Abstract
Bacteriophages T7, λ, P22, and P2/P4 (from Escherichia coli), as well as ϕ29 (from Bacillus subtilis), are among the best-studied bacterial viruses. This chapter summarizes published protein interaction data of intraviral protein interactions, as well as known phage–host protein interactions of these phages retrieved from the literature. We also review the published results of comprehensive protein interaction analyses of Pneumococcus phages Dp-1 and Cp-1, as well as coliphages λ and T7. For example, the ≈55 proteins encoded by the T7 genome are connected by ≈43 interactions with another ≈15 between the phage and its host. The chapter compiles published interactions for the well-studied phages λ (33 intra-phage/22 phage-host), P22 (38/9), P2/P4 (14/3), and ϕ29 (20/2). We discuss whether different interaction patterns reflect different phage lifestyles or whether they may be artifacts of sampling. Phages that infect the same host can interact with different host target proteins, as exemplified by E. coli phage λ and T7. Despite decades of intensive investigation, only a fraction of these phage interactomes are known. Technical limitations and a lack of depth in many studies explain the gaps in our knowledge. Strategies to complete current interactome maps are described. Although limited space precludes detailed overviews of phage molecular biology, this compilation will allow future studies to put interaction data into the context of phage biology.
I. INTRODUCTION
Bacteriophages have been extremely important model systems in molecular biology. They were critical to many breakthrough discoveries, such as the nature of the genetic material (Hershey, 1952), the structure of genes and genomes and the first genome sequence (Sanger et al., 1977), recombination (Kaiser and Jacob, 1957), transcriptional regulation (e.g., Roberts, 1969), and many others. In addition, phages have become important technical tools in molecular genetics and nanobiology, but these aspects are not reviewed here.
A great deal of work has been invested in learning the mechanistic details of the life cycles of a number of phages, including those described in this chapter. Although phages are tiny biological systems, surprisingly, not even the biology of main “model system phages” is completely understood. Despite a plethora of papers, few comprehensive summaries exist of the molecular interaction events that occur in both phage gene regulation and virion assembly. As in all organisms, protein–protein interactions (PPIs) play a fundamental role in phage biology. PPIs are important in all virus-related processes, including host infection, transcriptional regulation, DNA replication, virion assembly, and lysis. Current protein interaction databases (Lehne and Schlitt, 2009) have consistently neglected phage interactions. For instance, Intact, one of the most comprehenisve databases for protein–protein interactions, listed only 22 interactions for Caudovirales, the only phage group represented at the time of this writing (April 2011). We have thus attempted to compile such information for several phages that infect Gram-negative hosts, including λ, T7, P22, and P2/P4, as well as others that infect Gram-positive bacteria, specifically ϕ29, Dp-1, and Cp-1 (Table I). Coliphage T4 is one of the best understood of all the obligate lytic phages. T4 has been an outstanding model for phage biology, particularly for DNA replication, control of transcription, and virion assembly. Unfortunately, this chapter cannot discuss the extensive T4 PPIs here for reasons of space. Many of the small DNA phages, such as ϕX174 or M13, have been very well studied, but because of their tiny proteomes they do not have many protein–protein interactions. It would be highly desirable to establish their PPIs so that more comprehensive analyses of phage systems biology would be possible. Nevertheless, it is hoped that this chapter proves useful to the phage community and encourages similar compilations for other phages. The interactions collected for this chapter are available in Intact (http://www.ebi.ac.uk/intact/) for easy access.
TABLE I.
Summary of phage interactomes described in this chaptera
| Phage | Proteins | Phage–phage PPIs | Phage–host PPIs |
|---|---|---|---|
| T7 | 55 | ~43 (including 7 HTSb) | 15 |
| λ | 73 | 33 (+ 81 HTS) | 26 |
| P22 | 64 | 38 | 9 |
| P2/P4 | 42/13 | 14 | 3 |
| ϕ29 | 30 | 20 | 3 |
| Dp-1 | 72 | 156 (HTS) | 38 (HTS) |
| Cp-1 | 28 | 17 (HTS) | 11 (HTS) |
Note that all interactions from this chapter are available as a downloadable supplement from http://www.uetz.us.
High-throughput data (see text for details).
II. METHODS USED TO DETECT PROTEIN–PROTEIN INTERACTIONS
A. Genetic detection of interactions
Methods used to study protein–protein interactions in phages have evolved over the decades. A few important methods are illustrated in Figure 1. In the beginnings of phage molecular biology, detection of genetic interactions dominated. For instance, certain mutants in the λ J gene abolished virion binding to host Escherichia coli cells. Similarly, mutants of the E. coli gene lamB made the cells resistant to λ infections. From such experiments it was concluded that J protein bound to LamB and that LamB was the λ receptor. This hypothesis has been confirmed by isolating compensatory mutations in both genes that regenerate the interaction (Werts et al., 1994).
FIGURE 1.
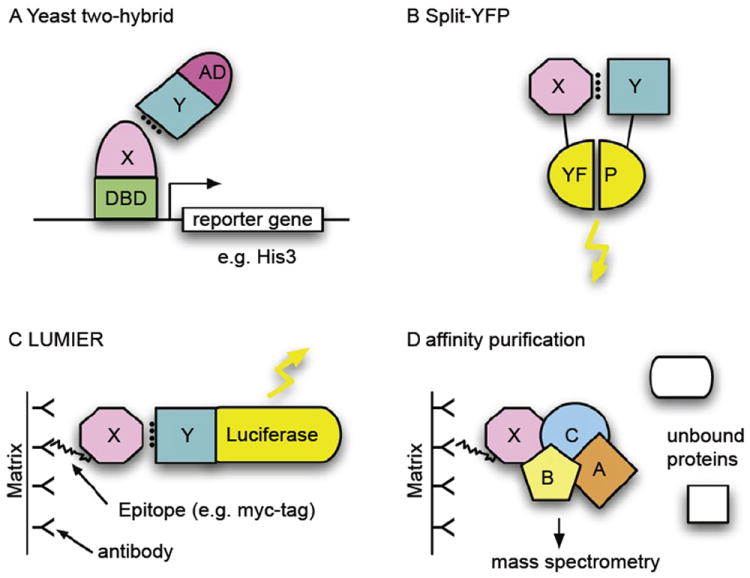
Selected methods for the study of protein–protein interactions. (A) The yeast two-hybrid (Y2H) system is based on the levels of two fusion proteins (typically in yeast but any cell type can be used). One of the proteins contains a DNA-binding domain (DBD), which can bind to the promoter of a reporter gene (here: HIS3), and a second protein X, the bait. The second fusion protein consists of a transcription-activation domain (AD) and a second protein, Y. If proteins X and Y interact, a transcription factor is formed and the reporter gene is activated. In this case, that means that the cell can grow on histidine-free medium. A yeast colony growing on such medium thus indicates an interaction of the two inserted proteins. (B) Protein complementation assay (PCA), e.g., split-YFP. As in the Y2H assay, two interacting proteins bring together two protein fragments that are inactive when separate but active when in close proximity. Here, fragments of yellow-fluorescent protein (YFP) reassociate and fluoresce when reassembled. Other fluorescent proteins, such as the green fluorescent protein (GFP), have been used in a similar way. (C) LUMIER (LUMInescence-based mammalian intERactome). Two fusion proteins are purified by means of an epitope tag (here: FLAG tag), usually on an antibody-coated matrix. Interactions between X and Y can be detected using Luciferase, whose gene is fused to Y and which emits light when luciferin is added. (D) Affinity purification. Protein complexes can be purified from cellular lysates using an affinity epitope, as in (C) with a FLAG tag. Components of the complex can then be identified using mass spectrometry, using the unique mass of peptides when the protein is digested by trypsin.
B. Electron microscopy
Another important method for the investigation of physical relationships between phage components is electron microscopy, especially in combination with genetic analysis. Mutants often have an aberrant morphology. If certain structures are missing in the mutant virion, it can be concluded that the mutated gene encodes this structure or is at least involved in its assembly. For instance, many laboratory strains of λ harbor a frameshift mutation in the stf gene, and these mutants lack side tail fibers (Hendrix and Duda, 1992). Many such studies not only showed which genes encode which proteins, but also indicated how the proteins interact (e.g., when particular structures could be “removed” from the virion by defined gene knockouts).
More recently, many bacteriophage capsids and phage protein complexes have been analyzed by cryoelectron microscopy and three-dimensional reconstructions. This technique gives resolutions approaching that of X-ray crystallography, especially for proteins present in symmetric arrays in the virion, and has proven ideal for analyzing protein–protein interactions in large complexes, especially in combination with high-resolution structural data from other techniques, such as X-ray crystallography.
C. Yeast two-hybrid screens
More recently, the yeast two-hybrid system (Y2H) has become an important method to detect pairwise protein interactions. Interaction screens of phage proteins can be carried out easily by cloning all open reading frames in appropriate vectors and then testing these in a systematic pairwise fashion. This has been done for several phages, including coliphages T7 (Bartel et al., 1996) and λ (Rajagopala et al., 2011), as well as Streptococcus pneumoniae phages Cp-1 and Dp-1 (Häuser et al., 2011; Sabri et al., 2010). The whole process can also be automated.
One disadvantage of the Y2H system is that a typical pairwise screen, also called an “array screen” because of the systematic arrangement of clones in microtiter plates, may only detect 20–30% of all interactions. That is, the rate of false negatives is on the order of 70–80%. This can, in theory, be overcome by using multiple vectors that differ in the structure of the fusion proteins, for instance in the production of N- or C-terminal fusion proteins (Stellberger et al., 2010). A combination of multiple vectors can achieve interaction detection rates of up to 80% (i.e., false negative rates as low as 20%) (Chen et al., 2010).
Y2H also suffers from false positives that can only be understood with additional experimentation. Nonetheless, a multivector Y2H strategy can help in the recognition of false positives. For instance, if multiple vector combinations are used, interactions found in all or most cases are more likely to be “true positives,” whereas interactions detected in a lower fraction of cases are less reliable and may be false positives.
D. Protein complex purification and mass spectrometry
Whole phage virions can be purified, typically by CsCl density gradient centrifugation. Proteins from purified phage can then be denatured and separated on polyacrylamide gels or subjected directly to trypsin digestion and subsequent liquid chromatography (LC) and mass spectrometry (MS). The mass of peptides determined by MS can usually identify the corresponding proteins unambiguously, and thus composition of the phage particles. Once all proteins in a virion are identified, at least some can be assigned easily to known structures, such as the icosahedral capsid or tail sheath. Typically, only a fraction of all phage-encoded proteins are represented in the mature virion (e.g., less than 20 of the ≈70 phage λ proteins). Especially for little-studied phages it can be extremely helpful to know which proteins are in the virion. For example, in an analysis of Streptococcus phage Dp-1, only 8 of the 72 predicted proteins were identified by LC/MS and were thus confirmed structural proteins (see later).
E. Biochemistry of protein complexes
Biochemical approaches often involve the purification of protein complexes and thus yield protein interaction information. This approach can be made technically easier if individual proteins are affinity tagged. For instance, phage-encoded proteins can often be fused to affinity column-binding proteins such as glutathione-S-transferase or oligohistidine without affecting their function. Such tagged proteins and proteins bound to them can then be purified using affinity reagents. The untagged proteins in the complex can then be identified by mass spectrometry or immunoblotting (if antibodies against specific proteins are available). A range of experimental techniques is available for the characterization of phage protein complexes and their activities; a comprehensive overview cannot be given here. For instance, chemical cross-linking of interacting proteins is used frequently to spatially locate physically associated proteins, and variable spacer lengths can determine the steric proximity of the binding partners. Other tests include activity assays (e.g., proteases or other modifying enzymes that may be required for phage assembly), replication, or other processes. Some examples of such activities are described here.
F. X-ray crystallography and nuclear magnetic resonance
Only atomic structures of all phage proteins and their interactions will give a mechanistic understanding of phage biology, and progress in this area has been accelerating. Individual proteins and protein complexes, if not the whole phage particle, can be crystallized and the structure of such proteins solved by X-ray analysis. Alternatively, protein structures can be solved by nuclear magnetic resonance (NMR) if high enough concentrations can be achieved. Unfortunately, size restrictions on NMR structural determinations limit the analysis of large protein complexes, but monomeric structures can be extremely useful in devising testable models for the interactions proteins may undergo. However, many phage proteins are refractory to structural analysis (especially those involved in virion assembly, as they often assemble only at high concentrations), and even in classical model systems such as phage λ less than a third of all phage-encoded proteins have been crystallized—most of these as pure proteins rather than as protein complexes (Rajagopala et al., 2011;Yura et al., 2006).
III. PROTEIN–PROTEIN INTERACTIONS OF BACTERIOPHAGES
A. Coliphage T7
Bacteriophage T7 was defined in 1945 as a member of the seven Type (“T”) phages that grow lytically on E. coli B (Demerec and Fano, 1945), although it is probably identical to phage δ, used earlier by Delbrück; a close relative of T7 was likely studied by d’Herelle in the 1920s (d’Herelle, 1926). Relatives of T7 exhibit high synteny across their entire genomes, with the closest relatives differing primarily only by the number and location of homing endonucleases or other selfish elements not known to have any role in phage development. These phages therefore will exhibit life cycles and PPIs that parallel T7 itself, modified only by the resources available in their host organisms.
1. Growth physiology
T7 grows on rough strains of E. coli (i.e., those without full-length O-antigen polysaccharide on their surface) and some other enteric bacteria, but close relatives have been described that infect smooth and even capsulated strains (Molineux, 2006a). In such phages, tail fibers present on T7 are replaced with tailspikes with enzymatic activity that degrades the O- or K-antigens on the cell surface. T7 has a short latent period of 17 min at 37°C, which has resulted in most physiological studies being conducted at 30°C where infected cells lyse after 30 min. However, adaptation of T7 to high fitness with an excess of an E. coli K-12 strain growing under optimal conditions at 37°C in rich media results in a phage with a latent period of only ≈11 min. This adapted phage can undergo an effective expansion of its population by more than 1013 in 1 hr of growth (Heineman and Bull, 2007). There is no obvious subtlety in the T7 developmental pathway: the early region of its genome is transcribed efficiently by the host RNA polymerase, leading to synthesis of a phage-specific RNA polymerase and the concurrent inhibition of the host enzyme. Thus, T7 rapidly takes control of the transcriptional capacity of the infected cell. As E. coli mRNAs are also intrinsically unstable, T7 therefore also gains control of the translational capacity of the cell. Further, because T7 uses nucleotides derived from the host chromosome for its own replication, more than 80% of the DNA in progeny is derived from the bacterial chromosome and only a small burst is observed after infection of DNA-free minicells. T7 development is not known to be dependent on host chaperones, although it does code for two proteins that have been suggested to have chaperone activity in capsid and tail assembly. Aside from host components used for phage adsorption, plus general biosynthetic processes and the translational apparatus, T7 development is remarkably independent of host proteins: only E. coli RNA polymerase, thioredoxin—the cofactor for T7 DNA polymerase—and (deoxy)cytidine monophosphate kinase are known to be essential.
2. Early steps in infection
T7 is a member of Podoviridae, possessing only a short stubby tail that needs to be extended functionally in order to span the cell envelope, which thus allows its genome to be translocated into the cell. Tail extension is affected through internal head proteins that are ejected into the cell at the initiation of infection prior to or concomitant with transport of the leading end of the genome (Chang et al., 2010; Kemp et al., 2005). A schematic of the T7 virion is shown in Figure 2. In T7, the core consists of several copies each of three proteins (gp14, gp15, and gp16), which must interact, albeit perhaps loosely, with themselves and each other, in the phage capsid. However, during their ejection into the cell they must at least partially dissociate and also partially unfold in order to pass through the narrow channel through the portal and tail (Molineux, 2006b). The N-terminal domain of gp16 refolds spontaneously in the periplasm of the infected cell to form an enzyme with lytic transglycosylase activity (Moak and Molineux, 2000, 2004). This muralytic activity is only conditionally essential for T7 infection (Moak and Molineux, 2000).
FIGURE 2.
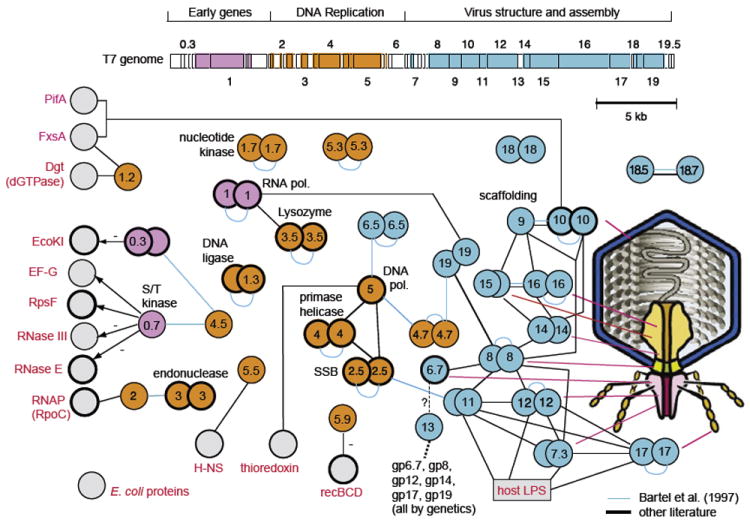
Phage T7: genome, virion, and interactome. ORFeome and protein interaction map of bacteriophage T7. Cloned open reading frames (ORFs) and random fragments were used to generate a two-dimensional interaction map. ORFeomes can also be used by structural genomics projects to derive three-dimensional structures. A combination of interaction maps and crystal structures often allows reconstruction of protein complexes as in viral particles or their subunits. (Top) T7 genome with each ORF represented as a box. ORFs indicated by integral numbers were identified originally as essential by genetic screens. Other genes have decimal numbers; of these, only 2.5 and 7.3 are essential. Proteins found to interact with other proteins are indicated by colored boxes; colors indicate a simplified assignment to functional classes as shown. Interactions between proteins are shown as lines, with self-interactions shown as dimers. Hatched lines indicate expected interactions that have not been found by two-hybrid analysis. Gray proteins correspond to host proteins (E. coli). Blue proteins are involved in virus assembly and structure, and, where known, their location in the virus particle is shown on the right. Proteins with thick borders have been crystallized and their structure determined. Genetic map modified after Dunn and Studier (1983). Figure modified after Uetz et al. (2004). Interactions based primarily on data in Table II (see references therein).
The C-terminal residues of gp16, likely together with a more central portion of gp15, also refold in the periplasm and then insert spontaneously into the cytoplasmic membrane, completing the channel across the cell envelope for DNA translocation into the cell. Likely the last of the core proteins to exit the phage head, gp14 is found exclusively within the outer membrane of the infected cell. Using energy derived from the membrane potential, these proteins then ratchet the leading 1 kb of the T7 genome into the cell (Chang et al., 2010; Kemp et al., 2005). In both T7 and T3, genome internalization by this process stops, and the remainder of the DNA is normally pulled into the cytoplasm as a consequence of host and then phage RNA polymerase-catalyzed transcription (Garcia and Molineux, 1995, 1996; Kemp et al., 2004). Several strong promoters lie on this leading 1-kb segment of the T7 genome.
3. Early genes
Transcription from T7 early promoters not only internalizes the remainder of the genome but also leads to expression of early T7 genes, some of which interact and inhibit or modulate the activity of host proteins (Molineux, 2006a). Of the early proteins, only T7 RNA polymerase is essential for phage growth. The first T7 protein to be synthesized is gp0.3, an anti-type I restriction protein (Studier, 1975). The structure of gp0.3 reveals that it resembles double-stranded DNA, and the protein binds to the specificity subunit of the heteropentameric EcoKI enzyme, preventing both it and the cognate modification enzyme from recognizing its target sequence (Atanasiu et al., 2001, 2002; Kennaway et al., 2009; Murray, 2000; Stephanou et al., 2009). T7 gp0.3 is active against all type I enzymes and thus T7 grows normally on B, C, and K-12 strains of E. coli regardless of its previous host. However, the protein affords no protection against type II and little, if any, against type III enzymes. Gp0.3 proteins of different T7-like phages fall into two distinct groups with no sequence similarity; both have anti-restriction activity but the group typified by T3 gp0.3 also has SAMase activity (Studier and Movva, 1976). The second major early protein whose function is known is gp0.7, a serine-threonine kinase (Rahmsdorf et al., 1974) that is also responsible for the shut off of host transcription. The two activities are separable (Simon and Studier, 1973). Gp0.7 phosphorylates several host proteins, including RNA polymerase (Severinova and Severinov, 2006; Zillig et al., 1975), the translational apparatus including EF-G and the small ribosomal subunit protein S6 (Robertson and Nicholson, 1990, 1992; Robertson et al., 1994), and both RNase III (Marchand et al., 2001) and RNase E (Mayer and Schweiger, 1983). T7 shuts down translation of host mRNAs but it remains unclear whether EF-G or S6 is involved directly. An unknown T7 late gene also modulates the specificity of RNase E (Savalia et al., 2010). Although it is likely that others do so, only 1 other of the 10 early T7 proteins is currently known to interact with host proteins. Gp1.2 binds to E. coli dGTPase, converting it into a rGTP-binding protein (Huber et al., 1988; Nakai and Richardson, 1990); gp1.2 also binds to the membrane protein FxsA and the F plasmid protein PifA (Cheng et al., 2004; Schmitt and Molineux, 1991; Wang et al., 1999).
4. T7 RNA polymerase
The hallmark of T7 is of course its phage-encoded RNA polymerase, which, together with its regulator T7 lysozyme, is responsible for most T7 gene expression. It is also essential for synthesizing RNA primers at the primary origin of replication (Fujiyama et al., 1981) and for initiation of DNA packaging (Zhang and Studier, 1997, 2004). In addition to several complexes containing a promoter and product RNAs (Cheetham and Steitz, 1999; Cheetham et al., 1999; Kennedy et al., 2007; Tahirov et al., 2002; Yin and Steitz, 2002, 2004), the crystal structure of the RNA polymerase–lysozyme complex has been determined (Jeruzalmi and Steitz, 1998). Many mutants in both RNA polymerase and lysozyme have been characterized that affect their functional interaction, but it is not obvious from the crystal structures how the mutations affect the regulation of transcription. Lysozyme is thought to target the initial transcribing complex in a promoter strength-dependent manner and have no effect on transcription elongation (Huang et al., 1999; Stano and Patel, 2004; Zhang and Studier, 1997, 2004). Lysozyme is, however, important in pausing or terminating transcription at the class II terminator CJ, where it probably interacts directly with the T7 terminase protein gp19 and thus directs the packaging machinery to create the DNA terminus (Lyakhov et al., 1997, 1998; Zhang and Studier, 2004).
After T7 RNA polymerase has been synthesized, there is no further requirement for the host polymerase and it actually becomes inhibitory to phage development. Inactivation is caused by the T7 gp2 protein, which binds to the β′ subunit and inhibits transcription by interfering with strand separation of the open complex at most promoters (Camara et al., 2010; Nechaev and Severinov, 1999). In vivo, the only T7 promoter that must be inhibited by gp2 is A3 (Savalia et al., 2010). Gp2 was found to interact with T7 endonuclease gp3 by a yeast two-hybrid screen (Bartel et al., 1996). An interaction between gp2 and gp3 (albeit not necessarily direct) was suggested earlier (Mooney et al., 1980), but the idea has not yet been critically evaluated experimentally.
5. DNA replication
T7 has been one of the primary model systems for understanding the biochemistry of DNA replication. Central to T7 replication is T7 DNA polymerase, which comprises a heterodimer of gp5 and the host thioredoxin. Binding of gp5 to thioredoxin is strong, with the latter serving as the processivity factor for DNA replication, increasing the length of DNA synthesized per binding event 100-fold. The interface between the two proteins also provides the docking site for binding of the other two proteins, SSB (gp2.5) and the primase–helicase gp4, which are central to T7 replication in vivo. The process of T7 replication in vitro has been reviewed recently (Hamdan and Richardson, 2009).
Two other proteins in the DNA replication cluster are known to interact with host proteins. The gene 5.5 product binds to the abundant nucleoid-associated protein H-NS (Ali et al., 2011) and in vitro relieves its inhibition of both E. coli and T7 RNA polymerase-catalyzed transcription (Studier, 1981). In vivo, T7 5.5 mutants direct reduced rates of DNA replication and exhibit a small plaque phenotype (Lin, 1992; Liu and Richardson, 1993; Studier, 1981). However, the phenotype persists in hns-defective strains, although an hns mutant that renders gene 5.5 essential has been isolated (Liu and Richardson, 1993). Gp5.5 also interacts with the λ proteins of RexAB system; it is not known whether this is a direct protein–protein interaction or whether gp5.5 interacts with a product of RexAB activity. A gene 5.5 missense, but not a null mutant, makes T7 sensitive to Rex-mediated exclusion. The product of gene 5.9 inhibits the E. coli RecBCD nuclease in a comparable manner to λ Gam protein; this interaction is also nonessential as mutants lacking gene 5.9 grow normally in rec(BCD)+ hosts (Lin, 1992).
The assembly of T7 virions involves several symmetry mismatches and many protein–protein interactions, some of which may be helped by other T7 proteins that potentially have a chaperone-like activity (Kemp et al., 2005). Note that, GenBank annotations notwithstanding, gp13 is not a structural component of the virion (Kemp et al., 2005). Assembly is thought to initiate with oligomerization of the portal protein gp8 to form a dodecameric ring, although both 12- and 13-mers are observed following overexpression of gene 8 (Cerritelli and Studier, 1996b; Kemp et al., 2005; Valpuesta et al., 1992, 2000). The pathway of prohead formation may then involve assembly of the internal core (gp14, gp15, and gp16) on the inner surface of the portal, perhaps concomitant with the recruitment of capsid protein gp10 hexamers bound to scaffolding protein gp9. Subsequent recruitment of gp10 pentamers and hexamers results in shell formation around a scaffold of gp9 (Cerritelli et al., 2003a). The 12-fold symmetrical portal protein faces a symmetry mismatch with the 5-fold vertex of the icosahedral capsid shell. The internal core of T7 contains 4 copies of gp16, 8 copies of gp15, and either 10 or 12 copies of gp14 (Agirrezabala et al., 2007; Cerritelli et al., 2003b; Kemp et al., 2005). These proteins are ejected into the infected cell in order to transport the phage genome into the cytoplasm, and the protein–protein interactions in the mature virion are likely quite different than those found in the infected cell. Gp14 lies at the base of the internal core and is therefore juxtaposed to the 12-fold symmetrical portal protein. Complicating our understanding of the T7 virion is the identification of ≈18 copies of the small gp6.7 protein, which is ejected into the outer membrane of the infected cell at the initiation of infection (Kemp et al., 2005). It is not known where gp6.7 is located; it likely forms part of the internal core or lies within the portal channel.
6. Tail
The outer face of the portal is the site of assembly of the sixfold symmetrical stubby tail. The tail is composed of three proteins—gp7.3, 11 and 12—to which six trimers of the tail fiber protein gp17 are attached. Each trimer consists of three domains: an N-terminal third that binds to the tail; this domain is conserved in phages related to T7 that have a different host range. This domain is followed by a triple-stranded coiled-coil of ≈120 residues and then a C-terminal half consisting of globular domains that interacts with the cell surface lipopolysaccharide (Steven et al., 1988). The body of the tail consists of 12 copies of gp11, 6 copies of gp12, and ≈30 copies of the small protein gp7.3 (Kemp et al., 2005). How these proteins are arranged is unknown. It has been suggested that gp7.3 functions as a tail assembly protein that remains with the mature virion; the protein is ejected into the outer membrane of the infected cell (Chang et al., 2010; Kemp et al, 2005). Host range mutants of T7 suggest that all three proteins interact directly with the lipopolysaccharide of the host cell.
7. Interactome screening
T7 was the focus of the first genome-wide screen for phage protein–protein interactions using the yeast two-hybrid assay (Bartel et al., 1996). As proof of principle, T7 was a good choice: the extensive genetic, molecular biology, and biochemical studies that had been performed on T7 provided a solid basis for evaluating the accuracy and completeness of the approach. Twenty-five interactions were identified in the screen, 15 of which were intramolecular. Many of these are to be expected as even monomeric polypeptides fold into a three-dimensional structure. Virion assembly clearly involves extensive protein–protein interactions, but as discussed by the authors, many of those that were known or plausibly expected from known structures were missed in the screen (Bartel et al., 1996). Conversely, screen-predicted interactions involving gp1.7 that were confirmed subsequently by its purification as an oligomer (Tran et al., 2010) and the predicted interactions between the spanin proteins gp18.5 and gp18.7 predated the discovery of a heterodimer even in phage λ (Casjens et al., 1989). Six intermolecular interactions were predicted that have not yet found confirmatory support, and our current understanding of T7 suggests that some are implausible. Improved screening procedures should help resolve these outstanding issues and, if more intermolecular interactions can be found that are consistent with, in particular, the pathways of virion assembly, the utility of those screening procedures in predicting the protein interactome of lesser-studied phages will be enhanced greatly. All published T7 interactions are summarized in Table II.
TABLE II.
Protein–protein interactions of phage T7
B. Coliphage λ
Phage λ may be the best-studied temperate phage and has been under investigation since the early 1950s. There are probably thousands of papers that have been published on phage λ and its closer relatives N15, HK97, and P22. For instance, there are more than 400 citations in a review of lambdoid phages that do not even focus on the genetic switch of λ, the paradigm for gene regulation (Hendrix and Casjens, 2006). The switch has been described in detail by many other papers and in its own book (Court et al., 2007; Ptashne, 2004). This section summarizes current knowledge of the interactions among phage λ proteins. Finally, a summary of interactions with its host, E. coli, is given. For a more detailed description of phage λ biology and regulation, the authors refer the reader to other reviews (Calendar, 2006; Court et al. 2007; Hendrix and Casjens, 2006; Hendrix et al., 1983; Oppenheim et al., 2005; Ptashne, 2004).
1. The λ virion
The phage λ genome and virion are shown in Figure 3. Note that the virion contains tail fibers that are not visible in most λ micrographs. The variant of λ used in most laboratories around the world, λPaPa, has a frameshift mutation in the structural stf gene and lacks the long “side tail fibers” of λ, Ur-λ, that comes directly from the original E. coli K-12 lysogen. It is still not entirely clear how many different proteins are in the λ particle but 12 are known with confidence and 2 more (gpM and gpL) are possibly in the particle in very low numbers. A current model of the particle with its known proteins is shown in Figure 3. Based on this model, we can predict at least 23 PPIs in the virion (Table III), although several are inferred from genetic experiments and remain to be detected by biochemical or biophysical methods. The virion adsorbs to sensitive cells through an interaction between the tail tip protein gpJ and the host LamB outer membrane protein. DNA is then released through the tail into the cytoplasm.
FIGURE 3.
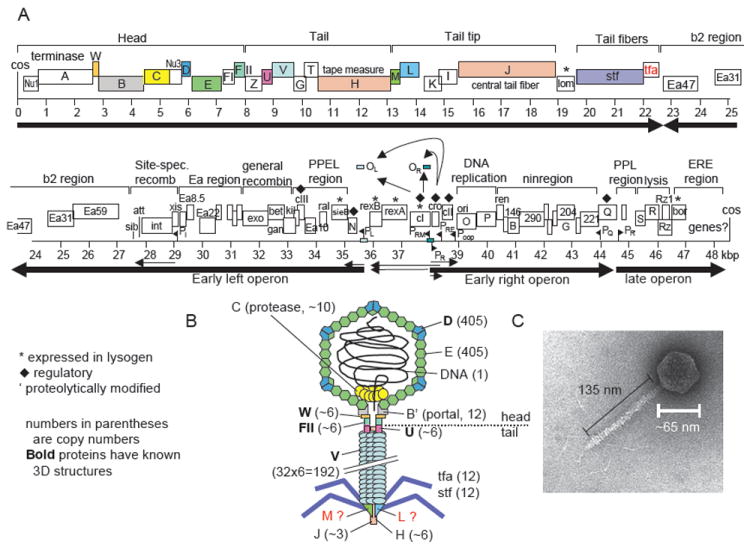
Genome and virion of phage λ. (A) Genome of phage λ. Colored ORFs correspond to colored proteins in (B). Main transcripts are shown as arrows. After Hendrix and Casjens in Calendar (2006). (B) Schematic model of λ virion. Numbers indicate the number of protein copies in the particle. It is unclear whether gpM and gpL proteins are in the final particle or only required for assembly. (C) Electron micrograph of phage λ. Modified after Hendrix and Casjens (2006).
TABLE III.
Published protein–protein interactions of phage λ
| λ 1 | λ 2 | Notes | References |
|---|---|---|---|
| gpA | gpNu1 | gpA (N-terminal) – gpNu1 (C-terminal) | Catalano, 2000; de Beer et al., 2002; Hang et al., 1999 |
| gpA | gpB | gpA (C-terminal) – gpB (= portal) | Catalano, 2000; Duffy and Feiss, 2002 |
| gpW | gpB | gpW (NMR, head–tail joining) | Casjens, 1974; Casjens et al., 1972 |
| gpW | gpFII | gpW required for gpFII binding | Casjens, 1974; Maxwell et al., 2001 |
| gpB | gpB | 12-mer (22 amino acids removed from gpB N-terminal) | Kochan, Carrascosa and Murialdo, 1984; Tsui and Hendrix, 1980 |
| gpC | gpE | Covalent PPI (in virion?) | Hendrix and Casjens, 1974;Hendrix and Duda, 1998 |
| gpC′ | gpB | Murialdo, 1979 | |
| gpC | gpNu3 | gpC may degrade gpNu3 (before DNA packaging) | Hendrix and Casjens, 1975; Hohn et al., 1975; Medina et al., 2010; Murialdo and Becker, 1978 |
| gpD | gpD | gpD forms trimers | Mikawa et al., 1996; Sternberg and Hoess, 1995; Yang et al., 2000 |
| gpE | gpE | Major capsid protein | Casjens and Hendrix, 1974; Dokland and Murialdo, 1993; Lander et al., 2008 |
| gpU | gpU | “Probably a hexamer” | Katsura and Tsugita, 1977 |
| gpD | gpE | Casjens and Hendrix, 1974; Dokland and Murialdo, 1993; Lander et al., 2008 | |
| gpU | gpV | gpU is head-proximal tail protein | Pell et al., 2009 |
| gpV | gpV | Buchwald et al., 1970a,b; Casjens and Hendrix, 1974; Katsura, 1983 | |
| gpO | gpP | Wickner and Zahn, 1986; Zahn and Blattner, 1985, 1987; Zahn and Landy, 1996) | |
| gpO | gpO | gpO–gpO interactions when bound to ori DNA | Alfano and McMacken, 1989 |
| gpB | gpA | Genetic evidence | Frackman et al., 1985; Sippy and Feiss, 1992 |
| gpFI | gpA | Genetic evidence | Catalano and Tomka, 1995 |
| gpFI | gpE | Genetic evidence | Murialdo and Tzamtzis, 1997 |
| gpH | gpG/gpGT | gpG/gpGT holds gpH in an extended fashion | Hendrix and Casjens, 2006 |
| gpV | gpGT | T domain binds soluble gpV | Hendrix and Casjens, 2006 |
| gpH | gpV | gpV probably assembles around gpH, displacing gpG/gpGT | Xu et al., 2004 |
| CI | CI | Forms octamer that links OR to OL | Bell and Lewis, 2001; Dodd et al., 2001 |
| Exo | Bet | Radding et al., 1971 | |
| SieB | Esc | Esc is encoded in frame in SieB and inhbits SieB | Ranade and Poteete, 1993a,b |
| CII | CII | Homotetramers | Ho and Rosenberg, 1982 |
| Xis | Int | Sam et al., 2002 | |
| Xis | Xis | Xis-Xis binding mediates cooperative DNA binding | Sam et al., 2004 |
| Int | Int | Dimer | Warren et al., 2003 |
| Stf | Stf | Likely homotrimer by homology with T4 fibers | |
| gpS | gpS | Large ring in inner membrane | Savva et al., 2008 |
| gpS | gpS′ | gpS′ inhibits gpS ring formation | Grundling et al., 2000 |
| gpB | gpE | Copurify in procapsid | Hendrix and Casjens, 1975 |
| gpNu3 | pNu3 | Monomer–dimer equilibrium | C. Catalano, personal communication |
| CIII | CIII | Dimer | Halder et al., 2007 |
| Cro | Cro | Dimer; X-ray str | Anderson et al., 1979 |
| gpRz | gpRz1 | Heteromultimer supposed to span the periplasm | Summer et al., 2007 |
| gpNu3 | gpB | gpNu3 required for gpB incorporation into procapsid | Ray and Murialdo, 1975 |
2. Assembly of virion head (Fig. 4)
FIGURE 4.
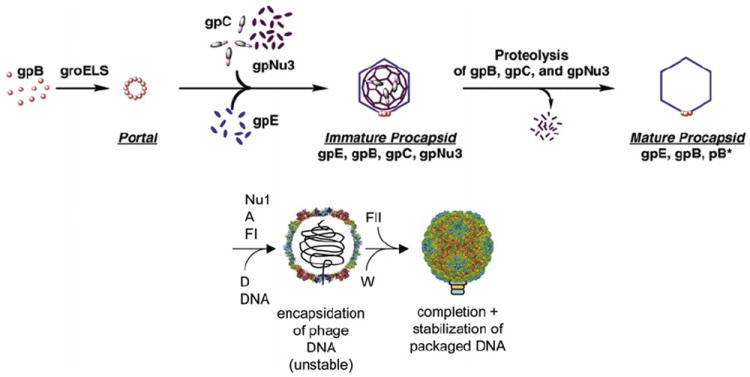
Assembly of the phage λ head. Head assembly has been subdivided into five steps, although most steps are not very well understood in mechanistic terms. Note that the tail is assembled independently. GpC protease, scaffolding protein gpNu3, and portal protein gpB form an ill-defined initiator structure. Protein gpE joins this complex in a step requiring the chaperonins GroES and GroEL. Proteins gpNu1, gpA, and gpFI are required for DNA packaging. GpD joins and stabilizes the capsid as a structural protein; gpFII and gpW are connecting the head to the tail that joins once the head is completed. Modified after Georgopoulos et al. (1983) and Lander et al. (2008).
λ encodes 10 proteins required for head assembly, of which 6 end up in the mature virion. Head assembly starts with assembly of the portal, which requires the host GroEL/S chaperone for folding. It is thought that the portal serves as the nucleus for assembling the remaining capsid. However, the mechanistic details of assembly are not well understood. GpC is involved as a head maturation protease that becomes attached covalently to E, the main capsid protein. The resulting fusion proteins subsequently get trimmed to make slightly shortened products named X1 and X2 (Hendrix and Casjens, 1974). The authors note, however, that attempts to identify X1 and X2 were unsuccessful and thus X1 and X2 may be artifacts (Medina et al., 2010). GpNu3 acts as putative scaffolding protein that aids coat shell assembly and is removed from the capsid by proteolysis before DNA is packaged. Structures of the procapsid and mature λ capsid have been solved by cryo-EM, showing the T=7 arrangement of capsid protein gpE clustered into hexamers and pentamers (Dokland and Murialdo; Lander et al., 2008). Trimers of the decoration protein gpD bind to the capsid at trivalent interaction points between the capsomers and stabilize the capsid (Sternberg and Weisberg, 1977). Comparison of the procapsid with the mature capsid reveals the considerable structural transitions that occur upon capsid maturation and DNA packaging, but this process is not well understood mechanistically.
3. Assembly of virion tail (Fig. 5)
FIGURE 5.
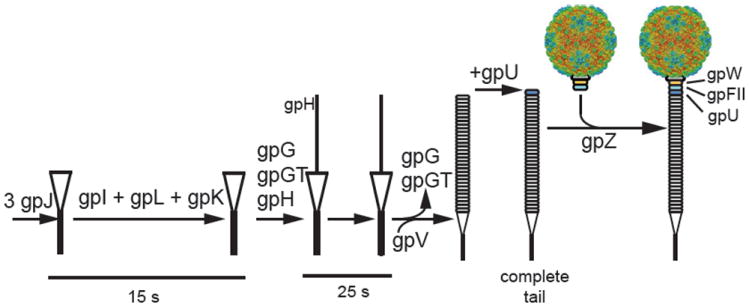
Assembly of the phage λ tail. The λ tail is made of at least six proteins (gpU, V, J, H, tfa, stf) with another seven required for assembly (gpI, M, L, K, G/T, Z). Assembly starts with protein gpJ, which then, in a poorly characterized fashion, requires proteins gpI, gpL, gpK, and gpG/T to add the tape measure protein H. GpM joins and then gpG and gpG/T leave the complex so that the main tail protein gpV can assemble on the gpJ/H scaffold. Finally, gpU is added to the head-proximal end of the tail. GpZ is required to connect the tail to the preassembled head. Protein gpH is cleaved by the action of gpU and gpZ (Tsui and Hendrix, 1983). It remains unclear if proteins gpM and gpL are part of the final particle (Hendrix and Casjens, 2006). From http://www.pitt.edu/~duda/lambdatail.html.
Tail assembly begins with the antireceptor protein gpJ. Proteins gpI, gpK, gpL, and gpM assemble around gpJ in that order, although no mechanistic or structural details are known about this process. In parallel events, gpH, gpG, and gpG-T (the frameshifted form of gpG including the T reading frame) form a complex that binds the tail tip and the major tail protein (gpV). The tape meassure protein gpH determines the length of the phage tail (Katsura, 1990). Once the correct length is achieved, gpV assembly stops and the proximal tail protein gpU is added. Protein gpZ somehow facilitates head–tail joining without being assembled into the final virion.
4. Lysogeny and induction
After phage DNA ejection, circularization, and ligation by the host DNA ligase, the earliest genes to be expressed include N, O, and P, as well as cII and cIII, which play a key role in the lysis/lysogeny decision (Echols, 1986). Both CII and CIII can interact with and be cleaved by the bacterial FtsH (HflB) protease directly, which in turn is complexed with the host HflK and HflC proteins at the cytoplasmic membrane. HflD, a cytoplasmic membrane protein, also binds CII, making it both more susceptible to FtsH and less able to bind DNA (Kihara et al., 2001; Parua et al., 2010). As a substrate for FtsH, CIII acts as a competitive inhibitor of CII degradation and, as the half-life of CII protein increases, leads to an increase in CI repressor synthesis (Datta et al., 2005a; Herman et al., 1997; Kobiler et al., 2007). Except when cIII is overexpressed, lysogeny is only established when the activity of these proteases (FtsH, Lon, and perhaps others) is low. Once established, lysogeny is a stable state until lysogens are exposed to ultraviolet light or other DNA-damaging agents that lead to the activation of RecA, which in turn accelerates autocleavage of the λ repressor CI. Loss of CI leads to prophage induction and the lytic cycle. The mechanism by which phage λ decides whether to grow lytically or become a lysogen is reviewed elsewhere (Court et al., 2007; Friedman and Court, 2001; Gottesman and Weisberg, 2004; Ptashne, 2006). However, Table IV contains the host–phage protein–protein interactions critical for gene expression and thus the genetic switch of phage λ.
TABLE IV.
Published phage λ–host interactionsa
| λ | Host | Description | References |
|---|---|---|---|
| Transcription | |||
| CI | RecA | RecA degrades CIb | Kobiler et al., 2004 |
| CI | RpoA | Kędzierska et al., 2007 | |
| CI | RpoD | Li et al., 1994; Nickels et al., 2002 | |
| CII | ClpYQ | ClpYQ degrades CII in vitro | Kobiler et al., 2004 |
| CII | ClpAP | ClpAP degrades CII in vitro | Kobiler et al., 2004 |
| CII | HflD | HflD makes CII more vulnerable to FtsH (hfl, high-frequency lysogenization) | Kihara et al., 2001 |
| CII | HflBc | HflB (= FtsH) protease degrades cII | Kobiler et al., 2002; Shotland et al., 2000 |
| CII | RpoA | Kędzierska et al., 2004; Marr et al., 2004 | |
| CII | RpoD | Kędzierska et al., 2004 | |
| CIII | HflB | CIII inhibits HflB; HflB degrades CIII | Herman et al., 1997; Kornitzer et al., 1991 |
| gpN | NusA | Transcriptional regulation | Friedman and Court, 2001 |
| gpN | Lon | Lon degrades gpN | Kobiler et al., 2004; Gottesman et al., 1981 |
| gpQ | σ70 | gpQ makes RNAP insensitive to cis terminal | Marr et al., 2001;Nickels et al., 2002; Roberts and Roberts, 1996; Roberts et al., 1998 |
| Head | |||
| gpB | GroE | genetic interaction; E. coli GroE | Ang et al., 2000; Georgopoulos et al., 1973; Murialdo, 1979 |
| gpE | GroE | genetic interaction | Georgopoulos et al., 1973 |
| Tail | |||
| gpJ | LamB | LamB is the E. coli receptor | Buchwald and Siminovitch, 1969; Clement et al., 1983; Mount et al., 1968; Wang et al., 2000a; Werts et al., 1994 |
| Stf | OmpC | OmpC is a secondary E. coli receptor | Hendrix and Duda, 1992 |
| Recombination | |||
| Xis | Lon | Xis is degraded by E. coli Lon protease | Leffers and Gottesman, 1998 |
| Xis | FtsH | Xis is degraded by E. coli FtsH protease | Leffers and Gottesman, 1998 |
| Xis | Fis | both required for excision | Ball and Johnson, 1991; Cho et al., 2002; Esposito and Gerard, 2003 |
| Int | IHF | Both catalyze recombination at attP/attB | Campbell et al., 2002; Crisona et al., 1999 |
| Gam | RecB | Gam inhibits RecBCD | Marsic et al., 1993 |
| NinB | SSB | NinB also binds ssDNA | Maxwell et al., 2005 |
| Replication | |||
| gpO | ClpXP | ClpXP degrades gpO | Kobiler et al., 2004 |
| gpP | DnaA | Datta et al., 2005b,c | |
| gpP | DnaB | Mallory et al., 1990 | |
CbpA (Wegrzyn et al., 1996) perhaps interacts with DnaA and SeqA (Potrykus et al., 2002; Wegrzyn and Wegrzyn, 2001, 2002).
After DNA damage, RecA bound to single-stranded DNA stimulates autocleavage of the CI repressor (Kobiler et al., 2004).
HflB = FtsH.
5. Anti-termination
Escherichia coli RNA polymerase transcribes λ DNA starting from the immediate early promoters PL and PR, which leads to expression of the N and cro genes, among others. As soon as the gpN-binding sites (nut sites) are transcribed, gpN modifies the transcription complex by recognizing a stem-loop structure in the mRNA and by binding to host Nus proteins [note that gpN interacts with nut RNA (not DNA)]. This modified complex is capable of overriding the termination signal that normally stops immediate early transcription. Because gpN is continually being degraded by the Lon protease, ongoing anti-termination requires continual transcription of the N gene (Fig. 6). In addition to the anti-terminator for early lytic genes (gpN), a second anti-terminator, gpQ, is required for late gene expression. GpQ anti-termination differs from that of gpN in that gpQ binds to the “Q binding element” (QBE, also known as the qut site) of the PR′ promotor (i.e., unlike the gpN protein, gpQ binds DNA). When RNA polymerase (RNAP) initiates transcription at the PR′ promotor, it almost immediately pauses at a pause-inducing sequence. Pausing allows gpQ to contact σ70 in the RNAP, which causes a conformational change in the enzyme that enables gpQ to bind the β subunit (RpoB). The gpQ-containing RNAP is then able to override the termination signal at tR′ (Deighan et al., 2008; Nickels et al., 2002).
FIGURE 6.
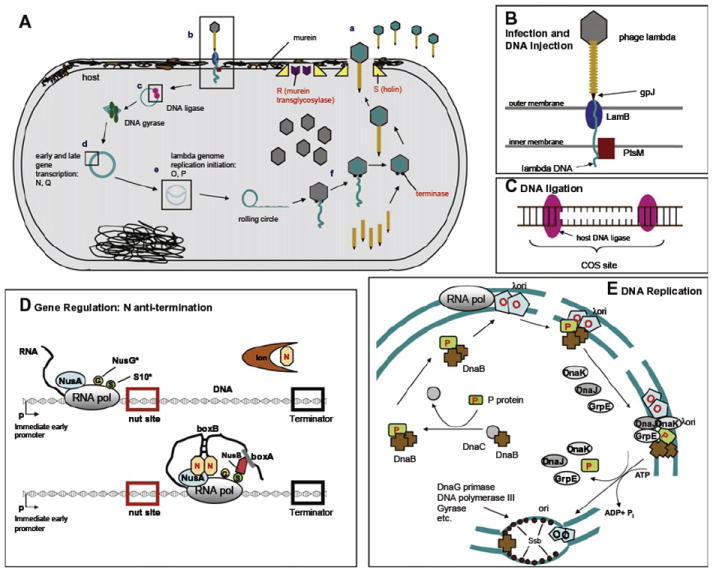
Interactions of phage λ with its host, E. coli. (A) Overview of λ activities in E. coli: (a) free phage, (b) binding and entry, (c) DNA circularization and supercoiling, (d) transcription, (e), replication, and (f) virion assembly. Details are shown in B–E. (B) Infection and DNA ejection, (C) DNA ligation, (D) gene regulation. An asterisk means weakly bound to the RNA polymerase. (E) DNA replication. Phage proteins are labeled in red, host proteins in black. Modified after Das (1992) and Mason and Greenblatt (1991).
6. Replication
After λ gene O and P protein synthesis, replication of the phage genome is initiated (Fig. 6). Replication initiation starts with dimeric gpO bound to the four 18-bp direct repeats of the λ origin, located within the sequence of the O gene. Each repeat binds a gpO dimer, leading to a total of eight monomers bound directly to the DNA, in addition to non-DNA-bound gpO that associates with the complex solely by PPIs (Dodson et al., 1986). This large nucleoprotein complex is called the O-some. GpP extracts the host DnaB helicase from its native complex with DnaC (Zylicz et al., 1989). The DnaB helicase is inactive as long as it is tightly associated with gpP. The gpP–DnaB pair is then added to the O-some through interactions between gpP and gpO. Three host heat shock chaperone proteins—DnaK, DnaJ, and GrpE—are subsequently recruited (DnaK through direct interaction with gpP and gpO) and activate the DnaB helicase by liberating it from its inhibitory interaction with gpP. Finally gpP and the chaperones are released, and replication starts with the help of the host DnaG primase, DNA polymerase III, gyrase, and single-stranded DNA-binding (SSB) proteins (Dodson et al., 1986; LeBowitz and McMacken, 1986). The RNA polymerase β subunit was shown to interact directly with gpO (Szambowska et al., 2010). Even though not yet fully understood, how E. coli RNA polymerase activates λ ori seems to be far more complex than the original idea that it simply changed DNA topology.
Studies on λ ori-dependent plasmids have revealed interactions with even more host proteins, including the DnaJ homolog CbpA (Wegrzyn et al., 1996), and through transcriptional activation of origin activity, perhaps also with DnaA and SeqA (Potrykus et al., 2002; Wegrzyn and Wegrzyn, 2001, 2002). As λ ori-dependent plasmid maintenance and copy number are very sensitive to changes in host physiology, interactions of the O-some with additional proteins may yet be revealed.
7. The λ interactome
A λ Y2H interactome analysis has been completed (Rajagopala et al., 2011). All λ open reading frames (ORFs) were cloned and tested in all possible pairs for interactions, using three different Y2H vectors. These screens identified 97 interactions among 68 tested λ proteins, of which 16 were known previously. Notably, because only 33 interactions had been reported previously, this screen found more than 50% of all previously published interactions in a single experiment! At least 18 of the newly observed Y2H interactions seem to be plausible, based on the functions of the interacting proteins, indicating that even in λ many new discoveries can be made. Interestingly, relatively few interactions involving morphogenetic proteins were revealed, and possible reasons for the still significant fraction of apparent false negatives are discussed later.
C. Salmonella phage P22
P22 infects Salmonella enterica serovar Typhimurium and was originally isolated by Zinder and Lederberg (1952). It is a temperate member of Podoviridae. Its virion proteins are not particularly closely related to other tailed phage groups, but its early genes and overall lifestyle are very closely related to phage λ. For this reason, it is often placed in the “lambdoid” phage group. Phage P22 is also a very important tool for those who use Salmonella genetics, as it is particularly easy to handle in the laboratory and is a very good generalized transducing phage (i.e., it packages a significant amount of host DNA by accident and can be used to move host alleles from one Salmonella strain to another).
1. P22 life cycle
The early operons of P22 are organizationally similar to those of λ and carry parallel prophage repressor, Cro, recombination, DNA replication, and transcription anti-termination functions (Susskind and Botstein, 1978). The P22 repressor has a different DNA operator target specificity from that of λ repressor and encodes nonhomologous types of recombination and replication proteins, but its late operon anti-termination protein is 97% identical to that of λ and has the same target specificity. Unlike λ, in addition to its prophage repressor (C2) and lysogeny control proteins (C1, C3, Cro), P22 carries an “immunity I” region that encodes an anti-repressor protein, Ant, that binds to the prophage repressor and blocks its ability to bind its operator. Two additional repressors, Mnt and Arc, regulate Ant synthesis (Susskind and Botstein, 1975; Vershon et al., 1987a,b). Also, instead of a protein that recruits the host DnaB protein to the replication origin, P22 encodes its own DnaB homologue (Backhaus and Petri, 1984; Wickner, 1984). The repressed P22 prophage encodes several “lysogenic conversion” genes, several of which encode a set of three Gtr (glucose transfer) proteins that modify the host O-antigen polysaccharide by glucosylation (Broadbent et al., 2010; Makela, 1973; Vander Byl and Kropinski, 2000). Finally, P22 has been studied as a prototypical headful DNA packaging phage, which is different from the packaging strategies used by the other phages discussed here; unlike these other phages that package DNA between two specific nucleotide sequences in the DNA substrate, P22 packages DNA until the head is full and the nucleotide sequence at the termination point does not matter (Casjens and Hayden, 1988; Jackson et al., 1978; Tye et al., 1974).
2. P22 virion assembly
The P22 virion is assembled by proteins encoded by 14 genes in its late operon (Fig. 7) (Botstein et al., 1973; Eppler et al., 1991; King et al., 1973; Poteete and King, 1977; Youderian and Susskind, 1980). Some P22-like phages, for example, phages ε34 and L, have a 15th virion assembly gene, dec, at the promoter proximal end of the cluster, whose encoded protein “decorates” the outside of the head and stabilizes it (Gilcrease et al., 2005; Tang et al., 2006; Villafane et al., 2008). The P22 late operon encodes lysis proteins very similar to those of λ; however, the virion assembly genes are not closely related to λ or to any other tailed phage type. P22 virion assembly is best known for the fact that its head-assembling scaffolding protein was the first to be discovered and studied (Cortines et al., 2011; King and Casjens, 1974; Prevelige et al., 1988; Weigele et al., 2005). The P22 procapsid assembles as a complex of 415 coat protein molecules, 12 portal ring subunits, and about 250 scaffolding protein molecules. At about the time when DNA is packaged, all 250 scaffolding proteins are released intact from the procapsid and are reused with newly synthesized coat protein in subsequent rounds of procapsid assembly. Thus, the P22 scaffolding protein acts catalytically in head assembly and individual molecules can participate in at least five rounds of procapsid assembly. A particularly high-resolution asymmetric cryo-EM reconstruction of the virion has been determined (Lander et al., 2006; Tang et al., 2011), and because assembly of the four proteins of its short tail has also been quite well studied [X-ray structures are known for three tail proteins and the portal protein (Olia et al., 2006, 2007; Steinbacher et al., 1997)], the P22 virion is perhaps the best understood of all tailed phage virions. The details of P22 virion assembly have been reviewed in light of its evolution and genome mosaicism (Casjens and Thuman-Commike, 2011), and the authors refer the reader to this review for further details. The published PPIs of phage P22 are summarized in Table V. Interactions between P22 and its host are much less well studied than those of λ and are mostly understood by homology arguments in the case of proteins that are similar in the two phages. Known P22–host interactions are listed in Table VI.
FIGURE 7.
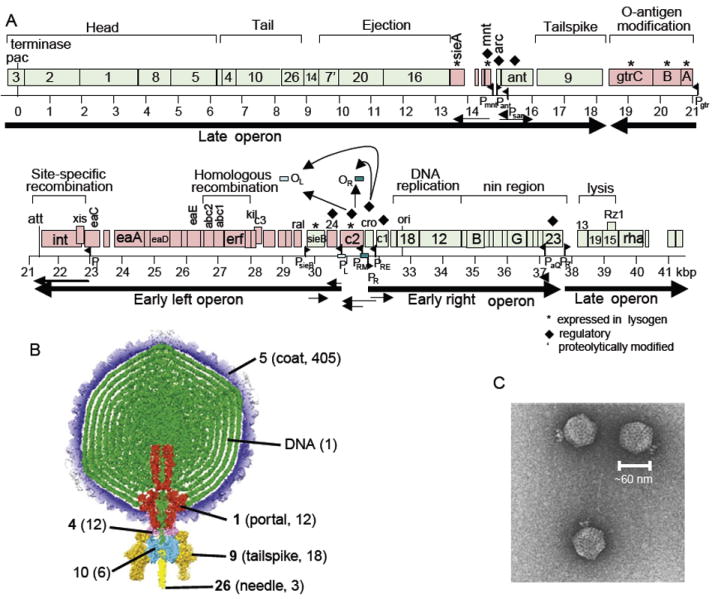
Genome and virion of phage P22. (A) Genome of phage P22 with a scale in kbp below. Green open reading frames (ORFs; rectangles) are transcribed left to right; red ORFs are transcribed right to left. Known RNA polymerase promoters and transcripts are shown as black flags and arrows, respectively. (B) The asymmetric (not icosahedrally averaged) P22 virion three-dimensional cryo-EM reconstruction from Tang et al. (2011). Numbers in parentheses indicate molecules/virion, and bold numbers indicate proteins for which X-ray structures are known. (C). Negatively stained electron micrograph of P22 virions.
TABLE V.
P22 intraphage protein interactions
| P22 1 | P22 2 | Description | References |
|---|---|---|---|
| gp3 | gp3 | Small terminase subunit; homononomer in solution | Nemecek et al., 2007, 2008 |
| gp3 | gp2 | Purify as complex; bind each other in vitro | Nemecek et al., 2007; Poteete and Botstein, 1979 |
| gp2 | gp1 | gp2 is large terminase subunit; assembles in packaging motor to gp1 by analogy with other phages (gp2 by itself is monomer in solution); evolutionary argument for P22 gp2–gp1 interaction | Casjens, 2011 |
| gp1 | gp1 | Portal protein; homododecamer; EM of 12-mer; EM of virion; X-ray structure | Bazinet et al., 1988; Lander et al., 2006; Olia et al., 2011 |
| gp1 | gp5 | Purify as complex (procapsids); three-dimensional (3D) reconstructions of virion and procapsid | Botstein et al., 1973; Chen et al., 2011; Lander et al., 2006 |
| gp1 | gp8 | gp8 required for assembly of gp1 into procapsid; interaction seen in procapsid 3D reconstruction | Chen et al., 1989; Greene and King, 1996; Weigele et al., 2005 |
| gp8 | gp8 | Scaffolding protein; forms homodimers and tetramers in solution | Parker et al., 1997 |
| gp8 | gp5 | Purify as complex (procapsid); bind in vitro | Fuller and King, 1982; Greene and King, 1994; Parker et al., 1998 |
| gp5 | gp5 | Coat (major capsid) protein; forms T=7 l icosahedral shell of virion | Botstein et al., 1973; Casjens, 1979; Lander et al., 2006 |
| gp5 | gp1 | Purify as complex (procapsids); interaction seen in 3D reconstruction | Chen et al., 2011; Lander et al., 2006 |
| gp4 | gp4 | Tail protein; monomer in solution (naturally unfolded?); gp4s barely touching each other in dodecamer ring in virion (i.e., contacts not large); EM, X-ray structure | Olia et al., 2011; Tang et al., 2011 |
| gp4 | gp5 | Contact in high-resolution 3D reconstruction of virion | Olia et al., 2006, 2011; Tang et al., 2011 |
| gp4 | gp1 | Order of assembly, EM, 3D reconstruction; X-ray cocrystal structure | Olia et al., 2006, 2011; Tang et al., 2011 |
| gp4 | gp10 | 12 gp4s contact 6 gp10s; order of assembly, EM, 3D reconstruction | Lander et al., 2006; Olia et al., 2007 |
| gp4 | gp9 | gp4 ring contacts 6 gp9 trimers; 3D reconstruction | Lander et al., 2006 |
| gp10 | gp10 | Tail protein; probably hexamer in solution; 3D reconstruction | Lander et al., 2006; Olia et al., 2007 |
| gp10 | gp9 | 6 gp4s contact 6 gp9 trimers; 3D reconstruction | Lander et al., 2006 |
| gp10 | gp26 | 6 gp10s contact one gp26 trimer | Landeret al., 2006 |
| gp26 | gp26 | Tail needle protein; trimer in solution - crystal structure; one trimer in virion | Olia et al., 2007 |
| gp16 | gp8 | Mutants of gp8 do not incorporate gp16 into virion | Chen et al., 1989; Greene and King, 1996; Weigele et al., 2005 |
| C1 | C1 | C1 is homologue of λ CII; homotetramer | Ho et al., 1992 |
| Mnt | Mnt | Repressor of antirepressor gene; tetramer | Nooren et al., 1999 |
| Arc | Arc | Repressor of antirepressor gene; dimer in solution, tetramer on DNA | Bowie and Sauer, 1989; Breg et al., 1990; Brown, Bowie and Sauer, 1990 |
| Ant | C2 | Antirepressor; binds to and inhibits C2 repressor | Susskind and Botstein, 1975 |
| gp9 | gp9 | Tailspike protein; trimer in solution, 6 trimers in virion; crystal structure; binds to polysaccharide on cell surface | Lander et al., 2006; Steinbacher et al., 1994, 1997 |
| Int | Xis | Form heteromultimer | Cho et al., 2002 |
| Xis | Xis | Excisionase; homomultimers | Mattis et al., 2008 |
| Erf | Erf | Recombination protein; homomultimeric ring of 10–14 subunits | Poteete et al., 1983 |
| C3 | C3 | C3 is homologue of λ CIII; λ CIII is dimer by disulfide bond; P22 interaction by homology with λ CIII | Halder et al., 2007 |
| SieB | Esc | Similar to λ but not studied in detail | Ranade and Poteete, 1993a |
| C2 | C2 | Prophage repressor homologue of λ CI; homodimer | De Anda et al., 1983 |
| Cro | Cro | Dimer; solution studies and crystal structure | Darling et al., 2000; Newlove et al., 2004 |
| gp18 | gp12 | Origin binding protein gp18 recruits DnaB-like gp12 to replication origin by analogy with λ (note that gp12 is not a homologue of λ P protein; gp18 is homologous to λ O protein only in N-terminal domain) | Backhaus and Petri, 1984 |
| Rz | Rz1 | Rz and Rz1 are supposed to bind to one another and form a bridge that spans the periplasm in phage λ; P22 gp15 and Rz1 should do the same by homology. | Summer et al., 2007 |
| gp13 | gp13 | Holin forms rings of large diameter in λ; by homology same should be true of P22 holin | Savva et al., 2008 |
| gp13 | gp13′ | Antiholin–holin interactions delay the formation of holin rings in λ; by homology same should be true in P22 | Rennell and Poteete, 1985 |
| Dec | Dec | Trimer; Note that P22 has no Dec, but its very close relative phage L does and L Dec binds to P22 capsids. | Tang et al., 2006 |
| Dec | gp5 | Dec occupies a subset of the quasi-equivalent local threefold positions on the shell | Tang et al., 2006 |
TABLE VI.
P22–host protein interactions
| P22 | Host | Description | References |
|---|---|---|---|
| gp5 | GroEL | Solution studies | de Beus et al., 2000 |
| gp24 | NusA? etc? | Homologue/analogue of λ gpN; likely binds same host factors as λ gpN | Franklin, 1985; Hendrix and Casjens, 2006 |
| C3 | FtsH(HflB) | C3 is homologue of λ CIII; inhibition of FstH by homology with λ CIII | Semerjian et al., 1989 |
| C1 | FtsH(HflB) | C1 is homologue of λ CII; CII is degraded by FtsH | Backhaus and Petri, 1984 |
| C2 | RecA | P22 repressor autocleavage stimulated by RecA binding like λ | Prell and Harvey, 1983; Tokuno and Gough, 1976 |
| Xis, gp18, gp24, C1 | Host protein degradation machinery | These may be degraded analogously to their λ homologues, but direct information is not available | Backhaus and Petri, 1984 |
| gp23 | RNA polymerase? | By homology to λ; gp23 is nearly identical to λ gpQ | Pedulla et al., 2003 |
| gp12 | Host replication machinery | Possible interaction with host DnaG primase | Wickner, 1984 |
| Abc2 | RecC | Solution studies | Murphy, 2000 |
3. Comparison of P22 and λ interactomes
As noted earlier, comparison of P22 and λ is a particularly informative way to understand the kinds of relationships among the proteins of different tailed phages. The “lambdoid” phages all encode similar functions and have similar gene organization, but these “similar functions” can be of several different types. Because these phages undergo a significant amount of horizontal transfer of genetic information (especially if they are “closely” related), their functions can be homologous or even nearly identical (Casjens, 2005; Hendrix, 2002). For example, the two “gpQ” late operon control anti-termination proteins of the two phages are nearly identical and can replace one another in function. A second kind of relationship is represented by the prophage repressors. These proteins from the two phages have weak but recognizable homology and similar folds, and probably interact with host proteins the same way, but have different operator-binding specificity. Thus, one repressor cannot functionally replace the other without additional changes in the DNA-binding sites of the protein. A third kind of relationship is that of the virion coat, terminase, and portal proteins, λ gpE, gpA, and gpB and P22 gp5, gp2, and gp1, respectively. These proteins have the same functions in the two phages—coat protein and DNA packaging nuclease/motor ATPase and procapsid DNA entry channel, respectively (summarized in Hendrix and Casjens, 2006). These cognate proteins are not recognizably similar in overall amino acid sequence between the two phages, although the two ATPase proteins have similar ATP-binding motifs. However, the two proteins almost certainly have the same basic fold. Thus, gpE and gp5 and gpA and gp2, as well as gpB and gp1, are all very likely ancient homologous pairs that have diverged beyond the point of having recognizable sequence similarity. Finally, similar functions are performed by nonhomologues in the two phages. Examples of this relationship are the (i) nonhomologous Exo (λ) and Erf (P22) proteins that catalyze homologous recombination in completely different ways (Little, 1967; Poteete and Fenton, 1983); (ii) the endolysin proteins gpR and gp19, where the λ protein is a transglycosylase and the P22 protein is a true lysozyme that both cleave the same peptidoglycan bond (Bienkowska-Szewczyk et al., 1981; Rennell and Poteete, 1989); (iii) the gpW and gp4 proteins, which bind between the portal ring of the head and the tail and yet have no recognizable homology or structural similarity; and (iv) the λ receptor binding “fiber” (gpJ and STF) and P22 tailspike (gp9), which have no homology, recognize different structures, and act in different ways [the P22 gp9 enzyme cleaves its polysaccharide receptor (Iwashita and Kanegasaki, 1976)]. Thus, there are potentially a number of different ways in which parallel λ and P22 functions might interact with the hosts of these two phages, and it will be interesting to understand these in detail in both cases. But are there very similar phage proteins that have different host interactions? Do very different phage proteins that have the same function have equally different host interactions? Although no such situations have been identified to date, it remains possible. A comparison of PPIs in λ and P22 is given in Tables VII and VIII.
TABLE VII.
Comparison of λ and P22 interactions
| λ 1 | λ 2 | P22 1 | P22 2 | Description |
|---|---|---|---|---|
| gpA | gpB | gp2 | gp1 | Likely similar contacts—by evolutionary argumenta |
| gpA | gpNu1 | gp2 | gp3 | Likely similar contacts—by evolutionary argument a |
| gpNu1 | gpNu1 | gp3 | gp3 | gpNu1 N-terminal fragment is dimer; gp3 is nonomer; not known if they have the same fold (i.e., are ancient homologues) |
| gpW | gpB | gp4 | gp1 | gpW and gp4 have no known homology, but are analogous in that they are the first proteins to bind the underside of the the portal ring; gp4 has no structural homology to λ gpFII or gpW (Cardarelli et al., 2010; Olia et al., 2011) |
| gpW | gpFII | No clear gpFII homologue in P22 (gp4 and gp10 of P22 have no sequence similarity) | ||
| gpB | gpB | gp1 | gp1 | Both form dodeamer portal ringsa |
| gpC | gpE | No gpC analogue in P22 | ||
| gpC′ | gpB | No gpC analogue in P22 | ||
| gpC | gpNu3 | No gpC analogue in P22 | ||
| gpNu3 | gpNu3 | gp8 | gp8 | Monomer/dimer/tetramer for gp8; monomer/dimer for gpNu3 |
| gpB | gpE | gp1 | gp5 | Seen in reconstructionsa |
| gpNu3 | gpB | gp8 | gp1 | Scaffolds required for portal assembly into procapsid |
| gp8 | gp5 | Scaffold–coat interaction not studied for λ | ||
| gpD | gpD | Dec | Dec | Both trimers; bind slightly different sites.a Note that P22 has no Dec, but its very close relative phage L does and L Dec binds to P22 capsids |
| gpE | gpE | gp5 | gp5 | Both make icosahedral T=7 head shellsa |
| gpU | gpU | No analogue in P22 | ||
| gpD | gpE | Dec | gp5 | Dec is more discriminatory than D, i.e., unlike D, Dec does not occupy all of the quasi-equivalent local threefold positions on the shell |
| gp4 | gp5 | Unkown in λ | ||
| gp4 | gp10 | No gp10 analogue in λ (but see gpFII above) | ||
| gp4 | gp9 | No gp9 analogue in λ (but see Stf below?) | ||
| gp10 | gp9 | No gp9 analogue in λ (but see Stf below?) | ||
| gp10 | gp10 | No gp10 analogue in λ (but see gpFII above) | ||
| gp10 | gp26 | No gp10 or gp26 (?) analogue in λ | ||
| gp26 | gp26 | No gp26 analogue in λ (maybe gpJ?) | ||
| Stf | Stf | gp9 | gp9 | Both trimers (Stf by homology to T4 fibers); analogues, not homologues |
| gpU | gpV | No analogues in P22 | ||
| gpV | gpV | No analogue in P22 | ||
| gpO | gpP | gp18 | gp12 | gpO and gp18 are homologous but gpP and gp12 are not; nonetheless, a good argument can be made that gp18 and gp12 must interact similarly to gpO and gpP to get replication started |
| gpW | gpW | gp4 | gp4 | Analogues—first proteins to bind below the portal ring (see gpW–gpB interaction above) |
| gpFI | gpA | No gpFI analogue in P22 | ||
| gpFI | gpE | No gpFI analogue in P22 | ||
| gpH | gpG/gpGT | No analogues in P22 | ||
| gpV | gpGT | No analogues in P22 | ||
| gpH | gpV | No analogues in P22 | ||
| Arc | Arc | No analogue in λ | ||
| Ant | C2 | No Ant in λ | ||
| Mnt | Mnt | No analogue in λ | ||
| CI | CI | C2 | C2 | Homologues (Sevilla-Sierra et al., 1994) |
| Exo | Bet | Erf | Erf | λ Exo and P22 Erf proteins are not homologues and function in basically different ways, but they are both essential for homologous recombination |
| SieB | Esc | SieB | Esc | Good homologues in P22 but they are unstudied |
| CII | CII | C1 | C1 | Homotetramers in both cases |
| Xis | Int | Xis | Int | Xis proteins are quite different |
| Xis? | Xis? | Xis | Xis | Monomer in λ, but binds cooperatively so could interact on DNA (Abbani et al., 2007); multimer in P22 |
| CIII | CIII | C3 | C3 | C3 is homologue of λ CIII; λ CIII is dimer (Halder et al., 2007); P22 interaction by homology with λ CIII |
| Cro | Cro | Cro | Cro | Both homodimers |
| gpRz | gpRz1 | gp15 | gp15 | Predicted for P22 by homology |
Not recognizably similar in amino acid sequence but very likely ancient homologues.
TABLE VIII.
Comparison of phage λ–host and P22–host interactions
| λ | Host | P22 | Host | Notes |
|---|---|---|---|---|
| CII | HflB | C1 | HflB | Should be true for P22 by homology |
| CIII | HflB | C3 | HflB | Should be true for P22 by homology |
| gpJ | LamB | P22 tails recognize polysaccharide so no analogue in P22 | ||
| Stf | OmpC | P22 tails recognize polysaccharide so no analogue in P22 | ||
| Xis | Lon | Xis | Lon? | Might be true for P22 by homology |
| Xis | FtsH | Xis | FtsH? | Might be true for P22 by homology |
| gpN | RNAP | gp24 | RNAP? | Likely true for P22 by similarity of function |
| Int | IHF | P22 does not interact with IHF (Cho et al., 1999) | ||
| Xis | Fis | Unknown for P22 | ||
| gpQ | σ70 | gp23 | σ70 | Likely true for P22 by similarity of function |
| P | DnaB | No analogue of P in P22 | ||
| gp12 | DnaG? | No analogue of 12 in λ | ||
| gpE | GroE | gp5 | GroE | |
| gpB | GroE | Not known for P22 | ||
| gpN | Lon | gp24 | Lon | Not known for P22 |
| CI | RecA | C2 | RecA | Might be true for P22 by homology and induction by ultraviolet light |
| CII | ClpYQ | C1 | ClpYQ | Might be true for P22 by homology |
| CII | ClpAP | C1 | ClpAP | Might be true for P22 by homology |
| gpO | ClpXP | gp18 | ClpXP | Might be true for P22 by homology |
| gpN | NusA | gp24 | NusA | Likely true by similarity of function |
| CII | HflD | C1 | HflD | Might be true for P22 by homology |
| Gam | RecB | No Gam in P22 | ||
| Abc2 | RecC | No Abc2 in λ |
D. Enterobacteriophage P2 and its satellite phage, P4
Bacteriophage P2 was originally isolated from the Lisbonne & Carrère strain of E. coli by Bertani in 1951 and is a member of the Myoviridae family of viruses. It has an icosahedral head and a contractile tail and is a 33.6-kb double-stranded DNA genome (Bertani, 1951; Bertani and Six, 1988; Nilsson and Haggård-Ljungquist, 2006). P2-like prophages are common in the environment (Breitbart et al., 2002) and are present in about 30% of strains in the E. coli reference collection (Nilsson et al., 2004).
Bacteriophage P2 has attracted much interest as a “helper” for bacteriophage P4, an 11.6-kb replicon that can exist either as a plasmid or integrated into the host genome as a prophage (Briani et al., 2001; Deho and Ghisotti, 2006; Lindqvist et al., 1993). P4 lacks genes encoding major structural proteins, but can utilize the structural gene products encoded by P2 for assembly of its own capsid (Christie and Calendar, 1990; Lindqvist et al., 1993; Six, 1975). An overview of the P2 structural biology and a summary of its interactions are shown in Figure 8 and Table IX.
FIGURE 8.
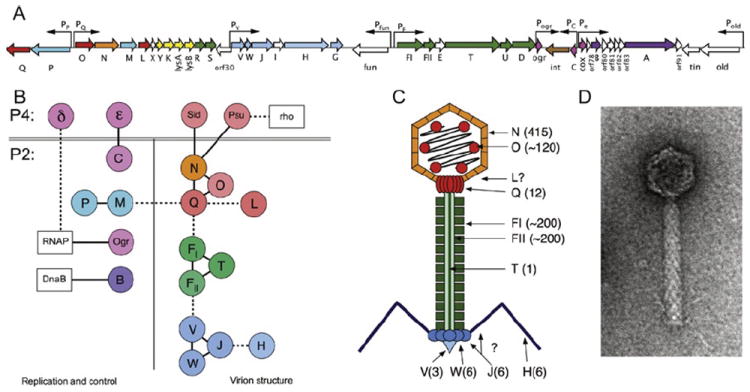
(A) Schematic diagram of the 33.6-kb P2 genome. Open reading frames (ORFs) are indicated by arrows that reflect their direction and size and are color coded as follow: red/orange, capsid-associated proteins (gpQ, O, N, L); blue, terminase proteins (gpP, M); yellow, lysis proteins (gpY, K, lysA, lysB); green, tail-related proteins (gpR, S, FI, FII, T, U, D); blue, base plate and tail fiber-related proteins (gpV, W, J, H, G); pink, transcriptional control proteins (Ogr, C, Cox); brown, integrase (Int); and purple, replication-related proteins (gpA,B). Nonessential genes and ORFs of unknown function are white. Promoters are indicated by arrows above ORFs. (B) Diagram of protein–protein interactions listed in Table IX. Each P2 and P4 protein is shown as a circle, colored as given earlier. Host proteins are shown as white boxes. (C) Schematic diagram of the P2 virion, color coded as given earlier. Copy numbers of proteins, when known, are indicated in parentheses. (D) Electron micrograph of a P2 virion, stained negatively with uranyl acetate.
TABLE IX.
Protein–protein interaction of phages P2 and P4
| Protein | Function | Interactions | Notes | References |
|---|---|---|---|---|
| P2–P2 proteins | ||||
| gpC | Immunity repressor | Dimer | Y2H 3D structure | Eriksson et al., 2000; Massad et al., 2010 |
| Cox | Transcriptional repressor, excisionase | Tetramer that can self-associate to octamers | In vitro; cross-linking and gel filtration | Eriksson and Haggard-Ljungquist, 2000 |
| Int | Integrase | Dimer | In vitro; cross-linking Y2H | Frumerie et al., 2005 |
| gpFI | Tail sheath | Polymer, six molecules per sheath striation | Electron microscopy | Lengyel et al., 1974 |
| gpFII | Tail tube | Polymer, six molecules per tube striation | Electron microscopy | Lengyel et al., 1974 |
| gpM | Small terminase subunit | Polymer, 8- to 10-fold ring | Biochemistry | Dokland, unpublished data |
| gpM | Small terminase subunit | gpP | Biochemistry | Bowden and Modrich, 1985 |
| gpN | Major capsid protein | T=7 icosahedron (415 copies) | Cryo-EM and 3D reconstruction | Dokland et al., 1992 |
| gpO | Biochemistry | Chang et al., 2008, 2009 | ||
| gpO | Internal scaffolding protein | Dimer | Biochemistry, chromatography | Chang et al., 2008, 2009 |
| gpN | ||||
| gpP | Large terminase subunit | gpP | Biochemistry | Bowden and Modrich, 1985 |
| gpQ | Connector, portal protein | Dodecameric ring | EM, crystallography | Doan and Dokland, 2007; Rishovd et al., 1994 |
| gpT | Tape measure | gpFII (gpFI ?) | Inside tail tube | |
| gpV | Tailspike | Tetramer | Sedimentation velocity and molecular mass determination | Haggard-Ljungquist et al., 1995; Kageyama et al., 2009 |
| gpV | Tailspike | gpW, base plate component. Belongs to the T4 gene 25-like superfamily | Copurification | Haggard-Ljungquist et al., 1995 |
| P2–other phage proteins | ||||
| Tin | Block superinfection of T-even phages | T4 SSB protein (gp32) | Genetic evidence | Mosig et al., 1997 |
| gpC | Immunity repressor | P4 anti-repressor ε | Genetic evidence. Y2H | Geisselsoder et al., 1981; Liu et al., 1998; Eriksson et al., 2000 |
| gpN | Major capsid protein | P4 Sid; size determinator, external scaffold | Cryo-EM and 3D reconstruction | Marvik et al., 1995; Wang et al., 2000; Dokland et al., 2002 |
| gpN | Major capsid protein | P4 Psu; polarity suppressor; decoration protein | Cryo-EM and 3D reconstruction | Dokland et al., 1993 |
| P2–host proteins | ||||
| gpB | Helicase loader. | E. coli helicase DnaB | Genetic evidence. | Sunshine et al., 1975; Funnell and Inman, 1983; Odegrip et al., 2000 |
| Required for lagging strand synthesis | Electron microscopy. | |||
| In vitro binding. | ||||
| Ogr | Transcriptional activator of late operons | E. coli RNA polymerase α C-terminal domain | Genetic evidence | Sunshine and Sauer, 1975; Fujiki et al., 1976; King et al., 1992; Ayers et al., 1994; Wood et al., 1997 |
| P4–host proteins | ||||
| Psu | Polarity suppressor; anti-terminator | Rho | Genetics; chemical cross-linking | Sauer et al., 1981; Pani et al., 2009 |
| Delta | Transcriptional activator, contains two Ogr domains | E. coli RNA polymerase | Genetics; homology to Ogr | Julien et al., 1997 |
1. Gene control circuits
The decision whether to grow lytically or form a lysogen after P2 infection is dependent on the levels of two repressors: the immunity repressor gpC, which blocks lytic growth, and Cox, which blocks the expression of the genes required for lysogenization. GpC acts as a symmetric dimer, and its three-dimensional structure has been determined (Eriksson et al., 2000; Massad et al., 2010). Cox is multifunctional and acts as a repressor, as well as an excisionase, thereby coupling the transcriptional switch that controls lytic growth versus formation of lysogeny with the integration/excision of the phage genome in or out of the host chromosome. Cox is a tetramer in solution that self-associates to octamers in the absence of DNA (Eriksson and Haggard-Ljungquist, 2000). The transcriptional activator Ogr is required to turn on late gene transcription. Ogr is a Zn finger-containing protein that interacts with the α subunit of the E. coli RNA polymerase, recruiting it to the late promoters (Ayers et al., 1994; Fujiki et al., 1976; King et al., 1992; Sunshine and Sauer, 1975; Wood et al., 1997).
Some P2 prophage genes are not blocked by the action of gpC and instead are expressed from the prophage providing the host cell with new properties. Among these “lysogenic conversion” genes, P2 has three genes, old, fun, and tin, that block superinfection of unrelated phages, but only gpTin is known to have a PPI. P2 gpTin excludes phage T4 by binding to and poisoning T4 gp32 (SSB), thereby inhibiting DNA replication (Mosig et al., 1997).
Satellite phage P4 has to get access to the morphogenetic genes of its helper P2 to grow lytically. This is accomplished by P4-encoded functions during coinfection with P2. The P4 antirepressor Epsilon derepresses the P2 prophage by interacting with gpC, leading to the expression of P2 lytic genes (Eriksson et al., 2000; Geisselsoder et al., 1981; Liu et al., 1998). P4 also has the capacity to trans-activate P2 late genes in the absence of Ogr using the transcriptional activator Delta. Delta contains two Ogr-like domains and recognizes the same DNA sequence as Ogr. Like Ogr, Delta interacts with the α subunit of E. coli RNA polymerase (Christie et al., 2003; Julien et al., 1997; Souza et al., 1977).
2. P2 DNA replication
After infection, the linear DNA molecule is circularized by ligation of the cohesive (cos) ends by the host DNA ligase, and if P2 enters the lytic pathway, DNA replication is initiated by the cis-acting gpA that creates a single-stranded cut at the origin. GpA remains linked covalently to the 5′ end of the cleaved strand (Liu and Haggard-Ljungquist, 1994). Replication occurs via a modified rolling-circle mechanism that generates a double-stranded monomeric circle that is the packaging substrate (Pruss et al., 1975). The cleavage/joining reactions are promoted by two tyrosine residues interspaced by three amino acids in gpA (Odegrip and Haggard-Ljungquist, 2001). The host Rep helicase is required for all DNA replication (Calendar et al., 1970), while the phage-encoded gpB is a helicase loader that interacts with the E. coli DnaB helicase required for lagging strand synthesis (Funnell and Inman, 1983; Odegrip et al., 2000).
3. Head structure and assembly
The main structural proteins involved in P2 capsid assembly are gpN, capsid protein; gpO, internal scaffolding protein; and gpQ, connector or portal protein (Chang et al., 2008; Lengyel et al., 1973; Linderoth et al., 1991). P2 capsids are assembled as empty precursor procapsids consisting of 415 copies of the gpN capsid protein arranged with T=7 icosahedral symmetry and clustered in hexamers and pentamers that make trivalent interactions (Dokland et al., 1992) (Fig. 9). GpQ forms a dodecameric ring that is incorporated into the capsid at one fivefold vertex and through which the DNA is subsequently packaged (Doan and Dokland, 2007; Rishovd et al., 1994).
FIGURE 9.
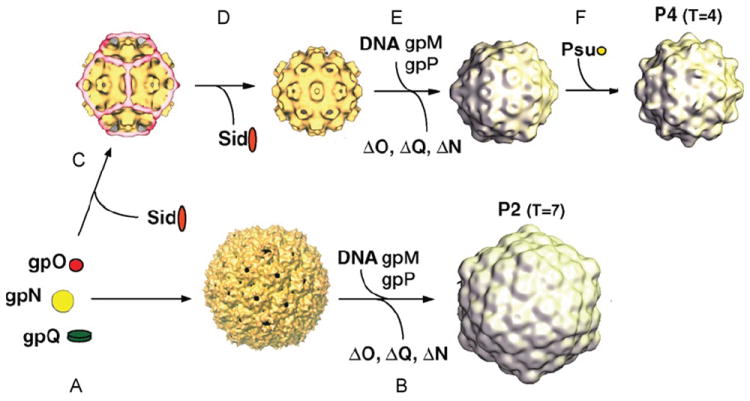
P2 assembly. Schematic diagram of the P2/P4 capsid assembly pathway. (A) Capsid protein gpN, scaffolding protein gpO, and connector (portal) protein gpQ are assembled into the T=7 P2 procapsid. (B) GpN, gpO, and gpQ are then processed to their mature forms, N*, O*, and Q*. DNA is packaged into the procapsid by the P2 terminase complex (gpM and gpP), accompanied by expansion into the mature, angular capsid (B). (C) In the presence of the P4-encoded external scaffolding protein Sid, P2 structural proteins are assembled into a small, T=4 P4 procapsid. Loss of Sid (D), protein processing, and DNA packaging lead to the mature, expanded P4 capsid (E). The decoration protein Psu is added to the mature capsid (F).
GpO plays a dual role in the assembly process as both a protease and a scaffolding protein (Chang et al., 2009; Dokland, 2011). The full-length, 284 residue gpO protein is associated with procapsids at early times (Marvik et al., 1994), but is cleaved autoproteolytically between residues 141 and 142, leaving an N-terminal fragment, gpO* (formerly known as h7), that remains inside the mature capsid (Chang et al., 2008; Lengyel et al., 1973). Concomitantly, gpN and gpQ are cleaved—presumably by gpO—to their mature forms, gpN* and gpQ*, by removal of 31 and 26 residues, respectively, from the N terminus (Rishovd and Lindqvist, 1992; Rishovd et al., 1994). The protease activity of gpO resides in its N-terminal domain, which binds tightly to gpN and constitutes a serine protease similar to that of the herpesviruses (Chang et al., 2009; Cheng et al., 2004; Dokland, 2011). The C-terminal 90 amino acids comprise a scaffolding domain, which is necessary and sufficient for promoting capsid assembly (Chang et al., 2008, 2009; Wang et al., 2006). It too presumably binds to gpN during assembly, but any interaction is transient (Chang et al., 2009). Both N- and C-terminal domains form dimers in solution (Chang et al., 2009).
Circular P2 genomic DNA substrates are packaged into empty procapsids by the terminase complex, which consists of the small and large terminase proteins (gpM and gpP, respectively). Terminase recognizes and cleaves P2 DNA at the cos sites (Bowden and Modrich, 1985; Ziermann and Calendar, 1990). DNA packaging is associated with major structural transitions that lead to a larger, more angular and thin-shelled, mature capsid (Chang et al., 2008; Dokland et al., 1992). A head completion protein, gpL, is added (Chang et al., 2008), and tails, which are assembled via a separate pathway, are attached to the unique gpQ-containing portal vertex (Lengyel et al., 1974).
4. Tail structure
The 135-nm-long tail is composed of an inner tube and a contractile sheath (Lengyel et al., 1974). The head-proximal end of the free tail has two disc-like structures of different thickness and diameter. In the complete phage, only the thinner disc is seen outside the head. The distal end of the tail contains a baseplate with a spike and six fibers. Sixteen essential tail genes have been identified located in three transcription units. GpFI and gpFII constitute the tail sheath and tail tube, respectively, and there are about 200 copies of each protein per tail (Temple et al., 1991). RpR and gpS are involved in tail completion, as amber mutations in the respective gene produce giant tails and giant tail sheaths (Linderoth et al., 1994). GpV is the tail spike, which forms a trimer in solution (Haggard-Ljungquist et al., 1995; Kageyama et al., 2009). GpW copurifies with gpV and is thus thought to be part of the baseplate, while gpJ localizes to the edge of the baseplate or proximal part of the tail fiber by immunoelectron microscopy (Haggard-Ljungquist et al., 1995). GpH is believed to be the distal part of the tail fiber because it contains regions similar to tail fiber proteins of phages Mu, P1, λ, K3, and T2 (Haggard-Ljungquist et al., 1992). A region of gene H is highly similar to the variable part of the S gene (Sv) of phage Mu and the variable part of gene 19 (19v) of phage P1, in both cases in the (+) orientation of the invertible segments. P2 does not have an invertible segment, but it contains a sequence identical to the left repeat of the Mu invertible G segment located within the H gene and upstream of the region with a high similarity to Mu Sv and P1 19v, indicating that the P2 H gene has been frozen in the (+) orientation, probably by a deletion covering the right inverted repeat and the invertase. This region is therefore inferred to constitute the receptor-binding tip of the tail fiber. By its similarities to gpU and gpU′ of phages Mu and P1, and gp38 of phages T4, Tula, and Tulb, gpG is probably required for fiber assembly. GpT is likely the tape measure, as it is the only protein long enough to span the length of the tail shaft (Christie et al., 2002). Function of the products of genes X, I, E, E′ (programmed -1 frameshift extension of E), U, and D remains unknown.
5. P4-induced redirection of P2 capsid assembly
Satellite phage P4 cannot form phage particles by itself, but in the presence of phage P2, P4 genomes are packaged into phage particles using structural proteins supplied by P2 (Six, 1975). However, the P4 capsid is smaller than P2 and contains only 235 copies of gpN, arranged on a T=4 icosahedral lattice (Dokland et al., 1992) (Fig. 9). This size control is affected by the P4 gene product Sid (size determination)(Barrett et al., 1976), which forms an dodecahedral cage-like scaffold around the outside of P4 procapsids (Marvik et al., 1995). Sid self-associates into trimers and dimers that interact with gpN only in two hexamer subunits near the twofold symmetry axes (Wang et al., 2000; Dokland et al., 2002). Gene N mutants, called sir (sid responsiveness), render the capsid protein unable to form small capsids even in the presence of Sid (Six et al., 1991). Conversely, the so-called super-sid or nms (N mutation sensitive) mutations in sid act as second-site suppressors of the sir mutations and enable the formation of small capsids (Kim et al., 2001). Although well-formed small procapsids can be assembled from Sid and gpN alone (Dokland et al., 2002), gpO is required for the formation of viable P4 phage (Six, 1975). Presumably this is because its protease activity is essential, and gpO may also facilitate incorporation of the portal. Indeed, sid complements the P2 Oam279 mutant, which produces a 237 residue N-terminal fragment of gpO that includes the protease domain but lacks scaffolding activity (Agarwal et al., 1990). The P4 genome contains P2 cos sites and is packaged into small capsids using the P2 terminase (Ziermann and Calendar, 1990). The P4-encoded protein Psu is a suppressor of Rho-dependent transcription termination (Pani et al., 2009; Sauer et al., 1981); it also functions as a decoration protein that binds to gpN hexamers and stabilizes the P4 capsid (Dokland et al., 1993; Isaksen et al., 1993).
E. Pneumococcus phage Dp-1 and Cp-1
The lytic bacteriophages Dp-1 and Cp-1 infect the Gram-positive bacterium Streptococcus pneumoniae, which colonizes the mucosal surface of the upper respiratory tract and causes serious invasive infections such as pneumonia, meningitis, and sepsis (van der Poll and Opal, 2009). Dp-1 was the first isolated Streptococcus phage and is an unassigned member of the Siphoviridae (Fig. 10) (McDonnell, Lain and Tomasz, 1975). It was reported that the Dp-1 virion surprisingly contains lipids that might be arranged as a bilayer surrounding the capsid (Lopez et al., 1977), a feature not reported for any other phage belonging to the Caudovirales. The lytic enzyme Pal of Dp-1 has been studied intensively because it kills virulent streptococci efficiently and has been used as an enzybiotic to cure pneumococcal infections in animals (Loeffler et al., 2001; Rodriguez-Cerrato et al., 2007). The podovirus Cp-1 lytic enzyme Cpl1 was also demonstrated to be an efficient enzybiotic (Loeffler et al., 2003). The Cp-1 genome is a linear, 20-kbp dsDNA predicted to encode 28 proteins and a packaging RNA (pRNA). Like ϕ29-like viruses, Cp-1 uses protein priming for the initiation of DNA replication (Martin et al., 1996). The biology of Dp-1 and Cp-1 is otherwise only poorly understood.
FIGURE 10.
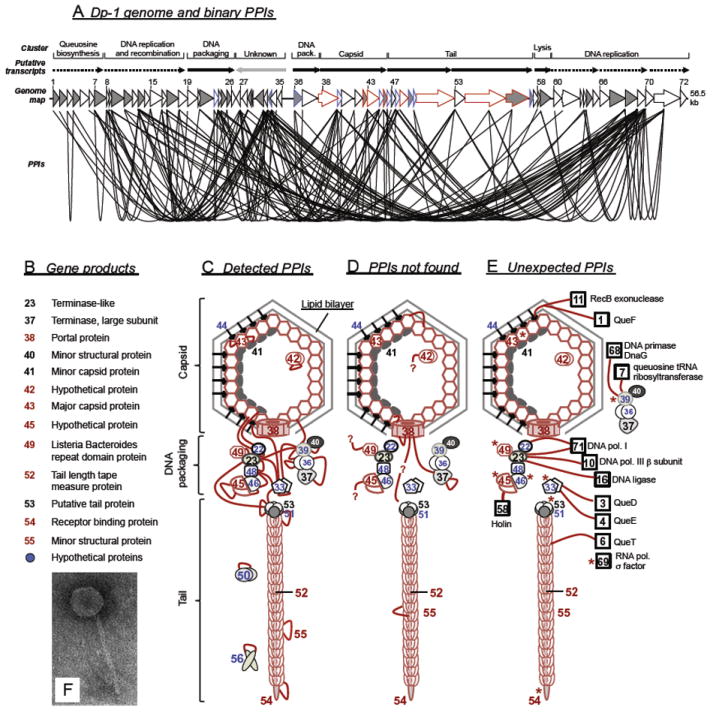
The Dp-1 genome and interactome. (A) Dp-1 genome map. Open reading frames (ORFs) corresponding to their transcription direction are shown as arrows (genome map). Predicted transcripts and functional gene clusters are indicated. All protein–protein interactions (PPIs) identified by systematic Y2H screens among Dp-1 proteins are shown as lines that connect the corresponding ORF symbols. Proteins that bind themselves (homomers) are highlighted by gray ORF symbols. Other ORF colors: refer to Figure C-E. (B–E) A Dp-1 virion model based on identified binary PPIs, mass spectrometry analysis (MS) of mature Dp-1 virions, and homology predictions. (B) Dp-1 virion proteins, including structural proteins identified by MS analysis (in red), or proteins with a homology-based annotation that indicates a virion-related function (in black). (C) Core model of the Dp-1 virion: proteins highlighted in red were identified by MS analysis as structural components. Proteins highlighted in blue represent hypothetical gene products that interact with structural components, thus presumably playing a transient role in virion assembly as they were not identified by MS analysis of Dp-1 virions. Red edges that connect the proteins were shown to interact in Y2H tests. (D) Y2H screens do not identify all expected PPIs. Red lines represent PPIs that were expected to occur among the corresponding proteins known from homologues but were not identified in the systematic Y2H screens. (E) Systematic Y2H screens reveal unexpected PPIs. Proteins are included that were shown to bind with structural proteins and putative morphogenetic factors, e.g., DNA metabolism (DNA polymerase subunits, RecB, DNA ligase, DnaG), queuosine biosynthesis (Que proteins), lysis (Holin), and transcription (sigma factor). Interactions are symbolized by edges that connect the corresponding proteins (in the case of sigma factor gp69, interaction partners are highlighted by an asterisk). (F) Electron micrograph of a mature Dp-1 virion. C–F from Sabri et al. (2010), with permission.
The 56.5-kbp genome of Dp-1 is predicted to encode 72 proteins (Fig. 10A) (Sabri et al., 2011). Approximately half of the gene products could be functionally annotated by homology predictions. However, in this and hundreds of other completely sequenced phage genomes, more than half of phage gene products could not be annotated. Proteome-wide Y2H screens, coupled with mass spectrometry analysis (MS) of mature Dp-1 virions, identified 156 unique PPIs among 57 Dp-1 proteins [a detailed list can be found in Sabri et al. (2011)], and MS revealed that the mature virion is composed of eight structural proteins. A Dp-1 virion model (Figs. 10B and 10C) physically links many proteins with unknown function to the virion. Because they bind structural proteins but are absent from the mature virion, these Y2H PPIs either indicate novel structural components (when found by MS of the virion particles) or virion assembly facilitators. For instance, homology comparisons of gp40, gp41, and gp53 predict that these proteins are structural (Fig. 10B). However, MS analysis did not detect them as components of the virion. Even though functional predictions by homology comparisons are extremely helpful in proteome annotation, a certain fraction of proteins may be wrongly annotated in the absence of other experimental data (i.e., whether a protein is annotated as a structural component or an assembly facilitator can have an important impact on future experiments).
Such a comprehensive analysis also reveals unexpected interactions that normally would not be tested in hypothesis-driven experiments (Fig. 10E). For example, Dp-1 proteins believed to be involved in DNA replication and recombination, transcriptional regulation, and other pathways exhibit interactions with structural components. This indicates that DNA packaging and DNA replication/repair might be coupled (as in phage T4), for example, by resolving intermediate DNA structures in parallel with packaging. Although the identified PPIs were helpful in obtaining the first hints about the function of the proteins, not all expected PPIs between the structural proteins were detected (Fig. 10D). Nevertheless, this study demonstrated that homology-based genome annotation complemented by proteomic approaches is a useful strategy to gain novel insights into the biology of poorly understood bacteriophages.
The same combination of genome annotation and proteomics was repeated for the 28 putative Cp-1 proteins (Häuser et al., 2011). They detected a total of 12 structural proteins by MS analysis. More than half the proteins showed at least one protein–protein interaction with 17 binary interactions in total. Notably, nearly all were detected between proteins encoded in the structural gene cluster, reflecting a highly modular organization of its interactome. In contrast, Dp-1, with its larger proteome/interactome, shows more cross-communication between proteins belonging to different pathways, although a functional enrichment of interactions between proteins belonging to related cellular processes was still detected (Sabri et al., 2011). It is likely that larger phage proteomes/interaction networks routinely exhibit a higher frequency of protein cross-communication at the protein interaction level due to regulation between various pathways (i.e., the larger the proteome, the more promiscuous the interaction network).
F. Bacillus subtilis phage ϕ29-like viruses
ϕ29-like phages belong to the Picovirinae subfamily of the Podoviridae. Members include ϕ29, ϕ15, PZA, BS32, B103, M2Y (M2), Nf, and GA-1. All are lytic; most infect B. subtilis, but some grow in B. pumilus, B. amyloliquefaciens, and B. licheniformis (Meijer et al., 2001a). The characteristics of their virions are a prolate capsid (e.g., ϕ29 is 41.5 by 31.5 nm) with attached head fibers, a connector, a short noncontractile tail (e.g., ϕ29 with 32.5 nm in length), and the presence of tail appendages (Anderson et al., 1966) (Fig. 11). GA-1 and M2Y lack the gene for head fibers (Yoshikawa et al., 1986), which may stabilize the capsid (Tao et al., 1998). The approximate 20-kbp genome of ϕ29-like phages is linear and encodes about 25 proteins. The 5′ ends of the genome are attached covalently to a terminal protein (TP). ϕ29-like viruses are among the smallest dsDNA phages isolated so far and thus represent minimal model systems (Meijer et al., 2001a).
FIGURE 11.
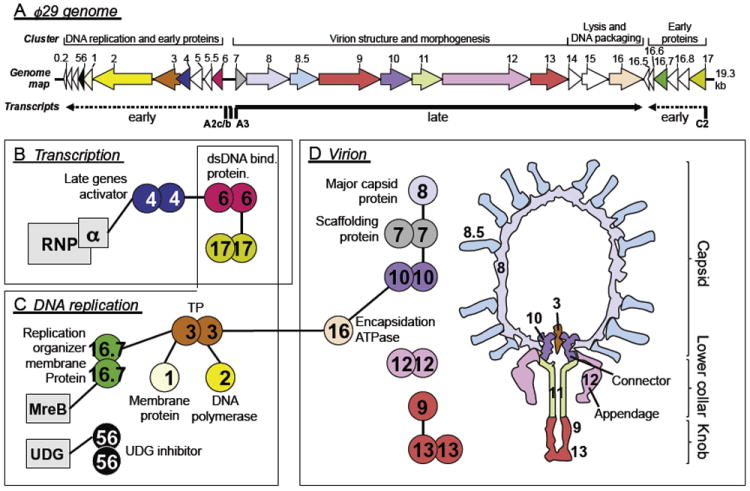
Genome map and literature curated ϕ29 interactome. (A) Genome and simplified transcriptional map of ϕ29 (after Meijer et al., 2001a). Major promoters are indicated by their names, and major transcripts are shown as dashed (early) and solid (late) arrows. Other promoters are not indicated for clarity. Open reading frame (ORF) arrows indicate transcription direction. The color of ORF symbols corresponds to the color used for protein symbols in B, C, and D. PPIs known for ϕ29, organized by their cellular function (transcription, DNA replication, or virion). B. subtilis proteins are shown as rectangles, phage proteins as circles. Equal protein symbols contacting each other indicate homomers. Heteromeric PPIs are highlighted by protein symbols that are connected by lines. (D) Schematic cross view of the mature ϕ29 virion (after Xiang et al., 2008). UDG, uracil–DNA glycosylase; RNAP, RNA polymerase; TP, terminal protein.
1. Protein–protein interactions
A literature and database search on ϕ29-like viruses retrieved 30 PPIs (Tables X and XI) with evidence mainly from directed experiments and structure determination studies. Twenty-three are attributed to phage ϕ29 itself, five to GA-1, and two to Nf, emphasizing the role of ϕ29 as a model for the whole group. The biology of other ϕ29-like phages is poorly understood, and only a few proteins have been characterized experimentally. In some cases, ϕ29 PPIs are conserved among other ϕ29-like homologues, supporting a close relationship of these phages. However, there are also differences that reveal interesting insights into their evolution. Known ϕ29 protein–protein interactions include proteins involved in transcriptional regulation, DNA replication, and virion structure. Only three PPIs between ϕ29 and B. subtilis proteins have been identified so far.
TABLE X.
Protein–protein interactions of phage ϕ29 including phage–host interactionsa
| ϕ29 1 | Function 1 | ϕ29 2 | Function 2 | References |
|---|---|---|---|---|
| gp2 | DNA polymerase | gp3 | DNA terminal protein TP | Kamtekar et al., 2006; Blanco et al., 1987; Dufour et al., 2000; González-Huici et al., 2000 |
| gp3 | DNA terminal protein TP | gp1 | Membrane-associated early protein | Bravo et al., 2000 |
| gp3 | DNA terminal protein TP | gp3 | DNA terminal protein TP | Serna-Rico et al., 2003 |
| gp3 | DNA terminal protein | gp16 | Packaging ATPase | Guo et al., 1987 Grimes and Anderson, 1997 |
| gp4 | Late gene activator | gp4 | Late gene activator | Badia et al., 2006 Mencía et al., 1996b |
| gp4 | Late gene activator | gp6 | Histone-like, nonspecific DNA-binding protein | Calles et al., 2002; Camacho and Salas, 2001 |
| gp6 | Histone-like, nonspecific dsDNA-binding protein | gp6 | Histone-like, nonspecific dsDNA-binding protein | Abril et al., 1999; Pastrana et al., 1985 |
| gp7 | Scaffolding protein | gp7 | Scaffolding protein | Morais et al., 2003 |
| gp7 | Scaffolding protein | gp8 | Major head protein | Lee and Guo, 1995 |
| gp7 | Scaffolding protein | gp10 | Upper collar/connector protein | Guo et al., 1991; Lee and Guo, 1995 |
| gp10 | Upper collar/connector protein | gp10 | Upper collar/connector protein | Guasch et al., 2002; Simpson et al., 2001 |
| gp12 | Preneck appendage protein (anti-receptor) | gp12 | Preneck appendage protein (anti-receptor) | Xiang et al., 2009 |
| gp13 | Tail-associated, peptidoglycan-degrading enzyme | gp9 | Tail knob protein | García et al., 1983 |
| gp13 | Tail-associated, peptidoglycan-degrading enzyme | gp13 | Tail-associated, peptidoglycan-degrading enzyme | Xiang et al., 2008 |
| gp16.7 | Replication organizer; DNA-binding membrane protein p16.7 | gp16.7 | Replication organizer; DNA-binding membrane protein p16.7 | Asensio et al., 2005; Albert et al., 2005; Crucitti et al., 1998; Serna-Rico et al., 2003; Meijer et al., 2001b |
| gp16.7 | Replication organizer; DNA-binding membrane protein p16.7 | gp3 | DNA terminal protein TP | Serna-Rico et al., 2003 |
| gp17 | Early protein involved in early DNA replication | gp6 | Histone-like, nonspecific dsDNA-binding protein | Crucitti et al. 2003 |
| gp17 | Early protein involved in early DNA replication | gp17 | Early protein involved in early DNA replication | Crucitti et al., 2003 |
| gp56 = gp0.8 | Host uracil-DNA glycosylase inhibitor | gp56= gp0.8 | Host uracil-DNA glycosylase inhibitor | Serrano-Heras et al., 2007) |
| ϕ29–Bacillus subtilis interactions | ||||
| gp4 | Late gene activator (3) | RpoA | RNA polymerase subunit α | Mencía et al., 1996a; Monsalve et al., 1996b |
| gp16.7 | Replication organizer, DNA-binding membrane protein gp16.7 (2) | MreB | Actin-like cytoskeleton protein | Muñoz-Espín et al., 2009 |
| gp56 = gp0.8 | UDG inhibitor (1) | UDG | Uracil-DNA glycosylase | Serrano-Heras et al., 2006 |
Data from BOND, IntACT, and PDB (ϕ29, Taxon id 186846). All proteins are termed “gp” (gene product), although they are also termed “p” (protein), e.g., gp4 = p4. In the phage–host interaction list, the first protein is from ϕ29, the second from B. subtilis. (1) Host RNAP binds via α subunit to p4 (i) bound at the late promoter PA3, thus activating late transcription and (ii) bound to the promoter PA2c, thus blocking transcription through promoter clearance inhibition. (2) PPI plays a role in the organization of ϕ29 DNA replication as peripheral helix-like structures. (3) Inhibition of host UDG; avoids removal of uracil from ϕ29 DNA via UDG-base excision repair.
TABLE XI.
Protein–protein interactions of ϕ29-related phagesa
| Phage | Protein 1
|
Protein 2
|
References | ||
|---|---|---|---|---|---|
| Function | Function | ||||
| GA-1 | gp6 | Histone-like, nonspecific dsDNA binding protein | gp6 | Histone-like, nonspecific dsDNA-binding protein | Freire et al., 1996 |
| Nf | gp6 | dsDNA binding protein | gp6 | dsDNA-binding protein | Freire et al., 1996 |
| GA-1 | gp2 | DNA polymerase | gp3 | DNA terminal protein | Illana et al., 1996; Longás et al., 2006 |
| Nf | gp2 | DNA polymerase | gp3 | DNA terminal protein | Longás et al., 2006; González-Huici et al., 2000 |
| GA-1 | gp5 | Single-stranded DNA-binding protein SSB (1) | gp5 | Single-stranded DNA-binding protein SSB | Gascón et al., 2000 |
| GA-1 | gp12 | Neck appendage protein (2) | gp12 | Neck appendage protein | Schulz et al., 2010 |
| GA-1 | gp4G | Transcriptional regulator (3) | gp4G | Transcriptional regulator | Horcajadas et al., 1999 |
Data from BOND, IntACT, and PDB (searched for ϕ29, Taxon id 186846). (1) ϕ29 and Nf SSB were shown to be stable monomers in solution, whereas GA-1 SSB forms homohexamers. (2) Homotrimer; autocatalytic cleavage similar to ϕ29 gp12. PDB: 3GUD. (3) Homodimer; negative regulator of early transcription; was shown not to interact with RNAP α subunit as the homologue gp4 of ϕ29.
2. Protein-primed DNA replication
Phage ϕ29 DNA polymerase (DNAP, gp2) interacts with terminal protein (TP, gp3) to initiate genome replication. Interactions have also been shown between the corresponding GA-1 and Nf orthologs (Table XI). The structure of the ϕ29 TP–DNAP complex has been determined and suggests that the priming domain of TP occupies the dsDNA-binding tunnel, allowing the DNAP to bind negatively charged amino acid residues of TP (Kamtekar et al., 2006).
Phage ϕ29 replication depends on the presence of a single dAMP residue linked covalently via its 5′-phosphoryl group to the hydroxyl group of Ser232 of TP. Formation of the phosphoester bond is catalyzed by the DNAP itself (Blanco and Salas, 1984; Hermoso et al., 1985); thus formation of a stable DNAP–TP heterodimeric protein complex is required (Blanco et al., 1987). The TP–dAMP is elongated by the same DNAP (Méndez et al., 1992). After the synthesis of six to nine nucleotides, DNAP dissociates from TP and starts continuous, linear synthesis (Méndez et al., 1997). This mechanism of protein-dependent DNA replication initiation was shown for Nf and GA-1 (Illana et al., 1996; Longás et al., 2008), S. pneumoniae phage Cp-1, E. coli phage PRD1, adenoviruses, and some plasmids and bacterial genomes (e.g., Streptomyces), although there may be differences in the various priming mechanisms (de Jong and van der Vliet, 1999; Martin et al., 1996; Salas, 1991).
The TP–DNAP interaction plays a central role in ϕ29 DNA replication initiation. However, other PPIs with TP are known, and thus protein-primed DNA replication may be more complicated than is currently known (Figs. 11C and Fig. 12). Chemical cross-linking shows that TP forms homodimers (Serna-Rico et al., 2003). Analysis of TP mutants suggests that the parental TP (TP bound to the parental genome) recruits the free TP–DNAP complex via a TP–TP interaction (Illana et al., 1999), but an additional interaction between parental TP and DNAP might be important for the specificity of replication initiation (González-Huici et al., 2000).
FIGURE 12.
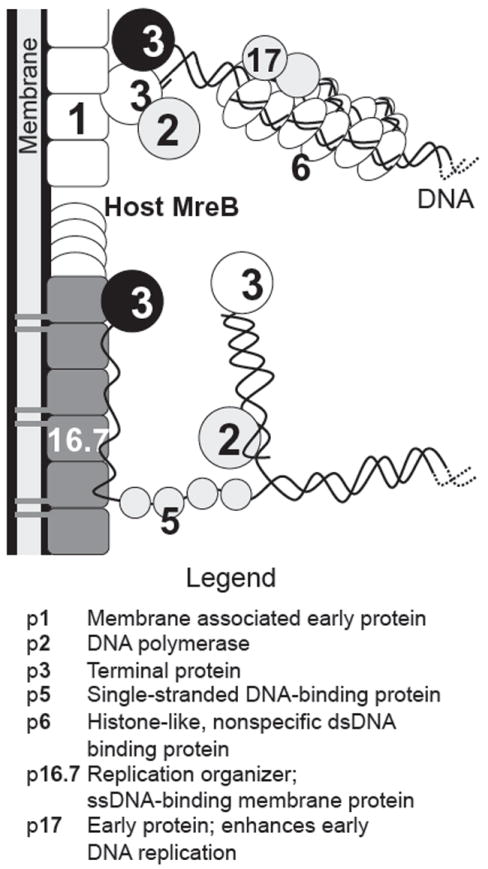
Interactions in ϕ29 genome replication and membrane localization. Proteins that contact each other have been shown to interact. For clarity, only one genome end is shown. Parental TP is highlighted by a black symbol, whereas primer TP is in white. The upper part of the figure includes PPIs that preferentially play a role in replication initiation. The lower part of the figure indicates elongated DNA with the associated ssDNA regions. Because p16.7 and p5 (SSB) both bind ssDNA, they could either redirect the parental ssDNA strand to the membrane via TP-p16.7 and p16.7–ssDNA interactions or stabilize it in the cytoplasm via SSB alone (Meijer et al., 2001b). After Bravo et al. (2000) and Serna-Rico et al. (2003).
TP interacts with the replication organizer gp16.7 (Serna-Rico et al., 2003) (Fig. 12). Gp16.7, which has the potential to bind to both ssDNA and dsDNA, is an integral membrane protein consisting of an N-terminal membrane anchor and a C-terminal cytoplasmic domain. It localizes ϕ29 DNA replication to the membrane (Meijer et al., 2000, 2001b; Serna-Rico et al., 2002, 2003). The C-terminal domain of gp16.7 dimerizes in solution but forms larger filaments in the presence of ssDNA (Asensio et al., 2005; Serna-Rico et al., 2003). Replication, which starts at both DNA ends, first produces a so-called type I replication intermediate consisting of dsDNA with ssDNA regions caused by displacement of the non-template DNA parental strand. Later, when the two replication forks have merged, the parental DNA strands become separated into two type II replication intermediates, still containing ssDNA regions in the parental template. A crystal structure of the gp16.7 DNA-binding C-terminal domain complexed with dsDNA reveals three gp16.7 dimers forming a deep, positively charged longitudinal cavity that interacts nonspecifically with the DNA phosphate backbone (Albert et al., 2005). Rp16.7 localizes in peripheral helix-like structures in a bacterial MreB cytoskeleton-dependent manner (Muñoz-Espín et al., 2010). In fact, protein gp16.7 interacts directly with MreB; this interaction is required for efficient ϕ29 DNA replication, allowing simultaneous replication of multiple templates at various peripheral sites.
Phage ϕ29 gp1 is another early protein that binds TP (Bravo et al., 2000). Rp1, like gp16.7, is membrane associated and self-associates into long filamentous structures (Bravo and Salas, 1997, 1998). Its membrane association and polymerization are mediated through a C-terminal hydrophobic sequence, whereas binding to TP involves an N-terminal region. In contrast to gp16.7, gp1 does not bind DNA (Fig. 12) and is thought to be another component that directs ϕ29 DNA replication to the membrane (Bravo et al., 2000). Any interplay between gp16.7 and gp1 in this process is unknown.
The ϕ29 histone-like, nonspecific dsDNA-binding protein gp6 forms a nucleoprotein complex with ϕ29 DNA that covers most of the genome, implicating it in genome organization (Serrano et al., 1994). Rp6 either unwinds the genome termini or interacts with TP directly, thus making a TP complex competent to bind the genome ends (Blanco et al., 1986; Serrano et al., 1994). Rp6 forms long oligomeric protein filaments thought to function as a protein backbone for DNA binding (Abril et al., 1999). GA-1 and Nf gp6 orthologues also self-associate (Freire et al., 1996). Rp6 also binds gp17, an early protein that stimulates the first viral replication round in vivo when other replication proteins are present at low levels (Crucitti et al., 1998, 2003). In contrast to gp6, gp17 does not bind DNA but its interaction with gp6 enhances gp6 binding to the replication origins, thereby stimulating early rounds of replication.
A homomeric interaction was demonstrated for the single-stranded DNA-binding protein of GA-1. GA-1 SSB was surprisingly found as a homohexamer in solution, whereas SSBs of Nf and ϕ29 (p5) appear to be stable monomers (Gascón et al., 2000). The homohexamer binds to ssDNA with higher affinity than the monomer and exhibits a higher stimulatory effect on DNA replication.
Bacteriophages have evolved different mechanisms to protect their genomes from degradation by host nucleases in the cell. One protective strategy includes the incorporation of noncanonical nucleotides into their DNA. For instance, B. subtilis phage PBS2 uses uracil instead of thymine (Takahashi and Marmur, 1963). PBS2 achieves this unusual goal by increasing the dUTP pool, relative to dTTP, after infection. In addition, PBS2 encodes Ugi, a protein that specifically inhibits the host uracil–DNA glycosylase (UDG)(Cone et al., 1980), which is involved in the first step of removing incorporated uracil residues via the base excision repair pathway. Normally, this takes place to avoid mutational effects in the host genome but it would impair the incorporation of uracil into the PBS2 genome. Cocrystallization of Ugi and UDG revealed that Ugi mimics uracil-containing duplex DNA, thereby blocking efficient DNA binding of UDG (Putnam et al., 1999). ϕ29 blocks the host UDG by an interaction with the early protein gp56 (gp0.8) that competes with the DNA-binding ability of UDG (Serrano-Heras et al., 2006, 2007, 2008). Although the ϕ29 genome does not contain uracil (Serrano-Heras et al., 2006), ϕ29 DNAP is able to incorporate dUMP with a catalytic activity only twofold lower than for that of dTMP. Because ϕ29 DNAP does not discriminate stringently between dTMP and dUMP, UDG inhibition via gp56 minimizes uracil incorporation.
3. Interactions during transcriptional regulation
Transcriptional regulation of the ϕ29 genome has been studied extensively (Meijer et al., 2001a; Rojo et al., 1998). The transition from early to late gene expression is controlled by gp4 (Fig. 11). Gp4 is a homodimer in solution (Mencía et al., 1996b); it forms a tetramer when bound to the intergenic region between gene 6 and gene 7, which contains the A2b, A2c, and A3 promoters (Fig. 11A). One binding site is upstream of A3, the late promoter that lacks a -35 box (Barthelemy and Salas, 1989). Once gp4 is bound, it recruits, via a PPI, the host RNAP, leading both to its stabilization on the promoter and transcriptional activation (Nuez et al., 1992). Although gp4 activates late gene expression, it represses early gene expression from the A2b and A2c promoters (Monsalve et al., 1997; Rojo and Salas, 1991). Interestingly, repression of A2c and activation of A3 involve the same residues of gp4 with the C-terminal domain of the RNAP α subunit (Mencía et al., 1996a; Monsalve et al., 1996a,b). Rp6 also interacts with gp4 in regulating the transcription of promoters A2b, A2c, and A3. Gp6 binds gp4 cooperatively to form a repression complex at promoters A2b and A2c while stimulating A3 (Calles et al., 2002; Elías-Arnanz and Salas, 1999).
4. Virion structure and morphogenesis
Cryo-EM analyses have revealed many details about the ϕ29 virion structure (Morais et al., 2003, 2005; Tao et al., 1998; Xiang et al., 2008, 2009), and many structures of virion proteins and factors are known (Table X). Assembly starts from a connector core particle consisting of 12 subunits of gp10 on which the scaffolding protein gp7 binds, followed by recruitment of the major capsid protein gp8 (Guo et al., 1991; Lee and Guo, 1995). The scaffolding protein is needed to create a lattice of concentric shells (Morais et al., 2003) and helps to organize the single capsomers built up by gp8 except for the basal connector capsomer. ϕ29 proheads were shown to consist of 235 copies of gp8 arranged as hexameric and pentameric capsomeres, indicating that direct interactions between gp8 monomers occur (Morais et al., 2005). The capsid fiber protein gp8.5, which is present in ϕ29 proheads, is presumably connected via gp8 to the prohead and is suggested to be organized as homotrimers via interactions of a C-terminal coiled-coil region (Morais et al., 2005). In addition to gp8, gp8.5, and gp10, ϕ29 proheads contain a homopentameric packaging RNA (pRNA) that binds to the connector, forming a basket-like complex (Simpson et al., 2000). pRNA is needed to translocate DNA into the prohead. Furthermore, the DNA translocation machinery needs the ATPase gp16 to drive the packaging reaction (see also chapter 9 of this volume). Rp16 was shown to bind to the connector protein (Ibarra et al., 2001). While other phages use specific DNA sequences to recognize and recruit the genome to the procapsids, ϕ29 DNA packaging initiation is dependent on the TP gp3 present at the genome ends, indicating that an interaction between gp16 and DNA-bound gp3 recruits the DNA to proheads and initiates packaging (Guo et al., 1987). Release of the scaffolding protein is associated with DNA packaging and pRNA and gp16 are released when there is no more DNA to package.
An interesting aspect of ϕ29 is that the virion contains several enzymes that are associated with structural proteins. For instance, gp12 is a homotrimer that participates in host cell recognition and entry. It also shows autocatalytic cleavage, although it remains unclear what the function of that reaction is. Similarly, gp13 is bound as a dimer at the distal end of the ϕ29 tail knob to gp9; gp13 cleaves both the polysaccharide backbone and peptide cross-links of the cell wall during infection.
IV. CONCLUSIONS
A. High-throughput data vs small-scale interactions
It took several decades to collect the interactions of phage λ, even though it has only a few dozen genes. These interactions have been studied in painstaking individual experiments. Since the mid-1990s, new strategies have been developed to identify and analyze PPIs on a larger scale, particularly two-hybrid-related methods and techniques involving protein complex purification and mass spectrometry. Despite the pioneering efforts of (Bartel et al., 1997) to map the interactome of phage T7 in a “single” experiment, their results remain somewhat contentious. Why is that? Studies show that each two-hybrid system produces system-specific data, and each system may only be able to identify as few as 20–30% of all interactions (Chen et al., 2010). It has been shown that the application of up to 10 different Y2H systems can identify up to 80% of all interactions, but there are still some that go undetected (Chen et al., 2010). This limited sensitivity can be explained by differences between the individual Y2H systems, expression levels, the way fusion proteins are constructed (e.g., N-terminal fusions vs C-terminal fusions), or linkers between fusion proteins. Some interactions may be completely undetectable by Y2H assays as these experiments are typically done in yeast, which may not have the necessary chaperones for bacterial or phage proteins to fold properly or the proteins may localize differently than in their normal organism.
Similar limitations apply to strategies that involve protein complex purification and mass spectrometry. Because many purifications are sensitive to small changes in protocols, even when a complex is purified within the same laboratory different preparations often result in different protein compositions in subsequent MS analyses (Gavin et al., 2006). If two different laboratories purify the same protein complexes using different methods, the results may differ even more dramatically (Hu et al., 2009).
Even if large-scale screens identify the majority of interactions, they will always also identify false positives. For instance, a screen of λPPIs yielded 97 interactions among λ proteins (Rajagopala et al., 2011). While the authors detected about half of the 33 published interactions listed in Table III, they also found many new interactions. Many do not seem plausible and are likely false positives. However, there is no way of knowing which ones are in fact false. We can only make educated guesses and rank all interactions by their plausibility, selecting only the best candidates for more detailed studies.
B. Phage of the same host has different interactomes
Where it is known, all bacteria are infected by numerous unrelated phages. While unrelated phages cannot have identical intraphage interactions, their proteins may interact with the same targets in their host cells. While interaction databases do not have enough information about phage–host interactions to draw general conclusions, several lines of evidence indicate that this is not generally true. For instance, the well-studied phages T7 and λ interact with different host proteins. They seem to share only one common interaction target, namely the RecBCD recombinase complex (see Tables II and IV). λ certainly has about twice as many host–phage interactions than T7, but this is certainly due to λ being temperate. T4 probably does interact somewhat more extensively with the host than T7, and one could argue that this is due to genome size. Similarly, in Streptococcus phage Dp-1 the authors found 38 interactions among the 72 phage proteins and its host, while the 28 (unrelated) proteins of phage Cp1 showed only 11 such interations (Häuser et al., 2011). Together, Dp-1 and Cp-1 target a total of 36 host proteins, of which only 2 are shared. More host–phage interactomes are needed to draw more general conclusions though.
C. Evolution of interactions [λ, P22]
We expect that related phage have similar interaction patterns, both among phage proteins and between phage and host proteins. This is certainly true for very closely related phages. However, phages evolve much faster than any other living system and thus may lose or gain interactions faster too. Unfortunately, there is not enough interaction data to make detailed comparisons, even among the well-studied lambdoid phages. However, many individual cases of proteins are not obviously homologous by sequence but which are believed to share a common ancestor, for example, the capsid proteins of λ and HK97. These proteins also have similar structures and make similar contacts in the capsid (Hendrix, 2005). Evolutionary relationships are also supported by the extensive similarities in parts of their genomes, that is, genome mosaicism (Casjens, 2011; Casjens and Thuman-Commike, 2011; Hendrix and Casjens, 2006). It remains to be seen how many interactions are conserved and how successful phages are in inventing new interactions (or discarding them). More systematic studies of multiple phages in parallel will be required to obtain comparable data sets in which differences in interaction patterns are not based on the use of noncomparable methods.
Acknowledgments
Work on this chapter was partially funded by NIH Grant R01GM79710 to P.U., the European Union (Grant HEALTH-F3-2009-223101)(P.U., R.H., and S.B.), NIH Grant RO1 AI074825 (S.R.C), and the Spanish Ministry of Science and Innovation (Grant BFU2008-00215) to M.S.
ABBREVIATIONS
- EM
electron microscopy
- LC
liquid chromatography
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- ORF
open reading frame
- PPI
protein–protein interaction
- Y2H
yeast two-hybrid
References
- Abbani MA, Papagiannis CV, Sam MD, Cascio D, Johnson RC, Clubb RT. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc Natl Acad Sci USA. 2007;104:2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abril AM, Marco S, Carrascosa JL, Salas M, Hermoso JM. Oligomeric structures of the phage phi29 histone-like protein p6. J Mol Biol. 1999;292:581–588. doi: 10.1006/jmbi.1999.3078. [DOI] [PubMed] [Google Scholar]
- Agarwal M, Arthur M, Arbeit RD, Goldstein R. Regulation of icosahedral virion capsid size by the in vivo activity of a cloned gene product. Proc Natl Acad Sci USA. 1990;87:2428–2432. doi: 10.1073/pnas.87.7.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005a;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: Structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005b;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Albert A, Munoz-Espin D, Jimenez M, Asensio JL, Hermoso JA, Salas M, Meijer WJ. Structural basis for membrane anchorage of viral phi29 DNA during replication. J Biol Chem. 2005;280:42486–42488. doi: 10.1074/jbc.C500429200. [DOI] [PubMed] [Google Scholar]
- Alfano C, McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem. 1989;264:10699–10708. [PubMed] [Google Scholar]
- Ali SS, Beckett E, Bae SJ, Navarre WW. The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain. J Bacteriol. 2011;193:4881–4892. doi: 10.1128/JB.05198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DL, Hickman DD, Reilly BE. Structure of Bacillus subtilis bacteriophage phi 29 and the length of phi 29 deoxyribonucleic acid. J Bacteriol. 1966;91:2081–2089. doi: 10.1128/jb.91.5.2081-2089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WF, Takeda Y, Echols H, Matthews BW. The structure of a repressor: Crystallographic data for the Cro regulatory protein of bacteriophage lambda. J Mol Biol. 1979;130:507–510. doi: 10.1016/0022-2836(79)90437-6. [DOI] [PubMed] [Google Scholar]
- Ang D, Keppel F, Klein G, Richardson A, Georgopoulos C. Genetic analysis of bacteriophage-encoded cochaperonins. Annu Rev Genet. 2000;34:439–456. doi: 10.1146/annurev.genet.34.1.439. [DOI] [PubMed] [Google Scholar]
- Asensio JL, Albert A, Muñoz-Espín D, González C, Hermoso J, Villar L, Jiménez-Barbero J, Salas M, Meijer WJ. Structure of the functional domain of phi29 replication organizer: Insights into oligomerization and DNA binding. J Biol Chem. 2005;280:20730–20739. doi: 10.1074/jbc.M501687200. [DOI] [PubMed] [Google Scholar]
- Atanasiu C, Byron O, McMiken H, Sturrock SS, Dryden DT. Characterisation of the structure of ocr, the gene 0.3 protein of bacteriophage T7. Nucleic Acids Res. 2001;29:3059–3068. doi: 10.1093/nar/29.14.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Su TJ, Sturrock SS, Dryden DT. Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–3944. doi: 10.1093/nar/gkf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers DJ, Sunshine MG, Six EW, Christie GE. Mutations affecting two adjacent amino acid residues in the alpha subunit of RNA polymerase block transcriptional activation by the bacteriophage P2 Ogr protein. J Bacteriol. 1994;176:7430–7438. doi: 10.1128/jb.176.24.7430-7438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus H, Petri JB. Sequence analysis of a region from the early right operon in phage P22 including the replication genes 18 and 12. Gene. 1984;32:289–303. doi: 10.1016/0378-1119(84)90004-0. [DOI] [PubMed] [Google Scholar]
- Badia D, Camacho A, Pérez-Lago L, Escandón C, Salas M, Coll M. The structure of phage phi29 transcription regulator p4-DNA complex reveals an N-hook motif for DNA. Mol Cell. 2006;22:73–81. doi: 10.1016/j.molcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Ball CA, Johnson RC. Efficient excision of phage lambda from the Escherichia coli chromosome requires the Fis protein. J Bacteriol. 1991;173:4027–4031. doi: 10.1128/jb.173.13.4027-4031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett KJ, Marsh ML, Calendar R. Interactions between a satellite bacteriophage and its helper. J Mol Biol. 1976;106:683–707. doi: 10.1016/0022-2836(76)90259-x. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- Barthelemy I, Salas M. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J Mol Biol. 1989;208:225–232. doi: 10.1016/0022-2836(89)90384-7. [DOI] [PubMed] [Google Scholar]
- Bazinet C, Benbasat J, King J, Carazo JM, Carrascosa JL. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- Bell CE, Lewis M. Crystal structure of the lambda repressor C-terminal domain octamer. J Mol Biol. 2001;314:1127–1136. doi: 10.1006/jmbi.2000.5196. [DOI] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani LE, Six E. In: The P2-like phages and their parasite, P4. Calendar R, editor. Plenum Press; New York: 1988. pp. 73–143. [Google Scholar]
- Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol Gen Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- Blanco L, Gutierrez J, Lazaro JM, Bernad A, Salas M. Replication of phage phi 29 DNA in vitro: Role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986;14:4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L, Prieto I, Gutierrez J, Bernad A, Lazaro JM, Hermoso JM, Salas M. Effect of NH4+ ions on phi 29 DNA-protein p3 replication: Formation of a complex between the terminal protein and the DNA polymerase. J Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L, Salas M. Characterization and purification of a phage phi 29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA. 1984;81:5325–5329. doi: 10.1073/pnas.81.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Bowden DW, Modrich P. In vitro maturation of circular bacteriophage P2 DNA: Purification of ter components and characterization of the reaction. J Biol Chem. 1985;260:6999–7007. [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Bravo A, Illana B, Salas M. Compartmentalization of phage phi29 DNA replication: Interaction between the primer terminal protein and the membrane-associated protein p1. EMBO J. 2000;19:5575–5584. doi: 10.1093/emboj/19.20.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Salas M. Initiation of bacteriophage phi29 DNA replication in vivo: assembly of a membrane-associated multiprotein complex. J Mol Biol. 1997;269:102–112. doi: 10.1006/jmbi.1997.1032. [DOI] [PubMed] [Google Scholar]
- Bravo A, Salas M. Polymerization of bacteriophage phi 29 replication protein p1 into protofilament sheets. EMBO J. 1998;17:6096–6105. doi: 10.1093/emboj/17.20.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breg JN, van Opheusden JH, Burgering MJ, Boelens R, Kaptein R. Structure of Arc repressor in solution: Evidence for a family of beta- sheet DNA-binding proteins. Nature. 1990;346:586–589. doi: 10.1038/346586a0. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briani F, Deho G, Forti F, Ghisotti D. The plasmid status of satellite bacteriophage P4. Plasmid. 2001;45:1–17. doi: 10.1006/plas.2000.1497. [DOI] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010;77:337–353. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BM, Bowie JU, Sauer RT. Arc repressor is tetrameric when bound to operator DNA. Biochemistry. 1990;29:11189–11195. doi: 10.1021/bi00503a006. [DOI] [PubMed] [Google Scholar]
- Buchwald M, Murialdo H, Siminovitch L. The morphogenesis of bacteriophage lambda. II. Identification of the principal structural proteins. Virology. 1970a;42:390–400. doi: 10.1016/0042-6822(70)90282-5. [DOI] [PubMed] [Google Scholar]
- Buchwald M, Siminovitch L. Production of serum-blocking material by mutants of the left arm of the lambda chromosome. Virology. 1969;38:1–7. doi: 10.1016/0042-6822(69)90121-4. [DOI] [PubMed] [Google Scholar]
- Buchwald M, Steed-Glaister P, Siminovitch L. The morphogenesis of bacteriophage lambda. I. Purification and characterization of lambda heads and lambda tails. Virology. 1970b;42:375–389. doi: 10.1016/0042-6822(70)90281-3. [DOI] [PubMed] [Google Scholar]
- Calendar R. The Bacteriophages. 2. Oxford University Press; Oxford: 2006. [Google Scholar]
- Calendar R, Lindqvist B, Sironi G, Clark A. Characterization of REP- mutants and their interaction with P2 phage. Virology. 1970;40:72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- Calles B, Salas M, Rojo F. The phi29 transcriptional regulator contacts the nucleoid protein p6 to organize a repression complex. EMBO J. 2002;21:6185–6194. doi: 10.1093/emboj/cdf623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho A, Salas M. Mechanism for the switch of phi29 DNA early to late transcription by regulatory protein p4 and histone-like protein p6. EMBO J. 2001;20:6060–6070. doi: 10.1093/emboj/20.21.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara B, Liu M, Reynolds J, Shadrin A, Liu B, Kwok K, Simpson P, Weinzierl R, Severinov K, Cota E, Matthews S, Wigneshweraraj SR. T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc Natl Acad Sci USA. 2010;107:2247–2252. doi: 10.1073/pnas.0907908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A, del-Campillo-Campbell A, Ginsberg ML. Specificity in DNA recognition by phage integrases. Gene. 2002;300:13–18. doi: 10.1016/s0378-1119(02)00846-6. [DOI] [PubMed] [Google Scholar]
- Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. The crystal structure of bacteriophage HK97 gp6: Defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- Casjens S. Bacteriophage lambda FII gene protein: Role in head assembly. J Mol Biol. 1974;90:1–20. doi: 10.1016/0022-2836(74)90252-6. [DOI] [PubMed] [Google Scholar]
- Casjens S. Molecular organization of the bacteriophage P22 coat protein shell. J Mol Biol. 1979;131:1–14. doi: 10.1016/0022-2836(79)90298-5. [DOI] [PubMed] [Google Scholar]
- Casjens S. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011 doi: 10.1016/j.virol.2010.12.046. in press. [DOI] [PubMed] [Google Scholar]
- Casjens S, Eppler K, Parr R, Poteete AR. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: Identification of a new gene required for lysis. Virology. 1989;171:588–598. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J Mol Biol. 1988;199:467–474. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- Casjens S, Horn T, Kaiser AD. Head assembly steps controlled by genes F and W in bacteriophage lambda. J Mol Biol. 1972;64:551–563. doi: 10.1016/0022-2836(72)90082-4. [DOI] [PubMed] [Google Scholar]
- Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–458. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Hendrix RW. Locations and amounts of major structural proteins in bacteriophage lambda. J Mol Biol. 1974;88:535–545. doi: 10.1016/0022-2836(74)90500-2. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Catalano CE. The terminase enzyme from bacteriophage lambda: A DNA-packaging machine. Cell Mol Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano CE, Tomka MA. Role of gpFI protein in DNA packaging by bacteriophage lambda. Biochemistry. 1995;34:10036–10042. doi: 10.1021/bi00031a027. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem. 2003a;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996a;258:286–298. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Studier FW. Purification and characterization of T7 head-tail connectors expressed from the cloned gene. J Mol Biol. 1996b;258:299–307. doi: 10.1006/jmbi.1996.0251. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Trus BL, Smith CS, Cheng N, Conway JF, Steven AC. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J Mol Biol. 2003b;327:1–6. doi: 10.1016/s0022-2836(03)00117-7. [DOI] [PubMed] [Google Scholar]
- Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology. 2008;370:352–361. doi: 10.1016/j.virol.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Spilman MS, Rodenburg CM, Dokland T. Functional domains of the bacteriophage P2 scaffolding protein: Identification of residues involved in assembly and protease activity. Virology. 2009;384:144–150. doi: 10.1016/j.virol.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci USA. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Vannais DB, Jones C, Patterson D, Davidson JN. Mapping of the gene encoding the multifunctional protein carrying out the first three steps of pyrimidine biosynthesis to human chromosome 2. Hum Genet. 1989;82:40–44. doi: 10.1007/BF00288269. [DOI] [PubMed] [Google Scholar]
- Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nature Methods. 2010;7:667–668. doi: 10.1038/nmeth0910-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang W, Molineux IJ. F exclusion of bacteriophage T7 occurs at the cell membrane. Virology. 2004;326:340–352. doi: 10.1016/j.virol.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Cho EH, Gumport RI, Gardner JF. Interactions between integrase and excisionase in the phage lambda excisive nucleoprotein complex. J Bacteriol. 2002;184:5200–5203. doi: 10.1128/JB.184.18.5200-5203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EH, Nam CE, Alcaraz R, Jr, Gardner JF. Site-specific recombination of bacteriophage P22 does not require integration host factor. J Bacteriol. 1999;181:4245–4249. doi: 10.1128/jb.181.14.4245-4249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Anders D, McAlister V, Goodwin T, Julien B, Calendar R. Identification of upstream sequences essential for activation of a bacteriophage P2 late promoter. J Bacteriol. 2003;185:4609–4614. doi: 10.1128/JB.185.15.4609-4614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Temple L, Bartlett B, Goodwin T. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J Bacteriol. 2002;184:6522–6531. doi: 10.1128/JB.184.23.6522-6531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Calendar R. Interactions between satellite bacteriophage P4 and its helpers. Annu Rev Genet. 1990;24:465–490. doi: 10.1146/annurev.ge.24.120190.002341. [DOI] [PubMed] [Google Scholar]
- Clement JM, Lepouce E, Marchal C, Hofnung M. Genetic study of a membrane protein: DNA sequence alterations due to 17 lamB point mutations affecting adsorption of phage lambda. EMBO J. 1983;2:77–80. doi: 10.1002/j.1460-2075.1983.tb01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone R, Bonura T, Friedberg EC. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2: Purification and preliminary characterization. J Biol Chem. 1980;255:10354–10358. [PubMed] [Google Scholar]
- Cortines JR, Weigele PR, Gilcrease EB, Casjens SR, Teschke CM. Decoding bacteriophage P22 assembly: Identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology. 2011;421:1–11. doi: 10.1016/j.virol.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court DL, Oppenheim AB, Adhya SL. A new look at bacteriophage lambda genetic networks. J Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisona NJ, Weinberg RL, Peter BJ, Sumners DW, Cozzarelli NR. The topological mechanism of phage lambda integrase. J Mol Biol. 1999;289:747–775. doi: 10.1006/jmbi.1999.2771. [DOI] [PubMed] [Google Scholar]
- Crucitti P, Abril AM, Salas M. Bacteriophage phi 29 early protein p17. Self-association and hetero-association with the viral histone-like protein p6. J Biol Chem. 2003;278:4906–4911. doi: 10.1074/jbc.M210289200. [DOI] [PubMed] [Google Scholar]
- Crucitti P, Lazaro JM, Benes V, Salas M. Bacteriophage phi29 early protein p17 is conditionally required for the first rounds of viral DNA replication. Gene. 1998;223:135–142. doi: 10.1016/s0378-1119(98)00167-x. [DOI] [PubMed] [Google Scholar]
- d’Herelle F. The Bacteriophage and Its Behavior. Williams & Wilkins; Baltimore, MD: 1926. pp. 490–497. [Google Scholar]
- Darling PJ, Holt JM, Ackers GK. Coupled energetics of lambda cro repressor self-assembly and site-specific DNA operator binding I: Analysis of cro dimerization from nanomolar to micromolar concentrations. Biochemistry. 2000;39:11500–11507. doi: 10.1021/bi000935s. [DOI] [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992;174:6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AB, Roy S, Parrack P. Role of C-terminal residues in oligomerization and stability of lambda CII: Implications for lysis-lysogeny decision of the phage. J Mol Biol. 2005a;345:315–324. doi: 10.1016/j.jmb.2004.09.098. [DOI] [PubMed] [Google Scholar]
- Datta I, Banik-Maiti S, Adhikari L, Sau S, Das N, Mandal NC. The mutation that makes Escherichia coli resistant to lambda P gene-mediated host lethality is located within the DNA initiator Gene dnaA of the bacterium. J Biochem Mol Biol. 2005b;38:89–96. doi: 10.5483/bmbrep.2005.38.1.089. [DOI] [PubMed] [Google Scholar]
- Datta I, Sau S, Sil AK, Mandal NC. The bacteriophage lambda DNA replication protein P inhibits the oriC DNA- and ATP-binding functions of the DNA replication initiator protein DnaA of Escherichia coli. J Biochem Mol Biol. 2005c;38:97–103. doi: 10.5483/bmbrep.2005.38.1.097. [DOI] [PubMed] [Google Scholar]
- De Anda J, Poteete AR, Sauer RT. P22 c2 repressor. Domain structure and function. J Biol Chem. 1983;258:10536–10542. [PubMed] [Google Scholar]
- de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- de Beus MD, Doyle SM, Teschke CM. GroEL binds a late folding intermediate of phage P22 coat protein. Cell Stress Chaperones. 2000;5:163–172. doi: 10.1379/1466-1268(2000)005<0163:gbalfi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong RN, van der Vliet PC. Mechanism of DNA replication in eukaryotic cells: Cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236:1–12. doi: 10.1016/s0378-1119(99)00249-8. [DOI] [PubMed] [Google Scholar]
- Deho G, Ghisotti D. In: The satellite phage P4. Calendar R, editor. Oxford University Press; New York: 2006. pp. 391–408. [Google Scholar]
- Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage lambda Q antiterminator protein contacts the beta-flap domain of RNA polymerase. Proc Natl Acad Sci USA. 2008;105:15305–15310. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M, Fano U. Bacteriophage-resistant mutants in Escherichia coli. Genetics. 1945;30:119–136. doi: 10.1093/genetics/30.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DNP, Dokland T. The gpQ portal protein of bacteriophage P2 forms dodecameric connctors in crystals. J Struct Biol. 2007;157:432–436. doi: 10.1016/j.jsb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M, Echols H, Wickner S, Alfano C, Mensa-Wilmot K, Gomes B, LeBowitz J, Roberts JD, McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: Localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci USA. 1986;83:7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T. gpO peptidase (Enterobacteria phage P2) In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Elsevier; Oxford: 2011. [Google Scholar]
- Dokland T, Isaksen ML, Fuller SD, Lindqvist BH. Capsid localization of the bacteriophage P4 Psu protein. Virology. 1993;194:682–687. doi: 10.1006/viro.1993.1308. [DOI] [PubMed] [Google Scholar]
- Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryoelectron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J Eur Mol Biol Organ J. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Dokland T, Wang S, Lindqvist BH. The structure of P4 procapsids produced by coexpression of capsid and external scaffolding proteins. Virology. 2002;298:224–231. doi: 10.1006/viro.2002.1485. [DOI] [PubMed] [Google Scholar]
- Duffy C, Feiss M. The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- Dufour E, Méndez J, Lázaro JM, de Vega M, Blanco L, Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J Mol Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986;233:1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Elías-Arnanz M, Salas M. Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of phi29 early-late transcriptional switch. Genes Dev. 1999;13:2502–2513. doi: 10.1101/gad.13.19.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- Eriksson JM, Haggard-Ljungquist E. The multifunctional bacteriophage P2 cox protein requires oligomerization for biological activity. J Bacteriol. 2000;182:6714–6723. doi: 10.1128/jb.182.23.6714-6723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson SK, Liu T, Haggard-Ljungquist E. Interacting interfaces of the P4 antirepressor E and the P2 immunity repressor C. Mol Microbiol. 2000;36:1148–1155. doi: 10.1046/j.1365-2958.2000.01937.x. [DOI] [PubMed] [Google Scholar]
- Esposito D, Gerard GF. The Escherichia coli Fis protein stimulates bacteriophage lambda integrative recombination in vitro. J Bacteriol. 2003;185:3076–3080. doi: 10.1128/JB.185.10.3076-3080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frackman S, Siegele DA, Feiss M. The terminase of bacteriophage lambda: Functional domains for cosB binding and multimer assembly. J Mol Biol. 1985;183:225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- Franklin NC. Conservation of genome form but not sequence in the transcription antitermination determinants of bacteriophages lambda, phi 21 and P22. J Mol Biol. 1985;181:75–84. doi: 10.1016/0022-2836(85)90325-0. [DOI] [PubMed] [Google Scholar]
- Freire R, Serrano M, Salas M, Hermoso JM. Activation of replication origins in phi29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J Biol Chem. 1996;271:31000–31007. doi: 10.1074/jbc.271.48.31000. [DOI] [PubMed] [Google Scholar]
- Friedman DI, Court DL. Bacteriophage lambda: Alive and well and still doing its thing. Curr Opin Microbiol. 2001;4:201–207. doi: 10.1016/s1369-5274(00)00189-2. [DOI] [PubMed] [Google Scholar]
- Frumerie CM, Eriksson J, Dugast M, Haggard-Ljungquist E. Dimerization of bacteriophage P2 integrase is not required for binding to its DNA target but for its biological activity. Gene. 2005;344:221–231. doi: 10.1016/j.gene.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Palm P, Zillig W, Calendar R, Sunshine M. Identification of a mutation within the structural gene for the a subunit of DNA-dependent RNA polymerase of E. coli. Mol Gen Genet. 1976;145:19–22. doi: 10.1007/BF00331552. [DOI] [PubMed] [Google Scholar]
- Fujisawa H, Shibata H, Kato H. Analysis of interactions among factors involved in the bacteriophage T3 DNA packaging reaction in a defined in vitro system. Virology. 1991;185:788–794. doi: 10.1016/0042-6822(91)90550-u. [DOI] [PubMed] [Google Scholar]
- Fujiyama A, Kohara Y, Okazaki T. Initiation sites for discontinuous DNA synthesis of bacteriophage T7. Proc Natl Acad Sci USA. 1981;78:903–907. doi: 10.1073/pnas.78.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J Mol Biol. 1982;156:633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- Funnell BE, Inman RB. Bacteriophage P2 DNA replication: Characterization of the requirement of the gene B protein in vivo. J Mol Biol. 1983;167:311–334. doi: 10.1016/s0022-2836(83)80338-6. [DOI] [PubMed] [Google Scholar]
- García JA, Carrascosa JL, Salas M. Assembly of the tail protein of the Bacillus subtilis phage phi 29. Virology. 1983;125:18–30. doi: 10.1016/0042-6822(83)90060-0. [DOI] [PubMed] [Google Scholar]
- Garcia LR, Molineux IJ. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J Bacteriol. 1995;177:4066–4076. doi: 10.1128/jb.177.14.4066-4076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia LR, Molineux IJ. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J Bacteriol. 1996;178:6921–6929. doi: 10.1128/jb.178.23.6921-6929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascón I, Gutiérrez C, Salas M. Structural and functional comparative study of the complexes formed by viral o29, Nf and GA-1 SSB proteins with DNA. J Mol Biol. 2000;296:989–999. doi: 10.1006/jmbi.2000.3521. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Geisselsoder J, Youdarian P, Deho G, Chidambaram M, Goldstein R, Ljungquist E. Mutants of satellite virus P4 that cannot derepress their bacteriophage P2 helper. J Mol Biol. 1981;148:1–19. doi: 10.1016/0022-2836(81)90232-1. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Tilly K, Casjens S. Lambdoid phage head assembly. In: Hendrix R, Roberts J, Stahl FW, Weisberg R, editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. pp. 279–304. [Google Scholar]
- Georgopoulos CP, Hendrix RW, Casjens SR, Kaiser AD. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973;76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hamdan SM, Cook TE, Richardson CC. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J Biol Chem. 2008;283:32077–32084. doi: 10.1074/jbc.M805062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Hamdan SM, Richardson CC. Two modes of interaction of the single-stranded DNA-binding protein of bacteriophage T7 with the DNA polymerase-thioredoxin complex. J Biol Chem. 2010;285:18103–18112. doi: 10.1074/jbc.M110.107656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, Casjens SR. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J Bacteriol. 2005;187:2050–2057. doi: 10.1128/JB.187.6.2050-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- Gottesman ME, Weisberg RA. Little lambda, who made thee? Microbiol Mol Biol Rev. 2004;68:796–813. doi: 10.1128/MMBR.68.4.796-813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Gottesman M, Shaw JE, Pearson ML. Protein degradation in E. coli: The lon mutation and bacteriophage lambda N and cII protein stability. Cell. 1981;24:225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- Grundling A, Smith DL, Blasi U, Young R. Dimerization between the holin and holin inhibitor of phage lambda. J Bacteriol. 2000;182:6075–6081. doi: 10.1128/jb.182.21.6075-6081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: The high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage phi 29 DNA-gp3. J Mol Biol. 1987;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- Guo PX, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden JM, Declais AC, Carr SB, Lilley DM, Phillips SE. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007;449:621–624. doi: 10.1038/nature06158. [DOI] [PubMed] [Google Scholar]
- Haggard-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggard-Ljungquist E, Jacobsen E, Rishovd S, Six EW, Nilssen O, Sunshine MG, Lindqvist BH, Kim KJ, Barreiro V, Koonin EV, et al. Bacteriophage P2: Genes involved in baseplate assembly. Virology. 1995;213:109–121. doi: 10.1006/viro.1995.1551. [DOI] [PubMed] [Google Scholar]
- Halder S, Datta AB, Parrack P. Probing the antiprotease activity of lambda-CIII, an inhibitor of the Escherichia coli metalloprotease HflB (FtsH) J Bacteriol. 2007;189:8130–8138. doi: 10.1128/JB.00820-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Fujisawa H, Minagawa T. Overproduction and purification of the products of bacteriophage T3 genes 18 and 19, two genes involved in DNA packaging. Virology. 1986;151:110–118. doi: 10.1016/0042-6822(86)90108-x. [DOI] [PubMed] [Google Scholar]
- Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci USA. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- Hang Q, Woods L, Feiss M, Catalano CE. Cloning, expression, and biochemical characterization of hexahistidine-tagged terminase proteins. J Biol Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
- Häuser R, Sabri M, Moineau S, Uetz P. The proteome and interactome of Streptococcus pneumoniae phage Cp-1. J Bacteriol. 2011;193:3135–3138. doi: 10.1128/JB.01481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J Biol Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- He ZG, Richardson CC. Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J Biol Chem. 2004;279:22190–22197. doi: 10.1074/jbc.M401100200. [DOI] [PubMed] [Google Scholar]
- Heineman RH, Bull JJ. Testing optimality with experimental evolution: Lysis time in a bacteriophage. Evolution. 2007;61:1695–1709. doi: 10.1111/j.1558-5646.2007.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R, Casjens S. Bacteriophage lambda and its genetic neighborhood. In: Calendar R, editor. The Bacteriophages. Chapter 27. Oxford University Press; Oxford: 2006. pp. 409–445. [Google Scholar]
- Hendrix R, Roberts J, Stahl FW, Weisberg R. Lambda II. CSHL Press; Cold Spring Harbor, NY: 1983. p. 694. [Google Scholar]
- Hendrix RW. Bacteriophages: Evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage HK97: Assembly of the capsid and evolutionary connections. Adv Virus Res. 2005;64:1–14. doi: 10.1016/S0065-3527(05)64001-8. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Casjens SR. Protein fusion: A novel reaction in bacteriophage lambda head assembly. Proc Natl Acad Sci USA. 1974;71:1451–1455. doi: 10.1073/pnas.71.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Casjens SR. Assembly of bacteriophage lambda heads: Protein processing and its genetic control in petit lambda assembly. J Mol Biol. 1975;91:187–199. doi: 10.1016/0022-2836(75)90159-x. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage lambda PaPa: Not the mother of all lambda phages. Science. 1992;258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: A protein ballet. Adv Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, D’Ari R, Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor lambda cIII. J Bacteriol. 1997;179:358–363. doi: 10.1128/jb.179.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoso JM, Méndez E, Soriano F, Salas M. Location of the serine residue involved in the linkage between the terminal protein and the DNA of phage phi 29. Nucleic Acids Res. 1985;13:7715–7728. doi: 10.1093/nar/13.21.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD. Inheritance in bacteriophage. Ann N Y Acad Sci. 1952;54:960–962. doi: 10.1111/j.1749-6632.1952.tb39971.x. [DOI] [PubMed] [Google Scholar]
- Ho Y, Rosenberg M. Characterization of the phage lambda regulatory protein cII. Ann Microbiol. 1982;133:215–218. [PubMed] [Google Scholar]
- Ho YS, Pfarr D, Strickler J, Rosenberg M. Characterization of the transcription activator protein C1 of bacteriophage P22. J Biol Chem. 1992;267:14388–14397. [PubMed] [Google Scholar]
- Hohn T, Flick H, Hohn B. Petit lambda, a family of particles from coliphage lambda infected cells. J Mol Biol. 1975;98:107–120. doi: 10.1016/s0022-2836(75)80104-5. [DOI] [PubMed] [Google Scholar]
- Horcajadas JA, Monsalve M, Rojo F, Salas M. The switch from early to late transcription in phage GA-1: Characterization of the regulatory protein p4G. J Mol Biol. 1999;290:917–928. doi: 10.1006/jmbi.1999.2932. [DOI] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, Chandran S, Christopoulos C, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Villemain J, Padilla R, Sousa R. Mechanisms by which T7 lysozyme specifically regulates T7 RNA polymerase during different phases of transcription. J Mol Biol. 1999;293:457–475. doi: 10.1006/jmbi.1999.3135. [DOI] [PubMed] [Google Scholar]
- Huber HE, Beauchamp BB, Richardson CC. Escherichia coli dGTP triphosphohydrolase is inhibited by gene 1.2 protein of bacteriophage T7. J Biol Chem. 1988;263:13549–13556. [PubMed] [Google Scholar]
- Ibarra B, Valpuesta JM, Carrascosa JL. Purification and functional characterization of p16, the ATPase of the bacteriophage phi29 packaging machinery. Nucleic Acids Res. 2001;29:4264–4273. doi: 10.1093/nar/29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1: Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J Mol Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- Illana B, Lázaro JM, Gutierrez C, Meijer WJ, Blanco L, Salas M. Phage phi29 terminal protein residues Asn80 and Tyr82 are recognition elements of the replication origins. J Biol Chem. 1999;274:15073–15079. doi: 10.1074/jbc.274.21.15073. [DOI] [PubMed] [Google Scholar]
- Isaksen ML, Dokland T, Lindqvist BH. Characterization of the capsid associating activity of bacteriophage P4’s Psu protein. Virology. 1993;194:647–681. doi: 10.1006/viro.1993.1307. [DOI] [PubMed] [Google Scholar]
- Iwashita S, Kanegasaki S. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur J Biochem. 1976;65:87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Steitz TA. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien B, Lefevre P, Calendar R. The two P2 Ogr-like domains of the delta protein from bacteriophage P4 are required for activity. Virology. 1997;230:292–299. doi: 10.1006/viro.1997.8487. [DOI] [PubMed] [Google Scholar]
- Kageyama Y, Murayama M, Onodera T, Yamada S, Fukada H, Kudou M, Tsumoto K, Toyama Y, Kado S, Kubota K, Takeda S. Observation of the membrane binding activity and domain structure of gpV, which comprises the tail spike of bacteriophage P2. Biochemistry. 2009;48:10129–10135. doi: 10.1021/bi900928n. [DOI] [PubMed] [Google Scholar]
- Kaiser AD, Jacob F. Recombination between related bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957;4:509–517. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura I. Tail assembly and injection. In: Hendrix R, Roberts J, Stahl FW, Weisberg R, editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. pp. 331–346. [Google Scholar]
- Katsura I. Mechanism of length determination in bacteriophage lambda tails. Adv Biophys. 1990;26:1–18. doi: 10.1016/0065-227x(90)90004-d. [DOI] [PubMed] [Google Scholar]
- Katsura I, Tsugita A. Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail. Virology. 1977;76:129–145. doi: 10.1016/0042-6822(77)90290-2. [DOI] [PubMed] [Google Scholar]
- Kędzierska B, Lee DJ, Węgrzyn G, Busby SJ, Thomas MS. Role of the RNA polymerase alpha subunits in CII-dependent activation of the bacteriophage lambda pE promoter: identification of important residues and positioning of the alpha C-terminal domains. Nucleic Acids Res. 2004;32:834–841. doi: 10.1093/nar/gkh230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kędzierska B, Szambowska A, Herman-Antosiewicz A, Lee DJ, Busby SJ, Węgrzyn G, Thomas MS. The C-terminal domain of the Escherichia coli RNA polymerase alpha subunit plays a role in the CI-dependent activation of the bacteriophage lambda pM promoter. Nucleic Acids Res. 2007;35:2311–2320. doi: 10.1093/nar/gkm123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DT. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy WP, Momand JR, Yin YW. Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase. J Mol Biol. 2007;370:256–268. doi: 10.1016/j.jmb.2007.03.041. [DOI] [PubMed] [Google Scholar]
- Kihara A, Akiyama Y, Ito K. Revisiting the lysogenization control of bacteriophage lambda: Identification and characterization of a new host component. HflD J Biol Chem. 2001;276:13695–13700. doi: 10.1074/jbc.M011699200. [DOI] [PubMed] [Google Scholar]
- Kim K-J, Sunshine MG, Lindqvist BH, Six EW. Capsid size determination in the P2–P4 bacteriophage system: suppression of sir mutations in P2’s capsid gene N by supersid mutations in P4’s external scaffold gene sid. Virology. 2001;283:49–58. doi: 10.1006/viro.2001.0853. [DOI] [PubMed] [Google Scholar]
- King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- King RA, Anders DL, Christie GE. Site-directed mutagenesis of an amino acid residue in the bacteriophage P2 ogr protein implicated in interaction with Escherichia coli RNA polymerase. Mol Microbiol. 1992;6:3313–3320. doi: 10.1111/j.1365-2958.1992.tb02199.x. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Koby S, Teff D, Court D, Oppenheim AB. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci USA. 2002;99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O, Oppenheim AB, Herman C. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J Struct Biol. 2004;146:72–78. doi: 10.1016/j.jsb.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Rokney A, Oppenheim AB. Phage lambda CIII: A protease inhibitor regulating the lysis-lysogeny decision. PLoS One. 2007;2:e363. doi: 10.1371/journal.pone.0000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan J, Carrascosa JL, Murialdo H. Bacteriophage lambda preconnectors. Purification and structure. J Mol Biol. 1984;174:433–447. doi: 10.1016/0022-2836(84)90330-9. [DOI] [PubMed] [Google Scholar]
- Kornitzer D, Altuvia S, Oppenheim AB. The activity of the CIII regulator of lambdoid bacteriophages resides within a 24-amino acid protein domain. Proc Natl Acad Sci USA. 1991;88:5217–5221. doi: 10.1073/pnas.88.12.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- Lee CS, Guo P. Sequential interactions of structural proteins in phage phi 29 procapsid assembly. J Virol. 1995;69:5024–5032. doi: 10.1128/jvi.69.8.5024-5032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers GG, Jr, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Schlitt T. Protein-protein interaction databases: Keeping up with growing interactomes. Hum Genom. 2009;3:291–297. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel JA, Goldstein RN, Marsh M, Calendar R. Structure of the bacteriophage P2 tail. Virology. 1974;62:161–174. doi: 10.1016/0042-6822(74)90312-2. [DOI] [PubMed] [Google Scholar]
- Lengyel JA, Goldstein RN, Marsh M, Sunshine MG, Calendar R. Bacteriophage P2 head morphogenesis: cleavage of the major capsid protein. Virology. 1973;53:1–23. doi: 10.1016/0042-6822(73)90461-3. [DOI] [PubMed] [Google Scholar]
- Li M, Moyle H, Susskind MM. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- Lin L. Study of Bacteriophage T7 Gene 5.9 and Gene 5.5. SUNY; Stonybrook, NY: 1992. [Google Scholar]
- Linderoth NA, Julien B, Flick KE, Calendar R, Christie G. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology. 1994;200:347–359. doi: 10.1006/viro.1994.1199. [DOI] [PubMed] [Google Scholar]
- Linderoth NA, Ziermann R, Haggård-Ljungquist E, Christie GE, Calendar R. Nucleotide sequence of the DNA packaging and capsid synthesis genes of bacteriophage P2. Nucl Acids Res. 1991;19:7207–7214. doi: 10.1093/nar/19.25.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist BH, Deho G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- Liu Q, Richardson CC. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Renberg SK, Haggard-Ljungquist E. The E protein of satellite phage P4 acts as an anti-repressor by binding to the C protein of helper phage P2. Mol Microbiol. 1998;30:1041–1050. doi: 10.1046/j.1365-2958.1998.01132.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Haggard-Ljungquist E. Studies of bacteriophage P2 DNA replication: localization of the cleavage site of the A protein. Nucleic Acids Res. 1994;22:5204–5210. doi: 10.1093/nar/22.24.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler JM, Djurkovic S, Fischetti VA. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect Immun. 2003;71:6199–6204. doi: 10.1128/IAI.71.11.6199-6204.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- Longás E, de Vega M, Lázaro JM, Salas M. Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res. 2006;34:6051–6063. doi: 10.1093/nar/gkl769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longás E, Villar L, Lázaro JM, de Vega M, Salas M. Phage phi29 and Nf terminal protein-priming domain specifies the internal template nucleotide to initiate DNA replication. Proc Natl Acad Sci USA. 2008;105:18290–18295. doi: 10.1073/pnas.0809882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R, Ronda C, Tomasz A, Portoles A. Properties of “diplophage”: A lipid-containing bacteriophage. J Virol. 1977;24:201–210. doi: 10.1128/jvi.24.1.201-210.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J Mol Biol. 1997;269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. Pausing and termination by bacteriophage T7 RNA polymerase. J Mol Biol. 1998;280:201–213. doi: 10.1006/jmbi.1998.1854. [DOI] [PubMed] [Google Scholar]
- Makela PH. Glucosylation of lipopolysaccharide in Salmonella: Mutants negative for O antigen factor 1221. J Bacteriol. 1973;116:847–856. doi: 10.1128/jb.116.2.847-856.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory JB, Alfano C, McMacken R. Host virus interactions in the initiation of bacteriophage lambda DNA replication: Recruitment of Escherichia coli DnaB helicase by lambda P replication protein. J Biol Chem. 1990;265:13297–13307. [PubMed] [Google Scholar]
- Marchand I, Nicholson AW, Dreyfus M. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol Microbiol. 2001;42:767–776. doi: 10.1046/j.1365-2958.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- Marintcheva B, Qimron U, Yu Y, Tabor S, Richardson CC. Mutations in the gene 5 DNA polymerase of bacteriophage T7 suppress the dominant lethal phenotype of gene 2.5 ssDNA binding protein lacking the C-terminal phenylalanine. Mol Microbiol. 2009;72:869–880. doi: 10.1111/j.1365-2958.2009.06682.x. [DOI] [PubMed] [Google Scholar]
- Marr MT, Datwyler SA, Meares CF, Roberts JW. Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Proc Natl Acad Sci USA. 2001;98:8972–8978. doi: 10.1073/pnas.161253298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, Roberts JW, Brown SE, Klee M, Gussin GN. Interactions among CII protein, RNA polymerase and the lambda PRE promoter: Contacts between RNA polymerase and the -35 region of PRE are identical in the presence and absence of CII protein. Nucleic Acids Res. 2004;32:1083–1090. doi: 10.1093/nar/gkh261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsic N, Roje S, Stojiljkovic I, Salaj-Smic E, Trgovcevic Z. In vivo studies on the interaction of RecBCD enzyme and lambda Gam protein. J Bacteriol. 1993;175:4738–4743. doi: 10.1128/jb.175.15.4738-4743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Blanco L, Garcia P, Salas M, Mendez J. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: A stepwise sliding-back mechanism. J Mol Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- Marvik OJ, Dokland T, Nøkling RH, Jacobsen E, Larsen T, Lindqvist BH. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- Marvik OJ, Jacobsen E, Dokland T, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: assembly precedes proteolytic processing of the capsid proteins. Virology. 1994;205:51–65. doi: 10.1006/viro.1994.1619. [DOI] [PubMed] [Google Scholar]
- Mason SW, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991;5:1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- Massad T, Skaar K, Nilsson H, Damberg P, Henriksson-Peltola P, Haggard-Ljungquist E, Hogbom M, Stenmark P. Crystal structure of the P2 C-repressor: A binder of non-palindromic direct DNA repeats. Nucleic Acids Res. 2010;38:7778–7790. doi: 10.1093/nar/gkq626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis AN, Gumport RI, Gardner JF. Purification and characterization of bacteriophage P22 Xis protein. J Bacteriol. 2008;190:5781–5796. doi: 10.1128/JB.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Reed P, Zhang RG, Beasley S, Walmsley AR, Curtis FA, Joachimiak A, Edwards AM, Sharples GJ. Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange. Proc Natl Acad Sci USA. 2005;102:11260–11265. doi: 10.1073/pnas.0503399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Yee AA, Booth V, Arrowsmith CH, Gold M, Davidson AR. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- Mayer JE, Schweiger M. RNase III is positively regulated by T7 protein kinase. J Biol Chem. 1983;258:5340–5343. [PubMed] [Google Scholar]
- McDonnell M, Lain R, Tomasz A. “Diplophage”: A bacteriophage of Diplococcus pneumoniae. Virology. 1975;63:577–582. doi: 10.1016/0042-6822(75)90329-3. [DOI] [PubMed] [Google Scholar]
- Medina E, Wieczorek D, Medina EM, Yang Q, Feiss M, Catalano CE. Assembly and maturation of the bacteriophage lambda procapsid: gpC is the viral protease. J Mol Biol. 2010;401:813–830. doi: 10.1016/j.jmb.2010.06.060. [DOI] [PubMed] [Google Scholar]
- Meijer WJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev. 2001a;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WJ, Lewis PJ, Errington J, Salas M. Dynamic relocalization of phage phi 29 DNA during replication and the role of the viral protein p16.7. EMBO J. 2000;19:4182–4190. doi: 10.1093/emboj/19.15.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WJ, Serna-Rico A, Salas M. Characterization of the bacteriophage phi29-encoded protein p16.7: A membrane protein involved in phage DNA replication. Mol Microbiol. 2001b;39:731–746. doi: 10.1046/j.1365-2958.2001.02260.x. [DOI] [PubMed] [Google Scholar]
- Mencía M, Monsalve M, Rojo F, Salas M. Transcription activation by phage phi29 protein p4 is mediated by interaction with the alpha subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996a;93:6616–6620. doi: 10.1073/pnas.93.13.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía M, Monsalve M, Salas M, Rojo F. Transcriptional activator of phage phi 29 late promoter: mapping of residues involved in interaction with RNA polymerase and in DNA bending. Mol Microbiol. 1996b;20:273–282. doi: 10.1111/j.1365-2958.1996.tb02616.x. [DOI] [PubMed] [Google Scholar]
- Méndez J, Blanco L, Esteban JA, Bernad A, Salas M. Initiation of phi 29 DNA replication occurs at the second 3′ nucleotide of the linear template: A sliding-back mechanism for protein-primed DNA replication. Proc Natl Acad Sci USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J, Blanco L, Salas M. Protein-primed DNA replication: A transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa YG, Maruyama IN, Brenner S. Surface display of proteins on bacteriophage lambda heads. J Mol Biol. 1996;262:21–30. doi: 10.1006/jmbi.1996.0495. [DOI] [PubMed] [Google Scholar]
- Moak M, Molineux IJ. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol Microbiol. 2000;37:345–355. doi: 10.1046/j.1365-2958.2000.01995.x. [DOI] [PubMed] [Google Scholar]
- Moak M, Molineux IJ. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol. 2004;51:1169–1183. doi: 10.1046/j.1365-2958.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- Molineux IJ. The T7 group. In: Calendar R, editor. The Bacteriophages. Chapter 20. Oxford University Press; Oxford: 2006a. pp. 277–301. [Google Scholar]
- Molineux IJ. Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology. 2006b;344:221–229. doi: 10.1016/j.virol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Calles B, Mencía M, Salas M, Rojo F. Transcription activation or repression by phage psi 29 protein p4 depends on the strength of the RNA polymerase-promoter interactions. Mol Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Mencía M, Rojo F, Salas M. Activation and repression of transcription at two different phage phi29 promoters are mediated by interaction of the same residues of regulatory protein p4 with RNA polymerase. EMBO J. 1996a;15:383–391. [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Mencía M, Salas M, Rojo F. Protein p4 represses phage phi 29 A2c promoter by interacting with the alpha subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996b;93:8913–8918. doi: 10.1073/pnas.93.17.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney PQ, North R, Molineux IJ. The role of bacteriophage T7 gene 2 protein in DNA replication. Nucleic Acids Res. 1980;8:3043–3053. doi: 10.1093/nar/8.13.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: The pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Analysis of functional domains of the packaging proteins of bacteriophage T3 by site-directed mutagenesis. J Mol Biol. 1994;235:248–259. doi: 10.1016/s0022-2836(05)80031-2. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: Importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- Mosig G, Yu S, Myung H, Haggard-Ljungquist E, Davenport L, Carlson K, Calendar R. A novel mechanism of virus-virus interactions: Bacteriophage P2 Tin protein inhibits phage T4 DNA synthesis by poisoning the T4 single-stranded DNA binding protein, gp32. Virology. 1997;230:72–81. doi: 10.1006/viro.1997.8464. [DOI] [PubMed] [Google Scholar]
- Mount DW, Harris AW, Fuerst CR, Siminovitch L. Mutations in bacteriophage lambda affecting particle morphogenesis. Virology. 1968;35:134–149. doi: 10.1016/0042-6822(68)90313-9. [DOI] [PubMed] [Google Scholar]
- Muñoz-Espín D, Daniel R, Kawai Y, Carballido-Lopez R, Castilla-Llorente V, Errington J, Meijer WJ, Salas M. The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0906465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Espín D, Holguera I, Ballesteros-Plaza D, Carballido-Lopez R, Salas M. Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc Natl Acad Sci USA. 2010;107:16548–16553. doi: 10.1073/pnas.1010530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H. Early intermediates in bacteriophage lambda prohead assembly. Virology. 1979;96:341–367. doi: 10.1016/0042-6822(79)90094-1. [DOI] [PubMed] [Google Scholar]
- Murialdo H, Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev. 1978;42:529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H, Tzamtzis D. Mutations of the coat protein gene of bacteriophage lambda that overcome the necessity for the Fl gene; the EFi domain. Mol Microbiol. 1997;24:341–353. doi: 10.1046/j.1365-2958.1997.3321698.x. [DOI] [PubMed] [Google Scholar]
- Murphy KC. Bacteriophage P22 Abc2 protein binds to RecC increases the 5′ strand nicking activity of RecBCD and together with lambda bet, promotes Chi- independent recombination. J Mol Biol. 2000;296:385–401. doi: 10.1006/jmbi.1999.3486. [DOI] [PubMed] [Google Scholar]
- Murray NE. Type I restriction systems: Sophisticated molecular machines (a legacy of Bertani and Weigle) MMBR. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Richardson CC. The gene 1.2 protein of bacteriophage T7 interacts with the Escherichia coli dGTP triphosphohydrolase to form a GTP-binding protein. J Biol Chem. 1990;265:4411–4419. [PubMed] [Google Scholar]
- Nechaev S, Severinov K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J Mol Biol. 1999;289:815–826. doi: 10.1006/jmbi.1999.2782. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Gilcrease EB, Kang S, Prevelige PE, Jr, Casjens S, Thomas GJ., Jr Subunit conformations and assembly states of a DNA-translocating motor: The terminase of bacteriophage P22. J Mol Biol. 2007;374:817–836. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlove T, Konieczka JH, Cordes MH. Secondary structure switching in Cro protein evolution. Structure (Camb) 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Nickels BE, Roberts CW, Sun H, Roberts JW, Hochschild A. The sigma (70) subunit of RNA polymerase is contacted by the (lambda)Q antiterminator during early elongation. Mol Cell. 2002;10:611–622. doi: 10.1016/s1097-2765(02)00648-2. [DOI] [PubMed] [Google Scholar]
- Nilsson AS, Haggård-Ljungquist E. In: The P2-like bacteriophages. Calendar R, editor. Oxford University Press; New York: 2006. pp. 365–390. [Google Scholar]
- Nilsson AS, Karlsson JL, Haggård-Ljungquist E. Site-specific recombination links the evolution of P2-like coliphages and pathogenic enterobacteria. Mol Biol Evol. 2004;21:1–13. doi: 10.1093/molbev/msg223. [DOI] [PubMed] [Google Scholar]
- Nooren IM, Kaptein R, Sauer RT, Boelens R. The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils. Nat Struct Biol. 1999;6:755–759. doi: 10.1038/11531. [DOI] [PubMed] [Google Scholar]
- Nuez B, Rojo F, Salas M. Phage phi 29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein-protein contacts. Proc Natl Acad Sci USA. 1992;89:11401–11405. doi: 10.1073/pnas.89.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegrip R, Haggard-Ljungquist E. The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage p2. J Mol Biol. 2001;308:147–163. doi: 10.1006/jmbi.2001.4607. [DOI] [PubMed] [Google Scholar]
- Odegrip R, Schoen S, Haggard-Ljungquist E, Park K, Chattoraj DK. The interaction of bacteriophage P2 B protein with Escherichia coli DnaB helicase. J Virol. 2000;74:4057–4063. doi: 10.1128/jvi.74.9.4057-4063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia A, Prevelige P, Jr, Johnson J, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Al-Bassam J, Winn-Stapley DA, Joss L, Casjens SR, Cingolani G. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J Mol Biol. 2006;363:558–576. doi: 10.1016/j.jmb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Olia AS, Bhardwaj A, Joss L, Casjens S, Cingolani G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry. 2007;46:8776–8784. doi: 10.1021/bi700186e. [DOI] [PubMed] [Google Scholar]
- Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- Pani B, Ranjan A, Sen R. Interaction surface of bacteriophage P4 protein Psu required for complex formation with the transcription terminator Rho. J Mol Biol. 2009;389:647–660. doi: 10.1016/j.jmb.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Parker MH, Casjens S, Prevelige PE., Jr Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- Parker MH, Stafford WF, 3rd, Prevelige PE., Jr Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- Parkinson MJ, Lilley DM. The junction-resolving enzyme T7 endonuclease I: Quaternary structure and interaction with DNA. J Mol Biol. 1997;270:169–178. doi: 10.1006/jmbi.1997.1128. [DOI] [PubMed] [Google Scholar]
- Parua PK, Mondal A, Parrack P. HflD, an Escherichia coli protein involved in the lambda lysis-lysogeny switch, impairs transcription activation by lambdaCII. Arch Biochem Biophys. 2010;493:175–183. doi: 10.1016/j.abb.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Pastrana R, Lazaro JM, Blanco L, Garcia JA, Mendez E, Salas M. Overproduction and purification of protein P6 of Bacillus subtilis phage phi 29: Role in the initiation of DNA replication. Nucleic Acids Res. 1985;13:3083–3100. doi: 10.1093/nar/13.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Karthikeyan T, Houtz JM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. Corrected sequence of the bacteriophage p22 genome. J Bacteriol. 2003;185:1475–1477. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell LG, Liu A, Edmonds L, Donaldson LW, Howell PL, Davidson AR. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol. 2009;389:938–951. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–573. doi: 10.1016/0042-6822(79)90509-9. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC. DNA-binding properties of the Erf protein of bacteriophage P22. J Mol Biol. 1983;163:257–275. doi: 10.1016/0022-2836(83)90006-2. [DOI] [PubMed] [Google Scholar]
- Poteete AR, King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977;76:725–739. doi: 10.1016/0042-6822(77)90254-9. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Baranska S, Wegrzyn A, Wegrzyn G. Composition of the lambda plasmid heritable replication complex. Biochem J. 2002;364:857–862. doi: 10.1042/BJ20011488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell HH, Harvey AM. P22 antirepressor protein prevents in vivo recA-dependent proteolysis of P22 repressor. Mol Gen Genet. 1983;190:427–431. doi: 10.1007/BF00331072. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- Pruss GJ, Wang JC, Calendar R. In vitro packaging of covalently-closed circular monomers of bacteriophage DNA. J Mol Biol. 1975;98:465–478. doi: 10.1016/s0022-2836(75)80080-5. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A Genetic Switch: Phage Lambda Revisited. Third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Ptashne M. A Genetic Switch. Phage Lambda Revisited. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J Mol Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- Qimron U, Marintcheva B, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci USA. 2006;103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding CM, Rosenzweig J, Richards J, Cassuto E. Appendix: Separation and characterization of exonuclease, β protein and a complex of both. J Biol Chem. 1971;146:2510–2512. [Google Scholar]
- Rahmsdorf HJ, Pai SH, Ponta H, Herrlich P, Roskoski R, Jr, Schweiger M, Studier FW. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc Natl Acad Sci USA. 1974;71:586–589. doi: 10.1073/pnas.71.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Casjens S, Uetz P. The protein interaction map of bacteriophage lambda. BMC Microbiol. 2011;11:213. doi: 10.1186/1471-2180-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade K, Poteete AR. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J Bacteriol. 1993a;175:4712–4718. doi: 10.1128/jb.175.15.4712-4718.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade K, Poteete AR. A switch in translation mediated by an antisense RNA. Genes Dev. 1993b;7:1498–1507. doi: 10.1101/gad.7.8.1498. [DOI] [PubMed] [Google Scholar]
- Ray P, Murialdo H. The role of gene Nu3 in bacteriophage lambda head morphogenesis. Virology. 1975;64:247–263. doi: 10.1016/0042-6822(75)90096-3. [DOI] [PubMed] [Google Scholar]
- Rennell D, Poteete AR. Phage P22 lysis genes: Nucleotide sequences and functional relationships with T4 and lambda genes. Virology. 1985;143:280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- Rennell D, Poteete AR. Genetic analysis of bacteriophage P22 lysozyme structure. Genetics. 1989;123:431–440. doi: 10.1093/genetics/123.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende LF, Hollis T, Ellenberger T, Richardson CC. Essential amino acid residues in the single-stranded DNA-binding protein of bacteriophage T7: Identification of the dimer interface. J Biol Chem. 2002;277:50643–50653. doi: 10.1074/jbc.M207359200. [DOI] [PubMed] [Google Scholar]
- Rishovd S, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: protein processing and capsid size determination. Virology. 1992;187:548–554. doi: 10.1016/0042-6822(92)90457-z. [DOI] [PubMed] [Google Scholar]
- Rishovd S, Marvik OJ, Jacobsen E, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: Identification and characterization of the portal protein. Virology. 1994;200:744–751. doi: 10.1006/viro.1994.1238. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Robertson ES, Aggison LA, Nicholson AW. Phosphorylation of elongation factor G and ribosomal protein S6 in bacteriophage T7-infected Escherichia coli. Mol Microbiol. 1994;11:1045–1057. doi: 10.1111/j.1365-2958.1994.tb00382.x. [DOI] [PubMed] [Google Scholar]
- Robertson ES, Nicholson AW. Protein kinase of bacteriophage T7 induces the phosphorylation of only a small number of proteins in the infected cell. Virology. 1990;175:525–534. doi: 10.1016/0042-6822(90)90437-v. [DOI] [PubMed] [Google Scholar]
- Robertson ES, Nicholson AW. Phosphorylation of Escherichia coli translation initiation factors by the bacteriophage T7 protein kinase. Biochemistry. 1992;31:4822–4827. doi: 10.1021/bi00135a012. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cerrato V, Garcia P, Del Prado G, Garcia E, Gracia M, Huelves L, Ponte C, Lopez R, Soriano F. In vitro interactions of LytA, the major pneumococcal autolysin, with two bacteriophage lytic enzymes (Cpl-1 and Pal), cefotaxime and moxi-floxacin against antibiotic-susceptible and -resistant Streptococcus pneumoniae strains. J Antimicrob Chemother. 2007;60:1159–1162. doi: 10.1093/jac/dkm342. [DOI] [PubMed] [Google Scholar]
- Rojo F, Mencia M, Monsalve M, Salas M. Transcription activation and repression by interaction of a regulator with the alpha subunit of RNA polymerase: The model of phage phi 29 protein p4. Prog Nucleic Acid Res Mol Biol. 1998;60:29–46. doi: 10.1016/s0079-6603(08)60888-0. [DOI] [PubMed] [Google Scholar]
- Rojo F, Salas M. A DNA curvature can substitute phage phi 29 regulatory protein p4 when acting as a transcriptional repressor. EMBO J. 1991;10:3429–3438. doi: 10.1002/j.1460-2075.1991.tb04907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri M, Häuser R, Ouellette M, Liu J, Dehbi M, Moeck G, Garcia E, Titz B, Uetz P, Moineau S. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent phage Dp-1. J Bacteriol. 2011;193:551–562. doi: 10.1128/JB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Richardson CC. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981;27:533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- Sam MD, Papagiannis CV, Connolly KM, Corselli L, Iwahara J, Lee J, Phillips M, Wojciak JM, Johnson RC, Clubb RT. Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein. J Mol Biol. 2002;324:791–805. doi: 10.1016/s0022-2836(02)01150-6. [DOI] [PubMed] [Google Scholar]
- Sam MD, Cascio D, Johnson RC, Clubb RT. Crystal structure of the excisionase-DNA complex from bacteriophage lambda. J Mol Biol. 2004;338:229–240. doi: 10.1016/j.jmb.2004.02.053. [DOI] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sauer B, Ow D, Ling L, Calendar R. Mutants of satellite bacteriophage P4 that are defective in the suppression of transcriptional polarity. J Mol Biol. 1981;145:29–46. doi: 10.1016/0022-2836(81)90333-8. [DOI] [PubMed] [Google Scholar]
- Savalia D, Robins W, Nechaev S, Molineux I, Severinov K. The role of the T7 Gp2 inhibitor of host RNA polymerase in phage development. J Mol Biol. 2010;402:118–126. doi: 10.1016/j.jmb.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva CG, Dewey JS, Deaton J, White RL, Struck DK, Holzenburg A, Young R. The holin of bacteriophage lambda forms rings with large diameter. Mol Microbiol. 2008;69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CK, Molineux IJ. Expression of gene 1.2 and gene 10 of bacteriophage T7 is lethal to F plasmid-containing Escherichia coli. J Bacteriol. 1991;173:1536–1543. doi: 10.1128/jb.173.4.1536-1543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EC, Dickmanns A, Urlaub H, Schmitt A, Muhlenhoff M, Stummeyer K, Schwarzer D, Gerardy-Schahn R, Ficner R. Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding. Nat Struct Mol Biol. 2010;17:210–215. doi: 10.1038/nsmb.1746. [DOI] [PubMed] [Google Scholar]
- Semerjian AV, Malloy DC, Poteete AR. Genetic structure of the bacteriophage P22 PL operon. J Mol Biol. 1989;207:1–13. doi: 10.1016/0022-2836(89)90437-3. [DOI] [PubMed] [Google Scholar]
- Serna-Rico A, Munoz-Espin D, Villar L, Salas M, Meijer WJ. The integral membrane protein p16.7 organizes in vivo phi29 DNA replication through interaction with both the terminal protein and ssDNA. EMBO J. 2003;22:2297–2306. doi: 10.1093/emboj/cdg221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna-Rico A, Salas M, Meijer WJ. The Bacillus subtilis phage phi 29 protein p16.7, involved in phi 29 DNA replication, is a membrane-localized single-stranded DNA-binding protein. J Biol Chem. 2002;277:6733–6742. doi: 10.1074/jbc.M109312200. [DOI] [PubMed] [Google Scholar]
- Serrano M, Gutierrez C, Freire R, Bravo A, Salas M, Hermoso JM. Phage phi 29 protein p6: A viral histone-like protein. Biochimie. 1994;76:981–991. doi: 10.1016/0300-9084(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Serrano-Heras G, Bravo A, Salas M. Phage phi29 protein p56 prevents viral DNA replication impairment caused by uracil excision activity of uracil-DNA glycosylase. Proc Natl Acad Sci USA. 2008;105:19044–19049. doi: 10.1073/pnas.0808797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Heras G, Ruiz-Maso JA, del Solar G, Espinosa M, Bravo A, Salas M. Protein p56 from the Bacillus subtilis phage phi29 inhibits DNA-binding ability of uracil-DNA glycosylase. Nucleic Acids Res. 2007;35:5393–5401. doi: 10.1093/nar/gkm584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Heras G, Salas M, Bravo A. A uracil-DNA glycosylase inhibitor encoded by a non-uracil containing viral DNA. J Biol Chem. 2006;281:7068–7074. doi: 10.1074/jbc.M511152200. [DOI] [PubMed] [Google Scholar]
- Severinova E, Severinov K. Localization of the Escherichia coli RNA polymerase beta’ subunit residue phosphorylated by bacteriophage T7 kinase Gp0.7. J Bacteriol. 2006;188:3470–3476. doi: 10.1128/JB.188.10.3470-3476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla-Sierra P, Otting G, Wuthrich K. Determination of the nuclear magnetic resonance structure of the DNA-binding domain of the P22 c2 repressor (1 to 76) in solution and comparison with the DNA-binding domain of the 434 repressor. J Mol Biol. 1994;235:1003–1020. doi: 10.1006/jmbi.1994.1053. [DOI] [PubMed] [Google Scholar]
- Shotland Y, Shifrin A, Ziv T, Teff D, Koby S, Kobiler O, Oppenheim AB. Proteolysis of bacteriophage lambda CII by Escherichia coli FtsH (HflB) J Bacteriol. 2000;182:3111–3116. doi: 10.1128/jb.182.11.3111-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MN, Studier FW. Physical mapping of the early region of bacteriophage T7 DNA. J Mol Biol. 1973;79:249–265. doi: 10.1016/0022-2836(73)90004-1. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2001;57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Sippy J, Feiss M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage lambda terminase. J Bacteriol. 1992;174:850–856. doi: 10.1128/jb.174.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six EW. The helper dependence of satellite bacteriophage P4: which gene functions of bacteriophage P2 are needed by P4? Virology. 1975;67:249–263. doi: 10.1016/0042-6822(75)90422-5. [DOI] [PubMed] [Google Scholar]
- Six EW, Sunshine MG, Williams J, Haggård-Ljungquist E, Lindqvist BH. Morphopoietic switch mutations of bacteriophage P2. Virology. 1991;182:34–46. doi: 10.1016/0042-6822(91)90645-r. [DOI] [PubMed] [Google Scholar]
- Souza L, Calendar R, Six E, Lindqvist B. A transactivation mutant of satellite phage P4. Virology. 1977;81:81–90. doi: 10.1016/0042-6822(77)90060-5. [DOI] [PubMed] [Google Scholar]
- Stano NM, Patel SS. T7 lysozyme represses T7 RNA polymerase transcription by destabilizing the open complex during initiation. J Biol Chem. 2004;279:16136–16143. doi: 10.1074/jbc.M400139200. [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: Crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike protein: Interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- Stellberger T, Häuser R, Baiker A, Pothineni VR, Haas J, Uetz P. Improving the yeast two-hybrid system with permutated fusions proteins: The Varicella Zoster Virus interactome. Proteome Sci. 2010;8:8. doi: 10.1186/1477-5956-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanou AS, Roberts GA, Cooper LP, Clarke DJ, Thomson AR, MacKay CL, Nutley M, Cooper A, Dryden DT. Dissection of the DNA mimicry of the bacteriophage T7 Ocr protein using chemical modification. J Mol Biol. 2009;391:565–576. doi: 10.1016/j.jmb.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage lambda. Proc Natl Acad Sci USA. 1995;92:1609–1613. doi: 10.1073/pnas.92.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J Mol Biol. 1977;117:733–759. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, Hainfeld JF, Studier FW. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- Stone JC, Miller RC., Jr Spontaneous temperature-sensitive mutations in bacteriophage T7. J Virol. 1985;54:886–888. doi: 10.1128/jvi.54.3.886-888.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975;94:283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- Studier FW, Movva NR. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol. 1976;19:136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Identification and mapping of five new genes in bacteriophage T7. J Mol Biol. 1981;153:493–502. doi: 10.1016/0022-2836(81)90404-6. [DOI] [PubMed] [Google Scholar]
- Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- Sunshine M, Usher D, Calendar R. Interaction of P2 bacteriophage with the dnaB gene of Escherichia coli. J Virol. 1975;16:284–289. doi: 10.1128/jvi.16.2.284-289.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine MG, Sauer B. A bacterial mutation blocking P2 phage late gene expression. Proc Natl Acad Sci USA. 1975;72:2770–2774. doi: 10.1073/pnas.72.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind MM, Botstein D. Mechanism of action of Salmonella phage P22 antirepressor. J Mol Biol. 1975;98:413–424. doi: 10.1016/s0022-2836(75)80127-6. [DOI] [PubMed] [Google Scholar]
- Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szambowska A, Pierechod M, Węrzyn G, Glinkowska M. Coupling of transcription and replication machineries in λ DNA replication initiation: Evidence for direct interaction of Escherichia coli RNA polymerase and the λ O protein. Nucleic Acids Res. 2010;39:168–177. doi: 10.1093/nar/gkq752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- Takahashi I, Marmur J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- Tang J, Lander G, Olia A, Li R, Casjens S, Prevelige P, Cingolani G, Baker T, Johnson J. Peering down the barrel of a bacteriophage portal: The genome packaging and release valve in P22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure. 2006;14:837–845. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple L, Forsburg S, Calendar R, Christie G. Nucleotide sequence of the genes encoding the major tail sheath and tail tube proteins of bacteriophage P2. Virology. 1991;181:353–358. doi: 10.1016/0042-6822(91)90502-3. [DOI] [PubMed] [Google Scholar]
- Tokuno SI, Gough M. UV sensitivity of a nonrepressor regulatory protein of bacteriophage P22. J Virol. 1976;18:65–70. doi: 10.1128/jvi.18.1.65-70.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NQ, Lee SJ, Richardson CC, Tabor S. A novel nucleotide kinase encoded by gene 1.7 of bacteriophage T7. Mol Microbiol. 2010;77:492–504. doi: 10.1111/j.1365-2958.2010.07221.x. [DOI] [PubMed] [Google Scholar]
- Tsui L, Hendrix RW. Head-tail connector of bacteriophage lambda. J Mol Biol. 1980;142:419–438. doi: 10.1016/0022-2836(80)90280-6. [DOI] [PubMed] [Google Scholar]
- Tsui LC, Hendrix RW. Proteolytic processing of phage lambda tail protein gpH: Timing of the cleavage. Virology. 1983;125:257–264. doi: 10.1016/0042-6822(83)90199-x. [DOI] [PubMed] [Google Scholar]
- Tye BK, Huberman JA, Botstein D. Non-random circular permutation of phage P22 DNA. J Mol Biol. 1974;85:501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- Uetz P, Rajagopala SV, Dong YA, Haas J. From ORFeomes to protein interaction maps in viruses. Genome Res. 2004;14:2029–2033. doi: 10.1101/gr.2583304. [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Fujisawa H, Marco S, Carazo JM, Carrascosa JL. Three-dimensional structure of T3 connector purified from overexpressing bacteria. J Mol Biol. 1992;224:103–112. doi: 10.1016/0022-2836(92)90579-9. [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Sousa N, Barthelemy I, Fernandez JJ, Fujisawa H, Ibarra B, Carrascosa JL. Structural analysis of the bacteriophage T3 head-to-tail connector. Journal of structural biology. 2000;131:146–155. doi: 10.1006/jsbi.2000.4281. [DOI] [PubMed] [Google Scholar]
- van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374:1543–1556. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- Vander Byl C, Kropinski AM. Sequence of the genome of Salmonella bacteriophage P22. J Bacteriol. 2000;182:6472–6481. doi: 10.1128/jb.182.22.6472-6481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon AK, Liao SM, McClure WR, Sauer RT. Bacteriophage P22 Mnt repressor: DNA binding and effects on transcription in vitro. J Mol Biol. 1987a;195:311–322. doi: 10.1016/0022-2836(87)90652-8. [DOI] [PubMed] [Google Scholar]
- Vershon AK, Liao SM, McClure WR, Sauer RT. Interaction of the bacteriophage P22 Arc repressor with operator DNA. J Mol Biol. 1987b;195:323–331. doi: 10.1016/0022-2836(87)90653-x. [DOI] [PubMed] [Google Scholar]
- Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. Genomic analysis of bacteriophage epsilon 34 of Salmonella enterica serovar Anatum (15+) BMC Microbiol. 2008;8:227. doi: 10.1186/1471-2180-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hofnung M, Charbit A. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J Bacteriol. 2000a;182:508–512. doi: 10.1128/jb.182.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Wang S, Palasingam P, Nøkling RH, Lindqvist BH, Dokland T. In vitro assembly of bacteriophage P4 procapsids from purified capsid and scaffolding proteins. Virology. 2000;275:133–144. doi: 10.1006/viro.2000.0521. [DOI] [PubMed] [Google Scholar]
- Wang WF, Cheng X, Molineux IJ. Isolation and identification of fxsA, an Escherichia coli gene that can suppress F exclusion of bacteriophage T7. J Mol Biol. 1999;292:485–499. doi: 10.1006/jmbi.1999.3087. [DOI] [PubMed] [Google Scholar]
- Warren D, Sam MD, Manley K, Sarkar D, Lee SY, Abbani M, Wojciak JM, Clubb RT, Landy A. Identification of the lambda integrase surface that interacts with Xis reveals a residue that is also critical for Int dimer formation. Proc Natl Acad Sci USA. 2003;100:8176–8181. doi: 10.1073/pnas.1033041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Węgrzyn A, Taylor K, Węrzyn G. The cbpA chaperone gene function compensates for dnaJ in lambda plasmid replication during amino acid starvation of Escherichia coli. J Bacteriol. 1996;178:5847–5849. doi: 10.1128/jb.178.19.5847-5849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Węrzyn A, Węrzyn G. Inheritance of the replication complex: A unique or common phenomenon in the control of DNA replication? Arch Microbiol. 2001;175:86–93. doi: 10.1007/s002030000245. [DOI] [PubMed] [Google Scholar]
- Węrzyn G, Węrzyn A. Stress responses and replication of plasmids in bacterial cells. Microb Cell Fact. 2002;1:2. doi: 10.1186/1475-2859-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J Mol Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Werts C, Michel V, Hofnung M, Charbit A. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J Bacteriol. 1994;176:941–947. doi: 10.1128/jb.176.4.941-947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Richardson CC. Gene 18 protein of bacteriophage T7: Overproduction, purification, and characterization. J Biol Chem. 1987;262:8845–8850. [PubMed] [Google Scholar]
- Wickner S. DNA-dependent ATPase activity associated with phage P22 gene 12 protein. J Biol Chem. 1984;259:14038–14043. [PubMed] [Google Scholar]
- Wickner SH, Zahn K. Characterization of the DNA binding domain of bacteriophage lambda O protein. J Biol Chem. 1986;261:7537–7543. [PubMed] [Google Scholar]
- Wood LF, Tszine NY, Christie GE. Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C terminus of the alpha subunit of Escherichia coli RNA polymerase. J Mol Biol. 1997;274:1–7. doi: 10.1006/jmbi.1997.1390. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Leiman PG, Li L, Grimes S, Anderson DL, Rossmann MG. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell. 2009;34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, Rossmann MG. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc Natl Acad Sci USA. 2008;105:9552–9557. doi: 10.1073/pnas.0803787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Elder JH, Ito J. Comparative studies on the small Bacillus bacteriophages. J Gen Appl Microbiol. 1986;155:392–401. [Google Scholar]
- Youderian P, Susskind MM. Identification of the products of bacteriophage P22 genes, including a new late gene. Virology. 1980;107:258–269. doi: 10.1016/0042-6822(80)90291-3. [DOI] [PubMed] [Google Scholar]
- Yura K, Yamaguchi A, Go M. Coverage of whole proteome by structural genomics observed through protein homology modeling database. J Struct Funct Genom. 2006;7:65–76. doi: 10.1007/s10969-006-9010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K, Blattner FR. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985;4:3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K, Blattner FR. Direct evidence for DNA bending at the lambda replication origin. Science. 1987;236:416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zahn K, Landy A. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol Microbiol. 1996;21:69–76. doi: 10.1046/j.1365-2958.1996.6201335.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Studier FW. Isolation of transcriptionally active mutants of T7 RNA polymerase that do not support phage growth. J Mol Biol. 1995;250:156–168. doi: 10.1006/jmbi.1995.0367. [DOI] [PubMed] [Google Scholar]
- Zhang X, Studier FW. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J Mol Biol. 1997;269:10–27. doi: 10.1006/jmbi.1997.1016. [DOI] [PubMed] [Google Scholar]
- Zhang X, Studier FW. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J Mol Biol. 2004;340:707–730. doi: 10.1016/j.jmb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Ziermann R, Calendar R. Characterization of the cos sites of bacteriophages P2 and P4. Gene. 1990;96:9–15. doi: 10.1016/0378-1119(90)90334-n. [DOI] [PubMed] [Google Scholar]
- Zillig W, Fujiki H, Blum W, Janekovic D, Schweiger M, Rahmsdorf H, Ponta H, Hirsch-Kauffmann M. In vivo and in vitro phosphorylation of DNA-dependent RNA polymerase of Escherichia coli by bacteriophage-T7-induced protein kinase. Proc Natl Acad Sci USA. 1975;72:2506–2510. doi: 10.1073/pnas.72.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: The role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


