An inducible dynein heavy chain 1b mutant reveals that robust retrograde intraflagellar transport is required for flagellar assembly and function but not the maintenance of flagellar length.
Abstract
The maintenance of flagellar length is believed to require both anterograde and retrograde intraflagellar transport (IFT). However, it is difficult to uncouple the functions of retrograde transport from anterograde, as null mutants in dynein heavy chain 1b (DHC1b) have stumpy flagella, demonstrating solely that retrograde IFT is required for flagellar assembly. We isolated a Chlamydomonas reinhardtii mutant (dhc1b-3) with a temperature-sensitive defect in DHC1b, enabling inducible inhibition of retrograde IFT in full-length flagella. Although dhc1b-3 flagella at the nonpermissive temperature (34°C) showed a dramatic reduction of retrograde IFT, they remained nearly full-length for many hours. However, dhc1b-3 cells at 34°C had strong defects in flagellar assembly after cell division or pH shock. Furthermore, dhc1b-3 cells displayed altered phototaxis and flagellar beat. Thus, robust retrograde IFT is required for flagellar assembly and function but is dispensable for the maintenance of flagellar length. Proteomic analysis of dhc1b-3 flagella revealed distinct classes of proteins that change in abundance when retrograde IFT is inhibited.
Introduction
Cilia and flagella are dynamic structures whose assembly and maintenance require an active transport process known as intraflagellar transport (IFT; Kozminski et al., 1993; Pedersen and Rosenbaum, 2008; Scholey, 2008; Ishikawa and Marshall, 2011). Intraflagellar transport is mediated by ∼20 IFT proteins (Piperno and Mead, 1997; Cole et al., 1998), which assemble into macromolecular complexes called IFT particles. The IFT particles then associate into linear arrays known as IFT trains (Pigino et al., 2009), which move processively from the base of the flagellum out to the tip. This anterograde transport is driven by kinesin-2, a heterotrimeric complex composed of the FLA10 and FLA8 motor subunits (Walther et al., 1994) and the kinesin-associated protein KAP (Cole et al., 1993; Mueller et al., 2005). After their anterograde motion to the flagellar tip, IFT particles rearrange into a new set of IFT trains that move back to the base of the flagellum. This retrograde transport is powered by cytoplasmic dynein 1b, a large complex composed of the heavy chain motor subunit DHC1b (Pazour et al., 1999a; Porter et al., 1999; Signor et al., 1999) and numerous smaller components including D1bLIC (Perrone et al., 2003; Schafer et al., 2003; Hou et al., 2004; Hao et al., 2011a), FAP133 (Rompolas et al., 2007), and LC8 (Pazour et al., 1998). Both the precursors for flagellar assembly and the breakdown products of flagellar turnover are thought to associate with IFT particles (Qin et al., 2004; Hao et al., 2011b).
The primary evidence that anterograde IFT supplies precursors for flagellar assembly to the growing tip of the flagellum comes from analysis of mutations in the anterograde motor kinesin-2. Using a fla10 temperature-sensitive (ts) allele, it was shown that kinesin activity is required for IFT train motion (Kozminski et al., 1995), flagellar assembly and length maintenance (Kozminski et al., 1995), and the incorporation of new tubulin into the flagellar tip during steady-state turnover (Marshall and Rosenbaum, 2001). Furthermore, changes in protein abundance in fla10 flagella were used for the initial identification of the IFT proteins (Piperno and Mead, 1997; Cole et al., 1998). Thus, our understanding of anterograde IFT has resulted in large part from the availability of a conditional mutation in the anterograde motor that allows inducible shutoff of motor function.
Compared with anterograde transport, our understanding of the functional role of retrograde transport is less well developed. Chlamydomonas mutant strains with null deletions in DHC1b (dhc1b-1, Pazour et al., 1999a; stf-1, stf-2, Porter et al., 1999) have stumpy flagella, swollen with IFT and axonemal proteins that have apparently been stranded at the flagellar tip in the absence of return transport. Based on this phenotype, it was concluded that an important function of retrograde transport is to carry back IFT particles and kinesin motors so they can execute a new cycle of anterograde transport. It is believed that flagellar length cannot be maintained without this recycling, as the anterograde transport machinery becomes sequestered in the flagellum.
Unfortunately, constitutive mutations in DHC1b only reveal endpoint phenotypes and do not provide information about the dynamics by which these phenotypes arise. Consequently, it is impossible to determine if the swollen stumpy flagella of dhc1b-1 reflect a role of dynein in the initial assembly of flagella or in their subsequent maintenance. In the case of anterograde transport, the ts fla10 mutant demonstrated the requirement of anterograde IFT for flagellar maintenance by dynamically shortening its flagella when the mutant was shifted to the nonpermissive temperature (Kozminski et al., 1995). However, the only ts dynein mutant that has been isolated up to this point (dhc1b-2, Pazour et al., 1999b; Witman, 2011) does not clearly reveal the role of retrograde IFT in flagellar maintenance, as even at the permissive temperature it has impaired dynein function and short flagella full of accumulated IFT material (Fig. S1).
Here, we describe a new conditional mutation in dynein, dhc1b-3, which enables the inducible decoupling of retrograde IFT from anterograde transport. Strains carrying this mutation have full-length flagella and nearly wild-type levels of IFT at the permissive temperature, but lose the majority of retrograde IFT events after a shift to the nonpermissive temperature. Anterograde IFT also becomes partially decreased upon temperature-shifting the dhc1b-3 mutant; however, flagellar length is maintained with only mild shortening for many hours until cell division, when new flagella are unable to assemble. This result is dramatically distinct from the immediate flagellar shortening seen in fla10, and suggests that robust retrograde IFT is necessary for flagellar assembly but not length maintenance. We further analyzed the role of retrograde IFT by comparative proteomics, which revealed complex patterns of protein accumulation and depletion in dhc1b-3 flagella. In addition, dhc1b-3 cells at the nonpermissive temperature displayed changes in phototaxis, swimming speed, and flagellar beat frequency. Taken together, our results indicate that retrograde IFT is not simply a means to recycle anterograde IFT trains, but plays a multifaceted role in maintaining flagellar composition and function.
Results
Isolation and cloning of dhc1b-3, a temperature-sensitive DHC1b mutation
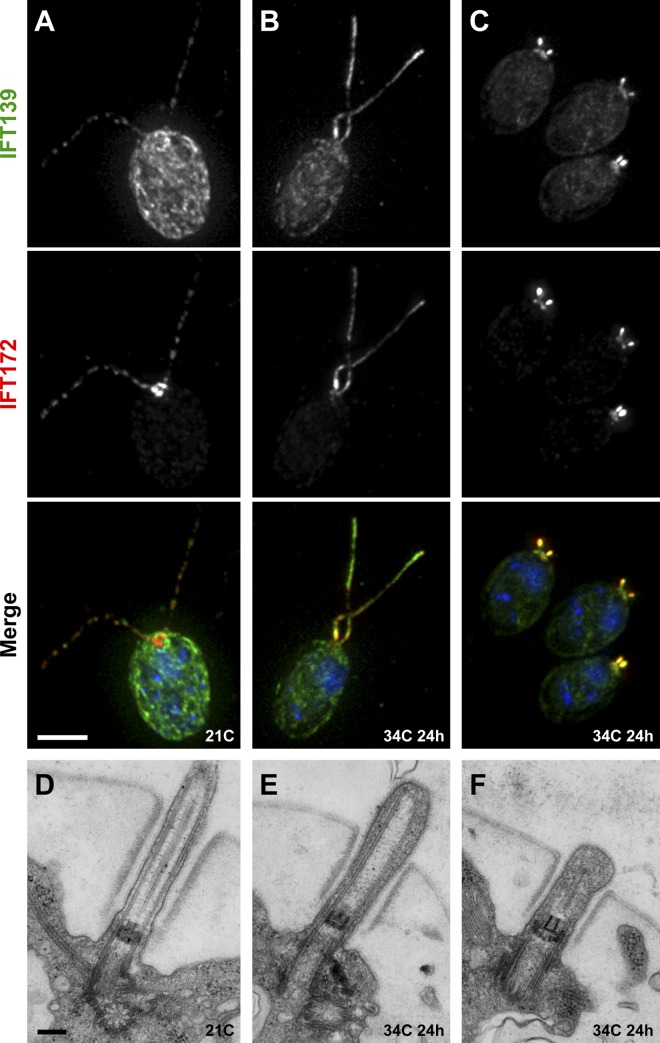
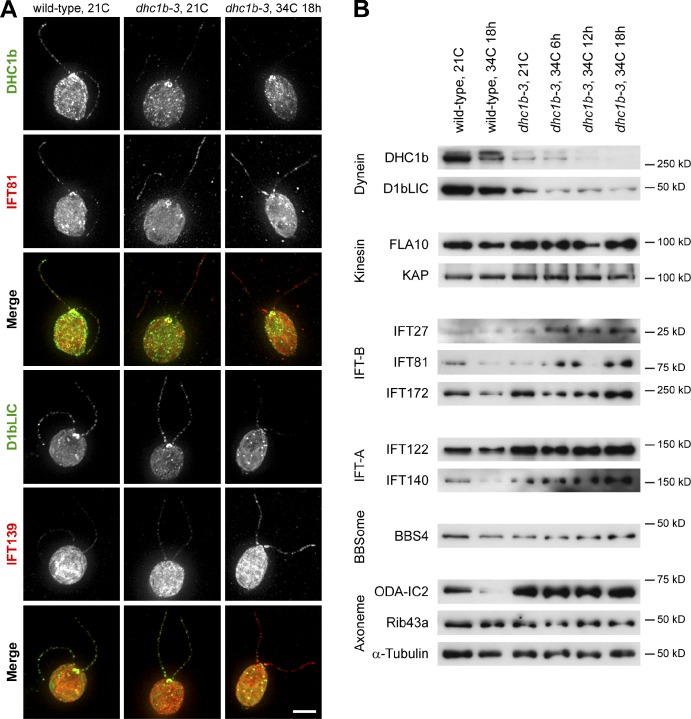
In an effort to obtain conditional mutations affecting retrograde IFT, we performed UV mutagenesis of Chlamydomonas cells, followed by a phenotypic screen for ts defects in cell motility (Fig. S2 E). Of the 122 punctate colonies isolated for further screening, 68 were bald, 13 had jerky swimming, 6 were paralyzed, 17 were sick, and 18 swam normally (screen false hits). Only 3 isolates exhibited ts motility defects. Two of these strains had conditional flagellar paralysis and were not further studied, whereas a single strain showed a robust and reversible defect in flagellar assembly. At 21°C, this mutant had full-length flagella containing a normal distribution of IFT proteins (Fig. 1 A) and appeared to have no ultrastructural defects in the axoneme or basal body (Fig. 1 D). However, after incubation for a day at 34°C, IFT proteins accumulated in flagella (Fig. 1, B and E) and flagella became stumpy and swollen with IFT material (Fig. 1, C and F). These stumpy flagella persisted until cells were shifted back to 21°C, at which temperature flagella regrew after several hours. The accumulation of IFT proteins in the mutant’s flagella at the nonpermissive temperature was typical of a retrograde IFT defect and was highly reminiscent of null mutants in DHC1b, the heavy chain of the retrograde dynein motor (dhc1b-1, Pazour et al., 1999a; stf-1, stf-2, Porter et al., 1999), which have stumpy swollen flagella.
Figure 1.
Temperature-sensitive “retrograde IFT” phenotype of isolated mutant strain dhc1b-3. (A–C) Immunofluorescence and (D–F) transmission electron microscopy (TEM) of the ts flagellar assembly mutant. Green, IFT139; red, IFT172; blue, DNA. At 21°C, the mutant appeared to have a regular distribution of IFT proteins along the flagella (A) and showed no discernible defects in axoneme or basal body ultrastructure (D). After 24 h at 34°C, flagella were either long (B) or stumpy (C) and in both cases showed a strong accumulation of IFT proteins. This accumulation of IFT proteins at 34°C was also apparent by TEM (E and F) as a buildup of electron-dense material between the axoneme and the flagellar membrane. Bars: (A–C) 5 µm; (D–F) 200 nm.
During initial analysis of tetrads obtained from backcrosses (Harris et al., 2009), we found that the mutant phenotype was tightly linked to the mating-type locus on chromosome 6. To perform higher resolution mapping, we crossed the mutant to the S1C5 polymorphic strain (Gross et al., 1988) and used PCR-based SNP analysis on the tetrad progeny to check for recombination between the mutation and several genetic markers (Fig. S2 A). This analysis tracked the mutation to a small region at the end of chromosome 6. Among the four potential flagellar genes in this region was DHC1b, our leading candidate based on phenotypic similarities to the null dhc1b-1 mutant.
Sequencing the entire length of the DHC1b cDNA from the mutant strain revealed a single base-pair mutation at 12313 bp in the coding sequence (Fig. S3 A). This A-to-T substitution results in a single amino acid switch from isoleucine to phenylalanine at position 4105 of the amino acid sequence (Fig. S3 B). The mutation occurs in the “C-sequence,” the C-terminal domain beyond the six ATPase domains (Kon et al., 2011, 2012). The C-sequence is present in the dynein heavy chains of many species (Numata et al., 2011), but is absent from budding yeast (Carter et al., 2011; Schmidt et al., 2012). ClustalX alignment (Larkin et al., 2007) of DHC1b C-sequences from several organisms revealed that the mutation occurs in a fully conserved hydrophobic residue in the middle of a small conserved hydrophobic motif (Fig. S3 C). Thus, the replacement of Ile with the bulkier Phe residue could presumably impede protein stability, a defect that would be exacerbated by the temperature shift. It is also possible that the mutation affects motor function, as the C-sequence is crucial for dynein processivity (Numata et al., 2011).
We named the mutant strain dhc1b-3 and developed a PCR assay to easily track the mutation, verifying that there is heterogeneity between dhc1b-3 and wild-type sequences at the mutated site (Fig. S2, B and C). To confirm that the dhc1b-3 phenotype is due to an impairment of the DHC1b gene, we created diploid strains by mating dhc1b-3 to the dhc1b-1 null mutant (Fig. S2 D). dhc1b-1 is a recessive mutation and would therefore show no phenotype if wild-type DHC1b function was present. The dhc1b-1 dhc1b-3 diploids exhibited conditional loss of flagella at 34°C with similar kinetics to the parental dhc1b-3 strain. Thus, the dhc1b-3 and dhc1b-1 strains do not complement, indicating that the dhc1b-3 phenotype is caused by a specific effect on the DHC1b gene.
Retrograde IFT is reduced in dhc1b-3 flagella
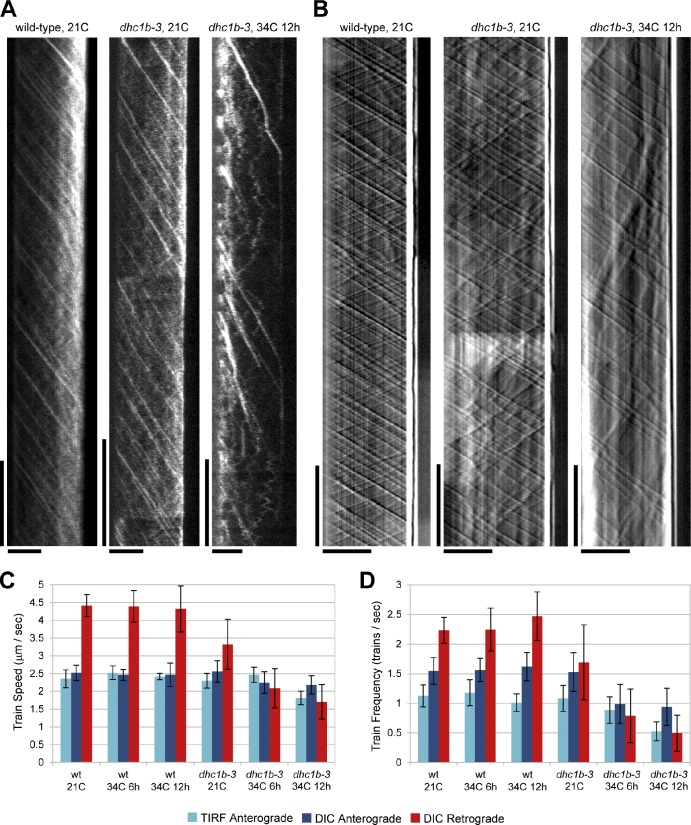
To determine how the dhc1b-3 mutation affects IFT, we imaged the flagella of live cells by two different light microscopy techniques. After crossing dhc1b-3 cells to a strain expressing the KAP subunit tagged with green fluorescent protein (KAP-GFP; Mueller et al., 2005), we visualized KAP-GFP movement using total internal reflection fluorescence (TIRF) microscopy (Fig. 2 A and Videos 1–3; for review of technique see Engel et al., 2009a). One distinct feature of KAP-GFP is that there is a high level of background fluorescence within flagella instead of clear retrograde traces (Engel et al., 2009b). Although dhc1b-3 KAP-GFP looked similar to wild-type at 21°C, anterograde IFT became less frequent at 34°C and the background fluorescence decreased. Interestingly, in the absence of strong background signal, we observed some KAP-GFP proteins undergoing bidirectional oscillatory movements, a behavior we have never witnessed in wild-type KAP-GFP flagella. To better understand how retrograde transport is affected in dhc1b-3 cells, we also visualized IFT using differential interference contrast (DIC) microscopy (Fig. 2 B and Videos 4–6; for reviews of technique see Kozminski, 1995; Dentler et al., 2009). At 21°C, dhc1b-3 IFT appeared similar to wild-type, though retrograde traces were slightly less processive. However, after 6–12 h at 34°C, retrograde transport events were dramatically slower and less frequent. Mirroring the KAP-GFP imaging, anterograde IFT frequency also decreased at 34°C.
Figure 2.
TIRF and DIC microscopy measurements of IFT in wild-type and dhc1b-3 cells at 21 and 34°C. (A) Kymographs generated from TIRF time series of wild-type and dhc1b-3 cells expressing KAP-GFP at 21 and 34°C, as indicated. (B) Kymographs generated from DIC time series of wild-type and dhc1b-3 cells at 21 and 34°C. For A and B, the x-axis is distance along the length of the flagellum (the flagellar base is on the left and the flagellar tip is on the right) and the y-axis is time (earliest times are at the top). Bars (vertical) 5 s; (horizontal) 5 µm. Quantification of IFT train speeds (C) and frequencies (D) from the TIRF and DIC datasets. Error bars indicate standard deviation. In dhc1b-3 flagella, retrograde IFT is moderately reduced at 21°C and strongly inhibited at 34°C. The large drop in dhc1b-3 retrograde IFT at 34°C is accompanied by a decrease in anterograde IFT frequency. The lower frequency of anterograde trains measured by TIRF vs. DIC likely means that the KAP-GFP fluorescence of some trains was not detected. Nflagella = 231 (TIRF), 109 (DIC).
After imaging, we quantified IFT speeds and frequencies by kymograph analysis (Fig. 2, C and D). Incubating wild-type cells at 34°C for 6 or 12 h had a negligible effect on IFT. Anterograde train speed was 2.5 µm/s, retrograde speed was 4.4 µm/s, anterograde frequency was 1.6 trains/s (1.1 trains/s for TIRF), and retrograde frequency was 2.3 trains/s. When dhc1b-3 cells were grown at 21°C, anterograde speed and frequency were unchanged from wild-type levels, but retrograde speed was decreased to 3.3 µm/s and retrograde frequency dropped to 1.7 trains/s. Retrograde train speed in dhc1b-3 flagella was further reduced to 2.1 µm/s after 6 h at 34°C and 1.7 µm/s after 12 h. Most dramatically, dhc1b-3 retrograde frequency dropped to 0.8 trains/s and 0.5 trains/s after 6 and 12 h incubation at 34°C, respectively. The impact of the dhc1b-3 mutation on anterograde IFT was less severe. Anterograde speed was only marginally affected, decreasing to 2.2 µm/s (1.8 µm/s for TIRF) after 12 h at 34°C. However, anterograde frequency experienced a significant reduction to 1.0 trains/s (0.9 trains/s for TIRF) after 6 h at 34°C and 0.9 trains/s (0.5 trains/s for TIRF) after 12 h. IFT train size (Engel et al., 2009b) was not measured in these experiments due to inadequate GFP expression in the dhc1b-3 KAP-GFP strain.
Unlike the ts kinesin mutant fla10, IFT does not completely stop in dhc1b-3 cells during the first 12 h at 34°C. Instead, retrograde IFT speed and frequency are greatly reduced, coupled with a moderate decrease in anterograde IFT frequency. How this IFT defect may be linked to the dhc1b-3 flagellar phenotype is explored further in the Discussion.
dhc1b-3 has distinct kinetics of flagellar loss
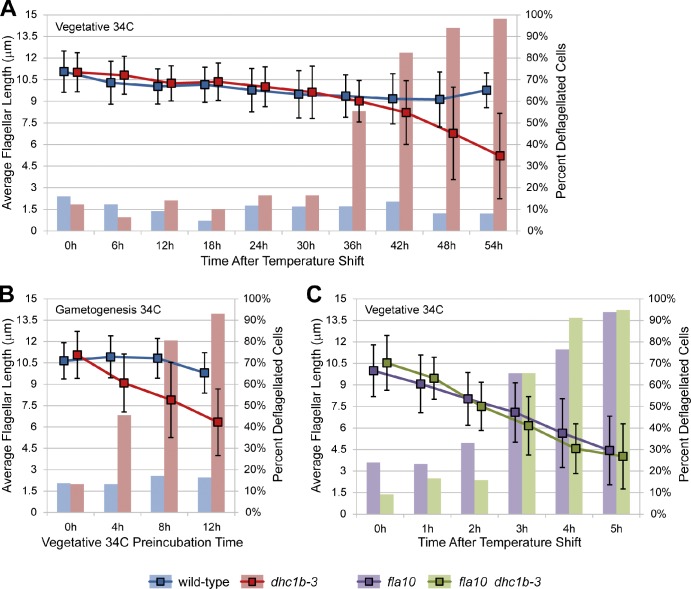
Although several characterized Chlamydomonas mutants have ts defects in flagellar assembly (most notably the fla mutants; Adams et al., 1982), the kinetics of flagellar loss in dhc1b-3 are strikingly different. Although fla10 flagella began shortening immediately at 34°C and were lost within 5 h (Fig. 3 C; Kozminski et al., 1995; Marshall and Rosenbaum, 2001), vegetative dhc1b-3 cells maintained nearly full-length flagella for over a day at 34°C before finally losing them (Fig. 3 A). Unlike fla10, only mild flagellar shortening was observed in dhc1b-3 cells until after the majority of the population had undergone deflagellation. The difference between fla10 and dhc1b-3 shortening kinetics suggests that although both anterograde and retrograde IFT are necessary for flagellar assembly, robust retrograde IFT may not be required for the maintenance of flagellar length.
Figure 3.
Kinetics of dhc1b-3 flagellar loss in asynchronous vegetative cells, during gametogenesis, and in fla10 dhc1b-3 double mutants. Bar graphs depict the percentage of deflagellated cells, and line graphs show the average flagellar length of flagellated cells. Error bars indicate standard deviation. (A) Vegetative wild-type and dhc1b-3 cells were grown in TAP media for 2 d at 21°C (dense, post-log phase cultures) and then shifted to 34°C. In this experiment, dhc1b-3 flagella were maintained with only mild shortening (10–20%) for 30 h. In less dense, rapidly dividing cultures, some dhc1b-3 cells lost flagella as early as 18 h at 34°C (not depicted). The cause of this variability is unknown, but it is likely related to the growth phase of the culture. It was not determined whether flagellar loss in these asynchronous cultures was due to cell division (as shown in Fig. S4 A) or other factors, such as retrograde IFT dropping below a critical threshold. (B) Vegetative cells were grown in TAP media at 21°C for 1 d, incubated for increasing periods of time at 34°C, and then induced to differentiate synchronously into gametes by diluting cells into M-N media followed by 18 h incubation at 34°C. Pre-incubation for 4–8 h at 34°C inhibited the assembly of new flagella in dhc1b-3 gametes. (C) Vegetative fla10 and fla10 dhc1b-3 double mutant strains were grown in TAP media for 2 d at 21°C and then shifted to 34°C. Both strains had the same kinetics of flagellar shortening and loss, which was far more rapid than flagellar loss in the dhc1b-3 single mutant. Nflagella = 1,849 (A), 833 (B), 940 (C) from single experiments.
In synchronized log growth-phase cultures, dhc1b-3 cells were flagellated at both 21 and 34°C until flagella were resorbed for cell division. After division, flagella regrew at 21°C but were unable to regrow at 34°C (Fig. S4 A). This implies that dhc1b-3 cells can maintain established flagella at 21 and 34°C, but can only assemble new flagella at 21°C. To further explore this hypothesis, we forced vegetative cells to undergo synchronous differentiation into gametes by nitrogen starvation (Harris et al., 2009). Incubation of vegetative dhc1b-3 cells for 4–8 h at 34°C before resuspending cells in nitrogen-free media inhibited flagellar assembly in the resulting gametes (Fig. 3 B). This again suggests that after several hours at 34°C, dhc1b-3 cells cannot assemble new flagella.
We created a fla10 dhc1b-3 double mutant via mating and found that this strain has identical kinetics of flagellar shortening and loss at 34°C as the fla10 single mutant (Fig. 3 C). Thus, kinesin inhibition is epistatic to dynein inhibition for the maintenance of flagellar length. This result is not surprising, as kinesin-powered anterograde transport brings the axonemal precursors required for length maintenance to the flagellar tip. One caveat is that the kinetics of kinesin inactivation in fla10 may be faster than the kinetics of dhc1b-3 dynein inhibition.
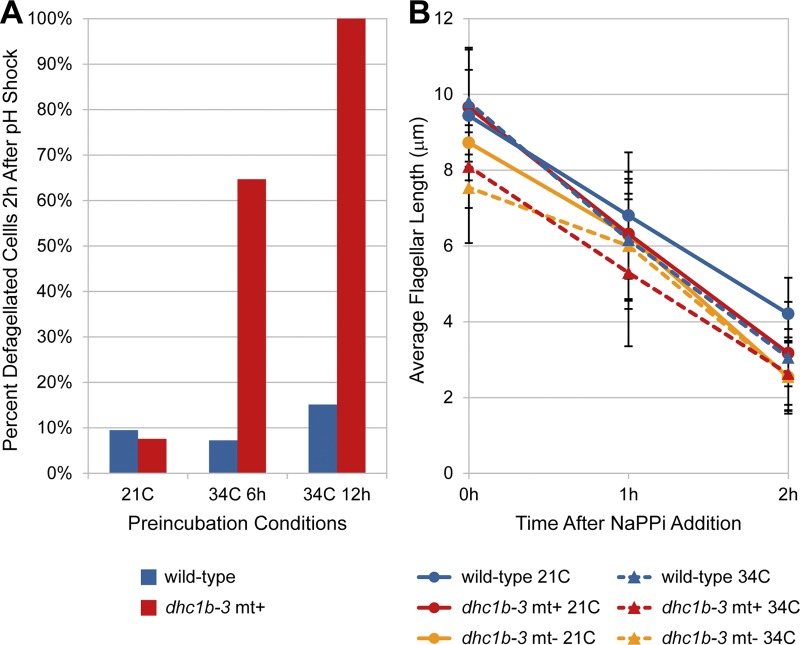
We next observed the effects of the dhc1b-3 mutation on flagellar assembly more directly by using pH shock regeneration. At 21°C, the extent (Fig. 4 A) and kinetics (Fig. S4 B) of dhc1b-3 flagellar regeneration were comparable to wild-type. In agreement with the gametogenesis measurements (Fig. 3 B), dhc1b-3 cells that were incubated at 34°C for 6 h before pH shock showed a strong flagellar assembly defect during 34°C regeneration, whereas preincubation for 12 h at 34°C completely inhibited regeneration (Fig. 4 A). Furthermore, when dhc1b-3 cells were incubated at 34°C for increasing periods of time before pH shock, and then allowed to recover at 21°C, flagellar assembly was delayed and proceeded at a slower rate (Fig. S4 B). These results again illustrate the strict requirement for dynein function in flagellar assembly.
Figure 4.
pH shock–induced flagellar regeneration and NaPPi-induced flagellar shortening in wild-type and dhc1b-3 cells at 21 and 34°C. (A) Wild-type and dhc1b-3 cells were incubated at 21 or 34°C (6 h, 12 h), deflagellated with pH shock, allowed to recover at 21 or 34°C, then scored after 2 h recovery for the percentage of deflagellated cells (regeneration failure). Although the extent of dhc1b-3 flagellar regeneration at 21°C was similar to wild-type, dhc1b-3 cells showed strong flagellar assembly defects at 34°C. Ncells = 2,335 from two experiments. (B) Cells were grown in M media, incubated for 12 h at 21 or 34°C, and then treated with 20 mM NaPPi. The flagella of wild-type and dhc1b-3 cells (mt+ and mt− strains) shortened with similar rates (∼3 µm/h) at both 21 and 34°C. Thus, NaPPi-induced flagellar shortening may be independent of retrograde IFT. The starting flagellar lengths of both mutant and wild-type strains were slightly shorter than normal, likely due to unknown variations in the culture conditions. Error bars indicate standard deviation. Nflagella = 540 from a single experiment.
Previous studies have indicated a role for IFT in active disassembly of flagella, as the rate of sodium pyrophosphate (NaPPi)–induced flagellar shortening is decreased in fla10 cells at the nonpermissive temperature (Pan and Snell, 2005). Because fla10 abrogates both anterograde and retrograde transport, this result does not reveal which phase of IFT is involved. One appealing hypothesis is that retrograde transport plays a key role in disassembly by facilitating the removal of flagellar proteins. However, we found that the rate and extent of NaPPi-induced flagellar shortening was not reduced in the dhc1b-3 mutant at 34°C compared with wild-type cells grown at 34°C (Fig. 4 B), indicating that retrograde IFT may not be required for active disassembly. Thus, the increased rate of flagellar shortening in the presence of IFT is likely due to anterograde transport, which has been shown to carry the microtubule-depolymerizing factor kinesin-13 into flagella in response to NaPPi treatment (Piao et al., 2009).
The depletion of dynein from dhc1b-3 flagella is correlated with the accumulation of IFT proteins, but not kinesin
Given the dramatic reduction in dhc1b-3 retrograde IFT, we decided to check the flagellar abundance of the retrograde dynein motor and IFT proteins by immunofluorescence microscopy (Fig. 5 A). Relative to wild-type, DHC1b fluorescence was markedly decreased in dhc1b-3 flagella at both 21°C and after 18 h at 34°C, though some protein remained localized around the basal bodies. This drop in DHC1b abundance was mirrored by the flagellar depletion of D1bLIC, a light intermediate chain of cytoplasmic dynein 1b (Hou et al., 2004). At 34°C, D1bLIC was also displaced from the basal bodies of many dhc1b-3 cells, localizing to punctate spots throughout the cytoplasm. As dynein fluorescence decreased, the intensities of IFT-B component IFT81 and IFT-A protein IFT139 increased. This result is not unexpected and indeed has been predicted based on the accumulation of IFT proteins in the stumpy flagella of DHC1b-null mutants (Pazour et al., 1999a; Porter et al., 1999) and the short flagella of fla14 (deletion of the dynein light chain LC8; Pazour et al., 1998). Nonetheless, to our knowledge this is the first direct demonstration that the induced depletion of cytoplasmic dynein 1b from full-length flagella (and subsequent drop in retrograde IFT, Fig. 2) results in the flagellar accumulation of IFT proteins.
Figure 5.
Accumulation and depletion of flagellar proteins in wild-type and dhc1b-3 cells at 21 and 34°C. (A) Immunofluorescence images of wild-type cells at 21°C (left column), dhc1b-3 cells at 21°C (middle column), and dhc1b-3 cells after 18 h at 34°C (right column), showing the relationship between dynein depletion and IFT protein accumulation. The top merged images are DHC1b (green) and IFT81 (red). The bottom merged images are D1bLIC (green) and IFT139 (red). Bar, 5 µm. (B) Western blots of flagella isolated from wild-type and dhc1b-3 cells at 21°C and increasing time at 34°C, as indicated. Dynein depletion correlated with the accumulation of IFT-B and IFT-A proteins, while the levels of kinesin, BBSome, and many axonemal proteins are unaffected (quantified in Table S2).
To expand our investigation to a wider scope of proteins and time points, we performed Western blot analysis of isolated flagella from wild-type and dhc1b-3 cells (Fig. 5 B). DHC1b and D1bLIC levels were greatly reduced in dhc1b-3 flagella at 21°C, and decreased even further after the mutant was incubated at 34°C. In agreement with the immunofluorescence data, this depletion of dynein was accompanied by an accumulation of IFT-B proteins IFT27, IFT81, and IFT172 (only at 34°C), as well as IFT-A proteins IFT122 and IFT140 (at both 21 and 34°C). We saw no change in BBS4, a component of the BBSome that is trafficked by IFT (Lechtreck et al., 2009). The levels of axonemal proteins α-tubulin and Rib43a (protofilament ribbons; Norrander et al., 2000) remained constant, although ODA-IC2 (intermediate chain of outer arm dynein, ODA6; Mitchell and Kang, 1991) appeared to accumulate in dhc1b-3 flagella.
Curiously, the depletion of dynein did not cause significant flagellar accumulation of kinesin proteins FLA10 and KAP. This result implies that, unlike IFT proteins, kinesin may not rely on dynein for transport out of flagella. Such a hypothesis is consistent with the absence of KAP-GFP retrograde traces in wild-type cells (Fig. 2 A; Engel et al., 2009a,b), immunofluorescence data showing that FLA10 remains concentrated at the peri-basal body region of dhc1b-1 cells in contrast to IFT protein redistribution into swollen stumpy flagella (Pazour et al., 1999a), and Western blotting evidence that FLA10 and KAP do not accumulate in flagella in the absence of D1bLIC (Pedersen et al., 2006). Conversely, in Caenorhabditis elegans KAP-GFP has been observed to undergo retrograde transport and to accumulate at the ciliary tips of DHC1b mutants (che-3; Signor et al., 1999).
It is also interesting to note that despite the greatly reduced levels of DHC1b protein in dhc1b-3 flagella at 21°C (Fig. 5 B), retrograde IFT speed and frequency were only moderately decreased (Fig. 2, C and D) and the kinetics of flagellar assembly remained comparable to wild type (Fig. S4). This discrepancy indicates that there is normally an excess of dynein in the flagellum, and begs the question of what function this extra dynein protein is performing. Perhaps it is simply surplus to ensure robust IFT, or perhaps it plays a role in specialized processes such as signal transduction, which our assays do not address.
2D-DIGE proteomic analysis of protein accumulation and depletion in dhc1b-3 flagella
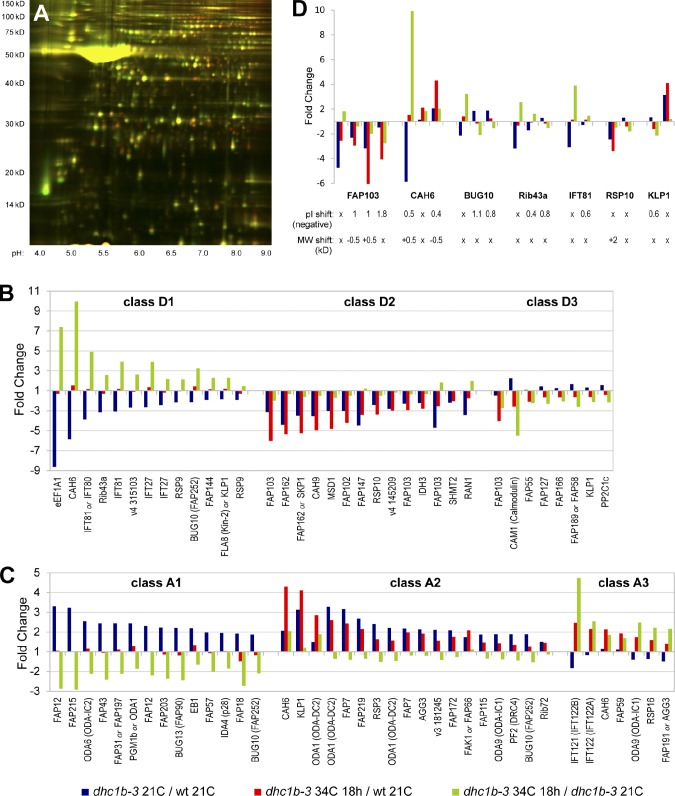
To get a more comprehensive and unbiased view of how defective retrograde IFT affects the abundance of flagellar proteins, we took a comparative proteomic approach using 2D-DIGE (two-dimensional difference in gel electrophoresis; Viswanathan et al., 2006). Isolated flagella from wild-type cells at 21°C, dhc1b-3 cells at 21°C, and dhc1b-3 cells after 18 h at 34°C were each labeled with a different fluorescent dye and run together on a 2D acrylamide gel (Figs. 6 A and S5 A). Spots with the greatest changes in abundance between the three samples were picked for identification via mass spectrometry. The majority of the proteins identified by this 2D-DIGE analysis proved to be known components of the flagellar proteome (Table S3; Pazour et al., 2005). Although numerous spots were proteins of known function, many other proteins were uncharacterized, including a wide assortment of FAPs (flagellar associated proteins; Pazour et al., 2005). The identified spots were clustered into classes based on their relative accumulation or depletion between the three samples (Fig. 6, B and C; Table 1). Proteins that group together in the same class may be transported together by IFT or share similar functions. For example, IFT complex B proteins cluster into class D1, IFT complex A proteins group into class A3, and class A2 is rich in axonemal proteins that may be accumulated turnover products. The potential relationships between the proteins in each class are further explored in the Discussion.
Figure 6.
2D-DIGE proteomic analysis of flagellar protein accumulation and depletion in dhc1b-3 cells at 21 and 34°C. (A) Fluorescence image of the 2D-DIGE analytical gel (dhc1b-3 21C = green; dhc1b-3 34C 18 h = red; for overlays of all three samples see Fig. S5). The large yellow spot at ∼50 kD is tubulin. (B and C) Depletion and accumulation profiles for every spot that was identified by mass spectrometry. Fold change is a spot’s relative fluorescence between two samples on the analytical gel (1 = equal abundance for both samples). Class D1 = depleted in only dhc1b-3 21C vs. wild-type 21C (accumulated in dhc1b-3 34C 18 h vs. dhc1b-3 21C); class D2 = depleted in both dhc1b-3 21C vs. wild-type 21C and dhc1b-3 34C 18 h vs. wild-type 21C; class D3 = depleted in only dhc1b-3 34C 18 h vs. wild-type 21C (depleted in dhc1b-3 34C 18 h vs. dhc1b-3 21C); class A1 = accumulated in only dhc1b-3 21C vs. wild-type 21C (depleted in dhc1b-3 34C 18 h vs. dhc1b-3 21C); class A2 = accumulated in both dhc1b-3 21C vs. wild-type 21C and dhc1b-3 34C 18 h vs. wild-type 21C; class A3 = accumulated in only dhc1b-3 34C 18 h vs. wild-type 21C (accumulated in dhc1b-3 34C 18 h vs. dhc1b-3 21C). (D) The profiles of proteins identified in multiple spots that group into more than one class, suggesting IFT-related protein modifications. Negative pI shifts (minus sign omitted) and MW shifts, both relative to the spot with the highest pI, are noted.
Table 1.
Flagellar proteins with changing abundance identified by 2D-DIGE analysis
| 21°C only | 21 and 34°C | 34°C only | |
| dhc1b-3 depletion vs. wt | Class D1 (mut-34A): eEF1A1, CAH6, IFT80, Rib43a, IFT81, v4 315103, IFT27, RSP9, BUG10 (FAP252), FAP144, FLA8 or KLP1 | Class D2 (mut-34NC): FAP103, FAP162, FAP102, FAP147, RSP10, v4 145209, RAN1 | Class D3 (mut-34D): FAP103, CAM1 (calmodulin), FAP55, FAP127, FAP166, FAP189 or FAP58, KLP1, PP2C1c |
| dhc1b-3 accumulation vs. wt | Class A1 (mut-34D): FAP12, FAP215, ODA6 (ODA-IC2), FAP43, FAP31 or FAP197, PGM1b or ODA1, FAP203, BUG13 (FAP90), EB1, FAP57, IDA4 (p28), FAP18, BUG10 (FAP252) | Class A2 (mut-34NC): CAH6, KLP1, ODA1 (ODA-DC2), FAP7, FAP219, RSP3, AGG3, FAP172, FAK1 or FAP66, FAP115, ODA9 (ODA-IC1), PF2 (DRC4), BUG10 (FAP252), Rib72 | Class A3 (mut-34A): IFT121, IFT122, CAH6, FAP59, ODA9 (ODA-IC1), RSP16, FAP191 or AGG3 |
| No change | RSP10, Rib43a, FAP164, ICL1, ODA13 (ODA-LC6), IDA5 (actin), IFT81, FAP161, IFT88, FAP52 | ||
Proteins are grouped by relative accumulation or depletion in dhc1b-3 flagella at 21 and 34°C vs. wild-type flagella at 21°C (as plotted by blue and red bars in Fig. 6). Duplicate hits, tubulin, and proteins not found in the flagellar proteome have been excluded from this list. For the complete catalogue and description of analyzed 2D-DIGE spots, see Fig. S5 and Table S3. Classes D1 and A3 are proteins accumulated in dhc1b-3 flagella after 18 h at 34°C vs. dhc1b-3 flagella at 21°C (as plotted by green bars in Fig. 6, indicated on the table by “mut-34A”). Classes D3 and A1 are proteins depleted in dhc1b-3 flagella after 18 h at 34°C vs. dhc1b-3 flagella at 21°C (indicated by “mut-34D”). Proteins in classes D2 and A2 experience no change in flagellar abundance when dhc1b-3 cells are shifted from 21 to 34°C (indicated by “mut-34NC”). Proteins are listed in descending order of fold accumulation or depletion (except the “No change” class).
Although many spots were identified as duplicate hits for the same proteins (Table S3), some proteins had spots that grouped into more than one class (Fig. 6 D), indicating that these proteins may receive modifications corresponding to different stages of transport or incorporation into the axoneme. BUG10, Rib43a, and IFT81 all have one spot that clusters with IFT-B proteins (class D1), but also have additional spots with acidic shifts to their isoelectric points (pI) that could indicate phosphorylation. It is plausible that such a modification could be added to these proteins after they have completed anterograde transport. As CAH6 has different spots that group with both IFT-B (class D1) and IFT-A (class A3) proteins, we may have identified forms of CAH6 that are trafficked by each phase of IFT. Interestingly, the transition from IFT-B to IFT-A grouping appears to be accompanied by a slight decrease in CAH6 molecular weight (MW). We identified four FAP103 spots that become increasingly acidic as the spots go from depletion in dhc1b-3 flagella primarily at 21°C, to depletion at both 21 and 34°C, and finally depletion at only 34°C. These changes in pI are likely related to the protein’s function as a nucleoside-diphosphate kinase. Although it is interesting that the activity of FAP103 could be linked to IFT, without further characterization of the protein’s role in flagella we cannot speculate about what this connection could be.
Phototaxis, swimming speed, and flagellar beat frequency are altered in dhc1b-3 cells
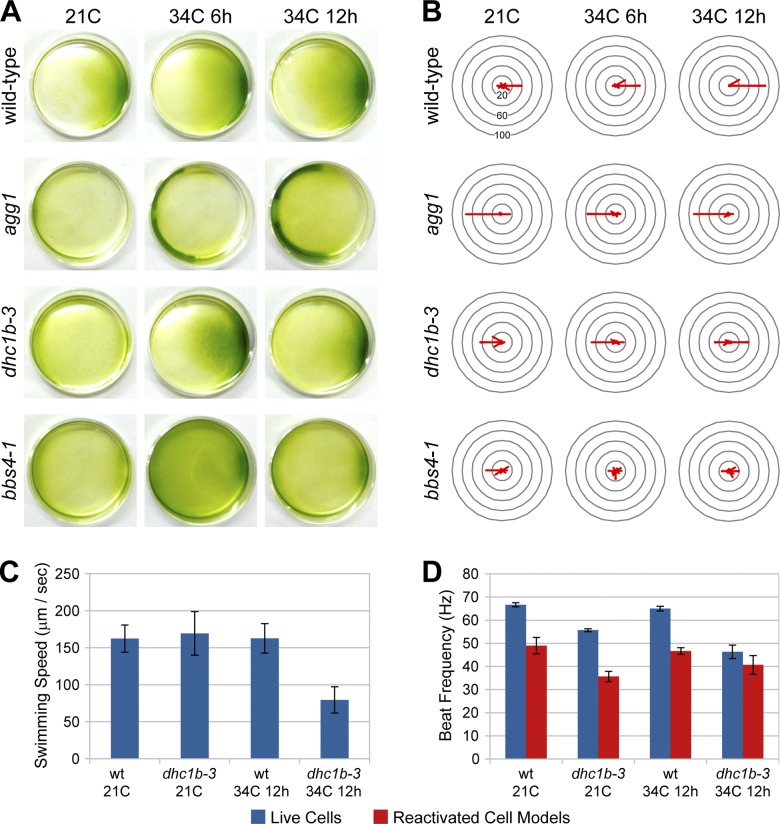
Although our results thus far indicate that fully functional retrograde IFT is not required for the maintenance of flagellar length, the changes in dhc1b-3 flagellar protein composition suggest that retrograde IFT may have an influence on flagellar function. We therefore tested several flagellar functions to see if they are affected by the reduction of retrograde IFT. In dish-based (Fig. 7 A) and single-cell (Fig. 7 B) phototaxis assays at both 21 and 34°C, wild-type cells swam toward the light source and agg1 cells (Smyth and Ebersold, 1985) swam away from the light. However, dhc1b-3 cells showed a switch from negative to positive phototaxis when shifted from 21 to 34°C (Fig. 7, A and B). This switch in phototactic sign was seen after 6 h at 34°C in the dish assays and after 12 h at 34°C in the single-cell measurements, a disparity that may be due to intrinsic differences between the assays. Bulk cell migration in the dish assay was the result of 15 min of continuous light exposure, whereas single-cell measurements were acquired 30 s after turning on the light source, before cells could accumulate at one side on the dish. Collectively, it may be inferred that dhc1b-3 cells incubated at 34°C for 6 h are at an intermediate stage in the switch from negative to positive swimming direction, and that by 12 h at 34°C they exhibit strong positive phototaxis. Because dhc1b-3 cells at 21°C showed the opposite phototaxis response to the wild-type strain from which they were generated, moderate reductions in retrograde IFT that do not affect anterograde IFT or flagellar length may still significantly influence cell signaling. Mutations in the BBSome component BBS4 have also been shown to affect phototaxis (bbs4-1; Lechtreck et al., 2009). Although reduced retrograde IFT in dhc1b-3 cells would also impair traffic of the BBSome, the dhc1b-3 and bbs4-1 phototaxis defects are distinct. Instead of switching the sign of phototaxis, bbs4-1 cells swam in random directions, irrespective of temperature. However, this difference may be due to the fact that the bbs4-1 mutant was generated from an agg1 strain.
Figure 7.
Effects of dhc1b-3 on phototaxis, swimming speed, and flagellar beat frequency. Phototaxis direction of wild-type, agg1, dhc1b-3, and bbs4-1 strains at 21 and 34°C, measured by the bulk migration of cells in dish assays (A) and the swimming trajectory of individual cells (B). For A and B, green light illumination is directed from the right. The compass plots in B indicate the percentage of cells swimming in a particular direction relative to the light source, grouped into 30-degree bins. Swimming speed (C) and flagellar beat frequency (D) of wild-type and dhc1b-3 cells at 21 and 34°C. Reactivated cell models are fixed permeabilized cells that have been treated with ATP, reinitiating flagellar beating (Kamiya and Witman, 1984). Error bars indicate standard deviation.
The swimming speed of dhc1b-3 cells was normal at 21°C, but was greatly diminished after 12 h at 34°C (Fig. 7 C). Furthermore, the beat frequency of dhc1b-3 flagella was decreased at both 21 and 34°C (Fig. 7 D), a telltale sign of an axonemal outer dynein arm (ODA) defect. Because this drop in beat frequency manifests in permeabilized ATP-reactivated cell models in addition to live cells, it is likely caused by a structural defect in the axoneme (although we did not identify an obvious deficiency in TEM cross sections of mutant axonemes; unpublished data). Thus, it appears that retrograde IFT helps maintain both physical (beat frequency) and signaling (phototaxis sign) functions of Chlamydomonas flagella. This is likely related to the turnover of flagellar proteins, a connection that we see some evidence for in the 2D-DIGE results (see Discussion). It will be interesting to determine whether additional flagellar functions, such as mating, are impacted by the dhc1b-3 mutation.
Discussion
2D-DIGE proteomics reveals distinct classes of IFT-related flagellar proteins
Class D1 proteins accumulate in dhc1b-3 flagella at 34°C vs. 21°C (Fig. 6, green bars), and include the IFT complex B proteins IFT81, IFT80, and IFT27. The increase in IFT81 and IFT27 abundance is consistent with our immunofluorescence and Western blot measurements (Fig. 5). Other proteins that group into the D1 profile may interact with IFT complex B as obligate cargos, including uncharacterized proteins FAP144, BUG10, and v4 315103. Of particular interest is eukaryotic translation elongation factor 1A1 (eEF1A1), a small GTPase that is involved in a diverse array of cellular processes in addition to its canonical function of delivering aa-tRNA to the ribosome (Mateyak and Kinzy, 2010). The 100-kD spot was double the expected MW for eEF1A1 and was also a 100% confidence hit for α-tubulin. Whereas tubulin hits for other spots in this study are less compelling because they are near tubulin’s expected 50 kD, this spot was well away from the tubulin region of the gel, at a MW and pI predicted for an eF1A1-tubulin dimer. Although it seems unlikely that such a dimer would survive denaturing electrophoresis, eEF1A does bind directly to microtubules (Moore and Cyr, 2000). eEF1A has been shown in different studies to bundle, stabilize, and sever microtubules (Durso and Cyr, 1994; Shiina et al., 1994; Moore et al., 1998). Although these diverse observations do not paint a clear picture of eEF1A1 in flagella, it is enticing for flagellar length control models that a small GTPase regulator of both translation and microtubule dynamics may be an obligate cargo of anterograde IFT.
The other group of proteins that accumulate in dhc1b-3 flagella at 34°C vs. 21°C is class A3, which includes the IFT complex A components IFT121 (IFT122B, IFTA-1) and IFT122 (IFT122A, DAF-10). Both proteins are required for retrograde transport and IFT122 has been implicated in the ciliary traffic of signaling proteins, including Gli2, Gli3, and TULP3 (Qin et al., 2001; Blacque et al., 2006; Cortellino et al., 2009; Mukhopadhyay et al., 2010; Ocbina et al., 2011; Qin et al., 2011). Other accumulated proteins in the A2 and A3 classes are likely cargoes that are ordinarily trafficked by retrograde IFT out of the flagella. These proteins include axonemal components such as ODA-DC2 (ODA1), ODA-IC1 (ODA9), KLP1, FAK1, RSP16, RSP3, DRC4 (PF2), FAP7, FAP219, as well as the “membrane plus matrix” proteins CAH6 and AGG3. The accumulation of ODA proteins and FAK1 could be indicative of outer dynein arm defects that may underlie the reduction in flagellar beat frequency (Fig. 7 D). CAH6 appears to be an abundant IFT cargo, with prominent spots in classes associated with IFT-B (D1), IFT-A (A3), and accumulated cargo (A2). As Chlamydomonas flagella are the only part of the cell membrane in direct contact with the extracellular environment, CAH6 may be using IFT to facilitate CO2 metabolism. The loss of ∼1 kD as CAH6 transitions from IFT-B– to IFT-A–associated forms could be related to this function. The accumulation of AGG3 is interesting as it may explain the change in dhc1b-3 phototaxis direction (Fig. 7, A and B). AGG3 RNAi induces the same phenotype as the agg1 mutant, where cells swim away from light (Iomini et al., 2006). Thus, it is logical that the accumulation of AGG3 in dhc1b-3 flagella could cause a switch to positive phototaxis.
Class A1 proteins are depleted from dhc1b-3 flagella at 34°C vs. 21°C, and have the opposite abundance as the IFT-B proteins in class D1. Most of the A1 proteins are uncharacterized (FAP12, FAP215, FAP31, FAP43, FAP203, BUG13, FAP57, BUG10, and FAP18) and many are in the membrane plus matrix fraction. One of the few well-characterized proteins in the group is EB1, which is found at the flagellar tip and stabilizes microtubules (Pedersen et al., 2003; Schrøder et al., 2007). As very few factors have been identified that regulate incorporation and removal of axonemal components, it would be of great interest if any of these uncharacterized proteins share EB1’s localization at the flagellar tip.
The second group of proteins depleted from dhc1b-3 flagella at 34°C vs. 21°C is class D3, which also primarily consists of uncharacterized proteins. Calmodulin has the opposite accumulation profile from IFT121, potentially providing a hint about how calmodulin is transported in flagella. FAP55, FAP127, FAP189/FAP58, and FAP166 are all axonemal proteins with very similar D3 profiles, so perhaps some of these structural proteins interact or even preassemble into a larger complex before being carried into flagella by IFT. FAP103 and FAP162 are repeatedly found among the top hits in classes D2 and D3. These two proteins are exclusively found in the membrane plus matrix fraction and may rely on DHC1b activity for entry into flagella.
Interestingly, IFT88 clusters into the “no change” class (Table 1). Although this could mean that there is something fundamentally different about the transport of IFT88, it should be noted that IFT81 also has one spot that sees no change in abundance, so we may have simply not picked the IFT88 spot that fits the D1 profile. Regardless, this observation warrants additional investigation.
The relationship between anterograde and retrograde IFT, and implications for flagellar length control
One clear conclusion that can be drawn from our data is that anterograde and retrograde IFT are coupled, but it is not a 1:1 relationship (Table 2). It may be easiest to understand the dhc1b-3 mutant’s flagellar phenotype in respect to this loose coupling. In dhc1b-3 cells at 21°C, anterograde IFT appears unaffected by a moderate ∼25% decrease in retrograde IFT speed and frequency relative to wild-type levels. It is perhaps no surprise then that dhc1b-3 flagellar assembly at 21°C is indistinguishable from wild type. After incubating dhc1b-3 cells for 12 h at 34°C, retrograde speed drops to ∼40% of wild-type levels and retrograde frequency drops to ∼20% of wild type. This strong retrograde IFT defect is coupled to less dramatic changes in anterograde speed and frequency, which decrease to ∼90% and ∼60% of wild type, respectively. Presumably, this level of anterograde IFT is sufficient to maintain flagella with little shortening, but is insufficient to drive flagellar assembly.
Table 2.
Summary of the dhc1b-3 IFT and flagellar phenotypes compared to wild-type cells at the same condition
| dhc1b-3 vs. wild type | Retrograde | Anterograde | Flagellar assembly | Length maintenance | Flagellar shortening | ||
| Speed | Freq. | Speed | Freq. | ||||
| % | % | % | % | ||||
| 21°C | 75 | 76 | 102 | 99 | + + | + + | + + |
| 34°C 6 h | 47 | 35 | 91 | 63 | − | + | ND |
| 34°C 12 h | 40 | 20 | 89 | 58 | − − | + | + + |
“+ +” = same phenotype as wt; “+” = close to wt; “−” = strong defect relative to wt; “− −” = complete defect; ND = no data.
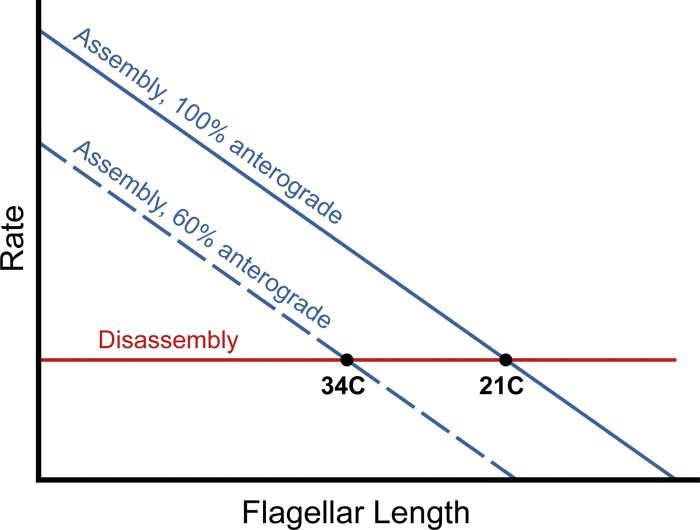
How well does the dhc1b-3 phenotype equate with the balance-point model of flagellar length control, which asserts that flagellar length is the resultant of opposing assembly and disassembly rates at the flagellar tip (Marshall and Rosenbaum, 2001; Marshall et al., 2005; Avasthi and Marshall, 2012)? The current iteration of the balance-point model predicts that, with a 40% reduction in anterograde IFT frequency at 34°C, both existing and newly assembling dhc1b-3 flagella should approach the same intermediate steady-state length (Fig. 8). Experimentally, however, dhc1b-3 flagellar length at 34°C is bi-stable. Existing flagella only slightly shorten, whereas new flagella experience strong assembly defects. Thus, there are likely uncharacterized intrinsic differences between established full-length flagella and short assembling flagella that are not yet factored into the balance-point model. One possibility is that stabilizing factors at the tips of full-length flagella, such as EB1 (Schrøder et al., 2007) or the distal filaments (Dentler and Rosenbaum, 1977; Dentler, 1980), could make the disassembly rate length dependent.
Figure 8.
Comparison of the dhc1b-3 phenotype with the current balance-point model of flagellar length control. The horizontal red line represents the length-independent disassembly rate and the blue lines are length-dependent assembly rates at 21°C (solid line) and 34°C (hashed line, 40% reduction in anterograde IFT frequency). The balance-point model predicts that both existing and assembling dhc1b-3 flagella should approach the same steady-state length (points indicated by “21C” and “34C”), where the assembly and disassembly rates are equal.
An important caveat about the nearly full-length dhc1b-3 flagella at 34°C is that we did not quantify IFT train size. If the drop in retrograde IFT leads to a reduction in anterograde train frequency, but also a commensurate increase in anterograde train size, then the effect on cargo delivery to the flagellar tip could potentially be very small. Future refinement of the balance-point model will require precise measurements of IFT train size, speed, and frequency, or perhaps even direct quantification of axonemal cargo delivery to the flagellar tip, coupled with single-cell measurements of flagellar length changes.
Finally, although our results clearly demonstrate the requirement for DHC1b in flagellar assembly, it remains to be shown why this is the case. The plausible default hypothesis is that continuous anterograde transport requires the recycling of IFT particles by retrograde IFT. However, it is possible that a signaling function of retrograde IFT is the true requirement for anterograde transport initiation. The unknown mechanism underlying the loose coupling between anterograde and retrograde IFT observed in the dhc1b-3 mutant may provide the key to understanding the reliance of flagellar assembly on retrograde transport.
Conclusion
The dhc1b-3 mutant’s unique kinetics of flagellar loss indicate that robust retrograde IFT is dispensable for the maintenance of flagellar length. However, it is clear that retrograde transport still plays key roles during flagellar assembly and in maintaining flagellar composition and function. Comparative proteomics of dhc1b-3 flagella revealed several distinct classes of known and uncharacterized IFT-related proteins that warrant further investigation. Like the fla10 mutant has done for kinesin-driven anterograde transport, the inducible and reversible inhibition of dynein function afforded by the dhc1b-3 mutant should prove exceptionally useful for future studies of retrograde IFT.
Materials and methods
Strains and culture conditions
Vegetative cells were grown in liquid Tris-acetate-phosphate (TAP) or minimal (M) media (Harris et al., 2009) at 21 or 34°C with constant aeration. Gametogenesis was induced by suspending cells in M-N media (M media without nitrogen). Wild-type mt+ (CC-125), wild-type agg1 mt− (CC-124), dhc1b-1 mt+ (CC-3711), and S1C5 mt− (CC-1952) strains were obtained from the Chlamydomonas Resource Center (University of Minnesota, Minneapolis, MN). The KAP-GFP fla3-1 (CC-4296) strain (Mueller et al., 2005) was provided by M. Porter (University of Minnesota, Minneapolis, MN). The bbs4-1 mt- (CC-4371) strain (Lechtreck et al., 2009) and the dhc1b-2 strain (Pazour et al., 1999b; Witman, 2011) were supplied by G. Witman (University of Massachusetts Medical School, Worcester, MA).
Isolation of the dhc1b-3 mt+ (CC-4422) strain is detailed in Fig. S2 E. Liquid cultures of wild-type CC-125 cells were mutagenized by exposure to UV radiation at a dose sufficient to kill 90% of the cells (LD90). Cells were then embedded in soft agar (0.35% agar in TAP) and grown for 1 wk at 34°C. Punctate colonies were picked out of the agar, grown in TAP at both 21 and 34°C, and checked by live-cell microscopy for ts conditional motility phenotypes. After isolation, the dhc1b-3 mt+ strain was backcrossed twice; the mutant phenotype consistently segregated 1:1. The dhc1b-3 mt− (CC-4423) strain was generated in the course of tetrad analysis for linkage mapping. The dhc1b-3 fla10 (CC-4424) double mutant was produced by mating dhc1b-3 mt+ (CC-4422) to fla10-1 mt− (CC-1919).
To produce diploid strains, electroporation was used to transform dhc1b-1 mt+ (CC-3711) cells with the pHyg3 hygromycin (hyg) resistance vector and dhc1b-3 mt− (CC-4423) cells with the pSI103 aphVIII paromomycin (par) resistance vector (plasmids were obtained from the Chlamydomonas Resource Center). The two drug-resistant strains were then mated, plated on 1.5% agar hyg/par TAP plates, and grown at room temperature in constant light until colonies appeared. Colonies were picked, struck onto 1.5% agar hyg/par TAP plates, and tested by PCR for mating type and the presence of the dhc1b-3 point mutation (Fig. S2 D). Positives were restruck onto new plates and retested by PCR.
Linkage mapping and sequencing the dhc1b-3 mutation
To locate the genomic region harboring the dhc1b-3 mutation, the mutant (mt+, CC-4422) was crossed to the S1C5 polymorphic strain (mt−, CC-1952; Gross et al., 1988) and tetrad progeny were isolated by dissection (Harris et al., 2009). The recombination frequencies between dhc1b-3 and the genetic markers SSR157-44 (5′-ACTGGGTTGTCACATACATGGTGG-3′, 5′-ACCTCAACCTTGGGGACGTAAAAT-3′), SSR75-62 (5′-CCCGCTTACCTACCCACTATGCTT-3′, 5′-CGACAGTGGATGTGTCCAGTAGGA-3′), FA1 (5′-ACGAGGAGGACATTCGGGAGCTGC-3′, 5′-GCTCAGCCGTTCCAAGGAAGCAATG-3′, 5′-GTCCAAGCAGGTCCAAGCCGTCAA-3′; Kathir et al., 2003), and the mating-type locus (MT; 5′-CGACGACTTGGCATCGACAGGTGG-3′, 5′-CTCGGCCAGAACCTTTCATAGGGTGG-3′, 5′-GCTGGCATTCCTGTATCCTTGACGC-3′, 5′-GCGGCGTAACATAAAGGAGGGTCG-3′; Zamora et al., 2004) were determined by detecting polymorphic PCR products in tetrad progeny with the dhc1b-3 flagellar assembly defect (Fig. S2 A). Recombinants were found for all markers other than SSR75-62, narrowing down the region where the mutation could be located.
To determine the precise location of the dhc1b-3 mutation, we sequenced cDNA of the DHC1b gene. Total RNA was isolated from dhc1b-3 and wild-type cells by TRIzol/Chloroform extraction, then cDNA was synthesized using Superscript III reverse transcription and random hexamer primers (Life Technologies). Using the primer pairs shown in Table S1, 29 overlapping PCR products were amplified from the dhc1b-3 cDNA, spanning the length of the coding sequence (CDS) and flanking untranslated regions (UTRs). PCR products were processed through a cleanup kit (QIAGEN) and then sequenced (Elim Biopharm). Only a single base pair differed from the wild-type sequence (12313 of the CDS). The mutation was confirmed by sequencing wild-type cDNA spanning the region, and through differential cutting of PCR products with restriction enzymes (Fig. S2, B and C). Noncomplementation in the dhc1b-1 dhc1b-3 diploid strain proved that the dhc1b-3 phenotype was due to a mutation in DHC1b (Fig. S2 D).
To examine the evolutionary conservation of the DHC1b C-sequence, the DHC1b amino acid sequences for the nine species displayed in Fig. S3 were aligned using ClustalX (Larkin et al., 2007). Aligned residues were colored to indicate conservation if they met the default thresholds that Clustal uses to assign Zappo colors, which are more stringent for some amino acid types than others. Instead of Zappo colors, the ClustalX “colprot.xml” file was modified to display a red-to-blue gradient corresponding to Kyte-Doolittle hydrophobicity values (Kyte and Doolittle, 1982). Dark blue indicates conserved hydrophilic residues, and dark red amino acids are conserved hydrophobic.
Flagellar regeneration and shortening
Flagellar regeneration was initiated by pH shock, as described previously (Lefebvre, 1995). TAP cell cultures were decreased to pH 4.5 (with 0.5 M acetic acid), incubated for 1.5 min to trigger flagellar abscission, then returned neutral pH (with 0.5 M KOH) to enable regeneration of flagella. To examine flagellar shortening, cells were grown in M media, incubated for 12 h at 21 or 34°C, and then treated with 20 mM NaPPi. Cells were incubated at 21 or 34°C after NaPPi treatment, fixed with glutaraldehyde, and imaged with a DeltaVision microscope (Applied Precision). Flagellar lengths were measured using ImageJ software (National Institutes of Health, Bethesda, MD).
Live-cell measurements of IFT by DIC and TIRF microscopy
Live-cell microscopy of IFT and kymograph analysis were performed as detailed previously (Engel et al., 2009a,b). For TIRF, 15 µl of GFP-expressing cells in TAP media were mounted on untreated cover glass (22 × 22 mm; Corning), inverted onto a slide (Gold Seal; Thermo Fisher Scientific) with a petroleum jelly spacer. Movies were acquired at ∼30 frames per second at room temperature (for <1 h after removing cells from 34°C) with NIS-Elements software (Nikon) and a cooled EM CCD camera (QuantEM:512SC; Photometrics) attached to a microscope (Eclipse Ti; Nikon) equipped with a 100×/1.49 NA TIRF oil immersion objective, a built-in perfect focus system, and through-the-objective TIRF with motorized control of the incident angle of illumination.
DIC imaging was performed essentially as described previously (Craige et al., 2010). Cells were immobilized between the cover glass and a 0.5% agarose pad. Movies were acquired at ∼24 frames per second at room temperature (for <1 h after removing cells from 34°C) with NIS-Elements software (Nikon) and a Clara Interline camera (Andor Technology) attached to a microscope (Eclipse Ti; Nikon) equipped with a 100×/1.4 NA TIRF oil immersion objective, a 1.4 NA oil immersion condenser, and a manually operated translating rotating stage, which enabled the specimen to be rotated so that flagella were perpendicular to the axis of DIC sheer before imaging.
Kymographs for both the TIRF and DIC movies were generated with MetaMorph software (Molecular Devices) by drawing a line along the flagella with a width slightly greater than the width of the flagella. The maximum intensity value (for TIRF) or average intensity (for DIC) for each point along the length of the line was plotted for each frame in the movie. IFT train speed and frequency were quantified from the kymographs by measuring the slope and number of traces, respectively.
Western blotting and immunofluorescence microscopy
Flagella for Western blots were isolated as described previously (Witman et al., 1972) by pH shock, pelleting the cell bodies (1,000 rcf; 5 min), centrifuging the supernatant onto an underlayed 25% sucrose cushion (2,500 rcf; 10 min, no brake), collecting the interface plus supernatant, and then pelleting the flagella (10,000 rcf; 20 min). Western blots were quantified by calculating the background-normalized integrated intensity using ImageJ. Immunofluorescence was performed essentially as described in Keller et al. (2010). In brief, cells were plated on polylysine-coated cover glass, fixed in methanol at −20°C, blocked in 5% BSA, 1% fish gelatin, and 10% normal goat serum in PBS, and incubated with primary antibodies overnight (1:100). After several washes with PBS, the cells were incubated with secondary antibodies (1:200; Jackson ImmunoResearch Laboratories, Inc.), washed with more PBS, incubated with 1 µg/ml DAPI, and mounted on slides with Vectashield mounting medium (Vector Laboratories). Cells were imaged using a DeltaVision deconvolution system (Applied Precision) with a 100×/1.4 NA oil immersion objective. Z-stacks were acquired, deconvolved, and then made into maximum intensity projections.
The primary antibodies used in this study were generously provided by D. Cole (rat α-KAP, mouse α-FLA10N, mouse α-IFT81, mouse α-IFT172, mouse α-IFT139, rabbit α-IFT140, and rabbit α-IFT122; Cole et al., 1998; Mueller et al., 2005; Behal et al., 2012), George Witman (rabbit α-DHC1b, rabbit α-D1bLIC, and rabbit α-BBS4; Pazour et al., 1999a; Hou et al., 2004; Lechtreck et al., 2009), Joel Rosenbaum (rabbit α-IFT27; Qin et al., 2007), and Richard Linck (rabbit α-Rib43a; Norrander et al., 2000). Antibodies to ODA-IC2 (IC69, clone D6168) and α-tubulin (DM1A, clone T6199) were obtained from Sigma-Aldrich.
2D-DIGE analysis
For each condition (wild-type 21C, dhc1b-3 21C, dhc1b-3 34C 18 h), two liters of cells were grown in TAP with constant light and aeration. Flagella were isolated as described previously (Witman et al., 1972), yielding 100–200 µg of flagellar protein for each sample. Protein concentrations were adjusted using Western blots with fluorescent secondary antibodies (measured by an Odyssey imager [LI-COR Biosciences]) to ensure equal α-tubulin loading between the samples. The samples were then sent to Applied Biomics, Inc. (Hayward, CA) for 2D-DIGE comparative proteomics.
The three samples were each labeled with a different cy-dye and then run together on a single 2D acrylamide gel (∼50 µg total protein), where they were separated by isoelectric focusing (isoelectric point, pI) followed by electrophoresis (molecular weight, MW). This “analytical” gel was scanned for the fluorescence signal of each cy-dye (Typhoon imager; GE Healthcare), and DeCyder software (GE Healthcare) was used to perform “in-gel” comparative analysis of every spot. Spots were chosen for mass spectrometry identification based on high relative accumulation or depletion between the three samples. A second “preparative” 2D gel was run with higher protein concentration (∼200 µg total protein) and spots were picked out of the gel using an automated Ettan Spot Picker (GE Healthcare). For spot identification, picked gel samples were washed, dried, and rehydrated in trypsin digestion buffer at 37°C. Digested peptides were extracted from the gel with trifluoroacetic acid buffer, desalted using C-18 Zip-tips (EMD Millipore), mixed with α-cyano-4-hydroxycinnamic acid matrix (CHCA), and spotted into wells of a matrix-assisted laser desorption ionization (MALDI) plate. The mass spectra (MS) of the peptides in each sample was determined by time of flight (MALDI-TOF) using an Applied Biosystems proteomics analyzer (Life Technologies). The most abundant peptides in each sample were subjected to further fragmentation and tandem mass spectrometry (MALDI-TOF/TOF, MS/MS). Protein identities were determined by searching the Chlamydomonas genome sequence (Joint Genome Institute, assembly v4.0) with the combined MS and MS/MS spectra using GPS Explorer software equipped with the MASCOT search engine (Matrix Science, Inc.). Only strong hits with close to 100% confidence were accepted.
Cell motility and phototaxis assays
The phototactic behavior of Chlamydomonas cells was observed and quantified as described previously (Wakabayashi et al., 2011). For dish assays, cells were washed with buffer (5 mM Hepes, pH 7.4, 0.2 mM EGTA, 1 mM KCl, and 0.3 mM CaCl2) and put in Petri dishes. Cell suspensions were illuminated from one side (from the right in Fig. 7) with a green LED (λ = 525 nm, ∼50 µmol photons m−2s−1) for 15 min and then photographed. For single-cell analysis, the Petri dishes were placed on the stage of an inverted microscope (model IX70; Olympus) and illuminated with the green LED. 30 s after the LED was turned on, the behavior of the cells was video-recorded using a CCD camera (model XC-75; Sony). The angle (θ) between the light direction and the swimming direction was measured from the tracks of swimming cells using Image Hyper software (Science-Eye).
Reactivation of demembranated cell models was performed by modifying the method described in Kamiya and Witman (1984). After cells were washed with a buffer containing 10 mM Hepes, pH 7.4, and 4% sucrose, cells were demembranated with HMDEKP buffer (30 mM Hepes, 5 mM MgSO4, 50 mM potassium acetate, and 1% polyethylene glycol Mr = 20,000) containing 0.1% Igepal CA-630. For reactivation, cell models were suspended in HMDEKP buffer containing 70 U/ml creatine kinase, 5 mM phosphocreatine, and 1 mM ATP.
Flagellar beat frequency was measured as described previously (Kamiya, 2000; Wakabayashi and King, 2006). In brief, a photomultiplier with a gradient filter was attached to a dark-field microscope (model BH-2; Olympus) and cells were observed with dim red light. Cell body vibration signals were transferred to a computer via an A/D converter (model SE-U55X; Onkyo Corporation) and put through a fast-Fourier transform (FFT) using SIGVIEW software (Signalab). Transformed signals were averaged for 30 s. The resulting power spectrum represents the beat frequency distribution, with the peak value indicating the median beat frequency. Swimming velocities were calculated from the tracks left by swimming cells during the extended exposure of images, obtained using Image Hyper software.
Online supplemental material
Fig. S1 shows the phenotype of dhc1b-2. Fig. S2 diagrams the scheme used to isolate ts motility mutants, and shows the linkage mapping of dhc1b-3, the PCR assay used to track the dhc1b-3 mutation, and noncomplementation in the dhc1b-1 dhc1b-3 diploid strain. Fig. S3 illustrates the location of the dhc1b-3 mutation and the conservation of the sequence where it is found. Fig. S4 graphs the kinetics of dhc1b-3 flagellar loss in synchronized dividing cells and the kinetics of flagellar regeneration in dhc1b-3 cells at 21°C after increasing preincubation times at 34°C. Together, Fig. S5 and Table S3 detail the complete results from the 2D-DIGE proteomic analysis. Table S1 lists the primers used to sequence the DHC1b cDNA. Table S2 shows the quantification of Fig. 5 B. Videos 1–6 were used to generate the kymographs in Fig. 2, A and B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206068/DC1.
Acknowledgments
We extend a special thanks to Ritsu Kamiya for hosting a two-month stay at the University of Tokyo and to all the members of the Kamiya laboratory for their help and gracious hospitality. We are grateful to Douglas Cole, Dennis Diener, Carlo Iomini, Karl Lechtreck, Richard Linck, and Joel Rosenbaum for generously sharing strains and antibodies. We thank Kurt Thorn and the Nikon Imaging Center at UC San Francisco for assistance with the TIRF microscope.
This work was funded by National Institutes of Health grant R01 GM 097017 (to W.F. Marshall), the Genentech Graduate Fellowship (to B.D. Engel), and the NSF East Asia and Pacific Summer Institutes Program (to B.D. Engel), co-funded by the Japan Society for the Promotion of Science.
Author contributions: B.D. Engel isolated the dhc1b-3 strain, sequenced the mutation, performed initial characterization of flagellar loss and regeneration, imaged and quantified IFT, and implemented the 2D-DIGE analysis with Applied Biomics, Inc. H. Ishikawa performed the Western blot analysis of flagellar protein levels. K.A. Wemmer created and characterized the dhc1b-1 dhc1b-3 diploid strain. S. Geimer performed the TEM sample preparation and imaging. K.-i. Wakabayashi measured phototaxis, cell swimming speed, and flagellar beat frequency. M. Hirono performed linkage mapping of the dhc1b-3 strain and isolated a mating-type minus recombinant used to produce the diploid. B. Craige helped with the DIC microscopy of IFT. G.J. Pazour isolated and characterized the dhc1b-2 strain. G.B. Witman provided DIC imaging facilities. R. Kamiya aided with TEM and dark-field microscopy, and provided facilities. W.F. Marshall provided facilities and measured flagellar regeneration and shortening. B.D. Engel and W.F. Marshall wrote the paper.
Footnotes
Abbreviations used in this paper:
- 2D-DIGE
- two-dimensional difference in gel electrophoresis
- CDS
- coding sequence
- DIC
- differential interference contrast
- IFT
- intraflagellar transport
- KAP
- kinesin-associated protein
- NaPPi
- sodium pyrophosphate
- TAP
- Tris-acetate-phosphate
- TEM
- transmission electron microscopy
- TIRF
- total internal reflection fluorescence
- ts
- temperature-sensitive
- UTR
- untranslated region
References
- Adams G.M., Huang B., Luck D.J. 1982. Temperature-sensitive, assembly defective flagellar mutants of Chlamydomonas reinhardtii. Genetics. 100:579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P., Marshall W.F. 2012. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 83:S30–S42 10.1016/j.diff.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal R.H., Miller M.S., Qin H., Lucker B.F., Jones A., Cole D.G. 2012. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J. Biol. Chem. 287:11689–11703 10.1074/jbc.M111.287102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Li C., Inglis P.N., Esmail M.A., Ou G., Mah A.K., Baillie D.L., Scholey J.M., Leroux M.R. 2006. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell. 17:5053–5062 10.1091/mbc.E06-06-0571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Cho C., Jin L., Vale R.D. 2011. Crystal structure of the dynein motor domain. Science. 331:1159–1165 10.1126/science.1202393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Chinn S.W., Wedaman K.P., Hall K., Vuong T., Scholey J.M. 1993. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 366:268–270 10.1038/366268a0 [DOI] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S., Wang C., Wang B., Bassi M.R., Caretti E., Champeval D., Calmont A., Jarnik M., Burch J., Zaret K.S., et al. 2009. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev. Biol. 325:225–237 10.1016/j.ydbio.2008.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190:927–940 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W.L. 1980. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J. Cell Sci. 42:207–220 [DOI] [PubMed] [Google Scholar]
- Dentler W.L., Rosenbaum J.L. 1977. Flagellar elongation and shortening in Chlamydomonas. III. structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 74:747–759 10.1083/jcb.74.3.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W.L., Vanderwaal K., Porter M.E. 2009. Recording and analyzing IFT in Chlamydomonas flagella. Methods Cell Biol. 93:145–155 10.1016/S0091-679X(08)93008-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso N.A., Cyr R.J. 1994. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1 alpha. Plant Cell. 6:893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Lechtreck K.F., Sakai T., Ikebe M., Witman G.B., Marshall W.F. 2009a. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 93:157–177 10.1016/S0091-679X(08)93009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Ludington W.B., Marshall W.F. 2009b. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J. Cell Biol. 187:81–89 10.1083/jcb.200812084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.H., Ranum L.P., Lefebvre P.A. 1988. Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13:503–508 10.1007/BF02427756 [DOI] [PubMed] [Google Scholar]
- Hao L., Efimenko E., Swoboda P., Scholey J.M. 2011a. The retrograde IFT machinery of C. elegans cilia: two IFT dynein complexes? PLoS ONE. 6:e20995 10.1371/journal.pone.0020995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Thein M., Brust-Mascher I., Civelekoglu-Scholey G., Lu Y., Acar S., Prevo B., Shaham S., Scholey J.M. 2011b. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat. Cell Biol. 13:790–798 10.1038/ncb2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Pazour G.J., Witman G.B. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 15:4382–4394 10.1091/mbc.E04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H., Stern D.B., Witman G.B. 2009. The Chlamydomonas Sourcebook (Second Edition). Academic Press/Elsevier, London.
- Iomini C., Li L., Mo W., Dutcher S.K., Piperno G. 2006. Two flagellar genes, AGG2 and AGG3, mediate orientation to light in Chlamydomonas. Curr. Biol. 16:1147–1153 10.1016/j.cub.2006.04.035 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W.F. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Kamiya R. 2000. Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods. 22:383–387 10.1006/meth.2000.1090 [DOI] [PubMed] [Google Scholar]
- Kamiya R., Witman G.B. 1984. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98:97–107 10.1083/jcb.98.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W.J., Haas N.A., Lefebvre P.A., Silflow C.D. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell. 2:362–379 10.1128/EC.2.2.362-379.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L.C., Wemmer K.A., Marshall W.F. 2010. Influence of centriole number on mitotic spindle length and symmetry. Cytoskeleton (Hoboken). 67:504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Sutoh K., Kurisu G. 2011. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 18:638–642 10.1038/nsmb.2074 [DOI] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., Kurisu G. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature. 484:345–350 10.1038/nature10955 [DOI] [PubMed] [Google Scholar]
- Kozminski K.G. 1995. High-resolution imaging of flagella. Methods Cell Biol. 47:263–271 10.1016/S0091-679X(08)60819-5 [DOI] [PubMed] [Google Scholar]
- Kozminski K.G., Beech P.L., Rosenbaum J.L. 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131:1517–1527 10.1083/jcb.131.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 90:5519–5523 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lechtreck K.F., Johnson E.C., Sakai T., Ikebe M., Witman G.B. 2009. The Chlamydomonas BBSome is transported by a subset of IFT particles and bbs mutants abnormally accumulate flagellar signaling proteins. J. Cell Biol. 187:1117–1132 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P.A. 1995. Flagellar amputation and regeneration in Chlamydomonas. Methods Cell Biol. 47:3–7 10.1016/S0091-679X(08)60782-7 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Rosenbaum J.L. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155:405–414 10.1083/jcb.200106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Qin H., Rodrigo Brenni M., Rosenbaum J.L. 2005. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol. Biol. Cell. 16:270–278 10.1091/mbc.E04-07-0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak M.K., Kinzy T.G. 2010. eEF1A: thinking outside the ribosome. J. Biol. Chem. 285:21209–21213 10.1074/jbc.R110.113795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.R., Kang Y. 1991. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 113:835–842 10.1083/jcb.113.4.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.C., Cyr R.J. 2000. Association between elongation factor-1alpha and microtubules in vivo is domain dependent and conditional. Cell Motil. Cytoskeleton. 45:279–292 [DOI] [PubMed] [Google Scholar]
- Moore R.C., Durso N.A., Cyr R.J. 1998. Elongation factor-1alpha stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil. Cytoskeleton. 41:168–180 [DOI] [PubMed] [Google Scholar]
- Mueller J., Perrone C.A., Bower R., Cole D.G., Porter M.E. 2005. The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell. 16:1341–1354 10.1091/mbc.E04-10-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., Jackson P.K. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J.M., deCathelineau A.M., Brown J.A., Porter M.E., Linck R.W. 2000. The Rib43a protein is associated with forming the specialized protofilament ribbons of flagellar microtubules in Chlamydomonas. Mol. Biol. Cell. 11:201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata N., Shima T., Ohkura R., Kon T., Sutoh K. 2011. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 585:1185–1190 10.1016/j.febslet.2011.03.036 [DOI] [PubMed] [Google Scholar]
- Ocbina P.J., Eggenschwiler J.T., Moskowitz I., Anderson K.V. 2011. Complex interactions between genes controlling trafficking in primary cilia. Nat. Genet. 43:547–553 10.1038/ng.832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Snell W.J. 2005. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev. Cell. 9:431–438 10.1016/j.devcel.2005.07.010 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Wilkerson C.G., Witman G.B. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979–992 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Witman G.B. 1999a. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144:473–481 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Witman G.B. 1999b. The DHC1b (DHC2) isoform of cytoplasmic dynein is necessary for flagellar maintenance as well as flagellar assembly. Mol. Biol. Cell. 10:369a (Supplement) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L.B., Rosenbaum J.L. 2008. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 85:23–61 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Pedersen L.B., Geimer S., Sloboda R.D., Rosenbaum J.L. 2003. The microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 13:1969–1974 10.1016/j.cub.2003.10.058 [DOI] [PubMed] [Google Scholar]
- Pedersen L.B., Geimer S., Rosenbaum J.L. 2006. Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr. Biol. 16:450–459 10.1016/j.cub.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Perrone C.A., Tritschler D., Taulman P., Bower R., Yoder B.K., Porter M.E. 2003. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell. 14:2041–2056 10.1091/mbc.E02-10-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao T., Luo M., Wang L., Guo Y., Li D., Li P., Snell W.J., Pan J. 2009. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 106:4713–4718 10.1073/pnas.0808671106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D.R., Rosenbaum J.L., Lupetti P. 2009. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187:135–148 10.1083/jcb.200905103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K. 1997. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 94:4457–4462 10.1073/pnas.94.9.4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M.E., Bower R., Knott J.A., Byrd P., Dentler W.L. 1999. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 10:693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Rosenbaum J.L., Barr M.M. 2001. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11:457–461 10.1016/S0960-9822(01)00122-1 [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Wang Z., Diener D.R., Rosenbaum J.L. 2007. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17:193–202 10.1016/j.cub.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Lin Y., Norman R.X., Ko H.W., Eggenschwiler J.T. 2011. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. USA. 108:1456–1461 10.1073/pnas.1011410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Pedersen L.B., Patel-King R.S., King S.M. 2007. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J. Cell Sci. 120:3653–3665 10.1242/jcs.012773 [DOI] [PubMed] [Google Scholar]
- Schafer J.C., Haycraft C.J., Thomas J.H., Yoder B.K., Swoboda P. 2003. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell. 14:2057–2070 10.1091/mbc.E02-10-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Gleave E.S., Carter A.P. 2012. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 19:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M. 2008. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J. Cell Biol. 180:23–29 10.1083/jcb.200709133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrøder J.M., Schneider L., Christensen S.T., Pedersen L.B. 2007. EB1 is required for primary cilia assembly in fibroblasts. Curr. Biol. 17:1134–1139 10.1016/j.cub.2007.05.055 [DOI] [PubMed] [Google Scholar]
- Shiina N., Gotoh Y., Kubomura N., Iwamatsu A., Nishida E. 1994. Microtubule severing by elongation factor 1 alpha. Science. 266:282–285 10.1126/science.7939665 [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K.P., Orozco J.T., Dwyer N.D., Bargmann C.I., Rose L.S., Scholey J.M. 1999. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147:519–530 10.1083/jcb.147.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth R.D., Ebersold W.T. 1985. Genetic investigation of a negatively phototactic strain of Chlamydomonas reinhardtii. Genet. Res. 46:133–148 10.1017/S001667230002262X [DOI] [PubMed] [Google Scholar]
- Viswanathan S., Unlü M., Minden J.S. 2006. Two-dimensional difference gel electrophoresis. Nat. Protoc. 1:1351–1358 10.1038/nprot.2006.234 [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., King S.M. 2006. Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J. Cell Biol. 173:743–754 10.1083/jcb.200603019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K., Misawa Y., Mochiji S., Kamiya R. 2011. Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 108:11280–11284 10.1073/pnas.1100592108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther Z., Vashishtha M., Hall J.L. 1994. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126:175–188 10.1083/jcb.126.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G.B. 2011. Dynein and intraflagellar transport. In Dyneins, Structure, Biology and Disease. First Edition. King SM, editor. Academic Press/Elsevier, London, Waltham, San Diego. 394–422.
- Witman G.B., Carlson K., Berliner J., Rosenbaum J.L. 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54:507–539 10.1083/jcb.54.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora I., Feldman J.L., Marshall W.F. 2004. PCR-based assay for mating type and diploidy in Chlamydomonas. Biotechniques. 37:534–536 [DOI] [PubMed] [Google Scholar]