Abstract
The Golgi apparatus is the central organelle in the secretory pathway and plays key roles in glycosylation, protein sorting, and secretion in plants. Enzymes involved in the biosynthesis of complex polysaccharides, glycoproteins, and glycolipids are located in this organelle, but the majority of them remain uncharacterized. Here, we studied the Arabidopsis (Arabidopsis thaliana) membrane proteome with a focus on the Golgi apparatus using localization of organelle proteins by isotope tagging. By applying multivariate data analysis to a combined data set of two new and two previously published localization of organelle proteins by isotope tagging experiments, we identified the subcellular localization of 1,110 proteins with high confidence. These include 197 Golgi apparatus proteins, 79 of which have not been localized previously by a high-confidence method, as well as the localization of 304 endoplasmic reticulum and 208 plasma membrane proteins. Comparison of the hydrophobic domains of the localized proteins showed that the single-span transmembrane domains have unique properties in each organelle. Many of the novel Golgi-localized proteins belong to uncharacterized protein families. Structure-based homology analysis identified 12 putative Golgi glycosyltransferase (GT) families that have no functionally characterized members and, therefore, are not yet assigned to a Carbohydrate-Active Enzymes database GT family. The substantial numbers of these putative GTs lead us to estimate that the true number of plant Golgi GTs might be one-third above those currently annotated. Other newly identified proteins are likely to be involved in the transport and interconversion of nucleotide sugar substrates as well as polysaccharide and protein modification.
The Golgi apparatus is the central organelle in the secretory pathway, and in higher plants it is involved in the biosynthesis and transport of cell wall matrix polysaccharides, glycoproteins, proteoglycans, and glycolipids as well as in protein trafficking to different subcellular compartments. The last decade has produced substantial findings on the function of the Golgi apparatus: insights into the protein trafficking at the endoplasmic reticulum (ER)/Golgi interface, Golgi structural maintenance, its involvement in endocytosis, and its behavior during cell division (for review, see Faso et al., 2009). However, despite its importance, only a small proportion of the Golgi proteome has been studied: relatively few Golgi proteins have been localized, and even fewer have been functionally characterized.
The Golgi apparatus is thought to contain a large and diverse group of membrane-bound glycosyltransferases (GTs). The current view is that different GT activities are required for synthesis of the linkage between different donor and acceptor sugars. Having in mind the diversity of linkage types found in cell wall polysaccharides, the number of different GTs involved is likely to be very large. For instance, it has been estimated that for the biosynthesis of pectin alone, the action of 65 different enzymatic activities is needed (Caffall and Mohnen, 2009). By the end of the year 2011, 468 Arabidopsis (Arabidopsis thaliana) sequences had been annotated in the Carbohydrate-Active EnZymes (CAZy) GT database (Cantarel et al., 2009; http://www.cazy.org). We estimate that two-thirds of these CAZy-classified GTs may be targeted to the Golgi. The remaining one-third are cytosolic or plastidic enzymes involved in processes including, secondary metabolism or starch synthesis. The reported sequences are classified into 43 CAZy families based on amino acid sequence similarities within which at least one member has been biochemically characterized. Each family is likely to have a common structural fold, and three-dimensional (3-D) structures have been resolved for 20 of these 43 families. These are divided mostly into two structural classes, having either a GT-A fold or a GT-B fold (Unligil and Rini, 2000; Bourne and Henrissat, 2001). Moreover, most of the structurally uncharacterized GT families are predicted to adopt either the GT-A or GT-B fold based on 3-D structural homology modeling (Coutinho et al., 2003; Lairson et al., 2008). Despite this conserved 3-D structure, different GT families have very low or undetectable sequence similarities. Consequently, predicting novel GTs based solely on their amino acid sequence similarities is not always achievable, and structural homology searches have also proven useful (Hansen et al., 2009).
The length and properties of the transmembrane domain (TMD) of endomembrane proteins appear to play a role in protein sorting and location within the secretory pathway and can be used to predict protein localization (Hanton et al., 2005; Sharpe et al., 2010). In order to perform such predictions, a high number of experimentally localized proteins is required, but only limited data sets have been available for plants to date.
In order to identify the most abundant CAZy-classified GTs as well as novel putative GTs, in this work we rigorously extended our proteomic studies of the Golgi apparatus. We have previously developed a high-throughput mass spectrometry (MS)-based quantitative proteomics technique for localization of organelle proteins by isotope tagging (LOPIT; Dunkley et al., 2004, 2006). Here, we report new LOPIT data sets and apply a new method of combining them with published LOPIT data sets, localizing an unprecedented number of plant organelle proteins. We have analyzed the TMD properties of the proteins assigned to the ER, Golgi, and plasma membrane (PM) and determined the organelle-specific features. Structural prediction analysis of the Golgi-localized proteins with unknown functions assessed the protein sequences for the potential to fold similarly to known GT structures. We found that the Golgi contains a substantial number of candidate GT families that have no characterized functions. These results yield a broader understanding of the Golgi function and its biochemical properties.
RESULTS
Integration of LOPIT Data Sets Increases Coverage of the Golgi Proteome
We have previously described the LOPIT approach for high-throughput determination of the steady-state localization of membrane proteins (Dunkley et al., 2004, 2006). In this quantitative proteomic technique, proteins can be confidently assigned to organelles according to the pattern of their distribution on density gradients (Dunkley et al., 2006). The main aim of the work described here was to increase the number of identified Golgi-localized proteins and to find proteins putatively involved in the biosynthesis of the plant cell wall. Earlier additional replicate experiments identified relatively few novel proteins (Sadowski et al., 2008). Moreover, the additional fractionation data from replicate experiments is not easily combined to strengthen localization predictions because there are often incomplete observations of proteins in each data set.
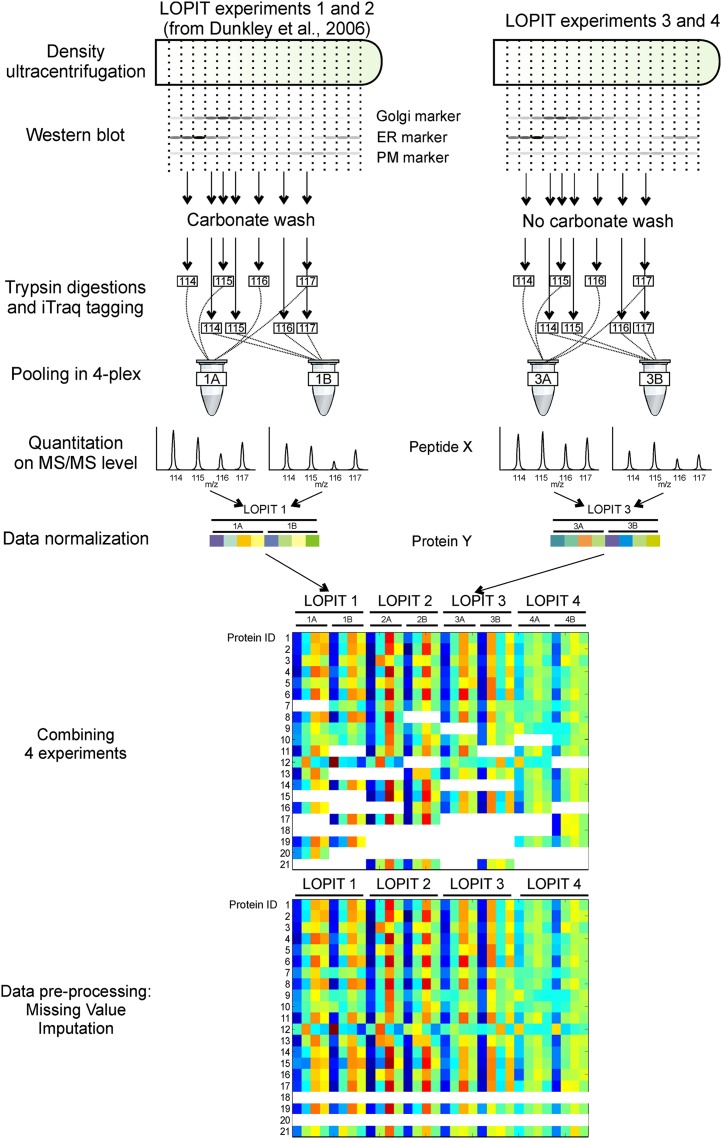
Many of the proteins involved in sugar nucleotide metabolism and membrane trafficking are peripheral. In the LOPIT experiments by Dunkley et al. (2006), these were removed by carbonate washing of the isolated membranes (Fig. 1, LOPIT experiments 1 and 2). To increase the likelihood of identifying novel proteins, we carried out two additional LOPIT experiments with samples enriched in peripheral and luminal proteins (Fig. 1, LOPIT experiments 3 and 4). Arabidopsis liquid-grown callus membrane fractions were prepared using conditions to preserve the association of the peripheral and luminal proteins. The distribution profiles of the proteins across the density gradient were estimated from their tryptic peptides labeled by four-plex iTRAQ followed by strong cation-exchange liquid chromatography-tandem mass spectrometry (MS/MS). To increase the number of fractions sampled, two four-plex labelings were performed per gradient. This allowed the quantitation of seven fractions, with one fraction common to the two labelings. The raw MS/MS spectral data sets from the two new LOPIT experiments and the two LOPIT experiments of Dunkley et al. (2006) were processed or reprocessed in parallel as described in “Materials and Methods” to produce protein abundance profiles. These analyses together identified and quantitated 2,205 proteins. Confident subcellular localization requires observations in two or more independent biological samples. A total of 1,385 proteins achieved this threshold of significance (Supplemental Table S1). The number of proteins identified and quantitated was not very different between LOPIT experiments 1 and 2 versus 3 and 4; however, as expected, almost all of the novel proteins found only in the experiments using non-carbonate-stripped membranes (LOPIT experiments 3 and 4) were without predicted TMDs (Supplemental Fig. S1).
Figure 1.
LOPIT experimental design and data preprocessing workflow. Organelle separation is obtained by density gradient ultracentrifugation. Four LOPIT experiments were performed, each consisting of two four-plex iTRAQ labelings from seven density gradient fractions. One fraction is common in both four-plex labelings. The peptide abundance values from the MS/MS quantitation of the iTRAQ reporter ions are collapsed to a protein level for each four-plex labeling and are presented as color-coded values. The missing value imputation was performed on protein profiles that were present in at least two experiments.
Our next aim was to develop an approach to classify the identified proteins into organelle categories using the incomplete protein fractionation information from all eight four-plex labelings. It has been demonstrated by Trotter et al. (2010) that combining LOPIT data sets from different experiments improves the subcellular clustering resolution of proteins. However, a known issue with quantitation is that proteins are not observed in every MS experiment, due to the intensity-dependent manner in which MS data are collected. Thus, we expected to see only a minor proportion of proteins present and quantitated in all the experiments. Indeed, only 423 of the 2,205 proteins identified and quantitated in these experiments were consistently detected in all eight iTRAQ labelings of the four experiments; the rest of the proteins had one or more missing measurements. These missing values can hinder subsequent multivariate analysis and classification algorithms, if not accounted for (Schafer, 1997). Imputation approaches have already been implemented in two-dimensional gel studies (Krogh et al., 2007; Pedreschi et al., 2008; Albrecht et al., 2010), and a few reports show the use of such algorithms in quantitative MS (Du et al., 2008; Karpievitch et al., 2009). The imputation of missing values prior to applying the classification algorithm exploits the covariance of all available protein data. Common approaches to missing data imputation include estimation of the joint probability distribution of all variables drawn from the relationship among replicates (using an expectation-maximization algorithm, for instance) and sampling missing values from this model (Rubin, 1987). In multivariate data sets, it can be difficult to specify a single joint distribution of all variables, so sequential sampling from a series of conditional predictive distributions is accepted as an approximation (Chen and Liu, 1996). A missing labeling in the LOPIT data consists of four values; therefore, it can be treated as a four-dimensional multivariate target variable, and in this case multivariate regression is a suitable choice for estimation. A partial least-squares (PLS) regression model was chosen, as it allows the use of multivariate predictors and target variables even when the number of variables exceeds the number of cases (Wold et al., 1984). In this respect, the multiple imputations procedure adopted in this study is closely related to a sequence-of-regression-models technique (Raghunathan et al., 2001). The choice of PLS regression as a predictive model was validated by a test on a subset of the data with complete observations in all eight four-plex labelings (consisting of profiles for 423 proteins). The number of latent variables of the model was chosen by a 7-fold cross-validation scheme (Geladi and Kowalski, 1986). The mean coefficient of determination for a target variable was 0.87 with se of 0.0023. The predictive power was measured by a score (cross-validated coefficient of determination [Q2]) where 0 is no better than chance and 1 is the theoretical maximum for a model that explains all the variation in the predicted data. The mean Q2 of all classes was 0.802 with se of 0.0212, showing the validity of the approach.
We applied the data imputation approach to estimate the missing values on a data matrix consisting of partial distribution profiles along the density gradient of the 1,385 proteins with two or more observations. The imputation scheme was validated by simulating random missing labelings and calculating the error of the imputation, and it demonstrated an expected relative error of the magnitude of 10% to 16% of the real data value (Fig. 2). The generation of multiple imputed data sets was then achieved by a bootstrap approach (Efron, 1994). One hundred imputed data sets were generated by predicting the missing values with a series of PLS regression models where the training set was sampled with replication from the complete and already imputed cases. This procedure is considered to give better estimation of uncertainty than a single imputation of missing data with the best-fit value or using a predictive model with a fixed set of parameters (Efron, 1994). For ease of presentation, the 100 data sets were averaged into one matrix that yielded complete subcellular fractionation profiles on 1,385 proteins in four experiments (Supplemental Table S2). This represents a 2-fold increase both in the amount of fractionation data for each protein and in the number of proteins over the experiments reported by Dunkley et al. (2006).
Figure 2.
Evaluation of the missing values imputation approach. The mean expected error of imputed data and its 95% confidence intervals (CI) are presented. The plot shows the results of 1,000 simulation runs of the imputation procedure.
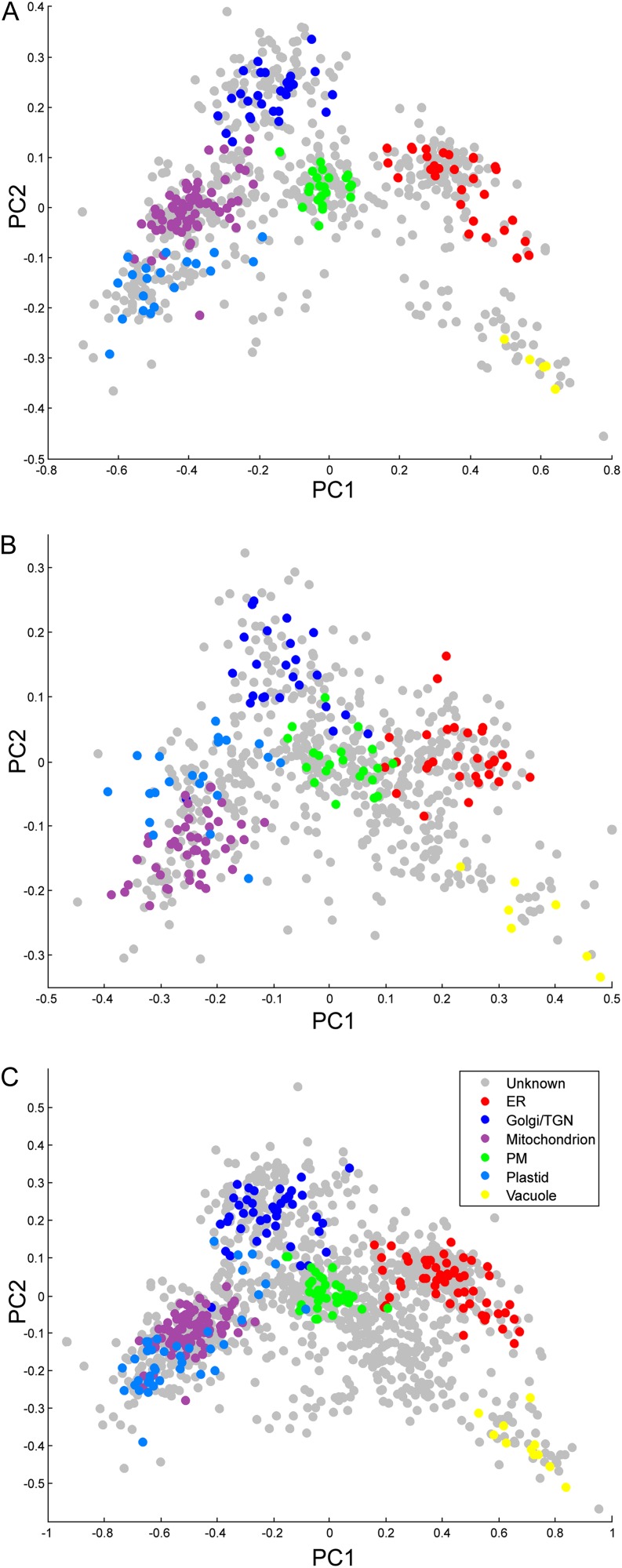
The averaged fractionation profiles were studied using principal component analysis in order to visualize the clustering of protein profiles by their organelle localization (Fig. 3). In the reanalyzed experiments 1 and 2, the plots show that marker proteins of the same organelle clustered together (Fig. 3A), as seen in the earlier analysis of the MS/MS data (Dunkley et al., 2006). The two new data sets with the modified sample preparation also showed clear clustering of marker proteins (experiments 3 and 4; Fig. 3B). The combined data from the four experiments using the imputed fractionation values showed clear clustering similar to the separate data sets, again supporting the view that the data imputation was successful (Fig. 3C).
Figure 3.
Multivariate analysis of the LOPIT data. A, Principal component analysis (PCA) plot of 728 protein profiles from the LOPIT experiments 1 and 2. B, PCA plot of 717 protein profiles from the LOPIT experiments 3 and 4. C, PCA plot of the 1,385 protein profiles of the combined data sets (LOPIT experiments 1–4) with imputed missing values. The profiles of marker proteins with previously reported subcellular localization (training set proteins) are colored in the PCA plots based on the organelle of residence.
Proteins in the LOPIT data sets previously localized to the Golgi (35 proteins) and six proteins localized to the trans-Golgi network (TGN) were used as a part of the training set for the classification analysis and prediction of further Golgi/TGN-associated proteins. The Golgi apparatus and the TGN proteins were combined into one class because they have a similar fractionation profile. The rest of the training set was composed of the proteins previously assigned to the PM (39 proteins), ER (53 proteins), vacuole (11 proteins), mitochondrion (92 proteins), and plastid (40 proteins) as well as a nonorganelle (cytosolic/ribosomal) class (72 proteins) used to constrict the classifier. The training set is presented in Supplemental Table S2.
To classify proteins into compartments, a multivariate naïve Bayesian classification model with a kernel density estimate was applied to each of the 100 imputed data sets. The choice of the naïve Bayesian model as a basis for multivariate classification was motivated by its particularly simple yet robust classification rule that uses a diagonal form of the covariance matrix. It has been reported to outperform a number of other popular classification models for multivariate biological data, including quadratic discriminant analysis and nearest neighbor classifier, and to yield better classification in situations where the sample values are correlated (Dudoit et al., 2002). Using the average posterior probability based on the naïve Bayesian classification models, 274 of the 275 membrane organelle training set proteins were assigned to the correct location. This indicates that the assignments using this approach are likely to be of high confidence.
The application of the classification procedure to the proteins with unknown localization resulted in a list of 197 proteins classified as Golgi/TGN localized (Supplemental Table S2). As expected, most of the proteins reported to be Golgi localized by Dunkley et al. (2006) lie within this set (85 out of 89). More than one-third (79 proteins) have not been previously identified to be Golgi localized by a high-confidence localization study (Supplemental Table S2; Supplemental Literature Cited S1). This represents an increase of 120% over the Golgi proteome reported by Dunkley et al. (2006). The same classification approach was applied to other organelle proteins, resulting in the assignment of 304 proteins to the ER, 208 to the PM, 213 to the mitochondrion, 151 to the plastid, and 37 to the vacuole (Supplemental Table S2).
The TMDs of Proteins Localized to Different Organelles Demonstrate Distinct Properties
TMDs of membrane proteins may harbor specific features that reflect the properties of the membrane in which they reside (Sharpe et al., 2010). Although in plants it has been shown that the TMD length has an effect on the localization of an artificial reporter protein (Brandizzi et al., 2002), there have been relatively few plant proteins localized, precluding detailed investigation of differences in the TMD properties of proteins in different organelles. The large number of proteins reliably localized by LOPIT here provides an opportunity to study whether plant proteins show organelle-specific TMD properties.
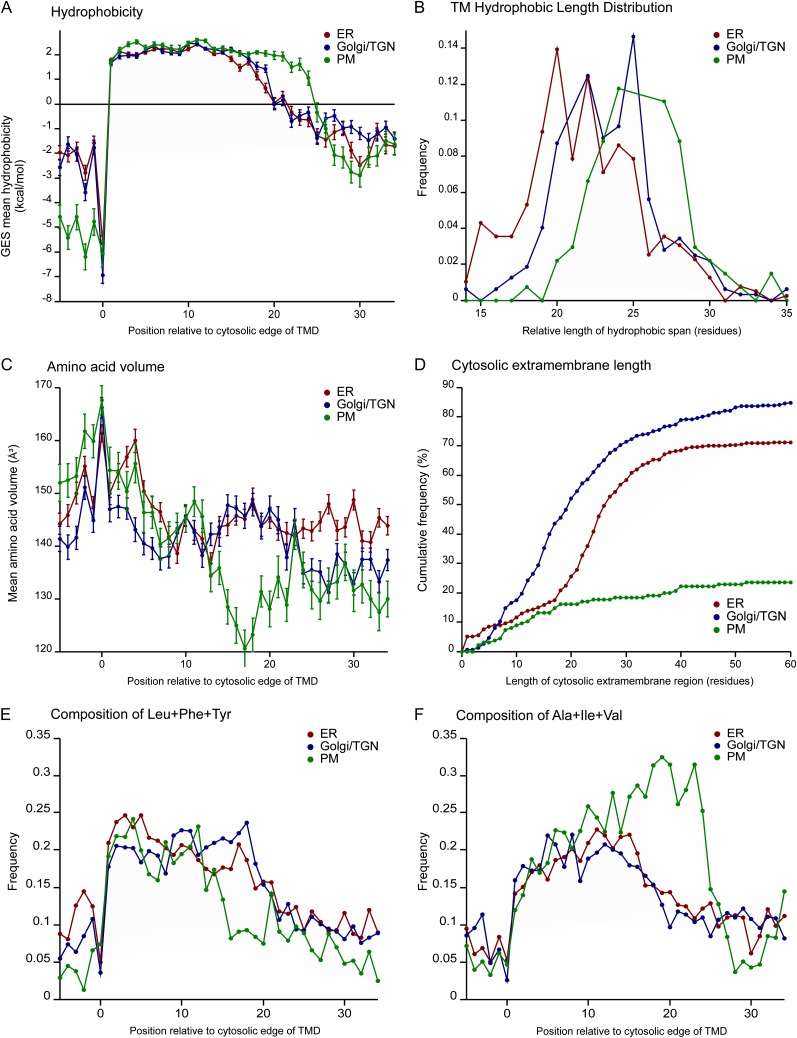
A subset of the proteins classified by the LOPIT approach was annotated as being putative single-span TMD proteins using the predictions from TMHMM (Table I; Krogh et al., 2001). The amino acid sequences of these proteins in and around their hydrophobic transmembrane span regions were analyzed similarly to Sharpe et al. (2010). This analysis refines the cytosolic hydrophobic start point of the TMD span according to a consistent heuristic function. The identification of hydrophobic start point allows aligning and computing different properties along the TMD sequence, such as average amino acid compositions, and corresponding derived parameters such as residue size and hydrophobicity. Also, measurements were made of the estimated extramembrane protein sequence length on the cytosolic side of the TMD from the same start point. To provide increased numbers of proteins for statistical analysis, the data set was augmented by the inclusion of plant orthologs of the organelle-classified Arabidopsis proteins (Table I). The method for incorporating close ortholog sequences and removing familial sample bias was done as described previously for fungi and vertebrates (Sharpe et al., 2010). The sequence database used for the ortholog search was the Streptophyta phylum subset of the UniProt database. The mean hydrophobicity of amino acid residues of all the putative TMD sequences, at positions relative to the cytosolic ends of their hydrophobic cores, was calculated using the Goldman, Engelman, and Steitz (GES) scale (Engelman et al., 1986) and plotted for the ER, Golgi, and PM organelle classifications (Fig. 4A). The hydrophobic regions of the TMDs from the Golgi and ER were of similar maximum hydrophobicity, with this maximum extending over the same number of residues. In contrast, for PM TMDs, the region of maximum hydrophobicity extended for a further five residues (Fig. 4A), similar to the situation observed in fungi and vertebrates (Sharpe et al., 2010). To demonstrate that positional averages did not obscure any differences, we plotted the frequency of hydrophobic length, defined with an intermediate GES scale threshold of 1 kcal mol−1 (Fig. 4B). The distribution plot shows distinct profiles for the three compartments, with the mean TMD hydrophobic span lengths of 21.5 residues for the ER, 22.7 for the Golgi, and 25.4 for the PM (Fig. 4B). We tested the dependence of the hydrophobic length output on the GES scale threshold used to define TMD ends and concluded that the difference between PM, ER, and Golgi is relatively unaffected by the threshold chosen, suggesting that the difference is robust (data not shown).
Table I. Summary of the number of organelle proteins used in the LOPIT approach and in the TMD analysis.
Figure 4.
Analysis of the TMD characteristics of single-span membrane proteins classified as localized to the ER, Golgi/TGN, and PM. A, Mean hydrophobicity (GES scale) of the residues at each position along the aligned TMDs relative to the cytosolic edge. The GES hydrophobicity values represent the free energy of partitioning from water into a hydrophobic environment. B, Distribution of TMD lengths. The exoplasmic ends of the TMD were defined using the hydrophobicity-scanning algorithm as for the cytosolic ends. C, Mean values for the residue volume at each position along the TMDs. D, Cumulative distribution of the cytosolic extramembrane lengths. E, Analysis of the cumulative abundance of Leu, Phe, and Tyr along the TMDs. F, Analysis of the cumulative abundance of Ala, Ile, and Val along the TMDs.
Since residue volumes are distributed asymmetrically in PM and Golgi TMDs of fungi and vertebrates (Sharpe et al., 2010), the mean residue volume was calculated along the plant TMD sequences (Fig. 4C). The TMD regions of different organelles showed no significant differences regarding the amino acid volume close to the cytosolic edge (positions 1–10). However, in the luminal/exoplasmic leaflet region, starting from position 12 there are clear differences: the PM TMDs have a significant drop in the mean residue volume, whereas Golgi and ER remain similarly high throughout the TMD (Fig. 4C). To investigate this in more detail, we analyzed the frequency of each amino acid at each position through the TMD. The amino acid compositions show differences between the TMDs from different organelles (Supplemental Fig. S2). Some of these differences are clearly associated with the differences in mean residue volume. In particular, the larger hydrophobic residues (Leu, Phe, and Tyr) are relatively depleted in the outer leaflet portion of the PM TMDs compared with Golgi and ER, while the smaller hydrophobic residues (Ala, Ile, and Val) show the opposite trend (Fig. 4, E and F). In addition to these differences in residue size, there appear to be other organelle-specific trends. For example, the PM TMDs have enriched abundance of the basic residues on the cytosolic side and increased occurrence of Cys in the interfacial region of the inner leaflet (perhaps reflecting increased palmitoylation). The Golgi TMDs are characterized by lower occurrence of Lys in the outer leaflet region, whereas the ER TMDs have increased occurrence of Met in the interfacial region of the outer leaflet (Supplemental Fig. S2).
The length of the cytosolic tail of the single-span TMD proteins also displayed specific variations for the three organelles. Over 80% of the Golgi apparatus proteins have a cytosolic tail of fewer than 45 amino acid residues. Almost as frequently, 70% of ER proteins have similarly short cytosolic tails; this observation holds for fewer than 22% of PM proteins (Fig. 4D). This is to be expected, since the transmembrane PM proteins (unlike the ER and Golgi proteins) are likely to perform more functions on the cytosolic side.
Novel Putative GTs Are Identified in the Golgi Proteome
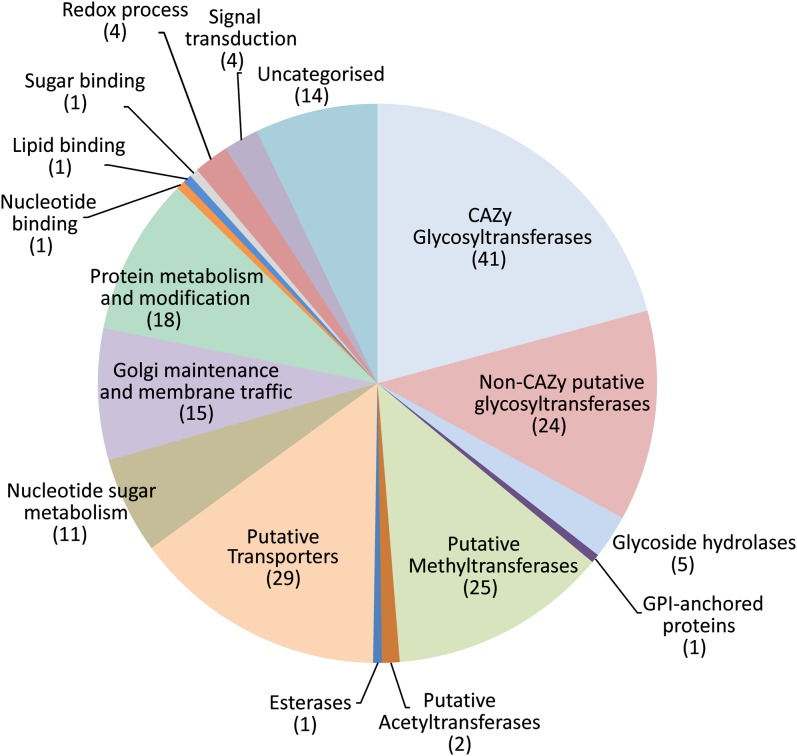
Many of the proteins (22%) identified as Golgi localized were annotated in The Arabidopsis Information Resource (TAIR) 10 database as proteins of unknown function. Since a large number of the proteins in the Golgi apparatus are likely to be involved in glycosylation (Dupree and Sherrier, 1998), we investigated whether any of these proteins show predicted structural homology to proteins of known function and structure. Currently, 3-D structural information is available for 39 of the 91 classified CAZy GT families. We analyzed the sequences of all 197 Golgi proteins using HHpred, an alignment tool based on the pairwise comparison of hidden Markov model profiles (Söding et al., 2005). HHpred builds a hidden Markov model profile from a query amino acid sequence and compares it with a database of hidden Markov models representing annotated protein families (e.g. Pfam, Simple Modular Architecture Research Tool [SMART], and Conserved Domains Database [CDD]) or domains with known structure (Protein Data Bank [PDB] and Structural Classification of Proteins [SCOP]) and provides greater sensitivity and selectivity in finding remote homologs than other sequence profile-based methods such as PSI-BLAST (Söding, 2005). The analysis of all the Golgi-localized proteins identified 65 of 197 as having a probable fold related to known GTs; of these, 41 were classified in CAZy and 24 proteins were identified as putative GTs not classified in CAZy families (Table II). In addition, the DUF579 family as well as three other unrelated proteins showed significant similarity to a conserved S-adenosylmethionine (SAM)-binding methyltransferase (MT) fold (Supplemental Fig. S3). To obtain an overview of the Golgi proteome, all 197 proteins were assigned to functional groups, and these are shown in Figure 5 and Supplemental Table S2.
Table II. The 12 putative GT families of the 24 proteins identified as Golgi localized and having predicted structural homology to CAZy-classified GTs.
For a full list of Arabidopsis GT-like proteins, see Supplemental Table S3.
| Gene Family Name | No. of Proteins Found in This Study | Gene Family Members | Structural Prediction and Remote Homology Detection (HHpred Results) |
|||
|---|---|---|---|---|---|---|
| Best PDB Hit (GT Family, Fold, Mechanism) | Probability | Best Arabidopsis CAZy-Classified GT Hit | Probability | |||
| % | % | |||||
| GT4R | 2 | 2 | 3c48 (GT4, GT-B, retaining) | 100 | At3g45100 (GT4) | 100 |
| GT8R-A | 1 | 8 | 1g9r (GT8, GT-A, retaining) | 84.2 | At3g50760 (GT8) | 83.6 |
| GT8R-B | 2 | 4 | 1g9r (GT8, GT-A, retaining) | 59.7 | At3g06260 (GT8) | 71.3 |
| GT13R | 1 | 1 | 1fo8 (GT13, GT-A, inverting) | 100 | At4g38240 (GT13), At2g35610 (GT77) | 100, 98.6 |
| GT14R | 1 | 23 | 2gak (GT14, GT-A, inverting) | 100 | At2g37585 (GT14) | 100 |
| GT27R | 2 | 11 | 2ffu (GT27, GT-A, retaining) | 95 | At1g20575 (GT2) | 96.4 |
| GT41R | 1 | 2 | 3tax (GT41, GT-B, inverting) | 100 | At3g11540 (GT41) | 100 |
| GT65R | 9 | 35 | 3zy2 (GT65, GT-B, inverting) | 100 | At3g05320 (GT65) | 99.9 |
| GT68R-A | 1 | 3 | 3zy2 (GT65, GT-B, inverting) | 99.6 | At1g52630 (GT68) | 98.8 |
| GT68R-B | 2 | 2 | 3zy2 (GT65, GT-B, inverting) | 99.8 | At1g53770 (GT68) | 99.8 |
| GT75R | 1 | 2 | 2d0j (GT43, GT-A, inverting) | 69.9 | At5g50750 (GT75) | 100 |
| GT92R | 1 | 3 | 1qg8 (GT2, GT-A, inverting) | 97.8 | At5g44670 (GT92) | 99.3 |
Figure 5.
Functional class assignments of the 197 Golgi-localized proteins. The classification is based on the TAIR annotation for the characterized proteins and the HHpred output predictions for the uncharacterized proteins, as indicated in Supplemental Table S2. The numbers of proteins per class are indicated in parentheses.
DISCUSSION
Replication of Quantitative Proteomic Experiments and Data Integration Allow Generation of Larger Quantitative Data Sets
The major difficulty in analyzing the Golgi proteome for many years was the purification of the organelle, because of its low abundance in the cell and its similar density to other cellular membranes (Mo et al., 2003). Furthermore, the Golgi apparatus contains proteins en route to other organelles because of anterograde and retrograde trafficking in the endomembrane system. Thus, even a pure Golgi preparation will not solely contain Golgi-resident proteins. The LOPIT method was developed to overcome these obstacles and is based on observations that a given organelle is not solely present in one particular density gradient fraction but rather forms a distribution profile along the gradient (De Duve, 1971; Dunkley et al., 2004). LOPIT describes the steady-state locations of proteins, thus allowing more permanent residents of the Golgi to be distinguished from cargo. Here, we reanalyzed previous data sets with an updated proteome database and combined it with two new LOPIT experiments. To overcome problems of multivariate statistical analysis of the incomplete observations across all these proteomic data sets, we introduced a statistical imputation approach. This allowed us to identify the subcellular localization of 1,110 proteins with high confidence. This is more than twice as many proteins as localized by Dunkley et al. (2006).
Characteristics of Plant ER, Golgi, and PM Protein Membrane Domains
Our collection of high-quality data sets of protein localization allowed us to investigate properties of the single-span TMDs in different organelles. TMDs of membrane proteins are likely to have features that reflect the properties of the membrane in which they reside (Sharpe et al., 2010), and these features may even influence their trafficking. It has been shown that TMDs in fungi and animals have organelle-specific properties, including a difference in TMD length between the early and late parts of the secretory pathway (Sharpe et al., 2010). A similar analysis of plant organelle proteins, based on our newly expanded LOPIT data, indicates that there are comparable differences in plant TMD length and amino acid composition (Fig. 4). The length of the TMD region showing the maximum average hydrophobicity is longer for the PM than for the Golgi and ER proteins by five residues. This difference is comparable to that observed for vertebrates (six residues), with fungi showing an even larger difference (nine residues; Sharpe et al., 2010). The average amino acid volume also shows a distinction between the organelles, with smaller residues being preferred in the exoplasmic leaflet of PM TMDs. Again, this is qualitatively similar to the situation in fungi and vertebrates, but this time the difference is more marked than in vertebrates, although again smaller than in fungi. The resulting plots and data of amino acid compositions, properties, and abundance were submitted to http://www.tmdsonline.org, and the differences in TMD properties described here could be used to improve the targeting prediction algorithm for single-span transmembrane proteins.
It has been suggested for fungi and vertebrates that these TMD differences reflect organelle-specific physical properties of the bilayer, with the PM being thicker with a particularly well-ordered outer leaflet (Sharpe et al., 2010). As in fungi and vertebrates, the PM of plants is enriched in sterols and sphingolipids (Hartmann, 1998; Pata et al., 2010). The precise structure of these lipids varies between the different clades, but the plant sterols and the glycosylinositol-phosphoceramides are believed to be abundant in plant PMs, perhaps even accounting for the large majority of the lipid molecules in the outer leaflet (Sperling et al., 2005). The ordering effect of the rigid sterols and the saturated, or trans-double-bonded, acyl chains of the sphingolipids would help exclude toxins and other undesirable molecules as well as having the effect of thickening the bilayer. Proteins would need to adapt to this different bilayer, and it may also be that the TMD differences could contribute to protein sorting in the secretory pathway in plants, as in the other two clades (Brandizzi et al., 2002; Wang et al., 2008).
Identification of the Cell Wall Synthesis Machinery in the Golgi Apparatus
Our rationale for extending the cataloging of the most abundant proteins in the Golgi apparatus is that within the identified Golgi proteins, most of the proteins involved in primary cell wall polysaccharide synthesis would be present, particularly proteins in pathways of synthesis of the more abundant polysaccharides. The Arabidopsis liquid-grown callus cell wall is a typical primary cell wall and is composed of abundant polygalacturonan, rhamnogalacturonan I, and xyloglucan with minor amounts of type II arabinogalactan, extensin, glucuronoarabinoxylan, rhamnogalacturonan II, and glucomannan (Goubet et al., 2002; Handford et al., 2003; Manfield et al., 2004; Barton et al., 2006; P. Dupree, unpublished data). Therefore, we expected to identify proteins involved in the synthesis of these polysaccharides.
One-fifth (41) of the 197 identified Golgi proteins are putative or confirmed GTs described in CAZy, strongly supporting the view that the callus Golgi apparatus is rich in GTs and is active in polysaccharide synthesis and protein and lipid glycosylation (Fig. 5). For many of these CAZy-classified GTs, we provide here, to our knowledge, the first evidence for Golgi localization, including the localization of two of the newly CAZy-classified GT90 enzymes. Xyloglucan is the main hemicellulose of dicot primary cell walls (Scheller and Ulvskov, 2010). We localized most of the GTs shown to be involved in xyloglucan synthesis to the Golgi (XT1, XT2, XXT5, MUR3, AtCslC4; Madson et al., 2003; Cocuron et al., 2007; Cavalier et al., 2008; Zabotina et al., 2008). Consistent with high pectin synthesis in the callus cells, we identified GAUT1, GAUT7, and many other GAUT family members in GT8 (Sterling et al., 2006; Atmodjo et al., 2011) as well as ARAD1 and XGD1 in GT47, involved in arabinan and xylogalacturonan synthesis, respectively (Harholt et al., 2006; Jensen et al., 2008). We also detected enzymes likely involved in primary cell wall glucuronoarabinoxylan synthesis: IRX10-L, IRX14, and two IRX15/DUF579 homologs (Brown et al., 2009, 2011; Wu et al., 2010; Jensen et al., 2011), and GUX3, a homolog of the GUX1 and GUX2 xylan glucuronosyltransferases (Mortimer et al., 2010). Proteins of extensin glycosylation (RRA3 and XEG113; Velasquez et al., 2011) and possibly glucomannan synthesis (CslD2 and CslD3; Yin et al., 2011) were identified as well, indicating that LOPIT also detects proteins synthesizing relatively minor cell wall components. Two subunits of the cellulose synthase were localized to the Golgi/TGN at steady state, in contrast to four callose synthases that were found in the plasma membrane. The remaining CAZy-classified Golgi GTs are involved in N-linked glycosylation or are of unknown function. A very recent paper from Heazlewood and colleagues (Parsons et al., 2012) describes a study of the Arabidopsis Golgi proteome using different membrane separation techniques and MS methods. Many of the proteins localized in the Golgi in this work were also found in their Golgi preparations.
Identification of Putative Golgi GTs
To address the question of whether additional identified Golgi proteins may be involved in glycosylation, we used comparative structural modeling to search for protein homologs with known functions. Since characterized sugar-nucleotide-binding Golgi GTs have one of two folds (GT-A or GT-B), unknown GTs are likely to have a similar 3-D structure (Unligil and Rini, 2000; Lairson et al., 2008). We applied HHpred, a sensitive structural prediction and remote homology detection algorithm, to the identified 197 Golgi proteins. In this way, we identified an additional 24 putative GTs in 12 sequence-related families (Table II). This bioinformatic prediction of GT-like folds is supported by our demonstration that they are Golgi residents. There are in total 96 proteins in these 12 putative Golgi GT families encoded in the Arabidopsis genome (Supplemental Table S3). CAZy GT families have at least one member biochemically characterized (Coutinho et al., 2003). We detected 41 putative or confirmed CAZy GTs by LOPIT (from 13 CAZy families, having a total of 230 members). Comparing the numbers of putative GTs in these two categories found in this study, we speculate that perhaps one-third of Arabidopsis Golgi GTs are not yet assigned to CAZy GT families because of the lack of biochemical characterization (Fig. 5). Egelund et al. (2004) and Hansen et al. (2009) also used structural bioinformatic approaches to identify some of these putative GTs. One of their identified families has since been shown to have GT activity and was added to the CAZy database as GT77 (Egelund et al., 2007; Gille et al., 2009).
KOBITO1 has earlier been reported to be necessary for cellulose synthesis in primary cell walls and localized to the PM of elongating root cells and in an internal compartment (Pagant et al., 2002). Here, we demonstrate the Golgi/TGN localization of this protein in callus (Supplemental Table S2). This is consistent with the similar finding that the cellulose synthases are also localized in the Golgi/TGN at steady state (Dunkley et al., 2006; Supplemental Table S2) but during cell elongation are targeted to the PM, where they are functionally active (Paredez et al., 2006; Crowell et al., 2009; Wightman et al., 2009). The HHpred alignment showed homology of KOBITO1, and its two homologs (At2g41451 and At3g57200), to the 3-D structures of GT2, and they are likely to have a GT-A fold. We name this group of three proteins GT92R because the putative GT domain is most closely related to GT92 (Table II; Supplemental Fig. S4). A recent study also identified KOBITO1 as a putative GT and proposed a function in a carbohydrate metabolic process essential for proper plasmodesmata closure (Kong et al., 2012).
The GT65R is a large family and consists of 35 members containing DUF246 domains in Arabidopsis, eight of which were shown in this study to be Golgi localized. This family has already been predicted to be a putative GT family (Egelund et al., 2004; Hansen et al., 2009), and in the Pfam database, this group has been recently fused into PF10250, which contains metazoan protein O-fucosyltransferases. Since the GT65R family shows structural homology to GT65, it is likely to have a GT-B fold. However, the sequence similarity to GT65 is low, suggesting that the activity may be different, and only one of its members (At1g52630) is currently in the CAZy database, in the group of “nonclassified” GTs. Two genes have been partially characterized: ROOT HAIR-SPECIFIC17 (RHS17) and EMBRYO SAC DEVELOPMENT ARREST30/TUBE GROWTH DEFECT1 (EDA30/TGD1). The RHS17 gene has a root hair-specific expression pattern and may play a role in root hair growth and morphogenesis (Won et al., 2009). The eda30 mutant fails in the fusion of polar nuclei in the embryo sac (Pagnussat et al., 2005), and another mutant in the same gene, tgd1, had defective pollen tube growth (Boavida et al., 2009).
The GT68R-A and GT68R-B families in Arabidopsis consist of three and two members, respectively. There are some distant similarities to GT65R (DUF246), since they show structural homology to the GT-B fold GT65 family. SUB1, a member of GT68R-A, has previously been suggested to be involved in the cryptochrome signaling pathway as a calcium-binding protein in the ER (Guo et al., 2001). Our results of SUB1 localization to the Golgi apparatus suggest that the protein fusion of SUB1 to GUS was possibly mislocalized.
The GT14R consists of 23 proteins with DUF266 domains in Arabidopsis, one of which was localized in our proteomic experiments. This family has already been predicted to be a putative GT family (Egelund et al., 2004; Hansen et al., 2009), and most of the members have been added to the CAZy nonclassified GTs. It was shown that this domain is distantly related to GT14, likely possessing the GT-A fold, but it lacks the DxD motif (Hansen et al., 2009). GT14 contains the characterized N-glycan β-1,6-N-acetylglucosaminyltransferase of animals, and there are 11 GT14s in Arabidopsis, but there are no characterized members in plants. In rice (Oryza sativa), the bc10 mutant with a loss of function of a DUF266 protein has an altered cell wall phenotype, suggesting that BC10 acts as a Golgi GT involved in cell wall synthesis (Zhou et al., 2009).
Two novel families have putative GT8 like structures: GT8R-A (DUF616) and GT8R-B. In Arabidopsis, there are eight GT8R-A proteins, containing the DUF616 domain, and one was found to be Golgi localized in this study. DUF616 is a domain present in bacterial and plant proteins. In several bacterial proteins, this domain is fused with GT2 domains (e.g. UniProt accessions Q2GAF2 and B3EF89). DUF616 shows highest structural homology to the GT-A fold GT43 family and to the GATL clade of GT8 in Arabidopsis (Table II). Several Arabidopsis GT8R-A genes are coexpressed with genes encoding pectin biosynthetic enzymes (http://atted.jp; Obayashi et al., 2007); hence, a possible function of this class of proteins could be in the synthesis of pectic polysaccharides. We have identified and localized two of the four GT8R-B family members in Arabidopsis. Similar to GT8R-A, the GT8R-B family shows homology to GT43 and to the GATL clade of GT8 in Arabidopsis (Table II).
The GT27R family in Arabidopsis comprises 11 proteins having a DUF707 domain and is restricted to plants and prokaryotes. Two GT27R proteins have been localized in our study to the Arabidopsis Golgi apparatus. The domain shows highest structural homology to the GT-A fold GT27 and to the GT2 family in Arabidopsis (Table II).
The GT13R protein (At5g12260) is a singleton. Its protein sequence suggested structural homology with two GT families: the N-terminal domain to GT13, and the C-terminal domain to GT77. Therefore, this protein appears to be a bimodular protein with two GT domains, similar to some heparan sulfate synthase proteins in animals. These fusions of GT47 and GT64 domains transfer alternately β-1,4-glucuronyl and α-1,4-N-acetylglucosaminyl residues (Sugahara and Kitagawa, 2002). It is possible that the Arabidopsis GT13R performs a similar alternating transfer of two different sugar residues (GT13 has an inverting and GT77 a retaining mechanism), and hence it could be involved in biosynthesis of a polymer with alternating sugar backbone, such as rhamnogalacturonan I.
The GT4R and GT75R proteins constitute two further putative GT families in Arabidopsis. We have identified and localized both GT4R and one of the two GT75R proteins encoded in the genome. Homology modeling shows that the DUF288 domains in GT75R are related to the reversibly glycosylated proteins/UDP-Ara mutase (GT75) and are structurally close to GT2, with a GT-A fold (Table II).
Several glycoside hydrolases were also detected in this Golgi proteome study: mannosidases, belonging to GH38 and GH47, presumably responsible for the maturation of N-glycans. However, we did not detect any Golgi-localized glycoside hydrolases that may be involved in the remodeling of cell wall polysaccharides.
Transporters and Other Multiple Membrane-Spanning Proteins
The Golgi apparatus transports various metabolites for luminal polysaccharide synthesis, including sugar nucleotides, SAM, and probably acetyl-CoA (Reyes and Orellana, 2008). Three families have been identified that fit the criteria of type III membrane proteins and are likely transporters: NUCLEOTIDE SUGAR TRANSPORTER/TRIOSE PHOSPHATE TRANSLOCATOR (NST/TPT), DUF791, and ENDOMEMBRANE PROTEIN70 (EMP70).
Six members of the NST/TPT family were identified (of 53 members proposed by Reyes and Orellana [2008]). The members of this family possess six to 10 transmembrane helices (Knappe et al., 2003), and most of them have a DUF250 domain. Four of the identified proteins in this study belong to the KD clade and one to the KT clade of phosphate translocator homologous proteins, as annotated by Knappe et al. (2003). Another nucleotide sugar transporter is the uncharacterized (At4g35335), which has the highest similarity to UTr6. One other uncharacterized transporter-like protein was identified, At4g39840, which contains five to six TMDs.
There are four DUF791 family members in Arabidopsis, and two of those were identified to be Golgi localized in this study. This family is part of the major facilitator superfamily, and they show homology to sugar proton symporters. Members of DUF791 are predicted to have 12 TMDs.
The EMP70 family is predicted to have a large luminal domain after the signal sequence and nine conserved putative TMDs, giving the name nonaspanin domain. Twelve proteins are encoded by the Arabidopsis genome, 10 of which were detected and localized to the Golgi by our approach. It has been shown that a yeast ortholog of EMP70 is important for early endosome-to-vacuole trafficking and vacuolar protein sorting (Aguilar et al., 2010). However, Arabidopsis EMP70 members have been shown to be localized exclusively to the Golgi (Dunkley et al., 2006; Gao et al., 2012). This family is quite conserved through all eukaryotes, and this suggests that members of EMP70 play important roles in secretion and/or Golgi functioning. The EMP70 proteins may function as channels or transporters.
Sugar Nucleotide Metabolism
Several enzymes catalyzing the nucleotide sugar interconversion reactions were identified with this approach as Golgi residents: GAE1, GAE5, UXS1, UXS2, and UXS4. The presence of UDP-Gal and UDP-Xyl synthesis enzymes is consistent with the high biosynthetic activity of xyloglucan and pectin. We also identified a previously undescribed putative epimerase/isomerase (At3g56820). The structure-based homology search against the CDD database showed that it is an rmlC-like protein. Proteins in this family have one or both of C3 and C5 epimerase activities, such as bacterial dTDP-4-keto-6-deoxyglucose-3,5-epimerase in dTDP-Rha synthesis. Another group of proteins that includes activities of nucleotide sugar interconversions is the Kelch motif-containing family; two such proteins were identified as Golgi residents (At1g51540 and At3g27220).
Putative SAM-Dependent MTs
There were a large number (24) of putative MTs detected in the Golgi by LOPIT. The functions of this type of Golgi enzyme are largely unknown. It is likely that most of these enzymes are involved in methylation of the glycans. MTs are an extensive class of enzymes that share little sequence identity but contain a well-conserved structural fold. Many crystal structures of SAM-dependent MTs have been determined, and they define the core SAM-MT fold as consisting of a central seven-stranded β-sheet that is flanked by three α-helices per side of the sheet. The SAM-binding region is localized to the N-terminal part and comprises the first three N-terminal β-strands separated by α-helices (Martin and McMillan, 2002). This conservation allowed us to discover distantly related proteins in the MT superfamily.
We identified 19 Golgi proteins containing the DUF248 domain. They have collectively been annotated previously as a putative MT family, with one member, QUA2, being involved in homogalacturonan methylation (Mouille et al., 2007; Ralet et al., 2008). The DUF579 domain shows high predicted structural similarity to several MTs (Supplemental Fig. S3). Two DUF579 family members have been shown to be involved in xylan biosynthesis, IRX15 and IRX15L (Brown et al., 2011; Jensen et al., 2011), but it is still unclear how they are involved. Three additional putative MTs assigned to the Golgi are encoded by the singleton At3g16200 and the two closely related At3g49720 and At5g65810. These are only distantly related to both DUF248 and DUF579.
CONCLUSION
The localization of over 1,100 plant membrane proteins will be an invaluable resource for the community and allow in-depth analysis of many organelle properties and functions. For example, the identification here of differences in TMD properties between the ER, Golgi, and PM proteins will allow better prediction of novel protein localization as well as facilitate studies of protein targeting. The currently identified GTs are insufficient to synthesize all the cell wall carbohydrate structures produced by plants. We identified 12 biochemically uncharacterized Golgi putative GT families, extending substantially the number of targets for functional studies that are now under way in our laboratory. We believe that these results will greatly contribute to the understanding of plant cell wall polysaccharide biosynthesis.
MATERIALS AND METHODS
Growth of Plant Material, Organelle Separation, Protein Digestion, Labeling, and MS Analysis
We used the established Arabidopsis (Arabidopsis thaliana) liquid-grown callus for the proteomic analysis. The conditions of growth were as described by Prime et al. (2000). Organelle separation was achieved from homogenized Arabidopsis cell culture using density centrifugation as described before (Sadowski et al., 2006). For the peripheral LOPIT experiment, the membrane fractions obtained from the density centrifugation were not treated with sodium carbonate. Proteolysis was achieved with trypsin, and the peptides were labeled with four-plex iTRAQ, separated by strong cation-exchange chromatography, and analyzed on an AB Sciex QSTAR Quadrupole Time-of-Flight liquid chromatography-MS system as described previously (Dunkley et al., 2004, 2006).
Data Processing: Integration of Four Data Sets and Missing Values Imputation
The raw MS/MS data files obtained from the QSTAR were processed using the wiff2DTA software to generate centroided and uncentroided peak lists (Boehm et al., 2004) containing mass-to-charge ratio and intensity information for each product ion spectrum. The centroided peak lists were analyzed by MASCOT 2.2 (Matrix Science) and queried against the TAIR 8 nonredundant protein database (27,234 sequences) using the following modifications: fixed, MMTS (Cys); variable, iTRAQ four-plex (Lys), iTRAQ four-plex (N-terminal), iTRAQ four-plex (Tyr), oxidation (Met). The ion score significance threshold was set to 0.05, MS tolerance was set to 1 D, and the MS/MS tolerance was set to 0.8 D. Each centroided peak list was also searched separately against a decoy reversed version of the database in order to determine the MASCOT peptide score resulting in a false identification rate of less than 1% in each LOPIT experiment. Hence, the minimum peptide MASCOT scores for proteins identified with two or more peptides were 32 for LOPIT experiment 1, 35 for LOPIT experiment 2, 33 for LOPIT experiment 3, and 33 for LOPIT experiment 4. The thresholds for identification of proteins with single peptides were 47, 51, 52, and 50 for experiments 1 to 4, respectively. Proteins identified by two or more distinct peptides in at least one experiment were retained. The XML files created from the integrated data set were exported to the PRIDE database (Vizcaíno et al., 2010). The uncentroided peak lists were processed with i-Tracker software in order to calculate the normalized iTRAQ reporter ion areas (Shadforth et al., 2005). The Genome Annotating Proteome Pipeline system (Shadforth et al., 2006) was used to link identification information with quantitation data in a MySQL database. Peptides were assigned to proteins only if at least three of their reporter ion peaks were above a threshold of 15 counts and if they had a MASCOT score of at least 20 (Dunkley et al., 2006). In addition, peptides matching multiple proteins were not used for protein quantitation. The quantitative data obtained from the four reporter ions of the iTRAQ label was corrected for the isotope impurities and normalized such that the four reporter ions sum to one (Fig. 1). Quantitation data from multiple peptides from the same protein were averaged to provide a fractionation profile for the identified proteins. Proteins identified and quantitated in two or more experiments were retained. The normalized four-plex data sets were concatenated in one matrix presented in Supplemental Table S1.
Multivariate Data Analysis
Protein profiles that were present in only one experiment were discarded (for data preprocessing, see Fig. 1). The protein profiles that had missing four-plex data (from one to six missing four-plex data points out of eight) were imputed by a PLS regression model (Geladi and Kowalski, 1986). The imputation was performed sequentially. First, the entire data set was divided into subgroups according to the number and positions of missing four-plex data. For each subgroup, the missing labeling was estimated by a PLS regression model built using proteins where all labelings were present. The estimated labeling was then considered “known” and used as a predictor for building subsequent regression models. The process was performed first for the protein profiles having one four-plex labeling missing and then repeated for those with two missing labelings. This process was iterated until all the missing values were imputed. The validation of this method was performed using a cross-validation approach. One nonmissing four-plex was randomly removed from each protein with up to five missing labelings, and the PLS regression method was applied to this new data set in order to predict known and unknown missing values. The predicted four-plexes were then compared with the known values, and the prediction relative error was calculated. This process was repeated 1,000 times, and the expected error (summarized by subgroups of proteins with the same number of missing values) was estimated by a mean error and its 95% confidence interval.
The list of protein identifiers with known subcellular localization was manually curated from published research papers, with the assistance of the TAIR (www.arabidopsis.org; Lamesch et al., 2012) and SUBA (http://suba.plantenergy.uwa.edu.au; Heazlewood et al., 2007) databases. The training set was composed of proteins localized by methods that were LOPIT orthogonal and considered of high confidence (fluorescence microscopy imaging, immunolocalization, or, in cases of proteins localized to the plastid or mitochondrion, sequence homology and protein import assays). This way, we gathered a list of 348 protein identifiers distributed in seven classes (41 proteins belonging to the Golgi/TGN class, 53 to the ER, 39 to the PM, 11 to the vacuole, 92 to the mitochondrion, 40 to the plastid, plus 72 to a decoy class consisting of ribosomal and cytosolic proteins; Supplemental Table S2). Their corresponding quantity profiles were used as a training set for the classification model. The multivariate classification rule with diagonal covariance matrices (known as naïve Bayesian classifier) was used to build a multivariate classification model. The prior probability for a particular protein to be assigned to one of six classes was set uniformly. The class posterior distributions were modeled in a nonparametric way using a Gaussian smoothing kernel.
In order to account for the uncertainty of classification originating from the missing value imputation step, a multiple imputation approach was undertaken as described previously (Rubin, 1987). Exactly 100 variants of an imputed data set were generated using the bootstrapped sequential imputation procedure described above in order to explore the variation in the prediction. Classification models were built for each of the 100 generated data sets. The average posterior probability was calculated to reflect the variability of the class probability distributions and the imputed data. The performance of the classifier was evaluated using a leave-one-out cross-validation scheme where the localization of one of the proteins from the training set was predicted using the remaining training set proteins. The proteins in the training set were correctly assigned in 98% of these models. Proteins were finally assigned using organelle-specific thresholds based on the average posterior probability of assignments of the training set proteins, and the predicted classes are presented in Supplemental Table S2. The data processing was done using the Matlab Statistical Toolbox (Mathworks).
TMD Analysis
The single-span TMD proteins from the LOPIT-classified set were identified using a combination of sequence filtering employing SignalP version 4.0 (Petersen et al., 2011), TMHMM (Krogh et al., 2001), and glycosylphosphatidylinositol (GPI) anchor prediction (Borner et al., 2002). In the first pass, only protein sequences were accepted, according to the TMHMM prediction of the transmembrane spans, if the protein appears to have only one transmembrane span or if it appears to have two but the first span is at the N terminus (and so could be a signal peptide). If a transmembrane span was at the N terminus and predicted to be a signal sequence, according to SignalP, then the protein was excluded if this was the only transmembrane span but accepted if this was the first of two TMHMM predictions. Any sequence that passed these tests was subject to the GPI-anchoring Prediction Tool method of GPI anchor prediction used by Borner et al. (2002), which excluded some putative GPI sequences on the basis of a C-terminal hydrophobic signal peptide.
In order to augment the organelle-classified Arabidopsis sequences, close orthologs in related organisms were identified in the Streptophyta clade within the UniProt database. The expanded lists of protein sequences were reduced for redundancy and analyzed for their cytosolic extra membrane length and TMD sequence properties: amino acid composition, hydrophobicity, and residue volume, as described by Sharpe et al. (2010).
Protein Structure Prediction
The remote homology detection server HHpred (www.toolkit.tuebingen.mpg.de/hhpred) was used to search for homologs to the uncharacterized protein families (Söding et al., 2005). All searches were conducted with the default settings: three HHblits iterations were performed in local alignment mode to build up a multiple sequence alignment from the input sequence. The algorithm was used to search the Protein Data Bank for 3-D structures, Pfam and CDD for primary structures, and the Arabidopsis protein database for determining the closest CAZy GT family to the query.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Number of proteins with and without predicted TMDs identified and quantitated in the four LOPIT experiments.
Supplemental Figure S2. Positional analysis of amino acid composition of TMDs from different organelles.
Supplemental Figure S3. HHpred alignment output of DUF579 query sequence against the PDB database.
Supplemental Figure S4. HHpred alignment output of KOBITO1 query sequence against the PDB database.
Supplemental Table S1. Summary of the 1385 proteins with normalized reporter ion intensities.
Supplemental Table S2. Functional annotations, classification results, and fractionation profiles of the 1385 proteins studied.
Supplemental Table S3. List of the 12 novel putative GT families with their members.
Acknowledgments
We thank Zhinong Zhang for maintaining the Arabidopsis callus lines, Svenja Hester and Julie Howard for assistance with MS, Hayley Sharpe for providing part of the python script, David Burke for useful discussions on protein structural prediction, and Bernard Henrissat, Pavel Shliaha, and Laurent Gatto for critical reading and comments on the manuscript.
Glossary
- ER
endoplasmic reticulum
- GT
glycosyltransferase
- CAZy
Carbohydrate-Active Enzymes
- 3-D
three-dimensional
- TMD
transmembrane domain
- MS
mass spectrometry
- LOPIT
localization of organelle proteins by isotope tagging
- PM
plasma membrane
- MS/MS
tandem mass spectrometry
- PLS
partial least-squares
- TGN
trans-Golgi network
- GES
Goldman, Engelman, and Steitz
- TAIR
The Arabidopsis Information Resource
- SAM
S-adenosylmethionine
- MT
methyltransferase
- GPI
glycosylphosphatidylinositol
References
- Aguilar PS, Fröhlich F, Rehman M, Shales M, Ulitsky I, Olivera-Couto A, Braberg H, Shamir R, Walter P, Mann M, et al. (2010) A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat Struct Mol Biol 17: 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D, Kniemeyer O, Brakhage AA, Guthke R. (2010) Missing values in gel-based proteomics. Proteomics 10: 1202–1211 [DOI] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton CJ, Tailford LE, Welchman H, Zhang Z, Gilbert HJ, Dupree P, Goubet F. (2006) Enzymatic fingerprinting of Arabidopsis pectic polysaccharides using polysaccharide analysis by carbohydrate gel electrophoresis (PACE). Planta 224: 163–174 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Shuai B, Yu HJ, Pagnussat GC, Sundaresan V, McCormick S. (2009) A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181: 1369–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AM, Galvin RP, Sickmann A. (2004) Extractor for ESI quadrupole TOF tandem MS data enabled for high throughput batch processing. BMC Bioinformatics 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Henrissat B. (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol 11: 593–600 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N. (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Wightman R, Zhang Z, Gomez LD, Atanassov I, Bukowski JP, Tryfona T, McQueen-Mason SJ, Dupree P, Turner S. (2011) Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J 66: 401–413 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Liu JS. (1996) Predictive updating methods with application to Bayesian classification. J R Stat Soc Ser B Stat Methodol 58: 397–415 [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C. (1971) Tissue fractionation: past and present. J Cell Biol 50: 20d–55d [PubMed] [Google Scholar]
- Du X, Callister SJ, Manes NP, Adkins JN, Alexandridis RA, Zeng X, Roh JH, Smith WE, Donohue TJ, Kaplan S, et al. (2008) A computational strategy to analyze label-free temporal bottom-up proteomics data. J Proteome Res 7: 2595–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J, Speed TP. (2002) Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc 97: 77–87 [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TPJ, Watson R, Griffin JL, Dupree P, Lilley KS. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol Cell Proteomics 3: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Dupree P, Sherrier DJ. (1998) The plant Golgi apparatus. Biochim Biophys Acta 1404: 259–270 [DOI] [PubMed] [Google Scholar]
- Efron B. (1994) Missing data, imputation, and the bootstrap. J Am Stat Assoc 89: 463–475 [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- Egelund J, Skjøt M, Geshi N, Ulvskov P, Petersen BL. (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136: 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA, Goldman A. (1986) Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem 15: 321–353 [DOI] [PubMed] [Google Scholar]
- Faso C, Boulaflous A, Brandizzi F. (2009) The plant Golgi apparatus: last 10 years of answered and open questions. FEBS Lett 583: 3752–3757 [DOI] [PubMed] [Google Scholar]
- Gao C, Yu CK, Qu S, San MW, Li KY, Lo SW, Jiang L. (2012) The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell 24: 2086–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geladi P, Kowalski BR. (1986) Partial least-squares regression: a tutorial. Anal Chim Acta 185: 1–17 [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Jackson P, Deery MJ, Dupree P. (2002) Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem 300: 53–68 [DOI] [PubMed] [Google Scholar]
- Guo H, Mockler T, Duong H, Lin C. (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291: 487–490 [DOI] [PubMed] [Google Scholar]
- Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P. (2003) Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218: 27–36 [DOI] [PubMed] [Google Scholar]
- Hansen SF, Bettler E, Wimmerová M, Imberty A, Lerouxel O, Breton C. (2009) Combination of several bioinformatics approaches for the identification of new putative glycosyltransferases in Arabidopsis. J Proteome Res 8: 743–753 [DOI] [PubMed] [Google Scholar]
- Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F. (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA. (1998) Plant sterols and the membrane environment. Trends Plant Sci 3: 170–175 [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron J-C, Orler R, Ralph J, Wilkerson CG. (2011) The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J 66: 387–400 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, et al. (2008) Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpievitch Y, Stanley J, Taverner T, Huang J, Adkins JN, Ansong C, Heffron F, Metz TO, Qian WJ, Yoon H, et al. (2009) A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinformatics 25: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S, Flügge UI, Fischer K. (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Karve R, Willet A, Chen MK, Oden J, Shpak ED. (2012) Regulation of plasmodesmatal permeability and stomatal patterning by the glycosyltransferase-like protein KOBITO1. Plant Physiol 159: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Krogh M, Fernandez C, Teilum M, Bengtsson S, James P. (2007) A probabilistic treatment of the missing spot problem in 2D gel electrophoresis experiments. J Proteome Res 6: 3335–3343 [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77: 521–555 [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WG. (2004) Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J 40: 260–275 [DOI] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol 12: 783–793 [DOI] [PubMed] [Google Scholar]
- Mo B, Tse YC, Jiang L. (2003) Organelle identification and proteomics in plant cells. Trends Biotechnol 21: 331–332 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al. (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Höfte H. (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Parsons HT, Christiansen K, Knierim B, Carroll A, Ito J, Batth TS, Smith-Moritz AM, Morrison S, McInerney P, Hadi MZ, et al. (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata MO, Hannun YA, Ng CK. (2010) Plant sphingolipids: decoding the enigma of the sphinx. New Phytol 185: 611–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedreschi R, Hertog ML, Carpentier SC, Lammertyn J, Robben J, Noben J-P, Panis B, Swennen R, Nicolaï BM. (2008) Treatment of missing values for multivariate statistical analysis of gel-based proteomics data. Proteomics 8: 1371–1383 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Prime TA, Sherrier DJ, Mahon P, Packman LC, Dupree P. (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Raghunathan TE, Lepkowski JML, Van Hoewyk J, Solenberger P. (2001) A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv Methodol 27: 85–95 [Google Scholar]
- Ralet MC, Crépeau MJ, Lefèbvre J, Mouille G, Höfte H, Thibault JF. (2008) Reduced number of homogalacturonan domains in pectins of an Arabidopsis mutant enhances the flexibility of the polymer. Biomacromolecules 9: 1454–1460 [DOI] [PubMed] [Google Scholar]
- Reyes F, Orellana A. (2008) Golgi transporters: opening the gate to cell wall polysaccharide biosynthesis. Curr Opin Plant Biol 11: 244–251 [DOI] [PubMed] [Google Scholar]
- Rubin DB. (1987) Multiple Imputation for Nonresponse in Surveys. John Wiley & Sons, New York
- Sadowski PG, Dunkley TPJ, Shadforth IP, Dupree P, Bessant C, Griffin JL, Lilley KS. (2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 1: 1778–1789 [DOI] [PubMed] [Google Scholar]
- Sadowski PG, Groen AJ, Dupree P, Lilley KS. (2008) Sub-cellular localization of membrane proteins. Proteomics 8: 3991–4011 [DOI] [PubMed] [Google Scholar]
- Schafer JL. (1997) Analysis of Incomplete Multivariate Data. Chapman & Hall, London
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Shadforth I, Xu W, Crowther D, Bessant C. (2006) GAPP: a fully automated software for the confident identification of human peptides from tandem mass spectra. J Proteome Res 5: 2849–2852 [DOI] [PubMed] [Google Scholar]
- Shadforth IP, Dunkley TP, Lilley KS, Bessant C. (2005) i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. (2010) A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960 [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Franke S, Lüthje S, Heinz E. (2005) Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol Biochem 43: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D. (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. (2002) Heparin and heparan sulfate biosynthesis. IUBMB Life 54: 163–175 [DOI] [PubMed] [Google Scholar]
- Trotter MW, Sadowski PG, Dunkley TP, Groen AJ, Lilley KS. (2010) Improved sub-cellular resolution via simultaneous analysis of organelle proteomics data across varied experimental conditions. Proteomics 10: 4213–4219 [DOI] [PubMed] [Google Scholar]
- Unligil UM, Rini JM. (2000) Glycosyltransferase structure and mechanism. Curr Opin Struct Biol 10: 510–517 [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Tangchaiburana S, Ndeh R, Markham JE, Tsegaye Y, Dunn TM, Wang GL, Bellizzi M, Parsons JF, et al. (2008) An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Marshall R, Turner SR. (2009) A cellulose synthase-containing compartment moves rapidly beneath sites of secondary wall synthesis. Plant Cell Physiol 50: 584–594 [DOI] [PubMed] [Google Scholar]
- Wold S, Ruhe A, Wold H, Dunn WJ. (1984) The collinearity problem in linear-regression: the partial least-squares (PLS) approach to generalized inverses. Siam J Sci Stat Comp 5: 735–743 [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. (2009) Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WG, et al. (2011) The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant 4: 1024–1037 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. (2008) Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li S, Qian Q, Zeng D, Zhang M, Guo L, Liu X, Zhang B, Deng L, Liu X, et al. (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57: 446–462 [DOI] [PubMed] [Google Scholar]