Abstract
The D1 protein of photosystem II in the thylakoid membrane of photosynthetic organisms is encoded by psbA genes, which in cyanobacteria occur in the form of a small gene family. Light-dependent up-regulation of psbA gene expression is crucial to ensure the proper replacement of the D1 protein. To gain a high level of gene expression, psbA transcription can be enhanced by several orders of magnitude. Recent transcriptome analyses demonstrated a high number of cis-encoded antisense RNAs (asRNAs) in bacteria, but very little is known about their possible functions. Here, we show the presence of two cis-encoded asRNAs (PsbA2R and PsbA3R) of psbA2 and psbA3 from Synechocystis sp. PCC 6803. These asRNAs are located in the 5′ untranslated region of psbA2 and psbA3 genes. Their expression becomes up-regulated by light and down-regulated by darkness, similar to their target mRNAs. In the PsbA2R-suppressing strain [PsbA2R(−)], the amount of psbA2 mRNA was only about 50% compared with the control strain. Likewise, we identified a 15% lowered activity of photosystem II and a reduced amount of the D1 protein in PsbA2R(−) compared with the control strain. The function of PsbA2R in the stabilization of psbA2 mRNA was shown from in vitro RNase E assay when the AU box and the ribosome-binding site in the 5′ untranslated region of psbA2 mRNA were both covered by PsbA2R. These results add another layer of complexity to the mechanisms that contribute to psbA gene expression and show PsbA2R as a positively acting factor to achieve a maximum level of D1 synthesis.
psbA genes encode the D1 reaction center protein of water-splitting PSII. The PSII structure from Thermosynechococcus vulcanus, a thermophilic cyanobacterium, has been resolved at 1.9 Å resolution (Umena et al., 2011). All the cofactors of PSII electron transfer are bound to the D1 protein and to a homologous D2 protein (Nanba and Satoh, 1987), forming together the reaction center core of PSII. The unique and highly oxidizing chemistry of PSII and the concomitant formation of reactive radicals induce lethal damage on the D1 protein, which needs to be replaced rapidly and properly by de novo synthesized D1 to avoid irreversible damage to the photosynthetic apparatus. Consequently, the D1 protein shows faster turnover than any other thylakoid protein, particularly under conditions where cells are exposed to light in excess of that safely used for photochemistry (Aro et al., 1993). In cyanobacteria, the major control of psbA gene expression has been identified to occur at the level of transcription (Golden et al., 1986; Mohamed and Jansson, 1989; Mulo et al., 2012).
Cyanobacteria in general have up to five psbA gene copies (Cyanobase; http://genome.kazusa.or.jp/cyanobase). These different copies respond differentially to changes in environmental conditions (Clarke et al., 1993; Mohamed et al., 1993; Máté et al., 1998; Kós et al., 2008; Summerfield et al., 2008; Sicora et al., 2009), emphasizing the importance of transcriptional regulation. Indeed, at high light intensities, at least one of the psbA genes belongs to the most highly expressed genes of the cyanobacterial cell. Besides the general transcriptional regulation whose components still remain unknown, there are several other molecular mechanisms that impact psbA gene expression. One such mechanism involves RNase E/G-type endoribonuclease-mediated cleavage at the “AU box” and the ribosome-binding site (RBS; Horie et al., 2007).
Recent transcriptome analyses have demonstrated a high number of noncoding transcripts in bacteria. In addition to the transacting regulatory sense RNAs, in many bacteria several hundreds of distinct cis-antisense transcripts have been reported (for review, see Georg and Hess, 2011). This matches a recent observation for plant chloroplasts, where antisense transcripts (asRNAs) were found for 35% of all genes (Zhelyazkova et al., 2012). Moreover, natural antisense transcripts are also abundant in the plant nuclear genome (Zhou et al., 2009; Zhang et al., 2012b).
In cyanobacteria, asRNAs amounting to 26% of all genes were reported for the unicellular Synechocystis sp. PCC 6803 (Georg et al., 2009; Mitschke et al., 2011a) and to 39% of all genes in the nitrogen-fixing Anabaena sp. PCC 7120 (Mitschke et al., 2011b). However, the functional relevance of specific antisense transcripts in plants, chloroplasts, as well as bacteria has scarcely been addressed. In Anabaena sp. PCC 7120, furA, the gene for the ferric uptake regulator, is covered by a long asRNA that originates by read through from the neighboring gene alr1690 (Hernández et al., 2006). In a Δalr1690 strain, increased levels of the Fur protein and an iron deficiency phenotype were observed, suggesting the biological relevance of this asRNA (Hernández et al., 2010). The 177-nucleotide asRNA IsrR in Synechocystis sp. PCC 6803 controls the expression of isiA, which encodes the iron stress-induced protein A, in a codegradation mechanism (Dühring et al., 2006). In this case, the transcript accumulations of IsrR and isiA mRNA are strictly inverse to each other. However, IsrR is transcribed from a constitutive promoter, whereas the isiA promoter is induced upon iron, redox, or light stress. When both transcripts are expressed simultaneously, they form an RNA duplex that is immediately degraded. Therefore, IsiA proteins are not observed before the number of isiA mRNA molecules has titrated out the number of IsrR molecules. As a result, IsrR filters short-time environmental stress signals out and causes a delay of isiA expression in the early stress phase but also a faster depletion during the recovery from stress, in a regulatory mode known as the threshold linear response (Legewie et al., 2008). Yet another cis-asRNA was recently characterized from the flv4-2 operon in Synechocystis sp. PCC 6803 and shown to be transiently expressed under inorganic carbon limitation (Eisenhut et al., 2012). Proteins encoded by the flv4-2 operon have been demonstrated as crucial in photoprotection of PSII under inorganic carbon limitation and high-light conditions (Zhang et al., 2009, 2012a), and the respective asRNA functions as a buffer mechanism to avoid premature expression of the flv4-2 operon when the stress conditions are only transient, thus avoiding the initiation of long-term acclimation processes upon short-term environment fluctuations.
Thus far, distinct functions have been identified for only a few and strongly expressed bacterial asRNAs, but many occur at relatively low abundance (for review, see Georg and Hess, 2011), raising the question of whether these low-abundance transcripts just constitute some kind of “transcriptional noise” or provide some selective advantage. In this study, we identified and characterized two short asRNAs, PsbA2R and PsbA3R, which originate from the 5′ untranslated region (UTR) of genes psbA2 and psbA3 in Synechocystis sp. PCC 6803. Despite low steady-state abundance, we show that PsbA2R acts as a positive regulator of the psbA2 expression and a positive effector of cell growth on solid medium.
RESULTS
Two asRNAs Originating in the 5′ UTR of psbA2 and psbA3
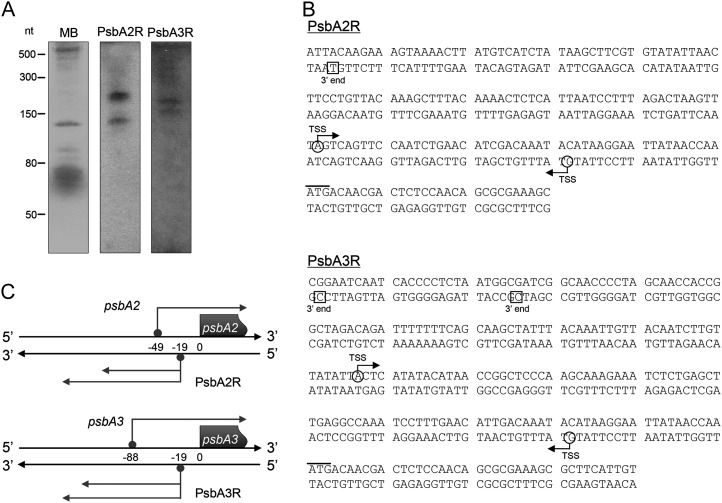
Analysis of the Synechocystis sp. PCC 6803 transcriptome by differential RNA sequencing revealed two transcription start sites immediately upstream of the psbA2 and psbA3 genes on the reverse complementary strand (Mitschke et al., 2011a). Both asRNAs (designated as PsbA2R and PsbA3R, respectively) were confirmed by northern analysis using strand-specific RNA probes (Fig. 1A) and by 5′ and 3′ RACE analyses to determine the full length of PsbA2R and PsbA3R (Fig. 1B). Two distinct signals with fragment lengths of approximately 130 and 220 nucleotides as well as approximately 160 and 180 nucleotides were detected for PsbA2R and PsbA3R, respectively (Fig. 1B). For both asRNAs, one transcription start site was found at position −19 counted from the ATG start codon of psbA mRNAs, corresponding to genome positions 7,210 and 1,820,016 on the reverse and forward strands, respectively (Fig. 1, B and C). This result is consistent with previous transcription start site mapping by differential RNA sequencing (Mitschke et al., 2011a). On the contrary, 3′ RACE analysis identified one reasonable 3′ end for PsbA2R at the position −147 and two 3′ ends for PsbA3R at positions −175 and −199 (Fig. 1B). However, no proper 3′ end was identified for the approximately 220-nucleotide signal of PsbA2R. Thus, there are two variants of PsbA2R (129 and about 220 nucleotides) and PsbA3R (157 and 181 nucleotides; Fig. 1C).
Figure 1.
Identification of the asRNAs PsbA2R and PsbA3R. A, Typical detection pattern of PsbA2R and PsbA3R by northern analysis. Five micrograms of total RNA was loaded on each lane. The blot was stained by methylene blue before being hybridized with RNA probes (MB). B, Determination of the 5′ and 3′ ends of the asRNAs by RACE analysis. mRNA and asRNAs are shown in the top and bottom DNA strands, respectively. Transcription start sites (TSS) and the 3′ ends of mRNAs or asRNAs are indicated by circles together with an arrow and by squares, respectively. The start codon of translation is indicated by a bar in the top DNA strand. C, Schematic presentation of PsbA2R and PsbA3R. Numbers within the two DNA strands indicate the positions counted from the ATG translation start codon of the mRNAs.
Light-Dependent Expression of PsbA2R and PsbA3R
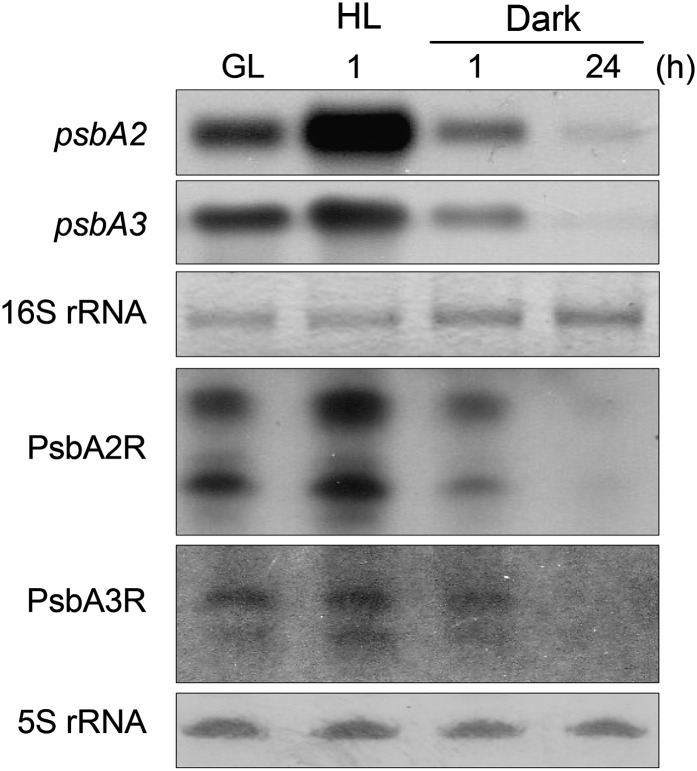
Since the transcription of the psbA genes is strictly regulated by light, we investigated whether PsbA2R and PsbA3R show any transcriptional response to changes in light intensity. As shown in Figure 2, the accumulation of PsbA2R was strongly enhanced upon a shift of cells from growth light (GL; 50 μmol photons m−2 s−1) to high light (HL; 1,000 μmol photons m−2 s−1), whereas the accumulation of PsbA3R transcripts was less drastic. This mimicked the expression patterns of the psbA2 and psbA3 genes, respectively. Likewise, the asRNAs were down-regulated in darkness similar to their target mRNAs.
Figure 2.
Response of the expression of psbA2 and psbA3 genes and their respective asRNAs to changes in light intensity. Total RNA was isolated from the cells grown under standard GL, 1 h after the shift to HL (1,000 μmol photons m−2 s−1), and 1 or 24 h after the shift to darkness. One microgram of total RNA was used for the detection of psbA2, and 5 μg of total RNA was applied to each lane in the case of PsbA2R, psbA3, and PsbA3R.
Molecular Ratio between mRNA and asRNA of psbA2 under Steady-State Conditions
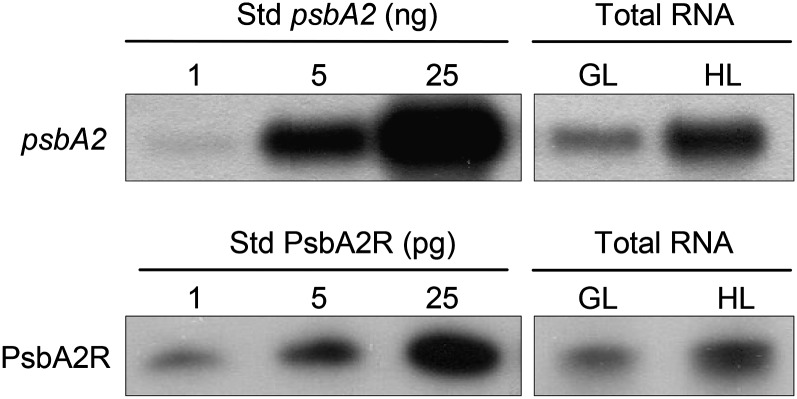
The molecular ratio between the target mRNA and the respective asRNA provides important information about the putative regulatory function of the asRNA on its target gene. To this end, we focused on the psbA2 gene, which is the most active one of the three psbA genes in Synechocystis sp. PCC 6803 at HL, and on the respective asRNA PsbA2R. The molecular ratio between mRNA and asRNA of psbA2 was estimated by comparing their amounts in the total RNA extract from Synechocystis sp. PCC 6803 cells with in vitro-synthesized RNA molecules as a reference. Such analyses revealed that 1 μg of total RNA isolated from the HL-treated cells contained about 3 ng of psbA2 mRNA and 1 pg of PsbA2R (Fig. 3). To estimate the molecular ratio between mRNA and asRNA of psbA2, the lengths of the psbA2 and PsbA2R transcripts, 1,083 and 129 nucleotides, respectively, were taken into account. This resulted in a steady-state molecular ratio between mRNA and asRNA of psbA2 at approximately 300:1.
Figure 3.
Molecular ratio of mRNA and asRNA of psbA2 in Synechocystis sp. PCC 6803 cells. RNA molecules synthesized by the in vitro transcription system were used as a standard series (Std; 1, 5, and 25 ng). Total RNA was isolated from cells grown under GL and from cells treated under HL for 30 min. One and 5 μg of total RNA were used for the detection of psbA2 mRNA and PsbA2R, respectively.
Rapid Turnover of asRNA PsbA2R Compared with psbA2 mRNA
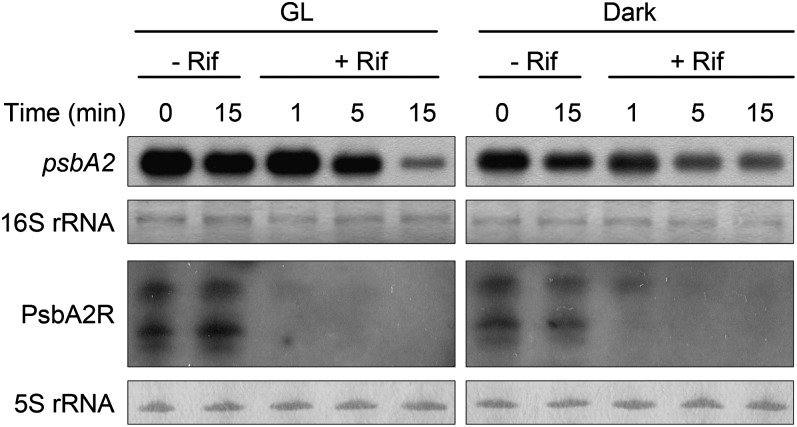
The low steady-state amount of PsbA2R prompted us to investigate the turnover rates of the asRNA and the target mRNA. To determine the half-lives of the psbA2 mRNA and the asRNA PsbA2R, the cell suspension was supplemented with rifampicin (Rif) to prevent de novo RNA synthesis. Furthermore, for practical reasons, the amounts of both psbA2 mRNA and PsbA2R were first left to increase by HL illumination for 30 min, and only thereafter were the measurements of psbA2 mRNA and PsbA2R amounts initiated (Fig. 4, 0 min). In the absence of Rif, the steady-state amount of psbA2 mRNA slightly decreased both in GL and in darkness, whereas the content of PsbA2R decreased only in darkness. On the contrary, in the presence of Rif, a fast RNA degradation occurred and distinct differences in the disappearance of psbA2 mRNA and PsbA2R were evident. Although the degradation of psbA2 mRNA was faster in GL compared with darkness, there was a clear signal of psbA2 mRNA still present after 15 min of Rif treatment in GL. In sharp contrast to the behavior of psbA2 mRNA, PsbA2R had nearly disappeared already within 1 min under GL conditions. The degradation of both the psbA2 mRNA and PsbA2R occurred more slowly in darkness, yet, again, there was a clear difference, with the asRNA disappearing faster. These results indicated that the turnover of PsbA2R is more than 1 order of magnitude faster, particularly in light, than that of the psbA2 mRNA (Fig. 4).
Figure 4.
Turnover of psbA2 mRNA and PsbA2R. Cells treated by HL for 30 min were transferred to GL or darkness in the absence or presence of Rif. Cells were harvested for RNA analysis at the designated time points. One and 5 μg of total RNA were used for the detection of psbA2 mRNA and PsbA2R, respectively.
Suppression of PsbA2R Decreases the Steady-State Amount of psbA2 mRNA
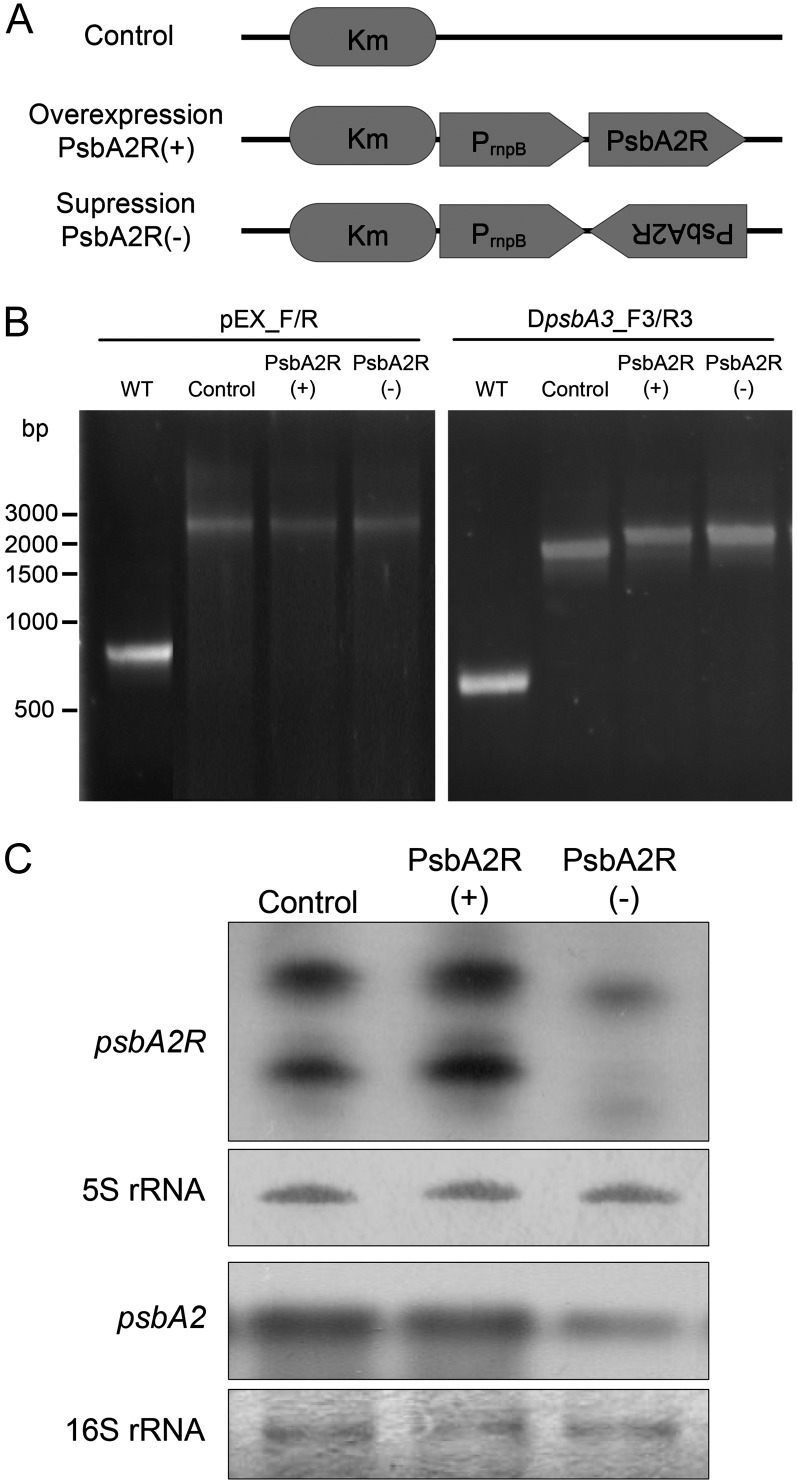
To further study the relationship between psbA gene expression and that of the respective asRNA, we prepared overexpressor and suppressor mutants of PsbA2R, referred to as PsbA2R(+) and PsbA2R(−), respectively (Fig. 5A). PsbA2R was selected for these analyses because a significant part, nearly 90% of the total psbA mRNA, is transcribed from the psbA2 gene, the putative target of PsbA2R (Mohamed et al., 1993). As mentioned above, the psbA3 gene also has a similar type of asRNA in the 5′ UTR as psbA2. Therefore, in order to eliminate the possibility that PsbA3R would compensate for the function of PsbA2R, the ΔpsbA3 strain that lacks PsbA3R was used as a parental strain to construct the PsbA2R(+) and PsbA2R(−) strains (Fig. 5B). Figure 5C shows the expression level of PsbA2R in each strain. PsbA2R(+) contained a slightly elevated amount of PsbA2R compared with the control strain. However, in the PsbA2R(−) strain, nearly complete suppression of PsbA2R occurred. To investigate the effect of PsbA2R on the expression of psbA2, the amount of psbA2 mRNA was also analyzed in the cells. As shown in Figure 5C, the PsbA2R(+) strain contained almost the same amount of psbA2 mRNA as the control strain. Intriguingly, however, the level of psbA2 mRNA in the PsbA2R(−) strain was significantly lower, being only about 50% of that in the control strain. These results provided strong evidence that PsbA2R, despite its low steady-state amount, is an important factor in controlling the cellular amount of psbA2 mRNA.
Figure 5.
Overexpression and suppression of PsbA2R. A, The sense and antisense fragments of PsbA2R were fused to the rnpB promoter, yielding overexpressor [PsbA2R(+)] and suppressor [PsbA2R(−)], respectively. The control ΔpsbA3 strain contains only the kanamycin (Km) resistance cassette. B, Segregation check of transformants. Disruption of the psbA3 gene and replacement of the slr2031 region were checked by PCR. WT, Wild type. C, Expression of PsbA2R and psbA2 mRNA in the overexpressor [PsbA2R(+)] and suppressor [PsbA2R(−)] mutant strains, respectively. One and 5 μg of total RNA isolated from the cells grown under GL were used for the detection of psbA2 mRNA and PsbA2R, respectively.
Effect of PsbA2R on the Stability of psbA2 mRNA
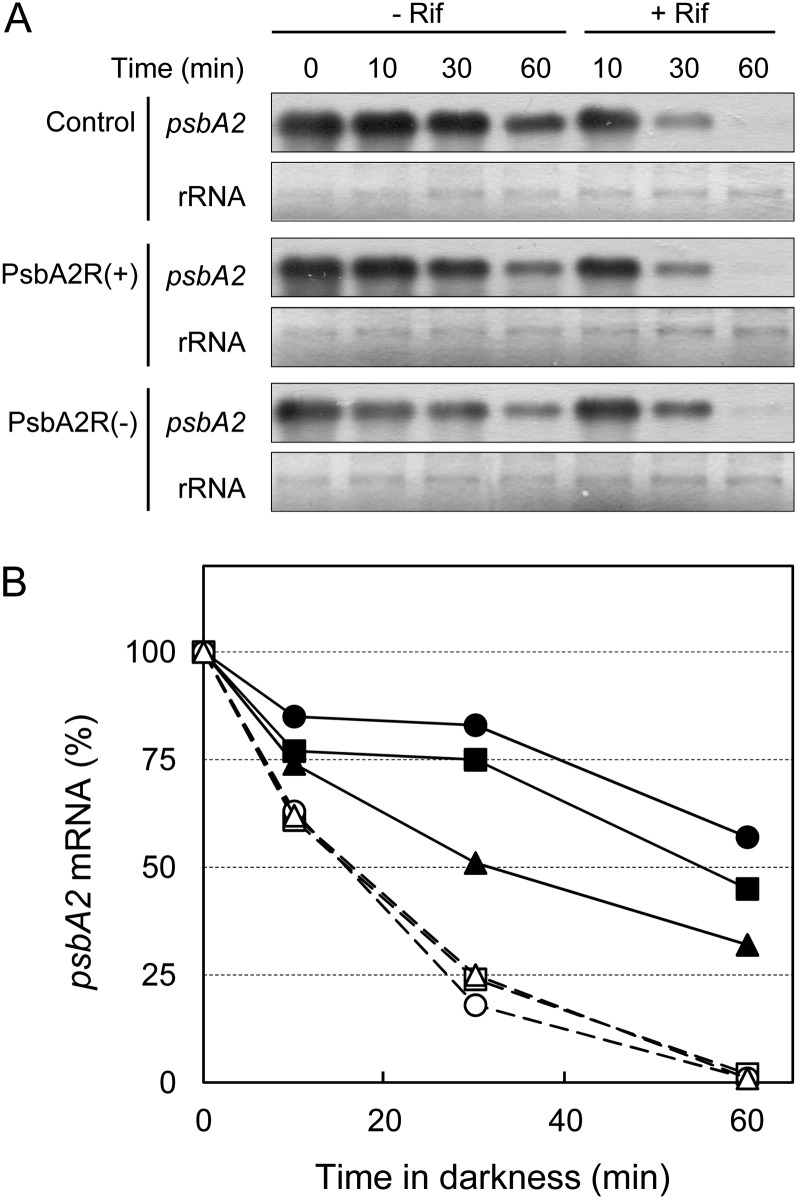
The balance between de novo synthesis and degradation determines the amount of mRNA. As the level of psbA2 mRNA was lower in the PsbA2R(−) strain as compared with the control strain, two possible functions of PsbA2R are conceivable. The asRNA PsbA2R either enhances the de novo synthesis of psbA2 mRNA or suppresses the degradation of psbA2 mRNA. To check the latter possibility, the stability of psbA2 mRNA was analyzed in the strains with modulated PsbA2R contents. Addition of Rif prevents the de novo synthesis of RNA, thus allowing the determination of transcript degradation rates. However, degradation of PsbA2R antisense transcripts is extremely fast in light as compared with the psbA2 mRNA (Fig. 4) and therefore would mask the putative effect of PsbA2R on the stability of psbA2 mRNA when the cells are incubated in light and transcription is blocked by Rif (dashed lines in Fig. 6B). Nevertheless, Figure 4 also showed that de novo transcription of psbA2 is somehow suppressed in darkness, where PsbA2R is likewise more stable. Consequently, the stability of psbA2 mRNA in PsbA2R(+) and PsbA2R(−) was analyzed in darkness in the absence of Rif. As shown in Figure 6, the degradation of psbA2 mRNA occurred more slowly in the PsbA2R(+) strain compared with the control strain. In contrast, the degradation rate of psbA2 mRNA was clearly faster in the PsbA2R(−) strain than in the control strain. These results provide strong evidence that PsbA2R functions in maintaining the stability of psbA2 mRNA.
Figure 6.
Effect of PsbA2R on the stability of psbA2 mRNA. A, Cells treated by HL for 1 h were transferred to darkness in the presence or absence of Rif. Cells were harvested for RNA analysis at the designated time points. One microgram of total RNA was used for the detection of psbA2 mRNA. B, Quantification of signal intensity. Signal intensity at 0 min (1 h of incubation of cells at HL) was set as 100%. Control, PsbA2R(+), and PsbA2R(−) are shown in squares, circles, and triangles, respectively, in the presence (closed symbols with solid lines) and absence (open symbols with dashed lines) of Rif.
Growth and Photosynthesis of PsbA2R(+) and PsbA2R(−) Strains
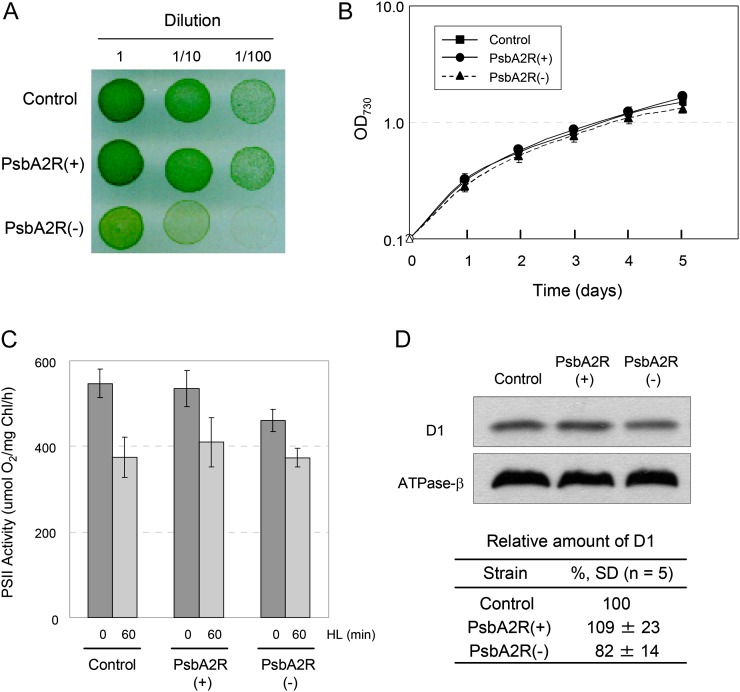
Next, we analyzed the physiological phenotypes of PsbA2R(+) and PsbA2R(−). On agar plates, the growth of PsbA2R(+) was similar to the control strain. On the contrary, PsbA2R(−) showed clearly slower growth compared with the control strain and PsbA2R(+) (Fig. 7A). However, the growth phenotype was lost when the PsbA2R(−) strain was grown in liquid medium (Fig. 7B).
Figure 7.
The effect of PsbA2R on growth and photosynthesis of Synechocystis sp. PCC 6803 cells. A, Growth of mutants on an agar plate. The cells were incubated for 4 d under GL conditions. B, Growth of mutants in liquid culture. OD730, Optical density at 730 nm. C, PSII-dependent oxygen-evolving activity. Oxygen evolution was measured in the presence of 2 mm 2,6-dimethyl-p-benzoquinone and 1 mm potassium ferricyanide at 30°C from the suspension culture grown under GL conditions and from cells illuminated under HL for 1 h prior. D, Amount of D1 protein in the mutants. Thylakoid membranes containing 0.5 μg of chlorophyll were solubilized, and the supernatant was applied to the gel. The same blot was used for the detection of the D1 protein and the β-subunit of ATPase. The table shows the relative amount of the D1 protein in each transformant. Data represent means ± sd from more than three independent experiments.
PSII activity of the PsbA2R(+) and PsbA2R(−) strains was analyzed using a Clark-type oxygen electrode (Fig. 7C). Under normal GL conditions, the control strain and PsbA2R(+) showed similar oxygen evolution activity of PSII, 550 and 535 μmol oxygen mg−1 chlorophyll h−1, respectively. PsbA2R(−) showed lower oxygen-evolving activity, approximately 85% of that measured in the control strain. In line with this result, PsbA2R(−) also contained a reduced amount of the D1 protein as compared with the control or the PsbA2R(+) strain (Fig. 7D).
In Vitro Indication for the Protection of psbA2 mRNA by PsbA2R against RNase E Attack
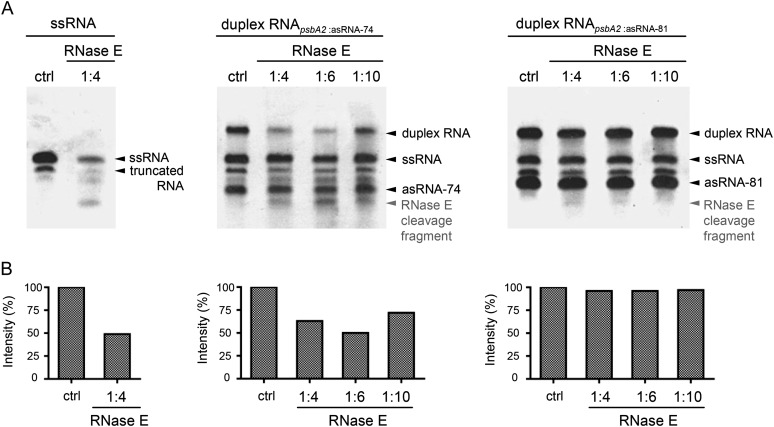
RNase E has an important role in the degradation of psbA2 mRNA, as shown previously (Horie et al., 2007). Since our experiments pointed to the fact that PsbA2R possibly functions in stabilizing the psbA2 mRNA (Fig. 6), we used an RNase E in vitro assay to gain further insights into the protective role of PsbA2R. Artificially synthesized psbA2 mRNA (110 nucleotides, positions −49 to +61, as described by Horie et al. [2007]) was incubated with purified RNase E both in the absence and presence of PsbA2R asRNA (74 nucleotides, positions −19 to −92). RNase E cleaved the single-stranded form of psbA2 mRNA into specific RNA fragments (Fig. 8A). Different from our expectations, however, psbA2 mRNA was degraded by RNase E also in the presence of PsbA2R (Fig. 8, middle panels). PsbA2R covers the AU box but not the RBS in the 5′ UTR of psbA2 mRNA (Horie et al., 2007). Both sites are recognized by RNase E, yet only the AU box can be masked by duplex formation. Nevertheless, Horie et al. (2007) demonstrated that psbA2 mRNA is successfully protected against RNase E also at the RBS, which would not be covered by PsbA2R. It is thus conceivable that in vivo binding of ribosomes protects psbA2 mRNA from RNase E attack at the latter site. To elucidate this question, we synthesized a “long and artificial” form of PsbA2R (81 nucleotides, positions −12 to −92), which covers both RNase E cleavage sites, and analyzed the protective effect of such an artificial PsbA2R. As shown in Figure 8 (right panels), the amount of the RNase E degradation products from psbA2 mRNA is substantially diminished in the presence of the long and artificial form of PsbA2R as compared with the presence of natural PsbA2R species. These data suggested that masking of both the AU box and the RBS is necessary to protect psbA2 mRNA from degradation by RNase E.
Figure 8.
Protection of psbA2 mRNA from RNase E attack by PsbA2R. A, Single-stranded psbA2-110 RNA (left panel) and duplex psbA2-110 RNA (middle and right panels) were treated without (ctrl) or with recombinant Synechocystis sp. PCC 6803 RNase E in a designated dilution. Duplex formation with psbA2 asRNA-74 (middle panel) and psbA2 asRNA-81 (right panel) served to mask one RNase E site (AU box) and two RNase E sites (AU box and RBS) in psbA2 mRNA (Horie et al., 2007), respectively. Following incubation with RNase E for 15 min, RNA cleavage patterns were visualized on ethidium bromide-stained 7 m urea-6% polyacrylamide gels. B, psbA2-110 RNA was quantified with Quantity One software (Bio-Rad). Signal intensities of each single-stranded RNA fraction and duplex fraction were combined to estimate total abundance of psbA2-110 RNA. Comparison of RNase E-treated samples and nontreated (ctrl) samples showed no decline in signal intensity for asRNA fractions. Thereby, the decline in signal intensity in duplex fractions exclusively accounts for the cleavage of psbA2-110 RNA.
DISCUSSION
Expression of the D1 protein of PSII is strictly regulated, and in general, no free D1 protein accumulates in the thylakoid membrane. A sufficient number of psbA mRNAs is expected to be always available, being tightly controlled by the intensity of light. In light, the psbA mRNAs reside in translation initiation complexes in the cytoplasm (Tyystjärvi et al., 2001), being ready for cotranslational insertion of the new D1 copy directly into the partially disassembled PSII complex whose D1 protein has been degraded. In cyanobacteria, regulation of psbA gene expression was postulated to occur mainly at the level of transcription (Golden et al., 1986; Mohamed and Jansson, 1989; Mulo et al., 2012). Yet, regulatory mechanisms at the molecular level are largely unresolved. Here, we provide novel data on psbA gene regulation in Synechocystis sp. PCC 6803 based on positive interaction between the psbA mRNAs and their respective asRNAs.
The asRNAs of the psbA2 and psbA3 genes were first discovered by deep sequencing of the Synechocystis sp. PCC 6803 transcriptome (Mitschke et al., 2011a). These asRNAs, named PsbA2R and PsbA3R, are located in the 5′ UTR of psbA2 and psbA3 genes, respectively, and thus belong to cis-type asRNAs (Waters and Storz, 2009). The expression of PsbA2R and PsbA3R is shown here to be regulated by light, the asRNAs accumulating with increasing light intensity, similar to the expression of their target psbA2 and psbA3 genes (Fig. 2). Corresponding responses of both the mRNA and asRNA upon changes in environmental conditions are so far unique for the psbA2 and psbA3 genes. Indeed, many asRNAs studied thus far have revealed an opposite expression pattern compared with their target mRNAs (Kawano et al., 2002; Silvaggi et al., 2005; Dühring et al., 2006). A clearly retarded growth of the PsbA2R(−) strain was evident on agar plates, which provides evidence for an important function of PsbA2R under some environmental conditions. Suppression of PsbA2R also induced a decrease in the abundance of psbA2 mRNA (Fig. 5C), which, based both on in vivo and in vitro experiments, was interpreted to result from a decrease in psbA2 mRNA stability (Fig. 6). From the cellular physiology point of view, such a decrease in the expression of the psbA2 gene was not detrimental for the growth of Synechocystis sp. PCC 6803 cells under standard growth conditions (Fig. 7B). Nevertheless, the down-regulation of psbA2 transcripts in the PsbA2R(−) strain resulted in lowering of the maximal PSII activity and in reduction in amount of the D1 protein (Fig. 7, C and D). Therefore, it is highly likely that the expression of PsbA2R has an important function in Synechocystis sp. PCC 6803 cells in maintaining the cellular capacity to rapidly respond to transient changes in environmental conditions that require higher performance of PSII than occurs under standard growth conditions.
It is also worth emphasizing that the steady-state amount of PsbA2R is much less than that of the psbA2 mRNA (1:300; Fig. 3). This is apparently compensated for by rapid turnover of the PsbA2R asRNA, which is at least 1 order of magnitude faster compared with the psbA2 mRNA. Our recent genome-wide analysis of the cyanobacterium Prochlorococcus sp. MED4 demonstrated the presence of many short-lived asRNAs (Steglich et al., 2010). Thus, a short lifetime appears as a common feature of many asRNAs, which further means that the molecular mechanism of regulation of the target mRNA depends on freshly synthesized antisense transcripts.
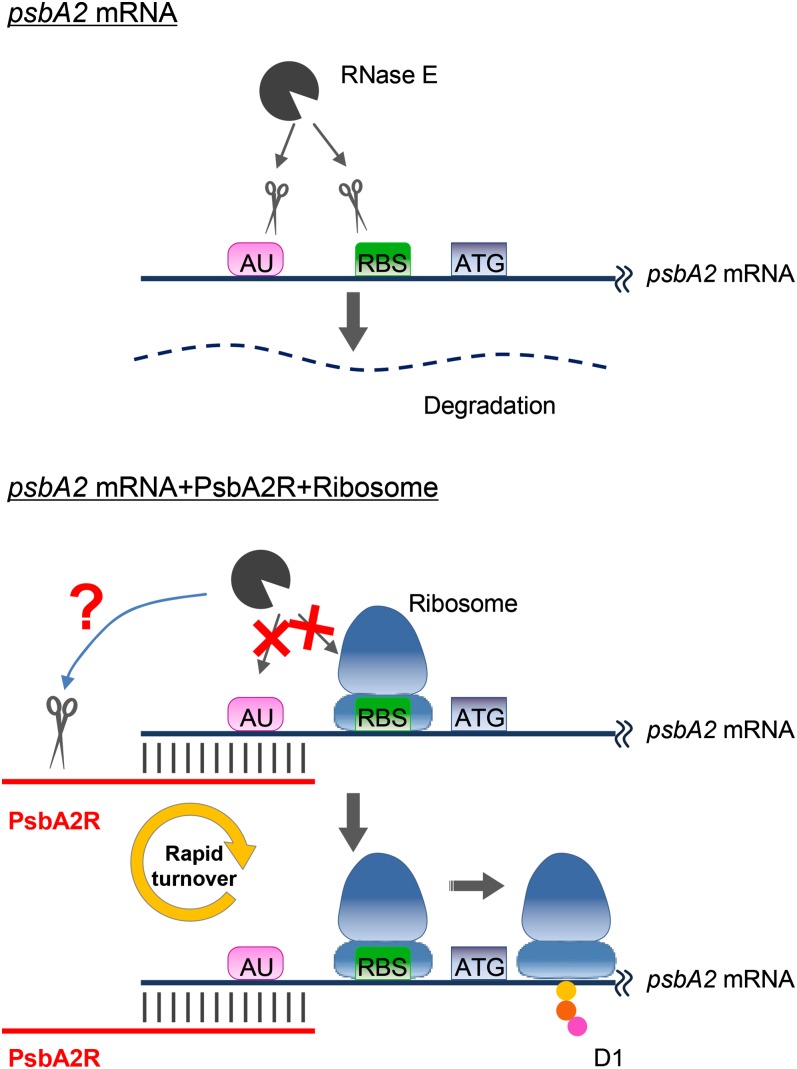
Elucidation of the mechanism for positive posttranscriptional regulation of the psbA2 gene by the cis-type PsbA2R transcript needs a closer sequence inspection of both the target gene psbA2 and the respective asRNA PsbA2R, their cleavage by RNases, as well as their interaction with ribosomes. In Figure 9, we have summarized the present understanding of the function of PsbA2R in the regulation of psbA2 gene expression.
Figure 9.
Hypothetical model on the regulation of psbA2 gene expression by PsbA2R. psbA2 mRNA is stabilized both by the annealing of PsbA2R and the binding of ribosomes. For more details, see text.
Single-stranded psbA2 mRNA is cleaved by RNase E at the AU box and RBS, both located in the 5′ UTR of the mRNA. Deduced from sequence information, PsbA2R has a capacity to protect the AU box by duplex formation with the psbA2 mRNA. Our in vitro RNase E assay, however, showed no protection of the psbA2 mRNA by the longest naturally occurring form of PsbA2R in cells. Only the long and artificial form of PsbA2R, covering both the AU box and RBS, had a protective effect against RNase E cleavage of the psbA2 mRNA. In the cellular environment in vivo, one would expect that the binding of the ribosome prevents cleavage at the RBS. It is thus conceivable that, in vivo, the psbA2 mRNA is stabilized by both PsbA2R and ribosome binding at the AU box and RBS, respectively. The double protection mechanism is likely important in stabilizing the initiation step of D1 protein translation. The requirement for PsbA2R drops sharply upon translational initiation, which is a very fast event, and would be consistent with the extremely short lifetime and low abundance of PsbA2R compared with the psbA2 mRNA, which then resides on polysomes during translation. Another feature of PsbA2R with possible mechanistic implications is that it is longer than the 49-nucleotide-long 5′ UTRs of the psbA2 mRNA and thus lacks an annealing partner. This raises the possibility that the single-stranded region of PsbA2R is subject to degradation in the cell by RNases more efficiently than the psbA2 mRNA during formation of the duplex structure and the subsequent translation event.
Besides clarification of the function of PsbA2R in regulation of the abundance of psbA2 transcripts, we also recognized the presence of PsbA3R. From the sequence similarity of these asRNAs, especially the 15 to 20 nucleotides at the 5′ end where the AU box was found, it is highly likely that the mechanism to regulate the expression of their target mRNAs is similar between PsbA2R and PsbA3R. On the other hand, PsbA2R and PsbA3R sequences are very different in other regions, providing evidence that both asRNAs specifically regulate the expression of their cis-encoded target gene.
In summary, asRNAs of psbA genes, PsbA2R and PsbA3R, were identified in the 5′ UTR of the psbA2 and psbA3 genes, respectively. These asRNA molecules are found in low abundance in the cell, yet they are shown here to act as important positive regulators to achieve a stable level of D1 protein synthesis that maintains the functionality of water-splitting PSII. Such an effect of the PsbA2R is reached by stabilizing the psbA2 mRNA. Collectively, our results strongly suggest that PsbA2R functions as a positive regulator in the expression of the psbA2 gene, which maintains photosynthesis in Synechocystis sp. PCC 6803 cells upon changes in environmental conditions, particularly at high light intensities. The results obtained in this study add a new layer of complexity to the mechanisms that contribute to the regulation of psbA gene expression. They also provide evidence that low-abundance asRNAs cannot be regarded as transcriptional noise but probably have important regulatory functions in cell physiology and indeed call for thorough investigation.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Synechocystis sp. PCC 6803 was grown photoautotrophically at 30°C in BG-11 medium (Allen, 1968) supplemented with 20 mm HEPES-NaOH (pH 7.5). The BG-11 medium for mutant strains (see below) was supplemented with 25 μg mL−1 kanamycin and 15 μg mL−1 spectinomycin. Cell growth was monitored by determination of optical density at 730 nm. As normal GL conditions, 50 μmol photons m−2 s−1 white light was provided from a fluorescent lamp, and 1,000 μmol photons m−2 s−1 white light provided from a slide projector was used as HL. For the dark treatment, the flask was covered with aluminum foil. Precultured cells were collected and resuspended in fresh BG-11 at the cell concentration of optical density at 730 nm = 0.6. These cultures were incubated under the normal GL conditions for 3 h before the experiments. Rif was used at 200 μg mL−1 to prevent de novo transcription.
RNA Extraction and Northern-Blot Analysis
Total RNA was isolated from 40 mL of culture using TriSure reagent (Bioline) according to the company’s instructions and treated with DNase (Ambion Turbo DNase kit) to remove the genomic DNA. psbA2 and psbA3 mRNAs were analyzed as described by Tyystjärvi et al. (2001), and PsbA2R and PsbA3R were analyzed as described by Axmann et al. (2005). All primers used to amplify the DNA templates for RNA probes and standard RNA are listed in Table I. RNA probe and standard RNA molecules were prepared with the MAXIscript kit (Ambion) according to the company’s instructions. [α-32P]UTP (Perkin-Elmer) was used for the labeling of probes.
Table I. Primers used in this work.
Restriction sites are underlined. fw, Forward; rev, reverse.
| Name | Sequence (5′→3′) | Experiment |
|---|---|---|
| psbA2_F-T7 | TAATACGACTCACTATAGGGAGCGCGCTGTTGGAGAGTCGTTGTC | psbA2 probe |
| psbA2_R | AGTCAGTTCCAATCTGAACATCGAC | psbA2 probe |
| PsbA2R_F-T7 | TAATACGACTCACTATAGGGAAAACTCTCATTAATCCTT | PsbA2R probe |
| PsbA2R _R | TGTATTTGTCGATGTTCAGA | PsbA2R probe |
| psbA3_F-T7 | TAATACGACTCACTATAGGGAGTATTTGTCAATGTTCAAAGGAT | psbA3 probe |
| psbA3_R | ATACATAACCGGCTCCCAAGC | psbA3 probe |
| PsbA3R_F-T7 | TAATACGACTCACTATAGGGAAACTCATATACATAACCGGCTCC | PsbA3R probe |
| PsbA3R _R | GTATTTGTCAATGTTCAAAGGATTTG | PsbA3R probe |
| 5′ RACE_RNA adaptor | GAUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA | 5′ RACE |
| 3′ RACE_RNA adaptor | UUCACUGUUCUUAGCGGCCGCAUGCUC | 3′ RACE |
| 5′ RACE_RNA Adaptor_comp_F | GAATTCCTGTAGAACGAACACTAG | 5′ RACE, PCR |
| 3′ RACE_RNA adaptor_comp_R | CATGCGGCCGCTAAGTTCAGTGAA | 3′ RACE, PCR |
| PsbA2R_5′ RACE_R1 | ACTTATGTCATCTATAAGCTTCG | 5′ RACE, first PCR |
| PsbA2R_5′ RACE_R2 | CGTGTATATTAACTTCCTGTTAC | 5′ RACE, second PCR |
| PsbA2R_3′ RACE_F | AGATTGGAACTGACTAAAC | 3′ RACE, PCR |
| PsbA3R_5′ RACE_R1 | TTCAGCAAGCTATTTACAAATTGT | 5′ RACE, first PCR |
| PsbA3R_5′ RACE_R2 | TTACTCATATACATAACCGGC | 5′ RACE, second PCR |
| PsbA3R_3′ RACE_F | AAAGGATTTGGCCTCAAGC | 3′ RACE, PCR |
| psbA2_Std_F-T7 | TAATACGACTCACTATAGGGAGTCAGTTCCAATCTGAACATCGA | psbA2 standard |
| psbA2_ Std_R | CCAAGGAATTAACCGTTGACAG | psbA2 standard |
| PsbA2R_Std_F-T7 | TAATACGACTCACTATAGGGAGGTATTTGTCGATGTTCAGATTGGA | PsbA2R standard |
| PsbA2R _ Std_R | ACAAGAAAGTAAAACTTATGTCATCTA | PsbA2R standard |
| ΔpsbA3_F1 | ACATGCATGCTAGTGGTGCTGTTGGCGTTG | Mutagenesis (psbA3, upstream) |
| ΔpsbA3_R1 | CCCAAGCTTATCTGTCTAGCCGGTGGTTG | Mutagenesis (psbA3, upstream) |
| ΔpsbA3_F2 | CCCAAGCTTTATCGGTTTGCACTTCTACC | Mutagenesis (psbA3, downstream) |
| ΔpsbA3_R2 | AACTGCAGTAACCGTTGACAGCAGGAGC | Mutagenesis (psbA3, downstream) |
| ΔpsbA3_F3 | TGCAGGATTAGTGGCTTTCC | Mutagenesis (psbA3, segregation check) |
| ΔpsbA3_R3 | CCATCAGAGAAGGAGCCTTGACCA | Mutagenesis (psbA3, segregation check) |
| PsbA2R (+)_F1 | GGCACACTAGAAAAAGTATTTGTCGATGTTCAGATTGG | Mutagenesis (two-step PCR) |
| PsbA2R (+)_R1 | ACGCGTCGACACAAGAAAGTAAAACTTATGTCATC | Mutagenesis (two-step PCR) |
| PsbA2R (−)_F1 | GGCACACTAGAAAAAACAAAGCTTTACAAAACTCTCAT | Mutagenesis (two-step PCR) |
| PsbA2R (−)_R1 | ACGCGTCGACGGAATAAAAAACGCCCGGCGGCAACCGAGCGTTGTATTTGTCGATGTTCAGATTGG | Mutagenesis (two-step PCR) |
| rnpB_F1 | AACTGCAGTTCAATGCGGTCCAATACCTCC | Mutagenesis (two-step PCR) |
| rnpB(+)_R1 | AACATCGACAAATACTTTTTCTAGTGTGCCATTGATTAG | Mutagenesis (two-step PCR) |
| rnpB(−)_R1 | TTTGTAAAGCTTTGTTTTTTCTAGTGTGCCATTGATTA | Mutagenesis (two-step PCR) |
| pEX_F1 | GGGCCCAGTGACTATTGGGGTCAG | Mutagenesis (slr2031, upstream) |
| pEX_R1 | GGTACCGGGTGTTATAACAGGCGATC | Mutagenesis (slr2031, upstream) |
| pEX_F2 | TCTAGAGATCAAACCGCTGTGGCG | Mutagenesis (slr2031, downstream) |
| pEX_R2 | ATGCATCAATGTCATATCGTCTCGGGC | Mutagenesis (slr2031, downstream) |
| pEX_F3 | GATGCCGATGGCAGTGC | Mutagenesis (slr2031, segregation check) |
| pEX_R3 | ACAATACTCCAGGGCACC | Mutagenesis (slr2031, segregation check) |
| T7-psbA2-110 fw | TAATACGACTCACTATAGGGAGTCAGTTCCAATCTGAACATC | In vitro RNase E assay |
| psbA2-110 rev | CCCACTGACAAAACTGTTCC | In vitro RNase E assay |
| T7-psbA2 asRNA-74 fw | TAATACGACTCACTATAGGGGTATTTGTCGATGTTCAGATTG | In vitro RNase E assay |
| psbA2 asRNA-74 rev | ACAAAGCTTTACAAAACTCTCATT | In vitro RNase E assay |
| T7-psbA2 asRNA-81 fw | TAATACGACTCACTATAGGGTCCTTATGTATTTGTCGATGTTC | In vitro RNase E assay |
| psbA2 asRNA-81 rev | ACAAAGCTTTACAAAACTCTCATT | In vitro RNase E assay |
5′ and 3′ RACE
The 5′ end and 3′ end RACE was performed according to Argaman et al. (2001). Sequences of RNA adaptors and DNA primers are listed in Table I.
Mutagenesis
Sequences of DNA primers used for mutagenesis are listed in Table I. The ΔpsbA3 mutant was prepared as a parental strain to generate PsbA2R(+) and PsbA2R(−) as well as the control strain. Two DNA fragments, psbA3-up and psbA3-down, were amplified by PCR with the primer sets ΔpsbA3_F1/ΔpsbA3_R1 and ΔpsbA3_F2/ΔpsbA3_R2, respectively. These fragments were treated with SphI/HindIII and HindIII/PstI, respectively. The spectinomycin resistance cassette was cut out from pHP45 by HindIII. pGEM-T vector was treated with SphI/PstI, and the fragment containing the ampicillin resistance cassette was extracted. These four fragments were ligated using the T4 DNA ligase (New England Biolabs), and the obtained plasmid was used to generate the ΔpsbA3 mutant by homologous recombination (Golden et al., 1987). The segregation of genomic DNA was confirmed by PCR using a ΔpsbA3_F3/ΔpsbA3_R3 primer set.
To generate PsbA2R(+) and PsbA2R(−) expression strains, sense or antisense versions of PsbA2R were amplified with the primer sets PsbA2R(+)_F1/PsbA2R(+)_R1 and PsbA2R(−)_F1/PsbA2R(−)_R1, respectively. PsbA2R(−)_R1 contains the sequence of the oop terminator of the bacteriophage λ. On the other hand, the rnpB promoter region was amplified with the primer sets rnpB_F1/rnpB(+)_R1 and rnpB_F1/rnpB(−)_R1 for PsbA2R(+) and PsbA2R(−), respectively. These fragments were fused in a two-step PCR with the primer sets rnpB_F1/PsbA2R(+)_R1 and rnpB_F1/PsbA2R (−)_R1. After digestion with PstI/SalI, these fragments were cloned into the PstI/SalI double-digested pEX6803 together with the kanamycin resistance cassette obtained by PstI digestion of pUC4K. The pEX6803 plasmid was prepared as below for homologous recombination at the slr2031 region. Segments upstream and downstream of slr2031 were amplified with the primer sets pEX_F1/pEX_R1 and pEX_F2/pEX_R2, respectively. These fragments were subcloned into pGEM-T vector and cut out by double digestion with ApaI/KpnI and XbaI/NsiI, respectively, for the following ligation. pDrive and pGEM-T were treated with KpnI/XbaI and ApaI/NsiI to get the multiple cloning site or ampicillin resistance cassette-containing fragments. Ligation of these four fragments gave pEX6803. The pEX6803 vector containing only the kanamycin resistance cassette was used for generation of the control strain. After homologous recombination, the segregation was checked by PCR with the pEX_F3/pEX_R3 primer set.
Thylakoid Membrane Isolation and Western-Blot Analysis
Thylakoid membranes were isolated from 30 mL of cell culture according to Kashino et al. (2002). Chlorophyll concentration was measured by the method of Arnon et al. (1974).
Western analysis was done by standard procedure using the thylakoid membranes containing 0.5 μg of chlorophyll. The D1 protein and β-subunit of ATPase were detected by the chemiluminescence method. The signal intensities of the D1 protein obtained from western analysis were quantified (NIH ImageJ software) and normalized by the amount of β-subunit of ATPase.
Measurement of PSII Activity
PSII activity was measured with a Clark-type oxygen electrode using 5 μg of chlorophyll-containing cells in the presence of 2 mm 2,6-dimethyl-p-benzoquinone and 1 mm potassium ferricyanide at 30°C.
Overexpression and Purification of Synechocystis sp. PCC 6803 Recombinant RNase E from Escherichia coli
pQESlr1129 (courtesy of M. Asayama; Horie et al., 2007) was transformed in E. coli M15 (pREP4) for the expression of recombinant Synechocystis sp. PCC 6803 RNase E containing a C-terminal hexa-His fusion. Strains were grown in Lysogeny broth supplemented with ampicillin (100 µg mL−1) and kanamycin (25 µg mL−1). Overnight cultures were diluted in Lysogeny broth and grown at 37°C to an optical density at 600 nm of 0.5 to 0.8. Following induction of heterologous expression with 1 mm isopropyl β-d-1-thiogalactopyranoside, incubation continued at 22°C for 24 h.
Recombinant Synechocystis sp. PCC 6803 RNase E was affinity purified under native conditions according to Stazic et al. (2011).
In Vitro RNase E Assay
In vitro cleavage assays were carried out according to Stazic et al. (2011) with the following modifications. The reaction mixture, containing 13 pmol of single-stranded RNA and 26 pmol of duplex RNA, was buffered with 25 mm Tris-HCl (pH 7.6). Transcripts for RNase E cleavage assays were synthesized with the MEGAshortscript system (Ambion) according to the manufacturer’s instructions. Removal of residual template DNA was conducted by incubating reaction mixtures with 2 units of TURBO DNase (Ambion) at 37°C for 15 min. Subsequently, RNA was purified by phenol-chloroform extraction and quantified with a NanoDrop ND-1000 spectrofluorometer (PEQLAB Biotechnology). The following in vitro RNAs were synthesized: psbA2-110, psbA2 asRNA-74, and psbA2 asRNA-81. Oligonucleotides used for template generation are listed in Table I.
Acknowledgments
We thank Munehiko Asayama (Ibaraki University) for providing plasmid pQESlr1129.
Glossary
- asRNA
antisense RNA
- UTR
untranslated region
- GL
growth light
- HL
high light
- Rif
rifampicin
- RBS
ribosome-binding site
References
- Allen M. (1968) Simple condition for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K. (1974) Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta 357: 231–245 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. (2005) Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol 6: R73.1–R73.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Hurry VM, Gustafsson P, Oquist G. (1993) Two functionally distinct forms of the photosystem II reaction-center protein D1 in the cyanobacterium Synechococcus sp. PCC 7942. Proc Natl Acad Sci USA 90: 11985–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring U, Axmann IM, Hess WR, Wilde A. (2006) An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA 103: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Georg J, Klaehn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro E-M. (2012) The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Hess WR. (2011) cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol 5: 305.1–305.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R. (1986) Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J 5: 2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R. (1987) Genetic engineering of the cyanobacterial chromosome. Methods Enzymol 153: 215–231 [DOI] [PubMed] [Google Scholar]
- Hernández JA, Alonso I, Pellicer S, Luisa Peleato M, Cases R, Strasser RJ, Barja F, Fillat MF. (2010) Mutants of Anabaena sp. PCC 7120 lacking alr1690 and alpha-furA antisense RNA show a pleiotropic phenotype and altered photosynthetic machinery. J Plant Physiol 167: 430–437 [DOI] [PubMed] [Google Scholar]
- Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. (2006) Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol 355: 325–334 [DOI] [PubMed] [Google Scholar]
- Horie Y, Ito Y, Ono M, Moriwaki N, Kato H, Hamakubo Y, Amano T, Wachi M, Shirai M, Asayama M. (2007) Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol Genet Genomics 278: 331–346 [DOI] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41: 8004–8012 [DOI] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. (2002) Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol 45: 333–349 [DOI] [PubMed] [Google Scholar]
- Kós PB, Deák Z, Cheregi O, Vass I. (2008) Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim Biophys Acta 1777: 74–83 [DOI] [PubMed] [Google Scholar]
- Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. (2008) Small RNAs establish delays and temporal thresholds in gene expression. Biophys J 95: 3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máté Z, Sass L, Szekeres M, Vass I, Nagy F. (1998) UV-B-induced differential transcription of psbA genes encoding the D1 protein of photosystem II in the cyanobacterium Synechocystis 6803. J Biol Chem 273: 17439–17444 [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. (2011a) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. (2011b) Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA 108: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Eriksson J, Osiewacz H, Jansson C. (1993) Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol Gen Genet 238: 161–168 [DOI] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. (1989) Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol 13: 693–700 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817: 247–257 [DOI] [PubMed] [Google Scholar]
- Nanba O, Satoh K. (1987) Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci USA 84: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicora CI, Ho FM, Salminen T, Styring S, Aro EM. (2009) Transcription of a “silent” cyanobacterial psbA gene is induced by microaerobic conditions. Biochim Biophys Acta 1787: 105–112 [DOI] [PubMed] [Google Scholar]
- Silvaggi JM, Perkins JB, Losick R. (2005) Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol 187: 6641–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stazic D, Lindell D, Steglich C. (2011) Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 39: 4890–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm SW. (2010) Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol 11: R54.1–R54.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield TC, Toepel J, Sherman LA. (2008) Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 47: 12939–12941 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi T, Herranen M, Aro EM. (2001) Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol Microbiol 40: 476–484 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. (2009) Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM. (2009) Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS ONE 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM. (2012a) Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, et al. (2012b) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol 13: R20.1–R20.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. (2012) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W. (2009) Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 19: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]