium bicarbonate (for a final concentration of 0.12 M sodium bicarbonate). Vortex briefly and then allow the labeling reaction to proceed for either 4 hours at room temperature or overnight at 4°C.
-
3For cell lysate samples, prepare a cell sample for lysis by centrifuging 10 million cells per chip to be analyzed at a speed of 1000 rpm for 10 minutes and discarding the supernatant.While 10 million cells is more than sufficient for immortalized cultured cells such as Jurkat T cells, primary cells tend to be smaller and larger numbers of cells may be necessary. Establishing the ideal concentration of your particular cells of interest may need to be done empirically.
-
4
For each chip to be analyzed, mix 500 µL of M-PER with 50 µL of 10× Complete Protease Inhibitor Cocktail and vortex briefly.
-
5Resuspend the pellet from Step 2 in the M-PER/Protease Inhibitor solution and incubate at room temperature for 15 minutes, vortexing frequently.If the proteins in your cell culture media are of concern, an additional wash step may be performed prior to this step by resuspending the pellet from Step 2 in PBS and centrifuging at a speed of 1000 rpm for 10 minutes.
-
6
Centrifuge this sample at 14,000 × g for 10 minutes. Transfer the supernatant to another tube, taking care not to disturb the pellet. Discard the pellet.
-
7
To this cell lysate sample, for each chip to be analyzed add 3.3 µL of Alexa Fluor 647 (prepared in Step 1) and 72.2 µL of 1 M sodium bicarbonate (for a final concentration of 0.12 M sodium bicarbonate). Vortex briefly and then allow the labeling reaction to proceed for either 4 hours at room temperature or overnight at 4°C.
Sample Purification
-
8Swell Sephadex G-100 beads by incubating a sufficient volume of beads to fill your column (beads swell to 15–20 mL/g of dry powder) in a volume of filter sterilized PBS equivalent to 130% the volume of the column. Incubate beads for 72 hours at room temperature or 5 hours at 90°C. Once swelling is complete, discard the supernatant and add sufficient PBS to create a 75% suspension. Degas this suspension and pack the column by pouring the Sephadex resin into the column in one motion.Following column usage, wash the column with at least two column volumes of filter sterilized buffer and recover the resin. Store in 20% ethanol or 0.2% sodium azide when not in use.
-
9
Wash the column with two column volumes of PBS and examine the column to ensure even packing and that no bubbles are present. Do not allow the column bed to dry.
-
10
When the PBS has just reached the bed of the column, add the labeled saliva or cell lysate solution from Step 2 or 7 to the top of the column. Once it has all entered into the bed of the column, carefully add PBS to the top of the column and continue to do so for the remainder of the fractionation procedures.
-
11Collect the column runoff as 0.5 mL fractions either by hand or using an automated fraction collector. Continue to collect fractions until there is no visible blue coloration remaining in the column.Labeled protein will run through the column faster than unconjugated dye, and therefore it is not necessary to collect the later fractions of the column except during pilot experiments or as required by individual assays.
-
12Add 100 µL aliquots of each fraction in triplicate to a black 96 well plate. Using a microplate reader, read the fluorescence values of each column fraction at 647 nm excitation 668 nm emission (665 nm auto-cutoff, when available). Plot fluorescence value (y-axis) versus fraction number (x-axis) to create a fluorescence fraction profile.For further confirmation that initial fluorescence peak fractions contain protein, perform a BCA or Bradford assay on these fractions and plot the resultant OD readings on a secondary axis on the same graph as the fluorescence fraction profile.
-
13Pool the three peak protein-containing fluorescence fractions and dilute to a final volume of 1.5 mL with PBS as needed.Once samples have been separated from free dye they must be used as quickly as possible, as size exclusion chromatography removes many reversibly bound protease inhibitors, resulting in an increase in protein degradation.
SPCE Assay
-
14
Complete Steps 3 and 6–9 of the Basic Protocol III with the exception that only 3 channels need to be primed (1.PBST 2. Blocking Buffer 3. Sample)
-
15
Wash chips with PBST for 10 minutes at a flow rate of 0.5 mL/min. Begin recording SPR angle data. Use this first wash step to generate a baseline SPR angle measurement for each individual ROI.
-
16
Recirculate the 1.5 mL sample from Step 13 over the sensor chips for 1 hour at a flow rate of 0.5 mL/min.
-
17
Wash chips with PBST for 10 minutes at a flow rate of 0.5 mL/min. Stop recording SPR data and save the SPR data file.
-
18Switch the laser to SPCE wavelength. Record an angle scan and determine the angle of maximum fluorescence (SPCE angle) for the chip. Set the camera angle to the maximum SPCE angle and record the fluorescence at each ROI. Save the fluorescent data file.Increasing the fluorescence exposure time will increase the fluorescence at each ROI. Set the exposure length so that the range of fluorescence values for the entire microarray falls between 0 and 1800 arbitrary fluorescence units
Alternate Protocol III – Detection of Viral Particles from Saliva on a microarray using GCSPRI
The general protocol for quantifying macromolecular biomarkers including viral particles and bacteria is similar to molecular analytes but special attention must be paid to the kinetic limitations of capture. Because particles do not diffuse, sample flow rates and array placement on the chip surface must be more carefully considered. In general, flow rates should be reduced compared to protein binding assays. Additionally, the distance between ROIs should be increased to prevent particle capture at one ROI altering the flow of fluid near another ROI. Finally, bacteria or viral particles may tend to build up more densely at the leading edge of ROIs facing the direction of flow. Therefore, interrogating the SPR angle shift only at the leading edge of an ROI may result in larger signal shifts, because the signal is not averaged over the entire ROI. This approach has been shown previously to be able to detect viral particle concentrations of 105 PFU/mL for M13 bacteriophage (Marusov et al.).
Additional Materials
Printed Gold Sensor Chip [see Basic Protocol II] (Ciencia)
Glass Coverslip Window with Inlet/Outlet Ports (Ciencia)
Double-sided adhesive gasket (Ciencia)
GCSPR/SPCE Instrument (Ciencia)
Sample containing viral particles
PBST
Blocking Buffer
Complete Steps 3 and 6–9 of the Basic Protocol III with the exception that only 3 channels need to be primed (1.PBST 2. Blocking Buffer 3. Sample)
Wash chips with PBST for 10 minutes at a flow rate of 0.5 mL/min. Begin recording SPR angle data. Use this first wash step to generate a baseline SPR angle measurement for each individual ROI.
Recirculate sample over the chip for 1 hour at a flow rate of 0.5 mL/min.
Wash the chip with PBST for 20 minutes at a flow rate of 0.5 mL/min to generate endpoint baseline. Stop recording SPR data and save the SPR data file.
Reagents and Solutions
Use ultrapure H2O (18 mΩ) or equivalent for all reagent preparations.
Phosphate Buffered Saline (PBS)
8 g/L NaCl
0.2 g/L KCl
1.15 g/L Na2HPO4
0.2 g/L KH2PO4
- pH to 7.2Filter (.22 µM) and store at 4°C
Phosphate Buffered Saline + Tween20 (PBST)
PBS (see above recipe)
0.1% (v/v)Tween 20
0.024% (w/v) NaN3
- pH to 7.2Filter (.22 µM) and store at 4°C
Blocking Buffer (BSA-Based; See Table 2)
PBST (see above recipe)
2% (w/v) Bovine Serum Albumin
0.1% ethanolamine *for DSP coated chips only
Reagent Buffer
PBST + 0.1% BSA
Commentary
Critical Parameters
Microspot Array Spacing –Adequate spacing between ROIs during printing is necessary to avoid fusion of capture ligand droplets during the adsorption to the chip surface. Spacing requirements depend on the volume deposited per spot and on the relative hydrophobicity of the printing surface. As a general rule, a minimum of 500 µm between the centers of each ROI should be maintained when using a 3 nL spot volume (7b pin) and a bare gold surface. This distance should be increased when using more hydrophilic surface (i.e. PDEC dextran) or a larger spot volume. More information on ROI spacing requirements can be found at http://arrayit.com/Products/Microarray_Printing/Microarray_Pins_Stealth_Techno/microarray_pins_stealth_techno.html. Additionally, replicate ROIs should be printed in the same row of the microarray. Therefore, the number of columns in the microarray will be a multiple of the number of replicates. For example, when using 4 replicates per ROI type, the microarray will have 4, 8, 12, 16, or 20 columns.
The position of the microarray on the sensor chip surface should be kept a minimum of 1.5 mm from each edge of the flow cell to minimize differences due to laminar flow.
Appropriate negative controls (e.g. Isotype matched antibodies, scrambled sequence peptides, etc) should be incorporated at the beginning and end of each printed matrix to ensure that residual capture ligand is not re-deposited after each pin washing step. Positive controls (e.g. Biotinylated proteins, fluorophore-conjugated proteins, etc) should be printed at the end of the matrix for the same reason. The concentration of positive controls for fluorescence-based assays should be carefully considered so that at the appropriate exposure length, the fluorescent signal from the positive control falls within the linear response range of the instrument. Alternatively, fluorescent spots can be placed in a checkerboard pattern interspersed between ROIs. In this configuration, the fluorescent signal from each ROI can be reported as a fraction of the four adjacent fluorescent positive control spots.
Statistical Analysis
The growing use of high-content screening (HCS) and analysis in drug discovery and toxicology is accompanied by a need to effectively manage and evaluate the massive amounts data. Statistical methodologies are required to transform HCS data into useful information that facilitates decision making in both an efficient and cost-effective manner. While the technology for generating high content data has advanced tremendously, accompanying statistical data analysis has lagged behind in theory and in practice. Data validation and tests for normality using multiple comparison Student t- tests are based on quantile-quantile tests. When parametric assumptions are not justifiable, one can use nonparametric tests, e.g., Wilcoxon and Komogorv-Smirnoff (KS) tests.
The most common approach to multiple comparison t-tests is the Significance Analysis of Microarrays (SAM). This statistical technique measures the strength of the relationship between analyte level and the experimental or response variable of interest. The use of this nonparametric, permutation-based analysis avoids assumptions of equal variance and/or interdependence of different analytes that are made when using ANOVA, which is essential when the data may not follow a normal distribution. SAM is available for download online at http://www-stat.stanford.edu/~tibs/SAM/ and can be run as a Microsoft Excel add-on. Once SAM has been run using Excel, the parameters can be adjusted for delta and sample size to identify significant differences in analyte levels. Other unbiased methods for dimension reduction usually rely on parameter correlation. Among these, factor analysis has been shown to derive a biologically meaningful set of parameters. Alternatively, principal component analysis (PCA) has been used. No comparative review of these techniques has been reported, and their impact on the information content has not been assessed.
A crucial step is how best to summarize values from replicate microspots. Often, the basic approach of computing one characteristic value (i.e., mean or median) for all spots on sensor chip surface is used. However, additional characteristics such as the value variation or percentile values can add information about the ROI population in the particular sensor chip surface. Alternatively, the Kolmogorov-Smirnoff (KS) statistic is used to compare the value distribution of each parameter. Hence, a range of possibilities on how to aggregate the single-spot information on a sensor chip surface exists that may capture different aspects of the biological effects being monitored.
Troubleshooting
In certain circumstances the amount of background fluorescence present on sensor chips in SPCE applications may be higher than the fluorescence of negative control spots. This happens most commonly during direct labeling assays, and can stem from a number of factors including an inadequate blocking step and the failure to fully remove all free amine-reactive dye before running the sample.
Increasing the protein or quenching agent concentration of the blocking buffer or the duration of the blocking step can often resolve this issue. Additional modulation of wash speeds/length can further enhance the signal to noise ratio. Alternatively, 5% Telostean fish gelatin in PBST may prove to be a more effective blocking agent for reducing nonspecific background fluorescence. For directly immunoassays employing direct labeling of sample, further separation of the labeled sample using a second round of size exclusion chromatography beads may allow for better removal of any free dye which may be increasing background fluorescence values, although this approach will further dilute the sample. The reactivity of amine-reactive dyes may alternatively be quenched by adding 0.1% hydroxylamine or 0.1% ethanolamine to the sample for 1 hour at room temperature prior to sample purification by size exclusion chromatography.
When viewing the fluorescent image of a chip, it is critical to inspect the image for optical variations on the chip surface. These anomalous spots of fluorescence and/or darkness can stem from defects in the chip or in the optics system, and can potentially alter your results. In order to limit these variations, each time a new batch of chips arrives the image calibration matrix in the GCSPR/SPCE instrument software must be recalibrated (refer to manual). Doing so will ensure that fluorescence stemming from optical anomalies will be properly normalized for each chip in a given batch. Regularly cleaning the optics with methanol and lens paper will further reduce variations stemming from poorly maintained optical systems.
Anticipated Results
Please refer to figures 2 and 3 for sample data of a salivary SPCE microarray and accompanying quantification.
Figure 2. SPCE Microarray Image using a Saliva Sample.
The SPCE microarray image was taken after exposure to human saliva doped with 14 recombinant human protein biomarkers of interest and a cocktail of fluorescently labeled secondary antibodies. Six cytokines were not included in the saliva sample to act as negative controls. Regions of interest (ROIs) are printed in groups of three for each biomarker. Isotype controls, diluent control (PBS), and positive control ROIs are included in the image.
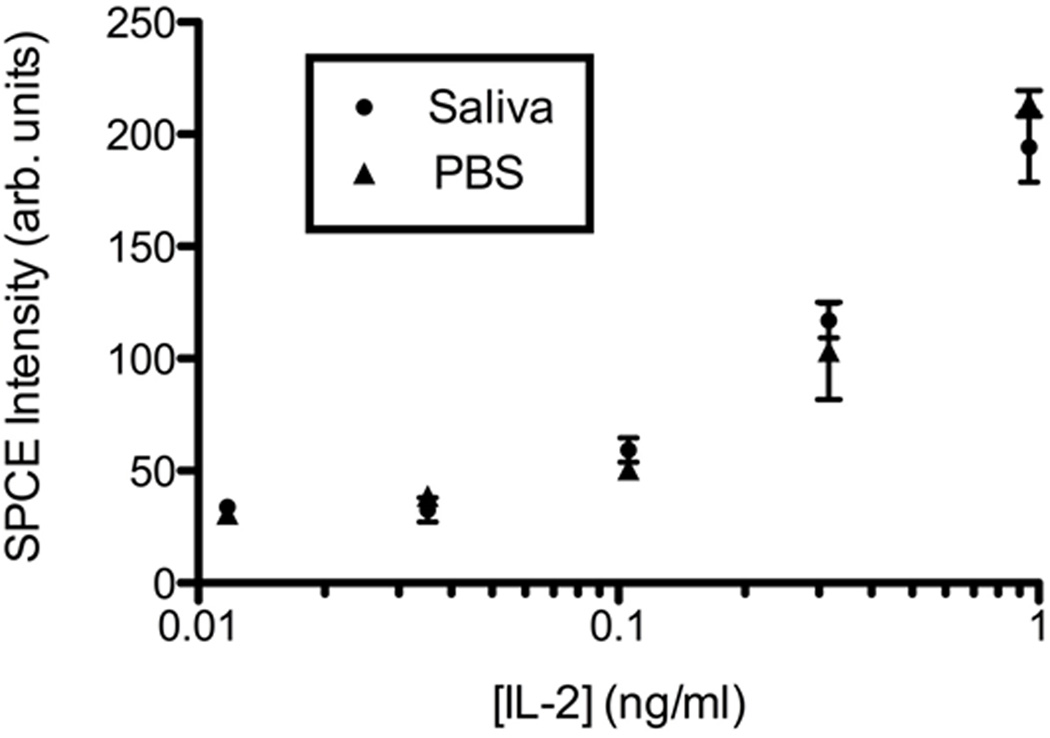
Figure 3. Saliva does not affect the IL-2 limit of detection by SPCE.
Recombinant human IL-2 was diluted in protease inhibitor treated saliva and PBS (1:1) (●) or PBS alone (▲) and detected using an indirect sandwich assay. Points indicate the mean fluorescence detected from 3 ROI microspots. Error bars indicate standard deviation.
Time Considerations
Sensor chip printing takes approximately two hours when factoring in the one hour humidified incubation period. This time can increase dramatically as the number of antibodies, chips, and spot replicates increases; however chips may be printed and stored for weeks so this step need not be completed on the day a given assay is to be completed.
Indirect SPCE assays typically take 4–5 hours to complete and analyze assuming the chips have already been printed and the samples have already been collected. Preparing for the ensuing steps during the intervening incubation periods can minimize this time.
Direct SPCE assay sample preparation and labeling can take 4.5–24 hours depending on the labeling technique chosen, not including the time it takes to obtain samples. The SPCE assay should only take 1.5–2 hours, however it must be conducted immediately after purification of labeled proteins as proteins should not be frozen or stored for extended periods of time.
Acknowledgments
This research was funded by grants for the National Institutes of Health (GM075407, AI066482, DK077291, and ES016014) and USDA (58-1940-007).
References
- Barnes AJ, Kim I, Schepers R, Moolchan ET, Wilson L, Cooper G, Reid C, Hand C, Huestis MA. Sensitivity, specificity, and efficiency in detecting opiates in oral fluid with the Cozart Opiate Microplate EIA and GC-MS following controlled codeine administration. J Anal Toxicol. 2003;27:402–407. doi: 10.1093/jat/27.7.402. [DOI] [PubMed] [Google Scholar]
- Borelli V, Marchioli A, Di Taranto R, Romano M, Chiandussi S, Di Lenarda R, Biasotto M, Zabucchi G. Neuropeptides in saliva of subjects with burning mouth syndrome: a pilot study. Oral Dis. 16:365–374. doi: 10.1111/j.1601-0825.2009.01648.x. [DOI] [PubMed] [Google Scholar]
- Bosker WM, Huestis MA. Oral fluid testing for drugs of abuse. Clin Chem. 2009;55:1910–1931. doi: 10.1373/clinchem.2008.108670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JK, Pemberton AD, Wright SH, Miller HR. Primary antibody-Fab fragment complexes: a flexible alternative to traditional direct and indirect immunolabeling techniques. J Histochem Cytochem. 2004;52:1219–1230. doi: 10.1369/jhc.3A6200.2004. [DOI] [PubMed] [Google Scholar]
- Carter CS, Pournajafi-Nazarloo H, Kramer KM, Ziegler TE, White-Traut R, Bello D, Schwertz D. Oxytocin: behavioral associations and potential as a salivary biomarker. Ann N Y Acad Sci. 2007;1098:312–322. doi: 10.1196/annals.1384.006. [DOI] [PubMed] [Google Scholar]
- Cone EJ, Presley L, Lehrer M, Seiter W, Smith M, Kardos KW, Fritch D, Salamone S, Niedbala RS. Oral fluid testing for drugs of abuse: positive prevalence rates by Intercept immunoassay screening and GC-MS-MS confirmation and suggested cutoff concentrations. J Anal Toxicol. 2002;26:541–546. doi: 10.1093/jat/26.8.541. [DOI] [PubMed] [Google Scholar]
- Cooper G, Wilson L, Reid C, Hand C, Spiehler V. Validation of the Cozart Amphetamine Microplate EIA for the analysis of amphetamines in oral fluid. Forensic Sci Int. 2006;159:104–112. doi: 10.1016/j.forsciint.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Dos-Santos MC, Matos-Gomes N, Makimoto FH, Katsurayama M, Santana LL, Becker MA, Paredes-Garcia E, Bertho AL. Cell phenotyping in saliva of individuals under psychological stress. Cell Immunol. 2009;260:39–43. doi: 10.1016/j.cellimm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Giampaolo B, Angelica M, Antonio S. Chromogranin 'A' in normal subjects, essential hypertensives and adrenalectomized patients. Clin Endocrinol (Oxf) 2002;57:41–50. doi: 10.1046/j.1365-2265.2002.01557.x. [DOI] [PubMed] [Google Scholar]
- Goldman ER, Balighian ED, Mattoussi H, Kuno MK, Mauro JM, Tran PT, Anderson GP. Avidin: a natural bridge for quantum dot-antibody conjugates. J Am Chem Soc. 2002;124:6378–6382. doi: 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- Granger DA, Schwartz EB, Booth A, Curran M, Zakaria D. Assessing dehydroepiandrosterone in saliva: a simple radioimmunoassay for use in studies of children, adolescents and adults. Psychoneuroendocrinology. 1999;24:567–579. doi: 10.1016/s0306-4530(99)00013-x. [DOI] [PubMed] [Google Scholar]
- Harden CJ, Perez-Carrion K, Babakordi Z, Plummer SF, Hepburn N, Barker ME, Wright PC, Evans CA, Corfe BM. Evaluation of the salivary proteome as a surrogate tissue for systems biology approaches to understanding appetite. J Proteomics. doi: 10.1016/j.jprot.2011.12.010. [DOI] [PubMed] [Google Scholar]
- He L, Li P, Sha YQ, Chen ZB, Luan QX, Wang XY. Levels of tumor necrosis factor-alpha and interleukin-1beta in saliva of metabolic syndrome patients. Zhonghua Kou Qiang Yi Xue Za Zhi. 45:269–273. [PubMed] [Google Scholar]
- Jang MU, Park JW, Kho HS, Chung SC, Chung JW. Plasma and saliva levels of nerve growth factor and neuropeptides in chronic migraine patients. Oral Dis. doi: 10.1111/j.1601-0825.2010.01717.x. [DOI] [PubMed] [Google Scholar]
- Jin GB, Unfricht DW, Fernandez SM, Lynes MA. Cytometry on a chip: cellular phenotypic and functional analysis using grating-coupled surface plasmon resonance. Biosens Bioelectron. 2006;22:200–206. doi: 10.1016/j.bios.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Kawas SA, Rahim ZH, Ferguson DB. Potential uses of human salivary protein and peptide analysis in the diagnosis of disease. Arch Oral Biol. 57:1–9. doi: 10.1016/j.archoralbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22:241–248. [PMC free article] [PubMed] [Google Scholar]
- Leigh JE, Steele C, Wormley F, Fidel PL., Jr Salivary cytokine profiles in the immunocompetent individual with Candida-associated denture stomatitis. Oral Microbiol Immunol. 2002;17:311–314. doi: 10.1034/j.1399-302x.2002.170508.x. [DOI] [PubMed] [Google Scholar]
- Li YW, Nath N, Reichert WM. Parallel comparison of sandwich and direct label assay protocols on cytokine detection protein arrays. Analytical Chemistry. 2003;75:5274–5281. [Google Scholar]
- Liedberg B, Lundstrom I, Stenberg E. Principles of Biosensing with an Extended Coupling Matrix and Surface-Plasmon Resonance. Sensors and Actuators B-Chemical. 1993;11:63–72. [Google Scholar]
- Liedberg B, Nylander C, Lundstrom I. Surface-Plasmon Resonance for Gas-Detection and Biosensing. Sensors and Actuators. 1983;4:299–304. [Google Scholar]
- Liu W, Dan H, Wang Z, Jiang L, Zhou Y, Zhao M, Chen Q, Zeng X. IFN-gamma and IL-4 in saliva of patients with oral lichen planus: a study in an ethnic Chinese population. Inflammation. 2009;32:176–181. doi: 10.1007/s10753-009-9118-2. [DOI] [PubMed] [Google Scholar]
- Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. J Dent Res. 89:1016–1023. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone GF, Paulpillai M, Evans RJ, Singleton EG, Heishman SJ. Breath carbon monoxide and semiquantitative saliva cotinine as biomarkers for smoking. Hum Psychopharmacol. 25:80–83. doi: 10.1002/hup.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusov G, Sweatt A, Pietrosimone K, Benson D, Geary SJ, Silbart LK, Challa S, Lagoy J, Lawrence DA, Lynes MA. A Microarray Biosensor for Multiplexed Detection of Microbes Using Grating-Coupled Surface Plasmon Resonance Imaging. Environ Sci Technol. doi: 10.1021/es201239f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicar AJ, Greenwood CR, Fewell F, D'Arcy V, Chandrasekharan S, Alldridge LC. Evaluation of anxiety, salivary cortisol and melatonin secretion following reflexology treatment: a pilot study in healthy individuals. Complement Ther Clin Pract. 2007;13:137–145. doi: 10.1016/j.ctcp.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Moulin AM, O'Shea SJ, Badley RA, Doyle P, Welland ME. Measuring surface-induced conformational changes in proteins. Langmuir. 1999;15:8776–8779. [Google Scholar]
- Nakane H, Asami O, Yamada Y, Ohira H. Effect of negative air ions on computer operation, anxiety and salivary chromogranin A-like immunoreactivity. Int J Psychophysiol. 2002;46:85–89. doi: 10.1016/s0167-8760(02)00067-3. [DOI] [PubMed] [Google Scholar]
- Ng V, Koh D, Mok B, Lim LP, Yang Y, Chia SE. Stressful life events of dental students and salivary immunoglobulin A. Int J Immunopathol Pharmacol. 2004;17:49–56. doi: 10.1177/03946320040170S209. [DOI] [PubMed] [Google Scholar]
- Ng V, Koh D, Mok BY, Chia SE, Lim LP. Salivary biomarkers associated with academic assessment stress among dental undergraduates. J Dent Educ. 2003;67:1091–1094. [PubMed] [Google Scholar]
- Noto Y, Sato T, Kudo M, Kurata K, Hirota K. The relationship between salivary biomarkers and state-trait anxiety inventory score under mental arithmetic stress: a pilot study. Anesth Analg. 2005;101:1873–1876. doi: 10.1213/01.ANE.0000184196.60838.8D. [DOI] [PubMed] [Google Scholar]
- Parahitiyawa NB, Scully C, Leung WK, Yam WC, Jin LJ, Samaranayake LP. Exploring the oral bacterial flora: current status and future directions. Oral Dis. 16:136–145. doi: 10.1111/j.1601-0825.2009.01607.x. [DOI] [PubMed] [Google Scholar]
- Rai B, Kaur J, Anand SC, Jacobs R. Salivary Stress Markers, Stress and Periodontitis: A Pilot Study. J Periodontol. doi: 10.1902/jop.2010.100319. [DOI] [PubMed] [Google Scholar]
- Randeva HS, Karteris E, Lewandowski KC, Sailesh S, O'Hare P, Hillhouse EW. Circadian rhythmicity of salivary leptin in healthy subjects. Mol Genet Metab. 2003;78:229–235. doi: 10.1016/s1096-7192(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Rehak NN, Cecco SA, Csako G. Biochemical composition and electrolyte balance of "unstimulated" whole human saliva. Clin Chem Lab Med. 2000;38:335–343. doi: 10.1515/CCLM.2000.049. [DOI] [PubMed] [Google Scholar]
- Reilly SMT, Nessing PA, Guignon EF, Lynes MA, Fernandez SM. Plasmonics in Biology and Medicine III. San Jose, CA, USA: 2006. SPR surface enhanced fluorescence with a gold-coated corrugated sensor chip. [Google Scholar]
- Rice JM, Stern LJ, Guignon EF, Lawrence DA, Lynes MA. Antigen-specific T cell phenotyping microarrays using grating coupled surface plasmon resonance imaging and surface plasmon coupled emission. Biosens Bioelectron. doi: 10.1016/j.bios.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Carbayo M. Antibody arrays: Technical considerations and clinical applications in cancer. Clinical Chemistry. 2006;52:1651–1659. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- Saruta J, Sato S, Tsukinoki K. The role of neurotrophins related to stress in saliva and salivary glands. Histol Histopathol. 25:1317–1330. doi: 10.14670/HH-25.1317. [DOI] [PubMed] [Google Scholar]
- Schwope DM, Milman G, Huestis MA. Validation of an enzyme immunoassay for detection and semiquantification of cannabinoids in oral fluid. Clin Chem. 56:1007–1014. doi: 10.1373/clinchem.2009.141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D, Vijay IK. A Method for the High-Efficiency of Water-Soluble Carbodiimide-Mediated Amidation. Analytical Biochemistry. 1994;218:87–91. doi: 10.1006/abio.1994.1144. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Kodama H. Relationship between circadian salivary melatonin levels and sleep-wake behavior in infants. Pediatr Int. doi: 10.1111/j.1442-200X.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- Shirotsuki K, Izawa S, Sugaya N, Yamada KC, Ogawa N, Ouchi Y, Nagano Y, Nomura S. Salivary cortisol and DHEA reactivity to psychosocial stress in socially anxious males. Int J Psychophysiol. 2009;72:198–203. doi: 10.1016/j.ijpsycho.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Simcic D, Pezelj-Ribaric S, Grzic R, Horvat J, Brumini G, Muhvic-Urek M. Detection of salivary interleukin 2 and interleukin 6 in patients with burning mouth syndrome. Mediators Inflamm. 2006;2006:54632. doi: 10.1155/MI/2006/54632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter JK, van der Beek AJ, Frings-Dresen MH. Medical staff in emergency situations: severity of patient status predicts stress hormone reactivity and recovery. Occup Environ Med. 2003;60:373–374. doi: 10.1136/oem.60.5.373. discussion 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo-Quee Koh D, Choon-Huat Koh G. The use of salivary biomarkers in occupational and environmental medicine. Occup Environ Med. 2007;64:202–210. doi: 10.1136/oem.2006.026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Yamaguchi M, Aragaki T, Eto K, Uchihashi K, Nishikawa Y. Gender-specific differences in salivary biomarker responses to acute psychological stress. Ann N Y Acad Sci. 2007;1098:510–515. doi: 10.1196/annals.1384.014. [DOI] [PubMed] [Google Scholar]
- Teles RP, Likhari V, Socransky SS, Haffajee AD. Salivary cytokine levels in subjects with chronic periodontitis and in periodontally healthy individuals: a cross-sectional study. J Periodontal Res. 2009;44:411–417. doi: 10.1111/j.1600-0765.2008.01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomadaki K, Helmerhorst EJ, Tian N, Sun X, Siqueira WL, Walt DR, Oppenheim FG. Whole-saliva proteolysis and its impact on salivary diagnostics. J Dent Res. 90:1325–1330. doi: 10.1177/0022034511420721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Koh D, Ng V, Lee CY, Chan G, Dong F, Goh SH, Anantharaman V, Chia SE. Self perceived work related stress and the relation with salivary IgA and lysozyme among emergency department nurses. Occup Environ Med. 2002;59:836–841. doi: 10.1136/oem.59.12.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelles T, Purushotham KR, Macauley SP, Oxford GE, Humphreys-Beher MG. Saliva and growth factors: the fountain of youth resides in us all. J Dent Res. 1995;74:1826–1832. doi: 10.1177/00220345950740120301. [DOI] [PubMed] [Google Scholar]