Summary
Rational design of functional enzymes with high turnovers is a significant challenge, especially those with complex active site and difficult reactions, such as in respiratory oxidases. Introducing 2 His and 1 Tyr into myoglobin resulted in designed enzymes that reduce O2 to H2O with > 1000 turnovers and minimal release of reactive oxygen species. This also showed that presence and positioning of Tyr, not Cu, are critical for activity.
Keywords: Heme Copper Oxidases, Metalloenzymes, Metalloprotein Design, Oxidoreductases, Protein Design
Much progress has been made in designing metalloproteins with structures similar to native enzymes[1] and advances in computational biology have allowed for rational design of function as well[2]. Despite these achievements, most designed enzymes have simple active site structures and low activities with limited turnover numbers. Designing artificial enzymes with higher complexity and numbers of turnovers is not only an important measure of success in the field, but can also reveal structural features responsible for tuning enzymatic activities and may result in artificial enzymes for practical applications. A primary example of a complex metalloenzyme with an important function is the family of terminal oxidases, such as heme-copper oxidases (HCOs)[3] and bd oxygen reductases[4], which catalyze the kinetically difficult reduction of O2 to water and, in doing so, generate the transmembrane proton gradient that drives important processes such as ATP synthesis. While many catalysts that reduce oxygen to water have been reported[5], a long-standing challenge is to carry out the reaction without the production of reactive oxygen species (ROS), such as superoxide and peroxide, under mild conditions. ROS not only damage biomolecules in cells and components in fuel cells, but also decrease energy efficiency, as ROS are a result of incomplete catalysis. In addition, efficient catalysts using non-precious metal ions such as iron or copper will greatly decrease costs in practical applications[6]. Therefore, designing enzymes that catalyse complex reactions, such as oxygen reduction, at mild pH that are based on small, stable, and easy to produce scaffold proteins, would yield ideal models for studying the fundamentals of this chemistry and produce useful catalysts for applications such as fuel cells.
In HCOs the active site responsible for O2 reduction is a heterobinuclear metal center containing a heme-Fe and a His-ligated Cu (CuB). We have previously reported the introduction of two histidines in the heme pocket of sperm whale myoglobin (swMb) through Leu29His and Phe43His mutations[7], which, together with the native distal His64 formed a copper binding site (this mutant is called CuBMb, see Supporting Information, Figure S1A). As purified, CuBMb has no metal in the CuB site (designated as E-CuBMb; i.e. empty). Introducing the copper binding site into myoglobin transformed it from a simple oxygen carrier into a copper-dependent heme oxygenase at pH 8[8], which degrades the heme cofactor to verdoheme. Since the active site Tyr in native HCOs has been shown to be critical to the function of these enzymes[9], we hypothesized that the lack of a tyrosine next to one of the histidine ligands in copper loaded CuBMb (Cu-CuBMb) may limit conversion of O2 to water. Herein, we report the introduction of a Tyr at two different positions close to the histidines of the putative copper binding site of CuBMb and demonstrate that these artificial enzymes are able to efficiently reduce O2 to water, one with over 1000 turnovers, and with minimal release of superoxide or peroxide. These results demonstrate that both the presence and positioning of the tyrosine is important for terminal oxidase activity.
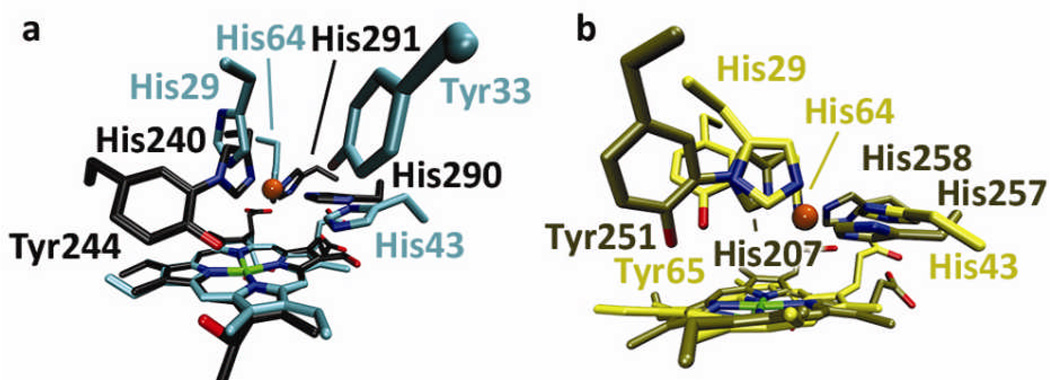
Since the design and activity of the CuBMb mutant were first reported[7–8], the crystal structure of E-CuBMb has been solved; the histidine ligands in this structure overlay well with those of bovine HCO (PDB 1V54) (see Supporting Information, Figure S1A) and this structure was used to guide the placement of Tyr in the active site pocket[11]. Since Tyr244 is four residues away from His240 in the primary sequence of bovine HCO, we first placed an analogous Tyr four residues from His29 via a Phe33Tyr mutation (called F33Y-CuBMb). The crystal structure of E-F33Y-CuBMb shows excellent agreement with the computer model (Figure S1B). A crystal structure of Cu-F33Y-CuBMb has also been obtained (Figures S1 and S2). However, although Tyr33 points into the Cu-binding site and is next to His29, Tyr33 does not overlay well with Tyr244 of bovine HCO (Figure 1A). To find a better structural overlay, a recent crystal structure of a cbb3 HCO[10] revealed that the Tyr can be near the His ligand spatially, while not necessarily close in the primary sequence. Therefore, we alternatively modelled Tyr next to His29 through a Gly65Tyr mutation (called G65Y-CuBMb). The energy-minimized computer model of E-G65Y-CuBMb overlays very well with that of cbb3 HCO (Figure 1B).
Figure 1.
Structures and computer models of native and designed oxidases. a) Overlay of the crystal structures of bovine CcO (grey) and E-F33Y-CuBMb (cyan); b) Overlay of the crystal structure of cbb3 HCO from Pseudomonas stutzeri[10] (tan) and E-G65Y-CuBMb computer model (yellow); The CuB copper in CcO is represented as an orange sphere.
The spectral properties of oxidized and reduced states of these variants are similar to those of wild type swMb (WTswMb)[12] (Supporting Information, Figure S4). However, upon exposure of the ferric- states of both E-F33Y-CuBMb and E-G65Y-CuBMb to excess reductant (ascorbate with N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) as a mediator) in air saturated buffer in a sealed cuvette, a transition from ferric through oxy to deoxy was observed (Supporting Information, Figure S4E–F), which is in contrast to WTswMb (Supporting Information, Figure S4D), and prompted an investigation of the product of this reduction.
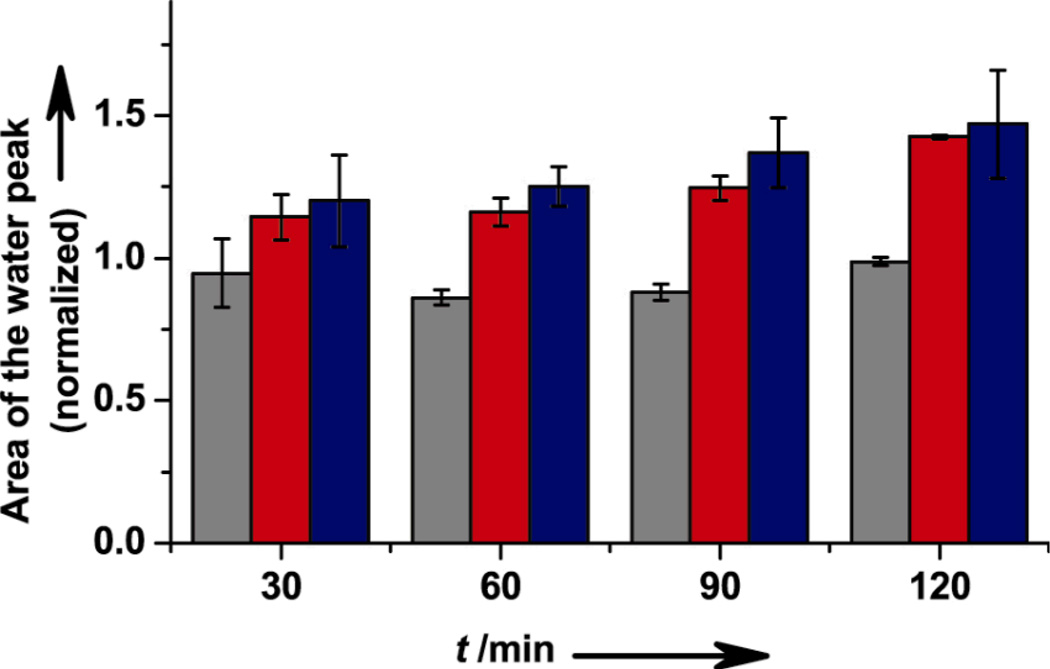
To confirm that the product of oxygen reduction is water, 17O NMR was carried out. Production of H217O above the background level (natural abundance) of H217O in water was monitored in a sealed vessel containing 17O2, the myoglobin variant, and excess reductant, using 17O-labeled Tyr as an external standard. A typical 17O NMR spectrum is shown in Supporting Information, Figure S5. Figure 2 shows the ratios of the H217O peak area to 17O-Tyr peak area at various time points after addition of 17O2 to initiate its reduction by WTswMb, E-F33Y-CuBMb, and E-G65Y-CuBMb, normalized to the area of the peak before initiating the reaction. The normalized ratio of water in the WTswMb sample remains close to 1, suggesting that the protein did not produce any new H217O. In contrast, E-F33Y-CuBMb and E-G65Y-CuBMb produced up to 10 mM H217O at 120 min, as indicated by an increase in the normalized ratio, confirming the production of water.
Figure 2.
Characterization of product by 17O NMR spectroscopy; normalized area of the H217O peak for WTswMb (gray), E-F33Y-CuBMb (red) and E-G65Y-CuBMb (blue) at 30, 60, 90, and 120 minutes. Error bars indicate standard deviation.
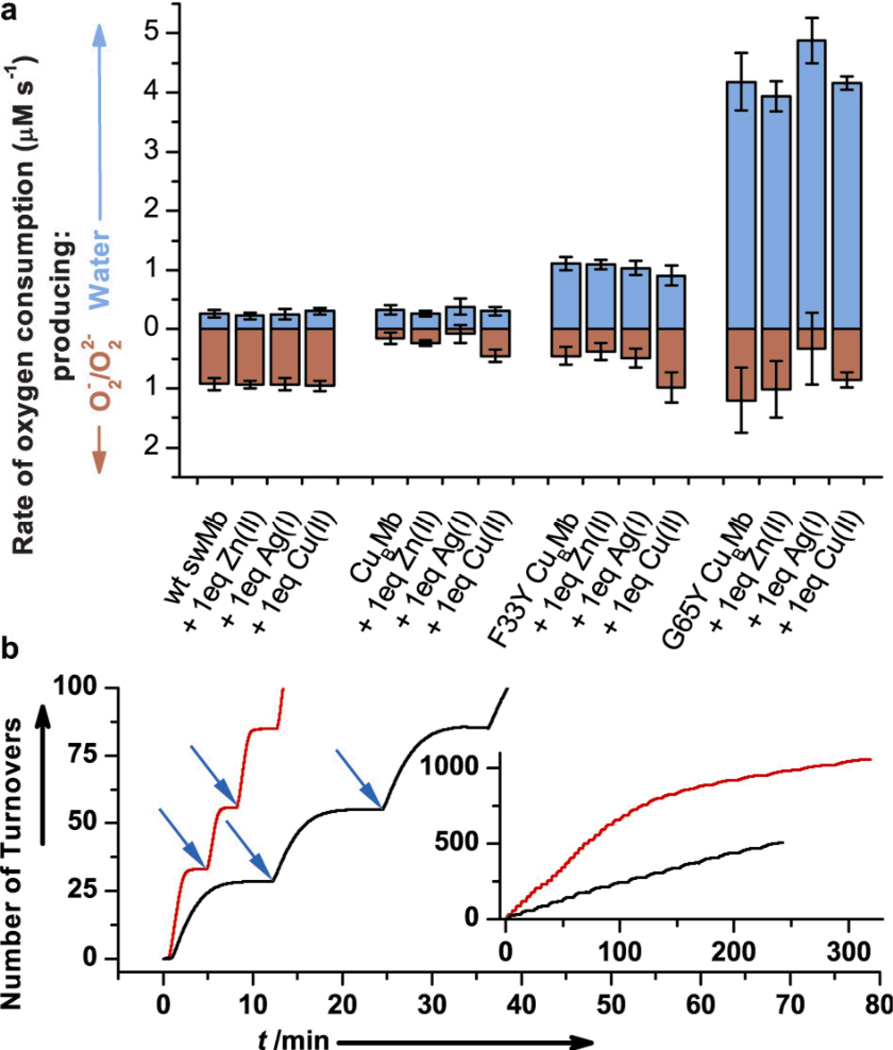
The rates of oxygen reduction were measured quantitatively using an O2 electrode to monitor the concentration of O2 over time in the presence of reductant, similar to protocols reported for native HCO[13]. The rate of O2 disappearance was measured for WTswMb and the CuBMb variants (Supporting Information, Figure S6). A hallmark of native terminal oxidases is the clean reduction of O2 to water with minimal release of superoxide or peroxide[14]. To identify the product (O2−, O22−, or H2O) we employed superoxide dismutase (SOD) and catalase which selectively react with superoxide and peroxide, respectively, producing O2 as one of the products, which would slow the apparent rate proportional to the amount of ROS released. By comparing the rates of reduction in the absence of and in the presence of SOD and catalase, the portion of O2 reduction due to water formation (in blue) and due to superoxide or peroxide formation (in red) can be calculated (see Figure 3A, and Supporting Information Table S5 and Supporting Text). Not surprisingly, most of the O2 consumption rate by WTswMb is due to superoxide or peroxide formation, consistent with autoxidation[15] and these rates remain unaffected in the presence of Cu2+, Zn2+ or Ag+. Interestingly, introducing two histidine residues into the distal pocket of WTswMb results in substantial inhibition of superoxide/peroxide formation in comparison to WTswMb, but does not significantly contribute to water formation, and therefore results in a decrease in the overall rate of O2 consumption. Similar to WTswMb, addition of Cu2+, Ag+ or Zn2+ to E-CuBMb does not perturb the rate and product of O2 reduction. In contrast to both WTswMb and CuBMb, introducing a Tyr next to the one of the His ligands at either position 33 or 65 results in a dramatic increase in water formation and overall rate of O2 consumption. The rate of water production increases up to 4.2 and 1.1 µM/s when using 18 µM E-G65Y-CuBMb and E-F33Y-CuBMb, respectively. To our knowledge, this is the first time that Tyr and its specific positioning has been shown to play an important role in directing the product of O2 reduction to water formation instead of release of superoxide or peroxide. Under further optimized conditions (with a higher concentration of reductant), the rate of O2 reduction to water by E-G65Y-CuBMb is 28 min-−1 (Supporting Information, Figure S10) – within ~150 fold of native HCOs[16]. Additionally, we show in Supporting Information, Figure S7 that 1.8 mM and 18 mM cyanide, a known inhibitor of HCO activity, inhibits the activity of E-G65Y-CuBMb and E-F33Y-CuBMb, respectively.
Figure 3.
Characterization of the O2 reduction; a) Rates of oxygen reduction to form either water (blue) or superoxide/peroxide (red) with 18 µM WTswMb, CuBMb, F33Y-CuBMb or G65Y-CuBMb; b) Number of turnovers of O2 reduction by E-F33Y-CuBMb (black) and E-G65Y-CuBMb (red); blue arrow indicates addition of ~28 equivalents O2. Inset, region of high number of turnovers.
Surprisingly, the O2 reduction activity is independent of the presence and identity of the metal in the engineered CuB site (Figure 3A). To further support this finding, we repeated the studies with varying equivalents of copper (0–2 equivalent) as well as a strong metal chelator, ethylenediaminetetraacetate (EDTA), but no further change was noticed (Supporting Information, Figure S8). As E-G65Y-CuBMb is still ~150 fold lower in activity than HCOs,[16] our results are not intended to rule out the role of copper ion native HCOs. Alternatively, these results do indicate that a copper center is not strictly necessary for oxygen reductase chemistry and are consistent with recent discovery of bd oxygen reductases[4] that lack the CuB center and yet can still perform the oxidase activity.
More importantly, this study demonstrates the importance of the presence and positioning of tyrosine in the active site. In both types of native oxidases the reaction is either known or proposed to proceed by the critical steps of two electron reduction of the oxygen to a peroxo intermediate, followed by rapid protonation and heterolytic O-O bond cleavage, leading to a transient ferryl intermediate; it is also proposed that the conserved tyrosine in HCOs is involved in donating an electron.[3–4] To elucidate the potential role of the Tyr, we have obtained the crystal structure of E-F33Y-CuBMb. When comparing with E-CuBMb and E-F33Y-CuBMb with WTswMb, we found that introducing two histidine residues and then an additional tyrosine residue has led to stabilization of two and three water molecules, respectively, in the distal pocket (Supporting Information, Figure S11), in comparison to one water molecule in the distal pocket of WTswMb. In addition, where WTswMb has only one hydrogen bond partner available to interact with the bound oxygen (H64), two and three more hydrogen bonding capable residues are available in E-CuBMb and E-F33Y-CuBMb, respectively. Studies of crystal structures of various states of HCOs have suggested a role for water molecules and an extended hydrogen bonding network in the oxygen reduction step[17], and computational studies[18] of HCOs have supported the role of similar interactions in oxidase activity. Based on these studies, we propose that tyrosine and its associated hydrogen-bonding network works to activate the ferric-superoxo state of our designed enzymes and this activation allows it to accept a second electron from the exogenous reductant towards complete reduction to water. Further studies are underway to elucidate the exact role of Tyr in our model protein system.
Finally, to test the extent of the functional activities, we have carried out multiple turnover reactions. To a solution containing E-F33Y-CuBMb or E-G65Y-CuBMb and excess reductant, ~500 µM (28 equivalents) O2 was added repeatedly and each time the total O2 consumption was monitored using an O2 electrode. These stepwise additions resulted in the multiple plateaus observed in Figure 3B. We calculate that E-F33Y-CuBMb and E-G65Y-CuBMb achieved >505 and > 1056 turnovers, respectively (Figure 3B, inset).
In summary, we have demonstrated the first successful design of a functional protein capable of cleanly reducing oxygen to water with minimal release of superoxide or peroxide, similar to the activity of terminal oxidases, with more than 1000 turnovers. Through these designed functional proteins, we have also shown that Tyr next to one of the copper coordinating His ligands plays a critical role in directing O2 reduction to water formation. Furthermore, the positioning of this Tyr is critical for affecting the rate of catalysis. Even though the designed proteins are still less active than terminal oxidases, it is remarkable that oxygen reduction to water was conferred to a much smaller protein, myoglobin, with only three mutations of the distal pocket. Given their high enzymatic turnovers, smaller size, higher stability, and higher expression yield, these designed enzymes will serve as ideal models for a more detailed understanding of terminal oxidases and allow for potential applications in biology and alternative energy.
Supplementary Material
Acknowledgments
We would like to thank Profs. Chad Rienstra and Robert Gennis for the kind use of their oxygen electrodes, Dr. Deborah Stoner-Ma of Brookhaven National Lab (BNL) for help with X-ray data collection of F33Y CuBMb, Dr. Ying-Wu Lin for helpful discussions, and Dr. Nicholas Marshall for help editing this manuscript. This work was supported by the US National Institute of Health (GM062211).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Kyle D. Miner, Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA
Arnab Mukherjee, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA.
Yi-Gui Gao, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA.
Eric L. Null, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA
Igor D. Petrik, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA
Xuan Zhao, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA.
Natasha Yeung, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA.
Howard Robinson, Department of Biology, Brookhaven National Laboratory, Upton, New York, 11973, USA.
Yi Lu, Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA, yi-lu@illinois.edu; Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois, 61801, USA.
References
- 1.a) Das R, Baker D. Annu. Rev. Biochem. 2008;77:363. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]; b) Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Calhoun JR, Nastri F, Maglio O, Pavone V, Lombardi A, DeGrado WF. Biopolymers. 2005;80:264. doi: 10.1002/bip.20230. [DOI] [PubMed] [Google Scholar]; d) Regan L, DeGrado WF. Science. 1988;241:976. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]; e) Bolon DN, Mayo SL. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14274. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Robertson DE, Farid RS, Moser CC, Urbauer JL, Mulholland SE, Pidikiti R, Lear JD, Wand AJ, DeGrado WF, Dutton PL. Nature. 1994;368:425. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]; g) Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 2.a) Korendovych IV, Senes A, Kim YH, Lear JD, Fry HC, Therien MJ, Blasie JK, Walker FA, DeGrado WF. J. Am. Chem. Soc. 2010;132:15516. doi: 10.1021/ja107487b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Nature. 2009;458:305. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. Nature. 2009;461:1300. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yeung N, Lin Y-W, Gao Y-G, Zhao X, Russell BS, Lei L, Miner KD, Robinson H, Lu Y. Nature. 2009;462:1079. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Watanabe Y, Hayashi T. Prog. Inorg. Chem. 2005;54:449. [Google Scholar]; g) Ghosh D, Pecoraro VL. Curr. Opin. Chem. Biol. 2005;9:97. doi: 10.1016/j.cbpa.2005.02.005. [DOI] [PubMed] [Google Scholar]; h) Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Nat Chem. 2012;4:118. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Namslauer A, Brzezinski P. FEBS Lett. 2004;567:103. doi: 10.1016/j.febslet.2004.04.027. [DOI] [PubMed] [Google Scholar]; b) Babcock GT, Wikstrom M. Nature. 1992;356:301. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]; c) Ferguson-Miller S, Babcock GT. Chem. Rev. 1996;96:2889. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 4.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. Biochim. Biophys. Acta. 2011;1807:1398. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Che CM, Chiang HJ, Margalit R, Gray HB. Catal. Lett. 1988;1:51. [Google Scholar]; b) Kim E, Chufan EE, Kamaraj K, Karlin KD. Chem. Rev. 2004;104:1077. doi: 10.1021/cr0206162. [DOI] [PubMed] [Google Scholar]; c) Collman JP, Devaraj NK, Decreau RA, Yang Y, Yan Y-L, Ebina W, Eberspacher TA, Chidsey CED. Science. 2007;315:1565. doi: 10.1126/science.1135844. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Collman JP, Decreau RA. Chem. Commun. 2008:5065. doi: 10.1039/b808070b. [DOI] [PubMed] [Google Scholar]; e) Halime Z, Kotani H, Li Y, Fukuzumi S, Karlin KD. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13990. doi: 10.1073/pnas.1104698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Bashyam R, Zelenay P. Nature. 2006;443:63. doi: 10.1038/nature05118. [DOI] [PubMed] [Google Scholar]; b) Lefevre M, Proietti E, Jaouen F, Dodelet J-P. Science. 2009;324:71. doi: 10.1126/science.1170051. [DOI] [PubMed] [Google Scholar]
- 7.Sigman JA, Kwok BC, Lu Y. J. Am. Chem. Soc. 2000;122:8192. [Google Scholar]
- 8.a) Sigman JA, Kim HK, Zhao X, Carey JR, Lu Y. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3629. doi: 10.1073/pnas.0737308100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang N, Zhao X, Lu Y. J. Am. Chem. Soc. 2005;127:16541. doi: 10.1021/ja052659g. [DOI] [PubMed] [Google Scholar]; c) Zhao X, Nilges MJ, Lu Y. Biochemsitry. 2005;44:6559–6564. doi: 10.1021/bi047465c. [DOI] [PubMed] [Google Scholar]; d) Zhao X, Yeung N, Wang Z, Lu Y. Biochemistry. 2005;44:1210–1214. doi: 10.1021/bi0479151. [DOI] [PubMed] [Google Scholar]
- 9.Kaila V, Verkhovsky MI, Wikstrom M. Chem. Rev. 2010;110:7062. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 10.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. Science. 2010;329:327. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 11.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15304. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonini E, Brunori M, Neuberger A, Tatum EL. Hemoglobin and Myoglobin in their Reactions with Ligands. Vol. 21. New York, NY: Elsevier; 1971. [Google Scholar]
- 13.Pawate AS, Morgan J, Namslauer A, Mills D, Brzezinski P, Ferguson-Miller S, Gennis RB. Biochemistry. 2002;41:13417. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- 14.a) Brzezinski P, Gennis RB. J. Bioenerg. Biomembr. 2008;40:521. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Koepke J, Olkhova E, Angerer H, Mueller H, Peng G, Michel H. Biochim. Biophys. Acta. 2009;1787:635. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]; c) Aoyama H, Muramoto K, Shinzawa-Itoh K, Hirata K, Yamashita E, Tsukihara T, Ogura T, Yoshikawa S. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2165. doi: 10.1073/pnas.0806391106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Egawa T, Lee HJ, Gennis RB, Yeh S-R, Rousseau DL. Biochim. Biophys. Acta. 2009;1787:1272. doi: 10.1016/j.bbabio.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brantley RE, Jr, Smerdon SJ, Wilkinson AJ, Singleton EW, Olson JS. J. Biol. Chem. 1993;268:6995. [PubMed] [Google Scholar]
- 16.Chang H-Y, Hemp J, Chen Y, Fee JA, Gennis RB. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16169. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshikawa S, Muramoto K, Shinzawa-Itoh K. Biochim. Biophys. Acta. 2011;1807:1279–1286. doi: 10.1016/j.bbabio.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Blomberg MRA, Siegbahn PEM, Wikstroem M. Inorg. Chem. 2003;42:5231. doi: 10.1021/ic034060s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.