Abstract
Here we report AWP28, an activity-based probe that can be used to biochemically monitor caspase-1 activation in response to pro-inflammatory stimuli. Using AWP28 we show that apoptosis is triggered upon bacterial infection in primary murine bone marrow macrophages lacking caspase-1. Furthermore we report that upon Salmonella infection, inflammasome-mediated caspase-1 activity is required to bypass apoptosis in favor of pro-inflammatory pyroptotic cell death.
Keywords: caspase, caspase-1, inflammasome, apoptosis, pyroptosis, activity-based probe
The cysteine protease caspase-1 initiates a pro-inflammatory cell death program termed pyroptosis in response to a variety of insults including metabolic stress and bacterial infection1–3. Pyroptosis is characterized by rapid loss of membrane integrity, resulting in cell lysis and release of pro-inflammatory components. Unlike apoptosis, pyroptosis is not immunologically silent and is regulated by formation of an intracellular multi-protein complex called the inflammasome1,3–5. Inflammasomes contain cytosolic pattern recognition receptors (PRRs), the pro-inflammatory cysteine protease caspase-1, and often the adapter protein ASC5. These receptors oligomerize in response to pathogen-or danger-associated molecular patterns (PAMPs or DAMPs), triggering autoproteolytic activation of the procaspase-1 zymogen into its active, tetrameric form.
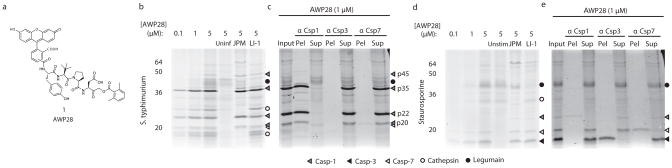
Because caspases are activated post-translationally, activity-based probes (ABPs) are powerful tools for studying their regulation and function6. In order to biochemically characterize inflammasome-mediated caspase-1 activation, we synthesized the fluorescently labeled probe AWP28 (Supplementary Methods), which contains an acyloxymethyl ketone electrophile linked to an optimized scaffold for caspase-1 recognition7 (Fig. 1a). When intact primary BMMs were infected with the Gram-negative intracellular bacterial pathogen Salmonella typhimurium, AWP28 primarily labeled multiple forms of caspase-1 as confirmed by immunoprecipitation (Fig. 1b,c). This included the p45 and p35 proforms8, revealing that higher molecular-weight forms of caspase-1 are active in response to S. typhimurium infection before being processed to the final catalytic p20 subunit in the p20/p10 heterotetramer. Interestingly, AWP28 labeled the p35 form of caspase-1 to a greater extent than the p20 and p22 forms, despite reports that these smaller forms are up to 130 fold more active8,9. Importantly, AWP28 showed significantly reduced non-specific background labeling compared to the commercially available caspase-1 probe FLICA, which contains the highly reactive fluoromethyl ketone electrophile10,11 (Supplementary Results, Supplementary Fig. 1). AWP28 also labeled a small amount of active executioner caspase-7 in response to S. typhimurium infection, consistent with its reported activation during pyroptosis12.
Figure 1.
AWP28 is an optimized caspase activity-based probe. (a) Structure of AWP28 (1). AWP28 labeling of intact primary murine bone marrow macrophages (BMMs) (b) infected with S. typhimurium (10:1) for 1 hour or (d) stimulated with 500 nM staurosporine for 4 hours. Cells were labeled with the indicated probe concentration for the final hour prior to harvest. Labeled proteins that are competed away upon pretreatment with 50 μM JPM-OEt (JPM) or 10 μM LI-1 reveal off-target lysosomal cysteine proteases (cathepsins and legumain, respectively). The identities of the labeled proteins as determined by immunopreciptiation in c and e are indicated with symbols that are defined in the key. (c,e) Labeled protein identification by immunoprecipitation. Stimulated BMMs were labeled with 1 μM AWP28 for the final hour, and subsequently labeled active caspases were immunoprecipitated using caspase-specific antibodies.
Because our caspase-1 probe was capable of labeling caspase-7, we wanted to determine if it could be used to visualize caspase activation upon initiation of classical apoptotic pathways. In BMMs undergoing staurosporine-induced intrinsic apoptosis, AWP28 labeled the executioner caspases-3 and -7, as confirmed by immunoprecipitation (Fig. 1d, e). Characterization by in vitro inhibition kinetics showed AWP28 is over 10-fold more potent against caspase-1 than other tested caspases (Supplementary Table 1), yet it can be used to simultaneously visualize activation of caspase-1 and downstream executioner caspases.
AWP28 can also be used in microscopy to show co-localization of active caspase-1 with the adapter protein ASC, which oligomerizes into large macromolecular inflammasome foci that serve as an activating platform for procaspase-113 (Supplementary Fig. 2). We found that the fraction of AWP28 labeled ASC foci saturated at concentrations as low as 100 nM whereas FLICA showed much higher levels of background and failed to completely label all ASC foci even at the highest concentration tested (Supplementary Fig. 2b). Importantly, despite this level of potency, AWP28 does not inhibit downstream physiological events of infection such as cell death or IL-1β processing and release (Supplementary Fig. 3).
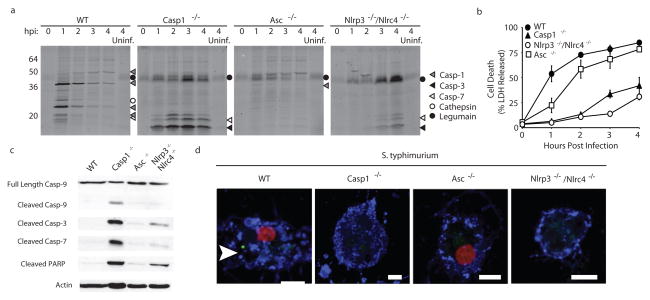
We next used AWP28 to dissect the kinetics of caspase-1 activation upon Salmonella infection in macrophages lacking different components of the inflammasome complex. Active forms of caspase-1 could be detected by AWP28 for several hours following infection of wild type BMMs (Fig. 2a). This activity was lost over time because the active protease is secreted and the cells are dying via pyroptosis14. Surprisingly, when the probe was used to visualize caspase activation in Casp1−/− BMMs, robust activation of the executioner caspases-3 and -7 was observed (Fig. 2a, Supplementary Fig. 4). We also found that activation of the executioner caspases was significantly delayed in infected BMMs lacking the receptors for S. typhimurium, NLRP3 and NLRC413 compared to Casp1−/− cells. In contrast, when ASC deficient BMMs were infected with S. typhimurium, none of the caspases were robustly activated (Fig. 2a). These cells undergo pyroptosis similar to wild type BMMs, but do not produce detectable levels of cleaved caspase-115. Activity of the p45 procaspase-1 has been shown to be essential for this cell death; however this activity appears to be below the detection limit of AWP28. S. typhimurium infected wild type and Asc−/− BMMs showed rapid release of LDH, which is a hallmark of pyroptosis1–3 (Fig. 2b). In contrast, infected cells lacking caspase-1 or the inflammasome receptors released LDH at a much slower rate, consistent with a primarily apoptotic cell death. Together these results suggest that caspase-3 and -7 are activated in response to S. typhimurium infection in the absence of inflammasome-mediated caspase-1 activation.
Figure 2.
S. typhimurium infection triggers apoptosis in the absence of inflammasome-mediated caspase-1 activation. (a) BMMs missing different inflammasome components as indicated were infected with S. typhimurium (10:1) and labeled with 1 μM AWP28 for one hour prior to each time point. The identities of the various labeled proteins are indicated with symbols that are defined in the key. (b) Cell death was measured by release of lactate dehydrogenase. Data represents the mean of 3 samples +/− one standard deviation. (c,d) BMMs were infected with S. typhimurium (10:1) for 2 hours. (c) Whole cell lysates were blotted for caspase-9 and cleaved caspase-3, -7, and cleaved poly (ADP-ribose) polymerase (PARP). β-actin serves as a loading control. Full blots are shown in Supplementary Fig. 8. (d) Cells were labeled with 1 μM AWP28 (green) for the last hour and stained with annexin V (blue) and propidium iodide (red). Uninfected and staurosporine controls are shown in Supplementary Fig. 9. Inflammasome focus is indicated with white arrowhead. Scale bars represent 5 μm.
We wanted to better characterize the cell death observed in the Casp1−/− and Nlrp3−/−Nlrc4−/− genotypes. Activation of executioner caspases-3 and -7 in Salmonella infected macrophages correlated with cleavage of apoptotic initiator caspase-9 as well as with cleavage of the apoptotic executioner caspase substrate poly (ADP-ribose) polymerase (PARP; Fig. 2c). These cells also retained membrane integrity two hours post infection as shown by lack of propidium iodide (PI) staining, but were stained by annexin V indicating a loss of membrane asymmetry characteristic of early apoptosis (Fig. 2d). In contrast, both infected wild type and Asc−/− BMMs were PI and annexin V positive due to membrane damage associated with pyroptosis2. Infected wild type cells also formed AWP28 labeled inflammasome foci (Fig. 2d). We did not detect cleaved caspase-3, -7, -9 or PARP in an equal amount of infected wild type BMMs undergoing pyroptosis (Fig. 2c). Taken together these results suggest that BMMs infected with Salmonella undergo apoptosis in the absence of inflammasome-mediated caspase-1 activation. This phenomenon was also observed upon infection with the Gram-negative coccobacillus Francisella novicida, consistent with recently published work16 (Supplementary Fig. 5).
We also examined mitochondrial cytochrome c release as a marker of apoptosis upon bacterial infection. Cytochrome c was detected in the cytosol of Casp1−/− BMMs and in a delayed manner in Nlrp3−/−Nlrc4−/− cells infected with S. typhimurium (Supplementary Fig. 6). Surprisingly, cytochrome c was also released at rates as fast or faster in wild type and Asc−/− macrophages, which undergo pyroptosis. Despite releasing cytochrome c, these pyroptotic cells were not able to effectively induce caspase-9 activation (Fig. 2c, Supplementary Fig. 5). Because active caspases are known to induce cytochrome c release17, it is possible that cytochrome c released during pyroptosis happens at a late stage of death and is not productive for apoptosome formation.
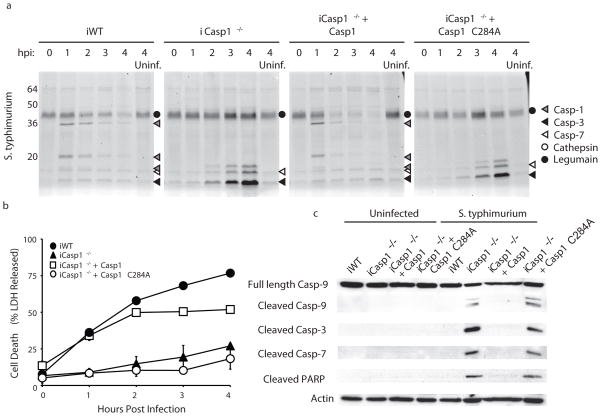
Because caspase-1 acts in a complex with other proteins, we wanted to determine if the presence of the protein alone was sufficient to block the apoptotic death observed in the casapse-1 deficient cells. Therefore, we infected immortalized Casp1−/− BMMs stably expressing active or catalytically dead (C284A) caspase-115 with S. typhimurium (Fig. 3a). We observed faint labeling of active caspase-3 and -7 by AWP28 in uninfected wild type and Casp1−/− control cells; however no significant background cytochrome c release was observed in these cells (Supplementary Fig. 7). Casp1−/− cells expressing wild type caspase-1 showed caspase-1 labeling by AWP28 upon infection and underwent pyroptosis with kinetics similar to immortalized wild type BMMs (Fig. 3a,b). However, expression of the catalytically dead mutant caspase-1 failed to block the robust activation of the apoptotic executioner caspases-3 and -7 seen in control Casp1−/− cells (Fig. 3a). Immortalized Casp1−/− BMMs expressing catalytically dead caspase-1 also retained markers of apoptosis including caspase-9 and PARP cleavage (Fig. 3c). Together these results indicate that caspase-1 proteolytic activity is required to bypass apoptosis in response to Salmonella infection. It was recently reported that Casp1−/− BMMs also lack caspase-1118. However, our ability to restore pyroptosis by expression of caspase-1 alone confirm that the interplay between apoptosis and pyroptosis is solely dependent on caspase-1 activity.
Figure 3.
Caspase-1 activity is required to bypass apoptosis upon S. typhimurium infection. Immortalized BMMs (indicated by “i”) stably transfected with wild type or catalytically dead (C284A) caspase-1 were infected with S. typhimurium (10:1). (a) Time course of caspase activation. BMMs were labeled with 1 μM AWP28 for 1 hour prior to each time point. The identities of the various labeled proteins are indicated with symbols that are defined in the key. (b) Time course of S. typhimurium induced cell death as measured by LDH release. (c) Characterization of cell death by blotting whole cell lysates for caspase-9 and cleaved caspase-3,-7, and cleaved PARP. β-actin serves as a loading control. Full blots are shown in Supplementary Fig. 10.
In conclusion, we used the activity-based caspase probe AWP28 to show that intracellular bacterial pathogens trigger apoptosis in the absence of caspase-1 activation. Pyroptosis provides an effective mechanism for the clearance of intracellular bacteria19. This explains why many intracellular pathogens have evolved strategies to prevent inflammasome-mediated caspase-1 activation20,21. It would therefore be beneficial for the host to evolve a backup cell death pathway to destroy the replicative niche for intracellular pathogens capable of inhibiting pyroptosis. The bacterial PAMPs that transduce the pro-apoptotic signal are unknown. However, we show that apoptosis triggered by Salmonella infection is significantly delayed in the absence of the inflammasome receptors NLRP3 and NLRC4, suggesting possible overlap between pro-apoptotic and pro-pyroptotic signals (Fig. 2, Supplementary Fig. 6). In some instances pro-inflammatory stimuli have been reported to trigger apoptosis in the absence of caspase-116,22,23. It is now clear from our results that caspase-1 activity is required to suppress apoptosis upon Salmonella infection. It has recently been shown that during embryogenesis and T-cell proliferation active pro-apoptotic caspase-8 blocks necroptosis by processing CYLD24,25, which is consistent with our findings of cross-regulation of death pathways by caspase-1 activity. It is not yet clear if caspase-1 is directly cleaving a substrate that prevents the activation of the apoptotic cascade. The identification of specific points of feedback between pro-inflammatory and immunologically silent cell death programs may enable innate immune responses to be therapeutically modulated in pathologies such as auto-inflammatory diseases or cancer.
Supplementary Material
Acknowledgments
We thank the Bogyo and Monack labs for useful discussions and help with macrophage harvesting. We thank S. Lynch for help with NMR. We thank B. Van Raam for apoptosis marker protocols, S. Snipas and E. Deu for helpful discussions about kinetics. Caspases-3, 4, 6, 7, and 8 and substrates were provided by the Salvesen Lab, Granzyme B and substrate was provided by the Craik Lab, anti-caspase-1 antibody was provided by Vandenabeele. This work was supported by NIH grants R01EB005011 and R01AI088541 to MB. A.W.P. was supported by an NSF Graduate Research Fellowship. P.B. is supported by a Stanford Institute for Immunity, Transplantation and Infection Young Investigator Award and was supported by the Swiss National Science Foundation and the Human Frontiers in Science Program. A.S. is supported by the NIH grant K99 GM092934. M.B. and D.M.M. are supported by the Burroughs Wellcome Foundation.
Footnotes
Author Contributions
A.W.P., P.B., A.S., D.M.M., and M.B. designed the experiments. A.W.P. and P.B. performed the experiments. A.W.P., P.B., A.S., D.M.M., and M.B. analyzed the data. A.W.P. and M.B. wrote the manuscript.
Competing Financial Interests Statement
The authors have no competing financial interests.
References
- 1.Bergsbaken T, Fink SL, Cookson BT. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fink SL, Cookson BT. Infect Immun. 2005;73:1907–16. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao EA, Rajan JV, Aderem A. Immunol Rev. 2011;243:206–14. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr JF, Wyllie AH, Currie AR. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder K, Tschopp J. Cell. 2010;140:821–32. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 6.Puri AW, Bogyo M. ACS Chem Biol. 2009;4:603–16. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wannamaker W, et al. J Pharmacol Exp Ther. 2007;321:509–16. doi: 10.1124/jpet.106.111344. [DOI] [PubMed] [Google Scholar]
- 8.Yamin TT, Ayala JM, Miller DK. J Biol Chem. 1996;271:13273–82. doi: 10.1074/jbc.271.22.13273. [DOI] [PubMed] [Google Scholar]
- 9.Elliott JM, Rouge L, Wiesmann C, Scheer JM. J Biol Chem. 2009;284:6546–53. doi: 10.1074/jbc.M806121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger AB, Sexton KB, Bogyo M. Cell Res. 2006;16:961–3. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- 11.Darzynkiewicz Z, Pozarowski P. Cytometry A. 2007;71:536–7. doi: 10.1002/cyto.a.20425. [DOI] [PubMed] [Google Scholar]
- 12.Lamkanfi M, et al. Mol Cell Proteomics. 2008;7:2350–63. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broz P, et al. J Exp Med. 2010;207:1745–55. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller M, Ruegg A, Werner S, Beer HD. Cell. 2008;132:818–31. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 15.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Cell Host Microbe. 2010;8:471–83. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierini R, et al. Cell Death Differ. 2012 doi: 10.1038/cdd.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bossy-Wetzel E, Green DR. J Biol Chem. 1999;274:17484–90. doi: 10.1074/jbc.274.25.17484. [DOI] [PubMed] [Google Scholar]
- 18.Kayagaki N, et al. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 19.Miao EA, et al. Nat Immunol. 2010;11:1136–42. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodsky IE, et al. Cell Host Microbe. 2010;7:376–87. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Master SS, et al. Cell Host Microbe. 2008;3:224–32. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jesenberger V, Procyk KJ, Yuan J, Reipert S, Baccarini M. J Exp Med. 2000;192:1035–46. doi: 10.1084/jem.192.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motani K, et al. J Biol Chem. 2011 [Google Scholar]
- 24.O’Donnell MA, et al. Nat Cell Biol. 2011;13:1437–42. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberst A, et al. Nature. 2011;471:363–7. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.