Abstract
Intracellular transport is essential for morphogenesis and functioning of the cell. The kinesin superfamily proteins (KIFs) have been shown to transport membranous organelles and protein complexes in a microtubule- and ATP-dependent manner. More than 30 KIFs have been reported in mice. However, the nomenclature of KIFs has not been clearly established, resulting in various designations and redundant names for a single KIF. Here, we report the identification and classification of all KIFs in mouse and human genome transcripts. Previously unidentified murine KIFs were found by a PCR-based search. The identification of all KIFs was confirmed by a database search of the total human genome. As a result, there are a total of 45 KIFs. The nomenclature of all KIFs is presented. To understand the function of KIFs in intracellular transport in a single tissue, we focused on the brain. The expression of 38 KIFs was detected in brain tissue by Northern blotting or PCR using cDNA. The brain, mainly composed of highly differentiated and polarized cells such as neurons and glia, requires a highly complex intracellular transport system as indicated by the increased number of KIFs for their sophisticated functions. It is becoming increasingly clear that the cell uses a number of KIFs and tightly controls the direction, destination, and velocity of transportation of various important functional molecules, including mRNA. This report will set the foundation of KIF and intracellular transport research.
Intracellular transport is essential for morphogenesis and functioning of the cell. After synthesis, proteins and lipids are sorted and transported to specific destinations within the cell as membranous organelles or protein complexes. The trafficking of proteins is tightly regulated and various different types of proteins are known to be involved. The kinesin superfamily proteins (KIFs) have been shown to transport organelles, protein complexes, and mRNAs to specific destinations in a microtubule- and ATP-dependent manner (1–3). KIFs are not only involved in the transport of organelles, protein complexes, and mRNAs, but also participate in chromosomal and spindle movements during mitosis and meiosis (4–6).
KIFs contain amino acid sequences that are highly conserved among all eukaryotic phyla studied thus far. Within the motor domain, there are two conserved sequences that are proximal to a Walker A ATP binding motif and a microtubule binding domain (2, 5, 7). Outside the motor domain, KIFs show few similarities. Interactions with cargo molecules have been shown to occur in regions outside the motor domain. Recently, it has been clearly shown that several KIFs attach to specific cargoes through interactions with adaptor proteins in these regions (8, 9).
Here, we report the identification of all KIFs in the mouse and human genomes. There are 45 members in total. Additional KIFs were identified by PCR cloning. The total number of KIFs was confirmed by a blast search of proteins in public and private genome databases. A unified nomenclature and phylogenic analysis also are presented to help categorize and understand functions of KIFs. This will set the foundation of KIF and intracellular transport research.
Materials and Methods
Identification of Additional KIFs by PCR Cloning.
To obtain sequences of mouse KIFs, PCR was conducted by using mouse cDNA and degenerate primers. Upstream primer sequences were derived from a putative ATP-binding motif and downstream primers from a conserved region 5′ to the second microtubule binding site (see Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). mRNA was isolated from 6- or 2-week-old or embryonic ICR mice (Oriental Yeast, Tokyo) tissue by the method of Okayama et al. (10) for reverse transcription (RT). RT was conducted by using the Choice cDNA synthesis system (Life Technologies, Rockville, MD). PCR was conducted for 40 cycles at 96°C for 30 sec, 55°C for 90 sec, and 72°C for 60 sec in a GeneAmp PCR system 9700 Thermal cycler (Perkin–Elmer). PCR products were blunted and subcloned into an EcoRV-digested pBluescript KS(+) vector (Toyobo, Osaka). Sequencing was performed by using the Dyenamic ET primer and Deza sequencing kit (Amersham Pharmacia) and an Applied Biosystems 377 DNA sequencer (Perkin–Elmer).
Northern Blotting.
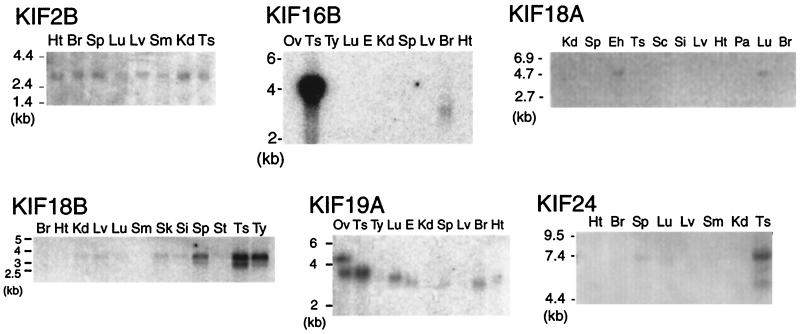
Obtained mRNAs also were used for Northern blotting whose results are shown in Fig. 1, KIF2B and KIF18A. Two micrograms of mRNA was run on a 1% formaldehyde agarose gel and transferred to Duralon UV uncharged nylon membranes (Stratagene). Semidried membranes were UV-crosslinked by using a Spectrolinker XL-1000 (Spectronic, Rochester, NY). The Northern blotting sheet used in Fig. 1 for KIF24 was purchased from CLONTECH. The sheet for KIF18B was purchased from Origene Technologies (Rockville, MD). KIF16B and KIF19A were analyzed by using Northern blotting sheets purchased from Ambion (Austin, TX). Random primed 32P-labeled probes were prepared by using the T7 Quick prime kit (Amersham Pharmacia). Hybridization was performed as described (11, 12). Radioactivity was visualized by using the Fuji Biological Analysis System BAS-2000. Membranes were exposed to a BAS-MS imaging plate, and the plate was processed through a BAS-2000.
Figure 1.
Northern blotting of additional KIFs. KIF2B is expressed ubiquitously in 2-week-old mouse tissue. KIF16B mRNA is detected in testis as a 4.1-kb band and a 3.3-kb band in brain. KIF18A was found in adult brain and embryonic head. KIF18B expression is dominant in testis. KIF19A is detected in testis, lung, and brain. KIF24 bands are seen in testis and spleen lanes. Ts, testis; Si, small intestine; Kd, kidney; Ht, heart; Br, brain; Sp, spleen; Sc, spinal cord; Lv, liver; Lu, lung; Pa, pancreas; Eh, embryo head; E, embryo; Sm, skeletal muscle; St, stomach; Ty, thymus; Ov, ovary.
Database Homology Search and Phylogenic Analysis.
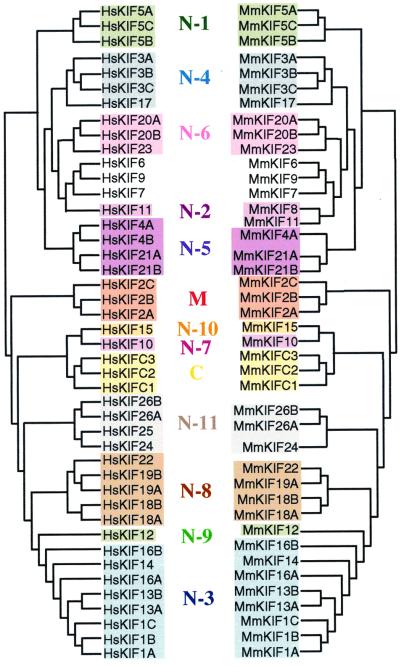
Full-length and partial amino acid sequences of KIFs obtained as described above were used for a database homology search through GenBank and the Celera Discovery System and Celera Genomics' associated databases (Celera Genomics, Rockville MD). An exhaustive tblastn search was conducted to detect KIF transcripts in the complete human genome. After obtaining all KIFs, the amino acid sequences between IFAY and LAGSE motifs were subjected to phylogenic analysis. In Fig. 2, human and mouse homologs were aligned with clustal w (13) software by the neighbor-joining method (14). The phylogenic tree was drawn with macvector software (Oxford Molecular, Cambridge, U.K.). For Fig. 3, maximum parsimony was calculated (15), and the phylogenic tree was drawn by using treeviewppc (16). Bootstrap values were assessed by 10,000 random samplings. Classification of all KIFs were carried out as described (2). Sequences used in this study can be obtained from our supplemental data or through the Celera database public segment (found on the web at www.celera.com).
Figure 2.
Phylogenic analysis of mouse and human orthologs. Forty-four of 45 murine KIFs have orthologs in humans. Sequences were analyzed by the neighbor-joining method.
Figure 3.
Phylogenic analysis of all KIFs expressed in mouse/humans, D. melanogaster, C. elegans, and S. cerevisiae. Amino acid sequences were aligned by using maximum parsimony. Sequences used for alignment are available as supplemental data or in the public segment of the Celera database (www.celera.com).
The KIFs presented here were identified by the following criteria: conservation of upstream Walker A ATP-binding motifs and a LAGSE or similar sequence ≈150–200 aa residues downstream, a YXXXXXDLL motif where X is any amino acid, and a SSRSH motif located between the Walker A and LAGSE sequences. Two predicted transcripts contained only the LAGSE consensus, and two transcripts had only IFAY sequences. In other organisms, a gene encoding only the LAGSE region and another encoding only IFAY were found in Drosophila. The prediction of KIFs using conventional software will automatically predict LAGSE-containing proteins to be KIFs. However, in a previous study it has been implied that the motor domain cannot be separated into modules (17). This indicates that IFAY and LAGSE sequences must be present at an appropriate order and spacing. Therefore, sequences lacking conserved motifs may not function as molecular motors and were excluded from this study. Additionally, genes from the same locus were considered to be splice variants and were omitted.
Results and Discussion
KIFs Previously Identified in Our Laboratory.
Previously, we have identified 25 KIFs in mice (11, 18- 22). Most were found by using molecular biological approaches. This study presents all KIFs in mouse and human and concludes the search for further unidentified KIFs.
Identification of 13 Additional KIFs.
We report 10 previously unidentified KIFs. KIF18B, KIF19A, KIF23, and KIF24 were identified in adult mouse brain, spinal cord, and small intestine cDNA by PCR. KIF23 has been reported in humans (23), but we have isolated it from mice by using PCR. KIF2B and KIF18A were found in embryonic cDNA by PCR. KIF4A and KIF4B, and KIF19A and KIF19B have a highly homologous motor domain. Therefore, it was difficult to discriminate the differences between them by PCR. These paralogs were found by using KIF4A and KIF19A amino acid sequences, respectively, as a template for blast searches. KIF26A and KIF26B also were discovered by blast searches using ScSMY1 as a template. SMY1 has amino acid motifs similar to KIF motor domains, although it may not be functional (24). With the exception of KIFs with motor domain sequences similar to that of SMY and therefore having low conservation in amino acid motifs, we were able to identify all KIFs by PCR.
KIF2C (25), KIF20B (26), and KIF25 (27) have been reported in humans and not in mice and were found in mice by our database search.
KIFs presented in this paper and previously identified KIFs were found by cross-hybridization methods or PCR using degenerate primers (11, 18, 28). These methods are laborious, hazardous, and time consuming. With this report there is no further need to search for new KIFs. However, the actual number of functional KIFs can only be determined after completing endogenous protein purification, peptide sequencing, and motility assays.
Tissue Distribution of Additional KIFs.
As part of our preliminary results concerning novel KIFs, Northern blotting results of KIF2B, KIF16B, KIF18A, KIF18B, KIF19A, and KIF24 are shown in Fig. 1. KIF2B is ubiquitously expressed in 2-week-old mice at 2.8 kb. KIF16B displays an intense 4-kb band in the testis lane and a 3.3-kb band in the brain lane. KIF18A is expressed in adult lung and embryonic head. The band migrates at 4.6 kb. The 3.9-kb band corresponding to KIF18B is most intense in adult testis. It is also highly expressed in the spleen and thymus and weakly in kidney, liver, lung, skin, small intestine, and stomach. No signal is detected in adult brain, heart, and muscle. KIF19A transcripts are found in adult testis, lung, and brain. A strong doublet band can be seen in the ovary lane. There are also bands in the embryo and spleen lanes. The higher band in ovary corresponds to a protein of 4.6 kb, the lower band to a protein of 3.8 kb. The lower band is found in testis and lung. The brain, embryo, and spleen lanes display a slightly lower band. KIF24 bands corresponding to proteins of 7.4 kb are found in the testis and spleen lanes.
Mouse and Human Similarity.
After obtaining all KIFs as described above, we next compared mouse and human KIFs. A phylogenic tree showing the analogy of murine and human KIFs is presented in Fig. 2. Forty-four of 45 mouse KIFs have orthologs in humans. The respective amino acid sequence conservation among murine and human KIFs exceeds 90%, indicating the significance of mice as a model of humans in KIF research.
Detection of KIFs in Mouse Brain.
There are eight KIFs reported to be expressed specifically or dominantly in mouse brain as demonstrated by Northern blotting, namely, KIF5A (18), KIF1A (18, 29), KIFC2 (21, 30), KIF3C (28), KIF5C (11), KIF21A and KIF21B (31), and KIF17 (8). Nineteen KIFs have been detected in the adult brain as intense bands by Northern blotting. PCR and KIF detection at various developmental stages reveal 11 KIFs expressed in brain. In total, the number of KIFs that have been detected at all stages of murine brain is 38. This number is much larger than the six KIFs reported in the single cell organism of Saccharomyces cerevisiae. This large number could mainly represent the necessity of delivering various functional molecules in highly polarized axons and dendrites for achieving complex functions of neurons.
Classification, Phylogenic Analysis, and Nomenclature of KIFs in Mouse and Human.
All KIFs shown or predicted to be transcribed in the human and mouse genomes are presented as a phylogenic tree along with all KIFs in S. cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans (Fig. 3). Sequences used are available as supplemental data. Using this occasion, a unified nomenclature is proposed (Table 1), which will abolish redundant designations that confuse researchers inside and outside this field of science.
Table 1.
Proposed and previous nomenclature of all KIFs
| Proposed | H. sapiens | D. melanogaster | C. elegans | S. cerevisiae | Others | Reference |
|---|---|---|---|---|---|---|
| KIF 1A | ATSV | unc104 | 18, GB | |||
| KIF 1B | CG8566 | (−) | 19, 57 | |||
| KIF 1C | (−) | RnKIF1D | 102 | |||
| KIF 2A | KIF 2 | CG1453 CG3219 | K11D9.1 | (−) | XIXKIF2 | 18, 103, 104 |
| KIF 2B | 0, GB, WB | |||||
| KIF 2C | CAKin/KNSL6 | CgMCAK XIKCM1 | 35 25, 105 | |||
| KIF 3A | Klp 64D | KRP85 | (−) | SpKRP85 | 18, 28, 106, 107 | |
| KIF 3B | Klp68D CG17461 | KRP 95 | SpKRP95 | |||
| KIF 3C | 11, 23, GB | |||||
| KIF 4A | KIF4 | Klp 3A | Y43F4B.6 | (−) | GgChrkin | 18, 80 |
| KIF 4B | 0, 78, WB | |||||
| KIF 5A | nKHC | KHC | Unc116 | (−) | RnnKHC | 18, 22, 108 109, 110 |
| KIF 5B | uKHC | |||||
| KIF 5C | xKHC | 11 | ||||
| KIF 6 | (−) | (−) | (−) | 11 | ||
| KIF 7 | (−) | (−) | (−) | 11 | ||
| KIF 8 | Klp 61F | BimC | Kip 1 Cin 8 | AnBimC | 11, 52, 54 56, 88, 111 | |
| KIF 11 | Eg5/KNSL1 | |||||
| KIF 9 | (−) | (−) | (−) | CrKlp1 | 11, 98 | |
| KIF 10 | CENP-E | Cmet CP15516 | (−) | Kip 2 | 11, 55 86, 88, GB | |
| KIF 12 | CG15844 | (−) | (−) | 11 | ||
| KIF 13A | Kin73 | Klp 4 | (−) | 9, 11, 112, 113 | ||
| KIF 13B | GAKIN | 11, 60 | ||||
| KIF 14 | HUMORFW | Neb | Klp 6 | (−) | 11, 114, ∗, GB | |
| KIF 15 | Hklp 2 | (−) | C06G3.2 C33H5.4 | (−) | 11, 91, WB | |
| KIF 16A | Klp98A | (−) | (−) | 11, 112 | ||
| KIF 16B | 11 | |||||
| KIF 17 | (−) | Osm3 | (−) | 8, 11, 73 | ||
| KIF 18A | Klp67A | (−) | Kip3 | 0, 112 | ||
| KIF 18B | ||||||
| KIF 19A | CG9913 | (−) | 0, 115, GB | |||
| KIF 19B | ||||||
| KIF 20A | Rab 6 Kin | CG12298 | (−) | (−) | MmKlp174 | 83 |
| KIF 20B | KlpMPP1 | 26, GB | ||||
| KIF 21A | CG5300 | T01G1.1 | (−) | 31, GB, WB | ||
| KIF 21B | ||||||
| KIF 22 | Kid/KNSL4 | Nod | (−) | (−) | 38, 116 | |
| KIF 23 | MKLP1/KNSL5 | Pav | Zen4A,B | (−) | CgCHO1 | 23, 81, 85, 117 |
| KIF 24 | CP38609 | (−) | (−) | 0, GB | ||
| KIF 25 | KNSL3 | (−) | (−) | (−) | 27 | |
| KIF 26A | (−) | Vab8 | (−) | 0, 94 | ||
| KIF 26B | ||||||
| KIF C1 | HSET/KNSL2 | Ncd | C41G7.2 M01E11.6 W02B12.7 | Kar3 | CgCHO2 | 36, 37,118, 119,120, WB |
| KIF C2 | (−) | Klp3 | (−) | XICTK1 | 21, 30 | |
| KIF C3 | 28, 112 |
GB, GenBank direct submission; WB, Worm Base; O, this paper.
Siddiqui, S. S., Hori, H., Mohammed, A. S. & Ali, M. Y., International C. elegans meeting, June 2–6, 1999, Madison, WI.
The number of KIFs is in accordance with the total number of genes in comparison with other phyla. The entire human genome is predicted to have 30,000 to 35,000 genes. The number of KIFs found in Drosophila and C. elegans are 23 and 21, respectively (Fig. 3). This is approximately half that of humans. The predicted numbers of genes in these two organisms are 13,601 and 18,424, respectively, likewise roughly half that of humans. S. cerevisiae has 6,241 proteins, of which six are KIFs. The total number of proteins is one-fifth of humans but there are less than a seventh of KIFs. This may be another example of the increased necessity of KIFs for higher cell function.
Three major types of KIFs have been identified according to the position of the motor domain: the NH2-terminal motor domain type (32, 33), middle motor domain type (18, 34, 35), and COOH-terminal motor domain type (21, 30, 36, 37) (referred to below as N-kinesins, M-kinesins, and C-kinesins, respectively). This study unexpectedly revealed abundant N-kinesins and few M- and C-kinesins (Fig. 3). Of the 45 KIFs, there are only three M-kinesins and C-kinesins each, leaving 39 N-kinesins. Of the 39 N-kinesins, two are monomeric and 37 seem to be multimeric.
There are 14 classes in total. C-kinesins are classified into two classes, C-1 kinesin and C-2 kinesin. M-kinesins make one class. N-kinesins are classified into 11 classes, comprising 16 families. Most classes consist of one family with the exception of N-3, N-4, and N-8 classes. N-3 kinesins consist of Unc104/KIF1, KIF13, and KIF16 families. Members of the Unc104/KIF1 family are mostly monomeric (29, 19), and amino acid sequences imply that those of the KIF16 family are multimeric (M.S., unpublished data). Members of the KIF13 family also have different characteristics (9). Thus, these families form subgroups within N-3 kinesins. N-4 kinesins consist of the KIF3 family and Osm3/KIF17 family. KIF3s are heterotrimeric, and Osm3/KIF17 form homodimers, indicating that these two families are distinct within this class. N-8 kinesins consist of the KIF18 family and Kid/KIF22 family. Due to the lack of similarity in the functional Kid domains, the KIF18 family was separated (38). This separation is supported by data obtained by using the neighbor-joining method (Fig. 2).
Most of the KIFs of other species including plants could be categorized into these 14 classes (data not shown). Below, we present a brief summary of the characterization of each kinesin class.
N-1 Kinesins.
This class consists of the kinesin heavy chain (KHC) family. KHC, the first KIF reported (39, 40), forms a heterotetramer made of two KHCs and two kinesin light chains (41). KHCs form a highly related family (KIF5A, KIF5B, and KIF5C). KIF5B is expressed ubiquitously in many tissues (42), whereas KIF5A and KIF5C are specific to nerve tissue (18, 22). Kinesin initially was characterized as a motor transporting membranous organelles anterogradely toward the plus end of microtubules and forming a crossbridge between membranous organelle and microtubules in nerve axons (32, 39, 40, 43, 44). However, recent studies have revealed various functions. In a wide variety of cells, kinesin works as a motor for transport of mitochondria, lysosomes, tubulin oligomer, and mRNA complex toward the plus end of microtubules (3, 45–48). The light chains of KIF5 have a role in binding these cargoes (49, 50). However, in the Ascomycete fungus, Neurospora crassa, kinesin light chains are lacking, implying that KHCs alone are sufficient by themselves for binding to some cargoes (51).
N-2 Kinesins.
This class consists of the BimC/Eg5/KIF11 family. This family contains KIF11 (Eg5), first found in Xenopus (52), a homotetrameric KIF (53) for bipolar spindle formation (54). Cell cycle-specific phosphorylation also has been reported in human (55).
BimC is a well-characterized KIF highly related to Eg5, functioning in cell division (56).
N-3 Kinesins.
This class is composed of three families, the Unc104/KIF1 family, the KIF13 family, and the KIF14 family.
The Unc104/KIF1 family.
There are three KIFs in this family: KIF1A, KIF1B, and KIF1C. C. elegans Unc104 is a homolog of mouse KIF1A (57). KIF1A is an anterograde motor transporting a subset of synaptic vesicle precursors and plays an important role in neuronal function and survival (29, 58). KIF1B is thought to convey mitochondria, sharing the role with KIF5B (19, 46). Interestingly, KIF1A and KIF1B are monomeric (19, 29). KIF1C is dimeric in vivo (59) and reported to have functions in endoplasmic reticulum-Golgi transport (60).
The KIF13 family.
In mice, this family consists of two proteins, KIF13A and KIF13B. KIF13A transports a cargo containing M6PR through direct interaction with the AP-1 complex (9). KIF13B (GAKIN) is reported to interact with hDLG and PSD95 in vitro with its proximal tail, which is highly conserved from C. elegans (61).
The KIF16 family.
KIF14, KIF16A, and KIF16B constitute a family with DmKlp98a. As the tail domains along with the expression patterns of KIF14, KIF16A, and KIF16B are different, these KIFs may have separate functions.
N-4 Kinesins.
This class consists of KIF3 and Osm3/KIF17 families.
The KIF3 family.
The KIF3 family is composed of KIF3A, KIF3B, and KIF3C in mice (18, 20, 62). A KIF3A-KIF3B heterodimer (KIF3A/3B) assembles with KAP3, forming a heterotrimeric complex (63, 64). The motor is expressed ubiquitously and is used for anterograde transport of membranous organelles containing fodrin in neurons (65). The KIF3 complex and its homolog were shown to transport protein complexes to form cilia (66–69). Gene-targeting studies showed that the nodal cilia, in which KIF3A/B are localized, rotate to generate a unidirectional flow of extraembryonic fluid (nodal flow), which could fundamentally control left-right determination (70, 71). Without KIF3, there is no nodal flow (70, 71). Thus, KIF3 is essential for development of the left-right axis determination in embryos (70–72).
The Osm3/KIF17 family.
Another molecule closely related to the KIF3 family is Osm3 in C. elegans (73). Mutations in Osm3 cause defects in chemosensory responses (74). Osm3 is necessary for sensory cilia growth in the dendrites of sensory neurons. Mutations in Osm-3 and Lin-10 result in a similar phenotype in osmotic avoidance, and Lin-10 has defect in glutamate receptor localization (75). Interestingly, KIF17, a member of this family, binds to mouse homolog of Lin-10 and transports vesicles containing a N-methyl-d-aspartate receptor subunit, NR2B, through the following interactions: KIF17-mLin10-mLin2-mLin7-NR2B (8). Human KIF17 also is reported to be highly expressed only in central nervous system, possessing a highly conserved Lin-10 binding domain (Kazusa DNA bank).
N-5 Kinesins.
This class is composed of the KIF4 family. A member of this family is KIF4A. KIF4A mRNA is expressed abundantly in juvenile tissues, including differentiated young neurons (76). KIF4A is a microtubule plus end-directed anterograde motor. Evidence has been shown of KIF4A transporting membranous organelles containing L1 in juvenile neurons (77). In this study, KIF4B, a transcript highly homologous to KIF4A was identified. Both genes have been assigned to respective loci (78). Chromokinesin, the chicken isolog of KIF4, is associated with chromosome arms and functions as a mitotic motor with DNA as its cargo (79). The other members of this family, KIF21A and KIF21B, are assumed to have some role in neurons (31). DmKlp3a is a critical component in the establishment or stabilization of the central spindle (80). Thus, members of this family may have multiple functions, including membrane trafficking and cell division.
N-6 Kinesins.
This class consists of the CHO1/KIF23 and KIF20/Rab6 kinesin families.
The CHO1/KIF23 family.
KIF23 (CHO1) is a KIF involved in mitosis, originally identified by a mAb raised against mitotic spindle components (81). It has been shown to be expressed in cultured neurons (82).
The KIF20/Rab6 kinesin family.
KIF20A (Rab6-KIF) is reported to have a fundamental role in Golgi-derived vesicle transport (83) and/or cell division (84). KIF20B (KlpMPP1) was isolated by using an antibody specific for M-phase phosphorylated proteins (26). The Drosophila homolog of KIF23, Pavarotti (Pav), is required for central spindle pole organization and cytokinesis (85).
N-7 Kinesins.
The CENP-E/KIF10 family forms N-7 kinesins. KIF10 (CENP-E) was first identified as a centromere-associated protein (86). It functions in chromosome segregation (87). Cenp-meta has a similar function in Drosophila (55). S. cerevisiae Kip2p, homolog of KIF10, functions in spindle positioning (88, 89).
N-8 Kinesins.
The KIF18 and Kid/KIF22 families constitute N-8 kinesins.
The KIF18 family.
This most recently discovered family of KIFs has not yet been well characterized. The tissue expression patterns of KIF18A and KIF18B are shown in Fig. 1. This family has a counterpart in Drosophila, but not in C. elegans.
The Kid/KIF22 family.
This family contains KIF22 (Kid), a KIF that colocalizes with mitotic chromosomes and may bind DNA (38). A highly homologous KIF, Nod is found in Drosophila (90). We report additional members of this family, KIF19A and KIF19B. KIF19A and KIF19B are highly homologous to each other, but have two loci (see Table 3, which is published as supplemental data, for accession numbers). We also have detected a splice variant of KIF19A. In the Northern blotting for KIF19A (Fig. 1), we detected bands at three different heights, in good agreement with the existence of three transcripts. However, only one loci is found in the Celera Discovery System. Further studies are necessary to clarify this inconsistency.
N-9 Kinesins.
The KIF12 family form this class. KIF12 has been an orphan KIF, not affiliated with any family previously reported (2). Here, it composes a family with its Dm counterpart, DmCG15844. KIF12 is highly expressed in kidney and may have a significant role in kidney cells (11).
N-10 Kinesins.
The KIF15 family forms this class. KIF15 was reported to be dominantly expressed in spleen and testis (11). Human KIF15 (Hklp2) is reported to associate with a cell proliferation marker protein (91). The two C. elegans proteins in this family, C06G3.2 and C33H5.4, seem to be paralogs (Worm Base).
N-11 Kinesins.
N-11 kinesins contain the KIF26 family. This family is related to DmCos2 and CeVab-8. Some members of this family have relatively low consensus motif conservation in comparison to other KIFs (see supplemental data). Localization of ScSMY1 is not affected by microtubule abolition (24). Costal2 (Cos2) has been shown to be part of the hedgehog signaling cascade with microtubule binding abilities (92, 93). Vab-8 is implicated in the regulation of cell and axon growth cone migration (94). KIF25 (KNSL3) has four splice variants and is expressed ubiquitously (27). Three additional KIFs, KIF24, KIF26A, and KIF26B, join this family. Their functions are yet to be determined.
M-Kinesins.
The KIF2 family forms M-kinesins. M-kinesins have a motor domain in the center of the molecule (18). KIF2A (formerly KIF2) forms a homodimer. KIF2A is a microtubule plus end-directed motor and is expressed ubiquitously (34). The cargo of KIF2A includes βgc, a β-subunit of the insulin-like growth factor-1 receptor (95). A splice variant of KIF2A has been reported (96). A Xenopus homolog of KIF2A, XKIF2, is reported to destabilize microtubules in vitro (104). KIF2C (MCAK) was identified as a mitotic-centromere-associated kinesin (35). KIF2B is a novel member of this family.
C-1 Kinesins.
This class contains the Ncd/Kar3/KIFC1 family. Several C-type motors, such as Ncd in Drosophila and Kar3 in S. cerevisiae, are motors for meiosis, mitosis, and karyogamy. These family members exhibit a microtubule minus end-directed motility (36, 37). Mammals have a counterpart, KIFC1 (21). It is noteworthy that C. elegans has developed many KIFC1 homologs. A highly related KIF also has been reported (97).
C-2 Kinesins.
The KIFC2/C3 family constitutes C-2 kinesins. Three C-type KIFs have been identified in mouse brain. KIFC2 forms a homodimer without associated polypeptides (21). It is a unique C-type motor that mainly functions in the dendritic transport of multivesicular body-like membranous organelles (21). KIFC3 is ubiquitously expressed (11).
A large number of C-kinesins were expected to function in the complex dendritic transport. Recently, it has been revealed that N-kinesins, plus end-directed motors, play important roles not only in axons but also in dendrites (8, 31). Indeed, the distal portions of developed dendrites have microtubule plus ends directed to the tips, having the same polarity as axons. The “mixed polarity” of microtubules exists only in proximal portions of dendrites. Future work should reveal the roles of KIFs in dendrites, which requires an accurate delivery regulation mechanism in comparison to axonal transport. Additionally, if the same motor for axonal transport is used for dendritic transport, how cargoes orient KIFs toward specific destinations will be a key question that needs to be answered in future studies.
Orphans.
These KIFs have no counterpart in Drosophila or C. elegans. KIF6 and KIF9 are localized near the BimC family. The full-length sequences of KIF6 and KIF9 are not similar with BimC. The phylogenic distance between KIF9 and KIF6 is large, implying that they do not form a family. These molecules may have evolved to attain other functions in mammals. KIF9 has a homolog in Chlamydomonas, Klp1 (98).
KIF7 has no evident homolog in Drosophila, C. elegans, or S. cerevisiae. Originally identified from murine brain by using PCR, it was found to be dominantly expressed in the testis by Northern blotting (11).
There have been reports of many proteins, RNA, and membranous organelles transported in a microtubule-dependent manner (99). Forty-five KIFs are insufficient to transport all organelles and vesicles. There must be a way to transport various cargoes using a limited number of KIFs. Other KIF members may arise through improved transcription algorithms. Alternative splicing also may increase the number of KIFs contributing to intracellular transport, and adaptor proteins also may contribute. The search for adaptor proteins other than kinesin light chains and kinectin (100) has begun (8, 9). The trend has extended to myosin research (101). To elucidate the mechanism of intracellular transport, the regulation of cargo binding is also an important problem. Currently, there are only a few reports clarifying this essential topic (102). Another question is how cargo and KIF dissociate. To enable cargo proteins to function properly, this dissociation is indispensable.
Concerning how each KIF recognizes and binds to their specific cargo molecules, one significant method is the formation of a receptor-adaptor (scaffold/scaffolding protein)-motor complex as in the case of KIF3, KIF17, and KIF13A (8, 9, 63). Alternatively, KIFs can bind to membrane proteins through light chains (49, 50). These adaptor proteins may contribute in increasing the variety of cargoes a KIF can convey. Thus, it is rapidly becoming clear that the cell uses a number of KIFs and tightly controls the direction, destination, and velocity of transports for various important functional molecules.
Supplementary Material
Acknowledgments
We are grateful to members of the Hirokawa laboratory, especially Dr. T. Nakagawa. This work was supported by a grant for Center of Excellence (COE) from the Ministry of Education, Science, and Culture (to N.H).
Abbreviations
- KIF
kinesin superfamily protein
- KHC
kinesin heavy chain
Footnotes
This paper was presented at the National Academy of Sciences colloquium, “Molecular Kinesis in Cellular Function and Plasticity,” held December 7–9, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Hirokawa N. Trends Cell Biol. 1996;6:135–141. doi: 10.1016/0962-8924(96)10003-9. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 3.Brendza R P, Serbus L R, Duffy J B, Saxton W M. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirokawa N, Noda Y, Okada Y. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 5.Vale R D, Fletterick R J. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 6.Sharp D J, Rogers G C, Scholey J M. Nature (London) 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 7.Kim A J, Endow S A. J Cell Sci. 2000;113:3681–3682. doi: 10.1242/jcs.113.21.3681. [DOI] [PubMed] [Google Scholar]
- 8.Setou M, Nakagawa T, Seog D H, Hirokawa N. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 10.Okayama H, Kawaichi M, Brownstein M, Lee F, Yokota T, Arai K. Methods Enzymol. 1987;154:3–28. doi: 10.1016/0076-6879(87)54067-8. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. Proc Natl Acad Sci USA. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y, Zhang Z, Hirokawa N. J Cell Sci. 1995;108:1883–1893. doi: 10.1242/jcs.108.5.1883. [DOI] [PubMed] [Google Scholar]
- 16.Page R D. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 17.Kikkawa M, Okada Y, Hirokawa N. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 18.Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. J Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki H, Nakata T, Okada Y, Hirokawa N. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N. Neuron. 1997;18:425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- 22.Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N. J Neurosci. 2000;20:6374–6384. doi: 10.1523/JNEUROSCI.20-17-06374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuriyama R, Dragas-Granoic S, Maekawa T, Vassilev A, Khodjakov A, Kobayashi H. J Cell Sci. 1994;107:3485–3499. doi: 10.1242/jcs.107.12.3485. [DOI] [PubMed] [Google Scholar]
- 24.Lillie S H, Brown S S. J Cell Biol. 1998;140:873–883. doi: 10.1083/jcb.140.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim I G, Jun D Y, Sohn U, Kim Y H. Biochim Biophys Acta. 1997;1359:181–186. doi: 10.1016/s0167-4889(97)00103-1. [DOI] [PubMed] [Google Scholar]
- 26.Westendorf J M, Rao P N, Gerace L. Proc Natl Acad Sci USA. 1994;91:714–718. doi: 10.1073/pnas.91.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto S, Matsushima M, Nakamura Y. Cytogenet Cell Genet. 1998;83:25–29. doi: 10.1159/000015159. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Hanlon D W, Marszalek J R, Goldstein L S. Genomics. 1997;45:123–131. doi: 10.1006/geno.1997.4901. [DOI] [PubMed] [Google Scholar]
- 29.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 30.Hanlon D W, Yang Z, Goldstein L S. Neuron. 1997;18:439–451. doi: 10.1016/s0896-6273(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 31.Marszalek J R, Weiner J A, Farlow S J, Chun J, Goldstein L S. J Cell Biol. 1999;145:469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirokawa N, Pfister K K, Yorifuji H, Wagner M C, Brady S T, Bloom G S. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 33.Yang J T, Laymon R A, Goldstein L S. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- 34.Noda Y, Sato-Yoshitake R, Kondo S, Nangaku M, Hirokawa N. J Cell Biol. 1995;129:157–167. doi: 10.1083/jcb.129.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wordeman L, Mitchison T J. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meluh P B, Rose M D. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 37.Endow S A, Henikoff S, Soler-Niedziela L. Nature (London) 1990;345:81–83. doi: 10.1038/345081a0. [DOI] [PubMed] [Google Scholar]
- 38.Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T. EMBO J. 1996;15:457–467. [PMC free article] [PubMed] [Google Scholar]
- 39.Vale R D, Reese T S, Sheetz M P. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brady S T. Nature (London) 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 41.Bloom G S, Wagner M C, Pfister K K, Brady S T. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- 42.Gudkov A V, Kazarov A R, Thimmapaya R, Axenovich S A, Mazo I A, Roninson I B. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirokawa N, Sato-Yoshitake R, Kobayashi N, Pfister K K, Bloom G S, Brady S T. J Cell Biol. 1991;114:295–302. doi: 10.1083/jcb.114.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirokawa N. J Cell Biol. 1982;94:129–142. doi: 10.1083/jcb.94.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollenbeck P J, Swanson J A. Nature (London) 1990;346:864–866. doi: 10.1038/346864a0. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 47.Nakata T, Hirokawa N. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terada S, Kinjo M, Hirokawa N. Cell. 2000;103:141–155. doi: 10.1016/s0092-8674(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 49.Kamal A, Stokin G B, Yang Z, Xia C H, Goldstein L S. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 50.Bowman A B, Kamal A, Ritchings B W, Philp A V, McGrail M, Gindhart J G, Goldstein L S. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 51.Steinberg G, Schliwa M. Mol Biol Cell. 1995;6:1605–1618. doi: 10.1091/mbc.6.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Guellec R, Paris J, Couturier A, Roghi C, Philippe M. Mol Cell Biol. 1991;11:3395–3398. doi: 10.1128/mcb.11.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole D G, Saxton W M, Sheehan K B, Scholey J M. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 54.Blangy A, Lane H A, d'Herin P, Harper M, Kress M, Nigg E A. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 55.Yucel J K, Marszalek J D, McIntosh J R, Goldstein L S, Cleveland D W, Philp A V. J Cell Biol. 2000;150:1–11. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enos A P, Morris N R. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- 57.Otsuka A J, Jeyaprakash A, Garcia-Anoveros J, Tang L Z, Fisk G, Hartshorne T, Franco R, Born T. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- 58.Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorner C, Ullrich A, Haring H U, Lammers R. J Biol Chem. 1999;274:33654–33660. doi: 10.1074/jbc.274.47.33654. [DOI] [PubMed] [Google Scholar]
- 60.Dorner C, Ciossek T, Muller S, Moller P H, Ullrich A, Lammers R. J Biol Chem. 1998;273:20267–20275. doi: 10.1074/jbc.273.32.20267. [DOI] [PubMed] [Google Scholar]
- 61.Hanada T, Lin L, Tibaldi E V, Reinherz E L, Chishti A H. J Biol Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- 62.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamazaki H, Nakata T, Okada Y, Hirokawa N. Proc Natl Acad Sci USA. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wedaman K P, Meyer D W, Rashid D J, Cole D G, Scholey J M. J Cell Biol. 1996;132:371–380. doi: 10.1083/jcb.132.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda S, Yamazaki H, Seog D H, Kanai Y, Terada S, Hirokawa N. J Cell Biol. 2000;148:1255–1265. doi: 10.1083/jcb.148.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cole D G, Cande W Z, Baskin R J, Skoufias D A, Hogan C J, Scholey J M. J Cell Sci. 1992;101:291–301. doi: 10.1242/jcs.101.2.291. [DOI] [PubMed] [Google Scholar]
- 67.Cole D G, Chinn S W, Wedaman K P, Hall K, Vuong T, Scholey J M. Nature (London) 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 68.Morris R L, Scholey J M. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cole D G, Diener D R, Himelblau A L, Beech P L, Fuster J C, Rosenbaum J L. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 71.Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marszalek J R, Ruiz-Lozano P, Roberts E, Chien K R, Goldstein L S. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shakir M A, Fukushige T, Yasuda H, Miwa J, Siddiqui S S. NeuroReport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- 74.Tabish M, Siddiqui Z K, Nishikawa K, Siddiqui S S. J Mol Biol. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- 75.Rongo C, Whitfield C W, Rodal A, Kim S K, Kaplan J M. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 76.Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peretti D, Peris L, Rosso S, Quiroga S, Caceres A. J Cell Biol. 2000;149:141–152. doi: 10.1083/jcb.149.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ha M J, Yoon J, Moon E, Lee Y M, Kim H J, Kim W. Cytogenet Cell Genet. 2000;88:41–42. doi: 10.1159/000015482. [DOI] [PubMed] [Google Scholar]
- 79.Wang S Z, Adler R. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams B C, Riedy M F, Williams E V, Gatti M, Goldberg M L. J Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nislow C, Sellitto C, Kuriyama R, McIntosh J R. J Cell Biol. 1990;111:511–522. doi: 10.1083/jcb.111.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferhat L, Kuriyama R, Lyons G E, Micales B, Baas P W. Eur J Neurosci. 1998;10:1383–1393. doi: 10.1046/j.1460-9568.1998.00159.x. [DOI] [PubMed] [Google Scholar]
- 83.Echard A, Jollivet F, Martinez O, Lacapere J J, Rousselet A, Janoueix-Lerosey I, Goud B. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 84.Hill E, Clarke M, Barr F A. EMBO J. 2000;19:5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adams R R, Tavares A A, Salzberg A, Bellen H J, Glover D M. Gene Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yen T J, Compton D A, Wise D, Zinkowski R P, Brinkley B R, Earnshaw W C, Cleveland D W. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yen T J, Li G, Schaar B T, Szilak I, Cleveland D W. Nature (London) 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- 88.Roof D M, Meluh P B, Rose M D. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cottingham F R, Hoyt M A. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang P, Knowles B A, Goldstein L S, Hawley R S. Cell. 1990;62:1053–1062. doi: 10.1016/0092-8674(90)90383-p. [DOI] [PubMed] [Google Scholar]
- 91.Sueishi M, Takagi M, Yoneda Y. J Biol Chem. 2000;275:28888–28892. doi: 10.1074/jbc.M003879200. [DOI] [PubMed] [Google Scholar]
- 92.Sisson J C, Ho K S, Suyama K, Scott M P. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 93.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Therond P P. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 94.Wolf F W, Hung M S, Wightman B, Way J, Garriga G. Neuron. 1998;20:655–666. doi: 10.1016/s0896-6273(00)81006-5. [DOI] [PubMed] [Google Scholar]
- 95.Morfini G, Quiroga S, Rosa A, Kosik K, Caceres A. J Cell Biol. 1997;138:657–669. doi: 10.1083/jcb.138.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santama N, Krijnse-Locker J, Griffiths G, Noda Y, Hirokawa N, Dotti C G. EMBO J. 1998;17:5855–5867. doi: 10.1093/emboj/17.20.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Navolanic P M, Sperry A O. Biol Reprod. 2000;62:1360–1369. doi: 10.1095/biolreprod62.5.1360. [DOI] [PubMed] [Google Scholar]
- 98.Bernstein M, Beech P L, Katz S G, Rosenbaum J L. J Cell Biol. 1994;125:1313–1326. doi: 10.1083/jcb.125.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Terada S, Hirokawa N. Curr Opin Neurobiol. 2000;10:566–573. doi: 10.1016/s0959-4388(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 100.Toyoshima I, Yu H, Steuer E R, Sheetz M P. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kussel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U, Petit C. EMBO J. 2000;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sato-Yoshitake R, Yorifuji H, Inagaki M, Hirokawa N. J Biol Chem. 1992;267:23930–23936. [PubMed] [Google Scholar]
- 103.Rogers S L, Tint I S, Fanapour P C, Gelfand V I. Proc Natl Acad Sci USA. 1997;94:3720–3725. doi: 10.1073/pnas.94.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Desai A, Verma S, Mitchison T J, Walczak C E. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 105.Walczak C E, Mitchison T J, Desai A. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 106.Rashid D J, Wedaman K P, Scholey J M. J Mol Biol. 1995;252:157–162. doi: 10.1006/jmbi.1995.0484. [DOI] [PubMed] [Google Scholar]
- 107.Cole D G, Diener D R, Himelblau A L, Beech P L, Fuster J C, Rosenbaum J L. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patel N, Thierry-Mieg D, Mancillas J R. Proc Natl Acad Sci USA. 1993;90:9181–9185. doi: 10.1073/pnas.90.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Niclas J, Navone F, Hom-Booher N, Vale R D. Neuron. 1994;12:1059–1072. doi: 10.1016/0896-6273(94)90314-x. [DOI] [PubMed] [Google Scholar]
- 110.Saxton W M, Hicks J, Goldstein L S, Raff E C. Cell. 1991;64:1093–1102. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- 111.Hoyt M A, He L, Loo K K, Saunders W S. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stewart R J, Pesavento P A, Woerpel D N, Goldstein L S. Proc Natl Acad Sci USA. 1991;88:8470–8474. doi: 10.1073/pnas.88.19.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li H P, Liu Z M, Nirenberg M. Proc Natl Acad Sci USA. 1997;94:1086–1091. doi: 10.1073/pnas.94.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Molina I, Baars S, Brill J A, Hales K G, Fuller M T, Ripoll P. J Cell Biol. 1997;139:1361–1371. doi: 10.1083/jcb.139.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.DeZwaan T M, Ellingson E, Pellman D, Roof D M. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Funabiki H, Murray A W. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 117.Raich W B, Moran A N, Rothman J H, Hardin J. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuriyama R, Kofron M, Essner R, Kato T, Dragas-Granoic S, Omoto C K, Khodjakov A. J Cell Biol. 1995;129:1049–1059. doi: 10.1083/jcb.129.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ando A, Kikuti Y Y, Kawata H, Okamoto N, Imai T, Eki T, Yokoyama K, Soeda E, Ikemura T, Abe K, et al. Immunogenetics. 1994;39:194–200. doi: 10.1007/BF00241260. [DOI] [PubMed] [Google Scholar]
- 120.McDonald H B, Goldstein L S. Cell. 1990;61:991–1000. doi: 10.1016/0092-8674(90)90064-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.