Abstract
Objective:
A better understanding of the processes influencing energy expenditure could provide new therapeutic strategies for reducing obesity. As the metabolic activity of the brown adipose tissue (BAT) and skeletal muscle is an important determinant of overall energy expenditure and adiposity, we investigated the role of genes that could influence cellular bioenergetics in these two tissues.
Design:
We screened for genes that are induced in both the BAT and skeletal muscle during acute adaptive thermogenesis in the mouse by microarray. We used C57BL/6J mice as well as the primary and immortalized brown adipocytes and C2C12 myocytes to validate the microarray data. Further characterization included gene expression, mitochondrial density, cellular respiration and substrate utilization. We also used a Hybrid Mouse Diversity Panel to assess in vivo effects on obesity and body fat content.
Results:
We identified the transcription factor Zbtb16 (also known as Plzf and Zfp14) as being induced in both the BAT and skeletal muscle during acute adaptive thermogenesis. Zbtb16 overexpression in brown adipocytes led to the induction of components of the thermogenic program, including genes involved in fatty acid oxidation, glycolysis and mitochondrial function. Enhanced Zbtb16 expression also increased mitochondrial number, as well as the respiratory capacity and uncoupling. These effects were accompanied by decreased triglyceride content and increased carbohydrate utilization in brown adipocytes. Natural variation in Zbtb16 mRNA levels in multiple tissues across a panel of >100 mouse strains was inversely correlated with body weight and body fat content.
Conclusion:
Our results implicate Zbtb16 as a novel determinant of substrate utilization in brown adipocytes and of adiposity in vivo.
Keywords: Zbtb16, Plzf, brown adipose tissue, adaptive thermogenesis, bioenergetics
Introduction
The increasing prevalence of obesity worldwide reflects changes in lifestyle, including a combination of increased food intake and reduced physical activity.1, 2, 3 This combination contributes to the accumulation of an energy surplus in the form of adipose tissue. Because obesity develops when energy intake exceeds energy expenditure, increasing energy expenditure is an attractive strategy to reduce body weight and fat storage. There has been considerable interest in modulating thermogenesis, particularly in the skeletal muscle and brown adipose tissue (BAT), as a target for the treatment of obesity.4, 5, 6
Skeletal muscle is one of the central tissues involved in shivering thermogenesis and also contributes to non-shivering thermogenesis.5, 7, 8 Skeletal muscle is responsible for at least 30% of the basal metabolic energy expenditure and 80% of whole-body glucose uptake.9 Because skeletal muscle accounts for approximately 30–40% of total body mass, it may contribute substantially to adaptive thermogenesis in humans.5 Recently, the existence of BAT in humans has been reappraised and there is good evidence that brown fat depots are active in adults and are capable of energy dissipation through adaptive thermogenesis.10, 11, 12, 13 Furthermore, body-mass index and percentage of body fat are inversely correlated with the BAT in humans12, suggesting that the BAT contributes to metabolic rate and is an important regulator of body fat accumulation. Further evidence for a shared physiological function between brown adipocytes and myocytes is the fact that they are derived from a common cell lineage.14, 15 Increased heat production through decreased coupling between fatty acid oxidation and ATP synthesis has been observed during both diet-induced and cold-induced adaptive thermogenesis.16, 17 During adaptive thermogenesis, a significant amount of energy stored as fat and glycogen is converted to heat instead of being efficiently converted to ATP, suggesting that induction of mitochondrial respiration or the level of uncoupling activity could promote caloric dissipation.
Because of its potential for energy dissipation, it is of great interest to identify the genes that orchestrate the changes occurring during adaptive thermogenesis and to determine their mechanisms of action. One key factor in adaptive thermogenesis of the BAT is the mitochondrial uncoupling protein-1 (Ucp1). Ucp1 is responsible for enabling the proton leak in mitochondria that dissipates energy produced through oxidative metabolism to generate heat.17, 18, 19 Previous systematic studies have identified genes that are induced in BAT after cold exposure in mouse, rat and squirrel,20, 21, 22, 23, 24, 25, 26 and a recent transcriptome-profiling study identified several genes that are induced in the skeletal muscle and BAT in mice.27
We hypothesized that understanding the function of genes promoting energy expenditure would reveal new targets for modulation of energy balance. Genes differentially expressed in both the BAT and skeletal muscle could be of great importance because they are indicative of a general thermogenic response not limited to the BAT-specific uncoupling or skeletal muscle-specific shivering. We performed transcriptional profiling of the BAT and skeletal muscle during acute cold exposure in mice, a model of adaptive thermogenesis. We identified genes that were induced in both the BAT and skeletal muscle following cold exposure. One of the significantly upregulated genes is the transcription factor Zbtb16, which belongs to the POZ/domain and Krüpel zinc finger family and generally acts as a repressor.28, 29, 30 Here, we demonstrate that Zbtb16 levels influence cellular bioenergetics in vitro and inversely correlate with adiposity in vivo.
Materials and methods
Mice
Animal studies were performed under approved UCLA animal research protocols and according to guidelines established in the Guide for Care and Use of Laboratory Animals. C57BL/6J mice were maintained in 12-h light/dark conditions and fed a regular chow diet (Purina 50010, Lab Diet, Richmond, IN, USA). Cold exposure was performed as described.31 After euthanasia, tissues were collected and frozen until use.
Microarray analysis
Total RNA was isolated from mouse tissues by extraction with TRIzol (Invitrogen, Carlsbad, CA, USA). Samples were then hybridized to Illumina 24K BeadChips using standard protocols (The Southern California Genotyping Consortium at UCLA). Differentially expressed genes were identified by applying a Student's t-test and fold-change thresholds. The microarray data can be accessed from the NCBI Gene Expression Omnibus (GEO) database.
Cell culture and over-expression
Primary brown adipocytes were cultured as described previously.31 C2C12 cells (ATCC, Manassas, VA, USA) were grown in complete DMEM (Dulbecco's modified Eagle's medium; 10% fetal bovine serum, 25 mℳ glucose, 1 mℳ pyruvate and 1 mℳ glutamine). For differentiation, serum was switched to 2% horse serum for 6 days. The immortalized brown adipocyte cell line was a gift from Bruce Spiegelman and was cultured as described.32
Mouse Zbtb16 cDNA was cloned into pcDNA3.1/V5-His vector (Invitrogen) and transfected with BioT (Bioland, La Palma, CA, USA). Empty vector was used as control. For transduction, an adenovirus was constructed from the Zbtb16 cDNA and the AdEasy XL Adenoviral Vector System (Stratagene, Santa Clara, CA, USA). Viral particles were purified with Adeno-X Maxi Purification Kit (Clontech, Mountain View, CA, USA) and titered with Adeno-X Rapid Titer Kit (Clontech). Transduction (MOI 50) was performed at day 3 post-confluence and gene expression analyzed at day 6.
Quantitative real-time (qRT-PCR) gene expression analyses
Total RNA was isolated from mouse tissues by extraction with TRIzol (Invitrogen). For human tissues, a cDNA panel was purchased from Clontech. cDNA synthesis, real-time PCR, and qRT-PCR were performed as described previously.33 Values are expressed in arbitrary units, normalized to housekeeping genes. All primer sequences used in this study are presented in Supplementary Table S1.
Immunoblotting
Cells were lysed in lysis buffer containing protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Proteins (50 μg/lane) were resolved by 7% Tris-Acetate pre-cast gel (Invitrogen), transferred to nitrocellulose membrane and incubated for 3 h with 5% milk TBS-T (Tris-buffered saline with Tween 20), overnight with primary antibody in 1% milk TBS-T, then 1 h with a horseradish peroxidase-conjugated secondary antibody in 1% milk TBS-T. ECL Plus (GE Healthcare, Piscataway, NJ, USA) was used for chemiluminescent detection. Antibodies included ZBTB16 (OP128, Calbiochem, Billerica, MA, USA), UCP1 (662045, Calbiochem), β-ACTIN (AC-15, Sigma, Dallas, TX, USA) and LAMIN A/C (sc-6215, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Triglyceride (TAG) contents and lipolysis assay
Cells were homogenized by sonification and TAG contents determined with L-Type triglyceride M kit (Waco Chemicals, Richmond, VA, USA). Lipolysis experiments from cell medium were performed after 10 h incubation with the Adipolysis Assay Kit (AB100, Millipore, Billerica, MA, USA). TAG contents and glycerol concentrations were normalized to protein.
Cellular oxygen consumption and extracellular acidification rates
Cellular metabolic rates were measured using a XF24 Analyzer (Seahorse Bioscience, Billerica, MA, USA). Immediately before the measurement, cells were washed with unbuffered DMEM as described.34 Plates were placed into the XF24 instrument for measurement of oxygen consumption and extracellular acidification (ECAR) rates. Mixing, waiting and measurement times were 3, 2, and 3 min for C2C12 and 5, 2 and 1.5 min for brown adipocyte cells, respectively. The measures were normalized per protein. Test compounds were obtained from Sigma and injected during the assay at the following final concentration: 0.75 or 0.5 μℳ oligomycin, 0.5 or 0.4 μℳ FCCP (carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone), 0.75 or 0.15 μℳ rotenone/myxothiazol (differentiated brown adipocyte cells or C2C12, respectively), 25 mℳ glucose, 100 μℳ 2,4-dinitrophenol, 50 mℳ Na-oxamate, and 200 mℳ 2-deoxyglucose. The basal and percentage of oxygen consumption rate and ECAR levels as well as area under the curve were obtained with the XF24 Analyzer software.
Mitochondrial DNA content
Total genomic DNA from the BAT tissue was isolated by phenol/chloroform/isoamyl alcohol extraction. Mitochondrial and nuclear DNA were amplified by RT-PCR with 25 ng of DNA and primers in the D-Loop region and Tert gene, respectively (sequences in Supplementary Table S1).
Substrate utilization with Biolog microplates
PM-M TOX1 microplates (Biolog, Hayward, CA, USA), consisting of eight different substrates in replicate, were used. Differentiated brown adipocyte cells were seeded at 60 000 cells per well in complete medium without pyruvate, glucose or phenol red. After 24 h, Biolog Redox Dye MA was added and rates measured every hour at 590 nm to detect the metabolic activity. Time 0 was subtracted for each time point. The reductase activity (NADH) is proportional to the energy produced from each substrate.
Hybrid Mouse Diversity Panel
Mouse strains, RNA isolation and expression profiling were described previously.35 Briefly, at 16 weeks of age, 4–6 mice per strain were assessed for body composition using an EchoMRI instrument (EchoMRI, Houston, TX, USA). Mice were fasted overnight before plasma and tissue collection. Tissues were weighed and then snap frozen in liquid nitrogen until RNA, cRNA and cDNA were prepared. Biotin-labeled cRNA was generated from the cDNA and used to probe by using Affymetrix Mouse Genome HT_MG430A arrays (4–6 mice per strain). Expression data (probe 1442025_a_at) and phenotypic traits are presented in Supplementary Table S2.
Statistical analysis
The data are expressed as the mean±s.d., except for the XF24 Seahorse Bioscience analysis where s.e. was used. Unpaired Student's t-test was used to compare the difference between the groups.
Results
Identifying genes differentially regulated in adaptive thermogenesis
We assayed the transcriptomes of mice exposed to cold using gene expression microarrays to identify genes differentially expressed during thermogenesis. In the BAT, 124 genes were upregulated and 176 were downregulated at least twofold in response to cold exposure. In the skeletal muscle (quadriceps), 69 genes were upregulated and 67 were downregulated at least twofold in response to cold exposure. We detected 11 genes that were altered by cold exposure in both the BAT and skeletal muscle, 5 of which were coordinately upregulated and 6 downregulated (Table 1). The transcription factor Zbtb16 (also known as Plzf and Zfp145) exhibited a robust induction in both the BAT (5.6-fold) and skeletal muscle (4.8-fold). Zbtb16 has been associated with white adipogensis36 but has not previously been implicated in the BAT or muscle metabolism. We selected this gene to further characterize its function.
Table 1. Common differential expression in BAT and muscle tissue (P-value<0.05 and change >twofold).
| Entrez ID | Gene |
Brown adipose tissue |
Muscle tissue |
||
|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | ||
| 235320 | Zbtb16 | 5.6 | 1.7 × 10−2 | 4.8 | 2.3 × 10−2 |
| 24082 | Gtlf3a | 3.7 | 3.6 × 10−2 | 6.6 | 4.5 × 10−2 |
| 20305 | Ccl6 | 2.3 | 3.3 × 10−3 | 4.5 | 2.8 × 10−3 |
| 227731 | Slc25a25 | 2.1 | 4.4 × 10−2 | 13.9 | 4.2 × 10−2 |
| 17750 | Mt2 | 2.1 | 2.2 × 10−2 | 2.5 | 2.0 × 10−2 |
| 20846 | Stat1 | −2.5 | 3.0 × 10−2 | −2.6 | 4.6 × 10−2 |
| 51789 | Tnk2 | −2.6 | 2.5 × 10−2 | −2.1 | 2.5 × 10−2 |
| 11481 | Acvr2b | −2.7 | 4.2 × 10−2 | −2.9 | 2.3 × 10−2 |
| 22341 | Vegf | −3.6 | 1.9 × 10−2 | −2.9 | 1.5 × 10−2 |
| 22418 | Wnt5a | −3.6 | 1.2 × 10−3 | −2.3 | 4.0 × 10−2 |
| 217198 | Plekhh3 | −4.8 | 1.1 × 10−2 | −2.2 | 1.9 × 10−3 |
Abbreviation: BAT, brown adipose tissue.
Validation of Zbtb16 induction in adaptive thermogenesis
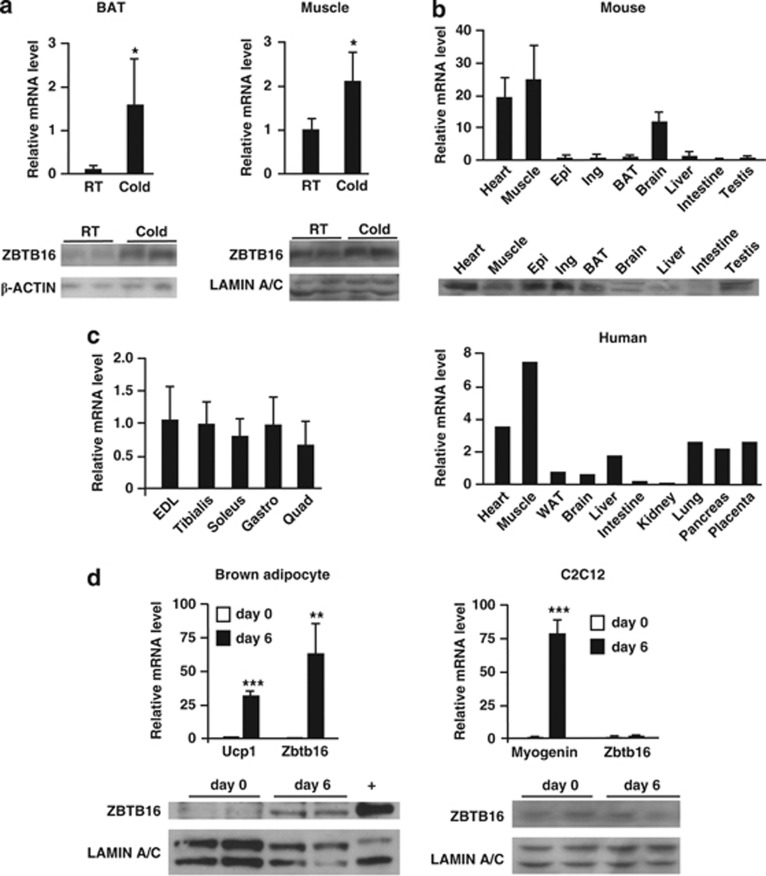
We first confirmed that Zbtb16 expression is consistently upregulated by cold exposure at the mRNA and protein levels in an independent group of mice. As shown in Figure 1a, Zbtb16 mRNA levels were increased 15.3-fold in the BAT and 2.1-fold in the muscle as detected by qPCR. Zbtb16 mRNA induction by cold was observed after 4 or 8 h and whether mice were fasted or not during the cold exposure (data not shown). Western blot analysis confirmed a concomitant increase in ZBTB16 protein levels (Figure 1a, bottom inserts). Thus, the upregulation of Zbtb16 during adaptive thermogenesis was validated at both the transcript and protein levels.
Figure 1.
Zbtb16 expression in tissues and cell lines. (a) Validation of Zbtb16 mRNA upregulation in BAT and skeletal muscle by qRT-PCR (mean±s.d.; n=4) and by immunoblot. (b) Zbtb16 mRNA and protein levels in mouse (mean±s.d.; n=3) and Zbtb16 mRNA in human (n=1) tissues. (c) Zbtb16 mRNA levels in different skeletal muscles (mean±s.d.; n=4). (d) Zbtb16 mRNA levels and protein levels during brown adipocyte and C2C12 differentiation (mean±s.d., n=4). Ucp1 and myogenin mRNA levels are differentiation controls for brown adipocyte and C2C12 cells, respectively. *P<0.05; **P<0.01; ***P<0.001 compared with control. EDL, extensor digitorum longus; Epi, epididymal fat; Gastro, gastrocnemius; Ing, inguinal fat; Quad, quadriceps; Tibialis, tibialis anterior; WAT, white adipose tissue.
Tissue expression pattern of Zbtb16 in mouse and human
We then investigated whether the BAT and skeletal muscle were the most biologically relevant tissues for studying Zbtb16 function by comparing mRNA expression levels in mouse and human tissue panels by qPCR. The highest Zbtb16 expression levels were detected in the skeletal muscle and heart for both mouse and human (Figure 1b). In contrast to what Mikkelsen, et al.36 reported, we detected a lower expression level in the mouse BAT than in the skeletal muscle. However, the levels of ZBTB16 protein measured by immunoblot in the white adipose tissue and BAT were similar to those of the heart and skeletal muscle (Figure 1b, insert). Within the muscle, the expression of Zbtb16 was similar in fast twitch (extensor digitorum longus and tibialis), slow twitch (soleus) and mixed (quadriceps and gastronecmius) muscles (Figure 1c), suggesting that Zbtb16 may function across muscle fiber types.
Expression of Zbtb16 during brown adipocyte and myocyte differentiation
We then studied Zbtb16 function in vitro using a mouse brown adipocyte cell line32 and the mouse C2C12 myocyte cell line. We first assessed Zbtb16 expression dynamics during differentiation of brown adipocyte precursors into mature brown adipocytes. Zbtb16 expression was dramatically upregulated 60-fold during brown adipocyte differentiation, whereas Ucp1 showed a 30-fold induction (Figure 1d). By contrast, Zbtb16 mRNA levels did not change during differentiation of C2C12 myoblasts to myotubes, whereas myogenin, used as control, was dramatically induced. The increased mRNA levels in brown adipocytes were mirrored by a substantial increase in ZBTB16 protein levels (Figure 1d, bottom inserts). The induction of Zbtb16 during brown adipogenesis is consistent with a role for this protein in mature brown adipocytes.
Zbtb16 expression activates the brown adipocyte gene expression program
To begin to elucidate the effect of Zbtb16 induction during thermogenesis, we overexpressed Zbtb16 in brown adipocyte cells using an adenoviral vector carrying Zbtb16 cDNA. We achieved roughly sixfold overexpression of Zbtb16 mRNA (Figure 2a), which led to alterations in expression of genes involved in brown adipocyte differentiation and mitochondrial function. Indeed, compared with cells infected with a control lacZ-containing adenovirus, cells overexpressing Zbtb16 exhibited significant upregulation of brown fat-enriched markers such as Ucp1, Ppargc1a, Ppara and Prdm16 (PR domain containing 16; Figure 2b). Enhanced Zbtb16 expression also stimulated the expression of genes promoting fatty acid oxidation, such as Fabp3 (fatty acid-binding protein 3), Fabp5 (also known as Mal1) and Cpt1b (carnitine palmitoyltransferase 1b). Zbtb16 expression also elevated expression levels of mitochondrial gene markers such as Tfam (mitochondrial transcription factor A), Elovl3 (elongation of very long chain fatty acids-like 3), Cidea (cell death-inducing DFFA-like effector a), Cox7a1 (cytochrome c oxidase, subunit VIIa 1) and Cox8b (cytochrome c oxidase, subunit VIIIb). To confirm these effects observed in an immortalized brown adipocyte cell line, we performed similar studies in primary brown adipocytes. Zbtb16 adenoviral expression induced UCP1 protein levels (Figure 2c) and increased expression of brown fat, fatty acid oxidation and mitochondrial genes (Figure 2d). Thus, Zbtb16 expression promotes a gene expression signature characteristic of active brown adipocytes and enhanced fatty acid oxidation.
Figure 2.
Zbtb16 expression increases the gene program of brown adipocyte cells. (a) Zbtb16 mRNA and protein levels after adenovirus transduction in differentiated immortalized brown adipocytes. (b) mRNA levels of brown adipocyte, fatty acid oxidation and mitochondrial markers in immortalized brown adipocytes (mean±s.d., n=4). (c) ZBTB16 and UCP1 protein levels after adenovirus transduction in differentiated primary brown adipocytes. (d) mRNA levels of brown adipocyte, fatty acid oxidation and mitochondrial markers in differentiated primary brown adipocytes (mean±s.d., n=4). *P<0.05; **P<0.01; ***P<0.001 compared with control.
Zbtb16 enhances mitochondrial respiration and mitochondrial biogenesis
The effect of Zbtb16 on mitochondrial gene expression described above suggested that it may promote changes in brown adipocyte energetics. To assess this, we measured cellular respiration in brown adipocytes by real-time detection of oxygen consumption. Using a XF24 Seahorse Bioscience instrument, we determined the level of different components of mitochondrial respiration contributing to the oxygen consumption using specific effectors. First, we determined the basal and coupling state of mitochondrial respiration with sequential injections of oligomycin and rotenone/myxothiazol. Oligomycin (F1F0-ATP synthase inhibitor) allows the measurement of ATP-linked oxygen consumption (ATP turnover) and rotenone/myxothiazol (complex I and III inhibitor, respectively) by blocking mitochondrial respiration infers the non-mitochondrial respiration and the mitochondrial uncoupling (difference between oligomycin and rotenone/myxothiazol injections). Cells were ‘activated' 2 h before the first measurement with 10 nℳ of the adrenoceptor agonist CL316,243. In both the immortalized and primary brown adipocytes, Zbtb16 overexpression did not alter basal mitochondrial respiration, but showed higher mitochondrial uncoupling at the expense of coupling respiration (Figures 3a and b). C2C12 cells can also be ‘activated' by beta-adrenergic stimulation when they are differentiated in myotubes.37 When we pretreated the myotubes for 1 h with 100 nℳ isoproterenol, we also observed a significant increase in mitochondrial uncoupling owing to Zbtb16 expression (Figure 3c). We also assessed mitochondrial reserve capacity by injecting the electron transport uncoupler FCCP. Brown adipocytes (primary and immortalized) and C2C12 cells over-expressing Zbtb16 exhibited a higher FCCP response, indicating more mitochondrial respiratory capacity (Figure 3d). Altogether, these results demonstrate that Zbtb16 is able to promote proton leak during mitochondrial respiration and increase mitochondrial respiration capacity.
Figure 3.
Zbtb16 expression increases mitochondrial respiration. Oxygen consumption rate (OCR) values were recorded in cells transduced with control or Zbtb16 adenovirus, before and after the sequential injection of oligomycin, and rotenone/myxothiazol, or FCCP alone. The measures are expressed as percentage of the baseline. (a) Brown adipocyte cells. (b) Primary brown adipocytes. (c) C2C12 myotubes. (mean±SE; n=8). (d) Percentage of change in OCR in immortalized and primary brown adipocytes, and C2C12 myotubes after injection of FCCP. (e) Relative mitochondrial DNA content measured by qRT-PCR. (f) Lipolysis assay after 10 h incubation. Forskolin treatment represents a positive control. (g) Triglyceride contents after 10 h of the lipolysis assay. *P<0.05; **P<0.01; ***P<0.001 compared with control. Mito, mitochondrial respiration.
The enhanced expression of mitochondrial gene markers and mitochondrial respiration capacity suggested a potential effect of Zbtb16 on mitochondrial biogenesis. To assess mitochondrial density, we measured the relative abundance of mitochondrial-encoded gene D-loop to the nuclear-encoded telomerase reverse transcriptase (Tert) by qPCR. Zbtb16 expression resulted in increased mitochondrial DNA by threefold in brown adipocytes and by 1.7-fold in C2C12 myotubes (Figure 3e).
We next asked whether the increased mitochondrial respiration owing to Zbtb16 overexpression affects lipolysis or lipid content in brown adipocytes. Lipolysis rates in brown adipocytes were not affected by Zbtb16 expression, whereas forskolin elicited the expected increase (Figure 3f). However, intracellular TAG content was decreased by 40% in Zbtb16 overexpressing cells (Figure 3g). Together, these data demonstrate that Zbtb16 promotes biogenesis and respiration of mitochondria, and that higher expression of Zbtb16 reduces cellular TAG levels in brown adipocytes.
Zbtb16 increases glucose oxidation
We then investigated whether Zbtb16 affects substrate utilization. Control brown adipocytes or Zbtb16 overexpressing cells were plated in Biolog microplates in which each well contains one of eight specific energy substrates. All the eight substrates were metabolized by brown adipocytes (Figure 4a and Supplementary Figure S1). Overexpression of Zbtb16 specifically increased utilization of glucose (glycolysis pathway) and xylitol (pentose phosphate pathway), while metabolism of other substrates was not affected by Zbtb16.
Figure 4.
Zbtb16 increases carbohydrate utilization. (a) Immortalized brown adipocyte cells transduced with control or Zbtb16 adenovirus were dispensed in PM-M TOX1 plate and incubated for 1 day. Then, MA dye was added into the wells and the change in reductase activity due to energy (NADH) production after 10 h was recorded at 590 nm. (mean±s.d.; n=4). (b) mRNA levels of glycolysis-related markers in immortalized brown adipocytes (mean±s.d.; n=4). *P<0.05; **P<0.01 compared with control. (c) ECAR values in immortalized brown adipocytes were recorded before and after glucose or 2,4-dinitrophenol (2,4-DNP) injection. (mean±SE; n=8). The adjacent bar graphs represent the average area under the curve. (d) Percentage of change in ECAR values after injection of oligomycin, 2-deoxyglucose (2-DG) or Na-oxamate injection (mean±SE; n=6). *P<0.05; **P<0.01 compared with control.
The preference in carbohydrate utilization suggests that Zbtb16 may influence glucose metabolism. Therefore, we assayed the expression of genes controlling glycolysis in response to Zbtb16 overexpression using qPCR. Levels of Glut4 mRNA were not changed, but Hexkin2 (hexokinase II) and Pkm2 (pyruvate kinase) expression levels were increased by Zbtb16 (Figure 4b). In addition, expression of Pdk4 was decreased. Increased glucose utilization and the differential expression of glycolysis-specific enzymes support a role for Zbtb16 in glucose consumption.
We then assessed the effects of Zbtb16 overexpression on glucose metabolism by quantitating a measure of glycolysis, the ECAR. Addition of glucose or the mitochondrial uncoupler 2,4-dinitrophenol increased ECAR in differentiated brown adipocytes. This response was amplified in cells with increased Zbtb16 expression, suggesting a greater ability to increase glycolysis (Figure 4c). We therefore exposed the brown adipocytes to different modulators of glycolysis and measured the ECAR response. Cells overexpressing Zbtb16 had a greater response to treatment with oligomycin which inhibits mitochondrial ATP production, suggesting an increased glycolytic capacity when ATP demand increases (Figure 4d). When treated with the glucose analog 2-deoxyglucose, which inhibits glycolysis, the effect was slightly less pronounced in cells expressing Zbtb16 (Figure 4d). We also performed experiments with Na-oxamate, an inhibitor of lactate dehydrogenase, which catalyzes the conversion of pyruvate to lactic acid. Na-oxamate did not significantly inhibit ECAR in brown adipocytes (∼7%), suggesting that the final step of glycolysis does not account for a substantial amount of glycolysis detected by the ECAR and that anerobic glycolysis is not very active in these cells. Nevertheless, this modest inhibition was abrogated when Zbtb16 was overexpressed (Figure 4d). Together, these results suggest that Zbtb16 promotes glucose oxidation, consistent with the increased mitochondrial respiration and lower Pdk4 expression.
Zbtb16 expression correlates with body weight, fat mass and diabetes in vivo
As described above, our data indicate that Zbtb16 is involved in the adaptive thermogenesis response and promotes mitochondrial respiration and carbohydrate utilization in brown adipocytes. To assess the role of Zbtb16 on energy balance in vivo, we assessed whether genetic variations in Zbtb16 gene expression levels are correlated with clinical traits. For these studies, we examined Zbtb16 expression levels in a Hybrid Mouse Diversity Panel.35 This resource consists of more than 100 highly diverse inbred and recombinant inbred mouse strains, which allow the analysis of association between gene expression levels and metabolic traits such as plasma lipids, plasma hormones and different body fat parameters.35, 38 We investigated the correlations between Zbtb16 expression levels and obesity traits in two tissues available in this panel that are most relevant to our study: as the BAT and skeletal muscle were not available in the hybrid mouse diversity panel, we examined the white adipose tissue and heart. The most robust associations for Zbtb16 were inverse correlations with body weight and fat mass (measured by nuclear magnetic resonance and weight) in both the tissues (Table 2 and Supplementary Figure S2). The inverse correlation for fat mass included gonadal, retroperitoneal, femoral and mesenteric fat pads. This finding demonstrates that common, natural variations in Zbtb16 expression levels are correlated with body fat content in the mouse.
Table 2. Correlation between Zbtb16 transcript levels and phenotypic traits.
|
Adipose |
Heart |
|||
|---|---|---|---|---|
| R | P-value | R | P-value | |
| Body weight | −0.18 | NS | −0.32 | 1.3E × 10−3 |
| NMR-total body fat | −0.20 | NS | −0.33 | 8.2 × 10−4 |
| NMR-% body fat | −0.30 | 3.5 × 10−3 | −0.38 | 1.1 × 10−4 |
| Total fat mass | −0.22 | 3.5 × 10−2 | −0.38 | 8.5 × 10−5 |
| Gonadal fat pad | −0.27 | 7.8 × 10−3 | −0.36 | 2.3 × 10−4 |
| % Gonadal fat pad | −0.31 | 2.5 × 10−3 | −0.33 | 8.1 × 10−4 |
| Retroperineal fat pad | −0.25 | 1.3 × 10−2 | −0.35 | 4.7 × 10−4 |
| % Retroperineal fat pad | −0.23 | 2.5 × 10−2 | −0.39 | 7.4 × 10−5 |
| Femoral fat pad | −0.23 | 2.7 × 10−2 | −0.39 | 8.3 × 10−5 |
| % Femoral fat pad | −0.27 | 7.2 × 10−3 | −0.42 | 1.5 × 10−5 |
| Mesenteric fat pad | −0.26 | NS | −0.42 | 1.2 × 10−5 |
| % Mesenteric fat pad | −0.09 | NS | −0.41 | 2.2 × 10−5 |
Abbreviations: NMR, nuclear magnetic resonance; NS, not significant.
R, correlation coefficient; % corresponds to total body weight.
Given the association of Zbtb16 mRNA levels with body weight and fat mass, we also assessed whether ZBTB16 expression levels are associated with fat mass in humans as well. Data mining from previously reported microarray analyses revealed that in visceral adipose tissue of obese diabetic women, ZBTB16 expression was significantly lower than age- and body-mass index-matched normal glucose-tolerant controls (t-test P-value=0.0056; GEO accession GDS3665). This result demonstrates that in humans, ZBTB16 expression levels are associated with glucose levels, suggesting a complex interrelationship between ZBTB16 expression, glucose and obesity.
Discussion
Manipulation of cellular bioenergetics is an attractive approach to increase cellular energy expenditure with the ultimate goal to decrease obesity at the whole body level. Mitochondrial respiration, primarily in the BAT and skeletal muscle, can be modified in order to respond to physical activity, diet or ambient temperature. In the current work, we identified Zbtb16 as a novel gene involved in the cold response in both the BAT and skeletal muscle. Zbtb16 was among a few genes that were transcriptionally induced by several-fold in both the BAT and skeletal muscle during acute cold exposure. Previous works on Zbtb16 have shown potential involvement in cancer and development. The human ZBTB16 gene has been implicated in acute promyelocytic leukemia, through chromosomal translocation and fusion to the retinoic receptor alpha.39, 40 Mice lacking Zbtb16 function exhibit limb defects and loss of spermatogonial stem cells.41, 42 Interestingly, Zbtb16 is induced during differentiation of 3T3-L1 preadipocytes.36 The role of Zbtb16 as a transcription factor provides a potential mechanism by which it may influence thermogenesis, through effects on expression of other genes. Indeed, we found that induction of Zbtb16 expression in brown adipocytes enhances the expression of many genes of the thermogenic program, such as genes involved in fatty acid oxidation and mitochondrial respiration. This effect was accompanied by an increase in mitochondrial content and maximal respiration capacity, and a decrease in intracellular TAG levels, consistent with a role for Zbtb16 in fatty acid metabolism in brown adipocytes. When differentiated brown adipocytes or myotubes were activated by adrenoceptor agonists, Zbtb16 increased mitochondrial uncoupling, in agreement with the increased Ucp1 gene expression.
Another aspect of the role of Zbtb16 is its function in glucose metabolism. Zbtb16 expression drives the expression of glycolysis-related genes (Hexkn2, Pkm2), and is able to increase glycolytic capacity. Consistent with this role, Zbtb16 expression increases carbohydrate utilization in vitro. The glucose utilization seems to be directed to mitochondrial oxidation as observed by the decrease in Pdk4 expression and lower Na-oxamate-ensitive ECAR levels. Altogether, these results indicate that Zbtb16 promotes oxidative metabolism.
Ultimately, the goal is to identify genes that influence energy metabolism in BAT that translates to whole-body effects on energy balance and adiposity. To evaluate the potential effect of variations in Zbtb16 expression levels in vivo, we assessed the relationship between Zbtb16 expression levels and body mass and fat pad mass across a set of >100 mouse strains for which gene expression and body fat traits have been determined.35 In this panel, we observed a striking association between Zbtb16 mRNA levels in two different tissues and body fat parameters. Higher levels of Zbtb16 expression were associated with a decrease in body fat storage in four major white adipose tissue depots and overall body weight. We also detected a correlation between decreased ZBTB16 expression levels in human visceral adipose tissue and diabetic state among obese women. These results are consistent with a role for Zbtb16 in increasing mitochondrial respiration and substrate utilization. Interestingly, a congenic rat strain harboring a mutation in Zbtb16 affected total body weight and adiposity, but it was not possible to rule out other mutations in the congenic strain.43 Although global loss of Zbtb16 function in mice41 and humans44, 45 causes skeletal defects, more subtle variation in Zbtb16 expression levels observed among individuals may be a determinant of adiposity. In the future, cell-type–specific knockout or transgenic models could be useful in determining further the role of Zbtb16 in adipose tissue function, energy balance and obesity.
Acknowledgments
We thank Dr Bruce Spiegelman for the kind gift of the mouse brown adipocyte cell line and Dr Andrea Hevener for providing cDNA for mouse skeletal muscle subtypes. This work was supported by the American Heart Association Grant 09BGIA2080363 (to LV), National Institutes of Health P01 HL28481 (to KR), and the National Center for research Resources Grant S10RR026744 (to KR).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Nutrition and Diabetes website (http://www.nature.com/nutd)
Supplementary Material
References
- Wyatt S, Winters KP, Dubbert PM. Overweight and obesity: prevalence, consequences, and causes of a growing public health problem. Am J Med Sci. 2006;331:166–174. doi: 10.1097/00000441-200604000-00002. [DOI] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- Hamilton MT, Hamilton DG, Zderic TW. Role of low energy expenditure and sitting in obesity, metabolic syndrome, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:2655–2667. doi: 10.2337/db07-0882. [DOI] [PubMed] [Google Scholar]
- Costford S, Gowing A, Harper ME. Mitochondrial uncoupling as a target in the treatment of obesity. Curr Opin Clin Nutr Metab Care. 2007;10:671–678. doi: 10.1097/MCO.0b013e3282f0dbe4. [DOI] [PubMed] [Google Scholar]
- Wijers SL, Saris WH, van Marken Lichtenbelt WD. Recent advances in adaptive thermogenesis: potential implications for the treatment of obesity. Obes Rev. 2009;10:218–226. doi: 10.1111/j.1467-789X.2008.00538.x. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke IJ, Henry BA. Targeting energy expenditure in muscle as a means of combating obesity. Clin Exp Pharmacol Physiol. 2010;37:121–124. doi: 10.1111/j.1440-1681.2009.05259.x. [DOI] [PubMed] [Google Scholar]
- van den Berg SA, van Marken Lichtenbelt W, Willems van Dijk K, Schrauwen P. Skeletal muscle mitochondrial uncoupling, adaptive thermogenesis and energy expenditure. Curr Opin Clin Nutr Metab Care. 2011;14:243–249. doi: 10.1097/MCO.0b013e3283455d7a. [DOI] [PubMed] [Google Scholar]
- de Lange P, Moreno M, Silvestri E, Lombardi A, Goglia F, Lanni A. Fuel economy in food-deprived skeletal muscle: signaling pathways and regulatory mechanisms. FASEB J. 2007;21:3431–3441. doi: 10.1096/fj.07-8527rev. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Metabolic consequences of the presence or absence of the thermogenic capacity of brown adipose tissue in mice (and probably in humans) Int J Obes (Lond) 2010;34 (Suppl 1:S7–16. doi: 10.1038/ijo.2010.177. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. A history of UCP1. Biochem Soc Trans. 2001;29:751–755. doi: 10.1042/bst0290751. [DOI] [PubMed] [Google Scholar]
- Klaus S, Casteilla L, Bouillaud F, Ricquier D. The uncoupling protein UCP: a membraneous mitochondrial ion carrier exclusively expressed in brown adipose tissue. Int J Biochem. 1991;23:791–801. doi: 10.1016/0020-711x(91)90062-r. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Daikoku T, Shinohara Y, Shima A, Yamazaki N, Terada H. Specific elevation of transcript levels of particular protein subtypes induced in brown adipose tissue by cold exposure. Biochim Biophys Acta. 2000;1457:263–272. doi: 10.1016/s0005-2728(00)00107-9. [DOI] [PubMed] [Google Scholar]
- Adams SH, Chui C, Schilbach SL, Yu XX, Goddard AD, Grimaldi JC, et al. BFIT, a unique acyl-CoA thioesterase induced in thermogenic brown adipose tissue: cloning, organization of the human gene and assessment of a potential link to obesity. Biochem J. 2001;360:135–142. doi: 10.1042/0264-6021:3600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XX, Lewin DA, Forrest W, Adams SH. Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 2002;16:155–168. doi: 10.1096/fj.01-0568com. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yamamoto T, Kakuhata R, Okada N, Kajimoto K, Yamazaki N, et al. Synchronized changes in transcript levels of genes activating cold exposure-induced thermogenesis in brown adipose tissue of experimental animals. Biochim Biophys Acta. 2008;1777:104–112. doi: 10.1016/j.bbabio.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Yan J, Burman A, Nichols C, Alila L, Showe LC, Showe MK, et al. Detection of differential gene expression in brown adipose tissue of hibernating arctic ground squirrels with mouse microarrays. Physiol Genomics. 2006;25:346–153. doi: 10.1152/physiolgenomics.00260.2005. [DOI] [PubMed] [Google Scholar]
- Navet R, Mathy G, Douette P, Dobson RL, Leprince P, De Pauw E, et al. Mitoproteome plasticity of rat brown adipocytes in response to cold acclimation. J Proteome Res. 2007;6:25–33. doi: 10.1021/pr060064u. [DOI] [PubMed] [Google Scholar]
- Anunciado-Koza RP, Zhang J, Ukropec J, Bajpeyi S, Koza RA, Rogers RC, et al. Inactivation of the mitochondrial carrier SLC25A25 (ATP-Mg2+/Pi transporter) reduces physical endurance and metabolic efficiency in mice. J Biol Chem. 2011;286:11659–111671. doi: 10.1074/jbc.M110.203000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipponi D, Hobbs RM, Ottolenghi S, Rossi P, Jannini EA, Pandolfi PP, et al. Repression of kit expression by Plzf in germ cells. Mol Cell Biol. 2007;27:6770–6781. doi: 10.1128/MCB.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe JH, Menk M, Reinemund J, Effertz K, Hobbs RM, Pandolfi PP, et al. A novel signal transduction cascade involving direct physical interaction of the renin/prorenin receptor with the transcription factor promyelocytic zinc finger protein. Circ Res. 2006;99:1355–1366. doi: 10.1161/01.RES.0000251700.00994.0d. [DOI] [PubMed] [Google Scholar]
- Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272:22447–2255. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Chin R, Young SG, Reue K. Heart-type fatty acid-binding protein is essential for efficient brown adipose tissue fatty acid oxidation and cold tolerance. J Biol Chem. 2011;286:380–390. doi: 10.1074/jbc.M110.184754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Phan J, Peterfy M, Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Ryall JG, Maxwell MA, Ohkura N, Lynch GS, Muscat GE. The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology. 2006;147:5217–5227. doi: 10.1210/en.2006-0447. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Licht JD. The PLZF gene of t (11;17)-associated APL. Curr Top Microbiol Immunol. 2007;313:31–48. doi: 10.1007/978-3-540-34594-7_3. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Pandolfi PP. The role of promyelocytic leukemia zinc finger and promyelocytic leukemia in leukemogenesis and development. Curr Opin Hematol. 2001;8:212–217. doi: 10.1097/00062752-200107000-00006. [DOI] [PubMed] [Google Scholar]
- Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Seda O, Liska F, Krenová D, Kazdová L, Sedová L, Zima T, et al. Differential linkage of triglyceride and glucose levels on rat chromosome 4 in two segregating rat populations. Folia Biol (Praha) 2003;49:223–226. [PubMed] [Google Scholar]
- Wieczorek D, Koster B, Gillessen-Kaesbach G. Absence of thumbs, A/hypoplasia of radius, hypoplasia of ulnae, retarded bone age, short stature, microcephaly, hypoplastic genitalia, and mental retardation. Am J Med Genet. 2002;108:209–213. doi: 10.1002/ajmg.10271. [DOI] [PubMed] [Google Scholar]
- Fischer S, Kohlhase J, Böhm D, Schweiger B, Hoffmann D, Heitmann M, et al. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet. 2008;45:731–737. doi: 10.1136/jmg.2008.059451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.