Abstract
Histidine kinase receptors are used ubiquitously by bacteria to monitor environmental changes, and they are also prevalent in plants, fungi and other protists. Typical histidine kinase receptors have an extracellular sensor portion to detect the signal, usually a chemical ligand, and an intracellular transmitter portion that includes both the kinase domain itself and the site for histidine phosphorylation. While the kinase domains are highly conserved, sensor domains are diverse. Histidine kinase receptors function as dimers, but the molecular mechanism for signal transduction across cell membranes remains obscure. In this study, eight crystal structures were determined from five sensor domains representative of the most populated family, Family HK1, found in a bioinformatic analysis of predicted sensor domains from transmembrane histidine kinases. Each structure contains an inserted repeat of PhoQ/DcuS/CitA (PDC) domains, and similarity between sequence and structure is correlated across these and other double-PDC sensor proteins. Three of the five sensors crystallize as dimers that appear to be physiologically relevant, and comparisons between ligated- and apo-state structures provide insights into signal transmission. Some HK1-family proteins prove to be sensors for chemotaxis proteins or for diguanylate cyclase receptors, which implies a combinatorial molecular evolution.
Keywords: crystal structure, molecular evolution, PDC domain, signal transduction, two-component system
Introduction
Two-component systems (TCSs) are critical for bacteria to sense and adapt to environmental changes1, 2. Although TCS signaling is also prevalent in plants, fungi and other protists, it is absent in metazoan animals; whereby, these systems are potential targets for novel antibiotics. The two components of a classical TCS system are a histidine kinase (HK) receptor and its cognate response regulator (RR). A typical HK contains an extracellular sensing domain in an N-terminal segment flanked by two transmembrane helices (TM1 and TM2), an intracellular dimerization-histidine-phosphorylation (DHp) domain and a C-terminal kinase domain, but there are many variations on that theme. Signals detected by the HK sensor domain are transmitted through the DHp domain to the kinase domain and can lead to phosphorylation of the conserved DHp histidine residue3. HK receptors are homodimeric proteins, and both trans and cis phosphorylation have been reported4, 5. Phosphotransfer to a conserved aspartate residue in the RR receiver domain then ensues, which affects the RR effector domain disposition. RR effectors typically bind to responsive elements to activate genes when the RR is phosphorylated. Indeed, a large majority of RRs serve as transcription factors for downstream cellular responses1. The concentration of phosphorylated RR determines the strength of output signals. Some HK receptors act constitutively as kinases when in the apo state, but become phophatases to reverse RR phosphorylation when bound to a stimulating ligand (e.g. LuxQ6). HK receptors that are stimulated to kinase activity by ligand binding may be apo-state phosphatases.
HK kinase and RR receiver domains are highly conserved in sequence and tertiary structure1, 7, 8, 9, 10, and it seem likely that basic elements of transduction mechanisms will be conserved as well, although many molecular details remain obscure. In contrast, HK sensor domains and RR effector domains are modular, having varied organizations reflecting the diversity of signals that are detected and outputs that are generated in TCS signaling. In an earlier study, we conducted a thorough bioinformatic analysis of HK sensor domains in all available sequences (J. Cheung, J. Liu, B. Rost and W.A. Hendrickson, unpublished) and found hundreds of different sequence families, including numerous singletons. We identified histidine kinase sensor domains as sequences lying between two N-terminal transmembrane helices annotated as putative histidine kinase receptors or found to contain a legitimate histidine kinase domain by sequence analysis. These putative sensor domain sequences were clustered at the level of E-value ≤ 1×10−3 as determined by PSI-BLAST11 and AGAPE12, and the largest resulting cluster, Family 1, was further divided into subfamilies at the level of E ≤ 1×10−5.
We and others have determined representative structures from a few of these sensor families, and there are fifteen histidine kinase sensor domains in the PDB at present. PhoQ13, DcuS14 and CitA15, 16 are founding members of the PAS-like PhoQ/DcuS/CitA (PDC) α+β fold; AbsF17 and PhoR18 are additional single PDC-domain sensors; LuxQ19, 20, DctB14, 21, KinD22, an uncharacterized sensor23, and an uncharacterized methyl-accepting chemotaxis protein domain (PDB ID: 3c8c)24 all contain inserted repeats of two PDC domains; HK29 25S has a periplasmic-binding-protein α/β fold; NarX26, Tar27 and an uncharacterized sensor (PDB ID: 3kkb)28 are all-α proteins, forming four-helix bundles; and TorS29 has an inserted-repeat double α-bundle fold; and RetS30 has β-sandwich sensor domain. Similarly, Family 3 sensors are predicted to have large, all-β structures. Families are identified in order of prevalence of members. Family 2 contains DcuS and CitA, Family 4 contains DctB, and Family 1 is likewise predicted to have an α+β fold. Among all Family 1 members, only KinD has been characterized. It is involved in sporulation regulation in Bacillus subtilis; however, the signaling ligand responsible for activation of KinD has not been identified31, 32, 33, 34.
Despite many biochemical analyses and the atomic structure information, the nature of signals generated upon ligand binding and transduced across lipid bilayer remains controversial. The idea of ligand induced dimerization is ruled out because histidine kinase receptors are constitutively homodimeric. A piston-like movement is proposed based on the crystallographic and biochemical studies on chemotactic sensor Tar35, 36, 37. Piston-like movements have also been suggested from the comparison of ligand-bound structures with ligand-free structures of citrate-binding protein CitA16 and nitrate-sensing protein NarX26 and from structural studies on the trimethylamine-N-oxide (TMAO) sensor TorS29. Neiditch et al. suggest that the signal transduction occurs through ligand-induced asymmetric rotation based on thorough structural and functional study on quorum sensing system LuxPQ19. Zhou et al. observe multiple dimerization states upon succinate binding to the C4-dicarboxylate sensor DctB and propose that differences at the dimerization interface initiate the signal transduction21.
We have undertaken a structural survey of the Family 1 sensor domains. Our results include eight crystal structures from five sensor domains in three subfamilies of Family 1. Two structures have an empty ligand binding site and six have ligands occupying a ligand binding site. We compare the structures with one another and with other double-PDC sensor domains. Structural deviations correlate with sequence variation. Three of the five sensors have structures similar to biological dimers, and conformational differences between ligated and apo states may have functional relevance. Moreover, we discovered that some HK1 proteins are sensors for chemotaxis proteins or for diguanylate cyclase receptors. Thus, our structural characterization of this large family of sensor proteins provides added insight into the molecular mechanism of signal transduction by histidine kinase receptors and it gives a framework for understanding the molecular evolution of these proteins.
Results
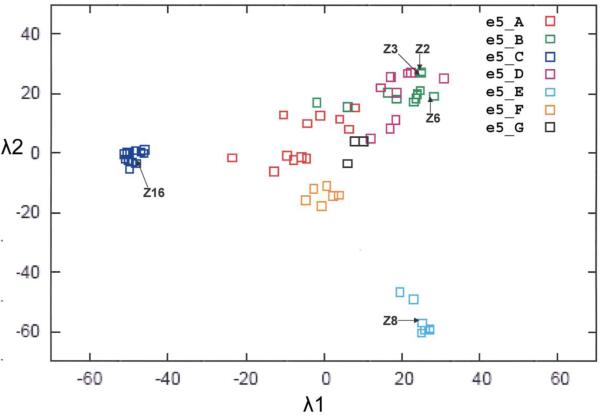
In this study we sought to analyze structures from representative Family 1 sensor domains, which were distinguished from other HK families at the PSI-BLAST level of E ≤ 1×10−3 and further divided into subfamilies at the level of E ≤ 1×10−5. Our original, restricted analysis had 65 sequences, which could be subdivided into seven sub-families and depicted as projections of positional coordinates deduced from inter-sequence distances (Figure 1). In a more recent PSI-BLAST expansion based solely on the sensor domains (unpublished results), we find 7933 homologs at the level of of E ≤ 10−3. We cloned and expressed several of these domains, seeking diversity in coverage of subfamilies, and we determined crystal structures from five purified proteins. None of these proteins has any genetic or functional characterization, and we name them for the species of origin (two initial letters) and the histidine kinase family (HK1). We further specify that these are sensor domain structures with subscript “s” and give each a unique laboratory label (Z number). Thus, we describe the sensor domains of five receptors: mmHK1S-Z2 and mmHK1S-Z3 from Methanosarcina mazei, soHK1S-Z6 from Shewanella oneidensis, vpHK1S-Z8 from Vibrio parahaemaolyticus and rpHK1S-Z16 from Rhodopseudomonas palustris (Table 1). Sensor domains Z2, Z3 and Z6 are from HK1 subfamily B, Z8 is from subfamily E, and Z16 is from subfamily C.
Figure 1.
Projection of sequence positions for Family 1 histidine kinase sensor domains (Cheung and Hendrickson, unpublished work). The matrix of all pairwise distances between sequences was transformed by singular value decomposition, based on an algorithm devised by Crippen and Havel66, into a set of multi-dimensional positions. The 2D projection onto the two principal eigenvectors (λ1 and λ2) yields this plot in which family members are represented by points distributed according to their relatedness. Family 1 members are distinguished from other families at the level of E ≤ 10−3; members of subfamilies A-G, distinguished at the level of E ≤ 10−5, are identified by distinctive colors as shown in the insert key. Structural and evolutionary relationships between sequence members can be quantitatively visualized. The five sensors reported in this work are identified in the projection by labeled arrows. Although the clusters of subfamilies A, B and D overlap somewhat in this 2D plot, they are separated when the third component (λ3) is visualized in 3D.
Table 1. Protein constructs and crystallization conditions.
| Protein | UniProt | Region | Source | Crystallization | Freezing |
|---|---|---|---|---|---|
| mmHK1s-Z2 | Q8PSW8 | 32-312 | M. mazei | 15% PEG3350 0.21 M NH4SO4 0.1M bistris (or cacodylate) pH 5.6 |
ethylene glycol |
| mmHK1s-Z3 | Q8PSW1 | 32-311 | M. mazei | 2M Na/K PO4 pH6.7, 0.2M NaCl |
ethylene glycol |
| soHK1s-Z6 | Q8EII0 | 36-308 | S. oneidensis | 5% PEG3350, 0.5M NH4SO4, 0.1M Na Citrate pH 5.8 |
glycerol |
| vpHK1s-Z8 | Q87SR8 | 28-312 |
V. parahaemolyticus |
5% PEG8K, 0.2M NaCl, 0.1M Na/K PO4 pH6.5 for PO4-bound 2M NaCl, 0.2M MgCl2, 0.1M Tris pH7 for apo |
ethylene glycol ethylene glycol |
| rpHK1S-Z16 | Q6N3S7 | 40-293 | R. palustris | 30% MPD, 0.2M NH4Ac, 0.1M Na citrate pH5.6 |
(MPD) |
Each of the five HK1 sensor proteins was crystallized (Table 1), diffraction data were measured, typically from selenomethionyl (SeMet) protein (Table 2), and a total of eight crystal structures were solved and refined (Table 3). Datasets are identified by the presence or not of SeMet, by the protein name, by a ligand if present, and by a wavelength if relevant (e.g. SeZ2-bis λ1 for SeMet Z2 complexed with bistris at wavelength λ1 and simply Z3 for apo Z3). These crystals are in seven different crystal lattices and they contain 21 distinct copies of HK1 sensor domains.
Table 2. Diffraction dataa.
| Dataset | dmin (Å) | Space Group |
Redundancy | Completeness | Rmergeb | <I>/δ | λ (Å) |
|---|---|---|---|---|---|---|---|
|
SeZ2-bis
λ1 |
1.70 | C2221 | 4.6 | 98.2 (93.5) | 4.8 (25.7) | 31.9 (4.8) | 0.97903 |
|
SeZ2-bis
λ2 |
1.70 | C2221 | 8.9 | 98.9 (93.2) | 5.1 (33.5) | 29.6 (3.8) | 0.97936 |
|
SeZ2-bis
λ3 |
1.70 | C2221 | 9.0 | 99.4 (95.8) | 6.2 (63.0) | 25.0 (2.2) | 0.97174 |
| SeZ2-cac | 1.75 | C2221 | 2.1 | 100 (99.8) | 9.4 (30.1) | 19.9 (6.6) | 0.96784 |
| Z2-bis | 2.00 | P21 | 3.8 | 99.8 (98.5) | 6.5 (25.1) | 21.4 (5.9) | 0.97157 |
| Z3 | 3.00 | P65 | 2.2 | 99.5(99.4) | 8.0 (43.1) | 15.9 (3.0) | 0.96788 |
| SeZ6 λ1 | 2.30 | P3121 | 13.3 | 100 (100) | 6.8 (22.5) | 28.6 (8.3) | 0.97927 |
| SeZ6 λ1 | 2.30 | P3121 | 13.2 | 100 (100) | 7.3 (28.5) | 26.4 (6.8) | 0.97937 |
| SeZ6 λ1 | 2.30 | P3121 | 13.3 | 100 (100) | 9.3 (48.2) | 21.0 (3.9) | 0.96789 |
|
SeZ8-PO4
λ1 |
2.00 | P212121 | 11.9 | 99.6 (99.3) | 10.2 (44.5) | 18.6 (4.4) | 0.97924 |
|
SeZ8-PO4
λ2 |
2.00 | P212121 | 11.9 | 99.6 (98.9) | 11.0 (48.2) | 17.9 (4.2) | 0.97929 |
|
SeZ8-PO4
λ3 |
2.00 | P212121 | 11.2 | 99.0 (96.5) | 19.1 (77.9) | 12.0 (2.7) | 0.96784 |
| Z8-PO4 | 1.76 | P212121 | 3.7 | 99.0 (95.7) | 5.4 (25.6) | 33.5 (6.5) | 0.97921 |
| Z8-apo | 2.60 | P21212 | 2.3 | 97.7 (95.9) | 7.7 (35.5) | 15.8 (3.6) | 0.97921 |
| SeZ16 λ1 | 2.70 | P4332 | 24.0 | 100 (100) | 6.5 (35.2) | 43.2 (8.4) | 0.97918 |
| SeZ16 λ2 | 2.70 | P4332 | 24.0 | 100 (100) | 6.5 (38.2) | 44.8 (7.9) | 0.97935 |
| SeZ16 λ3 | 2.70 | P4332 | 24.0 | 100 (100) | 8.5 (59.5) | 34.3 (4.9) | 0.96788 |
Values in the outermost shell are given in parentheses.
Rmerge = (Σ |li − < li > |) / Σ |li|, where li is the integrated intensity of a given reflection.
Table 3. Refinement statistics.
| Proteins | Z2-bis | Z2-cac | Z2 | Z3 | Z6 | Z8-PO4 | Z8-apo | Z16 | |
|---|---|---|---|---|---|---|---|---|---|
| Label | SeMet | SeMet | Native | Native | SeMet | Native | Native | SeMet | |
| Resolution (Å) | 1.70 | 1.75 | 2.00 | 3.00 | 2.30 | 1.76 | 2.60 | 2.70 | |
| Space Group | C2221 | C2221 | P21 | P65 | P3121 | P212121 | P21212 | P4332 | |
| Unit cell | a (Å) | 67.3 | 67.5 | 46.5 | 129.5 | 133.7 | 74.1 | 78.0 | 184.9 |
| b (Å) | 88.5 | 87.7 | 98.4 | 129.5 | 133.7 | 79.4 | 115.0 | 184.9 | |
| c (Å) | 99.2 | 99.0 | 71.0 | 404.7 | 32.5 | 123.1 | 69.6 | 184.9 | |
| β (°) | 87.5 | ||||||||
| Za a | 1 | 1 | 2 | 10 | 1 | 2 | 2 | 2 | |
| Solvent content | 46% | 45% | 52% | 59% | 54% | 53% | 46% | 72% | |
| Unique | 31298 | 28471 | 41368 | 72589 | 14044 | 68773 | 18510 | 28663 | |
| reflections | |||||||||
| Total atoms | 2306 | 2275 | 4643 | 21176 | 2229 | 5184 | 4317 | 3984 | |
| Protein atoms | 2062 | 2066 | 4139 | 21117 | 2114 | 4552 | 4285 | 3849 | |
| Water | 230 | 183 | 456 | 49 | 111 | 584 | 30 | 93 | |
| Ligand | bistris | SO4 | bistris | EDO | PO4 | MPD | |||
| Rwork b | 18.1 | 18.9 | 17.8 | 23.2 | 18.1 | 18.2 | 25.3 | 20.5 | |
| Rfree c | 21.1 | 23.4 | 24.6 | 27.7 | 24.5 | 22.2 | 32.4 | 25.6 | |
| RMS Bond (Å) | 0.027 | 0.027 | 0.024 | 0.018 | 0.022 | 0.026 | 0.013 | 0.021 | |
| RMS angle (Å) | 2.079 | 1.950 | 1.958 | 1.892 | 1.956 | 2.117 | 1.431 | 2.417 | |
| Bfactor (Å2) | 22.8 | 21.4 | 22.9 | 75.5 | 27.2 | 24.9 | 58.6 | 41.8 | |
| PDB code | 3LI9 | 3LI8 | 3LIA | 3LIB | 3LIC | 3LID | 3LIE | 3LIF |
Za stands for number of molecules per asymmetric unit.
Rwork = (Σ ∥Fo | − |Fc∥) / Σ|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
Rfree was calculated using 5% of data excluded from refinement.
Structural descriptions
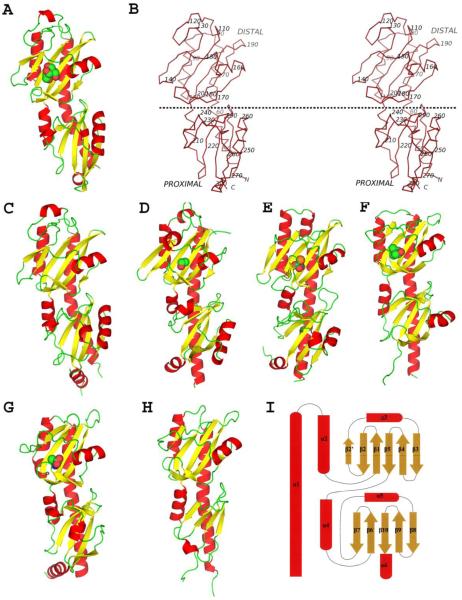
All five HK1 sensor domains are α+β structures containing inserted repeats of PDC domains as in other double PDC sensor proteins. The structures are obviously similar to one another (Figures 2A-F). Top hits from Dali searches38 with these double-PDC structures as templates are other PDC proteins with the best Z-scores close to 20 followed by PAS-domain proteins with the best Z-score smaller than 10. Scores for searches with individual HK1 domains give best hits at Z-scores of 10-15 for PDC domains. Each protein begins with helix α1, which participates in both PDC domains such that its C-terminal half and the rest of the membrane-distal, “upper” domain is inserted before returning at α4 to complete the membrane-proximal, “lower” domain and, in most cases, terminating with a short inclined helix, α6 (Figure 2I). In this sense, the membrane-proximal PDC domain of HK1 sensors is like the HK2 sensors DcuS14 and CitA15, 16. Helix α1 is poised to be a continuation of TM1 in the intact receptor, emerging perpendicular to the membrane, but although the C-terminal end is positioned proximate to the TM2 segment, the juncture between helix α6 and TM2 must necessarily be kinked. The PAS-like β-sheet at the core of PDC domains is typically five-stranded, but here all but Z16 include a sixth strand (β2′) in their membrane-distal domains (Figure 2I).
Figure 2.
Overall structures of Family sensor proteins. (A) Ribbon diagram of Z2-bis. (B) Stereo plot of a Cα trace of Z2. Every tenth C position is labeled. A dotted horizontal line, hypothetically parallel to the membrane surface, separates the distal and proximal domains. (C, D, E, F, G and H) Ribbon diagrams of Z3, Z6, Z8-PO4, Z16, McpX24 and KinD22, respectively. Ligands are shown in spacie-filling models for (A, D, E, F and G) where ligands were modeled into the structures. (I) Topology diagram of the Family 1 double-PDC sensor proteins. The figures were generated using PyMol67.
Structure of Z2
SeMet Z2 was crystallized in bistris buffer (SeZ2-bis) and the structure was solved by MAD at 1.70Å resolution in space group C2221 (Table 3). There is one protein molecule in an asymmetric unit with 261 well-ordered residues (42-302) and 230 water molecules. Native Z2-bis was crystallized in space group P21 and the structure was solved by molecular replacement using SeZ2-bis as the search model. Two well-ordered protein molecules and 460 water molecules were found in the asymmetric unit. In both structures, there is clear electron density in the putative ligand binding site of the protein, which could be best modeled as a bistris molecule. The binding pocket in the structure of SeMet Z2 crystallized in cacodylate buffer (SeZ2-cac) also contains an extra density, but it has a completely different shape and size from bistris, indicating that bistris is correctly identified in SeZ2-bis. The extra features in SeZ2-cac were modeled as a sulfate ion and an ethylene glycol molecule. The three structures, with or without bistris, are nearly identical to one another with root-mean-squared deviation (RMSD) less than 0.5Å for all atoms, implying that bistris is not the physiological substrate of Z2.
Structure of Z3
The native crystal structure of Z3 was solved by molecular replacement at 3Å resolution using SeZ2-bis structure as template (Table 3). The crystal has P65 symmetry. There are ten protein molecules, 53 water molecules and 10 potassium ions in an asymmetric unit. The ten Z3 molecules form five equivalent dimers. The protomers inside a dimer are related by a non-crystallographic 2-fold axis, which is parallel to the long N-terminal α-helix. Three dimers pack in one cluster forming a three-layer sandwich with the 108, 108, and 142 degree rotations along a common axis perpendicular to the three 2-fold axes. The other two dimers make another cluster with a 108 degree rotation between one another. There are extensive contacts within dimers and clusters, while only a few contacts exist between the two clusters.
Structure of Z6
The crystal structure of SeZ6 was solved by MAD at 2.30Å resolution (Table 3). This crystal is in space group P3121 and has one protein molecule, 140 water molecules and one ethylene glycol molecule per asymmetric unit. The loop between 238 and 241 was not modeled in the structure for the missing electron density.
Structure of Z8
The crystal structure of Z8 was solved in two states: the apo state (Z8-apo) and the phosphate-bound state (Z8-PO4) (Table 3). Z8-PO4 was crystallized in the presence of phosphate as buffer and was able to diffract to 1.75Å spacings. The structure was solved by MAD and refined against a native dataset to Rwork and Rfree of 18.2% and 22.2%. There are two protein molecules, two phosphate groups and 594 water molecules in each asymmetric unit. The two protein molecules are related by a two-fold rotation axis, which, unlike that in Z3, is perpendicular to the long N-terminal α-helix, meaning that the two N-terminal ends point into opposite directions. Z8-apo was crystallized without phosphate during purification or crystallization. It was able to diffract to 2.6Å spacings, albeit with a high mosaicity at 1.0°, and the structure was solved by molecular replacement using the Z8-PO4 structure as the search model. There are two protein molecules per asymmetric unit, which are again related by a two-fold axis perpendicular to the long N-terminal helix. After we had completed our structure determinations, a structure of the same protein was deposited (PDB ID: 2p7j). Although the construct and crystallization conditions differ, the lattice for this structure is quasi-isomorphous with that of Z8-PO4, but here a sulfate ion occupies the phosphate site.
Structure of Z16
The crystal structure of SeZ16 was solved by MAD at 2.70Å resolution (Table 3). There are two protein molecules, 2 MPD molecules in the putative binding pockets, 2 citrate molecules and 69 water molecules in each asymmetric unit of a P4332 lattice. In contrast to the other structures, Z16 does not have an ordered C-terminal helix. In addition, the central β-sheet in the membrane-distal domain appears special in that β3 is one single continuous strand instead of two separate strands linked by a bulge and there is a loop at the position corresponding to β2′ in other structures. Both citrate molecules are at the surface of the proximal domain of Z16, near the α1 helix, but without perfect occupancy.
Sequence and structural comparisons
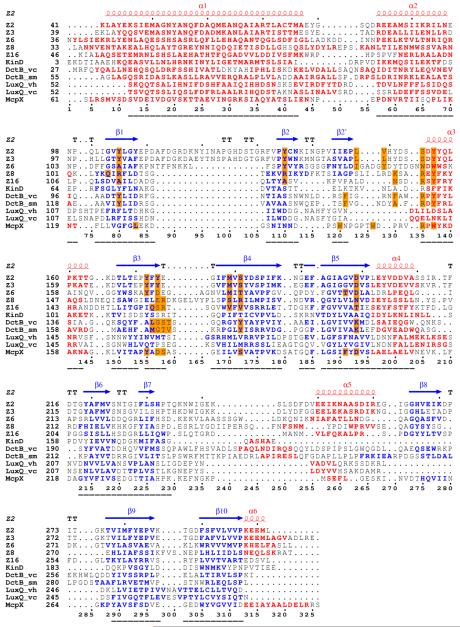
All double-sized PDC sensors with available structures have been compared based on structure-based sequence alignments. These double-PDC sensors include the five structures presented in this work, other histidine kinase sensors KinD from B. subtilis (PDB ID: 3fos)22, DctB from Vibrio cholerae (vcDctB, PDB ID: 3BY9), DctB from Sinorhizobium meliloti (smDctB, PDB ID: 3E4P), LuxQ from V. harveyi (vhLuxQ, PDB ID: 2HJE) and LuxQ from V. cholerae (vcLuxQ, 3C38), and an uncharacterized methyl-accepting chemotaxis domain (PDB ID: 3c8c) from V. cholerae, which we call McpX24. The structure-based sequence alignment (Figure 3A) indicates that core features of the PDC domains align well with one another despite having no positions of overall identity. Structural similarity despite sequence diversity has been noted previously for other histidine kinase sensors13, 14, 18. Although all of these double-PDC sensors are similar structurally, only KinD and McpX conform to sequence family HK1, whereas the DctBs are in family HK4 and the LuxQs are in family HK50.
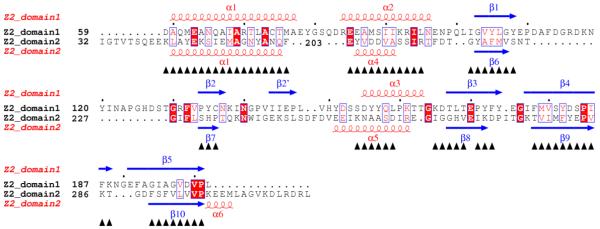
Figure 3.
Structure-based sequence alignments. (A) Alignment of double-PDC sensor domains. Secondary structure elements are specified at the top of the alignment based on the DSSP calculation for Z2. Sequences are shown for the sensor domains of Z2, Z3, Z6, Z8, Z16, KinD, vcDctB, smDctB, McpX, vhLuxQ and vcLuxQ with starting residue numbers given on each line. Helix residues are shown in red and strand residues are shown in blue. Ligand binding site residues are identified by orange-filled boxes. Numbers at the bottom specify positions used in comparisons. Sequence-aligned core residues used for RMSD calculations (Table 4) are identified by underlining. (B) Alignment of the two domains of Z2. Starting residue numbers are shown for each sequence segment. Superposed residues are identified by black triangles at the bottom. Identical residues are identified by red boxes and similar residues are identified by empty boxes. The pictures are made using ESPript68, 69, 70.
Sequence motif RXYF
The most conservative feature among these proteins is the consensus RXYF motif at the beginning of helix α3, in all but the LuxQ proteins. The X residue in this motif is typically polar and most often negatively charged, and the YF pair are always aromatic residues (Figure 3A). This region is located near the ligand-binding site (see below) and the first aromatic residue always points into the ligand-binding site, implying that this residue may be important on shaping the ligand binding site. The idea that the RXYF motif may be critical for substrate recognition is consistent with its absence from the LuxQ sensors; LuxQs interact indirectly with their quorum-sensing autoinducer ligands through LuxP periplasmic binding proteins.
Sequence relationships overall
These proteins show a wide range of pairwise sequence similarities. Z2, Z3, Z6, Z8 and Z16 all belong to family HK1 based on our previous bioinformatic analysis but, as this classification is at Psi-BLAST level E≤10−3, even these are diverse. Z2 and Z3 are closely related with 69% sequence identity; whereas Z6 shares 19% sequence identity with Z2 and 18% with Z3. These three proteins all belong to subfamily B. McpX, which is similar to the asparagine-responsive McpB chemotaxis protein from B. subtilis39 also belongs to subfamily B and is close to Z3 at 23% identity. B. subtilis KinD is also an HK1 protein, but in subfamily A. Similarities among representatives from different HK1 subfamilies are lower, often barely detectable by sequence identity alone; KinD (A), Z2 (B), Z16 (C) and Z8 (E) compare in the range of 10-17% identity. All seven HK1 proteins have less than 19% sequence identity with DctBs and less than 14% with LuxQs.
At this point, having discovered that the sensor domain of the McpX chemotaxis receptor is a member of HK1 subfamily and noticing that the PDB entry (ID: 2p7j) for an isomorph of our Z8 structure is annotated as from a “GGDEF protein”, named for a motif in diguanylate cyclases, we questioned the annotation-based assumption that our database of HK1 sequences really pertained to histidine kinase receptors. We found that all database members of subfamilies B, D and G did indeed contain HK catalytic domains, but that some members of subfamilies A, C and E, including our Z16 and Z8 proteins, contain diguanylate cyclase (DGC) domains but no HK domains. In addition, the McpX sensor conforms to HK1 subfamily B but has a chemotaxis-like cytoplasmic sequence, consistent with the PDB annotation. Each HK1 subfamily does have members with legitimate HK domains (11 of 14 for subfamily A, all 10 for B, 3 of 12 for C, all 10 for D, 3 of 7 for E, all 6 for F, all 3 for G, and all 3 subfamily singletons) and HK members do predominate (49 of 65). All Family 2 members are legitimate HK proteins, and with 51 members it technically becomes the largest true HK sensor family in our dataset. Moreover, the surprising discovery that clearly similar sensor domains are associated with cytoplasmic output domains of three very different classes opens new questions for molecular evolution of microbial signaling. Superpositions show members of the HK1 family of sensor domains are even more closely related in structure than in sequence.
Correlation of structure superpositions with sequence comparisons
It was obvious from even a superficial inspection that the juncture between domains of the double-PDC sensors has some flexibility. Thus, we separately superimposed the respective membrane-distal and membrane-proximal domains from each pair of sensors. Superpositions were based on Cα atoms with a criterion for superposition that required stretches of at least three contiguous residues should match within 3Å. Superposition results comparing the domains of our HK1 proteins with one another and with other PDC proteins are given in Supplementary Table 1. We also compared all double-PDC structures with one another. From the resulting structure-based alignment (Figure 3A), we defined a core of residues in common to all sequences. This core comprises 98 distal residues and 57 proximal residues, and the combined RMSD values are reported in Table 4. Pair-wise sequence comparisons are reported as Blastp Z-scores.
Table 4. Structure and sequence comparisons of sensors having a double-PDC fold.
| Z2 | Z3 | Z6 | Z8 | Z16 | KinD | DctB (vc) |
DctB (sm) |
LuxQ (vh) |
LuxQ (vc) |
McpX | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Z2 | 0.98 | 1.96 | 2.86 | 2.19 | 2.28 | 2.33 | 2.18 | 3.72 | 3.82 | 2.10 | |
| Z3 | 424 | 1.83 | 2.86 | 2.16 | 2.30 | 2.43 | 2.27 | 3.58 | 3.68 | 1.97 | |
| Z6 | 40.4 | 40 | 2.86 | 2.15 | 2.35 | 2.31 | 2.13 | 3.92 | 4.01 | 2.26 | |
| Z8 | 14.6 | 18.1 | 17.7 | 2.91 | 2.82 | 2.72 | 2.84 | 4.07 | 4.14 | 2.87 | |
| Z16 | 17.3 | 18.5 | 17.3 | 25 | 1.86 | 2.01 | 1.83 | 3.94 | 4.09 | 2.25 | |
| KinD | 16.9 | 15.8 | 14.2 | 26.9 | 38.1 | 1.92 | 1.79 | 3.84 | 3.89 | 2.23 | |
| vcDctB | 16.9 | 10 | 15.8 | 18.5 | 14.2 | 17.3 | 1.35 | 3.67 | 3.71 | 2.36 | |
| smDctB | 14.6 | 15.8 | 16.9 | 15.0 | 18.5 | 13.5 | 119 | 3.69 | 3.80 | 2.18 | |
| vhLuxQ | 13.9 | 21.6 | 11.9 | 15 | 10.8 | 15.4 | 13.9 | 16.9 | 1.15 | 2.60 | |
| vcLuxQ | 13.9 | 21.6 | 10.8 | 15.0 | 12.3 | 15.0 | 13.9 | 18.5 | 174 | 3.69 | |
| McpX | 50.4 | 55.1 | 20.8 | 11.9 | 26.6 | 16.9 | 15.4 | 12.7 | 9.6 | 9.6 |
RMSD values (upper triangle) and Blastp Z-scores (lower triangle) are shown for pair-wise comparisons of all sensors with double-PDC fold in the PDB. The PDB code of Z2, Z3, Z6, Z8, Z16, KinD, vcDctB, smDctB, vhLuxQ, vcLuxQ and McpX are 3LI9, 3LIB, 3LIC, 3LID, 3LIF, 3fos, 3by9, 3e4p, 2hje, 3c38, and 3c8c, respectively.
We found earlier that sequence similarity and structural deviation are anti-correlated among hemoglobins40, even when sequence relationships are remote. Here, the correlation coefficient between the primary sequence relatedness (Z-score) and 3D structural deviation (RMSD) is −0.46, indicating a mild negative correlation; i.e. as Z-score increases, RMSD tends to decrease. The correlation coefficient is stronger at −0.61 if both LuxQ molecules are removed from the calculation, whereas the removal of other sequences did not significantly affect the number. This distinction implies that LuxQs have taken a different path through evolution; in fact, the residues making LuxQs an outlier are mainly located around α3, which is an important ligand binding determinant in all of these double-PDC proteins except for LuxQ. Interestingly, LuxQ proteins do not use the common ligand binding site to sense their ligands. Instead, LuxQ proteins recruit LuxP to interact with substrates. Thus, the α3 region in LuxQ proteins is under different selection pressure from the others. This region retains a helical structure in LuxQ, but it swings away from an HK1-like place in the 3D structure. Indeed, the correlation coefficient does not change much with or without LuxQ molecules if it is calculated by the structure-invariant core method, which does not include the α3 region for the calculation of RMSD.
Domain duplication
The two PDC domains of all five of our HK1 proteins can be aligned with one another (Figure 3B) with a modest RMSD values (1.4-1.7Å) and more than 50% of all residues superposed (Supplementary Table 2), as is also true for DctB and LuxQ, consistent with these proteins having evolved by gene duplication. The RXYF motif is not conserved in the membrane-proximal domains, however, which may be a reason why the membrane-proximal domains lose the ability to recognize substrates.
Ligand binding site composition
The physiological substrates of these HK1 sensors have not been identified. Other PDC proteins do have well characterized ligands, however; and in the cases of DcuS, CitA and DctB these ligands are bound in a ligand binding site on the side of the β-sheet opposite from N-terminal helix α1. We find ligands, derived from the crystallization medium, to be bound in four crystal structures of the five sensor proteins reported in this paper. Interestingly, as for succinate with DctB, the ligand is always located at a site in the membrane-distal domain. In each case, the distal β-sheet forms the back of the ligand binding site, as viewed in Figures 2 and 4, and loops embrace the ligand. A major loop from between β2 and α3, including β2”, runs across the top of the ligand binding site from the other side of the β-sheet, that is, the front of the ligand binding site. A minor loop, which is between β3 and β4, turns over to contact the ligand from the bottom due to a kink in both β3 and β4. In the cases of Z2, Z3, Z6 and Z8, the kink is so sharp that β3 separates into two segments.
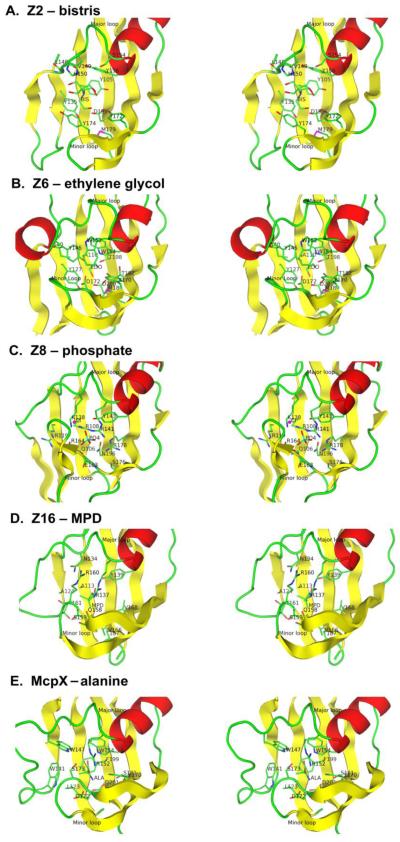
Figure 4.
Stereodiagrams of ligand-binding interactions in Family 1 sensors. (A) Z2. (B) Z6. (C) Z8. (D) Z16. (E) McpX24. The distal domain from each structure is superimposed onto that of Z2. The central β-sheet is shown in yellow, helix α3 is shown in red, and loop regions are shown in green. The ligand and the contacting residues are shown in ball-and-stick representation with carbon atoms green, oxygen atoms red and nitrogen atoms blue. The picture was made using PyMol67.
The residues making contact with ligands are very consistent across the HK1 family based on the structure-based sequence alignment (Figure 3A). Contacting residues include one from β1 (position 80, Figure 3A), one from β2 (position 110), two from the major loop, two from the beginning of α3 (positions 135 and 137), a few from β3 and the minor loop (mainly positions 155 and 157), two from β4 (positions 170 and 172), and finally one from β5 (position 193). We did not find any ligand in the structure of Z3, so residues in this binding site are imputed based on structure superimposition onto Z2. The major loop (β2 - α3) is variable in length and sequence and therefore its ligand binding site residues cannot be superimposed; thus that they do not appear at the same position in the structure-based sequence alignment. The RXYF motif (positions 135-138) has been identified as the most conserved region among the aligned sensors, except for LuxQs, and the third RXYF residue, always an aromatic amino acid, points into the ligand binding site as does the first. Except for the RXYF conservation, Family 1 binding site residues are quite variable as is true for these sensors overall. Besides the identified ligand-contacting residues, others are also in the binding pocket and may be involved in the binding of authentic ligands. The lack of conservation would seem to support great diversity in potential ligands.
A bistris molecule has been identified in the crystal structures of SeZ2-bis and Z2-bis (Figure 4A). The most notable feature of the binding pocket is the presence of five tyrosine residues: Y105, Y135, Y156, Y172, and Y174. The side chains of D199, S181, L148 and backbone atoms of L148 and V149 are also within 4Å distance from bistris (Figure 4). Even though S154 is located too far away to make contact with bistris (4.9Å), its side chain points to bistris and may make contribution to the binding of another ligand. S154 is located in the major loop right before α3, and the residue at this position (135) is in the ligand binding site of all double PDC sensors with known structure (Figure 3A and Figure 4). The ligand binding site of Z3 is empty but otherwise almost identical to that of Z2. The only difference is that major loop residue V149 in Z2, which makes contact with bistris using main chain atoms, becomes L148 in Z3.
An ethylene glycol molecule, presumably from the cryoprotectant, was modeled into the binding pocket of Z6 (Figure 4B). The residues surrounding the Z6 binding pocket are A110, Y127, I140, Y145, W154, D172, T182, T198, D200, which are similar to those of Z2 and Z3, but with fewer aromatics and more polar and negatively charged residues. The ethylene glycol molecule is coordinated well with B factors (~15Å2) similar to those of surrounding residues, suggesting that this adventitious ligand may relate to the natural one. Negatively charged residues D172 and D200 make contacts at either end of the ethylene glycol ligand; thus, replacement of the terminal hydroxyl groups with amines could yield attractive candidates for authentic ligands for Z6. Both ethanolamine and ethylenediamine can serve as nitrogen sources and ethylene glycol can be a carbon source for some microorganisms.
A phosphate ion was modeled into the crystal structure of Z8-PO4 at its ligand binding site (Figure 4C). The Z8 binding pocket is similar to that of Z2, but it has a longer minor loop, which flaps over and covers the front side of the pocket. The phosphate ion is coordinated by mainly positively charged residues including R108, R141, R178 and K138. Other residues within a 4Å contact radius include Q106, Y143, E163, N196 and the backbone of R164. We have modeled a chloride ion just outside the binding pocket, coordinated by the charged group of K138 and R141 and main chain atoms of R164, K166 and G167. This seemingly closes a phosphate exit channel to solution. A sulfate ion was found in another crystal structure of the same protein (PDB ID: 2p7j). Z8 is from bacterium V. parahaemolyticus, which inhabits coast-line water where both sulfate and phosphate are present, but with sulfate at greater abundance. Thus, SO4 may possibly be a natural signal for Z8. Z8-apo, crystallized in the absence of phosphate, diffracted only to 2.6Å with high mosaicity, indicating that Z8 is more flexible without the ligand.
An MPD molecule was modeled into extra density located in the ligand binding site of Z16, which was surrounded by residues A113, A124, N134, R137, Y139, Q158, S159, R160, T161, V166, V168 and T187 (Figure 4D). The MPD has slightly higher B factors than for surrounding residues. A citrate molecule, which is also present in the medium, could also fit the density, but with lower occupancy and higher B factors. There is no indication that either MPD or citrate is the physiological ligand for the sensor.
The structure of McpX (PDB ID: 3c8c)24, which is a member of HK1 subfamily B and closest to Z3 (Table 4), contains an alanine molecule at its ligand binding site (Figure 4E). A structural description is not yet published, but alanine is not a component of the PDB-reported crystallization solution. Thus, we presume that the alanine derives from the expression culture, as also happened for succinate with vcDctB14, and that McpX is likely to be an alanine sensor. The ligand binding site of McpX is similar to that of Z2 and Z3 in that aromatic residues predominate, but here it is rich in tryptophans. Interestingly, R152 (position 135) from the RXYF motif is as a hydrogen-bonded counter-ion to the carboxyl group of the alanine ligand and D201 (position 193, Figure 3A), which is also aspartate in Z2 and Z3, forms a hydrogen-bonded charge pair with the amino group of the alanine ligand.
Dimer interface
Histidine kinases are believed to function as homodimers3 and structures of cytoplasmic domains from these receptors are dimeric2, 7. Each of our HK1 sensor domains behaves as a monomer in solution based on size exclusion chromatography and static light scattering (data not shown), however, which is consistent with many other histidine kinase sensor domains. We do expect that the sensor domains will be functionally dimeric in intact receptors since membrane tethering will increase the intrinsic binding affinity of associated domains by orders of magnitude41, 42. Thus, we examined potential dimerization interfaces in detail and found various interfaces in the crystal packing of these structures. Based on previous studies13, 16, interfaces involving the N-terminal helix are most likely function-related. Four dimer interfaces of this kind have been identified and these are shown in Figure 5.
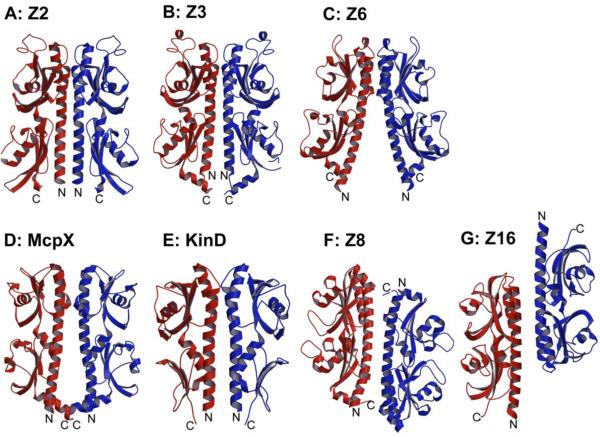
Figure 5.
Dimeric organization of Family 1 sensors. (A) Z2-bis. (B) Z3. (C) Z6. (D) McpX24. (E) KinD22. (F) Z8-PO4. (G) Z16. In each case, one protomer is colored red and the other is colored blue. N and C termini are labeled. Ribbon diagrams were drawn by Molscript71 and Raster3D72.
Both Z2 and Z3 dimers (Figures 5A and 5B) are related by two-fold rotation axes that are roughly parallel to the α1 helices. In the membrane-distal domain, the two α1 helices are joined by two α2 helices to form four-helix bundles. The α4 helices, which are membrane-proximal equivalents of α2, rotate away and do not contribute to the interface. Even though the C-terminal α6 helices are not part of the interface, they do point toward the N-terminal end of the sensors, poised to attach to the transmembrane domain, and they may be involved in the interface of the intact receptors. These interfaces bury 3263 Å2 and 2786 Å2 surface area for Z2 and Z3, respectively, and they are dominated by charged and hydrophilic residues. The Z6 dimer (Figure 5C) has the same diad-related four-helix bundle configuration as in Z2 and Z3 at the membrane-distal domain; however, the Z6 dimer interface breaks up in the membrane-proximal domain and the N-terminal ends are splayed apart, perhaps reflecting only a partially natural association. The buried surface area (1931 Å2) is much smaller than for Z2 and Z3, but interface residues in the Z6 dimer are also mainly charged and hydrophilic residues.
The McpX sensor, which is close to Z3 in sequence (Table 4), also forms a Z3-like dimer although the proximal domains are slightly separated and the buried interface (1888 Å2) is thereby closer to that of the Z6 dimer. This symmetric McpX dimer (Figure 5D) has a bound ligand and its organization appears to be physiologically relevant. KinD, which is from subfamily A, also forms a dimer (Figure 5E) that is consistent with physiological relevance. As for McpX, its buried surface area (1885 Å2) is like that of Z6 even though its appearance is more like Z2 and Z3..
Diad-symmetric, but seemingly just crystallographic, “dimers” are also found in the Z8 and Z16 crystal structures; but in these cases the membrane-proximal ends of the two protomers point in opposite directions because the two-fold axes are perpendicular to the α1 helices. In the resulting Z8 “dimer” (Figure 5F), the membrane-distal domain of one protomer makes contact with the membrane-proximal domain of the other protomer. In the Z16 “dimer” (Figure 5G), oppositely oriented membrane-distal domains interact with one another and the membrane-proximal domains extend away. These crystal “dimers” do not appear to have direct physiological relevance, but the surfaces of contact are similar to those in Z2 and Z3 dimers. Indeed, these same association-prone surfaces are used to form dimer interfaces in all Family 1 sensors (Figure 5).
Conformational change upon ligand binding
The sensors must undergo some kind of conformational change upon ligand binding in order to generate a signal that can be transduced across the cell membrane to trigger the downstream events. One major goal of this research is to look for such conformational changes and to deduce models for mechanisms of signal transmission. Since we do not have validated ligands for these receptors, we cannot be certain that differences observed here between apo and ligand-bound states are authentic. Moreover, unnatural ligands might not impart sufficient binding energy to compete with lattice interactions. Indeed, the structures of Z2 with and without bistris are almost identical with RMSD of 0.5Å for all atoms, and structures of the Z2-bistris complex in two different lattices are also the same. There is no significant change in protein conformation or in dimer configuration among any of these structures. It is possible that both bistris and sulfate plus ethylene glycol have elicited the same conformational change relative to the apo state.
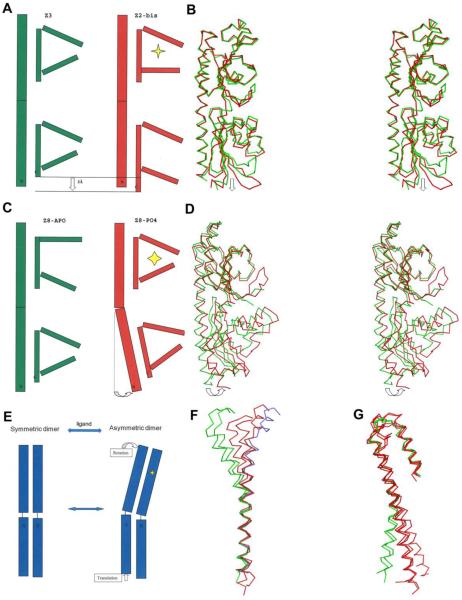
Piston-like translation movement
Since Z2 and Z3 are very similar, both in sequence (69% identity) and in structure, domain-by-domain (0.65Å distal and 0.96Å proximal, Supplementary Table 1), we consider Z3-apo as a candidate for the apo state of Z2. The ligand binding site of Z3 is nearly identical to that of Z2-bistris, but it is empty of any structured features. Thus, we compared the conformations of this apo and ligated pair (Figure 6A and 6B). In order to identify the structural difference between the two structures, we used Escet43 to identify the conformationally invariant core region, which was input to LSQMAN to align the two structures. The superposition reveals that the central β-sheet of the membrane-proximal domain moves toward the cell membrane relative to the N-terminal helix upon ligand-binding. Most interestingly, the C-terminal helix, which links to the intracellular kinase domain through TM2, dislocates by about 3Å relative to the N-terminal α1 helix next to TM1, implying that a piston-like movement may occur upon ligand binding. Both the Z2-bistris and Z3-apo sensors are symmetric or quasi-symmetric dimers here; but asymmetry, such as occurs for Tar35, may arise for signal transduction in the functional receptors.
Figure 6.
Conformational changes associated with ligand binding. (A) Cartoon illustration of the piston-like movement seen in comparing apo Z3 (green) and bistris-complexed Z2 (red). The ligand is labeled as a yellow star. (B) Stereo plot of superimposed apo Z3 (green) and Z2-bis (red) and showing the piston-like movement. (C) Cartoon illustration of the domain rotation seen in comparing apo Z8 and phosphate-complexed Z8 (red). The ligand is labeled as a yellow star. (D) Stereo plot of superimposed Z8-apo (green) and Z8-PO4 (red) showing the domain rotation movement. (E) Cartoon illustration of a rotation-induced translation due to asymmetry. (F) Cα traces of the two N-terminal helices of all five sensors when the membrane-distal domains are superimposed. Z2 and Z3 are colored green; Z6, Z8-apo and Z16 are colored red; Z8-PO4 is colored cyan. (G) Cα traces as in (F) except that the membrane-proximal domains are superimposed. Kinking or curvature is evident at the juncture between domains.
Domain rotation movement
The superimposition of Z8-apo onto Z8-PO4 reveals a conformational change within and beyond the ligand binding pocket (Figure 6C and 6D). The pocket is relatively open in the apo state, with the major loop turned away from the ligand binding site and the minor loop disordered, whereas these loops close down onto the ligand in the PO4-bound state. When the membrane-distal domains of both states are superposed, the membrane-proximal domains need to be rotated 12 degrees to fit to each other (Table 5). This conformational change may possibly be relevant to transmembrane signal transduction.
Table 5. Domain movement.
| χ (°) | tχ (Å) | |
|---|---|---|
| Z2-Z3 | 6.59 | −0.06 |
| Z2-Z6 | 32.39 | −0.41 |
| Z2-Z8-apo | 23.94 | 0.53 |
| Z2-Z8-PO4 | 33.67 | 2.46 |
| Z2-Z16 | 23.01 | −1.35 |
| Z8-apo-Z8-PO4 | 11.95 | 0.74 |
Parameters are shown for the transformation required to superimpose two proximal domains having already superimposed the corresponding distal domains. The value χ is the required rotation angle and the value tχ is the required translation along the unique axis of rotation.
Discussion
All five HK1 sensor domain structures have inserted repeats of PDC domains (Figure 1) organized in a manner similar to DctB and LuxQ structures. DALI searches38 find PDC proteins as the best matches, followed by PAS-containing proteins, consistent with previous results13, 14. The structure-based sequence alignment (Figure 3A) shows that there are no identically conserved residues across the Family 1 proteins, but there is a striking similarity in structure nevertheless; and sequence similarity is correlated with structural similarity across all double-PDC sensor domain structures (Table 4). Moreover, these proteins appear to have evolved by gene duplication (Figure 3B, Supplementary Table S2). In addition, as also observed for other sensor domains44, HK1-like domains clearly serve as evolutionary modules supporting a variety of receptor systems. We find that some HK1 family members actually have diguanylate cyclase output domains and that the HK1-like McpX protein has a chemotaxis output domain. Since closely related extracellular sensors are used by widely disparate cytoplasmic transmitters, this suggests relatively recent events of modular molecular evolution. Similar modular associations of sensor domains with alternative transmitter domains also occur for four-helix bundle sensors26 and for PDC-type sensors18.
There have been several previous attempts to classify extracellular sensor domains. Both an MCP_N helical domain and a Cache domain were defined based on sequence analyses of McpABC homologs45. Conserved-domain searches find the sequence motifs corresponding to the MCP_N and Cache domains in both Z2 and Z3, which is as expected since these subfamily B proteins and McpX are homologs of McpABC; however, a conserved-domain search does not detect either MCP_N or Cache motifs in Z6, Z8 and Z16 despite the fact that these sensor domains are clearly related both in structure and sequence (Table 4, Figure 3B). Moreover, the HK1 crystal structures show that neither MCP_N nor Cache can serve as an independent domain, but rather that both are inseparably integral components of Family 1 double-PDC domains. Thus, although the Cache motif does capture some elements of the HK1 family, it is not sufficiently comprehensive to define the entire domain or the full and yet limited scope of the family. CHASE domains were defined in the similar manner as for the so-called Cache domain44, 45, 46, and similar difficulties can be expected in finding all sensors within the respective sensor-domain families.
The sensor domains of histidine kinase receptors and of chemotactic receptors are diverse in structural organization47 and they recognize diverse ligands. There is no obvious relationship between structural organization and the mode of ligand recognition, however, as is clear from many examples: (a) Both DctB and LuxQ have similar double-PDC structures; but whereas DctB binds succinate directly, LuxQ binds to the periplasmic-binding protein LuxR, which in turn binds the AI-2 autoinducer ligand. (b) The chemotactic receptors Tar, McpB and McpX all bind amino-acid ligands (aspartate, asparagine and alanine, respectively), but whereas Tar is a four-helix bundle protein, we now know that McpX is a double-PDC protein24 as is now also clear for McpB48. (c) DcuS and DctB both bind C4-dicarboxylate ligands14, but whereas DcuS binds malate in a PDC domain, DctB binds succinate in the distal domain of a double-PDC sensor protein. It is in this context that we have examined the ligand binding possibilities for HK1 proteins, which were completely uncharacterized hitherto.
Although natural ligands are not known for any of our HK1 sensor proteins, we do find adventitious ligands bound to four of the five crystallized proteins. Each of these ligands binds to the distal PDC domain at a site that overlaps with that to which succinate binds in DctB14, 21 and to which malate and citrate bind in the single-PDC sensor domains of DcuS14 and CitA16, respectively. In addition, HK1 subfamily B homolog McpX binds alanine at the very same site. Thus, these ligands undoubtedly mark the ligand binding site of HK1 sensor proteins. We find no evidence of ligand binding to the proximal PDC domains of any of the HK1 sensors, which as in DctB are much less encapsulating. Each of our HK1 ligands is identified as an ingredient of the crystallization medium, but while these are not likely to be physiological ligands they likely do mimic features of the natural ligands. The only HK1 protein for which a physiological function has been identified is KinD, which is involved in the regulation of sporulation in B. subtilis31, 34 through phosphorylation of the phosphorelay protein spo0F. Clearly, Family 1 members are not all specialized in sporulation control, however, since many members come from bacteria that do not sporulate. It is probable that McpX is an alanine sensor since in this case the ligand must have come from the cellular milieu and homologous B. subtilis McpB is a sensor for another amino acid, asparagine48, 49, 50. On the other hand, three other MCPs, namely PctA, PctB and PctC, which are closer to Z2 and Z3 (~20% identity) than is McpB (~10%), have been identified as trichloroethylene sensors51, 52. Thus, it is very likely that the HK1 family members do not share a common class of substrates.
In contrast with the receptor tyrosine kinases of eukaryotic animals, which typically undergo ligand-mediated dimerization, it is well accepted that histidine kinase receptors remain constitutively homodimeric, independent of ligand-binding. Thus, the signal transduction event is not triggered by dimerization3. Moreover, many histidine kinase receptors can serve both as kinases or phosphatases, suggesting that they act more as either/or switches than as on/off switches. Despite the expected dimeric organization for the intact receptors, all of the HK1 sensor domains characterized in this paper appear to be monomeric in solution as found by size exclusion chromatography and static light scattering (data not shown). Among all characterized extracellular PDC sensors of histidine kinases, only DcuS and CitA are confirmed to have weak dimerization in solution14, 15. Nevertheless, at the high protein concentrations in crystal lattices, we do observe dimeric associations that appear to be physiologically meaningful. The weak intrinsic dimer affinity of these domains is expected to be enhanced by orders of magnitude when restricted to two-dimensional diffusion by membrane-tethering in the intact protein41, 42, and the weakness of intrinsic association may be essential for permitting the ligand-directed plasticity that can serve for signal transmission. The largely hydrophilic nature of HK1 dimer interfaces is consistent with the observed weak intrinsic associations. In keeping with such lability, sensor domains can also form swapped dimers18.
Signal transmission through histidine kinase dimers must involve some kind of ligand-induced conformational change. Here, in this work, we identify two types of possible changes associated with ligand binding. We see a piston-like translation between Z2-bis and Z3 and a domain rotation between Z8-apo and Z8-PO4 (Figure 6). Piston-like movements have been proposed from structural and biochemical studies of Tar35, TorS29, CitA16, and NarX26 while a disulfide crosslinking experiment on McpB shows no sign of relative movement between TM1 and TM253. An interdomain rotation upon ligand binding was also reported before in the crystallographic study of DctB21. Although it is possible that the observed rotations are artifactual due to the lack of membrane tethering or because of the crystallization medium, we believe the flexibility between the two PDC domains is an intrinsic feature and may be involved in the signal transduction.
Interdomain movement is a common feature for double-PDC sensors. Among all eight HK1 structures, three distinct curvatures of the N-terminal helix have been observed (Figures 6F and 6G). How one might integrate these findings and the two types of ligand-induced movements into a common system is quite puzzling, however. Intrigued by the idea of asymmetry-induced signal transduction proposed for LuxQ, a coupling of domain rotation to piston-like movement might be explained in the context of dimer configuration as exemplified in Figure 6E. Ligand binding induces rotation of the membrane-distal domain relative to membrane-proximal domain in protomer A, which causes the membrane-distal domain of protomer B to rotate in a similar direction. Then, the membrane-proximal domain of protomer B must slide relative to protomer A in order to maintain the dimeric state. Thereby, a ligand-induced rotation could lead to a piston-like movement through transmembrane helices. Further experiments are required to test this hypothesis.
Materials and Methods
Materials
The DH5α and Bl21(DE3) competent cells were purchased from Invitrogen. pET22b+ was purchased from Novagen. Endonucleases were purchased from NEB. Primers were purchased from Operon. Genomic DNAs were purchased from ATCC. Chemicals were from Sigma, Fisher or Hampton Research.
Cloning
Many genes from three well-defined HK1 subfamilies, namely subgroup B, C, and E (Figure 1), were amplified from genomic DNA (ATCC) by PCR using appropriate primers at both ends flanking the DNA fragments encoding the periplasmic domains of the histidine kinases, as predicted by PredictProtein at http://www.predictprotein.org/54. The amplified DNA fragments were inserted into pET22b+ between NdeI and XhoI cutting sites in frame with the C-terminal His-tag. The constructs were tested for expression and solubility in small scale. All failed constructs were cloned into pSMT3 between BamH1 and Xho1, which attaches a His-SUMO tag in the N-termini of the target proteins in order to improve expression and solubility. In the end, Z2, Z3 and Z8 were cloned into pET22b+, and Z6 and Z16 were produced from pSMT3 (Table 1).
Protein expression
Large scale protein production was carried out from a 1L culture of BL21(DE3) cells containing either ampicillin for pET22b+ constructs or kanamycin for pSMT3 constructs. The culture was inoculated with 10ml from an overnight culture and left shaking at 37 °C. The temperature was lowered to 20 °C when OD600 reached 0.4-0.6 and the induction was begun with the addition of final 0.5mM IPTG 30min after that. Cells were collected by centrifugation after overnight growth and stored in −80 °C until use. Selenomethionyl (SeMet) proteins were expressed in the same manner except that minimal media containing SeMet was used.
Purification
For each protein, the cell pellet from a 1L culture was melted at RT and resuspended by 50 ml lysis buffer (20mM Tris 150 mM NaCl pH8.5). The cells were broken by sonication and cell debris was removed by centrifugation. The supernatant was applied to 5 ml chelating column charged with nickel. Impurities were washed out by step-wise increases of imidazole concentration in lysis buffer at 0, 10, 30, 50, 100 and 250 mM. Both OD280 and SDS-PAGE were used to identify where the target protein was eluted. The fractions were pooled, concentrated and applied to a sizing column running in lysis buffer. Native gel electrophoresis was used to test the purity and only those fractions with a single band were pooled and concentrated for crystallization. For the two SUMO-tagged constructs, Z6 and Z16, an overnight digestion step by 1:1000 (w/w) sumo protease ULP at 4 °C was employed immediately following immobilized metal affinity chromatography. The digested protein solution was diluted to final 10mM imidazole and applied again to a nickel affinity column. The target protein flowed through the column while SUMO tag and ULP protease were bound to the column since both are His-tagged. The flowthrough was concentrated and applied to sizing column. The fractions with single bands in native gels were pooled and concentrated for crystallization.
Crystallization
The initial crystallization screening with Crystal Screen HT and Index kit from Hampton Research and Crystal Screen Wizard kit from Emerald Biosystems was carried out at two protein concentrations (OD280=10 and highest possible concentration) using a Mosquito crystallization robot. Crystal optimization was done by manual screening of the surrounding conditions of the initial hit. The final crystallization conditions are listed in Table 1.
Structure determination and refinement
Se-MAD was used to solve the structure of Z2, Z6, Z8 and Z16. In each case, a three-wavelength MAD experiment at the Se K-edge was collected on a single frozen SeMet crystal at the X4A beamline of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. HKL2000 package55 was used to process the datasets and the statistics are listed in Table 2. SOLVE56 was used to determine the positions of the Se sites for Z2, Z6 and Z8 and ShelxCD57 was used for Z16. Arp/Warp58 was used to build the initial models after density modification performed with DM59 from the CCP4 package60. The models were completed by manual building in Coot61 and refined using refmac562 from the CCP4 package60. Waters were added after models were refined well.
Phaser63 was used to solve the structure of Z3 using Z2 as the template because the two proteins share 67% identity. Eight copies were found with strong correlation. The initial model was refined by rigid-body refinement in the CCP4 package and the density map was examined in Coot61. There were clear electron densities for two more copies. The ten-copy model was mutated to the sequence of Z3 from Z2 in Coot and refined using Refmac5. The native Z2 protein was crystallized in a different space group from the SeMet protein, and this native structure was solved by molecular replacement using Phaser. Two protein molecules were found in one asymmetric unit. The SeMet residues were mutated to Met and the whole structure was refined using refmac5. The structures of SeMet Z2 crystallized in the absence of bistris buffer and Z8 crystallized without phosphate were solved in the same procedure.
Structure superposition and structure-based sequence alignment
Structure superposition was done using LSQMAN64 from the Uppsala Software Factory with the following restriction. Two residues are considered superimposed only when at least three contiguous Cα positions of one structure are within 3.0 Å of the counterparts in the other structure. The superimposition was done domain by domain instead of for the whole protein in order to remove the effect of domain movement. The sequence alignment by ClustalW65 was used as initial template, which was manually adjusted according to structure superposition.
RMSD calculation
RMSD values were computed as the sum of squared deviations from the two separately superimposed domains. To compare all structures in the same context, we needed to define a structure-invariant core. Two methods were tried. One uses the common sequence-aligned core of residues, for which overall structural similarity prevails although individual pairs may violate the rule of three consecutive Cα positions being within 3.0 Å. Resulting RMSD values should have minimal bias because a relatively large number of residues are included, but this value is sensitive to accuracy of the alignment. The alternative method uses a structure-invariant core that matches, in common, for all pair-wise superpositions. RMSD values from this method are robust with respect to the accuracy of alignment, but being biased toward the so-called structure-invariant core region they may underestimate real distances between the two structures. The structure-invariant core had few residues based on the 3Å criterion, but a larger basis could be obtained with a 4Å criterion. RMSD values reported in Table 4 were calculated using the former method based on a sequence-aligned core. The alignment has 98 residues from the distal domain and 57 residues from the proximal domain, but residues absent in the individual PDB files were excluded.
Supplementary Material
Acknowledgements
We thank John Schwanof and Randy Abramowitz of NSLS X4 for help with synchrotron data collection, Dr. Erik Martinez-Hackert and Dr. Jonah Cheung at Columbia University for aid in the structure determinations, and Dr. Jinfeng Liu of the Rost laboratory for the bioinformatic analysis that laid the foundation for this work. We also thank Dr. Masoud Vedadi and Dr. Aled Edwards of Canada SGC for their effort in Stargazer screening for natural ligands of the sensors, even though the results turned out negative. This work was supported in part by NIH grant GM34102 (W.A.H.). Beamline X4A at the National Synchrotron Light Source (NSLS) of Brookhaven National Laboratory is supported by the New York Structural Biology Center.
Abbreviations
- HK
histidine kinase
- IPTG
isopropyl β-D-thiogalactopyranoside
- PAS
Per/ARNT/Sim
- PCR
polymerase chain reaction
- PDC
PhoQ/DcuS/CitA
- PEG
polyethylene glycol
- RMSD
root-mean-squared deviation
- RR
response regulator
- SeMet
selenomethionine
- TCS
two-component system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes
Structural data depositions for crystallographic results have PDB IDs: 3LI8, 3LI9, 3LIA, 3LIB, 3LIC, 3LID, 3LIE and 3LIF with associations to individual structures as designated in Table 3.
References
- 1.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 7.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO. J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada S, Shiro Y. Structural basis of the signal transduction in the two-component system. Adv. Exp. Med. Biol. 2008;631:22–39. doi: 10.1007/978-0-387-78885-2_3. [DOI] [PubMed] [Google Scholar]
- 9.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada S, Sugimoto H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Przybylski D, Rost B. Improving fold recognition without folds. J. Mol. Biol. 2004;341:255–269. doi: 10.1016/j.jmb.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 13.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domain. J. Biol. Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 16.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Emami K, Topakas E, Nagy T, Henshaw J, Jackson KA, Nelson KE, Mongodin EF, Murray JW, Lewis RJ, Gilbert HJ. Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J. Biol. Chem. 2009;284:1086–1096. doi: 10.1074/jbc.M805100200. [DOI] [PubMed] [Google Scholar]
- 18.Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Pokkuluri PR, Szurmant H, Schiffer M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J. Bacteriol. 2010;192:1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slama B, Hendrickson W. Crystal structure of the periplasmic domain of Vibrio cholerae LuxQ. 2008 http://dx.doi.org/10.2210/pdb3c38/pdb.
- 21.Zhou YF, Nan B, Nan J, Ma Q, Panjikar S, Liang YH, Wang Y, Su XD. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Wu R, Schiffer M, Gu M, Joachimiak A, Midwest Center for Structural Genomics (MCSG) The crystal structure of two-component sensor histidine kinase domain from Bacillus subtilis. 2008 http://dx.doi.org/10.2210/pdb3fos/pdb.
- 23.Wu R, James A, Joachimiak A, Midwest Center for Structural Genomics (MCSG) The crystal structure of the domain of putative sensory box/GGDEF family protein from Vibrio parahaemolyticus. 2007 http://dx.doi.org/10.2210/pdb2p7j/pdb.
- 24.Patskovsky Y, Ozyurt S, Freeman J, Hu S, Smith D, Bain K, Wasserman SR, Sauder JM, Burley SK, Almo SC, New York SGX Research Center for Structural Genomics (NYSGXRC) Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae. 2008 http://dx.doi.org/10.2210/pdb3c8c/pdb.
- 25.Cheung J, Le-Khac M, Hendrickson WA. Crystal structure of a histidine kinase sensor domain with similarity to periplasmic binding proteins. Proteins. 2009;77:235–241. doi: 10.1002/prot.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE, Jr., Kim SH. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 28.Tan K, Chhor G, Buck K, Joachimiak A, Midwest Center for Structural Genomics The crystal structure of a two-component sensor domain from Pseudomonas aeruginosa PA01. 2009 http://dx.doi.org/10.2210/pdb3kkb/pdb.
- 29.Moore JO, Hendrickson WA. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure. 2009;17:1195–1204. doi: 10.1016/j.str.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Jing X, Jaw J, Robinson HH, Schubot FD. Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins. 2010;78:1631–1640. doi: 10.1002/prot.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castilla-Llorente V, Salas M, Meijer WJ. KinC/D-mediated heterogeneous expression of spo0A during logarithmical growth in Bacillus subtilis is responsible for partial suppression of phi 29 development. Mol. Microbiol. 2008;68:1406–1417. doi: 10.1111/j.1365-2958.2008.06234.x. [DOI] [PubMed] [Google Scholar]
- 32.Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 33.Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol. Microbiol. 1999;33:389–395. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- 34.Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 2002;46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- 35.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 38.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrity LF, Ordal GW. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology. 1997;143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- 40.Aronson HE, Royer WE, Jr., Hendrickson WA. Quantification of tertiary structural conservation despite primary sequence drift in the globin fold. Protein Sci. 1994;3:1706–1711. doi: 10.1002/pro.5560031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grasberger B, Minton AP, DeLisi C, Metzger H. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. USA. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger H. Transmembrane signaling: the joy of aggregation. J. Immunol. 1992;149:1477–1487. [PubMed] [Google Scholar]
- 43.Schneider TR. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr. D. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- 44.Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 2003;185:285–294. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anantharaman V, Aravind L. Cache - a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 2000;25:535–537. doi: 10.1016/s0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- 46.Mougel C, Zhulin IB. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 2001;26:582–584. doi: 10.1016/s0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- 47.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 2010;13:116–123. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. A PAS domain binds asparagine in the chemotaxis receptor MCPB in Bacillus subtilis. J. Biol. Chem. 2009;285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirby JR, Saulmon MM, Kristich CJ, Ordal GW. CheY-dependent methylation of the asparagine receptor, McpB, during chemotaxis in Bacillus subtilis. J. Biol. Chem. 1999;274:11092–11100. doi: 10.1074/jbc.274.16.11092. [DOI] [PubMed] [Google Scholar]
- 50.Zimmer MA, Szurmant H, Saulmon MM, Collins MA, Bant JS, Ordal GW. The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol. Microbiol. 2002;45:555–568. doi: 10.1046/j.1365-2958.2002.03035.x. [DOI] [PubMed] [Google Scholar]
- 51.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Ohtake H, Kato J. Identification and characterization of the chemotactic transducer in Pseudomonas aeruginosa PAO1 for positive chemotaxis to trichloroethylene. J. Bacteriol. 2006;188:6700–6702. doi: 10.1128/JB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shitashiro M, Tanaka H, Hong CS, Kuroda A, Takiguchi N, Ohtake H, Kato J. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J. Biosci. Bioeng. 2005;99:396–402. doi: 10.1263/jbb.99.396. [DOI] [PubMed] [Google Scholar]
- 53.Szurmant H, Bunn MW, Cho SH, Ordal GW. Ligand-induced conformational changes in the Bacillus subtilis chemoreceptor McpB determined by disulfide crosslinking in vivo. J. Mol. Biol. 2004;344:919–928. doi: 10.1016/j.jmb.2004.09.093. [DOI] [PubMed] [Google Scholar]
- 54.Rost B, Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 58.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 59.Cowtan K. ‘DM’: An automated procedure for phase improvement by density modification. CCP4/ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 60.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 65.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 66.Crippen GM, Havel TF. Stable calculation of coordinates from distance information. Acta Crystallogr. A. 1978;34:282–284. [Google Scholar]
- 67.DeLano WL. The PyMOL Molecular Graphics System DeLano Scientific. Palo Alto, CA, USA: 2002. [Google Scholar]
- 68.Gouet P, Courcelle E. ENDscript: a workflow to display sequence and structure information. Bioinformatics. 2002;18:767–768. doi: 10.1093/bioinformatics/18.5.767. [DOI] [PubMed] [Google Scholar]
- 69.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 70.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 72.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.