Abstract
CD8+ T cells respond to signals mediated through a specific interaction between the T-cell receptor (TCR) and a composite antigen in the form of an epitopic peptide bound between the polymorphic α1 and α2 helices of an MHC class I (MHCI) molecule. The CD8 glycoprotein ‘co-receives’ antigen by binding to an invariant region of the MHCI molecule and can enhance ligand recognition by up to 1 million-fold. In recent years, a number of structural and biophysical investigations have shed light on the role of the CD8 co-receptor during T-cell antigen recognition. Here, we provide a collated resource for these data, and discuss how the structural and biophysical parameters governing CD8 co-receptor function further our understanding of T-cell cross-reactivity and the productive engagement of low-affinity antigenic ligands.
Keywords: CD8, co-receptor, biophysics; crystal structure; peptide–major histocompatibility complex; T-cell activation; T-cell receptor
Introduction
Overview of the cellular expression pattern and function of CD8
T-cell antigen recognition and subsequent T-cell activation are governed by the interaction between the T-cell receptor (TCR) and peptide–major histocompatibility complex (pMHC) molecules.1 In a unique bipartite recognition mechanism TCR–pMHC-mediated T-cell activation is enhanced through the activities of co-receptor molecules that bind independently from the TCR to an invariant region of the pMHC (Fig. 1). The CD8 co-receptor exists as an αα homodimer (Fig. 2a) on the surface of many different cell types within the lymphoid system, including natural killer cells, γδ T cells2 and intestinal intra-epithelial T lymphocytes3; it is also expressed in this form on certain dendritic cell subsets.4 In the alternative αβ heterodimeric form (Fig. 2b), CD8 is found on ∼ 90% of cytotoxic T lymphocytes.5 The functional role of the CD8αα homodimer has not been formally identified, although a regulatory role has been proposed in the case of intestinal intra-epithelial T lymphocytes.6 In contrast, the CD8αβ co-receptor plays a major role in CD8+ T-cell activation by increasing antigen sensitivity7,8 and by stabilizing the TCR–pMHC class I (pMHCI) interaction at the cell surface.9–11 The pMHCI–CD8 interaction is central to these functional roles.
Figure 1.
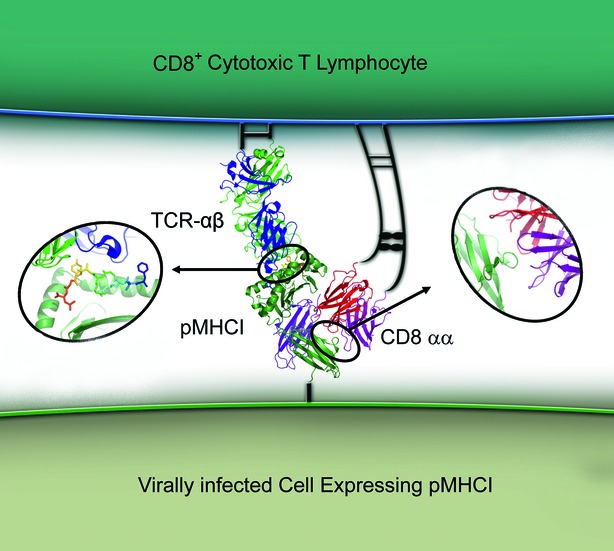
The tripartite T-cell receptor–peptide–MHC class I complex (TCR–pMHCI)–CD8 interaction. A representation of the tripartite TCR–pMHCI–CD8 interaction that forms part of the polyvalent interface between a CD8+ cytotoxic T lymphocyte and a target cell during the process of antigen recognition. This interaction governs T-cell activation and triggers target cell killing through the release of cytotoxic granules and lymphokines, and through the induction of apoptotic signals.
Figure 2.
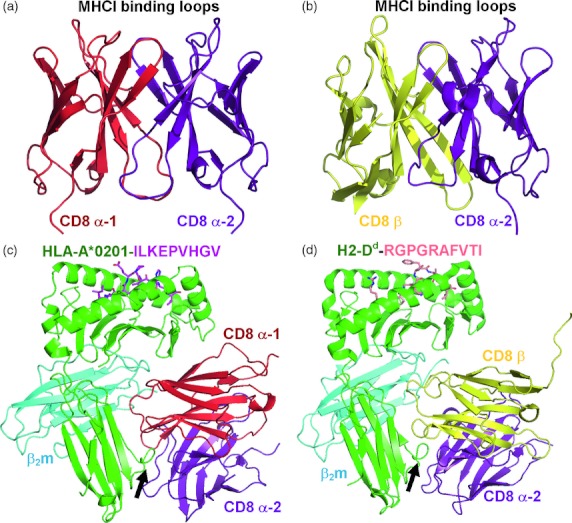
Crystal structures of CD8αα and CD8αβ in complex with peptide–MHC class I complex (pMHCI). (a) Crystal structure of the human CD8αα homodimer (PDB: 1CD8) with the α1-chain shown in red and the α2-chain shown in purple. (b) Crystal structure of the human CD8αβ heterodimer (PDB: 2ATP) with the α-chain shown in purple and the β-chain shown in yellow. (c) The co-crystal complex between human CD8αα and HLA-A2*0201-ILKEPVHGV (PDB: 1AKJ). CD8 is shown in red (α1) and purple (α2) binding mainly to the α3 domain of MHCI. The complementarity-determining region (CDR)-like loops of the CD8 molecule bind to a finger-like loop formed by residues 223–227 of the MHCI α3 domain; this interaction comprises the main binding interface between CD8 and MHCI (indicated by an arrow). (d) The co-crystal complex between murine CD8αβ and H2-Dd-RGPGRAFVTI (PDB: 3DMM). CD8 is shown in purple (α2) and yellow (β) binding mainly to the α3 domain of MHCI (indicated by an arrow). Although the amino acid sequence of CD8β is distinct from that of CD8α, the CD8αβ heterodimer adopts a virtually identical conformation to that of the CD8αα homodimer and binds to pMHCI in a similar overall manner.
Co-receptor functions of CD8
CD8 acts as a co-receptor during T-cell antigen engagement.8 The dominant molecular basis for this functional role in antigen recognition centres on the association of the CD8 α-chain with p56Lck, via two vicinal cysteines, which interact through a zinc chelate complex to produce a co-activation signal.12,13 This interaction leads to a signalling cascade that recruits ZAP-70 to the TCR–CD3 complex, leading to the amplification or enhancement of T-cell activation signals.14,15 The signalling role of the CD8 α-chain can be enhanced by palmitoylation of the CD8 β-chain at a membrane-proximal cysteine.16 Palmitoylation at this site allows the recruitment of the tripartite TCR–CD3–CD8 signalling complex to detergent-insoluble membrane domains, or lipid rafts.17,18 Lipid rafts are made up of ordered microdomains, enriched with sphingolipids and cholesterol, that exclude molecules such as phosphatases (CD45) but recruit molecules that are critical for T-cell activation, such as p56Lck and the linker for activation of T cells. Accordingly, these membrane microdomains represent privileged sites for TCR-mediated signal transduction.19–21 Hence, the tripartite extracellular interaction between TCR, pMHCI and CD8 (Fig. 1) has important consequences in terms of intracellular signalling.22 Although it is now generally accepted that CD8 enhances antigen sensitivity, recent studies have shown that certain CD8+ T-cell responses can occur independently of the CD8 co-receptor.23 This review will cover newly reported molecular aspects of the pMHCI–CD8 interaction and the role of the co-receptor during CD8+ T-cell antigen surveillance.
Molecular basis of the pMHCI–CD8 interaction
Structural basis of the pMHCI–CD8αα and pMHCI–CD8αβ interactions
The CD8 co-receptor binds to a largely invariant region of MHCI that is spatially distinct from the TCR binding platform, allowing the potential for tripartite (TCR–pMHCI–CD8) complex formation (Fig. 1). In an analogous fashion to the TCR, the soluble domain of CD8 contains a number of flexible complementarity-determining region-like (CDR) loops that are involved in MHCI binding. The interaction between the CDR-like loops of human CD8αα (residues 51–55) and a finger-like loop in the α3 domain of HLA-A*0201 (residues 223–229) forms the main contact zone of the complex. The CDR-like loops of CD8αα ‘clamp’ onto this flexible finger-like loop asymmetrically, with each molecule in the dimer contributing differently to the overall binding (Fig. 2c). Additionally, CD8αα contacts the α2 and β2m domains of HLA-A*0201, compounding the overall stability of the complex.24,25 These findings have been confirmed recently by another study that reported the co-crystal structure of CD8αα in complex with HLA-A*2402.26 In this structure, CD8αα bound primarily to the flexible α3 domain of HLA-A*2402 in a virtually identical conformation to that observed with HLA-A*0201.26 Although murine CD8αα bound to H2-Kb in a similar fashion compared with the human HLA-A*0201-CD8αα complex,27 there were some key differences in fine specificity between these two interactions. For example, in the murine system, more contacts were made between CD8 and the MHCI α3 domain, fewer contacts existed between CD8 and the MHCI α2 domain, and a number of unique bonds were formed at the interface between CD8 and β2m. These differences probably explain the higher binding affinity of murine CD8 compared with human CD8 for their corresponding species-specific MHCIs.15
Until recently, the orientation of the CD8αβ heterodimer in complex with pMHCI remained speculative.28 The atomic structure of murine CD8αβ in complex with H-2Dd29 revealed that the binding mode of the CD8αβ heterodimer was largely homologous to that of the CD8αα homodimer.24,27 Accordingly, the CDR-like loops of CD8αβ bound predominantly to the conserved finger-like loop in the H-2Dd α3 domain (Fig. 2d). Moreover, CD8αβ adopted a single orientation in the H-2Dd–CD8αβ co-complex, with the β-chain in the equivalent position to the CD8 α1-chain in the pMHCI–CD8αα complex, proximal to the T-cell membrane, in opposition to the original structural conformation predicted previously24 (Fig. 2d). Nonetheless, there were also some notable differences between the murine pMHCI–CD8αα and pMHCI–CD8αβ complex structures. For example, CD8αβ did not contact the α2 and β2m domains of H-2Dd, which reduced the buried surface area of this complex compared with murine pMHCI–CD8αα.
The T-cell co-receptors govern TCR binding orientation and MHC restriction
Accumulated structural evidence of TCR–pMHC interactions has shown that the TCR binds with a conserved general topology, with the TCR α-chain positioned over the N-terminus of the peptide and the TCR β-chain over the C-terminus.30 It has been postulated that this binding mode is essential to allow co-receptor binding to the same pMHC as the TCR at the cell surface (Fig. 1).31 Hence, the CD8 co-receptor (and CD4 co-receptor) may have a role in governing the conserved binding mode of the TCR to allow the formation of a functional signalling complex at the T-cell surface.32 Indeed, Kuhns and Davis33 have shown that the ectodomains of CD3εδ and CD3εγ, that constitute an important part of the TCR signalling complex, associate with the Cα DE and Cβ CC' loops, respectively, within the constant domain of the TCR (Fig. 3a). In this study, mutation of these conserved loops disrupted the formation of the TCR–CD3 signalling domain and subsequent T-cell activation. So it seems that these CD3 subunits, that contain intracellular tyrosine kinase activation motifs and play an important role in providing T-cell activation signals, form specific interactions with the TCR, fixing their position at the cell surface with respect to the TCR. Yin et al.32 showed that the structure of the tripartite TCR–pMHCII–CD4 complex is compatible with this notion. Assuming that the TCR and co-receptor co-engage the same pMHC at the cell surface, the fixed polarity of the TCR–pMHC interaction could orientate the co-receptor in such a way as to allow the CD3 molecules to lie between the TCR and co-receptor (Fig. 3a,b). This would position the intracellular signalling domains of CD3 and the co-receptor in close proximity to enable cross-signalling during antigen engagement. If the TCR bound in the reverse polarity, with the TCR β-chain over the peptide N-terminus and the TCR α-chain over the C-terminus, the CD3 complex would lie distal from the co-receptor, and this could presumably reduce the efficiency of the T-cell activation signal between the co-receptor and the CD3 complex (Fig. 3c,d). Adding further support to the idea that the T-cell co-receptors can influence the nature of TCR antigen recognition, Van Laethem et al.34 have shown that the CD4 and CD8 co-receptors impose MHC-restriction on T cells by preventing Lck availability during TCR interactions with non-MHC antigens. Indeed, in the absence of the co-receptors T cells develop with antibody-like specificities that can respond to other cell surface molecules, such as CD155.35 Taken together, it seems probable that the ability of the CD8 co-receptor to interact with the MHCI α3 domain enables the formation of an orientationally correct TCR–CD3 signalling complex essential for positive selection in the thymus, and subsequent efficient recognition of antigen in the periphery.
Figure 3.
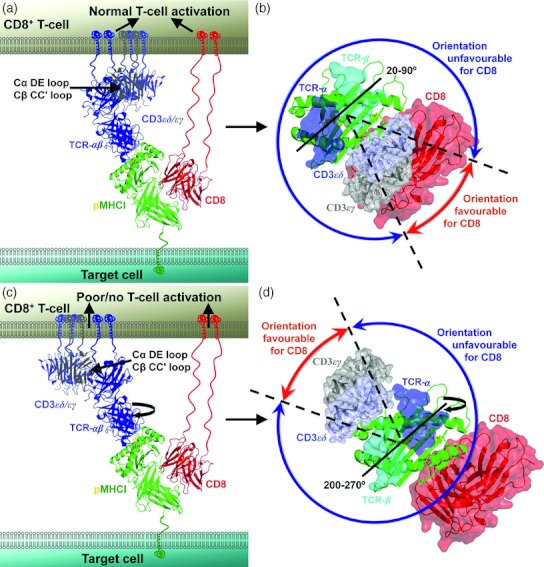
The CD8 co-receptor governs T-cell receptor (TCR) binding orientation. The CD3 co-signalling complex is shown in mauve (CD3εδ), or grey (CD3εγ), the TCR-αβ is shown in blue, peptide–MHC class I complex (pMHCI) is shown in green (peptide in yellow), and the CD8αα co-receptor is shown in red. Note that the vast majority of CD8+ T cells express the CD8αβ heterodimer, but the structure of human CD8αβ is not available for representation. (a) Proposed orientation of the TCR, CD3 and CD8 at the cell surface when the TCR binds with the α chain over the N-terminus of the peptide and the β chain over the C-terminus. In this orientation, CD3 is located adjacent to the TCR and CD8 to allow effective T-cell signalling. (b) Proposed orientation of the TCR, CD3 and CD8 as in (a) looking down the pMHC surface. In this orientation (which takes into account the range of binding angles published for TCR–pMHC complexes), CD8 is positioned optimally to and induce T-cell signalling (red arrow). (c) Proposed orientation of the TCR, CD3 and CD8 at the cell surface when the TCR binds with the reverse polarity (β-chain over the N-terminus of the peptide and the α-chain over the C-terminus). In this orientation, CD3 is located adjacent to the TCR, but distal from CD8, resulting in ineffective T-cell signalling. (d) Proposed orientation of the TCR, CD3 and CD8 as in (c) looking down the pMHC surface. In this TCR–pMHC orientation, CD8 is positioned unfavourably to induce T-cell signalling (blue arrow). Hence, the fixed polarity of the TCR–pMHC interaction is likely to be determined during thymic selection based on the orientation of the CD8 co-receptor in a mode in which it can effectively co-receive antigen.
Low solution binding affinity of the pMHCI–CD8 interaction maintains CD8+ T-cell specificity
The binding affinity of the pMHCI–CD8 interaction, measured by surface plasmon resonance, is largely conserved across the majority of MHCI allotypes studied to date (Tables 1a–c). Notably, the average human pMHCI–CD8αα interaction exhibits very low solution binding affinities (average KD = 145 μm) in a relatively tight range (KD = 100–220 μm) (Table 1a) and is characterized by extremely rapid kinetics (Koff > 18 s−1).36,37 There are, however, some exceptions to this overall uniformity. For example, HLA-A*6801 and HLA-B*4801 contain A245V and A245T mutations, respectively, in their α3 domains that substantially reduce CD8 binding (KD ∼ 1000 μm) (Table 1a).38 The biology that underlies these anomalies remains poorly defined, although the fact that CD8 can still bind, albeit with very low binding affinity, is likely to be important to impose MHCI restriction upon T cells restricted by these alleles.34 Furthermore, the extremely weak binding affinity of CD8 to HLA-A*6801 still allows most of the benefits, in terms of antigen recognition, that are seen with the wild-type interaction.38
Table 1.
Binding affinities of (a) human CD8αα to peptide–MHC class I complex (pMHCI); (b) murine CD8αα to pMHCI; and (c) murine CD8αβ to pMHCI
| KD (μm) | |
|---|---|
| (a) Human pMHCI–CD8 αα | |
| HLA-A*0201-GILGFVFTL37,44 | 1661 |
| HLA-A*0201-FIDSYICQV37 | 173 |
| HLA-A*0201-VLHDDLLEA37 | 107 |
| HLA-A*0201-ILKEPVHGV37 | 126 |
| HLA-A*0201-ILAKFLHWL44 | 183 |
| HLA-A*0201-SLLMWITQC44 | 125 |
| HLA-A*0201-YLEPGPVTV44 | 144 |
| HLA-A*0201-ELAGIGILTV 44 | 127 |
| HLA-A*0201-LLFGYPVYV44,78 | 1491 |
| HLA-A*0201-SLYNTVATL 38 | 128 |
| HLA-A*1101-AIFQSSMTK36 | 100 |
| HLA-A*2402-PYLFWLAAI44,78 | 1541 |
| HLA-A*6801-ITKGLGISYGR 38 | ∼9802 |
| HLA-A*6801-KTGGGPIYK 36 | > 10002 |
| HLA-B*0801-FLRGRAYGL44 | 135 |
| HLA-B*2702-KRWIILGLNK 79 | 130 |
| HLA-B*3501-TPEGIIPTL36 | 130 |
| HLA-B*4801-KQSTLHLV 36 | > 10002 |
| HLA-C*0702-KYFDEHYEY36 | 220 |
| HLA-G-RIIPRHLQL36 | > 10002 |
| HLA-E-VMAPRTVL36 | 160 |
| Average CD8αα KD (μm) | 145 |
| (b) Murine pMHCI–CD8αα | |
| H-2Kb-SIINFEKL3 | 91·6 |
| H-2Kb-SIINFEKL39 | 30·4 |
| H-2Kb-IFSK829 | 34·7 |
| H-2Kb-RGYVYQGL71 | 643 |
| H-2Kb-RGYVYQGL39 | 39·3 |
| H-2Kb-SIYRYYGL40 | 78·83 |
| H-2Dd-P18I1029 | 6·7 |
| H-2Db-KAVYNFATM40 | 210 |
| Average CD8αα KD (μm) | 69 |
| (c) Murine pMHCI–CD8αβ | |
| H-2Kb-SIINFEKL3 | 135 |
| H-2Kb-SIINFEKL39 | 14 |
| H-2Kb-IFSK829 | 38·4 |
| H-2Kb-RGYVYQGL71 | 403 |
| H-2Kb-RGYVYQGL39 | 11·8 |
| H-2Kb-SIYRYYGL40 | 122·33 |
| H-2Kd-SYIPSAEK17 | 99 |
| H-2Dd-P18I1029 | 8·2 |
| H-2Db-FAGHNLDLI39 | 14·1 |
| H-2Db-KAVYNFATM40 | > 10002 |
| H-2Ld-p2Ca39 | 11·2 |
| Average CD8αβ KD | 49 |
Values are averages from cited studies.
Values excluded from the average binding affinity.
Value is an average from multiple measurements in cited study.
In the murine system, affinity measurements have been reported for CD8αα and CD8αβ binding to a range of different MHCI alleles (Table 1b,c). The average binding affinity for CD8αα (KD = 69 μm) is similar to that of CD8αβ (KD = 49 μm) despite the small structural differences reported for pMHCI–CD8αα and pMHCI–CD8αβ,29 but the range of affinity measurements is somewhat larger than in the human system (CD8αα KD = 6·7–210 μm and CD8αβ KD = 14·1–135 μm). Hence, unlike in the human system, there seems to be some substantial differences in binding affinity between alleles. However, this observation should be considered with caution as there are inconsistencies for some measurements. For example, the interaction between CD8αβ and H2-Db has been measured by one group as KD = 14·1 μm 39 and by another group as KD > 1000 μm.40 The H2-Db molecules used in these separate experiments were complexed to different peptides, raising the possibility that peptide-induced modulation of CD8 binding could be at play. However, there has been no evidence in any other MHCI system to suggest that the bound peptide can affect CD8 binding, hence it is possible that differences in protein synthesis and experimental design may have had some impact on these disparate findings. Nonetheless, it is clear that CD8 operates at a very weak binding affinity compared with the TCR in both the human and murine systems.
Monomeric TCR–pMHCI binding does not alter the monomeric pMHCI–CD8 interaction
Although pMHCI–CD8 binding affinity measurements have shown that the interaction is weak, there is potential for CD8 to bind to pMHCI simultaneously with the TCR. This begs the question of whether the TCR, or CD8, binds more strongly to pMHCI during TCR–pMHCI–CD8 tripartite complex formation compared with the dipartite interactions. Consequently, a number of studies have investigated whether soluble CD8 binding can modify the TCR–pMHCI interaction. One investigation, using surface plasmon resonance analysis, indicated that pMHCI–CD8 binding occurred independently of the TCR–pMHCI interaction during antigen engagement.37 However, recent fluorescence resonance energy transfer-based examinations of the TCR–pMHCI–CD8 antigen recognition complex have shown that the TCR binds initially to pMHCI, satisfying the antigen-specific portion of the interaction. CD8 then binds to the same pMHCI as the TCR, fulfilling its role as a co-receptor.41 This ‘order’ of antigen engagement, which is also observed in the CD4+ T helper cell TCR–pMHCII–CD4 antigen recognition system,42,43 is likely to be important in ensuring that the specific interaction between the TCR and pMHC dominate T-cell recognition. Consequently, it is more reasonable to assume that, if binding modifications do occur, it is the initial TCR–pMHCI interaction that alters subsequent pMHCI–CD8 binding affinity. To confirm that CD8 binding occurred independently of TCR binding to pMHCI, we recently performed a study to investigate pMHCI–CD8 binding before and during TCR–pMHCI docking.44 We engineered a high affinity TCR with a half-life of many hours to overcome experimental limitations associated with the extremely rapid kinetics of natural TCR binding to pMHC. This development enabled us to measure the binding affinity of soluble CD8 to both unligated pMHCI and to TCR–pMHCI complex. The ensuing data demonstrated that dipartite CD8 binding was unaffected by TCR–pMHCI docking, thereby excluding the possibility that TCR modulation of the pMHCI–CD8 binding domain could influence CD8 interactions (Fig. 4).
Figure 4.
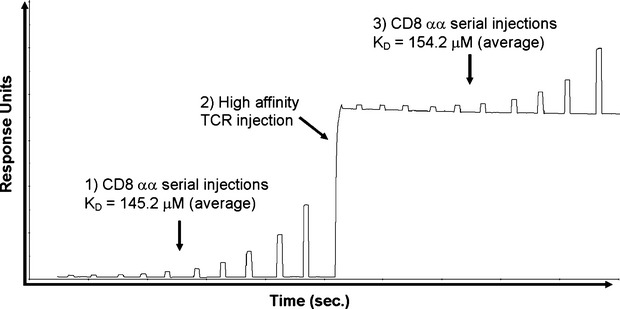
Monomeric T-cell receptor–peptide–MHC class I complex (TCR–pMHCI) binding does not alter the monomeric pMHCI–CD8 interaction. Binding affinity analysis of CD8αα using surface plasmon resonance. Ten serial dilutions of soluble CD8αα were injected over HLA-A*0201-ILAKFLHWL immobilized onto the surface of a CM5 sensor chip to determine the equilibrium binding constant for the pMHCI–CD8 interaction. The experiment was then repeated following a single injection of a high-affinity TCR (c13), which formed a complex with HLA-A*0201-ILAKFLHWL for the duration of the second CD8αα equilibrium binding measurement. No significant difference in CD8αα binding affinity was observed in the presence or absence of TCR–pMHCI engagement, indicating that TCR docking did not modify the CD8 binding domain of pMHCI.
Low pMHCI–CD8 binding affinity ensures TCR-dependent T-cell activation
In contrast to pMHCI–CD8, the affinity of the TCR–pMHCI interaction can be > 100-fold stronger and can exhibit considerably slower kinetics.23,30,44–48 It seems unlikely that the striking biophysical characteristics of the pMHCI–CD8 interaction have occurred by accident. In addition, the observation that the pMHCI–CD8 interaction is capable of exerting the vast majority of its biological function when weakened even further38 suggests that CD8 has specifically evolved to operate at very weak binding affinities. In a recent study, we generated pMHCI molecules with super-enhanced CD8 binding properties. Using these reagents, we demonstrated that pMHCI molecules with affinities for CD8 that lie within the typical range for agonist TCR–pMHCI interactions (KD = 10 μm) were able to activate CD8+ T cells in the absence of an antigen-specific TCR–pMHCI interaction.49 Hence, the weak binding affinity of the pMHCI–CD8 interaction is essential for the maintenance of CD8+ T-cell antigen specificity. It seems likely that MHCI molecules with a super-enhanced affinity for CD8 are capable of cross-linking CD8 at the cell surface in an ‘antibody-like’ manner. Indeed, this is consistent with other studies showing that antibody-mediated CD8 cross-linking can induce heterogeneous multimeric pMHCI binding,50–53 T-cell signalling can elicit downstream effector functions such as chemokine release and cytotoxicity in the absence of cognate antigen binding to the TCR.53,54 It is interesting to note that the average murine pMHCI–CD8 interaction is substantially stronger (KD = 49–69 μm) (Table 1b,c) than the equivalent human interaction (KD = 145 μm) (Table 1a) 15 but does not result in non-cognate CD8+ T-cell activation. Despite differences in TCR and CD8 binding (the average murine TCR–pMHCI and pMHCI–CD8 binding affinities are KD = 3·3 μm17,55–59 and KD = 59 μm, respectively, compared with the average human TCR–pMHCI and pMHCI–CD8 binding affinities of KD = 8·7 μm45,59–65 KD = 145 μm did, respectively37,45,66) the ratio of TCR and CD8 binding affinity is maintained between the two species (murine = 1 : 17, human = 1 : 18), so that the TCR binds with around 17–18 times stronger affinity than CD8. Therefore, the relationship between the binding affinity of the CD8 co-receptor compared with the TCR could represent a fundamental mechanism by which T cells maintain peptide antigen specificity through the TCR while retaining the required level of antigen sensitivity via CD8. Thus, pMHCI–CD8 interactions may have evolved in a highly constrained manner dictated by the need to balance high levels of T-cell cross-reactivity with non-specific T-cell activation, of which the latter could instigate auto-immunity. It should also be noted that the ratio of TCR : CD8 binding affinity may be different in the thymus because positively selecting pMHC ligands have been shown to have a very weak binding affinity for cognate TCRs.55,67 Hence, CD8 has been implicated as an important player during thymic selection of immature thymocytes.19
Disruption of the pMHCI–CD8 interaction can modulate TCR–pMHCI stability
Although the weak binding affinity of the pMHCI–CD8 interaction excludes the possibility that CD8 plays a major role during T-cell/target cell adhesion, experiments using mutated pMHCI tetramers with altered CD8 binding properties have shown that CD8 can profoundly affect TCR–pMHCI avidity.11,23,53,68 Accordingly, mutations in the α3 domain of HLA-A*0201 (D227K/T228A) that abolish CD8 binding (CD8-null) decreased both tetramer association rate and tetramer half-life compared with wild-type HLA-A*0201 tetramers23 (Fig. 5a,b). Furthermore, the shift in mean fluorescence intensity (MFI) using weakly binding pMHCI variants was substantially reduced using CD8-null tetramers compared with wild-type reagents (Fig. 5c,d). These data show that, although the interaction is weak, pMHCI–CD8 binding has an important role in stabilizing the TCR–pMHCI complex at the cell surface. In support of this notion, two-dimensional binding affinity measurements have shown that the TCR and CD8 bind pMHCI co-operatively to modulate T-cell antigen discrimination.69
Figure 5.
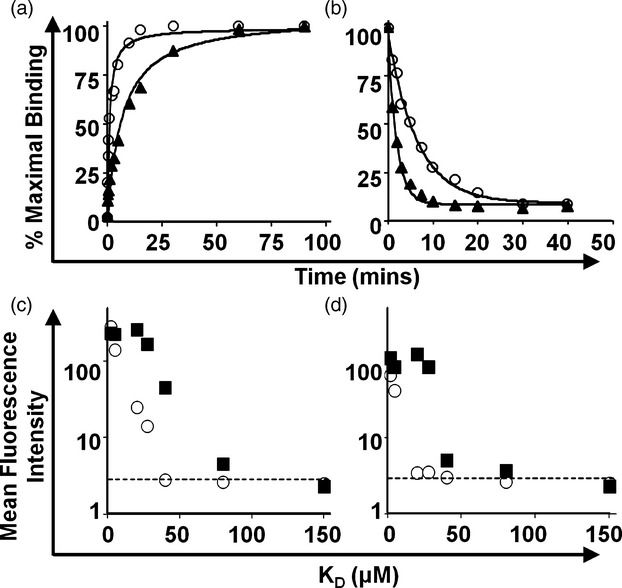
The effect of CD8 binding on peptide–MHC class I complex (pMHCI) multimer kinetics and staining using the ILA1 CD8+ T-cell clone. (a) Association kinetics of soluble multimeric pMHCI complexes with the ILA1 CD8+ T-cell clone expressing the cognate ILA1 T-cell receptor (TCR). Wild-type HLA-A*0201 molecules (open circles) that interact with CD8 (KD = 183 μm) reach binding equilibrium faster than mutated HLA-A*0201 molecules that fail to engage CD8 (triangles). (b) Dissociation kinetics of pMHCI complexes as a function of CD8 engagement. The dissociation of pMHCI complexes under conditions that prevent TCR re-binding is accelerated in the absence of a CD8 interaction (triangles) compared with wild-type HLA-A*0201 molecules (circles). (c, d) The ILA1 CD8+ T-cell clone was stained with seven different hTERT540-548 variants refolded in multimeric form with HLA-A*0201 wild-type (squares) or HLA-A*0201 that fail to engage CD8 (open circles) molecules at 37° (c) or 4° (d). In all cases, lack of CD8 binding reduced multimer sensitivity. Dot plots represent mean fluorescence intensity (MFI) versus TCR–pMHC interaction (KD). Background staining of with an irrelevant HLA-A*0201 wild-type HIV-1 p17 Gag multimer is represented by the horizontal dotted line in each case.
CD8 affects T-cell specificity
Tuning T-cell cross-reactivity through modulation of pMHCI–CD8 binding affinity
Disrupting the pMHCI–CD8 interaction clearly impacts the ability of T cells to recognize antigen. Accordingly, it has been shown that T-cell activation can be abolished if the pMHCI–CD8 interaction is blocked.70–72 However, recent evidence suggests that the requirements for CD8 co-activation may vary according to antigen potency and TCR–pMHCI affinity. Indeed, we and others7,23,73 have demonstrated that CD8-dependence during T-cell activation can be linked directly to the affinity of the TCR for pMHCI. In our study, pMHCI molecules with compromised CD8 binding were used to demonstrate that T-cell activation could not occur in the presence of weaker agonist antigens without CD8 co-activation, whereas T-cell activation by strong agonists was only partially impaired by the loss of CD8 engagement.23 Therefore, in instances where antigen potency is low, CD8 appears to play a greater role in increasing T-cell antigen sensitivity. In contrast, for stronger agonists, the contribution of CD8 to T-cell activation may be less.23 By extension, it might be predicted that the CD8 co-receptor acts to increase T-cell cross-reactivity by facilitating responses to a wider range of agonist ligands. To test this idea, we conducted a comprehensive evaluation of clonal CD8+ T-cell degeneracy using combinatorial peptide libraries and antigen-presenting cells expressing mutant HLA-A*0201 molecules with the following CD8 binding affinities: enhanced (KD = 85 μm),74 normal (KD ∼ 145 μm), decreased (KD = 500 μm) 38 or abrogated (KD < 10 000 μm). Using this approach, we were able to show a direct positive association between pMHCI–CD8 binding affinity and the number of ligands that elicited T-cell activation.75 Furthermore, in agreement with our previous findings, increasing the affinity of CD8 for HLA-A*0201 by more than one order of magnitude (KD = 10 μm) resulted in the loss of cognate antigen specificity and indiscriminate killing of HLA A2+ target cells.49,75 Hence, CD8 extends the range of pMHCI ligands that can be recognized by an individual cell surface-bound TCR, a feature that is essential for effective immune coverage.76 These findings suggest that the pMHCI–CD8 interaction is necessary to regulate the balance between optimal T-cell cross-reactivity and T-cell antigen specificity. This ‘CD8 effect’ (Fig. 6) can be controlled to optimize the degree of cross-reactivity and antigen sensitivity of CD8+ T cells at various stages of their development.
Figure 6.
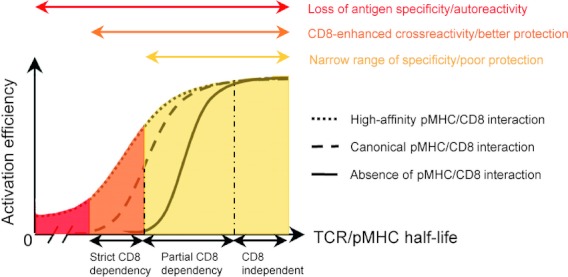
The influence of CD8 on the specificity of antigen recognition. The ‘CD8 effect’ broadens the range of ligands, represented by the curves, that can be productively engaged by the T-cell receptor (TCR). In the absence of a peptide–MHC class I complex (pMHCI) –CD8 interaction, only strong (CD8-independent) ligands can be recognized (plain line). This low level of cross-reactivity is likely to result in poor immunity because of insufficient coverage against all potential foreign peptides. The canonical pMHCI–CD8 interaction (dashed line) enhances the recognition efficiency of partially CD8-dependent ligands and broadens the spectrum of CD8+ T-cell antigen degeneracy by conferring a degree of reactivity against strictly CD8-dependent antigens. Developmentally regulated or activation-induced mechanisms that enhance or decrease CD8 co-receptor functions dynamically tune the overall pattern of CD8+ T-cell antigen specificity, which oscillates between minimal functional cross-reactivity afforded by TCR binding plasticity (plain line) and maximal antigen degeneracy licensed by full co-receptor activity (dashed line). High-affinity pMHCI–CD8 binding (dotted line) can trigger CD8+ T cells irrespective of the MHC-bound peptide or, it seems, the MHC molecule. The associated loss of specificity in this scenario would be deleterious to the organism.
Conclusions
The CD8 co-receptor plays an important and diverse role as a regulator of CD8+ T-cell immunity. Structural investigations have shown that CD8αα binds to an invariant domain of pMHCI independently from the TCR.24,25 The interaction between CD8αβ and pMHCI is similar, with the β-chain proximal to the T-cell surface.28,29 CD8, and indeed the CD4 co-receptor, may govern T-cell MHC restriction and TCR binding orientation to pMHC by enabling the formation of a functional signalling complex at the T-cell surface.32–34 Modulation of CD8 binding to pMHCI during TCR–pMHCI engagement does not occur, ensuring a fixed interaction across antigen specificities.44 Furthermore, the weak binding affinity of the pMHCI–CD8 interaction safeguards the role of TCR-mediated pMHCI engagement as the primary determinant of CD8+ T-cell activation in response to antigen.37,44,45,66 Indeed, increasing the affinity of the pMHCI–CD8 interaction into the range typically observed for TCR–pMHCI interactions can lead to CD8+ T-cell activation that does not require cognate antigen.49 From a therapeutic perspective, it is notable that CD8+ T cells with low-affinity TCR–pMHCI interactions are more dependent on the CD8 co-receptor for antigen-specific activation compared with CD8+ T cells with high-affinity TCR–pMHCI interactions. Consequently, therapeutic blockade of CD8 may be desirable for systems in which the TCR–pMHC interaction is weak, as typified by autoreactive CD8+ T cells.23,77 Finally, modulation of the pMHCI–CD8 interaction can affect CD8+ T-cell cross-reactivity.75 CD8 therefore appears to play a role in ‘tuning’ the sensitivity and specificity of CD8+ T-cell activation to ensure both effective and appropriately constrained behaviour during the continuous process of antigen surveillance.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Moebius U, Kober G, Griscelli AL, Hercend T, Meuer SC. Expression of different CD8 isoforms on distinct human lymphocyte subpopulations. Eur J Immunol. 1991;21:1793–800. doi: 10.1002/eji.1830210803. [DOI] [PubMed] [Google Scholar]
- 3.Leishman AJ, Naidenko OV, Attinger A, et al. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–9. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 4.Vremec D, Pooley J, Hochrein H, Wu L, Shortman K. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J Immunol. 2000;164:2978–86. doi: 10.4049/jimmunol.164.6.2978. [DOI] [PubMed] [Google Scholar]
- 5.Norment AM, Littman DR. A second subunit of CD8 is expressed in human T cells. EMBO J. 1988;7:3433–9. doi: 10.1002/j.1460-2075.1988.tb03217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayday A, Theodoridis E, Ramsburg E, Shires J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat Immunol. 2001;2:997–1003. doi: 10.1038/ni1101-997. [DOI] [PubMed] [Google Scholar]
- 7.Holler PD, Kranz DM. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–64. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 8.Janeway CA., Jr The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu Rev Immunol. 1992;10:645–74. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 9.Luescher IF, Vivier E, Layer A, Mahiou J, Godeau F, Malissen B, Romero P. CD8 modulation of T-cell antigen receptor–ligand interactions on living cytotoxic T lymphocytes. Nature. 1995;373:353–6. doi: 10.1038/373353a0. [DOI] [PubMed] [Google Scholar]
- 10.Norment AM, Salter RD, Parham P, Engelhard VH, Littman DR. Cell–cell adhesion mediated by CD8 and MHC class I molecules. Nature. 1988;336:79–81. doi: 10.1038/336079a0. [DOI] [PubMed] [Google Scholar]
- 11.Wooldridge L, van den Berg HA, Glick M, et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor–antigen complexes at the cell surface. J Biol Chem. 2005;280:27491–501. doi: 10.1074/jbc.M500555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner JM, Brodsky MH, Irving BA, Levin SD, Perlmutter RM, Littman DR. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60:755–65. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 13.Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–8. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 14.Laugel B, Price DA, Milicic A, Sewell AK. CD8 exerts differential effects on the deployment of cytotoxic T lymphocyte effector functions. Eur J Immunol. 2007;37:905–13. doi: 10.1002/eji.200636718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purbhoo MA, Boulter JM, Price DA, et al. The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor ζ chain. J Biol Chem. 2001;276:32786–92. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- 16.Rybakin V, Clamme JP, Ampudia J, Yachi PP, Gascoigne NR. CD8αα and -αβ isotypes are equally recruited to the immunological synapse through their ability to bind to MHC class I. EMBO Rep. 2011;12:1251–6. doi: 10.1038/embor.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arcaro A, Gregoire C, Bakker TR, et al. CD8β endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56(lck) complexes. J Exp Med. 2001;194:1485–95. doi: 10.1084/jem.194.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF. Essential role of CD8 palmitoylation in CD8 coreceptor function. J Immunol. 2000;165:2068–76. doi: 10.4049/jimmunol.165.4.2068. [DOI] [PubMed] [Google Scholar]
- 19.Bosselut R, Kubo S, Guinter T, Kopacz JL, Altman JD, Feigenbaum L, Singer A. Role of CD8β domains in CD8 coreceptor function: importance for MHC I binding, signaling, and positive selection of CD8+ T cells in the thymus. Immunity. 2000;12:409–18. doi: 10.1016/s1074-7613(00)80193-4. [DOI] [PubMed] [Google Scholar]
- 20.Bosselut R, Zhang W, Ashe JM, Kopacz JL, Samelson LE, Singer A. Association of the adaptor molecule LAT with CD4 and CD8 coreceptors identifies a new coreceptor function in T cell receptor signal transduction. J Exp Med. 1999;190:1517–26. doi: 10.1084/jem.190.10.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Trible RP, Samelson LE. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9:239–46. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 22.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 23.Laugel B, van den Berg HA, Gostick E, et al. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J Biol Chem. 2007;282:23799–810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 24.Gao GF, Tormo J, Gerth UC, et al. Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature. 1997;387:630–4. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 25.Leahy DJ, Axel R, Hendrickson WA. Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6 Å resolution. Cell. 1992;68:1145–62. doi: 10.1016/0092-8674(92)90085-q. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Qi J, Iwamoto A, Gao GF. Plasticity of human CD8αα binding to peptide-HLA-A*2402. Mol Immunol. 2011;48:2198–202. doi: 10.1016/j.molimm.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Kern PS, Teng MK, Smolyar A, et al. Structural basis of CD8 coreceptor function revealed by crystallographic analysis of a murine CD8αα ectodomain fragment in complex with H-2Kb. Immunity. 1998;9:519–30. doi: 10.1016/s1074-7613(00)80635-4. [DOI] [PubMed] [Google Scholar]
- 28.Chang HC, Tan K, Ouyang J, et al. Structural and mutational analyses of a CD8αβ heterodimer and comparison with the CD8αα homodimer. Immunity. 2005;23:661–71. doi: 10.1016/j.immuni.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Natarajan K, Margulies DH. Structural basis of the CD8αβ/MHC class I interaction: focused recognition orients CD8β to a T cell proximal position. J Immunol. 2009;183:2554–64. doi: 10.4049/jimmunol.0901276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridgeman JS, Sewell AK, Miles JJ, Price DA, Cole DK. Structural and biophysical determinants of αβ T-cell antigen recognition. Immunology. 2012;135:9–18. doi: 10.1111/j.1365-2567.2011.03515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Vα docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 32.Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci U S A. 2012;109:5405–10. doi: 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26:357–69. doi: 10.1016/j.immuni.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Van Laethem F, Sarafova SD, Park JH, et al. Deletion of CD4 and CD8 coreceptors permits generation of αβ T cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–50. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Tikhonova AN, Van Laethem F, Hanada K, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao GF, Willcox BE, Wyer JR, et al. Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8αα. J Biol Chem. 2000;275:15232–8. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 37.Wyer JR, Willcox BE, Gao GF, Gerth UC, Davis SJ, Bell JI, van der Merwe PA, Jakobsen BK. T cell receptor and coreceptor CD8αα bind peptide-MHC independently and with distinct kinetics. Immunity. 1999;10:219–25. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 38.Hutchinson SL, Wooldridge L, Tafuro S, et al. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J Biol Chem. 2003;278:24285–93. doi: 10.1074/jbc.M300633200. [DOI] [PubMed] [Google Scholar]
- 39.Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–81. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 40.Moody AM, Xiong Y, Chang HC, Reinherz EL. The CD8αβ co-receptor on double-positive thymocytes binds with differing affinities to the products of distinct class I MHC loci. Eur J Immunol. 2001;31:2791–9. doi: 10.1002/1521-4141(200109)31:9<2791::aid-immu2791>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 41.Yachi PP, Ampudia J, Zal T, Gascoigne NR. Altered peptide ligands induce delayed CD8-T cell receptor interaction–a role for CD8 in distinguishing antigen quality. Immunity. 2006;25:203–11. doi: 10.1016/j.immuni.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Hampl J, Chien YH, Davis MM. CD4 augments the response of a T cell to agonist but not to antagonist ligands. Immunity. 1997;7:379–85. doi: 10.1016/s1074-7613(00)80359-3. [DOI] [PubMed] [Google Scholar]
- 43.Madrenas J, Chau LA, Smith J, Bluestone JA, Germain RN. The efficiency of CD4 recruitment to ligand-engaged TCR controls the agonist/partial agonist properties of peptide-MHC molecule ligands. J Exp Med. 1997;185:219–29. doi: 10.1084/jem.185.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole DK, Dunn SM, Sami M, Boulter JM, Jakobsen BK, Sewell AK. T cell receptor engagement of peptide-major histocompatibility complex class I does not modify CD8 binding. Mol Immunol. 2008;45:2700–9. doi: 10.1016/j.molimm.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Cole DK, Pumphrey NJ, Boulter JM, et al. Human TCR-binding affinity is governed by MHC Class restriction. J Immunol. 2007;178:5727–34. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- 46.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by αβ T cell receptors. Annu Rev Immunol. 1998;16:523–44. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 47.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–76. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varela-Rohena A, Molloy PE, Dunn SM, et al. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat Med. 2008;14:1390–5. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wooldridge L, Clement M, Lissina A, et al. MHC class I molecules with Superenhanced CD8 binding properties bypass the requirement for cognate TCR recognition and nonspecifically activate CTLs. J Immunol. 2010;184:3357–66. doi: 10.4049/jimmunol.0902398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campanelli R, Palermo B, Garbelli S, Mantovani S, Lucchi P, Necker A, Lantelme E, Giachino C. Human CD8 co-receptor is strictly involved in MHC-peptide tetramer-TCR binding and T cell activation. Int Immunol. 2002;14:39–44. doi: 10.1093/intimm/14.1.39. [DOI] [PubMed] [Google Scholar]
- 51.Daniels MA, Devine L, Miller JD, et al. CD8 binding to MHC class I molecules is influenced by T cell maturation and glycosylation. Immunity. 2001;15:1051–61. doi: 10.1016/s1074-7613(01)00252-7. [DOI] [PubMed] [Google Scholar]
- 52.Denkberg G, Cohen CJ, Reiter Y. Critical role for CD8 in binding of MHC tetramers to TCR: CD8 antibodies block specific binding of human tumor-specific MHC-peptide tetramers to TCR. J Immunol. 2001;167:270–6. doi: 10.4049/jimmunol.167.1.270. [DOI] [PubMed] [Google Scholar]
- 53.Wooldridge L, Hutchinson SL, Choi EM, et al. Anti-CD8 antibodies can inhibit or enhance peptide-MHC class I (pMHCI) multimer binding: this is paralleled by their effects on CTL activation and occurs in the absence of an interaction between pMHCI and CD8 on the cell surface. J Immunol. 2003;171:6650–60. doi: 10.4049/jimmunol.171.12.6650. [DOI] [PubMed] [Google Scholar]
- 54.Clement M, Ladell K, Ekeruche-Makinde J, et al. Anti-CD8 antibodies can trigger CD8+ T cell effector function in the absence of TCR engagement and improve peptide-MHCI tetramer staining. J Immunol. 2011;187:654–63. doi: 10.4049/jimmunol.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–20. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 56.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–46. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 57.Garcia KC, Tallquist MD, Pease LR, et al. αβ T cell receptor interactions with syngeneic and allogeneic ligands: affinity measurements and crystallization. Proc Natl Acad Sci U S A. 1997;94:13838–43. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiser JB, Darnault C, Guimezanes A, et al. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–7. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 59.Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, van der Merwe PA. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 1999;10:357–65. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 60.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–96. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borg NA, Ely LK, Beddoe T, et al. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat Immunol. 2005;6:171–80. doi: 10.1038/ni1155. [DOI] [PubMed] [Google Scholar]
- 62.Davis-Harrison RL, Armstrong KM, Baker BM. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol. 2005;346:533–50. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 63.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 64.Gakamsky DM, Luescher IF, Pecht I. T cell receptor-ligand interactions: a conformational preequilibrium or an induced fit. Proc Natl Acad Sci U S A. 2004;101:9063–6. doi: 10.1073/pnas.0402840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laugel B, Boulter JM, Lissin N, et al. Design of soluble recombinant T cell receptors for antigen targeting and T cell inhibition. J Biol Chem. 2005;280:1882–92. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- 66.van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–84. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 67.Naeher D, Daniels MA, Hausmann B, Guillaume P, Luescher I, Palmer E. A constant affinity threshold for T cell tolerance. J Exp Med. 2007;204:2553–9. doi: 10.1084/jem.20070254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wooldridge L, Scriba TJ, Milicic A, Laugel B, Gostick E, Price DA, Phillips RE, Sewell AK. Anti-coreceptor antibodies profoundly affect staining with peptide-MHC class I and class II tetramers. Eur J Immunol. 2006;36:1847–55. doi: 10.1002/eji.200635886. [DOI] [PubMed] [Google Scholar]
- 69.Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choksi S, Jameson BA, Korngold R. A structure-based approach to designing synthetic CD8α peptides that can inhibit cytotoxic T-lymphocyte responses. Nat Med. 1998;4:309–14. doi: 10.1038/nm0398-309. [DOI] [PubMed] [Google Scholar]
- 71.Kern P, Hussey RE, Spoerl R, Reinherz EL, Chang HC. Expression, purification, and functional analysis of murine ectodomain fragments of CD8αα and CD8αβ dimers. J Biol Chem. 1999;274:27237–43. doi: 10.1074/jbc.274.38.27237. [DOI] [PubMed] [Google Scholar]
- 72.Sewell AK, Gerth UC, Price DA, et al. Antagonism of cytotoxic T-lymphocyte activation by soluble CD8. Nat Med. 1999;5:399–404. doi: 10.1038/7398. [DOI] [PubMed] [Google Scholar]
- 73.Holler PD, Lim AR, Cho BK, Rund LA, Kranz DM. CD8– T cell transfectants that express a high affinity T cell receptor exhibit enhanced peptide-dependent activation. J Exp Med. 2001;194:1043–52. doi: 10.1084/jem.194.8.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wooldridge L, Lissina A, Vernazza J, et al. Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur J Immunol. 2007;37:1323–33. doi: 10.1002/eji.200636765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wooldridge L, Laugel B, Ekeruche J, Clement M, van den Berg HA, Price DA, Sewell AK. CD8 controls T cell cross-reactivity. J Immunol. 2010;185:4625–32. doi: 10.4049/jimmunol.1001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.iSewell AK. Why must T cells be cross-reactive. Nat Rev Immunol. 2002;12 doi: 10.1038/nri3279. doi: 10.1038/nri327910.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laugel B, Cole DK, Clement M, Wooldridge L, Price DA, Sewell AK. The multiple roles of the CD8 coreceptor in T cell biology: opportunities for the selective modulation of self-reactive cytotoxic T cells. J Leukoc Biol. 2011;90:1089–99. doi: 10.1189/jlb.0611316. [DOI] [PubMed] [Google Scholar]
- 78.Cole DK, Rizkallah PJ, Boulter JM, et al. Computational design and crystal structure of an enhanced affinity mutant human CD8αα coreceptor. Proteins. 2007;67:65–74. doi: 10.1002/prot.21176. [DOI] [PubMed] [Google Scholar]
- 79.Iglesias MC, Almeida JR, Fastenackels S, et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–49. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]


