Abstract
Background
Activating point mutations of GNAS at codon 201 have been detected in approximately two thirds of intraductal papillary mucinous neoplasms (IPMNs) of the pancreas. Intraductal papillary neoplasms of the bile ducts (IPNBs) morphologically resemble pancreatic IPMNs. This study sought to assess the mutational status of GNAS at codon 201 in IPNBs.
Methods
Thirty-four patients were included. DNA from microdissected IPNBs was subjected to a polymerase chain reaction and ligation method for the detection of GNAS mutations at codon 201 and of KRAS mutations at codon 12. Mutational status was compared with clinical and pathologic data.
Results
The IPNBs had a median diameter of 3.5 cm and were located intrahepatically (n= 6), extrahepatically (n= 13), both intra- and extrahepatically (n= 4) or in the gallbladder (intracystic papillary neoplasms, n= 11). Most exhibited pancreatobiliary differentiation (n= 20), high-grade dysplasia (n= 26) and an associated adenocarcinoma (n= 20). Analysis of GNAS codon 201 identified only one mutant sample in a multifocal intestinal subtype intrahepatic IPNB with high-grade dysplasia. Six lesions harboured a KRAS codon 12 mutation.
Conclusions
GNAS codon 201 mutations are uncommon in IPNBs, by contrast with pancreatic IPMNs. More comprehensive molecular profiling is needed to uncover the pathways involved in IPNB development.
Introduction
Intraductal papillary neoplasms of the bile duct (IPNBs) are uncommon neoplasms arising in the biliary tree, which were first described more than a century ago.1 In 1959, Caroli provided a rigorous anatomical description of this entity.2 These neoplasms can arise as either solitary or multifocal lesions within the biliary tree; in the latter scenario they are termed in classical parlance as ‘biliary papillomatosis’.3–6 Intraductal papillary neoplasms of the bile duct show considerable morphologic similarities to intraductal papillary mucinous neoplasms (IPMNs) of the pancreas. Both entities present with a prominent intraductal papillary component within larger-diameter ducts, both are mucin-producing, both harbour varying grades of dysplasia in the lining epithelium, and both exhibit distinct patterns of differentiation, such as intestinal and pancreatobiliary manifestations.3 Finally, both IPMNs and IPNBs are bona fide precursor lesions of invasive adenocarcinomas at their respective anatomic sites. In view of these similarities, it has been proposed that IPNBs may be the biliary counterpart of pancreatic IPMNs,7 although there are also significant differences between IPMNs and IPNBs in incidence of mucin hypersecretion, relative prevalence of different papilla phenotypes and the frequency of high-grade dysplasia. Reports of IPNBs in the Asian literature also cite aetiologic associations (lithiasis, flukes) that differ from those of IPMNs.
Intraductal papillary neoplasms of the bile duct are predominantly diagnosed in the elderly population and show a slight male predominance. Characteristically, patients suffer from abdominal pain and recurrent cholangitis. The accompanying jaundice arises as a result of the physical obstruction of bile outflow by the neoplasm itself or the viscous inspissated mucin.5 There are no evident risk factors for IPNBs, although some studies have reported an association with longstanding biliary inflammation.5,8 Because of the underlying risk for malignant transformation to invasive adenocarcinomas (cholangiocarcinomas)5 and the obstructive nature of the lesions, resection with clear margins is the therapy of choice for IPNBs. Unfortunately, multifocal IPNBs may recur even after apparently curative resection and, in rare cases with diffuse intrahepatic disease, orthotopic liver transplantation may be considered.4,6
Recently, Wu et al. analysed a large sample of 132 IPMNs, including an admixture of tissue specimens and cyst fluid samples for somatic alterations in 169 cancer-associated genes.9 They identified mutations of GNAS at codon 201 in two thirds (66%) of analysed cases.9GNAS, located on chromosome 20q, encodes for the alpha subunit of a stimulatory G-protein, which activates a cyclic 3′-5′-cyclic adenosine monophosphate (cAMP)-mediated intracellular cascade, culminating in cellular proliferation and growth.10 Although this ‘hotspot’ codon 201 mutation has been previously described in endocrine neoplasms and, rarely, in colorectal carcinomas, this was the first report of involvement of this oncogenic pathway in pancreatic neoplasia. The high prevalence of GNAS mutations in IPMNs has been confirmed by others,11 and a subsequent study demonstrated that amongst cystic neoplasms of the pancreas, GNAS mutations are restricted to IPMNs and are not observed in solid pseudopapillary neoplasms or mucinous cystic neoplasms.12 In light of the morphologic similarities between IPMNs and IPNBs, and the shared embryologic origins of the pancreas and biliary tree,13 the present authors investigated the frequency of GNAS alterations in the latter using a sensitive polymerase chain reaction (PCR)/ligation methodology. Notably, the study found that GNAS codon 201 mutations are uncommon in IPNBs, which suggests that alternative mechanisms for the development of these neoplasms exist.
Materials and methods
Patients and tissues
The present study was approved by the Johns Hopkins University Institutional Review Board and the review boards of collaborating institutions, Memorial Sloan–Kettering Cancer Center, New York, NY, and Emory University, Atlanta, GA, USA. Papillary lesions located in the ampullary region were not included because it is sometimes difficult to group them according to anatomical origin (pancreatic vs. biliary vs. ampullary). The neoplasms were classified according to recent international guidelines.14 Reference slides of all eligible cases were reviewed by pathologists at the participating institutes, who were experts in pancreatobiliary pathology (RHH, AM, NVA, NK and DSK) to confirm the diagnosis of IPNB, and corresponding formalin-fixed and paraffin-embedded (FFPE) blocks for the most suitable specimens were selected for subsequent molecular studies. A total of 34 IPNB specimens were collated from the three collaborating institutions. Detailed information about the patients included is shown in Table S1 (online).
Microdissection and DNA preparation
Twenty sections measuring 7 µm in thickness were cut from every FFPE block in PCR clean conditions and mounted on plus-charged glass slides, after which they were stained with hematoxylin and eosin (H&E). The first and last sections were used as references for needle microdissection. Thereafter, manual microdissection of the neoplastic epithelium was performed using a sterile needle on a stereoscopic zoom microscope SMZ1500 (Nikon Corp., Tokyo, Japan). Approximately 10 000 cells were harvested from each lesion with an estimated tumour cellularity of >80% and subjected to genomic DNA extraction using a QIAamp DNA FFPE Tissue Kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol.
PCR/ligation assay for the detection of GNAS and KRAS mutations
Polymerase chain reaction products containing codon 12 of KRAS and codon 201 of GNAS were amplified using the primers described in Table S2. Each 10-µl PCR contained 200 template molecules in 5 µl of 2× Phusion Flash PCR Master Mix (New England Biolabs, Inc., Ipswich, MA, USA), and final concentrations of 0.25 µM forward and 1.5 µM reverse primers. Note that the mutant-specific probes in some cases included locked nucleic acid residues (Exiqon Life Sciences A/S, Vedbaek, Denmark). The following cycling conditions were used: 98 °C for 2 min; three cycles of 98 °C for 10 s, 69 °C for 15 s, 72 °C for 15 s; three cycles of 98 °C for 10 s, 66 °C for 15 s, 72 °C for 15 s; three cycles of 98 °C for 10 s, 63 °C for 15 s, 72 °C for 15 s, and 41 cycles of 98 °C for 10 s and 60 °C for 60 s. Reactions were performed in at least quadruplicate and each was evaluated independently. Five µl of a solution containing 0.5 µl of Proteinase K, (18.8 mg/ml; Roche AG, Basel, Switzerland) and 4.5 µl of dH2O was added to each well and incubated at 60 °C for 30 min to inactivate the Phusion polymerase and then for 10 min at 98 °C to inactivate the Proteinase K.
The ligation assay was based on techniques described previously, using thermotolerant DNA ligases.15–18 Each 10-µl reaction contained 2 µl of PCR product (unpurified), 1 µl of 10× Ampligase buffer (Epicentre Biotechnologies, Inc., Madison, WI, USA), 0.5 µl of Ampligase (5 U/µl; Epicentre Biotechnologies, Inc.), anchoring primer (final concentration 2 µM), wild-type specific primer (final concentration 0.1 µM), and mutant-specific primer (final concentration 0.025 µM). The sequences of these primers are listed in Table S2. The following cycling conditions were used: 95 °C for 3 min, and 35 cycles of 95 °C for 10 s, 37 °C for 30 s and 45 °C for 60 s. Five µl of each reaction was added to 5 µl of formamide and the ligation products were separated on a 10% Urea-Tris-Borate-EDTA gel (Life Technology, Inc., Grand Island, NY, USA) and imaged with an Amersham-GE Typhoon instrument (GE Healthcare Ltd, Waukesha, WI, USA).
Results
Clinical and pathologic characteristics of IPNBs
This study was conducted using archival IPNB cases collected from three large hepatopancreatobiliary surgery centres in the USA. The clinical and pathologic characteristics associated with each of the 34 individual IPNB samples are detailed in Table S1 and a summary tabulation is presented in Table 1. The median age of the 34 patients was 65 years (range: 26–88 years) and the sample included 20 men and 14 women. The non-invasive component of the IPNBs had a median diameter of 3.5 cm (range: 0.7–21.1 cm), and the lesions were located throughout the biliary tree, including in the intrahepatic ducts (n= 6), extrahepatic ducts (n= 13), both intra- and extrahepatic ducts (n= 4) and in the gallbladder (n= 11). An associated invasive adenocarcinoma was observed in 20 cases. Tumour–node–metastasis (TNM) staging showed most of the associated invasive cancers were T3 (n= 9) and had not yet spread to local lymph nodes (n= 13). Upon histopathologic examination, the non-invasive components uniformly exhibited an intraductal papillary growth pattern (Fig. 1). Based on the maximum grade of epithelial dysplasia, the lesions were classified into 26 high-grade, five intermediate-grade and three low-grade IPNBs.
Table 1.
Summary of clinical and pathologic data for 34 patients with intraductal papillary neoplasms of the bile duct
| Age, years, median (range) | 65 (54–76) | |
| Gender, n | Male | 20 |
| Female | 14 | |
| Surgery, n | Hepatectomy | 7 |
| Segmental bile duct resection | 3 | |
| Hepatectomy and segmental bile duct resection | 5 | |
| Tumour cholecystectomy | 11 | |
| Orthotopic liver transplantation | 1 | |
| Pancreatoduodenectomy (Whipple procedure) | 7 | |
| Resection status, n | 0 | 28 |
| 1 | 2 | |
| Unknown | 4 | |
| Location, n | Intrahepatic bile ducts | 6 |
| Extrahepatic bile ducts | 13 | |
| Intra- and extrahepatic bile ducts | 4 | |
| Gallbladder | 11 | |
| Size, cm, median (range) | 3.5 (0.7–21.1) | |
| Subtype, n | Gastric | 10 |
| Intestinal | 2 | |
| Pancreatobiliary | 20 | |
| Oncocytic | 2 | |
| Grade of dysplasia, n | Low | 3 |
| Intermediate | 5 | |
| High | 26 | |
| Associated invasive cancer, n | Yes | 20 |
| No | 14 | |
| Tumour stage | T1 | 4 |
| T2 | 6 | |
| T3 | 9 | |
| T4 | 1 | |
| N/A | 14 | |
| Lymph node metastases, n | N0 | 13 |
| N1 | 3 | |
| NX | 4 | |
| KRAS | Mutation detected, n | 6 |
| GNAS | Mutation detected, n | 1 |
Figure 1.
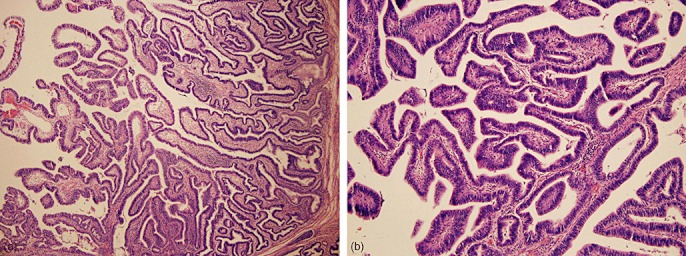
(a) Low- and (b) high-power photomicrographs obtained from an intraductal papillary neoplasms of the bile duct with high-grade dysplasia. [Haematoxylin and eosin stain; original magnification (a) ×10, (b) ×20]
Assessment of mutational status and clinicopathologic correlation
A sensitive PCR/ligation method was used to detect GNAS and KRAS mutations in DNA samples isolated from the 34 IPNBs. Only one case harboured a GNAS codon 201 mutation. The GNAS mutation was detected in a diffuse intrahepatic IPNB, of intestinal subtype and exhibiting high-grade dysplasia. The 65-year-old male patient in whom this neoplasm arose underwent a left lateral hepatectomy and was alive without evident disease at 60 months after resection.
KRAS mutations were found in six patients. When the cohort of patients with KRAS mutations in their IPNBs were compared with those without KRAS mutations for patient characteristics and clinical and pathologic features (age, gender, location, subtype, grade, maximum size, presence of invasive carcinoma and outcome), no significant differences between the groups were found (chi-squared test, P > 0.05).
Discussion
Invasive adenocarcinomas of the pancreas are postulated to arise via two major dichotomous pathways involving non-invasive precursor lesions: pancreatic intraepithelial neoplasia (panIN), and IPMNs.19 These two pathways are clinically, pathologically and genetically distinct. Analogous to the pancreas, two precursor pathways have been pathologically identified in the biliary tree, comprising non-papillary biliary intra-epithelial neoplasia (bilIN) lesions, and IPBNs.20 Although molecular analyses of either of the two biliary precursor lesions are limited, the available information suggests underlying differences in terms of genetic alterations, as well as clinical distinctions in the invasive carcinomas arising in the respective contexts.21–23
Emerging evidence suggests a number of similarities between IPMNs and IPNBs. For example, Zen et al. analysed both entities with respect to clinicopathologic features, expression of lineage-associated proteins and prognostic parameters.7 They compared a series of 32 IPNBs, including 22 with invasive cholangiocarcinomas, with a series of 31 pancreatic IPMNs (15 with associated invasive cancer); 15 non-IPNB-associated cholangiocarcinomas served as controls.7 The authors found that, similarly to those in IPMN, invasive adenocarcinomas arising from IPNBs were often of the mucinous type (five of 14, 35.7%). In addition, as seen in the pancreas, three predominant epithelial subtypes were observed in the biliary lesions, including gastric, intestinal and pancreatobiliary phenotypes. Immunohistochemically, IPNBs were characterized by the expression of apomucin MUC2, the intestinal differentiation factor CDX2, and the intermediate filament cytokeratin CK20, all of which are features prevalent in IPMNs, especially those of the intestinal subtype. Notably, these expression patterns were uncommon in cholangioadenocarcinomas not associated with IPNBs. The authors concluded that biliary papillary lesions may be regarded as the counterpart of IPMNs of the pancreas.7
Thus far, only limited information exists on the prevalent molecular alterations in IPNBs. Abraham et al. reported that IPNBs can have low levels of microsatellite instability in up to a third of cases, although these changes are not associated with loss of hMLH1 function.24 Furthermore, the same group of investigators assessed a panel of IPNBs for somatic alterations of KRAS and CTNNB1 hotspot regions, and found KRAS codon 12 mutations in four of 14 (28.6%) IPNBs, whereas no CTNNB1 mutations were detected.25Table 2 provides an overview of the available mutational data on IPNBs. The present study also suggests that the rate of KRAS mutations in IPNBs is lower (six of 34, 17.6%) than that previously reported in IPMNs (∼80%),9 but comparable with that described previously in IPNBs.25,26 Notably, a recent genetically engineered mouse model of cholangiocarcinoma, generated by expression of mutant KRAS and Tp53 under an albumin promoter during development, also demonstrated non-invasive papillary lesions of the biliary tree histologically similar to the cognate precursor lesions in humans, underscoring the potential importance of mutant KRAS to the pathogenesis of some IPNBs.27
Table 2.
Studies investigating the presence of distinct DNA mutations in intraductal papillary neoplasms of the bile duct (IPNB) located in the gallbladder and biliary ductal system
| Author | Year | Location of IPNB | Gene locus | Involvement |
|---|---|---|---|---|
| Ohta et al.46 | 1993 | Bile duct | KRAS codon 12 | 1 of 1 |
| Yanagisawa et al.43 | 2001 | Gallbladder | β-catenin exon 3 | 10 of 16 |
| Kim et al.44 | 2001 | Gallbladder | KRAS codon 12 | 0 of 3 |
| p53 | 0 of 3 | |||
| Iwasaki et al.45 | 2002 | Bile duct | KRAS codon 12 | 1 of 1 |
| Abraham et al.25 | 2003 | Bile duct | KRAS codon 12 | 4 of 14 |
| β-catenin exon 3 | 0 of 14 | |||
| Pai & Mojtahed26 | 2011 | Gallbladder | KRAS codons 12 and 13 | 5 of 16 |
| BRAF codon 600 | 1 of 16 | |||
The availability of massively parallel sequencing technologies has facilitated the identification of novel cancer-associated genes in several cancer types.12,28–30 Using a massively parallel approach, the present group and others identified activating mutations of GNAS at codon 201 in about two thirds of IPMNs.9,11 Point mutations of GNAS codon 227 have been reported in other tumour types, although not in IPMNs. Mutations at both codons 201 and 227 constitutively activate the encoded G-protein alpha subunit.31 The functional impact of these activating mutations is to confer cells with high adenyl cyclase activity and cAMP levels. GNAS mutations have predominantly been observed in association with endocrine disorders. For example, patients with McCune–Albright syndrome harbour germline GNAS mutations and present with a characteristic compendium of findings, including acromegaly, fibrous dysplasia and café au lait discoloration.32–34 Approximately 40% of pituitary somatotroph (growth hormone secreting) adenomas demonstrate GNAS mutations at either codon 201 or codon 227.35GNAS mutations are uncommon in epithelial malignancies, occurring only in minor subsets of colorectal, prostate and breast cancers, renal cell carcinomas and small cell carcinomas of the lung, and these mutations are almost always restricted to codon 201.36–40 In the hepatobiliary region, GNAS mutations have been reported in only two of 245 (∼1%) hepatocellular carcinomas (HCCs) and in four of 164 (2.4%) hepatocellular adenomas.41 Again, the mutations were all clustered at codon 201 and the corresponding HCCs were associated with an inflammatory infiltrate, postulated to reflect the activating of underlying proinflammatory pathways [interleukin-6 and STAT-3 (signal transducer and activator of transcription 3)] by the constitutively activated G-protein. Of note, GNAS mutations have not been identified in ‘usual’ pancreatic ductal adenocarcinomas arising outwith the context of IPMNs.9,30
In the present study, GNAS mutations were detected only in a single intrahepatic IPNB and the remaining 33 lesions were wild-type at codon 201. These results are by stark contrast with the high prevalence (66%) previously reported in non-invasive IPMNs by Wu et al.9 In their study, GNAS mutations were observed at all grades of epithelial dysplasia, suggesting it was an ‘early’ genetic alteration. However, there was a tendency for subtype-specific prevalence, with 100% of intestinal IPMNs and only ∼50% of gastric and pancreatobiliary IPMNs demonstrating GNAS codon 201 mutations. Interestingly, the single IPNB with a GNAS mutation in the present series was of the intestinal subtype. Overall, only two IPNBs showed intestinal differentiation in the present series, and one of the two was positive for GNAS mutation. Thus, the low prevalence of GNAS mutations in IPNBs may reflect the low prevalence of intestinal differentiation in these neoplasms. Nonetheless, it should be noted that ∼50% of gastric and pancreatobiliary type IPMNs also harboured GNAS mutations,9 and all of the 31 IPNBs comprising these two subtypes were wild-type, suggesting a truly lower rate in the biliary counterparts.
There are several limitations to the present study. In this series, a subset of the IPNBs presented as multifocal lesions (biliary papillomatosis), but only the largest neoplastic foci were analysed. The present authors have previously shown that IPMNs can be heterogeneous with regard to GNAS mutations,9 and this study's failure to examine multiple synchronous lesions may have produced false negatives. The authors find this possibility less likely as GNAS mutations would confer a growth advantage to the index lesion and thus there is a high likelihood that the index lesion will be the most prominent neoplastic focus in a multifocal IPNB. Another caveat may be that the activating hotspot mutation in IPNBs is not at GNAS codon 201, but, rather, at codon 227, which was not examined in the current study. However, based on the data that GNAS mutations in epithelial neoplasms (including IPMNs) are always restricted to codon 201,9,35,42 the authors find this possibility unlikely. Finally, the issue of assay sensitivity, which is always a concern in a ‘negative’ study of this nature, should be addressed. The PCR/ligation assay used herein can detect a single mutant GNAS molecule amongst 200 wild-type templates (0.5% lower limit of detection)9 and has been successfully used for identifying mutant GNAS in highly biologically heterogeneous pancreatic cyst fluid samples. Thus, the present authors consider it quite unlikely that false negative results have been obtained using microdissected tumour tissue (with neoplastic cellularity of >80%).
In conclusion, GNAS codon 201 mutation, a newly discovered ‘mountain’ in the genetic landscape of IPMNs, is an uncommon alteration in IPNBs, despite the morphologic similarities between the two lesions arising at embryologically related sites. Additionally, KRAS mutations are also less common in IPNBs than in IPMNs. Based on the present series, it can be assumed that the signalling pathways involved in the pathogenesis of IPNBs are distinct from those in IPMNs. Larger, more comprehensive sequencing studies should help to elucidate the genetic landscape of IPNBs.
Acknowledgments
This study was supported by the National Institutes of Health (P50CA062924, P01CA134292), the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation for Pancreatic Cancer Research and by a fellowship grant from German Cancer Aid (Deutsche Krebshilfe eV) to HM.
Conflicts of interest
None declared.
Supporting information
Additional supporting information may be found in the online version of this article:
Table S1. Detailed clinical and histopathologic data for the study population.
Table S2. Oligonucleotides used for sensitivepolymerase chain reaction ligation method for detecting GNASand KRAS mutations.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Chappet V. Cancer epithelial primitif du canal cholédoque. Lyon Med. 1894;76:145–157. [Google Scholar]
- 2.Caroli J. [Papillomas and papillomatoses of the common bile duct.] Rev Med Chir Mal Foie. 1959;34:191–230. [PubMed] [Google Scholar]
- 3.Amaya S, Sasaki M, Watanabe Y, Tsui WM, Tsuneyama K, Harada K, et al. Expression of MUC1 and MUC2 and carbohydrate antigen Tn change during malignant transformation of biliary papillomatosis. Histopathology. 2001;38:550–560. doi: 10.1046/j.1365-2559.2001.01103.x. [DOI] [PubMed] [Google Scholar]
- 4.Imvrios G, Papanikolaou V, Lalountas M, Patsiaoura K, Giakoustidis D, Fouzas I, et al. Papillomatosis of intra- and extrahepatic biliary tree: successful treatment with liver transplantation. Liver Transpl. 2007;13:1045–1048. doi: 10.1002/lt.21207. [DOI] [PubMed] [Google Scholar]
- 5.Lee SS, Kim MH, Lee SK, Jang SJ, Song MH, Kim KP, et al. Clinicopathologic review of 58 patients with biliary papillomatosis. Cancer. 2004;100:783–793. doi: 10.1002/cncr.20031. [DOI] [PubMed] [Google Scholar]
- 6.Marion-Audibert AM, Guillet M, Rode A, Barnoud R, Mesnil A, Ducerf C, et al. [Diffuse biliary papillomatosis: a rare indication for liver transplantation.] Gastroenterol Clin Biol. 2009;33(1 Pt 1):82–85. doi: 10.1016/j.gcb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Zen Y, Fujii T, Itatsu K, Nakamura K, Minato H, Kasashima S, et al. Biliary papillary tumours share pathological features with intraductal papillary mucinous neoplasm of the pancreas. Hepatology. 2006;44:1333–1343. doi: 10.1002/hep.21387. [DOI] [PubMed] [Google Scholar]
- 8.Nakanuma Y, Sasaki M, Ishikawa A, Tsui W, Chen TC, Huang SF. Biliary papillary neoplasm of the liver. Histol Histopathol. 2002;17:851–861. doi: 10.14670/HH-17.851. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lania AG, Mantovani G, Spada A. Mechanisms of disease: mutations of G proteins and G-protein-coupled receptors in endocrine diseases. Nat Clin Pract Endocrinol Metab. 2006;2:681–693. doi: 10.1038/ncpendmet0324. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, Yamamoto M, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1(161):1–7. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lammert E, Cleaver O, Melton D. Role of endothelial cells in early pancreas and liver development. Mech Dev. 2003;120:59–64. doi: 10.1016/s0925-4773(02)00332-5. [DOI] [PubMed] [Google Scholar]
- 14.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. Lyon: IARC Press; 2010. [Google Scholar]
- 15.Fouquet C, Antoine M, Tisserand P, Favis R, Wislez M, Commo F, et al. Rapid and sensitive p53 alteration analysis in biopsies from lung cancer patients using a functional assay and a universal oligonucleotide array: a prospective study. Clin Cancer Res. 2004;10:3479–3489. doi: 10.1158/1078-0432.CCR-0994-03. [DOI] [PubMed] [Google Scholar]
- 16.Dong SM, Traverso G, Johnson C, Geng L, Favis R, Boynton K, et al. Detecting colorectal cancer in stool with the use of multiple genetic targets. J Natl Cancer Inst. 2001;93:858–865. doi: 10.1093/jnci/93.11.858. [DOI] [PubMed] [Google Scholar]
- 17.Luo J, Bergstrom DE, Barany F. Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res. 1996;24:3071–3078. doi: 10.1093/nar/24.15.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, et al. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 19.Basturk O, Khayyata S, Klimstra DS, Hruban RH, Zamboni G, Coban I, et al. Preferential expression of MUC6 in oncocytic and pancreatobiliary types of intraductal papillary neoplasms highlights a pyloropancreatic pathway, distinct from the intestinal pathway, in pancreatic carcinogenesis. Am J Surg Pathol. 2010;34:364–370. doi: 10.1097/PAS.0b013e3181cf8bb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bickenbach K, Galka E, Roggin KK. Molecular mechanisms of cholangiocarcinogenesis: are biliary intraepithelial neoplasia and intraductal papillary neoplasms of the bile duct precursors to cholangiocarcinoma? Surg Oncol Clin N Am. 2009;18:215–224. doi: 10.1016/j.soc.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Zen Y, Quaglia A, Heaton N, Rela M, Portmann B. Two distinct pathways of carcinogenesis in primary sclerosing cholangitis. Histopathology. 2011;59:1100–1110. doi: 10.1111/j.1365-2559.2011.04048.x. [DOI] [PubMed] [Google Scholar]
- 22.Yonezawa S, Higashi M, Yamada N, Yokoyama S, Goto M. Significance of mucin expression in pancreatobiliary neoplasms. J Hepatobiliary Pancreat Sci. 2010;17:108–124. doi: 10.1007/s00534-009-0174-7. [DOI] [PubMed] [Google Scholar]
- 23.Nakanuma Y, Zen Y, Harada K, Ikeda H, Sato Y, Uehara T, et al. Tumorigenesis and phenotypic characteristics of mucin-producing bile duct tumours: an immunohistochemical approach. J Hepatobiliary Pancreat Sci. 2010;17:211–222. doi: 10.1007/s00534-009-0158-7. [DOI] [PubMed] [Google Scholar]
- 24.Abraham SC, Lee JH, Boitnott JK, Argani P, Furth EE, Wu TT. Microsatellite instability in intraductal papillary neoplasms of the biliary tract. Mod Pathol. 2002;15:1309–1317. doi: 10.1097/01.MP.0000038461.80167.34. [DOI] [PubMed] [Google Scholar]
- 25.Abraham SC, Lee JH, Hruban RH, Argani P, Furth EE, Wu TT. Molecular and immunohistochemical analysis of intraductal papillary neoplasms of the biliary tract. Hum Pathol. 2003;34:902–910. doi: 10.1016/s0046-8177(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 26.Pai RK, Mojtahed K. Mutations in the RAS/RAF/MAP kinase pathway commonly occur in gallbladder adenomas but are uncommon in gallbladder adenocarcinomas. Appl Immunohistochem Mol Morphol. 2011;19:133–140. doi: 10.1097/PAI.0b013e3181f09179. [DOI] [PubMed] [Google Scholar]
- 27.O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Gross V, et al. KrasG12D and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, EN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumours. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signalling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340:692–696. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 32.Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N Engl J Med. 1991;325:1688–1695. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- 33.Lumbroso S, Paris F, Sultan C. Activating Gsa mutations: analysis of 113 patients with signs of McCune–Albright syndrome – a European collaborative study. J Clin Endocrinol Metab. 2004;89:2107–2113. doi: 10.1210/jc.2003-031225. [DOI] [PubMed] [Google Scholar]
- 34.Imanaka M, Iida K, Nishizawa H, Fukuoka H, Takeno R, Takahashi K, et al. McCune–Albright syndrome with acromegaly and fibrous dysplasia associated with the GNAS gene mutation identified by sensitive PNA-clamping method. Intern Med. 2007;46:1577–1583. doi: 10.2169/internalmedicine.46.0048. [DOI] [PubMed] [Google Scholar]
- 35.Freda PU, Chung WK, Matsuoka N, Walsh JE, Kanibir MN, Kleinman G, et al. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumours: correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary. 2007;10:275–282. doi: 10.1007/s11102-007-0058-2. [DOI] [PubMed] [Google Scholar]
- 36.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 37.Kalfa N, Lumbroso S, Boulle N, Guiter J, Soustelle L, Costa P, et al. Activating mutations of Gsa in kidney cancer. J Urol. 2006;176:891–895. doi: 10.1016/j.juro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 39.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 40.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nault JC, Fabre M, Couchy G, Pilati C, Jeannot E, Tran Van Nhieu J, et al. GNAS-activating mutations define a rare subgroup of inflammatory liver tumours characterized by STAT3 activation. J Hepatol. 2012;56:184–191. doi: 10.1016/j.jhep.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Kalfa N, Lumbroso S, Boulle N, Guiter J, Soustelle L, Costa P, et al. Activating mutations of Gsalpha in kidney cancer. J Urol. 2006;176:891–895. doi: 10.1016/j.juro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 43.Yanagisawa N, Mikami T, Saegusa M, Okayasu I. More frequent beta-catenin exon 3 mutations in gallbladder adenomas than in carcinomas indicate different lineages. Cancer Res. 2001;61:19–22. [PubMed] [Google Scholar]
- 44.Kim YT, Kim J, Jang YH, Lee WJ, Ryu JK, Park YK, et al. Genetic alterations in gallbladder adenoma, dysplasia and carcinoma. Cancer Lett. 2001;169:59–68. doi: 10.1016/s0304-3835(01)00562-6. [DOI] [PubMed] [Google Scholar]
- 45.Iwasaki Y, Shimoda M, Furihata T, Rokkaku K, Sakuma A, Ichikawa K, et al. Biliary papillomatosis arising in a congenital choledochal cyst: report of a case. Surg Today. 2002;32:1019–1022. doi: 10.1007/s005950200206. [DOI] [PubMed] [Google Scholar]
- 46.Ohta H, Yamaguchi Y, Yamakawa O, Watanabe H, Satomura Y, Motoo Y, et al. Biliary papillomatosis with the point mutation of K-ras gene arising in congenital choledochal cyst. Gastroenterology. 1993;105:1209–1212. doi: 10.1016/0016-5085(93)90969-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


